ABSTRACT
Cell differentiation is associated with changes in chromatin organization and gene expression. In this study, we examine chromatin structure following differentiation of the human myeloid leukemia cell line (HL-60/S4) into granulocytes with retinoic acid (RA) or into macrophage with phorbol ester (TPA). We performed ChIP-seq of histone H3 and its modifications, analyzing changes in nucleosome occupancy, nucleosome repeat length, eu-/heterochromatin redistribution and properties of epichromatin (surface chromatin adjacent to the nuclear envelope). Nucleosome positions changed genome-wide, exhibiting a specific class of alterations involving nucleosome loss in extended (∼1kb) regions, pronounced in enhancers and promoters. Genes that lost nucleosomes at their promoters showed a tendency to be upregulated. On the other hand, nucleosome gain did not show simple effects on transcript levels. The average genome-wide nucleosome repeat length (NRL) did not change significantly with differentiation. However, we detected an approximate 10 bp NRL decrease around the haematopoietic transcription factor (TF) PU.1 and the architectural protein CTCF, suggesting an effect on NRL proximal to TF binding sites. Nucleosome occupancy changed in regions associated with active promoters in differentiated cells, compared with untreated HL-60/S4 cells. Epichromatin regions revealed an increased GC content and high nucleosome density compared with surrounding chromatin. Epichromatin showed depletion of major histone modifications and revealed enrichment with PML body-associated genes. In general, chromatin changes during HL-60/S4 differentiation appeared to be more localized to regulatory regions, compared with genome-wide changes among diverse cell types studied elsewhere.
KEYWORDS: CTCF: Epichromatin, histone modifications, nucleosome positioning, nucleosome repeat length, transcription factor binding
Introduction
Cell differentiation is known to be accompanied by chromatin changes and, in particular, nucleosome repositioning,1-6 since only about 37.5% of human nucleosomes have DNA sequence-determined positions.7 The magnitude of chromatin changes and their effect on gene expression depend upon the particular biologic system. Here, we consider 2 differentiation pathways of the human cell line HL-60/S4.
The immortalized myeloid leukemia cell line (HL-60), isolated from a female patient, was originally described to be an acute promyelocytic leukemia (APL);8,9 but was subsequently reclassified as an acute myeloblastic leukemia (AML) with maturation.10 HL-60 cells are multipotential and can be readily differentiated into granulocytes with DMSO9 or retinoic acid (RA),10 into monocytes with vitamin D3,11,12 or into macrophage with phorbol ester (TPA).12,13 The multipotential character of HL-60 cells to differentiate along various myeloid directions was summarized in an early review by one of the original discoverers of this important cell line.14 The mutagenically-derived subline HL-60/S4 available through ATCC as #CRL-3306, exhibits distinctly faster differentiation than the parent HL-60 cell line.15 HL-60/S4 cells develop granulocytic nuclear segmentation in 4 d, whereas the parent HL-60 line requires at least 6 d for the same level of differentiation.16 We have previously studied HL-60/S4, exploring nuclear shape, chromatin structure and cytoskeletal changes during differentiation induced by RA, TPA and vitamin D3.17-23 Furthermore, we have recently introduced the concept of “epichromatin,” which involves a small percentage (∼5%) of the HL-60/S4 genome, is highly enriched with retrotransposon Alu, and is reproducibly located adjacent to the nuclear envelope in undifferentiated, RA and TPA treated HL-60/S4 cells.23 While being adjacent to the nuclear envelope, this type of regions should be distinguished from lamina-associated domains (LADs) defined elsewhere,24-26 because epichromatin regions are identified after short formaldehyde fixation times (“snapshots”); whereas LADs are defined after longer transfection times, yielding 30–40% of the genome. Most recently, we have also described the transcriptomes of the undifferentiated, granulocytic and macrophage forms of HL-60/S4.27
Differential gene expression, the basic mechanism underlying cell differentiation, depends upon a complex interplay between nucleosome positioning, histone and DNA covalent modifications and chromatin binding proteins.3,28 Integrating these various changes at individual promoters represents a current frontier in understanding the molecular biology of cell differentiation and cancer formation. In recent studies, epigenetic profiling has been performed in several blood cells types to study the differentiation-induced changes of the epigenome. For example, the BLUEPRINT consortium studied the differentiation of purified circulating monocytes from healthy volunteers into macrophages, and quantified this process in terms of the correlation of changes of H3K4me1, H3K4me3, H3K27ac, and DNase I accessibility with changes of gene expression.29 A more recent study has followed up this analysis, including 3 additional histone modifications and DNA methylation data, as well as performing NOME-seq to study the relation of nucleosome positioning with DNA methylation.5 However, the change of the chromatin density per se was not systematically analyzed in these works. Perhaps, one of the reasons for this is the intrinsic variability of nucleosome positions between healthy individuals – this problem can be overcome by using well-defined cell lines rather than primary cells. For example, in single-cell studies it is possible to distinguish quite nicely between HL-60 and GM12878 cells just by the characteristic differences of the chromatin accessibilities30
In the present study, we have performed ChIP-seq with antibodies against histone H3 and 6 histone H3 modifications, combining these data with the RNA-seq to characterize the differentiation-specific chromatin changes in the HL-60/S4 cell system.
Results
Nucleosome repositioning at promoters and enhancers
We have determined nucleosome occupancy profiles using Micrococcal Nuclease (MNase)-assisted ChIP-seq with H3 antibody in untreated, RA-treated and TPA-treated HL-60/S4 cells. Complex scenarios of nucleosome gain or loss and nucleosome repositioning take place upon HL-60/S4 cells differentiation into granulocyte (RA-treated) or macrophage (TPA-treated) cell forms. Using a “sliding window” approach with the help of NucTools software,31 we defined 1000-bp genomic regions which are characterized by significant loss or gain of nucleosome occupancy upon differentiation with RA or TPA (see Methods). We have identified 22,024 regions with nucleosome loss and 224,140 regions with nucleosome gain upon RA treatment. Similarly, there were 28,732 regions with nucleosome loss and 237,850 regions with nucleosome gain upon TPA treatment. This is different from the previous NOME-seq study in primary monocyte cells differentiating into macrophages, which claimed preferential nucleosome depletions upon differentiation.5 These data are not directly comparable since we were calling fixed-size 1000-bp windows of differential histone H3 occupancy, while the NOME-seq study was calling single-nucleosome peaks, but the difference is interesting anyway.
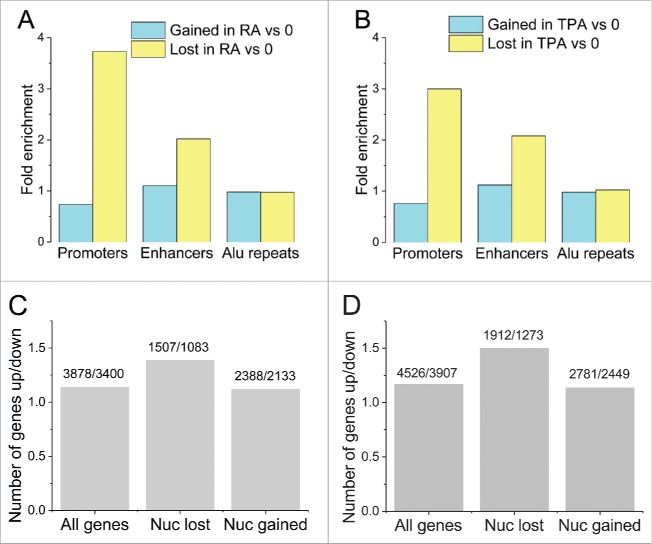
Figure 1A and B present the statistics of differential gain/loss of nucleosomes in 3 types of HL-60/S4 genomic regions (promoters, enhancers and retrotransposon Alu elements) as a consequence of RA or TPA treatment. Promoters and enhancers were enriched within regions that lost nucleosomes, but not enriched within regions that gained nucleosomes. Alu elements showed no significant gain or loss of nucleosomes. Promoters of upregulated genes that lost nucleosomes upon RA treatment were enriched within a very large number of KEGG pathways including among others “B-cell receptor signaling pathway” (P = 9.5e-5) and “T cell receptor signaling” (P = 1.4e-3). Promoters of downregulated genes that lost nucleosomes upon RA treatment were enriched only with “DNA replication” (P = 2.7e-3) and “mismatch repair” pathways (P = 6.6e-3). Promoters of upregulated genes that lost nucleosomes upon TPA treatment were most significantly enriched for “cGMP-PKG signaling” (P = 4.4e-5) and “cAMP signaling” pathways (P = 2.8e-3). Promoters of downregulated genes that lost nucleosomes upon TPA treatment were enriched with “propanoate metabolism” (P = 1.3e-4) and “butanoate metabolism” pathways (P = 6.8e-4).
Figure 1.
Statistics of the distribution of regions that gained or lost nucleosomes upon HL-60/S4 differentiation by RA (A) or TPA (B), among different functional elements (enhancers, promoters, Alu repeats). Bar color code: light blue, nucleosomes gained, relative to 0 (undifferentiated) cells; yellow, nucleosomes lost, relative to 0. Fold enrichment equal to 1.0 signifies that a given genomic feature exhibits no net change in nucleosome numbers. Following RA or TPA treatment, more nucleosomes are lost, than are gained, in promoters and enhancers; but this is not observed with Alu repeat sequences. (C-D) Relation between changes of gene expression and nucleosome repositioning upon RA treatment (C) and TPA treatment (D). “Number of genes up/down,” the number of up or downregulated genes for 3 groups: “All genes,” all significantly expressed genes; “Nuc lost,” genes that lost nucleosomes at their promoters; “Nuc gained,” genes that gained nucleosomes at their promoters. The group of genes that lost nucleosomes at their promoters contains a larger proportion of upregulated genes, than the group of genes that gained nucleosomes at the promoter.
A typical parameter of chromatin structural changes occurring during cell differentiation is “nucleosome occupancy,” which can be interpreted as the fraction of cells from the population in which a given region of DNA is occupied by a histone octamer.32 Figure 1C and D show that when comparing the numbers of up/downregulated genes with lost nucleosomes, the genes which have lost nucleosomes were preferentially upregulated (i.e., increased transcript levels) compared with those which gained nucleosomes or to all genes. In particular, the ratio of the number of upregulated versus downregulated genes was around 1.14, both for “all genes” and for genes that gained nucleosomes at their promoters. On the other hand, this ratio (up/down gene regulation) increased to 1.39 and 1.5 for genes that lost nucleosomes at their promoters upon treatment by RA and TPA, respectively. Fisher's exact test confirmed that nucleosome loss at promoters upon RA treatment correlates with a significant increase in the number of upregulated genes (χ2-test, P = 1.7e-5); but the difference for genes that gained nucleosomes was not significant (χ2-test, P = 0.64). The same effect was observed for TPA-treated genes.
Figure 2 presents nucleosome occupancy changes for specific genes belonging to haematopoietic differentiation pathways that have lost nucleosomes in promoter regions, comparing undifferentiated (O) to differentiated (RA and TPA) cell forms. This set of genes generally exhibited large increases in transcript levels after both RA and TPA treatment (with the exception of CD38, which showed a decrease after TPA). At our H3 ChIP-seq resolution, we mostly observe distinct changes that can be localized to regions enclosing 1–5 nucleosomes. The loss of nucleosomes and corresponding increase of gene expression is particularly apparent in the CD38 (RA), IRF7 (RA and TPA), EGR2 (RA) and EGR3 (RA and TPA) gene examples. On the other hand, nucleosome gain does not seem to correlate with changes of gene expression, as exemplified in Supplementary Figure S1. Thus, nucleosome loss in promoter regions, but not nucleosome gain, appears to be a particular functional mechanism during HL-60/S4 differentiation by RA and TPA.
Figure 2.
Examples of nucleosome repositioning in genes that exhibit significant changes in mRNA levels in the differentiated state, compared with the undifferentiated HL-60/S4 cell form. A blue-green bar under the X-axis shows the gene body and the direction of transcription is indicated by an arrow. Promoters are defined as (+/−1000 bp) centered around the TSS (Transcription Start Site). Plot line colors: O (undifferentiated), black; RA treated, red; TPA treated, blue. Images available in color online.
Nucleosome repeat length during cell differentiation
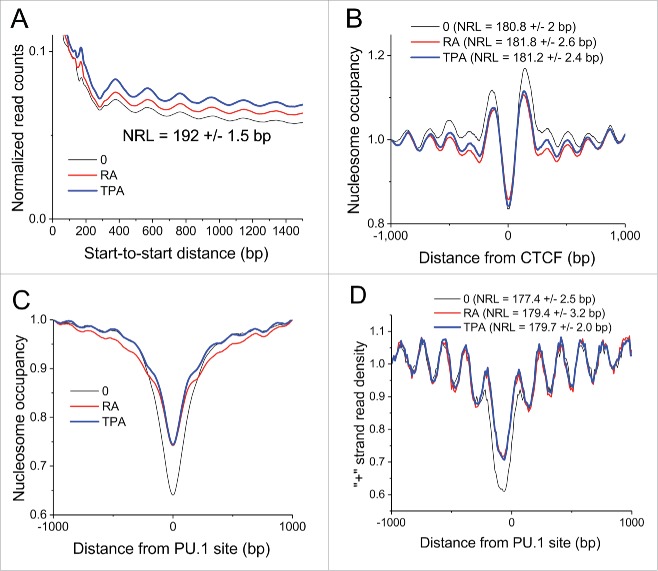
Global nucleosome repeat length (NRL) was calculated for undifferentiated, RA and TPA-treated HL-60/S4 cells, following the calculation procedure described previously.2,3,33 Fig. 3A demonstrates that there are no significant differences in global NRL (192 +/−1.5 bp) among the 3 cell states. This apparent constancy of NRL within the HL-60/S4 cell states is interesting, because, for example, the genome-wide NRL values measured for mouse embryonic stem cells differentiation varied by 5–7 bp.3
Figure 3.
Nucleosome repeat length (NRL) in different cell states and genomic regions. (A) Genome-wide average NRL is 192 bp without significant difference upon HL-60/S4 differentiation by RA or TPA. (B) Average nucleosome occupancy profile around bound CTCF. The corresponding NRL values are indicated on the figure. (C) Average nucleosome occupancy around bound PU.1 transcription factor. (D) The same as (C), but only the density of “+” strand reads is shown. The NRL determined for nucleosomes periodically arranged near PU.1 sites is indicated on this panel. The locations of CTCF and PU.1 binding sites were obtained from ENCODE ChIP-seq data on undifferentiated HL-60 cells; nucleosome positions were calculated from present data on undifferentiated and differentiated HL-60/S4 cells. Plot line colors: O (undifferentiated), black; RA treated, red; TPA treated, blue.
As pointed out in the earlier publications,3,33 specific genomic regions may display local variations and marked differences from the global NRL values. One such example is shown in Fig. 3B, which illustrates the global nucleosome occupancy surrounding CTCF binding sites in HL-60/S4 cells (undifferentiated, 180.8 +/−2.0 bp; RA, 181.8 +/−2.6 bp; TPA, 181.2 +/−2.4 bp), which is ∼10 bp smaller than the global average NRL. The center of the CTCF binding site exhibits a clear deficiency of bound nucleosomes, compared with the adjacent chromatin environment. Another specific example is the nucleosome occupancy around bound PU.1 transcription factors in the 3 HL-60/S4 cell states (Fig. 3C and D). The NRL values change by ∼2 bp during cell differentiation (undifferentiated, 177.4 +/−2.5 bp; RA, 179.4 +/−3.2 bp; TPA, 179.7 +/−2.0 bp). The chromatin region around bound PU.1 has ∼12 bp shorter NRL than the global NRL, and is also slightly smaller than that for CTCF. It remains to be demonstrated how general is the change in NRL around transcription factor (TF) binding sites, and whether such changes depend upon TF binding. In the examples above, we used CTCF and PU.1 binding sites determined by ChIP-seq in HL-60 cells by the ENCODE consortium.34
Nucleosome occupancy changes associated with histone modifications
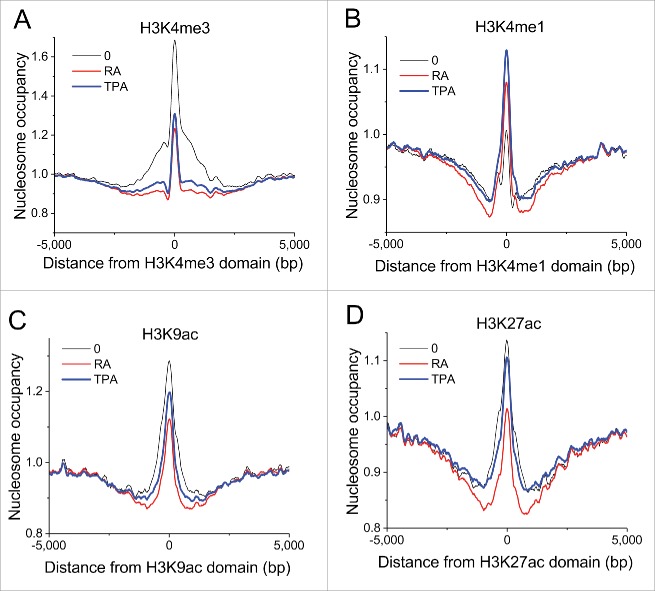
We have determined histone modifications using ChIP-seq in untreated, RA-treated and TPA-treated HL-60/S4 cells. As shown previously for other differentiation systems, nucleosome occupancy signatures of different histone modification domains vary significantly.3 The differentiation of HL-60/S4 is no exception. Figure 4 illustrates the nucleosome occupancy profiles of chromatin domains of 4 different histone modifications that are related to chromatin functions, comparing nucleosome occupancy in undifferentiated, RA- and TPA-differentiated cell states. Sites that were enriched with H3K4me3 (a marker for active promoters) in untreated cells reveal reduced (but still apparent) nucleosome occupancy in the center of their domain following treatment with RA or TPA (Fig. 4A). Sites that were enriched with H3K4me1, H3K9ac and H3K27ac in untreated cells reveal less dramatic changes in nucleosome occupancy following treatment (Fig. 4B, C and D). In contrast to activating marks, the sites enriched with H3K4me1 (which can be associated with repression)35 in untreated cells appear to show an increased nucleosome occupancy following either treatment. The finding that major decrease of the nucleosome density peak occurs at H3K4me3 sites in undifferentiated HL-60 cells is consistent with our statistics shown in Fig. 1A and B, which suggests significant nucleosome loss at some promoters upon HL-60/S4 differentiation.
Figure 4.
Nucleosome occupancies at different histone modification domains exhibit changes upon HL-60/S4 differentiation with RA or TPA. Panels: (A) H3K4me3; (B) H3K4me1; (C) H3K9ac; (D) H3K27ac. The largest changes occur in H3K4me3 domains, indicating major nucleosome loss at promoters upon HL-60/S4 differentiation (consistent with Fig. 1A and B). Coordinates of H3K4me1, H3K9ac and H3K27ac domains are obtained from the corresponding ChIP-seq experiments performed in this study. Coordinates of H3K4me3 domains are based upon ENCODE ChIP-seq data on undifferentiated HL-60 cells; nucleosome occupancy was calculated from present data on undifferentiated and differentiated HL-60/S4 cells. Plot line colors: O (undifferentiated), black; RA treated, red; TPA treated, blue.
Nucleosome features in HL-60/S4 epichromatin
Using the new data on nucleosome positioning and histone modifications during HL-60/S4 differentiation obtained in this study, we attempt to deepen our understanding of “epichromatin” regions, previously mapped in undifferentiated, RA and TPA treated HL-60/S4 cells.23 The concept of epichromatin (i.e., the surface of chromatin adjacent to the interphase nuclear envelope and at the “outer” surface of mitotic chromosomes) is based upon specifically localized immunostaining experiments.36,37 Two mouse monoclonal antibodies of divergent origin, PL2–638 and 1H6,39 yield identical staining patterns on fixed and permeabilized cells of considerable phylogenetic diversity, spanning from human to plant cells,36,37 implying the existence of a conserved chromatin epitope. Recently, 2 of us (ALO and DEO) enriched and characterized epichromatin from fixed and sonicated HL-60/S4 cells (0, RA and TPA cell states) by using a modified immunoprecipitation method (“xxChIP-seq”) using PL2–6 and 1H6.23 Some of the previously described characteristics of HL-60/S4 epichromatin regions include: 1) Representation of only ∼5% of the human genome. 2) Enrichment of retrotransposon Alu, ∼10-fold more concentrated than average for the human genome. 3) Epichromatin regions averaged ∼1 kb in size. 4) Epichromatin-containing DNA sequences, derived from 0, RA and TPA cells reacted with PL2–6 and 1H6 revealed a discontinuous distribution along each chromosome, which appeared to be relatively constant in chromosome location, comparing the 3 cell states. Still, the question of what exactly is epichromatin remains unanswered, and therefore we investigated the relationship between epichromatin regions and nucleosome occupancy changes during HL-60/S4 differentiation.
Since ChIP-seq is inherently a very noisy technique, we computed a list of common epichromatic regions, defined as the intersection of all replicate experiments between untreated and RA-treated cells. The intersection of 2 genomic regions is defined in the following way: 2 regions intersect if they share at least one base pair (based on their genomic coordinates). As input for these calculations, we took the coordinates of epichromatic regions for each replicate experiment reported earlier.23 Defined in this way, replicate PL2–6 immunoprecipitates of undifferentiated HL-60/S4 cells exhibited 40,288 epichromatin regions; replicate RA-treated cells, 77,897 regions. Defining only common regions based on all replicates, both antibodies (PL2–6 and 1H6) and 2 cell states (undifferentiated and RA-treated) resulted in 6541 common epichromatin regions.
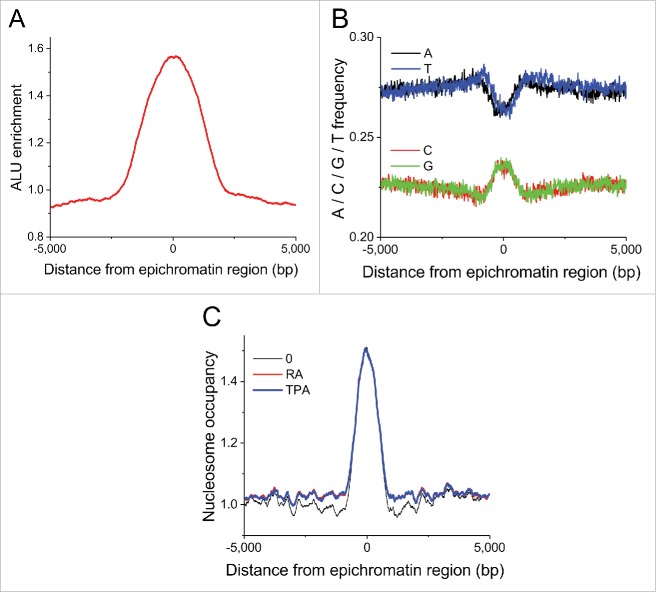
We have found the following 3 properties of the common HL-60/S4 epichromatin regions: (1) Alu enrichment, compared with average genomic background levels, is about 2-fold at the center of epichromatin, falling to background at +/−2.5 kb from the center of the distribution (Fig. 5A). (2) The average nucleotide frequency reveals increased GC nucleotide content at the center of epichromatin (Fig. 5B). (3) Nucleosome occupancy (i.e., histone H3 read density) in common epichromatin regions exhibits a similar spatial distribution (comparing O, RA and TPA), showing highest occupancy in the center, declining to background levels at +/− ∼2 kb (Fig. 5C). This latter analysis is consistent with the notion that the epichromatin epitope is highly exposed within the epichromatin regions. In addition, ∼12% of the epichromatin data set from PL2–6 and 1H6 antibody experiments on untreated cells intersect with broad DNase I sensitive regions (identified in ENCODE for HL-60 cells).34 These results imply a significant enrichment of “open” chromatin within epichromatin regions. On the other hand, the calculated NRL, based on all candidate epichromatin regions, was not different from the genome-wide NRL, while the number of confirmed common epichromatin regions was too small to quantify NRL.
Figure 5.
Average feature distribution of common HL-60/S4 epichromatin regions as a function of the distance from the center of these regions. (A) Enrichment of retrotransposon Alu repeats. (B) Nucleotide frequencies. (C) Nucleosome occupancy profiles. Plot line colors for panel C: O (undifferentiated), black; RA treated, red; TPA treated, blue.
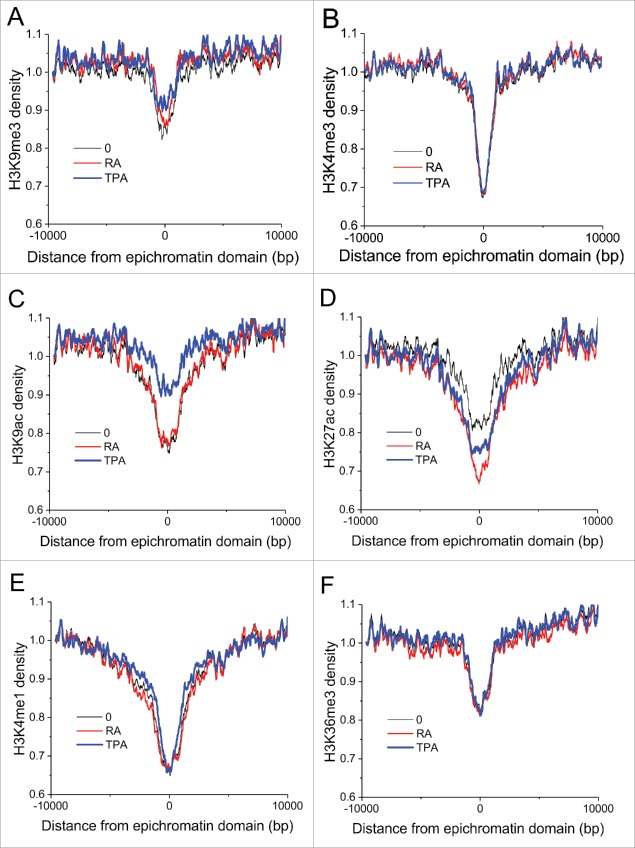
Signatures of histone post-translational modifications within common epichromatin regions are presented in Fig. 6. In all cases, modifications show a reduced presence in the middle of epichromatin: H3K9me3, a marker for constitutive heterochromatin (Fig. 6A); H3K4me3 (Fig. 6B) and H3K9ac (Fig. 6C), both markers for active promoters (although H3K9ac appears to be less depressed in TPA-treated cells); H3K27ac (Fig. 6D) and H3K4me1 (Fig. 6E), both markers for active enhancers; and H3K36me3 (Fig. 6F), a marker for actively transcribing genes. In summary, from the perspective of histone post-translational modifications, epichromatin is not clearly transcriptionally repressed, nor transcriptionally active.
Figure 6.
Signatures of histone modification marks around common epichromatin regions. Panels: (A) H3K9me3; (B) H3K4me3; (C) H3K9ac; (D) H3K27ac; (E) H3K4me1; (F) H3K36me3. Plot line colors: O (undifferentiated), black; RA treated, red; TPA treated, blue.
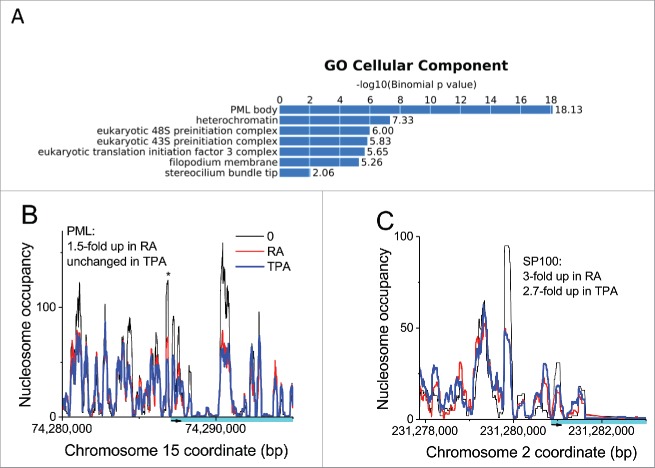
Finally, our analysis of the common epichromatin regions revealed a significant increase in gene promoters, compared with the average promoter density in the total human genome. Six percent of epichromatin regions contain promoters, corresponding to a 2.3-fold enrichment with respect to the genome average. Gene ontology (GO) analysis of these enriched promoters, unexpectedly, revealed enrichment for genes related to PML bodies (Fig. 7A and Supplementary Table 1). It is not clear what is the functional significance of having so many (i.e., 41) PML-associated genes in proximity to the interphase nuclear envelope (by virtue of their close continuity to epichromatin). The changes in transcript levels, compared with undifferentiated cells, for the specific PML-associated genes are frequently not the same for RA and TPA treated cells, implying that the PML bodies likely have different compositions in the 3 cell states. Figure 7B and C present nucleosome occupancy and transcript level changes for 2 important proteins of PML bodies (PML and SP100), also implying differential gene regulation between the 3 cell states. Because PML bodies appear to have multiple connections to stress responses40 the compositional heterogeneity of PML bodies suggests functional heterogeneity in the different cell states.
Figure 7.
(A) Gene Ontology (GO) analysis of common epichromatin promoter regions, indicating that these domains are enriched in genomic regions containing genes related to PML bodies. (B and C) Illustrating that PML bodies may have variable composition within the differentiated states: (B) Nucleosome repositioning in the PML gene, comparing 0, RA and TPA cell states; (C) Nucleosome repositioning in the SP100 gene, comparing 0, RA and TPA cell states. Plot line colors: O (undifferentiated), black; RA treated, red; TPA treated, blue.
Discussion
The present study used antibodies against histone H3 and several histone modifications in a ChIP-seq study of various nuclear elements (promoters, enhancers, TF binding sites and epichromatin) on undifferentiated (0) and differentiated (RA and TPA treated) forms of the myeloid leukemic cell line HL-60/S4. This yielded information about the relation of nucleosome loss to gene expression changes, the effects on nucleosome repeat length (NRL), nucleosome positions and nucleosome occupancy in and around CTCF and PU.1 binding sites and within common epichromatin regions. We have found changes in nucleosome occupancy surrounding various histone modification domains and revealed the distribution of histone modifications within and surrounding common epichromatin regions.
Our previous work on HL-60/S4 cells since 199817 was facilitated by their robust growth ability in the undifferentiated state and their rapid and reproducible cell differentiation in response to RA or TPA treatment. Furthermore, we recently obtained the transcriptomes of these 3 cell states (0, RA and TPA), and demonstrated that these cells have a very stable (abnormal) karyotype.27 Transcription factor binding measurements in HL-60/S4 are still missing, but we can extrapolate, to some extent, from the ENCODE ChIP-seq data for a related HL-60 cell line.34 All these data complemented the experimental data obtained in the current work, allowing us to perform the computational analyses.
Some of the most interesting conclusions from this study are the following: 1) During RA and TPA induced differentiation, regions that lost (but not regions that gained) nucleosomes are enriched at functional genomic elements such as promoters and enhancers (Fig. 1A and B). The number of genes upregulated upon nucleosome loss at the corresponding promoters was significantly elevated in comparison with genes that gained nucleosomes or in comparison with all genes (Fig. 1C and D). Documenting this observation, we presented a set of specific genes that show increased transcript levels and decreased promoter nucleosome occupancy in the differentiated state (Fig. 2). This observation has an intriguing parallel in the aforementioned transcriptome study,27 which demonstrated that following differentiation, more genes contributed more mRNA transcripts than genes that contributed decreased transcripts. 2) The global nucleosome repeat length (NRL) of HL-60/S4 cells appears essentially unchanged during cell differentiation, in contrast to other cell differentiation systems,3 such as stem cells differentiation into neuronal precursor cells and embryonic fibroblasts. 3) The NRLs around CTCF and PU.1 binding sites in undifferentiated and differentiated HL-60/S4 cells are about 10 bp shorter than the global NRL, arguing that transcription factor binding can lead to local changes of NRL. 4) Nucleosome occupancies can also change during HL-60/S4 differentiation in regions of specific histone modifications; this is, for example, well revealed around H3K4me3 domains. In the latter case, we observed a significant loss of nucleosomes following RA or TPA treatment (consistent with conclusion #1 above). 5) Epichromatin regions in undifferentiated and differentiated HL-60/S4 cells maintain their unusual characteristics, consistent with earlier observations.23 Alu and nucleosomes exhibit their peak densities at the “center” of the common epichromatin regions, which also shows a “spike” in G/C richness, compared with the surrounding regions. Common epichromatin regions also reveal a depletion of histone post-translational modifications considered indicative of either active or repressed transcription. In general, epichromatin is not within the gene body, but rather, is away from genes. If we assume that gene regulatory regions can extend up to 1000 kb from the promoter (unless it intersects another gene; the default parameter in the software GREAT41 used in this analysis), then it appears that the regulatory regions responsible for PML-associated genes (Supplementary Table 1) are enriched in epichromatin. At least 2 of these “close” promoters (i.e., for SP100 and DAXX) regulate transcription for proteins that are key components of ALT-associated PML bodies, believed to play a role in telomere elongation independent of telomerase.42 The large number of common chromosomal locations of epichromatin, comparing the 3 cell states, suggests stability of nuclear architecture during the lineage differentiation of HL-60/S4 cells. It is important to emphasize that epichromatin is identified by a conserved epitope (involving histones H2A, H2B and DNA) that remains to be defined, but likely represents a highly conserved structural feature of nucleosomes.36,37 A recent study43 presents evidence that etoposide-induced DNA damage of an embryonic teratocarcinoma cell line results in a disappearance of the peripheral epichromatin epitope in damaged nuclei, but not in undamaged nuclei. The epitope disappearance may represent histone-DNA loss, histone modification or masking by other proteins, an intriguing probe for changes at the nuclear periphery.
Finally, the present manuscript argues that in HL-60/S4 cells, there is a statistically significant association between the loss of nucleosomes within gene promoters (in differentiated cells) and increased levels of gene transcripts. Nucleosome depletion at promoters in many systems is quantitatively linked to the level of gene expression.3,28,44,45 However, it is also clear that there are promoters that require the presence of nucleosomes to initiate the binding of transcription factors, leading to subsequent transcription.46,47 Our observation that, during HL-60/S4 differentiation, nucleosome loss and nucleosome gain affect gene expression asymmetrically (i.e., a clear association with nucleosome loss and a more complicated response to nucleosome gain) agrees with the concept that there are several different mechanisms by which nucleosome positioning can affect gene expression. Interestingly, these changes in nucleosome occupancy involve extended regions of one to several kb, which is different from previously considered single-nucleosome repositioning scenarios and parallels a similar effect recently found at the V(D)J recombination domains48 The molecular mechanisms of the change of chromatin density in extended regions that we report here may be linked to chromatin compartmentalization in domains such as TADs (topologically associated domains),49,50 which have been recently shown to be correlated with regions of differential chromatin accessibility.51 An exploration of this idea may provide a direction for new studies.
Materials and methods
Cell culture and cell preparation
HL-60/S4 cells were maintained in RPMI 1640 medium, plus 10% fetal calf serum and 1% Pen/Strep/Glutamine. This cell line is available from ATCC (www.atcc.org, #CRL-3306). For the current experiments, cells were maintained in duplicate T-75 flasks, each flask containing 30 ml. Undifferentiated HL-60/S4 cells were seeded at a concentration of 0.83×105. RA treated cells were seeded at a concentration of 1.67×105, with RA (Sigma-Aldrich R2625) added to 1 µM. TPA treated cells were seeded at a concentration of 4 × 105, with TPA (Sigma-Aldrich P1585) added to 16 nM. After 4 d, approximately 107 were removed from each flask.
Harvested cells were centrifuged at 300xg for 5 min, resuspended in PBS and centrifuged again. The cell pellets were suspended in 20 ml PBS, made 1% HCHO (fresh ampule of 16% HCHO, methanol-free) and incubated 10 min at RT on a rotator. Fixation was stopped by addition of 2.5 M glycine to a final concentration of 125 mM and incubated 5 min at RT on a rotator. Fixed cells were centrifuged at 1000xg for 5 min and washed twice in PBS plus 50mM PMSF. Cell pellets were suspended in 10 ml of ice-cold “swelling buffer” (10 mM KCl, 1 mM MgCl2, 0.1% NP-40, 1 mM DTT, 25 mM Hepes pH 7.8, made 0.5 mM PMSF and containing Roche COMPLETE), incubated 10 min on ice, then centrifuged at 1000xg followed by plunging the cell pellets in liquid N2.
ChIP-seq protocol
The frozen cell pellets were resuspended in MNase (Micrococcal Nuclease) buffer (25 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 50 mM Tris/HCl pH 7.4) and 10 U MNase per 1×106 cells were added. After 15 min, incubation at 37°C MNase was stopped by adding 10x Covaris buffer (100 mM Tris pH 8.0, 2 M NaCl, 10 mM EDTA, 5% N-lauroylsarcosine, 1% Na-deoxycholate, supplemented with protease inhibitors). The samples were sonicated for 15 min with the following parameters with a Covaris S2 system: burst 200, cycle 20%, and intensity 8. Following centrifugation the supernatant was collected and directly used for IP.
After IgG preclearance the sheared chromatin was incubated overnight with protein G magnetic beads (Cell signaling, 9006) and 4 μg of anti-pan H3 (Abcam, ab1791), H3K4me1 (Abcam, ab8895), H3K4me3 (Abcam, ab8580), H3K9ac (Active Motif, 39917), H3K9me3 (Abcam, ab8898), H3K27ac (Abcam, ab4729) or H3K27me3 (Abcam, ab6002), H3K36me3 (Abcam, ab9050). After washes with 1x Covaris buffer (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% N-lauroylsarcosine, 0.1% Na–deoxycholate), high-salt-buffer (50 mM HEPES pH 7.9, 500 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Na–deoxycholate, 0.1% SDS), lithium buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na–deoxycholate) and 10 mM Tris–HCl, chromatin was eluted from the magnetic beads (elution buffer: 50 mM Tris pH 8.0, 1 mM EDTA, 1% SDS, 50 mM NaHCO3) and the crosslink was reversed overnight. After RNase A and proteinase K digestion, DNA was purified and cloned in a barcoded sequencing library for the Illumina sequencing platform. In brief, after DNA repair and A-addition NEBNext adapters (NEB, E7335) were ligated and digested with the USER enzyme. Barcodes (NEB, E7335) were introduced via PCR with a maximum of 14 cycles by the NEBNext polymerase (NEB, M0541). Size selection for mononucleosomal insert fragments was done with Ampure XP beads (Agencourt, A63880). Each ChIP-seq library was sequenced on the Illumina HiSeq 2000 with 50bp single-end in 2 replicates, whereas the H3-chip samples were sequenced with 50 bp paired-ends in 2 replicates.
Analysis of H3 ChIP-seq
H3 ChIP-seq reads were mapped using Bowtie52 allowing up to 1 mismatch and counting only unique hits, obtaining the following number of mapped 100-bp paired-end reads: 248 million for undifferentiated HL-60/S4 cells, 319 million for RA-treated HL-60/S4, and 338 million reads for TPA-treated HL-60/S4. The average DNA fragments lengths were correspondingly 160 bp, 159 bp and 159 bp, reflecting a moderate chromatin digestion (in a strongly digested chromatin DNA fragments are closer to the 147bp nucleosome DNA length). Mapped reads were processed using NucTools31 to generate genome-wide nucleosome occupancy landscapes, extract individual genomic regions and calculate NRL, as described previously.3,33,53 Aggregate nucleosome occupancy profiles around genomic features were calculated using HOMER54 and visualized using OriginPro 2016 (OriginLab Corporation). For the analysis performed in this manuscripts, we used the corresponding RefSeq definitions of promoters55 (extended +/−1,000 from TSS), FANTOM definitions of enhancers56 and Dfam definitions of ALU repeats.57 The coordinates of transcription factor binding sites and DNaseI-sensitive regions in HL-60 cells determined by the ENCODE consortium were obtained from the following GEO entries: GSM749688 (CTCF), GSM1010843 (PU.1) and GSM736626 (DNaseI hypersensitivity).
Analysis of ChIP-seq of histone modifications
The basic data processing was performed similarly to H3 ChIp-seq, with a difference that these were single-end reads, and the sequencing was to a smaller depth (30–40 million reads for each of 6 histone modification for each of the 3 cell conditions). Peak calling was performed using HOMER and repeated with MACS58 and SICER59 to confirm that the number of peaks did not differ significantly for different peak calling procedures. In the case of H3K4me3, the data from the ENCODE consortium obtained for HL-60 cells (GSM945222) had higher genomic coverage than our H3K4me3 ChIP-seq in HL-60/S4, but the aggregate profiles around genomic features were qualitatively similar.
Finding genomic regions which lost/gained nucleosomes
1000-bp regions which lost or gained nucleosome were determined genome-wide using NucTools31 with the following criteria applied to the pairs of cell states (e.g. RA vs 0): If the average nucleosome occupancy in RA-treated cells (PRA) in a given genomic window was higher than that in the control state (P0), i.e., if 2x (PRA – P0)/(PRA + P0) > 0.99 this was assigned as a nucleosome gain region. If 2x (PRA – P0)/(PRA + P0) < −0.99 the corresponding region was considered to have a nucleosome loss.
Analysis of RNA-seq data
We used RNA-seq data for undifferentiated HL-60/S4 and RA- and TPA-differentiated counterparts (4 replicates for each state) reported in the accompanying paper (Mark Welch et al, Nucleus, in press; deposited at the Short Read Archive (SRA), bioproject PRJNA303179). Because different methods can produce inconsistent results60-62 we measured gene-level differential expression using DESeq2,63 EBSeq,64 EdgeR,65 and limma-voom.66 For our data, EBSeq proved the most conservative, with the fewest genes called as significantly differentially expressed and the fewest genes called, that were not called by other methods (Supplemental Table 2). Therefore, only genes that EBSeq determined to have posterior probability of differential expression (PPDE) >0.95 were considered in the analysis (Supplemental Table 3).
Epichromatin coordinates
Coordinates of epichromatin domains in undifferentiated HL-60/S4 and RA-differentiated cells were determined in our previous publications using modified immunoprecipitation method (“xxChIP-seq”) with PL2–6 and 1H6 antibodies.23 Here we have defined a subset of these regions that was reproduced in all replicate experiments with both antibodies, both in undifferentiated HL-60/S4 and RA-differentiated cells. Defined in this way, replicate PL2–6 immunoprecipitates of undifferentiated HL-60/S4 cells exhibited 40,288 epichromatin regions; replicate RA-treated cells, 77,897 regions. Defining only common regions based on all replicates, both antibodies (PL2–6 and 1H6) and both cell states (undifferentiated and RA-treated) resulted in 6,541 epichromatin regions.
Enrichment analysis
Enrichment of genomic regions which lost or gained nucleosomes at functional genomic elements (promoters, enhancers, Alu repeats) was calculated as a ratio of several regions falling to a given class of functional elements vs. the number of random regions (generated for a data set of the same and with the same region length distribution) falling to a given class of functional elements. Random regions were generated using BedTools.67 Gene Ontology enrichment at promoters with differential nucleosome occupancy was calculated using DAVID68 (2016 update) with default parameters. The enrichment of epichromatin regions in functional regulatory elements was calculated using GREAT41 with the following parameters: Each gene was assigned a basal regulatory domain of 5kb upstream and 1kb downstream of the TSS (regardless of other nearby genes). The gene regulatory domain is extended in both directions to the nearest gene's basal domain but no more than 1000kb in one direction.
Permission
No human patients or experimental animals were used. Approval by the Ethics Committee of Human Experimentation was not required.
Supplementary Material
Availability of materials and data
The data has been deposited to the GEO database, accession number GSE90992.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors express their appreciation to Naveed Ishaque (DKFZ, Heidelberg) for advice on interpreting ChIP-seq data.
Funding
The Guest Scientist Program at the DKFZ supported ALO and DEO while working in Heidelberg. VBT is supported by the Wellcome Trust grant 200733/Z/16/Z. Open Access fees were paid by the Wellcome Trust.
References
- [1].Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016; 164:57-68; PMID:26771485; http://dx.doi.org/ 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature 2011; 474:516-20; PMID:21602827; http://dx.doi.org/ 10.1038/nature10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 2012; 19:1185-92; PMID:23085715; http://dx.doi.org/ 10.1038/nsmb.2419 [DOI] [PubMed] [Google Scholar]
- [4].Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008; 132:887-98; PMID:18329373; http://dx.doi.org/ 10.1016/j.cell.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wallner S, Schroder C, Leitao E, Berulava T, Haak C, Beisser D, Rahmann S, Richter AS, Manke T, Bönisch U, et al.. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenetics Chromatin 2016; 9:33; PMID:27478504; http://dx.doi.org/ 10.1186/s13072-016-0079-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang W, Li Y, Kulik M, Tiedemann RL, Robertson KD, Dalton S, Zhao S. Nucleosome positioning changes during human embryonic stem cell differentiation. Epigenetics 2016; 11:426-37; PMID:27088311; http://dx.doi.org/ 10.1080/15592294.2016.1176649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drillon G, Audit B, Argoul F, Arneodo A. Evidence of selection for an accessible nucleosomal array in human. BMC Genomics 2016; 17:526; PMID:27472913; http://dx.doi.org/ 10.1186/s12864-016-2880-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Collins SJ, Gallo RC, Gallagher RE. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature 1977; 270:347-9; PMID:271272; http://dx.doi.org/ 10.1038/270347a0 [DOI] [PubMed] [Google Scholar]
- [9].Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci U S A 1978; 75:2458-62; PMID:276884; http://dx.doi.org/ 10.1073/pnas.75.5.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci U S A 1980; 77:2936-40; PMID:6930676; http://dx.doi.org/ 10.1073/pnas.77.5.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dalton WT Jr., Ahearn MJ, McCredie KB, Freireich EJ, Stass SA, Trujillo JM. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood 1988; 71:242-7; PMID:3422031 [PubMed] [Google Scholar]
- [12].Rovera G, Santoli D, Damsky C. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc Natl Acad Sci U S A 1979; 76:2779-83; PMID:288066; http://dx.doi.org/ 10.1073/pnas.76.6.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rovera G, O'Brien TG, Diamond L. Induction of differentiation in human promyelocytic leukemia cells by tumor promoters. Science (New York, NY) 1979; 204:868-70; http://dx.doi.org/ 10.1126/science.286421 [DOI] [PubMed] [Google Scholar]
- [14].Collins SJ. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood 1987; 70:1233-44; PMID:3311197 [PubMed] [Google Scholar]
- [15].Leung MF, Sokoloski JA, Sartorelli AC. Changes in microtubules, microtubule-associated proteins, and intermediate filaments during the differentiation of HL-60 leukemia cells. Cancer Res 1992; 52:949-54; PMID:1737356 [PubMed] [Google Scholar]
- [16].Campbell MS, Lovell MA, Gorbsky GJ. Stability of nuclear segments in human neutrophils and evidence against a role for microfilaments or microtubules in their genesis during differentiation of HL60 myelocytes. J Leukocyte Biol 1995; 58:659-66; PMID:7499963 [DOI] [PubMed] [Google Scholar]
- [17].Olins AL, Buendia B, Herrmann H, Lichter P, Olins DE. Retinoic acid induction of nuclear envelope-limited chromatin sheets in HL-60. Exp Cell Res 1998; 245:91-104; PMID:9828104; http://dx.doi.org/ 10.1006/excr.1998.4210 [DOI] [PubMed] [Google Scholar]
- [18].Olins AL, Herrmann H, Lichter P, Olins DE. Retinoic acid differentiation of HL-60 cells promotes cytoskeletal polarization. Exp Cell Res 2000; 254:130-42; PMID:10623473; http://dx.doi.org/ 10.1006/excr.1999.4727 [DOI] [PubMed] [Google Scholar]
- [19].Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res 2001; 268:115-27; PMID:11478838; http://dx.doi.org/ 10.1006/excr.2001.5269 [DOI] [PubMed] [Google Scholar]
- [20].Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. The LINC-less granulocyte nucleus. Euro J Cell Biol 2009; 88:203-14; http://dx.doi.org/ 10.1016/j.ejcb.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Olins AL, Ernst A, Zwerger M, Herrmann H, Olins DE. An in vitro model for Pelger-Huet anomaly: stable knockdown of lamin B receptor in HL-60 cells. Nucleus (Austin, Tex) 2010; 1:506-12; PMID:21327094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem 2013; 288:8610-8; PMID:23355469; http://dx.doi.org/ 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Olins AL, Ishaque N, Chotewutmontri S, Langowski J, Olins DE. Retrotransposon Alu is enriched in the epichromatin of HL-60 cells. Nucleus 2014; 5:237-46; PMID:24824428; http://dx.doi.org/ 10.4161/nucl.29141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gonzalez-Sandoval A, Gasser SM. On TADs and LADs: Spatial Control Over Gene Expression. Trends Genet 2016; 32:485-95; PMID:27312344; http://dx.doi.org/ 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- [25].Yanez-Cuna JO, van Steensel B. Genome-nuclear lamina interactions: from cell populations to single cells. Curr Opin Genet Dev 2017; 43:67-72; PMID:28107752; http://dx.doi.org/ 10.1016/j.gde.2016.12.005 [DOI] [PubMed] [Google Scholar]
- [26].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948-51; PMID:18463634; http://dx.doi.org/ 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- [27].Mark Welch DB, Jauch A, Langowski J, Olins AL, Olins DE Nucleus 2017; PMID:28152343; http://dx.doi.org/ 10.1080/19491034.2017.1285989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Teif VB, Erdel F, Beshnova DA, Vainshtein Y, Mallm JP, Rippe K. Taking into account nucleosomes for predicting gene expression. Methods (San Diego, Calif) 2013; 62:26-38; PMID:23523656; http://dx.doi.org/ 10.1016/j.ymeth.2013.03.011 [DOI] [PubMed] [Google Scholar]
- [29].Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, et al.. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014; 345:1251086; PMID:25258085; http://dx.doi.org/ 10.1126/science.1251086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015; 348:910-4; PMID:25953818; http://dx.doi.org/ 10.1126/science.aab1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vainshtein Y, Rippe K, Teif VB. NucTools: analysis of chromatin feature occupancy profiles from high-throughput sequencing data. BMC Genomics Submitted 2017; 18(1):158; PMID:28196481; http://dx.doi.org/ 10.1186/s12864-017-3580-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol 2013; 20:267-73; PMID:23463311; http://dx.doi.org/ 10.1038/nsmb.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Beshnova DA, Cherstvy AG, Vainshtein Y, Teif VB. Regulation of the nucleosome repeat length in vivo by the DNA sequence, protein concentrations and long-range interactions. PLoS Comput Biol 2014; 10:e1003698; PMID:24992723; http://dx.doi.org/ 10.1371/journal.pcbi.1003698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].de Souza N. The ENCODE project. Nat Methods 2012; 9:1046; PMID:23281567; http://dx.doi.org/ 10.1038/nmeth.2238 [DOI] [PubMed] [Google Scholar]
- [35].Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell 2014; 53:979-92; PMID:24656132; http://dx.doi.org/ 10.1016/j.molcel.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Olins AL, Langhans M, Monestier M, Schlotterer A, Robinson DG, Viotti C, Zentgraf H, Zwerger M, Olins DE. An epichromatin epitope: persistence in the cell cycle and conservation in evolution. Nucleus (Austin, Tex) 2011; 2:47-60; PMID:21647299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Prudovsky I, Vary CP, Markaki Y, Olins AL, Olins DE. Phosphatidylserine colocalizes with epichromatin in interphase nuclei and mitotic chromosomes. Nucleus (Austin, Tex) 2012; 3:200-10; PMID:22555604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Losman MJ, Fasy TM, Novick KE, Monestier M. Monoclonal autoantibodies to subnucleosomes from a MRL/Mp(−)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B, and DNA. J Immunol 1992; 148:1561-9; PMID:1371530 [PubMed] [Google Scholar]
- [39].Mandinov L, Mandinova A, Kyurkchiev S, Kyurkchiev D, Kehayov I, Kolev V, Soldi R, Bagala C, de Muinck ED, Lindner V, et al.. Copper chelation represses the vascular response to injury. Proc Natl Acad Sci U S A 2003; 100:6700-5; PMID:12754378; http://dx.doi.org/ 10.1073/pnas.1231994100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sahin U, de The H, Lallemand-Breitenbach V. PML nuclear bodies: assembly and oxidative stress-sensitive sumoylation. Nucleus (Austin, Tex) 2014; 5:499-507; PMID:25482067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G et al.. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28:495-501; PMID:20436461; http://dx.doi.org/ 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang Y, Liang P, Liu D, Huang J, Songyang Z. Telomere regulation in pluripotent stem cells. Protein Cell 2014; 5:194-202; PMID:24563217; http://dx.doi.org/ 10.1007/s13238-014-0028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huna A, Salmina K, Erenpreisa J, Vazquez-Martin A, Krigerts J, Inashkina I, Gerashchenko BI, Townsend PA, Cragg MS, Jackson TR. Role of stress-activated OCT4A in the cell fate decisions of embryonal carcinoma cells treated with etoposide. Cell Cycle 2015; 14:2969-84; PMID:26102294; http://dx.doi.org/ 10.1080/15384101.2015.1056948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wight A, Yang D, Ioshikhes I, Makrigiannis AP. Nucleosome presence at AML-1 binding sites inversely correlates with Ly49 expression: revelations from an informatics analysis of nucleosomes and immune cell transcription factors. PLoS Comput Biol 2016; 12:e1004894; PMID:27124577; http://dx.doi.org/ 10.1371/journal.pcbi.1004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet 2010; 26:476-83; PMID:20832136; http://dx.doi.org/ 10.1016/j.tig.2010.08.003 [DOI] [PubMed] [Google Scholar]
- [46].Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP, et al.. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 2013; 49:67-79; PMID:23177737;http://dx.doi.org/ 10.1016/j.molcel.2012.10.019 [DOI] [PubMed] [Google Scholar]
- [47].Ballare C, Zaurin R, Vicent GP, Beato M. More help than hindrance: nucleosomes aid transcriptional regulation. Nucleus (Austin, Tex) 2013; 4:189-94; PMID:23756349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pulivarthy SR, Lion M, Kuzu G, Matthews AG, Borowsky ML, Morris J, Kingston RE, Dennis JH, Tolstorukov MY, Oettinger MA. Regulated large-scale nucleosome density patterns and precise nucleosome positioning correlate with V(D)J recombination. Proc Natl Acad Sci U S A 2016; 113(42):E6427-36; PMID:27698124; http://dx.doi.org/ 10.1073/pnas.1605543113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; http://dx.doi.org/ 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376-80; PMID:22495300; http://dx.doi.org/ 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schreiber J, Libbrecht M, Bilmes J, Noble W. Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture. bioRxiv 103614; http://dx.doi.org/ https://doi.org/ 10.1101/103614 [DOI] [Google Scholar]
- [52].Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/ 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Teif VB, Beshnova DA, Vainshtein Y, Marth C, Mallm JP, Hofer T, Rippe K. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res 2014; 24:1285-95; PMID:24812327; http://dx.doi.org/ 10.1101/gr.164418.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576-89; PMID:20513432; http://dx.doi.org/ 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016; 44:D733-45; PMID:26553804; http://dx.doi.org/ 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al.. An atlas of active enhancers across human cell types and tissues. Nature 2014; 507:455-61; PMID:24670763; http://dx.doi.org/ 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AF, Wheeler TJ. The Dfam database of repetitive DNA families. Nucleic Acids Res 2016; 44:D81-9; PMID:26612867; http://dx.doi.org/ 10.1093/nar/gkv1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al.. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137; PMID:18798982; http://dx.doi.org/ 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xu S, Grullon S, Ge K, Peng W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 2014; 1150:97-111; PMID:24743992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lin Y, Golovnina K, Chen ZX, Lee HN, Negron YL, Sultana H, Oliver B, Harbison ST. Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genomics 2016; 17:28; PMID:26732976; http://dx.doi.org/ 10.1186/s12864-015-2353-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rajkumar AP, Qvist P, Lazarus R, Lescai F, Ju J, Nyegaard M, Mors O, Børglum AD, Li Q, Christensen JH. Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genomics 2015; 16:548; PMID:26208977; http://dx.doi.org/ 10.1186/s12864-015-1767-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform 2015; 16:59-70; PMID:24300110; http://dx.doi.org/ 10.1093/bib/bbt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550; PMID:25516281; http://dx.doi.org/ 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013; 29:1035-43; PMID:23428641; http://dx.doi.org/ 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139-40; PMID:19910308; http://dx.doi.org/ 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15:R29; PMID:24485249; http://dx.doi.org/ 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 2014; 47:11 2 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4:P3; PMID:12734009; http://dx.doi.org/ 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.