Summary
Astrocytes exist throughout the nervous system and are proposed to affect neural circuits and behavior. However, studying astrocytes has proven difficult because of the lack of tools permitting astrocyte selective genetic manipulations. Here, we report the generation of Aldh1l1-Cre/ERT2 transgenic mice to selectively target astrocytes in vivo. We characterised Aldh1l1-Cre/ERT2 mice using imaging, immunohistochemistry, AAV-FLEX-GFP microinjections and crosses to RiboTag, Ai95 and new Cre-dependent membrane tethered Lck-GCaMP6f knock-in mice that we also generated. Two-to-three weeks after tamoxifen induction, Aldh1l1-Cre/ERT2 selectively targeted essentially all adult (P80) brain astrocytes with no detectable neuronal contamination, resulting in expression of cytosolic and Lck-GCaMP6f and permitting subcellular astrocyte calcium imaging during startle responses in vivo. Crosses with RiboTag mice allowed sequencing of actively translated mRNAs and determination of the adult cortical astrocyte transcriptome. Thus, we provide well characterised, easy-to-use resources with which to selectively study astrocytes in situ and in vivo in multiple experimental scenarios.
Keywords: astrocyte, calcium, GCaMP, Lck-GCaMP, Aldh1l1, Cre/ERT2
ETOC/“In brief”
The Khakh laboratory developed and characterized new optical and genetic reagents to explore astrocyte functions in neural circuits. The approaches permit inducible genetic targeting of astrocytes in vivo and the selective monitoring of calcium signals in astrocyte processes.
Introduction
Astrocytes tile the entire central nervous system, serve essential support functions, and contribute to disease. In addition, astrocytes regulate synapse formation, removal and function, as well as regulate blood flow (Allen, 2014; Bazargani and Attwell, 2015). Astrocytes also respond with intracellular calcium elevations during distinct responses in vivo such as locomotion and startle (Shigetomi et al., 2016) and they are implicated in the regulation of complex mouse behaviours (Halassa and Haydon, 2010).
A key necessity to study astrocytes in vivo is the ability to manipulate them selectively without concomitantly impacting other cells. From this perspective much attention has focussed on genetic methods to selectively target astrocytes (Davila et al., 2013; Xie et al., 2015). Cell type specific expression of Cre recombinase (Cre) is frequently used to achieve gene expression and deletion by exploiting the Cre-loxP system (Sauer, 1994; Tsien, 2016). However, existing mouse lines expressing Cre under the control of astrocyte promoters are neither selective for astrocytes, nor are they pan-astrocytic (Khakh and Sofroniew, 2015; Xie et al., 2015; Zhang and Barres, 2010). Furthermore, several of the commonly used mouse lines express Cre from birth, which vitiates inducible gene expression/deletion in adult mice and complicates analyses of complex behaviors and disease mechanisms. In addition, several available Cre lines are known to target neural stem cells, which compromises interpretation of behavior in adults. Overall, existing astrocyte Cre lines display some, several or all of these shortcomings, and thus fall short of minimal experimental requirements for cell specificity (Xie et al., 2015). For example, widely used hGfap-Cre lines target large populations of neurons (Sun et al., 2016; Zhuo et al., 2001) and several mGfap-Cre and Slc1a3-Cre/ERT2 lines target neural progenitors that give rise to neurons in the olfactory bulb and hippocampus (Garcia et al., 2004; Gregorian et al., 2009; Niu et al., 2013; Slezak et al., 2007) and smaller populations of neurons elsewhere (Xie et al., 2015). This problem is shared with mice constitutively expressing Cre under the control of the Aldh1l1 promoter (Foo and Dougherty, 2013). Hence, the field lacks a reliable method to achieve pan-astrocytic, specific and inducible genetic manipulations in order to explore astrocyte physiology in vivo. The limitations of Gfap based mouse lines have been discussed (Sloan and Barres, 2014; Su et al., 2004).
The GENSAT project (Gong et al., 2003) found that the aldehyde dehydrogenase 1 family member L1 (Aldh1l1) gene marked astrocytes, a finding confirmed by subsequent RNA (Cahoy et al., 2008; Molofsky and Deneen, 2015; Zhang et al., 2014; Zhang et al., 2016), lineage (Molofsky et al., 2013) and expression analyses (Cahoy et al., 2008; Foo and Dougherty, 2013). This raised the possibility that Aldh1l1 could be used as a genetic locus to express tamoxifen inducible Cre/ERT2 and thus achieve temporally specific pan-astrocytic genetic manipulations. In the present study, we explored this possibility using BAC transgenesis (Yang and Gong, 2005). In order to characterise the Aldh1l1-Cre/ERT2 mice, we used immunohistochemistry (IHC) and Cre-dependent FLEX-GFP adeno-associated viruses (AAV) (Atasoy et al., 2008). We also used genetic crosses of the Aldh1l1-Cre/ERT2 mice with Ai95 mice that express cytosolic GCaMP6f (cyto-GCaMP6f) in a Cre-dependent manner (Chen et al., 2013; Madisen et al., 2015). Furthermore, in light of studies showing that membrane targeted Lck-GCaMPs reliably report calcium signals in astrocyte processes (Haustein et al., 2014; Shigetomi et al., 2013; Shigetomi et al., 2010), we generated knock-in mice at the ROSA26 locus for Cre-dependent Lck-GCaMP6f, crossed these to Aldh1l1-Cre/ERT2 mice and characterised the offspring using in situ and in vivo imaging. Finally, we crossed the Aldh1l1-Cre/ERT2 mice with RiboTag mice that express the ribosomal protein Rpl22HA in a Cre-dependent manner (Sanz et al., 2009). We used this strategy to determine the cortical astrocyte transcriptome in adult mice, which we compared to data from P7 mice (Zhang et al., 2014). Our studies provide well characterized, much needed and easy-to-use resources.
Results and Discussion
Generation of BAC transgenic Aldh1l1-Cre/ERT2 mice
The Cre lines used are named according to the gene (Table 1). Cre/ERT2 was inserted at the Aldh1l1 gene (Aldh1l1) start codon within the BAC DNA (Figure 1A). Insertion was confirmed by PCR using flanking and insert specific primers and with restriction analyses. The Cre/ERT2 sequence harbors a rare AsiS1 restriction site and cutting with AsiS1 linearized the modified BAC (Supp Figure 1A). The modified Aldh1l1-Cre/ERT2 BAC clone was used for pronuclear injections after detailed characterization with PCR over regions of Aldh1l1 and Cre/ERT2. Following injection of ~200 embryos, two founder lines of transgenic mice were generated (Aldh1l1-Cre/ERT2; lines 1 and 2). Both resulted in germ-line transmission and were identical in initial experiments that are described below for line 1. The insertion of Cre/ERT2 was confirmed by PCR of tail biopsy genomic DNA and by characterization with PCR over regions of Aldh1l1 (Supp Figure 1B). Identical methods were used to make Slc6a11-Cre/ERT2 mice at the GABA transporter type 3 (GAT3) locus. Both the Aldh1l1- and Slc6a11-Cre/ERT2 mice reproduced, provided offspring in expected numbers and displayed no obvious behavioral alterations. This was expected, because the BAC method leaves both copies of the endogenous genes intact while achieving expression from the full length locus within the BAC (Yang and Gong, 2005).
Table 1.
Summary of the Cre mice (with regards to astrocyte and neuronal expression of GFP from AAV FLEX-GFP microinjections).
| Gene name | Protein name | Cre type | Tamoxifen inducible | Expression in astrocytes | Expression in neurons |
|---|---|---|---|---|---|
| Gfap | GFAP | Cre | No | High | High |
| Slc1a3 | GLAST | Cre/ERT | Yes | Moderate | Very high |
| Gjb6 | Cx30 | Cre/ERT2 | Yes | Very high | Moderate |
| Slc6a11 | GAT3 | Cre/ERT2 | Yes | Moderate | Moderate |
| ALDH1L1 | Aldh1l1 | Cre/ERT2 | Yes | Very high | Not detected |
Note to table: See also Figure 1, Supp Figure 1 and the associated text in the Results.
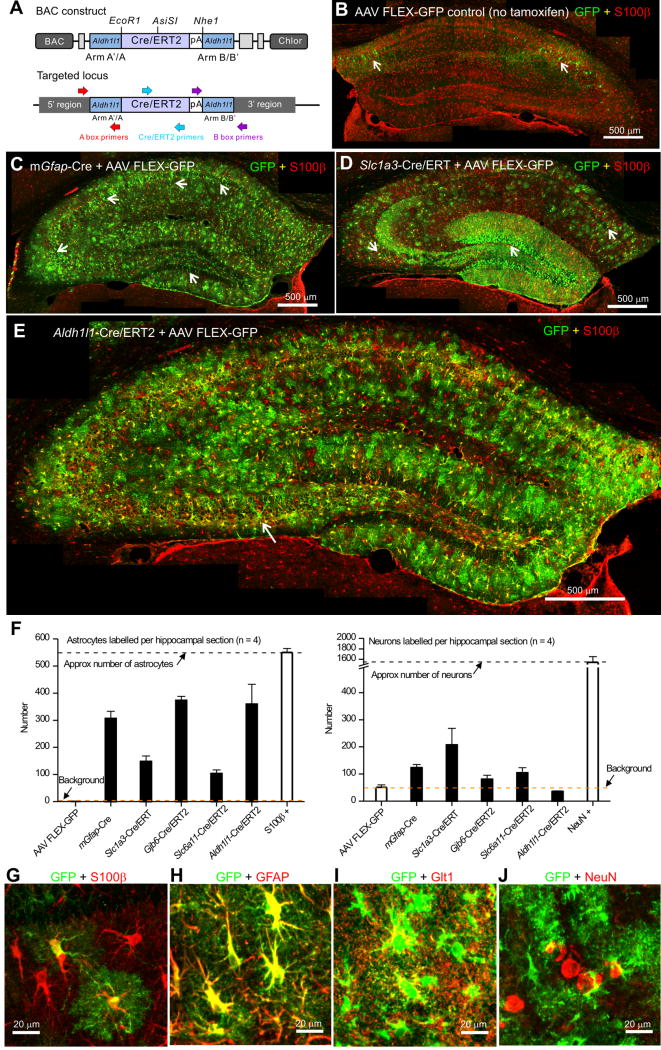
Figure 1. Creation and characterization of Aldh1l1-Cre/ERT2 BAC transgenic mice.
A. Schematic showing the BAC targeting construct used to create the Aldh1l1-Cre/ERT2 transgenic mice (key primer locations are also shown). B. Representative montage of the hippocampus from the Aldh1l1-Cre/ERT2 transgenic mouse injected with AAV FLEX-GFP virus with no tamoxifen injection. The white arrows show leaky expression of GFP in neurons. C. Representative montage of the hippocampus from the mGfap-Cre transgenic mouse injected with AAV FLEX-GFP virus stained for GFP (green) and S100β (red). There was expression of GFP in astrocytes; the white arrows show expression of GFP in neurons within the CA1, CA3 and dentate gyrus. D. Representative montage of the hippocampus from a Slc1a3-Cre/ERT2 transgenic mouse injected with AAV FLEX-GFP virus, followed by 75 mg/kg i.p. tamoxifen for 5 consecutive days. E. Representative montage of the hippocampus from an Aldh1l1-Cre/ERT2 transgenic mouse injected with AAV FLEX-GFP virus, followed by 75 mg/kg i.p. tamoxifen for 5 days. There was abundant, widespread expression in astrocytes; the white arrow shows GFP expression in one neuron. F. Bar graphs showing the number of astrocytes and neurons in hippocampal montages from each transgenic line following AAV FLEX-GFP virus injection. White bars show the total number of S100β positive astrocytes, the number of neurons observed in controls without tamoxifen injection and the total number of NeuN positive neurons in hippocampal montages. G–J. Representative high magnification images of CA1 astrocytes in the Aldh1l1-Cre/ERT2 mouse showing co-staining for S100β (G), GFAP (H), Glt1 (I), but not NeuN (J). In B–G, the sections were stained for GFP (green) and S100β (red).
Testing of Aldh1l1-Cre/ERT2 mice in relation to other commonly used Cre lines
We initially tested for Cre/ERT2 mediated expression of GFP within adult Aldh1l1-Cre/ERT2 mice using IHC 14-21 days after microinjections (i.e. at ~P80) of Cre-dependent AAVs FLEX-GFP, followed by tamoxifen induction (Atasoy et al., 2008). With this strategy, GFP is expressed in cells that express Cre. As a control, we injected AAV FLEX-GFP into the hippocampus of Aldh1l1-Cre/ERT2 mice, and in the absence of tamoxifen we detected no GFP expression in astrocytes based on cell morphology or colocalisation with the astrocyte marker S100β in 40 μm hippocampal sections (Figure 1B; n = 4 mice). However, we did detect GFP expression in a small number of neurons (~50), which represents leaky expression from the AAV FLEX-GFP (Figure 1B,F; n = 4 mice). Later, we report no detectable leakiness from the Aldh1l1-Cre/ERT2 mouse line. However, with the AAV FLEX-GFP controls, we microinjected AAV FLEX-GFP into the hippocampi of mGfap-Cre 77.6 mice (Gregorian et al., 2009) that constitutively express Cre in GFAP positive cells. We observed expression of GFP in 306 ± 27 astrocytes, but also in a substantial population of neurons (123 ± 12) throughout the hippocampus (arrows in Figure 1C,F; n = 4 mice). We repeated these experiments with Slc1a3-Cre/ERT2 mice at the GLAST glutamate transporter locus (Slezak et al., 2007), and again noted clear GFP expression in astrocytes (148 ± 21) and large numbers of neurons (207 ± 61; arrows in Figure 1D,F; n = 4 mice). Similar experiments were performed for Gjb6-Cre/ERT2 (Slezak et al., 2007) and Slc6a11-Cre/ERT2 mice (Supp Figure 1; Figure 1F; n = 4 mice) the numbers of labeled astrocytes and neurons were 374 ± 14 and 81 ± 14, and 104 ± 14 and 104 ± 19 for Gjb6-Cre/ERT2 and Slc6a11-Cre/ERT2 mice, respectively. Thus in the case of mGfap-Cre, Slc1a3-Cre/ERT2, Gjb6-Cre/ERT2 and Slc6a11-Cre/ERT2 the number of GFP expressing neurons as a percentage of GFP expressing astrocytes in the hippocampus was 40, 140, 21 and 101%, respectively (n = 4 mice of each genotype). In contrast, with Aldh1l1-Cre/ERT2 mice we detected GFP expression in large numbers of astrocytes (360 ± 73) and in essentially no neurons (36 ± 2 versus the AAV control of 50 ± 9; Figure 1E,F; Table 1; n = 4 mice). We emphasize that the comparison of different Cre lines shown in Figure 1 and Table 1 relates to the hippocampus at P80. We do not rule out judicious use of existing Cre lines or the need for further controls.
The astrocytes labeled with GFP from AAV FLEX-GFP in the Aldh1l1-Cre/ERT2 mice displayed typical, unambiguous, complex bushy astrocyte morphologies and colocalized with S100β, GFAP and Glt1, but not with NeuN (Figure 1G–J; n = 4 mice each; Supp Figure 1E). A single pan-astrocyte antibody marker does not exist, but the bushy morphologies and colocalisation with S100β, GFAP and Glt1 provides strong evidence that the GFP expressing cells were astrocytes.
Generation of Lck-GCaMP6fflox mice at the ROSA26 locus
Genetically-encoded calcium indicators (GECIs) can be expressed in specific cells and offer advantages over organic calcium indicators for studying astrocyte calcium signaling. GCaMP6f has fast kinetics and higher signal-to-noise than its predecessors (Chen et al., 2013), and Cre-dependent knock-in mice at the ROSA26 locus are available (Madisen et al., 2015). Plasma membrane targeting of GCaMP6f improves its ability to reveal calcium signals in near membrane regions and fine processes of astrocytes (Shigetomi et al., 2016). In light of this, we made knock-in Lck-GCaMP6f mice at the ROSA26 locus (Lck-GCaMP6fflox) in which GCaMP6f is targeted to the plasma membrane by a Lck tag and expressed in a Cre-dependent manner (Figure 2A–C).
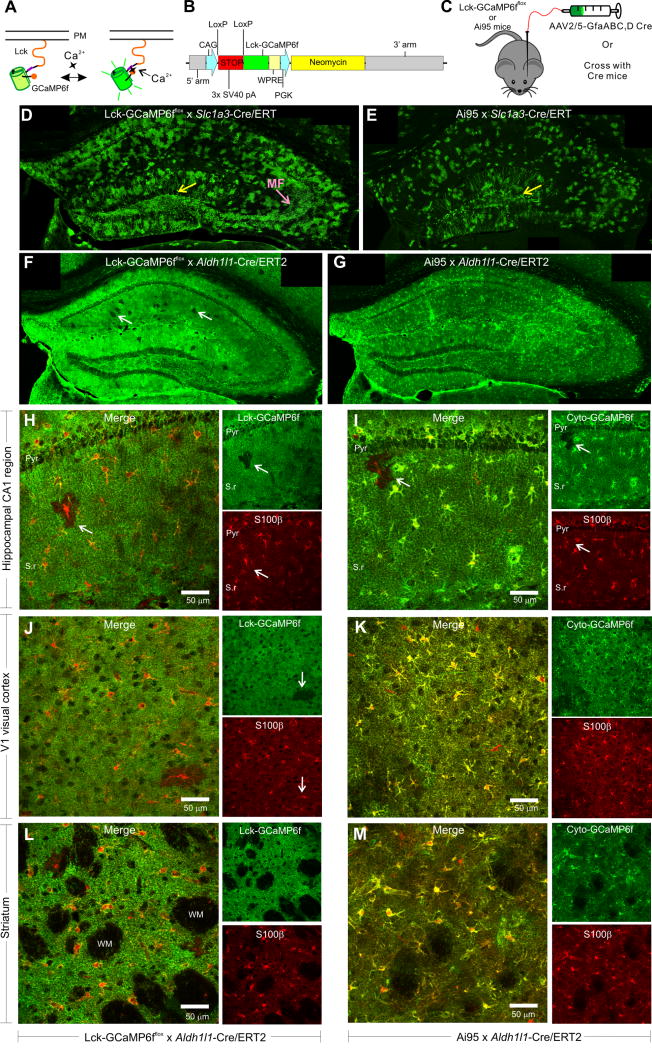
Figure 2. Creation of Lck-GCaMP6fflox knock-in mice and characterization of Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice.
A. Lck-GCaMP6f localizes to the plasma membrane and detects near membrane calcium signals. B. Schematic of the targeting construct used to create Lck-GCaMP6fflox knock-in mice. C. For initial characterization, Lck-GCaMP6fflox knock-in mice and Ai95 mice were injected with AAV2/5-GfaABC1D Cre AAVs or crossed with Cre/ERT2 transgenic mice. D. Representative montage of Lck-GCaMP6fflox x Slc1a3-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained for Lck-GCaMP6f. The yellow arrow indicates expression of Lck-GCaMP6f and cyto-GCaMP6f in granule cells. The pink arrow shows expression of Lck-GCaMP6f in the mossy fiber pathway. E. Representative montage of Ai95 x Slc1a3-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained for cyto-GCaMP6f. F. Representative hippocampal montage of Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained for Lck-GCaMP6f. The white arrows point to astrocytes that lack Lck-GCaMP6f expression. G. Representative hippocampal montage of Ai95 x Aldh1l1-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained with a GFP antibody shows GCaMP6f expression in nearly all astrocytes of the hippocampus. H. Representative image of the hippocampal CA1 region from a Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained for Lck-GCaMP6f (green) and S100β (red); the pyramidal cell layer (Pyr) and Stratum radiatum (S.r) are indicated. The white arrow shows an astrocyte with S100β that lacks Lck-GCaMP6f expression. The panels to the right separately show Lck-GCaMP6f staining using a GFP antibody and S100β staining. I. Representative image of the hippocampal CA1 region from a Ai95 x Aldh1l1-Cre/ERT2 double transgenic mouse injected with tamoxifen and stained for cyto-GCaMP6f (green) and S100β (red). The white arrow shows an astrocyte with S100β lacking cyto-GCaMP6f expression. The panels to the right separately show cyto-GCaMP6f staining using a GFP antibody and S100β staining. J–K and L–M. As in H–I, but for V1 of the visual cortex and striatum, respectively.
We initially tested Lck-GCaMP6fflox mice by injecting AAV2/5 GfaABC1D Cre into the hippocampus of adult mice. We also used the same AAV to express cyto-GCaMP6f in Ai95 mice. In both cases we noted expression of Lck-GCaMP6f and cyto-GCaMP6f in S100β positive astrocytes and neurons in the hippocampus (Supp Figure 2A–F; n = 4 mice; Supp Figure 3A,B; n = 4 mice). Our data show that AAV2/5 GfaABC1D Cre is not selective for Cre-dependent astrocyte gene deletion/expression in the hippocampus, however, it was convenient for testing/validating the knock-in mice. Lck-GCaMP6f revealed entire bushy astrocytes, whereas cyto-GCaMP6f revealed the somata, major branches and branchlets (Supp Figure 3A,B; n = 4 mice). Moreover, with live slice imaging we could readily measure 100 μM ATP-evoked and spontaneous Ca2+ signals from Lck-GCaMP6f and cyto-GCaMP6f expressing astrocytes (Supp Figure 3C–E and G). The ATP-evoked responses measured with Lck-GCaMP6f were significantly faster than those measured with cyto-GCaMP6f (Supp Figure 3C,D & E,G; n = 46 & 84 cells from 4 mice each). In the case of spontaneous Ca2+ signals, Lck-GCaMP6f revealed greater numbers of ROIs per-cell and greater numbers of events per-cell per-minute in relation to cyto-GCaMP6f (Supp Figure 3F,H; n = 7 & 8 cells, from 4 mice each). The kinetics of the Ca2+ signals were significantly faster with Lck-GCaMP6f (Supp Figure 3H). These initial experiments demonstrate the potential utility of Lck-GCaMP6fflox mice in relation to Ai95 mice.
Validating Aldh1l1-Cre/ERT2 mice by crosses with Lck-GCaMP6fflox and Ai95 mice
We crossed both Ai95 and Lck-GCaMP6fflox mice with Slc1a3-Cre/ERT2 and Aldh1l1-Cre/ERT2 mice. We induced gene expression with tamoxifen (75 mg/kg once per day for five days at ~P56) in the offspring that were heterozygous for both alleles and studied GCaMP6f expression two to three weeks later with IHC (i.e. at ~P80, Figure 2). We did not examine reporter expression in neonatal mice or in adult mice greater than 3 weeks after tamoxifen.
Cyto-GCaMP6f and Lck-GCaMP6f expression was sparse with Slc1a3-Cre/ERT2 mice, but expression of both GECIs was ubiquitous and high with Aldh1l1-Cre/ERT2 mice (Figure 2D–G; n = 4 mice each). Indeed, the entire brain was green when either Lck-GCaMP6f or cyto-GCaMP6f was driven with the Aldh1l1-cre/ERT2 mouse line (Figure 2F,G). Occasional astrocytes that did not express GCaMP6f appeared as dark patches in an otherwise uniformly green neuropil (white arrows; Figure 2F). This was clear from higher magnification images; the dark patches contained S100β positive astrocytes (Figure 2H–I). It was clear that Lck-GCaMP6f and cyto-GCaMP6f were expressed in the vast majority of S100β positive astrocytes in the hippocampus, visual cortex and striatum, with occasional patches of S100β positive cells that lacked the GECIs (Figure 2H–M). Furthermore, examination of example brain areas in sagittal sections showed that Aldh1l1-Cre/ERT2 resulted in astrocyte GCaMP6f expression throughout the brain (Supp Movie 1).
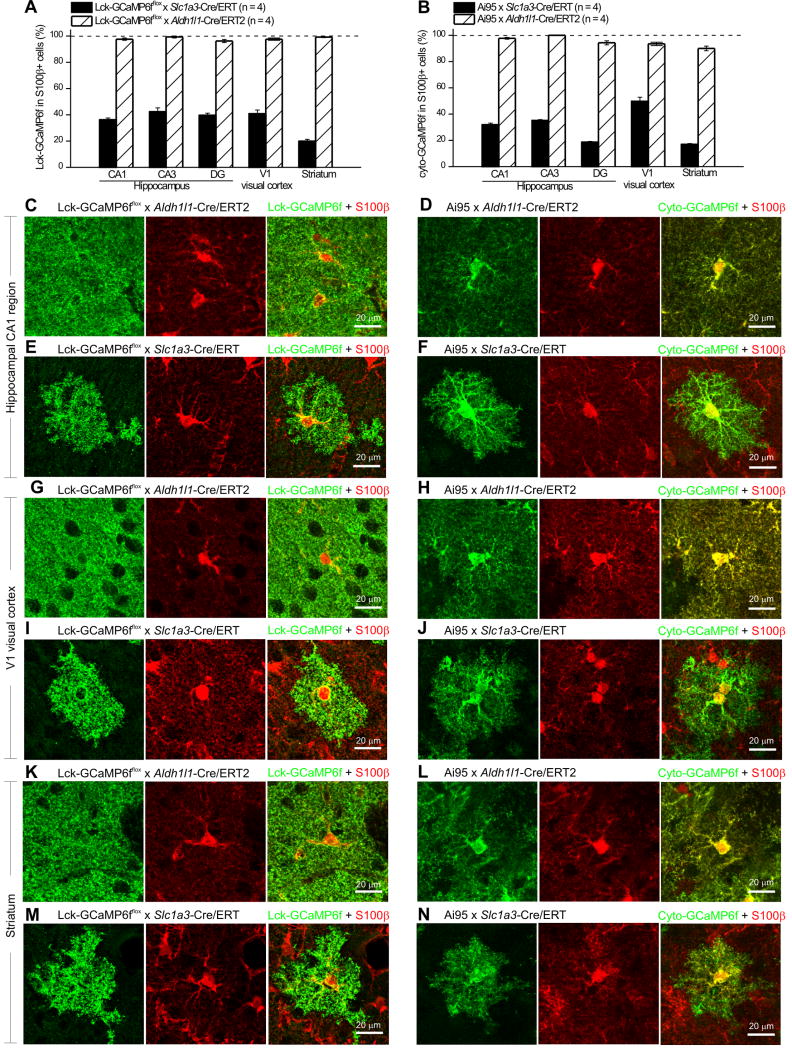
Overall, Aldh1l1-Cre/ERT2 mice resulted in GECI expression in >90% of astrocytes in the hippocampal CA1, CA3 and DG regions, in V1 of the visual cortex and in the striatum (Figure 3A,B; n = 4 mice for each case). In contrast Slc1a3-Cre/ERT2 resulted in expression of Lck-GCaMP6f or cyto-GCaMP6f in ~20–40% of astrocytes in the same areas (Figure 3A,B; n = 4 mice each).
Figure 3. Comparison of Slc1a3-Cre/ERT2 and Aldh1l1-Cre/ERT2 transgenic mice.
A. Bar graph showing the percentage of S100β+ astrocytes that express Lck-GCaMP6f in the Lck-GCaMP6fflox x Slc1a3-Cre/ERT2 and the Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice in the hippocampal CA1, CA3 and DG areas, in the V1 visual cortex and dorsolateral striatum. B. As in A, but for Ai95 x Slc1a3-Cre/ERT2 and Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice. C. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the hippocampal CA1 region of Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice. D. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the hippocampal CA1 region of Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice. E. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the hippocampal CA1 region of Lck-GCaMP6fflox x Slc1a3-Cre/ERT2 double transgenic mice. F. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the hippocampal CA1 region of Ai95 x Slc1a3-Cre/ERT2 double transgenic mice. G. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the V1 visual cortex of Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice. H. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the V1 visual cortex of Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice. I. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the V1 visual cortex of Lck-GCaMP6fflox x Slc1a3-Cre/ERT2 double transgenic mice. J. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the V1 visual cortex of Ai95 x Slc1a3-Cre/ERT2 double transgenic mice. K. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the striatum of Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice. L. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the striatum of Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice. M. Representative staining for Lck-GCaMP6f (green) and S100β (red) in the striatum of Lck-GCaMP6fflox x Slc1a3-Cre/ERT2 double transgenic mice. N. Representative staining for cyto-GCaMP6f (green) and S100β (red) in the striatum of Ai95 x Slc1a3-Cre/ERT2 double transgenic mice.
As expected (Slezak et al., 2007), Slc1a3-Cre/ERT2 resulted in GECI expression in granule cells of the dentate gyrus (yellow arrows, Figure 2D,E; n = 4), but expression in the mossy fiber pathway was only observed for Lck-GCaMP6f (pink arrow; Figure 2D), which implies that Lck-GCaMP6f traffics more efficiently than cyto-GCaMP6f to axons and nerve terminals. No similar neuronal expression was observed with Aldh1l1-Cre/ERT2 mice (n = 4 mice; Figure 2F,G). We also performed a specific set of experiments to evaluate GCaMP6f expression in neurogenic zones of the brain by examining colocalisation with NeuN, which is one of several markers that label newly born neurons (Garcia et al., 2004). Thus, we found no significant colocalisation between NeuN and GCaMP6f driven by Aldh1l1-Cre/ERT2 in the subgranular zone (SGZ), dentate gyrus (DG), subventricular zone (SVZ), rostral migratory stream (RMS) or the granule cell layer of the olfactory bulb (GCL OB; Supp Figure 4; n = 4 mice each). The data illustrate the pan-astrocyte and specific nature of Cre-dependent gene expression driven by Aldh1l1-Cre/ERT2 in adult mice at ~P56 when evaluated ~3 weeks after the final tamoxifen injection with a standard five-day induction protocol.
We acquired higher magnification images of Lck- and cyto-GCaMP6f expressing astrocytes from the CA1 region of the hippocampus, V1 area of the visual cortex and striatum when Cre was expressed either from the Slc1a3-Cre/ERT2 or the Aldh1l1-Cre/ERT2 mouse lines (Figure 3C–N). In all three areas, we noted that Lck-GCaMP6f was expressed within entire bushy astrocytes (Figure 3C–N). Because expression driven by Aldh1l1-Cre/ERT2 occurred in virtually all astrocytes, individual astrocyte territories were hard to delineate (Figure 3C,D & G, H & K,L). In contrast, individual astrocyte territories could easily be delineated in mice in which expression was driven by Slc1a3-Cre/ERT2 mice (Figure 3E,F, I,J & M,N). Moreover, by comparing the expression of Lck-GCaMP6f to that of cyto-GCaMP6f, irrespective of the Cre/ERT2 line used or the brain area, it was apparent that the membrane tethered Lck-GCaMP6f labelled the surface area of astrocytes whereas cyto-GCaMP6f mostly revealed internal volumes (Figure 3). Thus, there were Cre-line-independent, precise differences in the subcellular pattern of GECI expression in astrocytes as revealed by Lck-GCaMP6f and cyto-GCaMP6f (Figure 3C,D, G,H & M,N). This speaks to the utility of both knock-in mouse lines expressing Lck-GCaMP6f and cyto-GCaMP6f to comprehensively study astrocyte calcium signals.
No detectable leaky reporter expression with Aldh1l1-Cre/ERT2 mice
We examined the possibility of leaky GCaMP6f reporter expression in the absence of tamoxifen in Aldh1l1-Cre/ERT2 mice. The injection of corn oil instead of tamoxifen resulted in no detectable GCaMP6f expression, whereas in parallel controls injection of tamoxifen clearly did (Supp Figure 5; n = 4 mice each). As reported in earlier sections, in these additional controls, we did not find any evidence for GCaMP6f colocalisation with NeuN when tamoxifen was administered (Supp Figure 5). In the absence of tamoxifen, there was no detectable GCaMP6f expression (Supp Figure 5; n = 4 mice). As far as we can ascertain, there was no leakiness in the Aldh1l1-Cre/ERT2 mouse line. As expected, the level of GCaMP6f expression in the hippocampus, V1 visual cortex or striatum was dependent on the tamoxifen dose (25–225 mg/kg, Supp Figure 6; n = 3 mice).
In situ calcium imaging: Aldh1l1-Cre/ERT2 mice x Lck-GCaMP6fflox or Ai95 mice
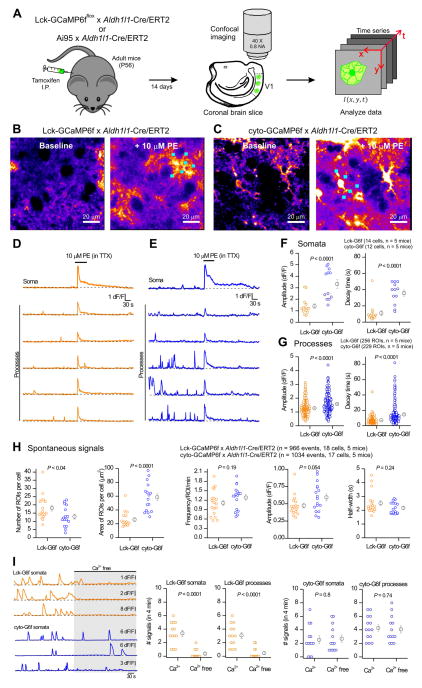
We next determined whether Aldh1l1-Cre/ERT2 mice would drive GECI expression sufficient for astrocyte calcium imaging. We studied astrocyte calcium signals in acute brain slices of V1 visual cortex following tamoxifen induction in adult mice (Figure 4A). Astrocytes respond to endogenously released noradrenaline with calcium elevations mediated by α1 adrenoceptors (Ding et al., 2013). We probed this process in situ with Lck-GCaMP6f and cyto-GCaMP6f mice by applying 10 μM phenylephrine, an α1 adrenoceptor agonist, in the continued presence of TTX (0.5 μM) to minimize secondary effects of neuronal activation. In astrocytes expressing Lck-GCaMP6f or cyto-GCaMP6f, PE-evoked robust calcium elevations (Figure 4B,C & D,E; n = 17–18 cells, 5 mice each). In addition, large amplitude spontaneous calcium signals could be detected with both GECIs (Figure 4D,E) driven by Aldh1l1-Cre/ERT2.
Figure 4. Visual cortex astrocytes from Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 and Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice display spontaneous and PE-evoked calcium signals.
A. Schematic of the approach/workflow for imaging of astrocyte calcium signals in brain slices. B. Representative images of baseline calcium signals in V1 visual cortex astrocytes, before and during 10 μM phenylephrine (PE) in Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mice. C. As in B, but for Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice expressing cyto-GCaMP6f. D. Representative traces of calcium signals in the soma and processes of the astrocyte from the Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 double transgenic mouse shown in B. E. Representative traces of Ca2+ signals in the soma and processes of the astrocyte from the Ai95 x Aldh1l1-Cre/ERT2 double transgenic mouse shown in C. F–G. Scatter plots of PE-evoked calcium signal amplitude and decay times from the somata (F) and processes (G) of visual cortex astrocytes of the Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 and Ai95 x Aldh1l1-Cre/ERT2 double transgenic mice, respectively. H. Scatter plots of spontaneous calcium signal properties in astrocyte processes from Lck-GCaMP6fflox x Aldh1l1-Cre/ERT2 (Lck-G6f) and Ai95 x Aldh1l1-Cre/ERT2 (cyto-G6f) double transgenic mice. I. Representative traces and average data for the effect of Ca2+ free buffers on Ca2+ signals measured with Lck- and cyto-GCaMP6f in astrocyte somata and processes. The data are from 5 mice in panels A–H and for 4 mice for panel I. Sometimes the bars representing the s.e.m. are smaller than the symbol used for the mean.
The PE-evoked calcium signals in cortical astrocytes were significantly smaller when detected with Lck-GCaMP6f as compared to cyto-GCaMP6f for both somata and processes (Figure 4D–G; n = 17–18 cells, n = 5 mice). However, they were also significantly faster in somata and processes, a finding that was clear from the raw traces and average data (Figure 4D–G). We interpret this to support the view that PE-evoked calcium signals are largely driven by intracellular release and hence are optimally detected in terms of amplitude and duration by cyto-GCaMP6f, rather than by Lck-GCaMP6f which may detect the PE-evoked signals only as they spread to the plasma membrane.
Spontaneous signals were also readily detected by Aldh1l1-Cre/ERT2-mediated GECI expression and their properties are reported in detail in Figure 4H. The most notable difference between cyto-GCaMP6f and Lck-GCaMP6f was that ~33% more ROIs were detected per cell with Lck-GCaMP6f than with cyto-GCaMP6f and that the calcium signals in these ROIs covered significantly smaller areas at 26 ± 3 and 59 ± 5 μm2 for Lck-GCaMP6f and cyto-GCaMP6f, respectively (Figure 4H; n = 17 & 18 cells, n = 5 mice each).
Application of Ca2+ free buffers significantly reduced the frequency of Ca2+ signals measured with Lck-GCaMP6f in somata and processes, but had no effect on Ca2+ signals measured with cyto-GCaMP6f (Figure 4I; n = 13 cells and 4 mice). These observations are consistent with studies suggesting that plasma membrane-targeted GECIs identify transmembrane calcium microdomains within astrocytes (Jackson and Robinson, 2015; Melom and Littleton, 2013; Shigetomi et al., 2013; Shigetomi et al., 2010). Overall, Aldh1l1-Cre/ERT2 mediated expression of Lck-GCaMP6f reveals aspects of astrocyte calcium signaling separable and distinct from that reported with cyto-GCaMP6f.
Cortical astrocyte Ca2+ signals in vivo measured with GECIs driven by Aldh1l1-cre/ERT2
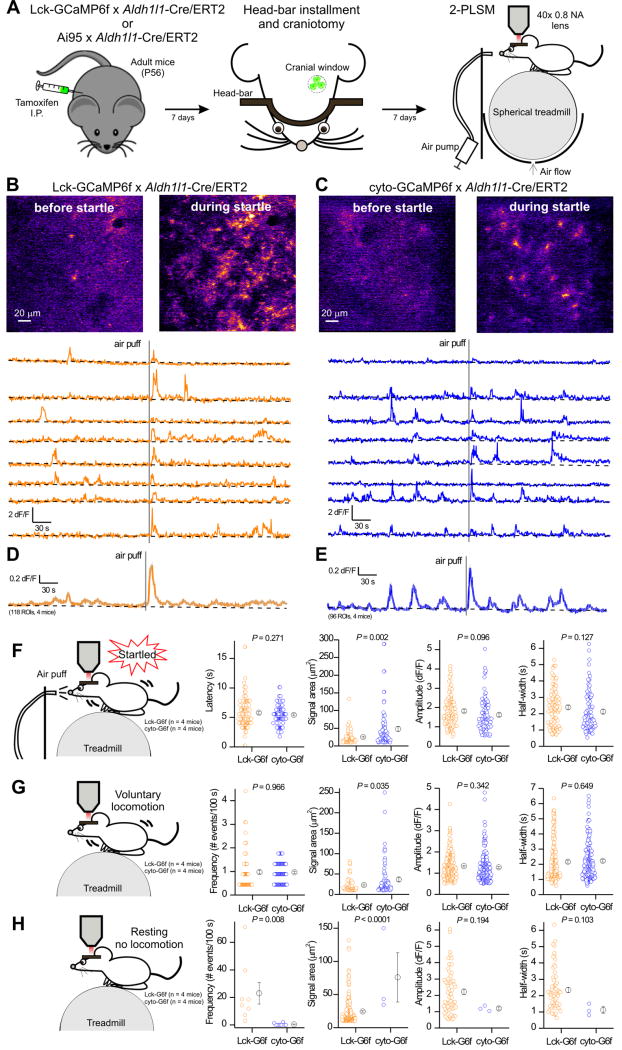
Astrocytes in the visual cortex respond with robust calcium elevations during arousal/startle due to the release of noradrenaline from noradrenergic projections via a mechanism likely involving α1 adrenoceptors on astrocytes (Shigetomi et al., 2016). We next tested if astrocyte GECI expression driven by Aldh1l1-Cre/ERT2 mice could detect calcium signals in vivo from non-anesthetized awake behaving mice. We measured spontaneous calcium signals and those triggered by startle, which was elicited by a gentle puff of air to the face (Figure 5A).
Figure 5. Comparison of in vivo astrocyte calcium signals measured with Lck-GCaMP6f and cyto-GCaMP6f driven by Aldh1l1-Cre/ERT2 mice.
A. Schematic drawings showing the workflow for in vivo 2PLSM in head-fixed, awake mice. In brief, after the mice were injected with tamoxifen for 7 days, a lightweight metal head bar was glued to their skull and a 3 mm cranial window was made above the visual cortex. Mice were then head-fixed onto a spherical treadmill where they were free to rest or run. A 40 x objective lens as part of a 2-photon laser scanning imaging microscope was positioned above the cranial window. An air pump outside the microscope enclosure was used to generate an unexpected air puff, which evoked startle response. B & C. Top: Representative pseudo-colored images showing the fluorescence increase of membrane-tethered Lck-GCaMP6f (B) and cyto-GCaMP6f (C) in astrocytes of mouse visual cortex before and during startle. Bottom: representative ΔF/F traces from 8 randomly selected ROIs (10 μm2 each) in Lck-GCaMP6f x Aldh1l1-Cre/ERT2 mice (orange) and cyto-GCaMP6f x Aldh1l1-Cre/ERT2 (blue). The gray vertical line indicates the air puff. The trend for the baseline is shown in black. D. The average F/F trace of 118 ROIs from four Lck-GCaMP6f x Aldh1l1-Cre/ERT2 mice (s.e.m. shown with gray lines for every 5th time point). E. The average F/F trace of 96 ROIs from four cyto-GCaMP6f x Aldh1l1-Cre/ERT2 mice (s.e.m. shown with gray lines for every 5th time point). F. Comparison of calcium signals detected by Lck-GCaMP6f (113 events, n = 4 mice) and cyto-GCaMP6f (94 events, n = 4 mice) during startle. G. Comparison of calcium signals detected by Lck-GCaMP6f and cyto-GCaMP6f during voluntary locomotion (without air puff). H. Comparisons of calcium signals detected by Lck-GCaMP6f and cyto-GCaMP6f during the resting phase (without air puff and locomotion).
The representative images and traces shown in Figure 5B,C and the average traces in Figure 5D,E demonstrate that Aldh1l1-Cre/ERT2 drove GECI expression in cortical astrocytes sufficiently to detect spontaneous and startle-evoked astrocyte calcium signals. Past work shows that cortical astrocytes display calcium elevations during voluntary locomotion (Paukert et al., 2014), which we tracked by recording the movement of the spherical treadmill on which the mice stood (Figure 5A). We next separated astrocyte calcium signals into those that were triggered by startle, those that occurred during voluntary locomotion and those that occurred independently of locomotion and startle, i.e. when the mouse was resting (Figure 5F–H). The main difference between Lck-GCaMP6f and cyto-GCaMP6f with respect to the startle-evoked calcium signals was in the areas that the calcium signals encompassed: the areas were significantly smaller for Lck-GCaMP6f (Figure 5F; 118 and 96 ROIs, n = 4 mice each). This was also observed for the calcium signals that occurred during voluntary locomotion (Figure 5G). Otherwise the startle-evoked and locomotion associated astrocyte calcium signals were similar in terms of amplitude and half-width (Figure 5F,G).
In the case of calcium signals that occurred during resting, we found that Lck-GCaMP6f revealed significantly more signals than cyto-GCaMP6f by a factor of ~60. Thus, the frequency (number of events in 100 s) was 23 ± 8 and 0.4 ± 0.3 for Lck-GCaMP6f and cyto-GCaMP6f, respectively (Figure 5H; P = 0.008). Moreover, the areas of these events were also significantly smaller in the case of Lck-GCaMP6f than those detected by cyto-GCaMP6f (Figure 5H; P < 0.0001). They tended to be larger in amplitude, but this did not reach statistical significance likely because few similar events existed in the cyto-GCaMP6f group (Figure 5H). Overall, these data show that Aldh1l1-Cre/ERT2 mice can drive Lck-GCaMP6f and cyto-GCaMP6f to permit measurement of calcium signals in awake behaving mice (Figure 5).
The signals measured in vivo with Lck-GCaMP6f were generally smaller in area and more microdomain-like, which recalls Figure 4. These data also emphasize that studying astrocytes in vivo with cytosolic and membrane targeted GECIs reveals distinct aspects of calcium signaling. This is relevant given that astrocyte calcium signals are recognised to be heterogeneous and to include significant transmembrane contributions (Jackson and Robinson, 2015; Khakh and Sofroniew, 2015; Melom and Littleton, 2013; Srinivasan et al., 2015). The P7 cortical astrocyte transcriptome (Zhang et al., 2014) also shows the presence of several channel and pump mRNAs that may underlie astrocyte calcium signals; we provide appropriate genetic tools to study these systematically.
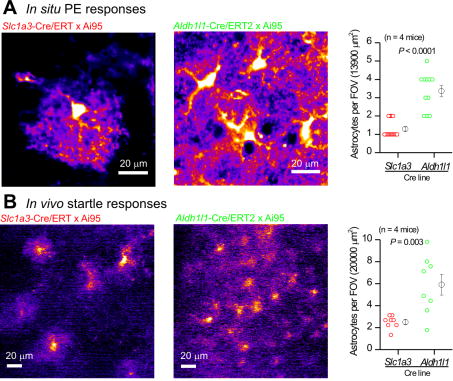
Aldh1l1-Cre/ERT2 mice permit high-density calcium imaging of astrocytes
IHC evaluations (Figures 1–3) showed that Aldh1l1-Cre/ERT2 targets most astrocytes. We next determined if this was also the case for live cell imaging in brain slices and in vivo. We repeated the key experiments shown in Figures 4 and 5 with mice in which cyto-GCaMP6f was driven by Slc1a3-Cre/ERT2 (n = 4 mice) and compared these data to those where cyto-GCaMP6f was driven by Aldh1l1-Cre/ERT2 (n = 4 mice). We found that many more astrocytes could be imaged with Aldh1l1-Cre/ERT2-driven GCaMP6f expression both for acute brain slices and in vivo during startle responses (Figure 6). These differences were highly significant for brain slices (Figure 6A; P < 0.0001) and for in vivo startle-evoked responses (Figure 6B; P = 0.003). The data demonstrate that the use of Aldh1l1-cre/ERT2 mice permits high-density imaging of astrocyte calcium signaling.
Figure 6. Aldh1l1-Cre/ERT2 mice permit high-density imaging of calcium signals in vivo and in brain slices.
A. Representative images and average data of 10 μM PE-evoked astrocyte calcium signals in visual cortex brain slices when cyto-GCaMP6f was driven by Slc1a3-Cre/ERT2 or by Aldh1l1-Cre/ERT2. B. As in A, but for in vivo startle-evoked response in the visual cortex. In both cases, many more astrocytes were detected when cyto-GCaMP6f was driven by the Aldh1l1-Cre/ERT2 mouse.
Aldh1l1-Cre/ERT2 x Ribotag mice reveal the adult cortical astrocyte transcriptome
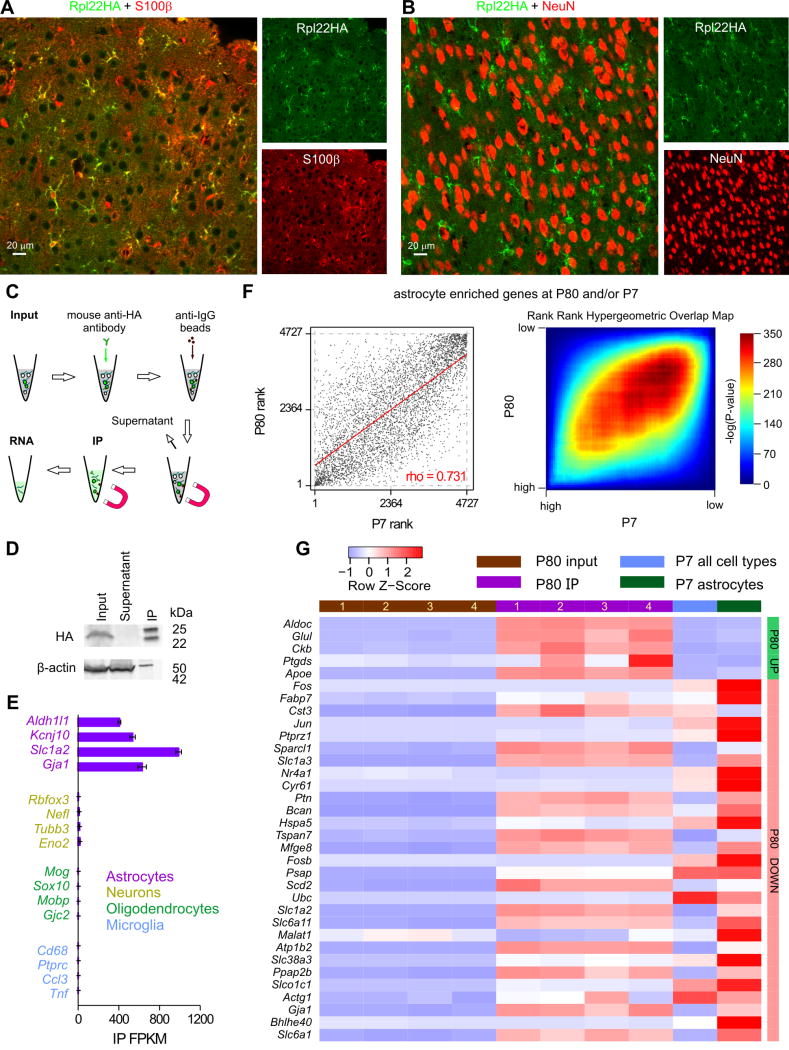
In order to determine the cortical astrocyte transcriptome from adult mice we crossed Aldh1l1-cre/ERT2 mice with RiboTag mice that express the ribosomal protein Rpl22HA in a Cre-dependent manner (Sanz et al., 2009). We found Rpl22HA was strongly expressed in S100β positive cortical astrocytes, but not in NeuN positive neurons (Figure 7A,B; n = 4 mice). We used standard methods (Figure 7C,D) to immunoprecipitate (IP) Rpl22HA containing ribosomes and their associated actively translated mRNAs (Sanz et al., 2009), and performed RNA sequencing to determine the transcriptome of cortical astrocytes at P80 (in quadruplicate). Table 1 of Supplementary Excel file 1 contains the results of the entire P80 RNAseq dataset for the astrocyte IP fraction as well as the input “soup” from all cortical cells (Figure 7C). We found the P80 astrocyte transcriptome (Figure 7E) replete with several known astrocyte markers (Zhang et al., 2014), but depleted of markers for neurons, oligodendrocytes and microglia (Figure 7E; Supplementary Fig 7).
Figure 7. Aldh1l1-Cre/ERT2 x RiboTag mice and the determination of the cortical astrocyte transcriptome at P80.
A. Representative photomicrographs of IHC data showing strong colocalisation between S100β and Rpl22HA in the visual cortex. B. Representative photomicrographs of IHC data showing no colocalisation between NeuN and Rpl22HA. C. Schematic of the workflow. D. The representative Western blot shows that Rpl22HA was preserved in the IP sample, whereas β-actin was depleted in relation to input. In contrast, there was no Rpl22HA in the supernatant. In the IP lane, the 25 and 50 kD bands are the light and heavy chains of the anti-HA antibody that was used in the IP. E. The RNAseq FPKM values of well-established markers of astrocytes, neurons, oligodendrocytes, and microglia in the IP samples are plotted as mean ± s.e.m. from four biological replicates (n = 4 mice). F. Graphs comparing expression of 4727 transcripts enriched in either P80 IP (2-fold enriched over input FDR < 0.05) or P7 astrocytes (2-fold enriched over average of all other cell types) ranked based on FPKM percentile. Genes that were not sequenced in both datasets were excluded from this list. Left: Scatter plot representing the rank of each gene in the P80 (x-axis) vs. the P7 (y-axis) dataset. Clustering along the diagonal indicates similar rank in both datasets. Right: Rank-rank hypergeometric overlap (RRHO) heatmap. Each pixel represents the significance of overlap between the two datasets (-log10(pvalue), hypergeometric test, bin size = 50). Red cells represent highly significant overlap. Color scale (right) represents a range between -log10(pvalue) = 0 (p = 1) and 350 (p = 10−350) G. A heatmap showing relative expression (row z-score) of the 32 genes whose percentile FPKM differ by at least 0.1 between highly expressed P80 and P7 cortical astrocytes determined by RRHO algorithm. These genes and their FPKM values are reported in Supp Table 2. All genes for the analysis in panel F and all the raw data are provided as part of Supplementary Excel file 1. The data used for P7 were from Zhang et al., (2014)
Supp Figure 7 plots the Row z-score values from our P80 data set for the top 50 genes that were astrocyte enriched in P7 cortical astrocytes (Zhang et al., 2014). Most of these are also enriched in the P80 data set, but there were exceptions indicating abundant astrocyte-enriched genes which were differentially expressed between IP and input at P80 (Supp Fig 7). In order to compare P7 and P80 across the whole dataset, we determined the 4727 transcripts enriched in astrocytes in either dataset and used the Rank-rank hypergeometric overlap (RRHO) method (Plaisier et al., 2010; Stein et al., 2014) to compare their relative rank according to FPKM percentile (Figure 7F). Most transcripts were significantly clustered along the diagonal, which indicates similarity in rank between P7 and P80 (Figure 7F). However, many transcripts did not cluster near the diagonal, indicating different expression between P7 and P80; these can be seen more clearly in the scatter graph in Figure 7F. Figure 7G plots heat maps for the top 34 differentially expressed genes between P7 and P80 (i.e. delta percentile > 0.1) and Supplemental Table 1 reports the results of Gene Ontology (GO) analyses for these 34, listed in Supplemental Table 2.
Examination of Figure 7G also reveals several genes that are markedly altered between P7 and P80: the genes encoding membrane proteins (Slc1a3) GLAST, (Slc1a2) Glt1, (Slc6a11) GAT3, (Slc6a1) GAT1 and (Gja1) Cx43, and the genes for a calcium binding protein and a chondroitin sulfate proteoglycan (Sparcl1 and Bcan). Similar changes are summarized in Supplemental Table 3 for several genes that are frequently invoked in astrocyte biology. S100b was highly expressed, but increased substantially at P80 (Supplemental Table 3). Moreover, as expected (Sun et al., 2013) mGluR2 receptor (Grm2) was increased in the P80 data set relative to P7, and mGluR5 (Grm5) was decreased at P80 relative to P7 (Supplemental Table 3). As expected, Grm2 was not astrocyte enriched at P7 or P80 (N/E in Supplemental Table 3). Of the known astrocyte secreted factors, HEVIN (Sparcl1) was higher at P80 and thrombospondins 1–3 were lower at P80 relative to P7. Vesicular neurotransmitter transporters for glutamate, ACh, nucleotides, GABA and monoamines were all very low in astrocytes at both P7 and P80, although vGlut1 (Slc17a7) was higher at P80 than at P7 (Supplemental Table 3). None of these transporters were astrocyte enriched (Supplemental Table 3). The role of astrocyte vesicular gliotransmission remains debated and our P80 dataset, along with Aldh1l1-Cre/ERT2 mice, will be useful to plan specific experiments in adult mice. Broadly, however, our data are in accord with those at P7 (Zhang et al., 2014) and show low mRNA for known vesicular neurotransmitter transporters (Supplemental Table 3). This brief analysis illustrates that astrocyte P80 RNAseq data provide validation of the Aldh1l1-Cre/ERT2 mouse and a resource to explore the functions of specific genes enriched in adult astrocytes (Figure 7) as well as those that are not enriched, but change in relative expression between P7 and P80 astrocytes (Supplementary Excel file).
Summary
We have developed much needed transgenic mouse resources that will advance our ability to explore astrocyte biology within neural circuits in brain slices and most importantly in vivo.
First, extending pioneering transcriptome studies (Cahoy et al., 2008), we provide well characterized Aldh1l1-Cre/ERT2 mice that allow for specific, regulated and pan-astrocytic gene expression under the conditions we report (induction at ~P56 and assessment at ~P80). These mice will be invaluable to study astrocyte biology at different developmental stages, during disease processes and in the context of injury and trauma. However, we believe the use of these mice will be particularly important to tease apart, through loss- and gain-of-function studies, the roles that astrocytes play at synapses, in neural circuits and their contributions to mouse behaviour (Allen, 2014). This has been a challenging problem to attack, especially with regards to gliotransmision (Sloan and Barres, 2014). In addition, several brain diseases are now considered to be non-cell autonomous and to include important astrocyte contributions (Barres, 2008). Aldh1l1-Cre/ERT2 mice will allow researchers to directly express and delete the disease causing genes specifically in astrocytes in a temporally controlled manner and thus tease apart disease mechanisms that may yield new therapeutic targets. This has been a critical issue that existing astrocyte specific genetic strategies have not conclusively addressed. With appropriate controls for age, disease settings and brain area in each case, we expect Aldhll1-Cre/ERT2 mice to fill this void.
Second, we provide knock-in Lck-GCaMP6fflox mice at the ROSA26 locus. These mice will be necessary to directly measure calcium signals in astrocyte processes. The use of these mice, along with Ai95 mice, will help to understand the molecular basis of the diverse calcium signals that astrocytes display and permit exploration of the rules for neuron-to-astrocyte calcium signaling (Shigetomi et al., 2016). Of note, the Lck-GCaMP6fflox mice represent robust tools to explore calcium signals in astrocyte processes, i.e. in locations where most physiological signaling may occur. The general nature of the Lck-GCaMP6fflox mice also means that the GECI could be expressed in any cell type for which a Cre line is available; they may be useful to study calcium signals in axons, nerve terminals and dendritic spines.
Third, we provide initial assessment of astrocyte calcium signals in V1 cortical slices and in vivo in awake-head fixed mice using both Lck-GCaMP6f and cyto-GCaMP6f, during startle, spontaneous locomotion and resting states. These data illustrate the similarities and key differences between measuring calcium signals in the cytosol and in near membrane regions and provide a rationale for future studies in vivo. The high density imaging enabled by the use of Aldh1l1-Cre/ERT2 mice opens up the possibility of imaging astrocyte calcium signals in freely behaving mice equipped with miniature wearable microscopes and perhaps even transcranially.
Fourth, we provide an RNAseq dataset for astrocyte-enriched genes at P80, which can be used in conjunction with a previously reported dataset for P7 to focus on individual genes and molecular pathways at these two ages. The combination of these two datasets will permit precise exploration of astrocyte signaling in vivo and allow for exploration of mechanisms based on knowledge of adult gene expression patterns in the cortex. Initial analyses already suggest several new mechanisms to explore (see Figure 7). The adult mouse cortical dataset is also valuable to compare with human mature astrocyte transcriptome datasets that have recently become available (Zhang et al., 2016). The Aldh1l1-Cre/ERT2 x RiboTag mice are the tools of choice to document astrocyte similarities and differences between distinct areas of the brain. Thus, our RNAseq data provide a basis to explore astrocyte diversity between specific adult neural circuits and brain areas (Khakh and Sofroniew, 2015; Zhang and Barres, 2010).
As exploration of astrocytes in mouse behavior and disease models advances, there will be necessity to perform astrocyte specific, temporally controlled and pan-astrocytic genetic manipulations, as well as study aspects of their signaling in physiologically relevant compartments such as processes. The resources we report are valuable in these regards. However, we emphasise that we did not examine tamoxifen inducible reporter expression in very young mice or in adult mice greater than 3 weeks after tamoxifen administration; we evaluated mice at ~P80. Hence, our results need to be interpreted with this in mind and future users of the Aldh1l1-Cre/ERT2 mice will need to perform their own controls: it is important not to extrapolate to all brain areas, all ages, all experimental settings and all reporter mice from any one study, including ours. Hence, our studies do not obviate the need for controls, especially in disease related research where gene expression patterns and cell fates may change significantly. We have specifically focused on adult mice and the expression of GECIs at levels appropriate for imaging in brain slices and in vivo. These analyses reveal Aldh1l1-Cre/ERT2 mice to be valuable tools to study astrocyte biology.
Experimental procedures
All animal experiments were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Chancellor’s Animal Research Committee at the University of California, Los Angeles. Detailed procedures are provided in the Supplemental information.
Generation of Aldh1l1-Cre/ERT2, Slc6a11-Cre/ERT2 and Lck-GCaMP6fflox mice
BAC transgenic mice expressing Cre/ERT2 from the Aldh1l1 and Slc6a11 locus were made using published methods (Yang and Gong, 2005). The Lck-GCaMP6fflox mice contain a CAG promoter, followed by a loxP-3xSV40pA-loxP cassette, followed by the Lck-GCaMP6f cDNA at the ROSA26 locus. In brief, ROSA26 targeting plasmid, called Ai39 was purchased from Addgene (plasmid #34884) and then modified to create the ROSA26 Lck-GCaMP6f targeting vector. Knock-in mice were then made using well established methods.
Our mouse lines will be available from The Jackson Laboratory: JAX Stock No. 029626: B6N.Cg-Gt(ROSA)26Sor<tm1(CAG-GCaMP6f)Khakh>/J with a common name of: “R26-Lck-GCaMP6fflox knock-in”, JAX Stock No. 029655: B6N.FVB-Tg(Aldh1l1-cre/ERT2)1Khakh/J with a common name of “Aldh1l1-Cre/ERT2 BAC transgenic” and JAX Stock No. 029656: B6N.FVB-Tg(Slc6a11-cre/ERT2)#Khakh/J with a common name of “Slc6a11-Cre/ERT2 BAC transgenic”.
Data analyses
Calcium signals were analyzed using MiniAnalysis program 6.0.7 (Synaptosoft) and Origin 8.5 (Origin Lab Corp.). Image analyses were performed using ImageJ v1.30 (NIH). In brief, the data were analyzed as previously reported (Srinivasan et al., 2015). Data for ATP-evoked responses were analyzed from circular 50 μm2 ROIs, encompassing astrocyte territories and data for PE-evoked responses were acquired from square 10 μm2 ROIs drawn within individual astrocyte territories. Time traces of fluorescence intensity were extracted from the ROIs, converted to dF/F values and the time-to-peak, response amplitude and decay time were measured using a data selector function in Origin 2015 (Origin labs). We analyzed spontaneous events that occurred in the first 250 s before agonist application. Events were identified based on amplitudes that were at least 2-fold above the baseline noise of the dF/F trace. Spontaneous events were manually marked and event amplitudes, half width and event frequency per ROI per min was measured using MiniAnalysis 6.0.07 (Synaptosoft).
To generate hippocampal montages (Figure 1), images of all hippocampal sub-regions (CA1, CA3 and DG) were obtained from 40 μm thick coronal brain sections using a Fluoview 1000 confocal microscope, equipped with a 20X 0.85 NA oil immersion lens (Olympus). Images were acquired at 1X digital zoom, such that immediately adjacent images had ~150 to 200 μm of overlapping cellular features. Individual images obtained in this way were manually stitched together and then exported as a single hippocampal montage. Numbers for GFP positive astrocytes and neurons were acquired by manually counting cells from hippocampal montages. Total numbers of astrocytes or neurons were obtained by counting the number of cells in the montage with S100β or NeuN staining, respectively.
Statistical tests were run in Origin 9 or GraphPad Instat 3. Data are presented as mean ± s.e.m. Note that in some of the graphs, the bars representing the s.e.m. are smaller than the symbols used to represent the mean. For each set of data to be compared we determined within GraphPad Instat whether the data were normally distributed or not. If they were normally distributed we used parametric tests, otherwise we used non-parametric tests. Paired and unpaired Student’s two-tailed t tests (as appropriate) and two tailed Mann–Whitney tests were used for most statistical analyses with significance declared at P < 0.05, but stated in each case with a precise a P value. When the P value was less than 0.0001, it is stated as P < 0.0001 to save space. N is defined as the numbers of cells and/or mice throughout on a case-by-case basis depending on the particular experiment.
Supplementary Material
Highlights.
Cre/ERT2 mice were made to achieve astrocyte specific genetic manipulations in vivo
Knock-in Lck-GCaMP6f mice were made to study astrocyte calcium signals in vivo
Mice were used to determine the adult cortical astrocyte transcriptome
New, well characterised and much needed in vivo genetic resources are provided
Acknowledgments
This work was mainly supported by the National Institutes of Health grant MH099559 and MH104069 (to BSK). We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics at UCLA (P30 NS062691). HC was supported by a NIH F30 Training Fellowship (MH106197). Thanks to Michael V. Sofroniew for mGfap-Cre 77.6 mice and for providing access to equipment. Thanks to Michael V. Sofroniew and Ben Novitch for guidance on studying neurogenic zones in tissue slices. Thanks to X. William Yang for use of a pulse-field gel apparatus, Fuying Gao for assistance with RNAseq data analysis, and to Xiao-Hong Lu for discussions. Thanks also to the UCLA Clinical Microarray Core. Thanks to Dr. Frank Pfrieger (Strasbourg, France) for Cx30-Cre/ERT2 mice. Many thanks to Dr. Amy Gleichman for help using the Nikon C2 confocal.
Footnotes
Supplementary information: 7 figures, 1 supplemental movie, 3 tables, Supplementary Excel file reporting the adult cortical astrocyte transcriptome, detailed methods.
Author contributions
RS and JX performed molecular biology for the BAC mice (Aldh1l and Slc6a11, respectively). RS performed cloning for the Lck-GCaMP6fflox mice. RS and T-YL performed IHC. RS performed brain slice imaging and AAV FLEX GFP experiments. T-YL performed 2PLSM. HC performed RNAseq. GC and HC carried out RNAseq data analyses. RS, JX, T-YL, HC and BSK performed some genotyping and mouse colony management. BSH shared expertise in cranial window surgery and trained T-YL. PG provided access to 2-PLSM equipment. BSK conceived, designed and directed the research project, helped with data analyses, made the final figures and wrote the paper with input from RS, T-YL, GC and HC. All authors commented on the final version. The data in main Figures 1, 2, 3, 4, 6A, Supp Fig 1, Supp Fig 2, Supp Fig 3, Supp Fig 4, Supp Fig 5, Supp Fig 6, Supplemental Movie 1 and Table 1 were gathered by RS. Also, two of the three mice were made by RS. Data in Figure 7C–G, Supp Fig 7, Supplemental Tables 1–3 and Tables 1–4 of the Supplemental Excel spreadsheet were gathered by HC. Data in Figures 5, 6B, 7A,B were gathered by T-YL.
Supplemental Information available online
Seven figures, a movie and three tables are submitted as supplementary information
A Supplementary Excel file containing the RNAseq data is included (4 tables in separate sheets)
The Supplementary information also contains all the methods
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen NJ. Astrocyte regulation of synaptic behavior. Annu Rev Cell Dev Biol. 2014;30:439–463. doi: 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Bazargani N, Attwell D. Astrocyte calcium signalling: the third wave. Nat Neurosci. 2015 doi: 10.1038/nn.4201. accepted. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila D, Thibault K, Fiacco TA, Agulhon C. Recent molecular approaches to understanding astrocyte function in vivo. Front Cell Neurosci. 2013 Dec 24;7:272. doi: 10.3389/fncel.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, O'Donnell J, Thrane AS, Zeppenfeld D, Kang H, Xie L, Wang F, Nedergaard M. alpha1-Adrenergic receptors mediate coordinated Ca2+ signaling of cortical astrocytes in awake, behaving mice. Cell Calcium. 2013;54:387–394. doi: 10.1016/j.ceca.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Dougherty JD. Aldh1L1 is expressed by postnatal neural stem cells in vivo. Glia. 2013;61:1533–1541. doi: 10.1002/glia.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gregorian C, Nakashima J, Le Belle J, Ohab J, Kim R, Liu A, Smith KB, Groszer M, Garcia AD, Sofroniew MV, et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci. 2009;29:1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Haydon PG. Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Annu Rev Physiol. 2010;72:335–355. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein MD, Kracun S, Lu XH, Shih T, Jackson-Weaver O, Tong X, Xu J, Yang XW, O'Dell TJ, Marvin JS, et al. Conditions and constraints for astrocyte calcium signaling in the hippocampal mossy fiber pathway. Neuron. 2014;82:413–429. doi: 10.1016/j.neuron.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JG, Robinson MB. Reciprocal Regulation of Mitochondrial Dynamics and Calcium Signaling in Astrocyte Processes. J Neurosci. 2015;35:15199–15213. doi: 10.1523/JNEUROSCI.2049-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melom JE, Littleton JT. Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J Neurosci. 2013;33:1169–1178. doi: 10.1523/JNEUROSCI.3920-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Deneen B. Astrocyte development: A Guide for the Perplexed. Glia. 2015;63:1320–1329. doi: 10.1002/glia.22836. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Glasgow SM, Chaboub LS, Tsai HH, Murnen AT, Kelley KW, Fancy SP, Yuen TJ, Madireddy L, Baranzini S, et al. Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia. 2013;61:1518–1532. doi: 10.1002/glia.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang CL. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukert M, Agarwal A, Cha J, Doze VA, Kang JU, Bergles DE. Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron. 2014;82:1263–1270. doi: 10.1016/j.neuron.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010 Sep;38(17):e169. doi: 10.1093/nar/gkq636. Epub 2010 Jul 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Site-specific recombination: developments and applications. Curr Opin Biotechnol. 1994;5:521–527. doi: 10.1016/0958-1669(94)90068-x. [DOI] [PubMed] [Google Scholar]
- Shigetomi E, Bushong EA, Haustein MD, Tong X, Jackson-Weaver O, Kracun S, Xu J, Sofroniew MV, Ellisman MH, Khakh BS. Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J Gen Physiol. 2013;141:633–647. doi: 10.1085/jgp.201210949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Kracun S, Sofroniew MV, Khakh BS. A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat Neurosci. 2010;13:759–766. doi: 10.1038/nn.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Patel S, Khakh BS. Probing the Complexities of Astrocyte Calcium Signaling. Trends Cell Biol. 2016;26:300–312. doi: 10.1016/j.tcb.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezak M, Göritz C, Niemiec A, Frisén J, Chambon P, Metzger D, Pfrieger FW. Transgenic mice for conditional gene manipulation in astroglial cells. Glia. 2007;55:1565–1576. doi: 10.1002/glia.20570. [DOI] [PubMed] [Google Scholar]
- Sloan SA, Barres BA. Looks can be deceiving: reconsidering the evidence for gliotransmission. Neuron. 2014;84:1112–1115. doi: 10.1016/j.neuron.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P, Khakh BS. Ca(2+) signaling in astrocytes from Ip3r2(-/-) mice in brain slices and during startle responses in vivo. Nat Neurosci. 2015;18:708–717. doi: 10.1038/nn.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernández IA, Marchetto MC, Baker DK, Lu D, Hinman CR, Lowe JK, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Hu H, Lee Y, d'Azzo A, Messing A, Brenner M. Expression specificity of GFAP transgenes. Neurochem Res. 2004;29:2075–2093. doi: 10.1007/s11064-004-6881-1. [DOI] [PubMed] [Google Scholar]
- Sun W, McConnell E, Pare JF, Xu Q, Chen M, Peng W, Lovatt D, Han X, Smith Y, Nedergaard M. Glutamate-dependent neuroglial calcium signaling differs between young and adult brain. Science. 2013;339:197–200. doi: 10.1126/science.1226740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XD, Li L, Liu F, Huang ZH, Bean JC, Jiao HF, Barik A, Kim SM, Wu H, Shen C, et al. Lrp4 in astrocytes modulates glutamatergic transmission. Nat Neurosci. 2016 Jun 13; doi: 10.1038/nn.4326. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ. Cre-Lox Neurogenetics: 20 Years of Versatile Applications in Brain Research and Counting…. Front Genet. 2016 Feb 19;7:19. doi: 10.3389/fgene.2016.00019. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie AX, Petravicz J, McCarthy KD. Molecular approaches for manipulating astrocytic signaling in vivo. Front Cell Neurosci. 2015 Apr 21;9:144. doi: 10.3389/fncel.2015.00144. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Gong S. An overview on the generation of BAC transgenic mice for neuroscience research. Curr Protoc Neurosci. 2005 May;Chapter 5(Unit 5.20) doi: 10.1002/0471142301.ns0520s31. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.