Abstract
Antibodies have traditionally served as the affinity reagents of choice in point-of-care diagnostic biosensors. However, this class of proteins is not ideally suited for this use, being poorly characterized and prone to thermal denaturation. Here, we present an activity-based assessment of an alternative engineered binding protein in a cellulose-based assay.
Immunoglobulins are the most commonly used binding proteins for a wide variety of diagnostic formats, including commercial ELISA kits and lateral flow assays, microfluidics-based tests,1 and paper-based diagnostics.2 However, while these antibodies undergo high-affinity binding interactions with their complementary antigens, they possess a number of physical and chemical attributes that are non-optimal for in vitro diagnostic applications.
Antibodies are known to be prone to thermal denaturation,3-6 and their production can be imprecise,7 leading to lot-to-lot variability in assay performance.8,9 These attributes make antibodies non-ideal for use in low-cost diagnostics, which are challenged with complex patient samples and used in settings where continuous cold-chain storage may not be guaranteed throughout the distribution network.10,11 Although efforts have been made to enhance the thermostability of antibodies via rational design,12–15 the task is complicated by a limited understanding of the structural determinants of protein stability,16,17 and how these features might be introduced without negatively impacting binding function. Thus, improved variants are often isolated by high-throughput screening of natural antibody repertoires18 and combinatorial mutagenesis libraries19–21. However, this process may be impractical for the routine development of binding proteins for applications that are not as commercially lucrative as therapeutics, as it requires intensive cloning, library manipulation, and protein characterization in addition to the involved workflows already associated with antibody production projects.
In recent years an alternative, bottom-up method has emerged, in which the desired binding functionality is imparted upon intrinsically stable biochemical species with structurally isolated paratopes, to yield functional binders with innate stability. Many potential binding proteins have been investigated over the past several decades for in vivo therapeutic uses and industrial applications,22,23 but relatively few binding species have been tested as replacements for antibodies in in vitro diagnostics; these include single-domain antibodies,24–27 aptamers,28,29 and slow off-rate aptamer derivatives.30 These candidates have historically suffered from a number of shortcomings: single-domain antibodies feature native disulfide bonds and have demonstrated low expression yields31 and poor solubility,32 and the in vitro aptamer selection process has proven inconsistent, frequently yielding binding molecules of low affinity or poor specificity.33
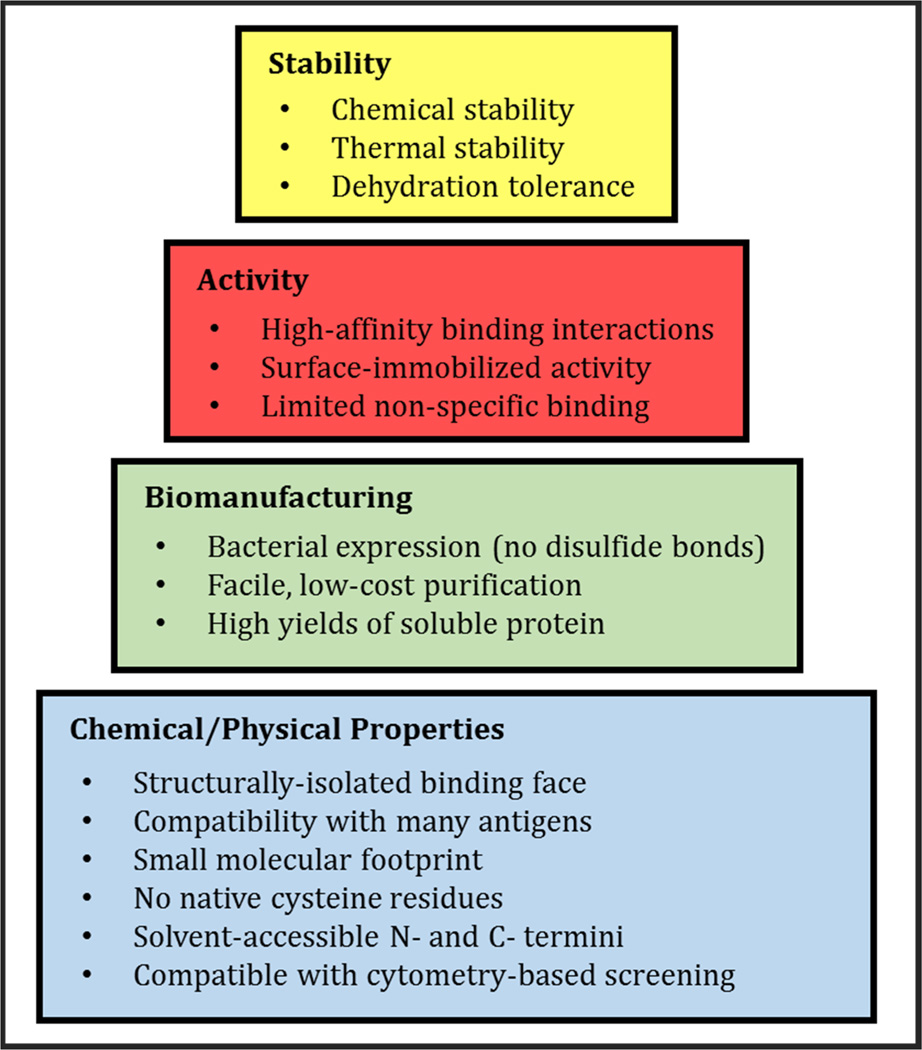
Here, we investigate the utility of the Sso7d protein from the hyperthermophilic archaeon Sulfolobus solfataricus as an alternative binding scaffold for in vitro point-of-care diagnostics. This 7 kDa DNA-binding protein features a well-defined, planar binding face which has proven to be a versatile scaffold for the development of novel, high-affinity binding functionalities, and which has been shown to be topologically compatible with a diverse range of target epitopes.34–36 Sso7d is also intrinsically stable, featuring a wild-type melting temperature of 98°C and demonstrated activity across a broad pH range. Sso7d lacks any native cysteine residues, allowing for scalable, facile recombinant expression in the reducing environment of the bacterial cytoplasm, and permitting the use of artificially-introduced cysteine residues as functional moieties for oriented surface immobilization and the conjugation of reporter species. Because the Sso7d scaffold is immuno-orthogonal, there is limited risk of cross-reaction with immunological factors found in patient samples. Sso7d has also demonstrated activity following surface immobilization,37 and features a small molecular footprint (rg = 1.31 nm), potentially allowing for higher molecular surface densities and enhanced substrate functionalization. Thus, the Sso7d scaffold meets many of the criteria required of a binding protein applied in point-of-care diagnostic biosensors (Figure 1).
Figure 1.
Characteristics of the ideal alternative binding protein for point-of-care diagnostic biosensors
To date, species from this family of hyperthermophilic oligonucleotide/oligosaccharide-binding (OB) fold proteins have been used to develop binding proteins for a range of applications, serving as purification ligands for the chromatographic separation of IgG antibodies37 and viral particles38, as capture agents in zirconium phosphonate-functionalized microarrays,39 and as polymerase fusion partners for stable DNA binding during polymerase chain reaction (PCR)40. Each of these applications takes advantage of the intrinsic chemical and thermal stability of Sso7d to perform functions otherwise inaccessible to traditional antibody-based affinity reagents. However, existing studies have not yet investigated the species’ fitness for in vitro point-of-care diagnostics such as paper-based assays, in which the surface-immobilized binding proteins must withstand harsh environmental conditions and capture dilute antigens from patient samples. In this dilute analyte regime, even small fractional losses in the binding activity of the immobilized capture molecule have important implications with respect to the sensitivity and reproducibility of the device; thus it is of critical importance that diagnostic capture agents be highly stable under thermal stress.
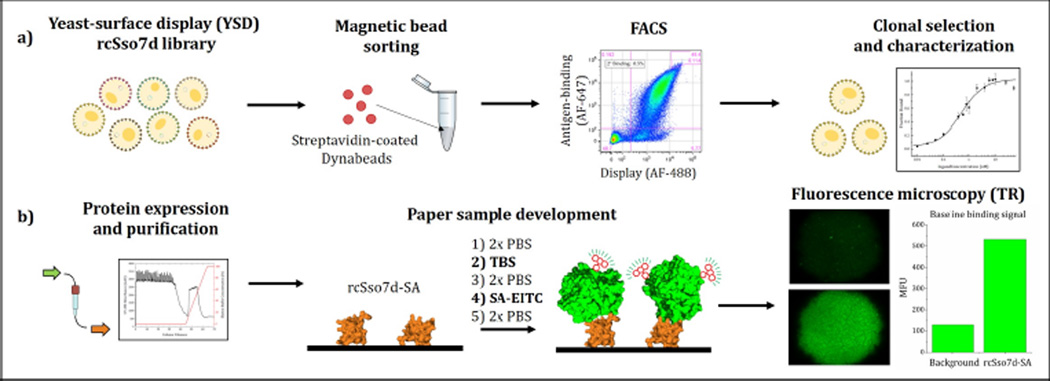
Given that the Sso7d scaffold possesses many favorable properties and has a strong record of successful technological integration in other applications, we assessed this species’ suitability for use in these challenging diagnostic contexts. We first developed and characterized a model binding species by screening a combinatorial library via yeast-surface display, and the activity of this species was assessed in a paper-based diagnostic format (Figure 2). We explored whether the immobilized Sso7d variant retained sufficient binding activity to capture a model analyte from a dilute, nanomolar solution, and compared this interfacial binding activity with that observed in the yeast surface-displayed format. We also conducted an activity-based stability measurement of this Sso7d variant when applied in a paper-based assay following thermal challenge.
Figure 2.
Schematic representation of experimental approach used in this study. (A) A combinatorial yeast library was screened via magnetic bead sorting and fluorescence-activated cell sorting were used to select a specific antigen-binding rcSso7d variant. (B) This species was then expressed in E. coli and purified via immobilized metal affinity chromatography, and its activity was assessed in a paper-based format using fluorescence microscopy.
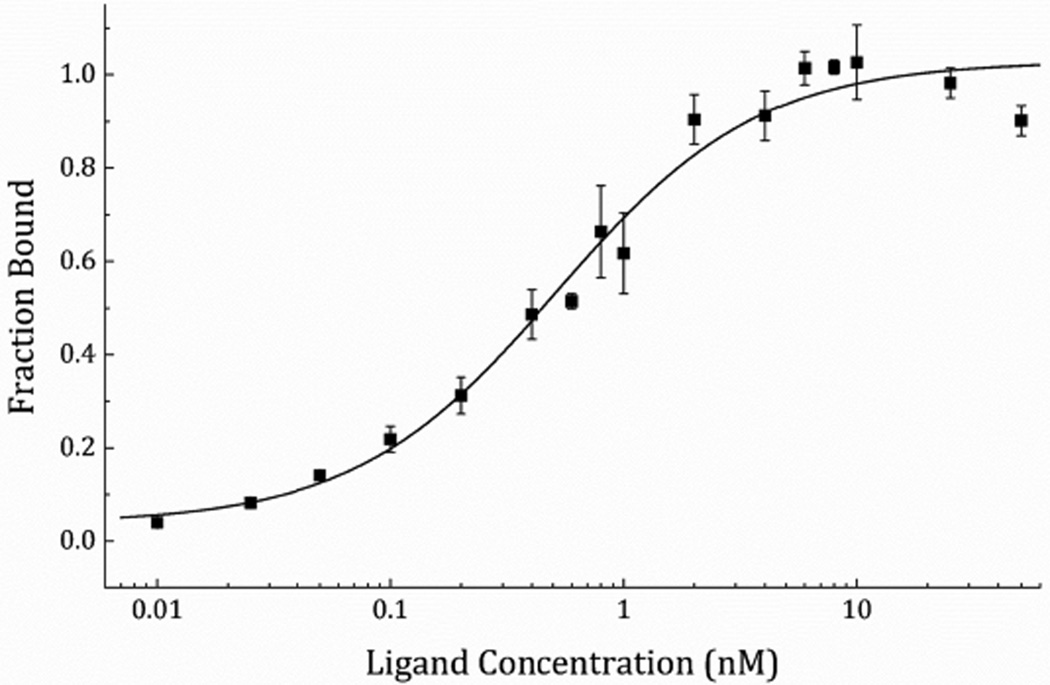
In order to select an engineered Sso7d variant which binds to the model analyte streptavidin, a high-quality saturation mutagenesis library of 1.4 × 109 unique yeast clones, referred to as reduced-charge Sso7d-11 (rcSso7d-11), was screened via yeast-surface display. This library was first refined over the course of two rounds of highly-avid magnetic bead sorting, using streptavidin-coated Dynabeads to isolate binders of even modest affinity, and to reduce the library size by approximately three orders of magnitude. This binder-enriched library was then further refined over the course of five rounds of fluorescence-activated cell sorting (FACS), during which selective pressure was applied to the clonal population both by progressively reducing the concentration of available ligand, and by designating increasingly stringent sort gates (Supporting Information (SI), Figure S1). A subset of the enriched population was sequenced, and a single clone, rcSso7d-SA, was selected for affinity characterization. This yeast surface-displayed clone was characterized by affinity titration using a serial dilution of streptavidin Alexa Fluor 647, and its affinity was determined to be 556 ± 136 pM (Figure 3).
Figure 3.
Equilibrium binding titration curve for determining the affinity of the yeast surface-displayed streptavidin binding protein, rcSso7d-SA. The data were collected over three biological replicates on separate days, and the mean fluorescence of each sample was normalized and averaged across the replicates.
The gene encoding this selected rcSso7d-SA species was then cloned into a pET-28b(+) bacterial expression plasmid, expressed in a BL21(DE3) strain of E. coli, purified using immobilized metal affinity chromatography (IMAC), and eluted via a 500 mM imidazole gradient. (SI, Table S2 and Figure S2) SDS-PAGE characterization of the purified fractions indicated that a single pass through a nickel column yielded electrophoretically pure recombinant product in a single band, and quantification of the purified protein via a bicinchoninic acid (BCA) assay indicated a single-pass yield of 42.2 mg/Lculture, achieved without any process optimization. Based on a per-test usage of 0.25 µg of rcSso7d, these results suggest that this process would support the production of over 170,000 diagnostic assays per single-liter culture, at a price that is nearly 100-fold more economical than the molar-equivalent cost of a streptavidin-binding polyclonal antibody blend (SI, Table S6).
We then sought to determine whether the purified protein would retain its activity following immobilization on a low-cost, field-relevant substrate, using aldehyde-functionalized chromatography paper. In a standard workflow for these paper-based assays, the capture protein is deposited on test zones defined using hydrophobic ink. Following protein immobilization via an overnight coupling reaction, the test zone substrate is inactivated in Tris-buffered saline and the samples are challenged with a solution containing the fluorescently-labeled protein analyte. The developed samples are then washed twice in 1× PBS and imaged using fluorescence microscopy (Figure 2), and the mean fluorescence intensity of each test zone is quantified using ImageJ software (US National Institutes of Health).
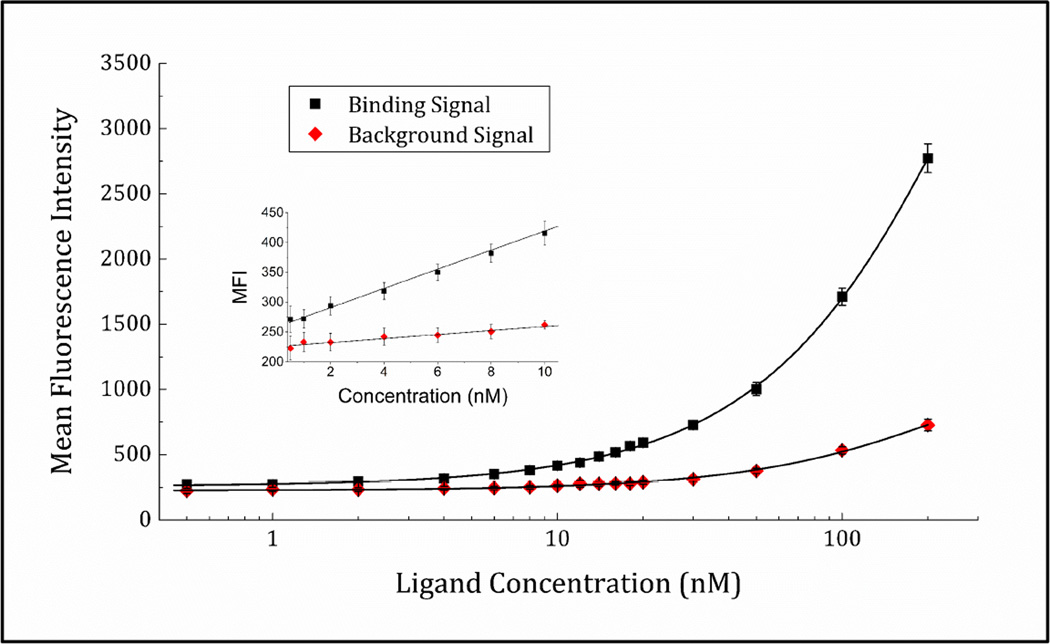
In order to test the antigen dose response and activity of surface-immobilized rcSso7d-SA, a serial dilution of streptavidin-eosin 5-isothiocyanate conjugate was prepared, and contacted with the paper-bound rcSso7d-SA for a period of time sufficient for the species to reach 95% of equilibrium binding. Comparison of binding-mediated fluorescence signal with the baseline fluorescence observed in BSA-passivated samples (Figure 4) indicates that rcSso7d-SA retains activity following surface immobilization, and yields binding signals that are perceptibly above baseline even at a low nanomolar analyte concentration.
Figure 4.
Titration of paper-immobilized rcSso7d-SA with streptavidin-eosin 5-isothiocyanate conjugate. Background signal was determined by challenging BSA-passivated test zones with the same concentration of antigen. The data were collected during independent experiments on two separate days, with four technical replicates on each day. The mean fluorescence observed for each experimental condition was averaged across all replicates.
However, while the plot does not reach saturation, it is clear that the surface-immobilized species exhibits an apparent KD higher than that observed in the context of yeast surface display, as evidenced by the right-shifted response curve. This is likely attributable to both a loss of activity upon expression and purification, and deleterious orientation effects following conjugation of the binder to the substrate. The introduction of N- or C- terminal residues or fusion peptides for site-specific, oriented surface immobilization may serve to improve this limit of detection.
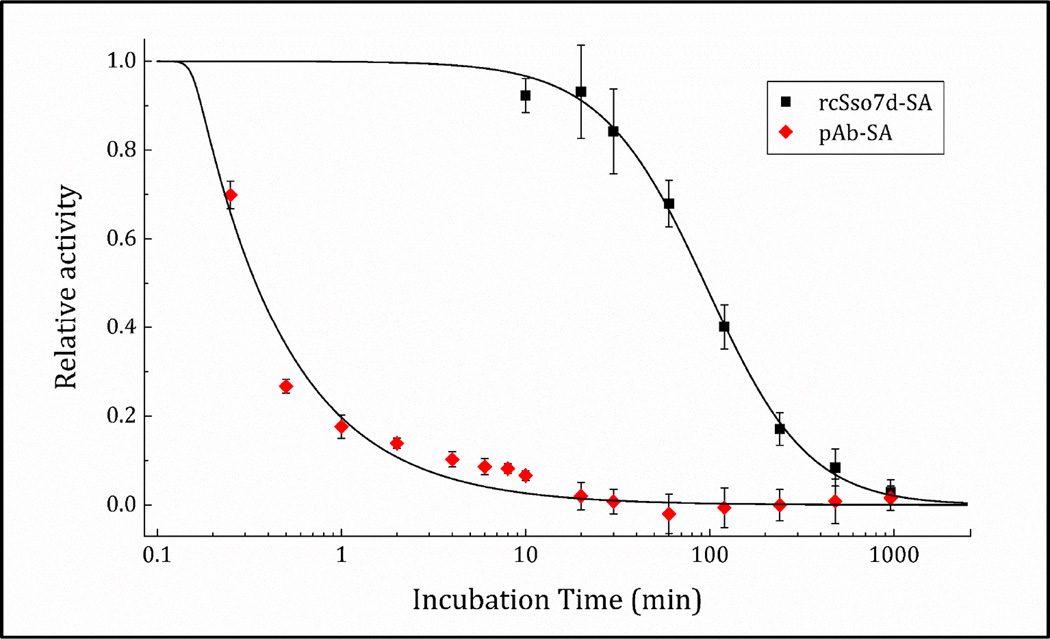
We also evaluated the binding protein’s activity retention under thermal challenge, in order to assess whether diagnostic test stability would be enhanced by the use of the rcSso7d scaffold in place of polyclonal antibodies. While standard practice is to report the biophysical melting temperature (Tm) of the species of interest, this parameter does not necessarily correlate with the temperature at which paratope perturbation and functional attenuation begins to occur at an appreciable rate.41 Thus, we employed an activity-based assay to test the relative stability of these species, subjecting both rcSso7d-SA and a commercially-available, polyclonal blend of anti-SA rabbit IgG to a thermal challenge antigen-binding assay.
Both binding species were diluted in their optimized buffer systems such that the monovalent rcSso7d-SA binding face was represented in equimolar proportion with that of the bivalent polyclonal antibodies. These samples were incubated in a thermocycler at 95°C for various time periods ranging up to 16 hours, and then applied to aldehyde-functionalized paper and challenged with the streptavidin-eosin 5-isothiocyanate conjugate according to the protocol outlined above. The relative activity observed for each sample was determined by dividing the background-subtracted fluorescence signal by the background-subtracted baseline fluorescence observed in the unheated sample.
The rcSso7d-SA species featured much slower unfolding kinetics, retaining binding activity sufficient for discerning a positive result after heating at 95°C for nearly 8 hours (Figure 5). The t1/2 (the time at which 50% of activity is lost) of rcSso7d was determined to be 94.0 minutes. In comparison, the polyclonal rabbit IgG lost nearly 100% of its activity within the initial 10 minutes of the thermal challenge. Studies at short timescales determined the approximate t1/2 of the pAbs to be 20.5 s, 275-fold faster than the denaturation of rcSso7d-SA.
Figure 5.
Activity-based accelerated degradation study for streptavidin-binding polyclonal antibodies (pAb) and rcSso7d-SA. The data were collected over three independent experiments on separate days, with four technical replicates on each day. The mean fluorescence observed for each experimental condition was averaged across all replicates.
This accelerated degradation study has established precedent in pharmaceutical formulation research,42 and captures the intrinsically higher kinetic stability of rcSso7d relative to polyclonal antibodies. These results suggest that incorporating Sso7d-based affinity reagents into point-of-care diagnostic tests may render these assays more thermally stable and better able to withstand the environments in which they are used. These improvements would likely be enhanced by the addition of small-molecule stabilizing agents such as trehalose, pullulan43, or silk44. Additional studies will investigate the relative long-term stability of lyophilized, surface-immobilized rcSso7d species formulated in commercial stabilization buffers, in order to assess whether the significantly higher stability of the rcSso7d scaffold in solution directly translates to kinetic stability in the interfacial context.
In summary, this study has provided further demonstration that the rcSso7d scaffold can be readily engineered to bind to a protein analyte, and that this binder can be inexpensively produced in bacterial culture and facilely purified in high yield. We have established that this binding protein retains activity following surface immobilization in a low-cost, paper-based format, and that rcSso7d drastically out-performs polyclonal antibodies in its retention of binding activity following thermal challenge. Future studies will seek to demonstrate the development of rcSso7d-based binders to disease-relevant antigens, and the incorporation of these binders into assay contexts which yield a colorimetric readout (e.g. via gold nanoparticles or polymerization-based amplification45), in order to further demonstrate the potential of this alternative binding scaffold as a suitable replacement for antibodies in point-of-care diagnostic tests.
Supplementary Material
Acknowledgments
EAM acknowledges support from NIH 2 T32 GM008334 26.
HDS acknowledges MIT’s Reed Fund. We thank Prof. Dane Wittrup, Prof. Balaji Rao, Shefali Lathwal, and Carlos Cruz for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available. See DOI:
Notes and references
- 1.Chin, et al. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 2.Yetisen AK, Akram MS, Lowe CR. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 3.Boscato M, Stuart MC. Clin. Chem. 1988;34:27–33. [PubMed] [Google Scholar]
- 4.World Health Organization. Malaria Rapid Diagnostic Test Performance: Summary results of WHO product testing of malaria RDTs: Round 1–5 (2008–2013) 2014
- 5.Lee, et al. J Clin. Microbiol. 2012;50:1397–1405. doi: 10.1128/JCM.06533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G, Srivastava J, Zaman MH. Anal. Biochem. 2014;449:147–154. doi: 10.1016/j.ab.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Wang, et al. Biomacromolecules. 2012;13:559–564. doi: 10.1021/bm2017405. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Gaza-Bulseco G, Sun J. J Chromatogr. B. 2006;837:35–43. doi: 10.1016/j.jchromb.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 9.Lipman, et al. ILAR J. 2005;46:258–268. doi: 10.1093/ilar.46.3.258. [DOI] [PubMed] [Google Scholar]
- 10.Bradbury ARM, Plückthun A. Protein Eng. Des. Sel. 2015;28:303–305. doi: 10.1093/protein/gzv051. [DOI] [PubMed] [Google Scholar]
- 11.Marx V. Nat. Methods. 2013;10:703–707. doi: 10.1038/nmeth.2570. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen, et al. Am. J. Trop. Med. Hyg. 2006;74:750–754. [PubMed] [Google Scholar]
- 13.Albertini, et al. Malar. J. 2012;11:1–9. [Google Scholar]
- 14.McConnell, et al. MAbs. 2014;6:37–41. [Google Scholar]
- 15.McConnell, et al. Protein Eng. Des. Sel. 2013;26:151–164. doi: 10.1093/protein/gzs090. [DOI] [PubMed] [Google Scholar]
- 16.Buchanan, et al. MAbs. 2013;5:255–262. doi: 10.4161/mabs.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano, et al. Anticancer. Drugs. 2010;21:907–916. doi: 10.1097/CAD.0b013e32833f5d68. [DOI] [PubMed] [Google Scholar]
- 18.Kovacic, et al. Protein Eng. Des. Sel. 2016;29:65–76. doi: 10.1093/protein/gzv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudgeon, et al. Proc. Natl. Acad. Sci. 2012;109:10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leow, et al. Malar. J. 2014;13:277. doi: 10.1186/1475-2875-13-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traxlmayr, et al. Biochim. Biophys. Acta. 2012;1824:542–549. doi: 10.1016/j.bbapap.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, et al. Protein Eng. Des. Sel. PEDS. 2010;23:549–557. doi: 10.1093/protein/gzq028. [DOI] [PubMed] [Google Scholar]
- 23.Christ D, Famm K, Winter G. Protein Eng. Des. Sel. 2007;20:413–416. doi: 10.1093/protein/gzm037. [DOI] [PubMed] [Google Scholar]
- 24.Goldman, et al. Anal. Chem. 2008;80:8583–8591. doi: 10.1021/ac8014774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fatima, et al. PLoS One. 2014;9:e95263. doi: 10.1371/journal.pone.0095263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths, et al. Antibodies. 2013;2:66–81. [Google Scholar]
- 27.Odongo, et al. PLoS Negl. Trop. Dis. 2016;10:e0004420. doi: 10.1371/journal.pntd.0004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang, et al. J Infect. 2014;69:569–580. doi: 10.1016/j.jinf.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Dirkzwager, et al. Chem. Commun. 2015;51:4697–4700. doi: 10.1039/c5cc00438a. [DOI] [PubMed] [Google Scholar]
- 30.Ochsner, et al. Biotechniques. 2014;56:125–128. 130, 132–133. doi: 10.2144/000114134. [DOI] [PubMed] [Google Scholar]
- 31.Liu, et al. Microb. Cell Fact. 2015;14:158. doi: 10.1186/s12934-015-0340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, et al. Biotechnol. Reports. 2015;6:27–35. doi: 10.1016/j.btre.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blind M, Blank M. Mol. Ther. Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gera, et al. J Mol. Biol. 2011;409:601–616. doi: 10.1016/j.jmb.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 35.Arcus V. Curr. Opin. Struct. Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, et al. FEBS. 2016;283:1351–1367. doi: 10.1111/febs.13674. [DOI] [PubMed] [Google Scholar]
- 37.Gera, et al. PLoS One. 2012;7:e48928. doi: 10.1371/journal.pone.0048928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain, et al. Biotechnol. Prog. 2012;29:237–246. doi: 10.1002/btpr.1655. [DOI] [PubMed] [Google Scholar]
- 39.Cinier, et al. Bioconjug. Chem. 2009;20:2270–2277. doi: 10.1021/bc9002597. [DOI] [PubMed] [Google Scholar]
- 40.Wang, et al. Nucleic Acids Res. 2004;32:1197–1207. doi: 10.1093/nar/gkh271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tischenko VM, Abramov VM, Zav’yalov VP. Biochemistry. 1998;37:5576–5581. doi: 10.1021/bi972647a. [DOI] [PubMed] [Google Scholar]
- 42.Alsante KM, Martin L, Baertschi SW. Pharm. Technol. 2003:60–72. [Google Scholar]
- 43.Kannan, et al. Anal. Chem. 2015;87:9288–9293. doi: 10.1021/acs.analchem.5b01923. [DOI] [PubMed] [Google Scholar]
- 44.Li, et al. J Control. Release. 2015;219:416–430. doi: 10.1016/j.jconrel.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badu-Tawiah Lab Chip. 2015;15:655–659. doi: 10.1039/c4lc01239a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.