Abstract
We have sequenced the genome of a second Drosophila species, Drosophila pseudoobscura, and compared this to the genome sequence of Drosophila melanogaster, a primary model organism. Throughout evolution the vast majority of Drosophila genes have remained on the same chromosome arm, but within each arm gene order has been extensively reshuffled, leading to a minimum of 921 syntenic blocks shared between the species. A repetitive sequence is found in the D. pseudoobscura genome at many junctions between adjacent syntenic blocks. Analysis of this novel repetitive element family suggests that recombination between offset elements may have given rise to many paracentric inversions, thereby contributing to the shuffling of gene order in the D. pseudoobscura lineage. Based on sequence similarity and synteny, 10,516 putative orthologs have been identified as a core gene set conserved over 25–55 million years (Myr) since the pseudoobscura/melanogaster divergence. Genes expressed in the testes had higher amino acid sequence divergence than the genome-wide average, consistent with the rapid evolution of sex-specific proteins. Cis-regulatory sequences are more conserved than random and nearby sequences between the species—but the difference is slight, suggesting that the evolution of cis-regulatory elements is flexible. Overall, a pattern of repeat-mediated chromosomal rearrangement, and high coadaptation of both male genes and cis-regulatory sequences emerges as important themes of genome divergence between these species of Drosophila.
Comparative genome sequencing is an important tool in the ongoing effort to annotate and analyze genes, cis-regulatory elements, and architectural features of genomes. A single genomic sequence provides a wealth of information about the number and location of genes, but the experimental confirmation of genetic function and regulation can be a painstaking process. The structure of the genetic code facilitates identification of conserved protein-coding regions (Nekrutenko et al. 2002), whereas approaches such as “phylogenetic footprinting” (Boffelli et al. 2003) may aid the identification of functional noncoding elements. A recent small-scale study of four Drosophila species (Bergman et al. 2002) suggested that the sequence divergence between Drosophila pseudoobscura and Drosophila melanogaster is appropriate for the identification of cis-regulatory regions. Such a comparison also provides support for gene predictions, allows conserved protein-coding sequences to be identified, and is a major rationale for the D. pseudoobscura genome sequencing project.
Comparative genomic sequencing can also provide insights into the evolutionary mechanisms of genome rearrangement, which is of special interest in these species. Selection may favor inversions, because they maintain epistatic combinations within the inverted segment (Dobzhansky 1949; Charlesworth and Charlesworth 1973; Wu and Beckenbach 1983; Otto and Barton 2001), or selection may favor rearrangements that reorganize genes into clusters of coordinately expressed genes (Roy et al. 2002; Spellman and Rubin 2002; Lercher et al. 2003). Once established in populations, chromosomal inversions may play a role in the formation of new species (Noor et al. 2001; Navarro and Barton 2003).
Drosophila has been a model system for studying the evolution of chromosomes and gene order (Sturtevant and Tan 1937; Sturtevant and Novitski 1941). Chromosomal rearrangements have negative fitness consequences in many organisms because of the deleterious effects of segmental aneuploidy resulting from chromosomal segregation (reciprocal translocations and transpositions) or recombination (pericentric inversions) (Swanson et al. 1981). In Drosophila, however, special features of meiosis avoid the negative fitness effect for one class of rearrangements— paracentric inversions (inversions with both breakpoints on the same chromosome arm). In male meiosis there is no crossing over and hence no recombinant aneuploid dicentric/acentric gametes. In female meiosis, where crossing over does occur, the dicentric/acentric recombinant chromosomes are directed into polar bodies rather than the functional gamete (Sturtevant and Beadle 1936). As a result, paracentric inversions are highly polymorphic within populations of most Drosophila species (Sperlich and Pfriem 1986), and some of these inversions become fixed during speciation.
Sturtevant and Dobzhansky discovered a wealth of naturally occurring chromosomal inversion polymorphisms in D. pseudoobscura, predominantly on the third and X-chromosomes (Sturtevant and Dobzhansky 1936), through an examination of salivary chromosomes (Painter 1934). Ten of these arrangements are widely distributed and abundant. Dobzhansky first used paracentric inversion events to reconstruct relationships among D. pseudoobscura and Drosophila persimilis third chromosomes (Dobzhansky and Epling 1944). Genes within the D. pseudoobscura chromosomal inversions are likely targets of selection as the polymorphic gene arrangements form stable geographic clines (Dobzhansky and Epling 1944), from altitudinal clines in certain populations (Dobzhansky 1948a), exhibit seasonal cycling (Dobzhansky 1948a) and exhibit high levels of linkage disequilibrium (Schaeffer et al. 2003). More than 300 inversions have been detected across the six chromosomal arms of D. melanogaster (Lemeunier and Aulard 1992), but only two arrangements per chromosome are widely distributed and abundant. An obvious question, therefore, is what is the mechanism responsible for differences in the distribution of inversions in different Drosophila genomes?
Random breakage (Ohno 1973; Nadeau and Taylor 1984), transposon-mediated recombination (Krimbas 1992; Caceres et al. 1999; Mathiopoulos et al. 1999; Evgen'ev et al. 2000; Casals et al. 2003), and fragile breakpoints (Novitski 1946; Pevzner and Tesler 2003) have been suggested as possible mechanisms for generating paracentric inversions in natural populations, but there is little definitive evidence. Our study provides a unique opportunity to explore the origin of these rearrangements by examining sequences at junctions of synteny blocks between the two species.
Genome sequence of D. pseudoobscura
We sequenced the genome of D. pseudoobscura using a whole genome shotgun method. In all, 2.6 million sequence reads were produced and assembled into a high-quality draft genome sequence (the numbers of reads from libraries of different insert sizes are summarized in Supplemental Table S1). The sequence is comprised of 8288 contigs (average length 16.3 kb, N50 51.9 kb) joined by paired end read information into 755 scaffolds with an N50 of 1.0 Mb, covering a total of 139 Mb (Supplemental Table S2). Although the total number of reads attempted suggests a 13× coverage, the actual read coverage within the assembly is 9.1×. A description of the N50 scaffold best reflects the quality of the sequence. Fifty percent of the sequence is in scaffolds longer than scaffold contig 2803–contig 3631, the N50 scaffold. The N50 scaffold is 994,609 bp in length and is comprised of 15,143 sequence reads in 26 contigs. Of the sequence reads 2927 are from the 2.7-kb library, 7565 from the 3.4-kb library, 4413 from the 6.3-kb library, 178 are fosmid end sequences, and 19 BAC end sequences. The average Phred (Ewing and Green 1998; Ewing et al. 1998) quality score of the consensus contig sequence is 86.8, with 908,521 bp (91.3%) having the highest Phred Score of 90 and only 1198 bp (0.1%) having a Phred score <20. The total estimated length of the gaps is 15,143 bp or 1.5% of the scaffold sequence length. The quality of the sequence has been further assessed by comparison with a small amount of finished sequence. From this comparison an error rate of 0.26 × 10-4 was estimated—see Methods for further information on this assessment.
The 755 scaffolds can be placed into 16 ultra-scaffolds anchored onto the six chromosomal arms or Muller elements (Muller 1940) (see Methods, `Anchoring sequence scaffolds to chromosomes'). These ultra-scaffolds have an N50 of ∼12 Mb, with Muller elements C and E covered by single ultra-scaffolds, Muller element B comprising four ultra-scaffolds, and Muller elements A and D having five and six ultra-scaffolds, respectively.
Genome size
The chromosome arms of D. pseudoobscura are ∼17% larger than those of D. melanogaster sequence version 3 (Celniker et al. 2002) with the exception of Muller element C, which is approximately the same size (the sizes of the chromosome arms are shown in Supplemental Table S3). The assembly contains 156 Mb of sequence in scaffolds with >1 contig of at least 1 kb, with 17 Mb of this in the form of reptigs (contigs produced from the separate assembly of highly repetitive sequence, then merged with the main assembly). We estimate the euchromatic portion to be ∼131 Mb, based on the extent of the genome covered by and between scaffolds >100 kb anchored to chromosomes. This set of sequence scaffolds is ∼18% longer than the finished D. melanogaster euchromatic sequence and forms the basis of the data in Supplemental Table S3. Much of the remaining sequence consists of small contigs resistant to scaffolding and anchoring. In D. melanogaster, many such small contigs mapped to heterochromatic regions of the genome, and we expect the same will be true of D. pseudoobscura.
To compare unique sequence between the two genomes, we identified distinct 16-mer sequences within the assemblies. We found 111.9 Mb and 102.3 Mb of unique sequence in the D. pseudoobscura and D. melanogaster assemblies, respectively. Thus, the additional sequence is not predominantly due to repeat expansion, unless such repeats are old enough to have significantly diverged from one another. To determine whether the relative additional sequence resides in a small number of large differences or a large number of smaller differences, we compared orthologous pairs of intergenic lengths in regions where syntenic order was conserved. The total length of these regions is 65 Mb in D. pseudoobscura and 59 Mb in D. melanogaster— ∼11% longer in D. pseudoobscura. The increase in length appears to be fairly evenly distributed over many intergenic regions and not due to a small number of large sequence insertions. The frequency of orthologous intergenic and intron length ratios is shown in graphical form in Supplemental Figure S1. The mean ratio of the orthologous intronic length pairs is very close to zero, indicating that intron length is not the source of the increased size of the genome. The orthologous intergenic pairs analyzed show an increased length of ∼17% in D. pseudoobscura relative to D. melanogaster.
Genome coverage
A biologically relevant measure of genome size is the gene content. In D. melanogaster release 3.1, 13,676 genes have been annotated in the euchromatic portion of the assembly (Misra et al. 2002). Others in the heterochromatic portion of the assembly bring the total gene content of D. melanogaster to ∼14,000. More than 90% of these genes can be putatively found using TBLASTN to search the D. pseudoobscura assembly. Of this number, 10,516 are likely orthologs (see annotation and gene prediction). The genome coverage of the assembly was additionally estimated by comparison with the 1.1 Mb total finished sequence: 96.3% was contained in the assembly. A search for a set of 22,347 D. pseudoobscura EST sequences found that 96.2% could be aligned to the assembly. Together, these data indicate that the draft sequence included >96% of the euchromatic genome in D. pseudoobscura.
Syntenic map
A map summarizing the syntenic regions between D. pseudoobscura and D. melanogaster is extremely useful in the identification of orthologous genes, for the identification of chromosomal rearrangements, and the seeding of genome alignments. As protein sequences provided more robust similarity signals than noncoding sequences, the initial syntenic map was based on TBLASTN comparison of release 3.1 D. melanogaster protein predictions (one chosen for each gene) to the D. pseudoobscura “freeze 1” genomic sequence. In most cases, the D. pseudoobscura sequence match was the best TBLASTN hit to a given melanogaster protein. Semiautomatic inspection was used to refine the initial set of matches. Occasionally, a less strong TBLASTN hit was selected as the “valid” match if it resided in the expected linkage location, closing a gap in a run of D. melanogaster–D. pseudoobscura syntenic conservation. Matches that were inconsistent with syntenic data, where other data were consistent with the true match falling into a sequence gap, were rejected as false positives. The syntenic relationships of the D. melanogaster protein to D. pseudoobscura sequence anchor points described by these BLAST hits were then manually inspected to define and refine the order of syntenic blocks. Synteny blocks were defined as runs of consecutive D. melanogaster protein sequence–D. pseudoobscura genomic sequence pairs. Within a syntenic block, gaps were permitted (since there are genes that fall into sequence gaps) and an occasional gene out of order was also permitted (if it fell within five genes of its expected location). Gene duplications in one species could be perceived as synteny breaks. Gene duplications were not considered in the derivation of synteny blocks.
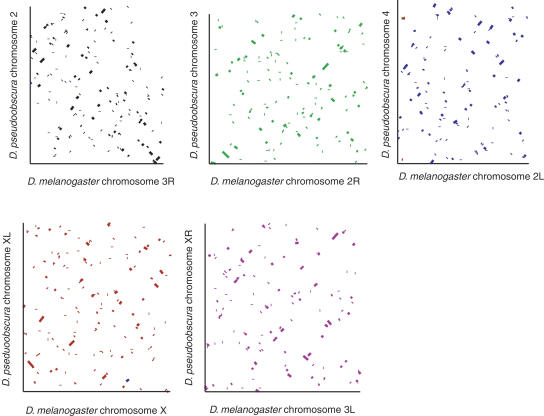
Figure 1 shows the synteny blocks of the chromosomes are short and extremely mixed, but the great majority of syntenic sequences are found on the same Muller element in D. pseudoobscura as they are in D. melanogaster. Thus, as expected, the majority of the chromosomal rearrangements between the D. pseudoobscura and D. melanogaster lineages have been confined to related chromosome arms. The average number of D. melanogaster genes in a syntenic block is 10.7, corresponding to ∼83 kb. The length distribution of syntenic blocks on different Muller elements is shown in Supplemental Figure S2.
Figure 1.
The syntenic relationship between D. pseudoobscura and D. melanogaster. Synteny dot-plots showing the shuffled syntenic relationships between D. pseudoobscura and D. melanogaster for the five chromosome arms. In each case the D. melanogaster chromosome is shown on the x-axis and the D. pseudoobscura chromosome on the y-axis. Note that lines within the graph are all of the same thickness, but are of varying length. Owing to the compression inherent in the figure, many of the lines are shorter than they are wide. Chromosomes have been color coded to allow identification of interchromosomal synteny blocks. For example, in the top left of the D. pseudoobscura Chromosome 4–D. melanogaster Chromosome 2L plot, a small region of sequence on D. pseudoobscura Chromosome 4 with similarity to D. melanogaster Chromosome X can be seen. Muller element F is not shown because of the lack of sequence anchoring data on this chromosome.
Alignment with D. melanogaster
We produced several alignments of D. pseudoobscura and D. melanogaster, and focused on a BLASTZ alignment filtered by comparison with our syntenic map (see Methods). The number of bases that could be aligned (alignability) of the different chromosome arms between D. pseudoobscura and D. melanogaster averages ∼48% and is shown for the five large chromosome arms in Supplemental Figure S3. The fraction of identical bases in the alignment along D. melanogaster chromosomes is quite variable, and is shown for both all sequence and aligned sequence only in Supplemental Figures S4 and S5. Notably, for Muller element A we are able to align only 34% of the bases, compared with between 46.5% and 51% of the bases on the other chromosome arms. The Muller element A is Chromosome XL in D. pseudoobscura and Chromosome X in D. melanogaster. Muller element D, which is the sex chromosome arm XR in D. pseudoobscura and the autosomal 3L in D. melanogaster, has 46.5% of its base pairs in alignments, the second lowest value. Further analysis showed that amino acid identities of orthologs between the two species on Muller elements A and D are not markedly different from the other chromosomes. Possibly, the haploid nature of the X-chromosome in males allows faster accumulation of differences than in the autosomes.
Annotation and gene prediction
Using TBLASTN we identified 12,179 regions of the D. pseudoobscura genome containing a putative ortholog of a D. melanogaster gene. These regions were compared to the locations of gene models produced by three separate gene prediction programs (see Methods). Of the 12,179 regions containing an ortholog of a D. melanogaster gene, 9946 overlapped a D. pseudoobscura gene model predicted by one or more of the prediction programs and for which the proteins in the two species were reciprocal best hits. These 9946 gene models are annotated in the GenBank submission, and the remaining 2233 gene loci have been annotated in the GenBank submission as gene loci identified by TBLASTN analysis but lacking gene models. Despite being a draft sequence, only 9.6% of the gene models contain sequence gaps. A total of 19.9% of the gene models align to the entire orthologous D. melanogaster gene model, while the remaining 80% of the D. pseudoobscura gene models align to an average of 92% of the orthologous D. melanogaster gene model. A frequency histogram of the percent identity within alignments of orthologous proteins is shown in Supplemental Figure S10. The mean amino acid identity for all of the gene pairs was 77%, with a mode around 85%. These gene models provide an excellent starting point for further annotation of the D. pseudoobscura genome. A modest amount of EST sequence is available in D. pseudoobscura. Of the 12,179 gene loci, 3859 overlap with one or more ESTs regardless of the strand, among them 3592 overlapping with one or more ESTs on the same strand. Unfortunately, the only cDNAs so far sequenced are from a non-normalized embryonic library; thus, without additional tissues or normalization, we expect little increase in this coverage.
This analysis of gene loci leaves two interesting sets of genes. First, there are 1485 (10.9% of all gene models) D. melanogaster gene models in FlyBase without associated D. pseudoobscura gene loci. We believe that the majority of these 1485 “orphaned” D. melanogaster genes have D. pseudoobscura orthologs, but they fail to be identified either because the D. pseudoobscura ortholog is located within a sequence gap in the current WGS assembly, or because there is no supporting synteny evidence. Of the remainder (which we expect are at most 500 genes), this set no doubt contains rapidly evolving genes whose sequence similarity is too low for our TBLASTN cutoffs, and possibly novel genes that have arisen since the D. pseudoobscura–D. melanogaster divergence. The reverse set of D. pseudoobscura gene predictions without D. melanogaster counterparts cannot currently be assessed. The three gene prediction programs produced 14,646 gene predictions (many of these are overlapping between the three programs) that did not correspond to a TBLASTN-identified putative ortholog to a D. melanogaster gene model. The majority of these are GENSCAN and TWINSCAN predictions as these do not require a D. melanogaster protein like GeneWise. We expect that most of these are invalid predictions, but that some will turn out to be novel genes not present in D. melanogaster, while some others will turn out to have D. melanogaster orthologs that have thus far escaped annotation by FlyBase. Additional data are required to distinguish between invalid predictions and true genes unique to D. pseudoobscura. Until such data are present, we are reluctant to speculate further on the gene set unique to D. pseudoobscura.
Chromosomal evolution
Comparison of the D. pseudoobscura and D. melanogaster genome sequences identifies conserved linkage blocks and the associated rearrangement breakpoints in the two lineages. Despite strong conservation of sequence blocks within the five orthologous chromosome arms, each chromosome arm has experienced extensive internal shuffling, much of which can be interpreted as the result of a sequential series of paracentric inversions (Fig. 1). No large interarm translocations were observed (with one possible exception), consistent with previous small-scale analyses (Ranz et al. 2001). D. pseudoobscura scaffold 7059_2327 had a mixture of best hits from genes located at the base of 2L and 2R in D. melanogaster. This exception may therefore reflect a class of pericentric inversions whose breaks are so proximal on each arm that recombination does not overlap the inversion, allowing them to be tolerated without loss of fitness. A similar pericentric inversion has been observed within the melanogaster species subgroup in Drosophila erecta, Drosophila teissieri, and Drosophila yakuba based on chromosome banding patterns (Lemeunier and Ashburner 1976), and it is possible that the D. melanogaster gene distribution between proximal 2L and 2R is not ancestral.
Single gene transpositions between Muller elements were observed, and in some cases a lack of introns in one ortholog indicates that these arose through retrotransposition events. Analysis of 27 well-defined retrotransposition events showed that 11 were from the D. melanogaster X-chromosome to a D. pseudoobscura autosome versus possible other directions (probability < 0.01, χ2 test), suggesting that gene movement away from the X-chromosome is favored, consistent with observations made by Betran et al. (2002). Thus far, transcripts from seven of the 11 D. melanogaster genes derived from the X to autosome transpositions have been found only in testis-derived EST libraries and absent from other EST collections derived from other tissues. This is consistent with the hypothesis that the selective pressure favors testis-specific gene movement to autosomes, ensuring gene expression despite X inactivation during spermatogenesis (Betran et al. 2002).
Chromosomal rearrangements
Transposable or repetitive elements may be involved in the genesis of rearrangements in Drosophila chromosomes through recombination between offset copies of an element in reverse orientation (Potter 1982; Collins and Rubin 1984; Engels and Preston 1984; Blackman et al. 1987; Lim 1988; Krimbas 1992; Lyttle and Haymer 1992; Sheen et al. 1993; Ladeveze et al. 1998; Caceres et al. 1999; Mathiopoulos et al. 1999; Evgen'ev et al. 2000; Casals et al. 2003). The differences in gene order observed between D. melanogaster and D. pseudoobscura reflect the rearrangement history since the two species diverged from a common ancestor (known rearrangements since the species diverged are depicted in Supplemental Fig. S6). Genes can move to different chromosome arms either through transpositions or pericentric inversions or can be shuffled within chromosomal arms via paracentric inversions. In all of these cases, junctions between adjacent syntenic blocks contain rearrangement breakpoints that have occurred in either the D. pseudoobscura or D. melanogaster lineage. Most of the rearrangement breakpoints are interspecific inversions long ago fixed in one or the other lineage, but eight breakpoints on Muller element C are the result of four inversion mutations that converted the ancestral D. pseudoobscura Tree Line arrangement into the Arrowhead arrangement (the chromosome arrangement in the D. pseudoobscura strain whose genome was sequenced).
A total of 921 rearrangement breakpoints were identified in the comparison of the D. pseudoobscura and D. melanogaster genomes. This number is likely an underestimate as breaks caused by scaffold ends were excluded if the map location of the next scaffold was not known. It has been estimated that 460 inversions have occurred in the two lineages (Ranz et al. 1997). We compared the sequences within the rearrangement breakpoints to determine (1) if breakpoints shared common sequence elements; (2) if shared sequences are similar to known transposable elements; (3) if the distribution of common sequence elements is correlated with the presence of inversion polymorphism; and (4) if sequences between breakpoints are similar between species. This analysis did not include a comparison to Anopheles gambiae because the intra- and interchromosomal rearrangements between D. melanogaster and the mosquito genomes have been too extensive (Zdobnov et al. 2002).
Identification of intraspecific inversion breakpoints
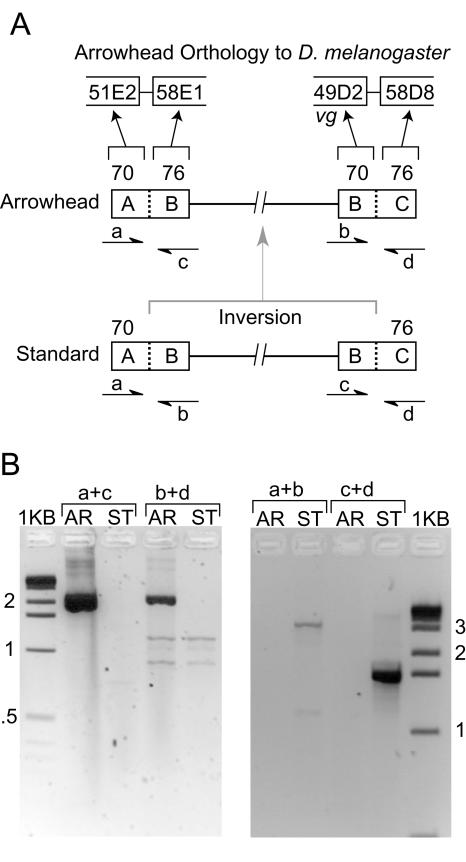
PCR was used to identify the two breakpoints for the inversion that converted the Standard gene arrangement into the Arrowhead gene arrangement. The vestigial gene is located near the distal Standard to Arrowhead breakpoint based on in situ hybridization (Fig. 2; Schaeffer et al. 2003). Using the synteny map described above, a break in conserved gene order between D. pseudoobscura and D. melanogaster, located 17 kb 3′ of the vestigial gene (Schaeffer et al. 2003) was confirmed as the distal Arrowhead breakpoint with a PCR amplification that spanned the breakpoint (Fig. 2). The proximal Arrowhead breakpoint was also mapped with conserved linkage information and verified with PCR analysis (Fig. 2).
Figure 2.
Mapping intraspecific inversion breakpoints. (A) Comparison of Muller element C between D. melanogaster and the Arrowhead arrangement of D. pseudoobscura revealed a junction in conserved linkage near vestigial (vg). The numbered sections 51E2, 58E1, 49D2, and 58D8 are the D. melanogaster cytological locations that are homologous to 70A, 76B, 70B, and 76C sections on the D. pseudoobscura cytological map, respectively. vg maps near the distal breakpoint of the inversion that converted the Standard arrangement into the Arrowhead arrangement (Schaeffer et al. 2003). The locations of four PCR primers, a, b, c, and d, are shown on the Standard and Arrowhead physical maps. Note that the two internal primers, b and c, are switched in the two chromosomes. (B) PCR results. The Arrowhead-specific primer combinations (a + c and b + d) only amplified Arrowhead DNA, while the Standard-specific primer combinations (a + b and c + d) only amplified breakpoints on Standard arrangements. Sequence analysis of the PCR products from the Standard and Arrowhead backgrounds verifies that PCR amplified the appropriate sequences.
If we reconstruct the Standard gene order by inverting the genes within the two breakpoints, then the two genes that flank the distal breakpoint are qkr58E-1 and qkr58E-2, which are also adjacent in D. melanogaster. The two genes flanking the proximal breakpoint are vestigial and the predicted gene CG11798, which are not adjacent in D. melanogaster. These data suggest that the distal breakpoint has been used a single time, while the proximal breakpoint has been used multiple times. The two breakpoints define a 6.0-Mb inverted region of Muller element C in D. pseudoobscura that is predicted to contain 775 genes with D. melanogaster putative orthologs. There is good evidence that natural selection modulates the frequencies of the D. pseudoobscura gene arrangements (Dobzhansky 1944, 1948b; Wright and Dobzhansky 1946; Schaeffer et al. 2003); however, it will require further work to identify which genes or combination of genes within the inverted regions are the targets of selection.
The junctions between syntenic blocks for the proximal and distal Arrowhead breakpoints, as defined by their flanking syntenic blocks, are 20 and 5 kb in length, respectively. Comparison of the two junctions revealed two, short repeat sequences of 128 and 315 bp (“breakpoint motifs”) (Fig. 3). The breakpoint motifs are in reverse orientation relative to each other, suggesting that pairing followed by ectopic exchange led to the Arrowhead gene arrangement. The breakage event between elements was staggered, at opposite ends of repeats p1F and d1B (Fig. 3). The similarity among the 13 copies of the 128-bp repeat vary between 49.6% and 96.9%, while similarity among the three copies of the 315-bp repeat 2 varies between 83.7% and 86% (alignments are shown in Supplemental Fig. S7). The sequences show no significant similarity to any known Drosophila transposable element sequence, and we have been unable to detect coding function for either a transposase or a reverse transcriptase near the breakpoint motifs.
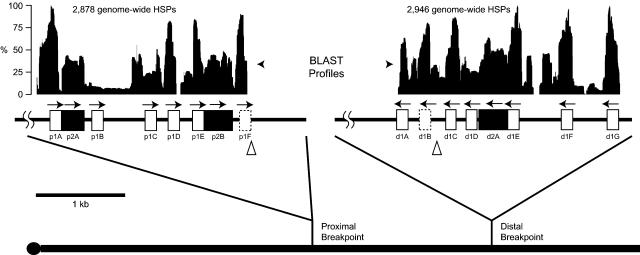
Figure 3.
Structure of the repeats within the breakpoints that converted the Standard gene arrangement into the Arrowhead arrangement. The heavy line at the bottom indicates Muller element C, and the tick marks indicate the locations of the proximal and distal breakpoints for the Arrowhead inversion. The black histograms at the top indicate the frequency that a BLAST High-scoring Segment Pair (HSP) included a particular nucleotide in BLASTN comparison of each breakpoint to the entire genome (E-value ≤1 × 10-5). Two repeat families of 128 and 315 bp (open and filled boxes, respectively) are shown within the two breakpoint regions within the detail regions at the top of the figure. The individual repeats were labeled with a three-letter designation, where the first letter indicates proximal or distal, the number indicates the repeat family, and the last letter indicates the distinct repeat copy. Larger repeats can be generated from the small repeats such as the 443-bp repeat created by the adjacent 128- and 315-bp repeats. The dashed box indicates the putative repeat unit involved in the rearrangement event, and the triangles indicate the approximate location of the DNA breaks with respect to the repeat motif.
Analysis of interspecific breakpoints
Junctions between syntenic blocks from the six Muller elements were extracted from D. pseudoobscura and D. melanogaster genomic sequence. The location and motif presence of each junction is listed in Supplemental Table S9. Junctions without inferred gaps have an average length of 5.6 kb, and tend to be A/T-rich sequences with a mean A+T content of 60%. These junction statistics are shown by Muller elements in Supplemental Table S4.
The breakpoint motif found at the two Arrowhead breakpoints of D. pseudoobscura is also found at other synteny breakpoints. A BLAST analysis of each breakpoint junction sequence against the set of all breakpoint junctions found that >60% of the sequences had at least one High-scoring Segment Pair (HSP) to one other breakpoint within the genome (E-value, 1 × 10-5) (Supplemental Table S5); this similarity is largely due to the breakpoint motif. Each chromosomal arm had at least one breakpoint that had an HSP to >40% of breakpoint sequences, supporting the idea that the breakpoint motif constitutes a single repetitive element family that has numerous degenerate copies in the D. pseudoobscura genome. The higher frequency of the breakpoint motif within the junction sequences on Muller elements C and E led to higher breakpoint similarity than Muller elements A, B, and D when a nonparametric Kruskal-Wallis test was used. The distribution of match fractions for breakpoints on the five major Muller elements is shown in Supplemental Figure S8. The interbreakpoint match frequency for Muller element F is not presented because only seven breakpoints were identified on this chromosome. This analysis shows that D. pseudoobscura breakpoints tend to have similar sequences; however, the repeat is not restricted to the two chromosomes segregating for inversions with D. pseudoobscura. A similar analysis of breakpoint junctions in D. melanogaster failed to detect an abundant repeat sequence (Supplemental Fig. S8).
Distribution of the breakpoint motif
The breakpoint motif is found at other locations in the genome, but the frequencies are much reduced (Table 1). The breakpoint motif is found at the highest frequencies at junctions between syntenic blocks (33.8%–42.6%), at moderate frequencies in noncoding sequences (10.3%–15.3%), and at minimal frequencies in coding regions (0.4%–0.8%). The motif frequencies in breakpoints and noncoding sequences of the Muller element F are high relative to the other chromosomal arms. This may reflect the small numbers of sequences on the Muller element F, the dot chromosome, or more likely, that the heterochromatic nature of the chromosome allows a greater accumulation of repetitive elements (Sun et al. 2000). These observed frequency differences are significantly different from each other with χ2 heterogeneity tests (S.W. Schaeffer, unpubl.). These data suggest that the breakpoint motif is nonrandomly distributed in the genome and is enriched in breakpoints.
Table 1.
Breakpoint sequence motif frequencies in three classes of sequence in six Muller elements in D. pseudoobscura
| Breakpoints
|
Noncoding
|
Coding
|
||||||
|---|---|---|---|---|---|---|---|---|
| Muller element | na | (% ± SD)b | n | (% ± SD) | χ2 | n | (% ± SD) | χ2 |
| A | 210 | 33.8 ± 3.3 | 1698 | 15.3 ± 0.9 | 45.5c | 1851 | 0.8 ± 0.2 | 513.6c |
| B | 135 | 43.0 ± 4.3 | 2031 | 12.9 ± 0.7 | 90.3c | 2124 | 0.8 ± 0.2 | 703.0c |
| C | 205 | 39.0 ± 3.4 | 2082 | 11.4 ± 0.7 | 119.4c | 2276 | 0.7 ± 0.2 | 733.4c |
| D | 141 | 42.6 ± 4.2 | 2068 | 14.3 ± 0.8 | 78.3c | 2159 | 0.6 ± 0.2 | 758.0c |
| E | 223 | 38.1 ± 3.3 | 2636 | 10.3 ± 0.6 | 146.1c | 2923 | 0.4 ± 0.1 | 985.8c |
| F | 7 | 57.1 ± 18.7 | 76 | 44.7 ± 5.7 | 0.4 | 63 | 17.5 ± 4.8 | 5.9c |
The total number of sequences within each category
The percentage of sequences within each category that matched the conserved sequence motif ± standard deviation. The three categories are breakpoints, sequences at the boundary of two conserved linkage groups; noncoding, sequences that are not breakpoints or coding; and coding, sequences of protein-coding genes including introns
Probability of the χ2 value for the heterogeneity test with one degree of freedom is ≤0.05 after applying a Bonferroni correction for multiple comparisons (Rice 1989). A χ2 heterogeneity test is used to determine if the frequency of the breakpoint motif is significantly different between either the noncoding or coding regions
We asked whether breakpoint motifs are associated with paracentric inversions on Muller element C (Fig. 4). In all, 80 junctions between syntenic blocks on Muller element C contain the motif. Of those, 18 motifs are within the boundaries of two ancestral syntenic blocks and can be hypothesized to generate simple two-break rearrangements. The other 62 motif-containing junctions were likely involved in multiple rearrangement events where exchanges involved a motif within an ancestral syntenic block and a motif at a pre-existing conserved linkage junction. The high frequency of motifs at pre-existing conserved linkage junctions suggests that reconstructing the rearrangement history between D. pseudoobscura and D. melanogaster will be difficult, because breakpoints have been used multiple times. In addition, this observation suggests that the estimate of 460 rearrangements is likely to be an underestimate of the true number of rearrangements that have occurred during the D. pseudoobscura and D. melanogaster lineages. Figure 4 also shows the orientations of the breakpoint motifs (indicated by open and filled triangles), which alternate more frequently than expected at random based on a runs test (ts = 2.20, P < 0.05) (Sokal and Rohlf 1981).
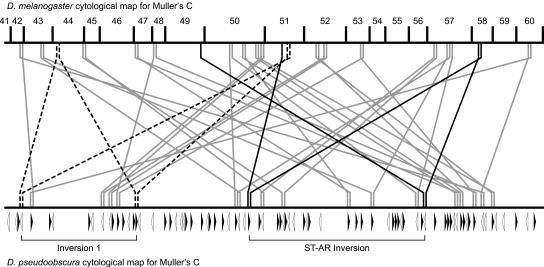
Figure 4.
Rearrangement of conserved linkage groups between D. melanogaster and D. pseudoobscura. The thick horizontal lines represent the chromosomal maps of the D. melanogaster and D. pseudoobscura Muller element C. Vertical lines drawn either down (D. melanogaster) or up (D. pseudoobscura) indicate conserved linkage groups. The locations and orientations of 80 breakpoint motifs are indicated with open and filled triangles at the junctions of conserved linkage groups. Diagonal lines connect homologous linkage groups in the two species where a single inversion event between breakpoint motifs will bring adjacent D. melanogaster genes together (dashed and gray lines). A second example that shows ectopic exchange between a pair of motifs where only one breakpoint brings adjacent D. melanogaster genes together is indicated with black solid lines.
The interbreakpoint sequence similarity is not solely due to the breakpoint motif seen in the Standard to Arrowhead breakpoints. Transposable elements and repetitive sequences were found in the junctions between syntenic blocks, but the junctions were not enriched for these known transposable elements. For example, some D. pseudoobscura breakpoints had sequences similar to the mini-me element (Wilder and Hollocher 2001), which uses reverse transcriptase for retrotransposition (S.W. Schaeffer, unpubl.). The mini-me element is found at a lower frequency at breakpoints than the breakpoint motif (3.4% vs. 38.9%) and is not found at significantly different frequencies between breakpoints and noncoding regions with χ2 heterogeneity tests.
A phylogeny of the breakpoint motifs, shown in Supplemental Figure S9, which are 85% identical on average, has many long terminal branches. This suggests that the breakpoint motif has rapidly radiated throughout the D. pseudoobscura genome. Breakpoint motifs fail to form monophyletic clusters by chromosome or region of origin, rejecting the idea that these elements are unique to a particular chromosome or have diversified based on their chromosome of origin. Also, breakpoint motifs from the same local genomic region are not more similar than sequences separated by longer distances. In fact, the two motifs that were the most similar in this subset of sequences are from different chromosomes.
Conservation of genes between D. pseudoobscura and D. melanogaster
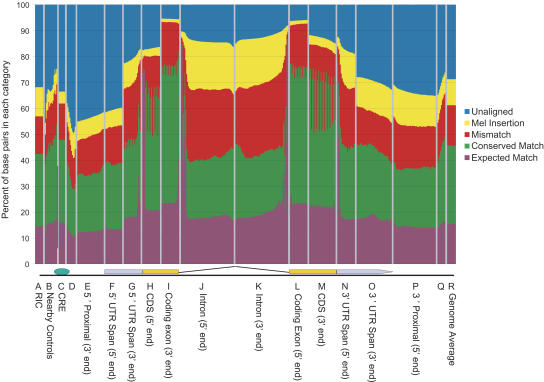
To investigate the conservation of genes between D. pseudoobscura and D. melanogaster, we examined both the nucleotide and amino acid sequences of orthologous genes. Using the filtered global BLASTZ alignment and D. melanogaster 3.1 gene model annotations (Misra et al. 2002), we were able to investigate the conservation of gene features between D. pseudoobscura and D. melanogaster. Figure 5 shows the degree of sequence conservation in promoter regions, upstream regions, untranslated regions (UTRs), coding regions, introns, and other gene features, averaged over a large number of orthologous D. pseudoobscura–D. melanogaster gene feature pairs. (The number of gene feature pairs analyzed for each category varies from 2300 to 43,000 as shown in Supplemental Fig. S11.) The average identity of coding sequence at the nucleotide level is ∼70% for the first and second base pair of the codon, and 49% for the wobble base. Intron sequences are ∼40% identical, UTRs 45%–50%, and protein-binding sites from the literature 63%. Within our genome alignment, 46% of total D. melanogaster base pairs are identical, and 71.3% of D. melanogaster base pairs are in aligned regions. We also examined sequence conservation at the protein level. Supplemental Figure S10 depicts the percent amino acid identity of aligned orthologous protein sequences as a frequency histogram for alignments for four sets of proteins—all, male-specific, transcription factors, and proteins with functions in the nervous system. The majority of protein sequences show >70% amino acid identity, with a mode around 85%.
Figure 5.
Averaged conservation of different segments of a “prototypical gene.” Conservation statistics were computed over thousands of aligned pairs of regions of various types, aligned at different reference points. At each position we compute the fraction of aligned pairs that have identical bases at that position (green + purple tiers), have mismatched bases (red), melanogaster bases aligned to deleted bases in pseudoobscura (yellow), or are unaligned in our synteny-filtered BLASTZ alignment (blue). The purple tier shows the fraction of bases that would be expected to match by chance given the base composition at that position in both species. The expected match is <25% because of the inclusion of unaligned and deleted sequences; if these are removed, the baseline is ∼28% because of the slight AT richness of the genome. The vertical panels correspond to different segments of a prototypical gene, indicated on the x-axis. A cartoon of the prototypical gene is represented under the panels. The segments are labeled by the segment of the gene followed in parentheses by the part of that segment by which the segment was aligned. For example, CDS (5′-end) represents the start of the coding sequence aligned by the ATG start sequence, whereas the coding exon (3′-end) is aligned at the 3′-end of the coding exon, and thus the sequences are not all in phase with each other. (A) RIC, random intergenic controls for CRE analysis; (B) nearby controls in order from -250 bp to +250 bp offset from CREs. The right-most nearby controls are closest to the gene start and therefore in a region that is on average more conserved. Some of the nearby controls have a higher match percent (green) as a result; however, CREs have the highest match percent of identical base pairs as a fraction of aligned bases (everything but blue). (C) 142 Cis-regulatory elements of 50 bp or less from literature; (D) compressed sampling of the 5′-proximal region every 50 bp from 50 to 500; (E) 50 bp proximal to the transcription start site (TS), aligned at TS; (F) genomic span of 5′-UTR, aligned at TS; (G) 5′-UTR span aligned at protein start site (PS); (H) 5′-end of protein-coding region aligned at PS; (I) 3′-end of coding exons aligned at donor site; (J) intron aligned at donor site; (K) introns aligned at acceptor; (L) 5′-end of internal coding exons aligned at acceptor site; (M) 3′-end of protein-coding region aligned at protein end site (PE); (N) 3′-UTR span aligned at PE; (O) 3′-UTR span aligned at transcript end; (P) 50 bp of 3′-proximal region aligned at transcript end; (Q) compressed sampling of 3′-proximal region every 50 bp from 50 to 500; and (R) genome-wide average.
Male-specific proteins are less conserved than others
In contrast to the overall mode of 85% amino acid identity, proteins with ESTs derived from testis-specific libraries had a mean amino acid identity of just 60%. This suggested that there might also be an excess of testis-specific genes for which orthologs might not be found because of overly rapid divergence. We searched for D. melanogaster genes for which no ortholog could be found in the entire D. pseudoobscura sequence set including unassembled sequence reads. We focused on cases in which the syntenic neighbors of the D. melanogaster orthologs of the missing D. pseudoobscura gene were present. We found 75 such genes, 20 of which contained no introns, suggesting they might be the result of a retrotransposition event. It is impossible to ascertain the origins of this class of genes without additional data, but of the 20 intronless D. melanogaster genes not found in D. pseudoobscura, 11 were male specific, based on representations in testis-derived EST libraries and absence from EST libraries derived from other tissues (χ2-value = 59.7, df = 1, p < 0.00001). Furthermore, in 761 cases in which putative orthologous genes with testis-specific derived ESTs could be identified, the mean identity was ∼15% more divergent than for other orthologs (p < e - 75) (Supplemental Fig. S10).
Evolutionary analysis of divergence of orthologous gene pairs
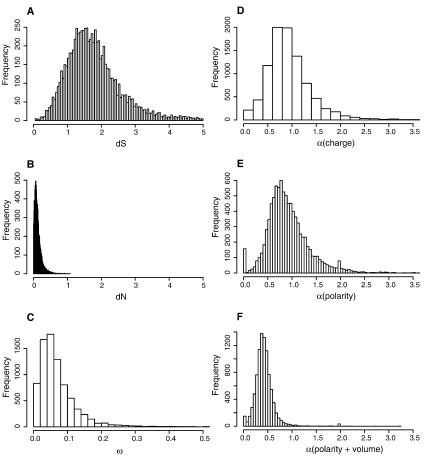
Coding regions of genomes have a built-in contrast between silent, synonymous sites and amino-acid-replacing nonsynonymous sites that allow a variety of evolutionary inferences. The median number of synonymous substitutions per synonymous site between D. pseudoobscura and D. melanogaster was dS = 1.79, and the median number of nonsynonymous substitutions per nonsynonymous site was dN = 0.14, with a skewed distribution around both values (Fig. 6). Estimates of median dS for XL, XR, and the autosomes were 1.82, 1.75, and 1.81, indicating that silent positions have suffered multiple hits per site. XR had a significantly lower dS by a nonparametric Kruskal-Wallis test (H = 14.39, P < 0.001). Median values of dN for XL, XR, and the autosomes were 0.118, 0.105, and 0.108, and XR is again significantly lower (H = 9.92, P = 0.007). This is a surprise, as one might expect the translocation of the Muller element D from an autosome to the X (to form XR) would result in an acceleration in evolution on this arm.
Figure 6.
Distributions of dN, dS, and radical and conservative amino acid changes. (A) Distribution of dS and (B) distribution of dN (numbers of synonymous substitutions per synonymous site and of nonsynonymous substitutions per nonsynonymous site). (C) Distribution of the ratio ω = dN/dS for the melanogaster–pseudoobscura comparison of 9184 inferred orthologous protein-coding genes. (D–F) Distributions of α, the ratio of rates of substitution that are radical to those that are conservative, based on 9184 alignments of orthologous protein-coding genes in D. pseudoobscura and D. melanogaster. Radical changes influence charge (D), polarity (E), or polarity and volume (F) to a greater degree than do conservative changes. A substitution model was fitted by maximum likelihood to estimate these rate parameters.
As the high level of synonymous divergence between D. pseudoobscura versus D. melanogaster gene sequences resulted in low power and low reliability to detect positive selection using the dN/dS ratio, an alternative test was required. We fitted substitution models that split the nonsynonymous substitution rate into two bins (radical vs. conservative as defined in Zhang 2000), each with its own rate parameter (see Methods). After controlling for several factors that can influence this test (Dagan et al. 2002; Smith 2003), we use a rate ratio of radical to conservative amino acid substitutions >1 to identify an accelerated rate of radical changes. The set of genes such that the probability a false positive is <5% (the 5% false discovery rate set) were identified from the P-values associated with the likelihood ratio test. There were 27 genes in the polarity 5% FDR set and 44 genes in the 5% FDR set for charge, providing rather conservative sets of genes showing excess rates of radical amino acid substitution (these genes are listed in Supplemental Tables S6 and S7). The list includes several transcription factors and activators, trithorax group genes, genes involved in innate immunity, cytochrome P450s, and chorion genes, reflecting a diverse set of biological functions that may have faced positive selection.
Conservation of known regulatory elements
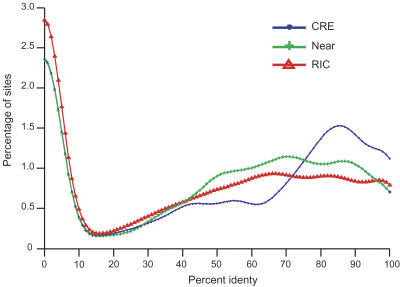
To investigate conservation of cis-regulatory elements (CREs), we collected a set of experimentally characterized regulatory sites curated by the FlyBase project from published papers. We restricted our attention to sites of length <50 bp that seemed likely to correspond to individual CREs. Our collection comprised 142 sites over 30 genes, characterized using a variety of experimental methods, ranging from in vitro binding assays to detection of a mutational phenotype. Of these sites, 63% were upstream of their respective gene, 25% were internal to the gene, and 6% were downstream of their respective gene; all of the sites were analyzed. The modal position of the sites was 2 kb from the putative transcription start site, and the median length was 14 bp. At least 83% of the sites were described as protein-binding sites and the remainder were characterized as regulatory sites, although many of these are likely to be protein-binding sites as well.16 In order to assess whether these regulatory elements were more conserved than expected by chance, we needed some way to estimate the expected sequence similarity in the absence of functional constraints. One difficulty is that since conservation statistics vary in different parts of the genome, and in different regions around genes (see Fig. 5), a test of CRE conservation must control for the effects of the local genomic region. We therefore created two sets of control sites for comparison with the CRE set: random intergenic control (RIC) sites, matched to the CREs for size but randomly positioned in noncoding intergenic sequence, and nearby sites, systematically offset from each CRE by offsets from -250 to +250 bp in increments of 50 bp. Our expectation was that the contrast with nearby sites might be overly conservative, because these may overlap other known or unknown functionally constrained sites, whereas the comparison with RICs might not be stringent enough, since it would detect neighborhood as well as CRE-specific effects. The three sets of sites enabled three pairwise contrasts between the distributions of percent identity values, for which we used the Kolmogorov-Smirnov test for comparison of distributions (see distribution in Fig. 7, results in Table 2). All three contrasts were statistically significant.17 Figure 7 shows that the CRE percent identity distribution has an excess of values >80% and a reduction of values around 50% compared to nearby sites, presumably as a result of stabilizing selection. It suggests that regions with 75%–100% conservation would be most promising for detecting regulatory elements, using conservation-directed motif search or discovery methods such as those described by Grad et al. (2004). However, it is worth noting that the mean conservation in aligned CREs of 72%18 amounts to ∼10 identical bases in 14, whereas the nearby sites, at ∼66% identical, would be expected to have 9.24 identical bases in 14. That difference, though statistically significant, amounts to <1 bp of excess conservation per site. Such a slight difference in conservation would appear to offer scant hope of identifying CREs through pairwise sequence conservation alone in these species. Additional information, such as knowledge of gene expression patterns and known motifs, as in Grad et al. (2004), or genome sequences of additional related species, as in Kellis et al. (2003), will be needed.
Figure 7.
Smoothed distributions of percent identity values for the three groups of cis-regulatory element sequences, excluding sequences with no aligned bases. The KS test can be viewed as answering the question “are these curves different?” All three curves are significantly different (see Table 2). The true CREs show a distinctive peak in the 80%–90% identity range, presumably a consequence of stabilizing selection. The rise on the left is due to unaligned or mostly deleted sequences.
Table 2.
Comparison of conservation of cis-regulatory elements (CREs) to two types of control sites
| Group 1 vs. group 2 | CRE vs. nearby | CRE vs. random intergenic | Nearby vs. random intergenic | |
|---|---|---|---|---|
| Per site analysis | Group 1 mean per site % identity | 51.3% | 51.3% | 47.8% |
| Group 2 mean per site % identity | 47.8% | 42.9% | 42.9% | |
| Difference of means (group 1 – group 2) | 3.6% | 8.4% | 4.9% | |
| Difference of means resampling p-value | 0.05 | 0.003 | 1E-5 | |
| Distribution comparison KS p-value | 0.026 | 0.0016 | 2E-6 | |
| Per base analysis | Group 1 mean per base % identity | 47.8% | 47.8% | 46.3% |
| Group 2 mean per base % identity | 46.3% | 42.4% | 42.4% | |
| Difference of means (group 1 – group 2) | 1.5% | 5.4% | 3.9% | |
| Difference of means resampling p-value | 0.24 | 0.05 | 5.8E-4 |
For each CRE 20 RICs were generated by randomly choosing sites of the same length as the CRE, on the same chromosome and strand, and rejecting any that overlapped a known gene. Then 10 nearby control sites were generated for each CRE by adding positive and negative (i.e., 3′ and 5′) offsets of 50, 100, 150, 200, and 250 bp to the coordinates of each true CRE. Percentage identities for all CRE and control sites were computed relative to reference alignment, on both a per site and per base basis. Unaligned bases, mismatchs, and D. melanogaster insertions contributed zeros to % identity results; D. pseudoobscura insertions were ignored. The distributions of % identity values were clearly not normal, thus we avoided using tests such as the t-test that assume normality. We compared the per site and per base mean % identities of each group using a resampling test, in which the p-value of the observed difference was estimated as the frequency (over a million trials) in which a value as large or larger than the observed CRE mean was observed in an equal-sized sample of control sites. Similarly, the p-value of the difference between the two control sets was estimated using a randomization test (over a million trials) in which the sets mixed and then repartitioned into corresponding mock control sets. We compared the distributions using the Kolmogorov-Smirnov test, which measures the likelihood that samples came from the same continuous distribution.
Discussion
Evolutionary model of genomic rearrangement
A striking feature of conservation between D. melanogaster and D. pseudoobscura is the overwhelming degree of conservation of gene synteny. This contrasts with the A. gambiae–D. melanogaster comparison, where there is a tendency for far weaker arm conservation. Thus, although the basic mechanism favoring paracentric rearrangements appears to be a dipteran-wide phenomenon, over longer evolutionary time (250–300 Mya since the divergence of Anopheles and Drosophila, compared with 25–55 Mya since the divergence of D. melanogaster and D. pseudoobscura) (Beckenbach et al. 1993; Russo et al. 1995; Tamura et al. 2003) there is clearly a breakdown of synteny (Zdobnov et al. 2002). Perhaps scaffold 7059_2327 with its mixture of proximally located genes from D. melanogaster 2L and 2R arms is a hint at one mechanism that can, over long evolutionary time, lead to extensive reshuffling of genes between arms of a chromosome.
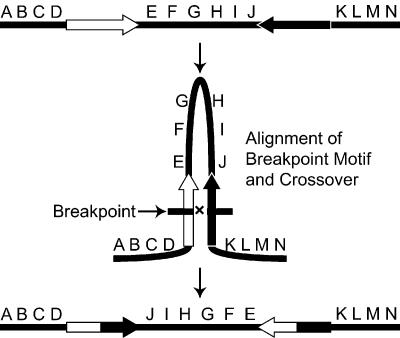
Previous investigations have identified specific cases of transposons and repetitive sequences at inversion breakpoints (Lyttle and Haymer 1992; Caceres et al. 1999; Mathiopoulos et al. 1999; Evgen'ev et al. 2000; Casals et al. 2003), and in other cases, repetitive elements have not been seen at inversion breakpoints (Wesley and Eanes 1994; Cirera et al. 1995). This study provides evidence that repetitive sequences can effect rearrangements on the genome scale, and may be the cause of the majority of inversions. Several pieces of evidence are consistent with the breakpoint motif being causal in the generation of chromosomal rearrangements in the D. pseudoobscura lineage. The breakpoint motifs at opposite ends of the Arrowhead inversion are in reverse orientation, consistent with a mechanism where ectopic exchange generates an inversion event (Fig. 8). The conserved sequence motif is virtually absent from intron and coding sequences. This suggests that strong purifying selection has acted to prevent the accumulation of this sequence within introns. An alternative explanation is that intron sequences are inaccessible either because of a nucleotide composition unfavorable for motif insertion, or because the introns are in an unfavorable location with respect to chromatin structure. If the conserved motif serves as the target for rearrangements, then inversions that use elements within a gene would cause loss-of-function mutations that would be quickly removed from populations (Charlesworth et al. 1992). Repeated sequences have also been detected at conserved linkage breakpoints among trypanosome species (Ghedin et al. 2004). The analysis of repeat structures within breakpoints should be viewed with caution. Each breakpoint sequence should be viewed as a composite of repetitive sequences. The breakpoint motif represents the largest family of repeats within the D. pseudoobscura genome detected to date, but other repeats within junctions of conserved lineage may contribute to the process of genomic rearrangement.
Figure 8.
Mechanism for chromosomal inversion with a repeated sequence motif. A hypothetical chromosome is shown with genes A through N and two repeated sequence motifs (open and black arrows) in a reverse orientation (top). Repeated motifs are shown pairing during meiosis with a recombination event occurring in the middle of the paired motifs (middle). Resolution of the recombination event between the repeated sequence motifs leading to the inversion of the central gene region (bottom).
One problem with the high frequency of the breakpoint motif is that ectopic exchange between elements in the same orientation would lead to deletion mutations. The lack of purifying selection on the breakpoint motif has allowed for its rapid decay through the accumulation of nucleotide and indel substitutions. These data are consistent with the “dead-on-arrival” elements of Drosophila virilis that preferentially delete sequence (Petrov et al. 1996; Petrov and Hartl 1998). As a consequence, few intact elements are capable of ectopic exchange. Molecular evolutionary studies of homologous breakpoint motifs will be necessary to test the element degradation hypothesis. The conclusion that the conserved sequence element causes paracentric inversions should be tempered as other possible explanations for the coincidence of the breakpoint repeat and inversion breakpoint may exist.
One can speculate about why breakpoint repeat elements are found only in D. pseudoobscura. Perhaps a new repetitive DNA element has been introduced in the obscura group lineage. Drosophila subobscura is a close relative of D. pseudoobscura, and five of the six chromosomal elements are segregating for paracentric inversions in European populations (Krimbas 1992). It will be interesting to determine if the repeat motif is present at the breakpoints of D. subobscura rearrangements.
The distribution of the breakpoint motif was not restricted to the D. pseudoobscura chromosomes with the major rearrangement polymorphisms. The genome-wide distribution of this repetitive element suggests that all chromosomes are capable of rearrangements, but has no bearing on the fixation of such inversions in the population. Rare inversions have been described on the other chromosomal arms both within D. pseudoobscura (Dobzhansky 1944) and between D. pseudoobscura and D. persimilis (Tan 1935).
Fixed inversion differences between the species may play a significant role in the formation of new species because inversions prevent the spread of incompatibility genes between different chromosomal backgrounds (Noor et al. 2001; Navarro and Barton 2003). By reducing rates of crossover, chromosomal inversions act as a barrier to gene flow, allowing Dobzhansky-Muller incompatibility genes to be fixed in different gene arrangement backgrounds, greatly enhancing the possibility of speciation (Noor et al. 2001; Navarro and Barton 2003). In Drosophila, hybrid male sterility genes appear to be involved in the process of speciation. In fact, we find that D. pseudoobscura genes with testis expression show a significant decrease in identity with their D. melanogaster orthologs. It will be interesting to determine if genes within inverted regions, and particularly those with male-specific expression, are associated with the sterility of male hybrids of D. pseudoobscura and D. persimilis.
Conservation of known cis-regulatory regions
D. pseudoobscura was chosen as the second fly species to be sequenced in part because it appeared to have the appropriate degree of sequence divergence from D. melanogaster to locate cis-regulatory sequences (Bergman et al. 2002). We were somewhat surprised at the overall low level of conservation of known cis-regulatory regions. Bergman et al. (2002) used clusters of these conserved noncoding sequences to identify enhancer sequences in the apterous gene. However, when known regulatory regions are examined, the conservation signal is not striking. Others have come to a similar conclusion using different alignment methods (Emberly et al. 2003). Alignment of Caenorhabditis elegans and Caenorhabditis briggsae has also suggested that many conserved noncoding regions will not be due to cis-regulatory sequences, increasing the noise in the conservation signal of these elements (Stein et al. 2003). Alignments of additional species of intermediate divergence may improve the detection of known regulatory elements as in Kellis et al. (2003), assuming the elements are conserved.
The lack of a clear conservation of cis-regulatory sequences suggests that simple models of sequence divergence in regulatory regions may be naive. Ludwig et al. (1998) observed that the D. pseudoobscura eve stripe 2 enhancer was functional in D. melanogaster despite significant differences between the regulatory protein-binding sites. In contrast, chimeric eve stripe 2 promoters had improper expression patterns, suggesting that stabilizing selection was acting on the enhancer (Ludwig et al. 2000), where “...selection can maintain functional conservation of gene expression for long periods of evolutionary time despite binding site turnover.” The D. pseudoobscura transcription factor proteins are 17% diverged from their D. melanogaster orthologs (Supplemental Fig. S5), different enough to allow variation of binding specificity. Evidence of cis-regulatory binding site conservation is encouraging; however, it is clear the D. pseudoobscura–D. melanogaster sequence comparisons will not identify binding sites alone. Instead, approaches like phylogenetic shadowing (Boffelli et al. 2003) that make use of a multiple alignment with species of intermediate divergence show more promise, owing to the reduced chance of binding site turnover between more recently diverged species. A recent paper (Berman et al. 2004) suggests the identification of binding-site clusters to reduce false positives when identifying regulatory regions.
Impact of the D. pseudoobscura sequence on the D. melanogaster annotation
The sequence of D. pseudoobscura will have a substantial impact on gene predictions in other species, most notably D. melanogaster. This will include direct conservation evidence for the validity of current predictions, modification of predictions based on conserved sequences with hallmarks of open reading frames such as third position variation, and support for ab initio predictions, which in previous annotation efforts were rejected as being too unreliable (Misra et al. 2002).
The first of these benefits from comparative analysis between these two flies, is the additional supporting evidence for the current D. melanogaster gene model set. FlyBase uses a simple confidence scoring system in which one point is given for a gene prediction being supported by at least one instance of each of the following four sources of evidence: full-length cDNA sequence, EST sequence, similarities to known proteins, and ab initio predictions. Thus, the most evidence-based gene models are assigned 4 points and the weakest accepted gene models have 1 (typically an ab initio prediction) (Misra et al. 2002). Of the 13,329 D. melanogaster Release_3.1 genes that were analyzed here and that remain in FlyBase, 11,989 have a putative ortholog in D. pseudoobscura (using a less stringent definition not requiring a gene model in D. pseudoobscura sequence). Table 3 describes the distribution of these putative orthologs by confidence value category. For the 1164 least supported gene models (confidence value of 1), 80.5% now have support based on orthology. For the confidence value 2 and 3 groups, the majority of gene models lack full-length cDNA evidence, and hence the conservations between the D. melanogaster and pseudoobscura genes are likely to permit significant improvement in the details of the gene models. Even for the most fully supported gene models (confidence value of 4), the conservation between D. melanogaster and pseudoobscura is likely to aid in the identification of the actual translation start and of alternative coding exons.
Table 3.
Number of D. melanogaster gene models having D. pseudoobscura orthologs
| Confidence value | D. metanogaster gene models | D. melanogaster gene models with D. pseudoobscura orthologsa (%) |
|---|---|---|
| 1 | 1194 | 962 (80.5) |
| 2 | 1961 | 1614 (82.3) |
| 3 | 2422 | 2137 (88.2) |
| 4 | 7752 | 7276 (93.8) |
| Total | 13,329 | 11,989 (89.9) |
Using a relaxed definition of orthology, not requiring a D. pseudoobscura gene prediction
Finally, it should be noted that identification of putative orthologs is 89.9% overall, and as expected goes up from 80.5% for the predictions with the least supporting evidence to 93.8% for those with the most. Hence, the majority of even D. melanogaster ab initio predictions accepted as valid gene models by FlyBase are likely to represent real expressed genes.
Methods
Strain selection
The availability of an isogenic or highly inbred strain is a critical factor to simplify the whole genome shotgun assembly problem. As the required balancer chromosomes are not available in D. pseudoobscura to produce an isogenic strain, an inbred strain was used. The sequenced strain was derived from a Mesa Verde, Colorado isolate collected in 1996 (W. Anderson, unpubl.). A population cage was set up in 1997 from eight iso-female lines. After ∼50 generations, inbred lines were established using a single virgin male and female for each line. A single brother–sister inbreeding procedure was repeated for an additional 14 generations, when a single line (MV-25) was selected on the basis of its viability. Cytological examination of the larval polytene chromosomes confirmed both the identity of the species and that the stock was homozygous for the Arrowhead inversion on the third chromosome. To avoid contamination with DNA from gut contents, and possible issues of unequal representation of the genome in larval polytene chromosomes, embryos were used for the isolation of genomic DNA for sequencing library production. The sequencing strain is available from the Tucson Drosophila Species Stock Center.
Library production
High-molecular-weight Genomic DNA was isolated from purified embryonic nuclei. pUC18 subclone libraries were constructed as described previously (Andersson et al. 1996). A BAC library (CHORI-226) and fosmid library (CHORI-1226) were prepared by and are available from BACPAC resources (Oakland, CA; http://bacpac.chori.org/). These large insert libraries were constructed from the same inbred MV-25 strain that was used for the preparation of the subclone libraries.
Sequencing and assembly
A total of 2.6 million high-quality sequence reads were produced from WGS sequencing libraries of ∼3 and 6 kb in pUC18 sub-clones, as well as additional reads from fosmids (40 kb) and BACs (130 kb) (Supplemental Table S1). DNA sequencing reactions were performed using BigDye version 3.1 (Applied Biosystems), and analyzed on ABI 3700 sequencing machines. These reads were assembled using the Atlas suite of assembly tools (Havlak et al. 2004). The Atlas suite identifies relatively small groups of reads that contain sequence overlap, assembles these groups individually, and uses paired-end information to join the resulting contigs into large scaffolds. All 2.6 million sequence reads were compared to each other for overlap using Atlas-overlapper. Putative overlaps were confirmed by banded dynamic programming alignment around the seed overlaps. Groups of sequence reads were then selected for local assembly by analysis of the sequence read overlaps using Atlas-binner, and individual assemblies were performed on the BCM-HGSC computer cluster. Paired-end sequence information was used with the Atlas-scaffolder program to generate larger scaffolds. This approach generated scaffolds with an N50 of 0.995 Mb and contigs within those scaffolds with an N50 of 51 kb (Supplemental Table S2). The total length of the sequence contained in scaffolds of this main assembly is ∼136 Mb. A complete description of the assembly process will be described elsewhere.
Certain sequence reads were resistant to assembly using this approach. Some lacked sufficient sequence overlap to be placed into a contig. Other sequence reads overlapped too many other sequences, and were assembled in a high-stringency repeat assembly. “Reptigs” from this repeat assembly were integrated into the main assembly on the basis of paired end sequence information. All of the sequence data are available on the BCM-HGSC Web site (http://hgsc.bcm.tmc.edu/projects/drosophila/). The annotated whole genome project has also been deposited into DDBJ/EMBL/GenBank under the project accession AADE00000000. The version described in this paper is the first version, AADE01000000.
Polymorphic sequence
Despite multiple generations of inbreeding the strain of D. pseudoobscura selected for sequencing still displayed a low level of sequence polymorphism. Because DNA was isolated from multiple D. pseudoobscura embryos, the assembly contained polymorphic sequences that do not assemble because of the presence of high-quality discrepancies including insertions and deletions. The sequence identity of these polymorphic regions ranges from 92% to 98% in short regions preventing assembly. In all, 6.4 Mb of sequence overlapped but did not assemble because of the presence of these discrepancies. It is possible to determine that a particular region contains strain polymorphisms, as opposed to repetitive regions in the genome, by using measures of sequence coverage and careful analysis of paired end sequence information. In cases in which similar sequences were identified in the assembly with high-quality discrepancies, further analysis suggested that the cause was polymorphism within the strain. These sequence reads were removed from the assembly, so that a single version of the polymorphic sequence was retained in the assembly. The fewest possible reads were removed that allowed proper assembly of the region, in order to keep the quality of the assembled sequence as high as possible. The sequence reads containing the putative polymorphisms are available from the BCM-HGSC Web site.
Anchoring sequence scaffolds to chromosomes
The lack of fine-resolution genetic maps, STS markers, and cytologically mapped sequences made the anchoring of draft sequences to the cytogenetic map a significant challenge. The genome of D. pseudoobscura is comprised of five chromosome arms (XL, XR, 2, 3, 4) and a dot chromosome (5). D. melanogaster has the same chromosome arm numerology although the arms are organized differently into chromosomes, presumably reflecting an evolutionary process of Robertsonian fusions and fissions. It has been recognized that within the genus Drosophila, there is a very strong tendency for the genes to remain on a single chromosome arm, although the relative gene order on the arm can be quite different (Segarra et al. 1995; Ranz et al. 2001). Drosophila evolutionary geneticists have thus identified six “Muller elements,” designated A–F to describe the conserved euchromatic chromosome arms. The correspondence between D. pseudoobscura, D. melanogaster, and Muller element nomenclature is shown in Supplemental Table S3. Thus, given that the D. melanogaster sequences are known, the identification of orthologous genes can be used to assign large scaffolds to D. pseudoobscura chromosome arms via Muller element conservation. In the vast majority of cases, this led to an unambiguous chromosome assignment. Of 234 large scaffolds covering >90% of the sequence, four aligned to more than one chromosome. Further investigation revealed erroneous joins between contigs based on paired-end reads assembling into repeat regions. In these four cases, the scaffolds were split at the gap erroneously joined by the paired-end reads. Since erroneous joins are more likely to occur between chromosome arms than within one (as any one arm is only ∼20% of the genome), we believe there are very few, if any, erroneous contig joins in the final assembly.
Despite the Muller element conservation, the D. pseudoobscura and D. melanogaster genomes differ by a succession of intrachromosomal arm inversions, such that only small stretches of synteny exist between the two species (see syntenic map). Therefore, chromosome arm assignment via Muller element conservation does not inform order and orientation of genes within arms. The order and orientation of the sequence scaffolds were aided by additional BAC end sequencing. BAC libraries were screened to identify new clones extending the ends of sequence scaffolds (in particular, probes were made at the ends of all scaffolds >50 kb). Local synteny information from the comparison with D. melanogaster was used to extend scaffold groups if it was consistent with other information, such as single BAC end sequence reads. This anchoring information was confirmed by comparison to a recombination map produced from microsatellite markers in all scaffolds >100 kb. This anchoring procedure yielded 16 groups of ordered and oriented scaffolds (ultra-scaffolds), containing ∼90% of the D. pseudoobscura sequence and all scaffolds >100 kb. Chromosomes 2 and 3 are represented by a single group of ordered and oriented scaffolds. The remaining three chromosome arms have a small number of groups: XL, four groups; XR, five groups; and Chromosome 4, five groups. Although scaffolds can be identified as belonging to the dot Chromosome 5, there was not enough data to reliably order and orient scaffolds on this chromosome. It is not surprising that less contiguity was obtained with XL and XR. Because embryos of both sexes (XX female and XY male) were used for the DNA preparation, the X portion of the WGS coverage is at 75% the coverage of the autosomes. The Y-chromosome sequence is the topic of a separate paper (Carvalho and Clark 2004).
Sequence quality assessment
We assessed the quality of the draft sequence at three levels of detail. At the finest level, the draft genome sequence was compared to 0.5 Mb of finished D. pseudoobscura sequences. Gap closure confirmed that the order and orientation of all 19 contigs in the finished part of this scaffold was correct, and estimates of gap size in the draft sequence were within expectations based on variation of subclone insert size. The only discrepancies observed in the alignment of the draft and finished sequences were at the single base level: a total of 13 mismatches were observed, an error rate of 0.26 × 10-4 per base. Thus the overall quality within the contigs of the draft sequence is comparable to that required for the finished human genome sequence (Felsenfeld et al. 1999).
To validate the assembly at the contig level, ten fosmids (GenBank accessions AC134177, AC131961, AC131959, AC131960, AC134174, AC132213, AC134175, AC132164, AC132165, AC132166) from a different D. pseudoobscura strain (Tucson Drosophila Species stock center strain 14011-0121.4, collected from Death Valley, CA; note that the assembly only contains Fosmid and BAC end sequences from libraries made from the same DNA as the plasmid libraries) were sequenced to the high-quality finished grade. Misassemblies at the contig level are observable when a portion of the contig aligns to the finished sequence, but the remaining contig sequence does not. Of 17 contigs aligned to the finished fosmids, no such cases were observed. In all cases, the order and orientation of the contigs were confirmed by comparison to the finished sequence. The alignment revealed 15 minor discrepancies between the assembly and the finished fosmid sequences, all of which were small insertion/deletions between 30 and 300 bp in length. These differences are likely due to sequence polymorphisms between the sequenced strain and the Death Valley strain from which the fosmid library was derived.
Genome alignment
We produced several alignments of D. pseudoobscura and D. melanogaster genomic sequences with BLASTZ (Schwartz et al. 2003), PARAGON (O. Couronne, unpubl.), and the local/global technique of the Berkeley Genome Pipeline (Couronne et al. 2003) with both AVID (Bray et al. 2003) and LAGAN (Brudno et al. 2003) global alignment programs. The AVID and LAGAN alignments are viewable via the VISTA Genome Browser at http://pipeline.lbl.gov/pseudo/. All these programs provide reasonable alignments of the two genomes. A higher-accuracy alignment was constructed based on the high-quality manually curated gene ortholog list. In theory, the high-quality alignments generated in the ortholog regions would provide anchor points for the genome alignment, and in the case of specific genes, we could be more certain that the orthologous sequences were being aligned. Two approaches to incorporating the curated ortholog data into the BLASTZ alignment were taken. The first was a filtering method: in cases in which conflicting high-scoring pairs (HSPs) overlapped a gene with a known ortholog, HSPs not in agreement with the known orthologous sequence were filtered out. The second was an “align and extend” method in which BLASTZ alignments were generated first around the known orthologous sequences and then extended as far as possible. The filtered BLASTZ alignment was the one used for the remainder of the analyses and is available at http://www.hgsc.bcm.tmc.edu/projects/drosophila/.
Gene prediction
In D. melanogaster, the majority of gene predictions are based on either full-length cDNA sequences or expressed sequence tag (EST) sequences (Misra et al. 2002) creating accurate gene models. Such a comprehensive biological data set is not available for D. pseudoobscura, thus a gene prediction method was designed to take advantage of the D. melanogaster annotations. Three gene prediction algorithms were used, GENSCAN (Burge and Karlin 1997), TWINSCAN (Korf et al. 2001), and GeneWise (Birney and Durbin 2000). These three were chosen because of their differing reliance on comparative sequence data. GENSCAN requires no input other than genomic sequence. TWINSCAN is based on GENSCAN, but makes use of a BLASTN comparison of the target sequence to a related sequence (in this case, D. melanogaster) to improve accuracy. GeneWise uses protein sequences from D. melanogaster as one of its inputs. A total of 48,000 gene predictions were produced by these three programs. The best predictions in this overlapping set were identified on the basis of similarity to D. melanogaster expressed sequence-based gene models—a similar process has been used for C. briggsae (Stein et al. 2003). Comparison of these gene predictions with D. melanogaster protein sequences was accomplished with the reciprocal use of BLASTP. D. pseudoobscura gene models with no similarity to any D. melanogaster protein sequence were removed. This reduced the number of independent gene predictions to 10,987. This set of gene predictions was further filtered for orthologous gene pairs using our synteny map to a total of 10,516 gene predictions. These 10,516 gene predictions were further screened for good gene models to annotate the D. pseudoobscura genome.
Identification of the Standard to Arrowhead breakpoint
Genomic DNA was prepared from D. pseudoobscura strains that were isochromosomal either for Arrowhead or Standard gene arrangements (Schaeffer et al. 2003). Four oligonucleotide primers were designed to amplify sequences that straddle the inversion breakpoints of the event that converted the Standard arrangement into the Arrowhead arrangement, a (5′-TCCTTGGAGCTGGTCTCGGA-3′), b (5′-CCAGAGGTAGTCGCAGTATG-3′), c (5′-TGGTGCGCTGCTGGTAGACA-3′), and d (5′-GCTGTGTCCTCGTTGTAGTC-3′). The following PCR conditions were used to amplify the proximal and distal breakpoints in the ancestral (Standard) and derived (Arrowhead) gene arrangements: 5.0 min at 94°C for 1 cycle; denature for 1.0 min at 94°C, anneal for 1.0 min at 65°C, extension for 2.0 min at 72°C for 30 cycles. The sequences of the proximal and distal Arrowhead breakpoints have been deposited in GenBank (accession nos. AY693425 and AY693426).
The proximal and distal breakpoints were compared using dot-plots and local alignment algorithms within the MEGALIGN program in the LASERGENE suite of DNA analysis software (DNAStar). In addition, BLAST analysis of the proximal and distal breakpoints was used to find regions that matched interspecific breakpoints in the genome.
Breakpoint sequence analysis
The conserved linkage groups and the breaks between groups were inferred from the orthologous gene calls between the two species in our synteny map. The junctions between homologous conserved linkage groups were assumed to be rearrangement breakpoints. The D. pseudoobscura breakpoints are inferred based on the ordered D. pseudoobscura sequence compared to the rearranged D. melanogaster sequence and the D. melanogaster breakpoints are inferred by the inverse process. The conserved linkage blocks within the scaffolds of each Muller element were numbered sequentially as a method of bookkeeping. Breakpoint sequences were extracted from scaffold sequences by taking the nucleotides defined by the end of one conserved linkage block and the beginning of the adjacent conserved linkage block. The breakpoint sequences were each labeled BP plus the left- and rightmost conserved linkage group numbers and the appropriate Muller element (Supplemental Table S9). For instance, BP_167_168_C is the breakpoint sequence at the boundary of conserved linkage groups 167 and 168 on Muller element C. The coordinates of all inferred interspecific breakpoints are shown in Supplemental Table S9. BLASTN (Altschul et al. 1990) was used to test each conserved linkage breakpoint for similarity to other breakpoint regions on the same chromosomal element. An E-value of 1 × 10-5 was used as the cutoff for the BLAST searches because the expected probability of a match given the size of the breakpoint database is <0.05. The breakpoint match distribution was determined by estimating the fraction of breakpoint sequences that each breakpoint matched on the chromosome. The number of interbreakpoint matches for an interspecific breakpoint are presented in Supplemental Table S5, and the distribution of interbreakpoint match fraction is shown in Supplemental Figure S8.
The discovery of a conserved repetitive element among breakpoints leads us to ask what the distribution of this sequence was in the genome. Sequences on each chromosomal element were partitioned into one of three categories, breakpoints, coding sequences, and noncoding/nonbreakpoint sequences. GeneWise (Birney and Durbin 2000) predictions (see below) of coding sequence locations in D. pseudoobscura were used to define a nonredundant set of coding regions for each chromosomal element. In cases in which two transcripts of the same gene or different genes overlapped, the intersection of the two transcripts was used to define a single coding segment of DNA. The noncoding/nonbreakpoint database was defined as all of the remaining sequences not found in either coding or breakpoint sequences. The frequency of conserved breakpoint motifs present in the three classes of sequence were determined with a BLASTN search of the three concatenated sequence databases using an E-value of 1 × 10-5. Seven conserved linkage breakpoints (BP_007_008_A, BP_081_082_B, BP_202_203_C, BP_104_105_D, BP_201_202_E, proximal and distal Arrowhead inversion breakpoints) were each used as query sequences for the BLASTN search for all chromosomal arms to detect the conserved motif in coding, noncoding, and breakpoint sequences. Multiple query sequences were used to survey the genome for the conserved breakpoint motif because the breakpoint sequence motifs are heterogeneous among breakpoints. We chose the breakpoint sequence from each of the five major chromosomal arms that had the maximum number of interbreakpoint matches as well as the proximal and distal Arrowhead inversion breakpoints. χ2 tests of homogeneity were used to determine if the distribution of the breakpoint motif was similar in the different classes of sequences. Supplemental Table S9 also indicates the presence and absence of the breakpoint motif sequence in each interspecific breakpoint.
Phylogenetic analysis of conserved sequence motifs
We wanted to determine the phylogenetic relationships among the breakpoint motifs to determine if copies from the same chromosome or local region were monophyletic. The breakpoint motifs are similar in sequence, but the different copies in the genome vary in length. We wanted to maximize the number of motifs and the number of base pairs in the phylogenetic analysis. Increasing the number of copies led to a reduction in the number of alignment bases, while increasing the number of aligned bases reduced the sample size. We chose to use a 443-bp motif sequence because this size provided an adequate sample of motif sequences from the major chromosomes, maximized the number of aligned bases, and tended to include adjacent copies of repeat 1 (128 bp) and repeat 2 (315 bp). A phylogenetic analysis was used to infer the relationships among the elements found in different categories of sequences and among different Muller elements. A total of 91 motifs that varied in length from 296 to 446 bp were aligned with CLUSTALX (Higgins et al. 1992), where 22, 3, and 66 sequences were found in breakpoints, coding regions, and noncoding regions, respectively. The 91 sequences shared the same 5′-end, but differed in their 3′-ends. The Kimura 2 parameter model was used to estimate the pairwise distances among breakpoint motifs because there is a slight bias in favor of transversions versus transitions. Deletions were excluded for pairwise estimates of distances. Phylogenetic trees of the breakpoint motifs were inferred with the neighbor-joining and maximum parsimony algorithms (Saitou and Nei 1987) as implemented in the Molecular Evolutionary Genetic Analysis package (Kumar et al. 2001).
Inference of positive selection from radical versus conservative amino acid substitutions
Each amino acid substitution was identified as being either a radical or a conservative change based on charge or on polarity, as defined by Zhang (2000). We tested for an excess of radical versus conservative substitutions using a likelihood model. A continuous time Markov chain was defined on the state space of all 20 amino acids and with transition rates derived from a codon-based model taking into account the observed codon frequencies and a transition/transversion bias as in Yang et al. (1998). Letting uij be the transition rate from amino acid i to amino acid j in the Yang et al. (1998) model, we defined the transition rates as
 |
The parameter α then measures the relative increase or decrease in the rate of evolution of radical amino acid substitutions compared to conservative amino acid substitutions. The maximum likelihood value assuming H0: α = 1, was compared to the maximum likelihood value assuming α ∈ [1, ∞). A p-value (p) calculated assuming two times the log likelihood ratio was distributed as a mixture between a  -distribution and a point mass at zero. Because the likelihood surface in a general model in which α ∈ [0, ∞) always had a single mode, maximum likelihood estimates of
-distribution and a point mass at zero. Because the likelihood surface in a general model in which α ∈ [0, ∞) always had a single mode, maximum likelihood estimates of  in this model imply that p = 1.
in this model imply that p = 1.
Acknowledgments
We thank Hugh Robertson for incisive comments and discussions. The sequencing of D. pseudoobscura was supported by NIH grant 1U01 HG02570 to R.G. Members of FlyBase were supported by Grant Numbers 5 P41 HG00739 and 5 R37 GM28669 from the National Institutes of Health (PHS). M.F.vB. was supported by grant BMI-050.50.201 from the Netherlands Organization for Scientific Research (NWO). H.J.B. was partly supported by NIH grant LM007276. A.G.C. was supported by NIH grants AI-45402 and GM-64590. K.T. was supported by an Alfred P. Sloan fellowship in Computational Biology. Finally, we thank the Center for Genomic Research at Harvard University for the use of computing resources, and Pieter J. de Jong and Kazutoyo Osoegawa of BAC-PAC at the Children's Hospital Oakland Research Institute for the construction of BAC and Fosmid libraries.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3059305. Freely available online through the Genome Research Immediate Open Access option.
Footnotes
[Supplemental material is available online at www.genome.org. The annotated whole genome project has been deposited into DDBJ/EMBL/GenBank under the project accession AADE00000000. The version described in this paper is the first version, AADE01000000. The sequences of the proximal and distal Arrowhead breakpoints have been deposited in GenBank with accession nos. AY693425 and AY693426. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: P.J. de Jong and K. Osoegawa.]
To the best of our knowledge, a comprehensive curated collection of experimentally determined cis-regulatory element information does not exist at the present time; such a resource would be of great value for analyses such as this.
The magnitude of the p-values is in part a function of the different set sizes and should not be viewed as an estimate of the magnitude of the effect.
This figure corresponds to 51.3% identity for all sites, because ∼70% of sites are at least partially aligned.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Andersson, B., Wentland, M.A., Ricafrente, J.Y., Liu, W., and Gibbs, R.A. 1996. A “double adaptor” method for improved shotgun library construction. Anal. Biochem. 236: 107-113. [DOI] [PubMed] [Google Scholar]
- Beckenbach, A.T., Wei, Y.W., and Liu, H. 1993. Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Mol. Biol. Evol. 10: 619-634. [DOI] [PubMed] [Google Scholar]
- Bergman, C.M., Pfeiffer, B.D., Rincon-Limas, D.E., Hoskins, R.A., Gnirke, A., Mungall, C.J., Wang, A.M., Kronmiller, B., Pacleb, J.M., Park, S., et al. 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3: RESEARCH0086. [DOI] [PMC free article] [PubMed]
- Berman, B.P., Pfeiffer, B.D., Laverty, T.R., Salzberg, S.L., Rubin, G.M., Eisen, M.B., and Celniker, S.E. 2004. Computational identification of developmental enhancers: Conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran, E., Thornton, K., and Long, M. 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. and Durbin, R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10: 547-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.K., Grimaila, R., Koehler, M.M., and Gelbart, W.M. 1987. Mobilization of hobo elements residing within the decapentaplegic gene complex: Suggestion of a new hybrid dysgenesis system in Drosophila melanogaster. Cell 49: 497-505. [DOI] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. [DOI] [PubMed] [Google Scholar]
- Bray, N., Dubchak, I., and Pachter, L. 2003. AVID: A global alignment program. Genome Res. 13: 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., and Batzoglou, S. 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Caceres, M., Ranz, J.M., Barbadilla, A., Long, M., and Ruiz, A. 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415-418. [DOI] [PubMed] [Google Scholar]
- Carvalho, A.B. and Clark, A.G. 2004. The Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science (in press). [DOI] [PubMed]
- Casals, F., Caceres, M., and Ruiz, A. 2003. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol. Biol. Evol. 20: 674-685. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed]
- Charlesworth, B. and Charlesworth, D. 1973. Selection of new inversion in multi-locus genetic systems. Genet. Res. 21: 167-183. [Google Scholar]
- Charlesworth, B., Lapid, A., and Canada, D. 1992. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. II. Inferences on the nature of selection against elements. Genet. Res. 60: 115-130. [DOI] [PubMed] [Google Scholar]
- Cirera, S., Martin-Campos, J.M., Segarra, C., and Aguade, M. 1995. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics 139: 321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, M. and Rubin, G.M. 1984. Structure of chromosomal rearrangements induced by the FB transposable element in Drosophila. Nature 308: 323-327. [DOI] [PubMed] [Google Scholar]
- Couronne, O., Poliakov, A., Bray, N., Ishkhanov, T., Ryaboy, D., Rubin, E., Pachter, L., and Dubchak, I. 2003. Strategies and tools for whole-genome alignments. Genome Res. 13: 73-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan, T., Talmor, Y., and Graur, D. 2002. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol. Biol. Evol. 19: 1022-1025. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T. 1944. Chromosomal races in Drosophila pseudoobscura and Drosophila persimilis. In Carnegie Institution of Washington Publication 554, pp. 47-144. Washington, DC.
- ———. 1948a. Genetics of natural populations XVI, altitudanal and seasonal changes in certain populations of Drosophila pseudoobscura and Drosophila persimilis. Genetics 33: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1948b. Genetics of natural populations. XVIII. Experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics 33: 588-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1949. Observations and experiments on natural selection in Drosophila. In Proc. Int. Congress. Genet., pp. 210-224.
- Dobzhansky, T. and Epling, C. 1944. Contributions to the genetics, taxonomy and ecology of Drosophila pseudoobscura and its relatives. In Carnegie Institution of Washington Publication 554, pp. 1-46. Washington, DC.
- Emberly, E.G., Rajewsky, N., and Siggia, E.D. 2003. Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics 4: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W.R. and Preston, C.R. 1984. Formation of chromosome rearrangements by P factors in Drosophila. Genetics 107: 657-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen'ev, M.B., Zelentsova, H., Poluectova, H., Lyozin, G.T., Veleikodvorskaja, V., Pyatkov, K.I., Zhivotovsky, L.A., and Kidwell, M.G. 2000. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc. Natl. Acad. Sci. 97: 11337-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Felsenfeld, A., Peterson, J., Schloss, J., and Guyer, M. 1999. Assessing the quality of the DNA sequence from the Human Genome Project. Genome Res. 9: 1-4. [PubMed] [Google Scholar]
- Ghedin, E., Bringaud, F., Peterson, J., Myler, P., Berriman, M., Ivens, A., Andersson, B., Bontempi, E., Eisen, J., Angiuoli, S., et al. 2004. Gene synteny and evolution of genome architecture in trypanosomatids. Mol. Biochem. Parasitol. 134: 183-191. [DOI] [PubMed] [Google Scholar]
- Grad, Y.H., Roth, F.P., Halfon, M.S., and Church, G.M. 2004. Prediction of similarly-acting cis-regulatory modules by subsequence profiling and comparative genomics in D. melanogaster and D. pseudoobscura. Bioinformatics May 14 [Epub ahead of print]. [DOI] [PubMed]
- Havlak, P., Chen, R., Durbin, K.J., Egan, A., Ren, Y., Song, X.Z., Weinstock, G.M., and Gibbs, R.A. 2004. The Atlas genome assembly system. Genome Res. 14: 721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D.G., Bleasby, A.J., and Fuchs, R. 1992. CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. Biosci. 8: 189-191. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241-254. [DOI] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17 Suppl 1: S140-S148. [DOI] [PubMed] [Google Scholar]
- Krimbas, C.B. 1992. In Drosophila inversion polymorphism (eds. C.B. Krimbas and J.R. Powell), pp. 127-220. CRC Press, Boca Raton, FL.
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244-1245. [DOI] [PubMed] [Google Scholar]
- Ladeveze, V., Aulard, S., Chaminade, N., Periquet, G., and Lemeunier, F. 1998. Hobo transposons causing chromosomal breakpoints. Proc. R Soc. Lond. B Biol. Sci. 265: 1157-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier, F. and Ashburner, M.A. 1976. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc. R Soc. Lond. B Biol. Sci. 193: 275-294. [DOI] [PubMed] [Google Scholar]
- Lemeunier, F. and Aulard, S. 1992. Inversion polymorphism in Drosophila melanogaster. In Drosophila inversion polymorphism (eds. C.B. Krimbas and J.R. Powell), pp. 339-405. CRC Press, Boca Raton, FL.
- Lercher, M.J., Blumenthal, T., and Hurst, L.D. 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13: 238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J.K. 1988. Intrachromosomal rearrangements mediated by hobo transposons in Drosophila melanogaster. Proc. Natl. Acad. Sci. 85: 9153-9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M.Z., Patel, N.H., and Kreitman, M. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: Rules governing conservation and change. Development 125: 949-958. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.Z., Bergman, C., Patel, N.H., and Kreitman, M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564-567. [DOI] [PubMed] [Google Scholar]
- Lyttle, T.W. and Haymer, D.S. 1992. The role of the transposable element hobo in the origin of endemic inversions in wild populations of Drosophila melanogaster. Genetica 86: 113-126. [DOI] [PubMed] [Google Scholar]
- Mathiopoulos, K.D., della Torre, A., Santolamazza, F., Predazzi, V., Petrarca, V., and Coluzzi, M. 1999. Are chromosomal inversions induced by transposable elements? A paradigm from the malaria mosquito Anopheles gambiae. Parassitologia 41: 119-123. [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed]
- Muller, H.J. 1940. In The new systematics (ed. J. Huxley), pp. 185-268. Clarendon Press, Oxford, UK.
- Nadeau, J.H. and Taylor, B.A. 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 81: 814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A. and Barton, N.H. 2003. Accumulating postzygotic isolation genes in parapatry: A new twist on chromosomal speciation. Evol. Int. J. Org. Evol. 57: 447-459. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A., Makova, K.D., and Li, W.H. 2002. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 12: 198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M.A., Grams, K.L., Bertucci, L.A., and Reiland, J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. 98: 12084-12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski, E. 1946. Chromosomal variation in Drosophila athabasca. Genetics 31: 508-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1973. Ancient linkage groups and frozen accidents. Nature 244: 259-262. [DOI] [PubMed] [Google Scholar]
- Otto, S.P. and Barton, N.H. 2001. Selection for recombination in small populations. Evol. Int. J. Org. Evol. 55: 1921-1931. [DOI] [PubMed] [Google Scholar]
- Painter, T.S. 1934. A new method for the study of chromosomal aberrations and the plotting of chromosomal maps in Drosophila melanogaster. Genetics 19: 175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D.A. and Hartl, D.L. 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol. Biol. Evol. 15: 293-302. [DOI] [PubMed] [Google Scholar]
- Petrov, D.A., Lozovskaya, E.R., and Hartl, D.L. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346-349. [DOI] [PubMed] [Google Scholar]
- Pevzner, P. and Tesler, G. 2003. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 100: 7672-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, S.S. 1982. DNA sequence analysis of a Drosophila foldback transposable element rearrangement. Mol. Gen. Genet. 188: 107-110. [DOI] [PubMed] [Google Scholar]
- Ranz, J.M., Segarra, C., and Ruiz, A. 1997. Chromosomal homology and molecular organization of Muller's elements D and E in the Drosophila repleta species group. Genetics 145: 281-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J.M., Casals, F., and Ruiz, A. 2001. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 11: 230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W.R. 1989. Analyzing tables of statistical tests. Evolution 43: 223-225. [DOI] [PubMed] [Google Scholar]
- Roy, P.J., Stuart, J.M., Lund, J., and Kim, S.K. 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418: 975-979. [DOI] [PubMed] [Google Scholar]
- Russo, C.A., Takezaki, N., and Nei, M. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391-404. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- Schaeffer, S.W., Goetting-Minesky, M.P., Kovacevic, M., Peoples, J.R., Graybill, J.L., Miller, J.M., Kim, K., Nelson, J.G., and Anderson, W.W. 2003. Evolutionary genomics of inversions in Drosophila pseudoobscura: Evidence for epistasis. Proc. Natl. Acad. Sci. 100: 8319-8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, C., Lozovskaya, E.R., Ribo, G., Aguade, M., and Hartl, D.L. 1995. P1 clones from Drosophila melanogaster as markers to study the chromosomal evolution of Muller's A element in two species of the obscura group of Drosophila. Chromosoma 104: 129-136. [DOI] [PubMed] [Google Scholar]
- Sheen, F., Lim, J.K., and Simmons, M.J. 1993. Genetic instability in Drosophila melanogaster mediated by hobo transposable elements. Genetics 133: 315-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N.G. 2003. Are radical and conservative substitution rates useful statistics in molecular evolution? J. Mol. Evol. 57: 467-478. [DOI] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1981. Biometry. W.H. Freeman, New York.
- Spellman, P.T. and Rubin, G.M. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlich, D. and Pfriem, P. 1986. Chromosomal polymorphism in natural and experimental populations. In The genetics and biology of Drosophila (eds. M. Ashburner et al.), pp. 257-309. Academic Press, New York.
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A.H. and Beadle, G.W. 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 544-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A.H. and Dobzhansky, T. 1936. Inversions in the third chromosome of wild race of Drosophila pseudoobscura, and their use in the study of the history of the species. Proc. Natl. Acad. Sci. 22: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A.H. and Novitski, E. 1941. The homologies of the chromosome elements in the genus Drosophila. Genetics 26: 517-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A.H. and Tan, C.C. 1937. The comparative genetics of Drosophila pseudoobscura and D. melanogaster. J. Genet. 34: 415-432. [Google Scholar]
- Sun, F.L., Cuaycong, M.H., Craig, C.A., Wallrath, L.L., Locke, J., and Elgin, S.C. 2000. The fourth chromosome of Drosophila melanogaster: Interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. 97: 5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, C.P., Merz, T., and Young, W.J. 1981. Cytogenetics: The chromosome in division, inheritance and evolution. Prentice-Hall, Upper Saddle River, NJ.
- Tamura, K., Subramanian, S., and Kumar, S. 2003. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36-44. [DOI] [PubMed] [Google Scholar]
- Tan, C.C. 1935. Salivary gland chromosomes in the two races of Drosophila pseudoobscura. Genetics 20: 392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, C.S. and Eanes, W.F. 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. 91: 3132-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, J. and Hollocher, H. 2001. Mobile elements and the genesis of microsatellites in Dipterans. Mol. Biol. Evol. 18: 384-392. [DOI] [PubMed] [Google Scholar]
- Wright, S. and Dobzhansky, T. 1946. Genetics of natural populations. XII. Experimental reproduction of some of the changes caused by natural selection in certain populations of Drosophila pseudoobscura. Genetics 31: 125-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-I. and Beckenbach, A.T. 1983. Evidence for extensive genetic differentiation between the sex ratio and the standard arrangement of Drosophila pseudoobscura and D. persimilis and identification of hybrid sterility factors. Genetics 105: 71-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., and Hasegawa, M. 1998. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol. Biol. Evol. 15: 1600-1611. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
- Zhang, J. 2000. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 50: 56-68. [DOI] [PubMed] [Google Scholar]
Web site references
- http://bacpac.chori.org/; BACPAC resources.
- http://hgsc.bcm.tmc.edu/projects/drosophila/; BCM-HGSC.
- http://pipeline.lbl.gov/pseudo/; VISTA Genome Browser.