Abstract
CHARMM-GUI, http://www.charmm-gui.org, is a web-based graphical user interface that prepares complex biomolecular systems for molecular simulations. CHARMM-GUI creates input files for a number of programs including CHARMM, NAMD, GROMACS, AMBER, GENESIS, LAMMPS, Desmond, OpenMM, and CHARMM/OpenMM. Since its original development in 2006, CHARMM-GUI has been widely adopted for various purposes and now contains a number of different modules designed to set up a broad range of simulations: (1) PDB Reader & Manipulator, Glycan Reader, and Ligand Reader & Modeler for reading and modifying molecules; (2) Quick MD Simulator, Membrane Builder, Nanodisc Builder, HMMM Builder, Monolayer Builder, Micelle Builder, and Hex Phase Builder for building all-atom simulation systems in various environments; (3) PACE CG Builder and Martini Maker for building coarse-grained simulation systems; (4) DEER Facilitator and MDFF/xMDFF Utilizer for experimentally guided simulations; (5) Implicit Solvent Modeler, PBEQ-Solver, and GCMC/BD Ion Simulator for implicit solvent related calculations; (6) Ligand Binder for ligand solvation and binding free energy simulations; and (7) Drude Prepper for preparation of simulations with the CHARMM Drude polarizable force field. Recently, new modules have been integrated into CHARMM-GUI, such as Glycolipid Modeler for generation of various glycolipid structures, and LPS Modeler for generation of lipopolysaccharide structures from various Gram-negative bacteria. These new features together with existing modules are expected to facilitate advanced molecular modeling and simulation thereby leading to an improved understanding of the molecular details of the structure and dynamics of complex biomolecular systems. Here, we briefly review these capabilities and discuss potential future directions in the CHARMM-GUI development project.
1. INTRODUCTION
Modeling and simulation of biologically important molecules have come a long way since their inception [1]. Various computational methodologies have been developed not only to elucidate the mysteries in biological phenomena [2–6], but also to predict the outcome when the systems or the conditions are changed. Nowadays, computations can be used to design new experiments to test the developed hypotheses [7, 8]. Combined with the tremendous increase in computing power, biomolecular modeling and simulation have become routine tools used in many research laboratories [9].
An issue that is not always fully appreciated is the difficulty to prepare and initiate the complex computational tasks needed to achieve a specific objective. Except for simple modeling and simulation tasks, the execution of most computational methods requires far more than a single “click”. Advanced biomolecular modeling programs rely on sophisticated scripts, and thus handling all the relevant information to prepare a complex simulation system correctly often incurs a considerable amount of human time. This is an important issue, for beginners and experts alike, that cannot be overstated. Various software tools have been produced to address this need and offer support to the users [10, 11]. Among these tools, the emergence of web-based modeling tools is notable. For example, CHARMMing [12], Glycam Biomolecule Builder [13], and MDWeb [14] provide graphical web interface for preparing chemical systems for molecular dynamics (MD) simulations. In addition, there is a battery of web-based tools for preparing custom force field (FF), such as ParamChem [15, 16], MATCH [17], SwissParam [18], GAAMP [19], and RED [20]. Compared to downloadable software, web-based tools do not require program installation or upgrades on the user end and can be accessed using a web browser, rendering them “platform-independent” and easy to use.
Among various web-based modeling tools, CHARMM-GUI [21], http://www.charmm-gui.org, sets out to simplify and generalize the protocols for building complex simulation systems and preparing simulation input files for widely used simulation packages, such as CHARMM [22], NAMD [23], GROMACS [24], AMBER [25], GENESIS [26], LAMMPS [27], Desmond [28], OpenMM [29], and CHARMM/OpenMM [30] to facilitate the usage of common and advanced simulation techniques. Molecular simulation systems are often comprised of multiple components, and in-depth knowledge of modeling software is often necessary to understand and build a sophisticated simulation system. In addition, generalizing the system building protocols is challenging because customization is often necessary. For example, building a realistic and physiologically relevant protein-membrane complex containing water, ions, protein, and various lipid types would require a considerable effort, even for an expert. CHARMM-GUI Membrane Builder successfully encapsulates such a sophisticated process into a well-defined workflow that can reproducibly generate a realistic protein-membrane complex or a membrane-only system within minutes to a few hours depending on the system size [21, 31–33].
Many online tools aim to provide a general modeling facility to users. One notable exception is the “What-If” server (http://swift.cmbi.ru.nl/whatif/) developed by Vriend and co-workers [34]. The server provides a simple and easy-to-use interface to explore consequences of structural changes to a protein; e.g., “what if a certain residue is mutated?” or “how would the solubility of the protein change?” By focusing on a task, instead of providing low-level functionality, e.g., introducing mutation, the server is able to simplify and streamline many aspects of interface design.
Inspired by such task-oriented design approach, our philosophy in developing CHARMM-GUI is less about providing the nuts and bolts of molecular modeling, but we instead focus on helping users to achieve a task, such as building a membrane system or solvating a protein, by providing a streamlined interface. This design principle helps our team to think of the workflow critically while designing the interface, which led CHARMM-GUI to be accessible to users with less experience in modeling tools and yet still useful to experts, especially for batch generation of systems. As with any design decision, there is a trade-off with the task-oriented approach that we have adopted in CHARMM-GUI. For example, a major limitation is that CHARMM-GUI contains many pre-determined parameters (e.g., number of steps for minimization or the specifics of how to solvate a protein), which are hidden from users, potentially making CHARMM-GUI less appealing to more advanced users for their specific customization needs. To address such issue, CHARMM-GUI provides raw CHARMM input files, allowing users to optimize our default protocols or adopt more sophisticated approaches without having to create an entire input file from scratch. This practice is particularly useful as the program CHARMM is now free for academic use (http://www.charmm.org).
Since its original development in 2006, CHARMM-GUI has been extended to a range of capabilities and now contains a number of different modules designed to set up a broad range of biological systems (Figure 1). Many of the original CHARMM-GUI modules were developed as an in-house effort, but we have undertaken a number of collaborative efforts with other groups to develop new modules. Many novel computational methodologies require complex system setup and the use of sensible parameters. In our co-development, we focus on a limited number of specific use cases and implement a clear and concise user-interface encapsulating a complex workflow with sensible, tested parameters. In the remainder of this review, we briefly discuss different modules in CHARMM-GUI that are currently available and our ongoing efforts in bringing more complex features to users.
Figure 1.
Schematic view of modules in CHARMM-GUI Input Generator.
2. Functionalities of CHARMM-GUI Input Generator
2.1 PDB reading and manipulation
Reading a PDB file containing multiple components into a simulation program is not straightforward and is generally considered the first hurdle in a simulation project. It is typically even harder to introduce mutations, different protonation states of titratable residues, disulfide bonds, other post-translational modifications (such as phosphorylation, glycosylation, and lipid-tail linkers), naturally modified nucleic acids, or experimental modifications (such as adding spin labels, fluorophores, and unnatural amino acids). PDB Reader & Manipulator provides a flexible web interface to handle these and converts a PDB file (downloaded from RCSB [35], http://www.rcsb.org, or uploaded by the user) into a protein structure file (PSF) that can be directly read by some simulation packages or readily converted to other formats, facilitating the setup and simulation of all-atom heterogeneous systems with the use of the comprehensive CHARMM additive force field that covers the full range of biomolecules [36–40]. In addition, since many PDB entries have missing protein residues, PDB Reader & Manipulator detects such missing residues and provides an option to model them using GalaxyFill [41]. If a user wants to only use some components of a PDB file, e.g., solvating a small drug-like molecule in water or embedding a lipid molecule in a lipid bilayer, PDB Reader & Manipulator provides options to model the ligand(s) using CGenFF (CHARMM Generalized force field) [42, 43] or ANTECHAMBER [44] if they do not exist already in the CHARMM additive force fields.
RCSB has been transitioning from PDB to PDBx/mmCIF format. Using mmCIF is advantageous over the conventional PDB format that has fixed column widths, causing issues in the number of atoms, residue names, and/or atom names when an entry is longer than the corresponding width. In addition, mmCIF format is more portable because the data format specifications are included in the same file. PDB Reader & Manipulator is able to handle both PDB and PDBx/mmCIF formats. While PDB Reader & Manipulator exists as an independent module, it is the first step in all other modules.
2.2 Visualization
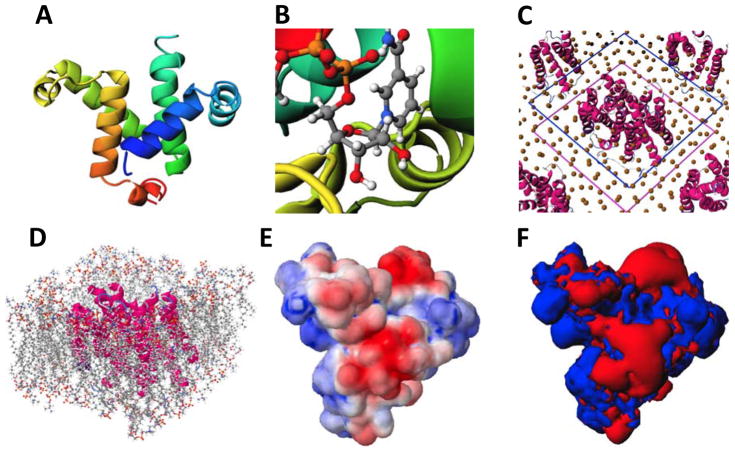
CHARMM-GUI uses JSmol [45] to visualize three-dimensional (3D) molecular structures, allowing users to check their system in each step. Currently, CHARMM-GUI provides a variety of viewing modes (Figure 2): (1) protein view for protein-only structures with ribbon diagram rendering; (2) protein/membrane (or ligand) complex view with lipids as CPK and protein as ribbon diagram; (3) orientation view for displaying both upper and lower planes indicating the bilayer hydrophobic core; (4) packing image view for lipid-like pseudo atoms with spheres (for both primary and neighboring image systems); and (5) electrostatic potential view for displaying electrostatic potential or contour maps obtained from the PBEQ Solver. Users do not need to install any additional program to display/render the structures in CHARMM-GUI, and the concise workflow with an appropriate mode in each building step increases the viewing speed.
Figure 2.
Molecular graphics views of (A) KIX domain and co-activator KID (PDB:1KDX), (B) DOX-P reductoisomerase with NADH (PDB:4ZQG), (C) lipid-like pseudo atom packing around multidrug transporter EmrD (PDB:2GFP) with the primary system indicated by lines, and (D) after replacement of lipid-like pseudo atoms in (C) with corresponding all-atom lipids (before equilibration), (E) electrostatic potential on the solvent-accessible surface representation of KIX domain, and (F) iso-electrostatic potential contour of KIX domain.
2.3 Solution simulation
Starting from PDB Reader & Manipulator, input files for standard MD (equilibration and production) simulations of globular proteins and/or other molecules in aqueous solvent environments can be quickly prepared using Quick MD Simulator. All the simulation input files are prepared to use periodic boundary conditions (PBC) with the particle-mesh Ewald (PME) method [46] for long-range electrostatic interactions. In general, all simulation protocols in CHARMM-GUI are optimized for each FF used in different modules [47], following the principles of the original FF developments.
2.4 Membrane simulation
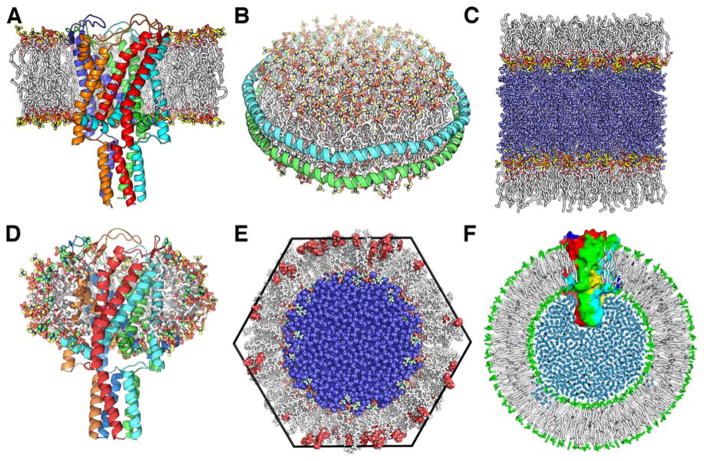
Biological membranes are complex in terms of lipid types and their compositions [48], and the lipid molecules may interact with proteins in a specific manner. To better represent a native environment, more fatty acid, lipid, and detergent types have been added to CHARMM-GUI. Although lipid bilayers are the most common in biological systems, other membrane environments such as monolayer, micelle, nanodisc, hexagonal lipid phase, and vesicle are commonly used in experiments and thus require computational tools to build them (Figure 3).
Figure 3.
Molecular graphics views of (A) bilayer, (B) nanodisc, (C) monolayer, (D), micelle, (E) inverse hexagonal lipid phase, and (F) vesicle. The mechanosensitive channel of large conductance (MscL) is embedded in a bilayer, a micelle, and a vesicle. Water molecules are shown as blue spheres only in (C), (E), and (F). The alkane molecules in the hexagonal phase system are shown as red spheres.
Membrane Builder [31–33] helps users to generate a protein-membrane complex with homogeneous or heterogeneous bilayers, as well as membrane-only bilayers. As of 2016, Membrane Builder has 295 lipid types in the context of CHARMM additive FF including phosphoinositides, cardiolipin, sphingolipids, bacterial lipids, sterols, and fatty acids, which makes it possible to build a realistic biological membrane system. Since RCSB PDB structures do not contain the orientation information of a membrane protein relative to lipid bilayers (whose normal is pre-defined to be parallel to the Z-axis and center is located at Z = 0), users can use the OPM database (http://opm.phar.umich.edu) [49] to obtain pre-oriented protein structures. Membrane Builder also provides several options to orient proteins. We have optimized the automated building process including system size determination as well as generation of lipid bilayer, water, and ions. Any complex (homogeneous or heterogeneous) bilayer system can be generated by the so-called “replacement method” [50, 51] that first packs the lipid-like pseudo atoms (Figure 2C), and then replaces them with lipid molecules one at a time by randomly selecting a lipid molecule from a lipid structural library [3, 32]. Using the replacement method, it generates nicely packed lipid molecules around a protein. For proteins with water-filled lumens, Membrane Builder estimates the pore’s solvent accessible area profile and hydrates the lumen with water. Careful checks of lipid membrane building are included to avoid unphysical structures, such as penetration of acyl chains into ring structures in sterols, aromatic residues, and carbohydrates [33]. The optimized equilibration scripts prevent any unwanted structural changes in lipids and embedded proteins, so that production simulations can probe physically realistic behavior of biomolecules.
Nanodisc Builder can be used to build a nanodisc system (with/without protein) in which two membrane scaffolding proteins (MSPs) wrap around a lipid bilayer to form a discoidal structure [52, 53]. Nanodiscs have been widely used in biophysical and biochemical studies of membrane proteins. As there is no structural information of the MSPs in nanodiscs, Nanodisc Builder uses pre-assembled MSP structures based on the double-belt model [54] by stacking two MSP monomers against each other. Eight different MSPs that form different sizes of nanodiscs are available. In addition to providing all-atom nanodisc simulation inputs, Nanodisc Builder is also available in PACE CG Builder [55] and Martini Maker [56] for coarse-grained (CG) simulations.
Monolayer Builder helps users to generate a series of CHARMM input files necessary to build a protein-monolayer (or monolayer-only) system. The building procedure of a monolayer system is the same as that of a bilayer system except that a bilayer system is converted to a monolayer system by manipulations of the PBC after assembling all the system components.
Micelle Builder [57] helps users to build homogeneous/heterogeneous micelle and protein-micelle systems. Currently, Micelle Builder provides 20 detergent types. Considering the size and shape of the transmembrane segments of the protein, Micelle Builder arranges detergents in a micelle of a torus shape. Therefore, protein-micelle complexes can be built for proteins with any shape [58].
Hex Phase Builder has been developed to probe a lipid phase that exists at high temperature or low water concentration regime (for a review, see [59]) and has been used in biophysical experiments and simulations to gather information on lipids at high curvature [60–65]. Hex Phase Builder uses a building procedure similar to Membrane Builder except that lipids are placed around a water cylinder instead of on a plane.
HMMM Builder [66] aids in studies that are hindered by the slow diffusion of lipids that exists in conventional all-atom membrane simulations, making it difficult to sample large rearrangement in a relatively short simulation time. HMMM Builder provides users with input files for simulating membrane systems (with/without proteins) using the highly mobile membrane-mimetic (HMMM) model [67], in which the lipid tails are replaced with small organic molecules (currently 1,1-dichloroethane, i.e., DCLE), resulting in acceleration of lipid diffusion by 1–2 orders of magnitude. HMMM Builder supports all lipid types available in Membrane Builder, and the user can choose the position to trim an acyl chain and lipid packing ratio. In addition to building HMMM systems, HMMM Builder also provides a conversion tool between HMMM and full-length lipid systems, allowing users to easily take advantage of both HMMM and full-length membranes.
2.5 Coarse-grained simulation
Coarse-graining is a popular approach to model large simulation systems. Martini Maker [56] and PACE CG Builder [55] provide simulation system preparations using the Martini FF [68] and the PACE FF [69], respectively. The Martini model is widely adopted to represent lipid molecules with on average four heavy atoms mapped to one CG atom. Martini Maker facilitates Martini system building for solution, micelle, nanodisc, bilayer, and vesicle simulations as well as simulations starting with randomly distributed detergent/lipid molecules. The supported versions of the Martini FF include standard Martini [68, 70, 71], Martini with polarizable water [72, 73], Dry Martini [74], and ElNeDyn using elastic networks for proteins [75]. The generated simulation input files are ready for use with the latest GROMACS simulation software in which Martini simulations have been optimized. The Martini model is suitable to CG lipid molecules, but proteins are not often suitable to model in the same manner. The PACE model has been developed to perform CG simulations of united-atom (UA) protein with the Martini water and lipid FF [68]. Compared to the corresponding all-atom simulations using a 2-fs time step, the PACE simulation reduces the number of atoms by a factor of 10, and allows an integration time step of 5 fs, which speeds up the simulation about 30 times. The PACE FF provides more details for the protein than the Martini FF, and could be a superior feature in applications for modeling/refining large, complicated biological systems.
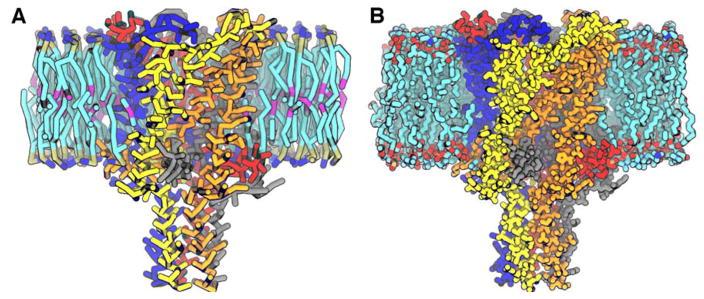
After performing CG simulations, it is sometimes desired to convert the CG system back to an all-atom representation. However, CG to all-atom mapping is a difficult problem. In this context, Martini Maker and PACE CG Builder provide back conversion of a simulation system to an all-atom system using a simple but effective method (Figure 4). The Martini protein is converted using the backward.py script [76] and the PACE protein is converted by adding the hydrogen atoms back using CHARMM HBUILD functionality. Different from other approaches [76–78], the CG lipids are converted into all-atom lipids using a lipid structure library (used in Membrane Builder) by aligning a randomly selected all-atom structure to the CG lipid. The CG waters are converted by mapping one CG water molecule back to four all-atom water molecules in a tetrahedron arrangement.
Figure 4.
Conversion of (A) a Martini bilayer system to (B) an all-atom system. The mechanosensitive channel MscL protein is embedded in a POPC bilayer. Water molecules and ions are not shown for clarity. Different subunits of the MscL are drawn with different colors.
2.6 Protein-ligand simulation
As computer-aided drug design becomes increasingly popular, the number of MD simulations of drug-like molecules is on the rise [79]. Having a well-validated FF is extremely important to obtain meaningful results. Ligand Reader & Modeler helps both beginners and experts to prepare FF parameters conveniently by searching for a ligand in the entire CHARMM FF or using CGenFF. Users are able to upload a ligand to Ligand Reader & Modeler in various ways, and the uploaded structure can be interactively modified on the sketchpad (powered by Marvin JS, http://www.chemaxon.com). In addition, it is possible to draw chemical substitution sites and substituents in each site to generate combinatorial structures and corresponding FF parameters. Finally, the output can be used in other CHARMM-GUI modules to build a simulation system.
Advanced free energy perturbation molecular dynamics (FEP/MD) simulation methods [80–85] can be used to calculate absolute binding free energies of protein–ligand complexes. Typically, setting up a FEP/MD simulation is very difficult because these methods rely on various biasing energy restraints to enhance the convergence of the calculations. Ligand Binder [86] provides the standardized CHARMM input files for calculations of absolute binding free energies using FEP/MD simulations. A number of features, such as implicit treatment of bulk water [87, 88], sophisticated decomposition scheme [89, 90], or Hamiltonian-tempering replica-exchange simulation [91, 92], are implemented to conveniently set up the FEP/MD simulations in a highly customizable manner, thereby permitting an accelerated throughput of this important class of computations while decreasing the possibility of human errors.
2.7 Glycoconjugate simulation
Carbohydrate moieties, referred to as glycans, can be covalently attached to proteins (glycoproteins) and lipids (glycolipids), or exist as free ligands (Figure 5). Understanding how glycosylation affects protein structure, dynamics, and function is an emerging and challenging problem in biology. Notably, about 30% of carbohydrate structures in the PDB contain at least one error regarding the sugar-type assignment [93]. As shown in Figure 6, Glycan Reader greatly simplifies the reading of PDB and PDBx/mmCIF structure files containing glycans using graph representations. Following considerable efforts, Glycan Reader is able to recognize all major carbohydrate chemical modifications in PDB and generate their structure models. While it exists as a separate module, Glycan Reader is linked to other functional modules as part of PDB Reader & Manipulator, allowing users to easily generate molecular simulation systems with carbohydrate or glycoprotein, and visualize the electrostatic potential on glycoprotein surfaces.
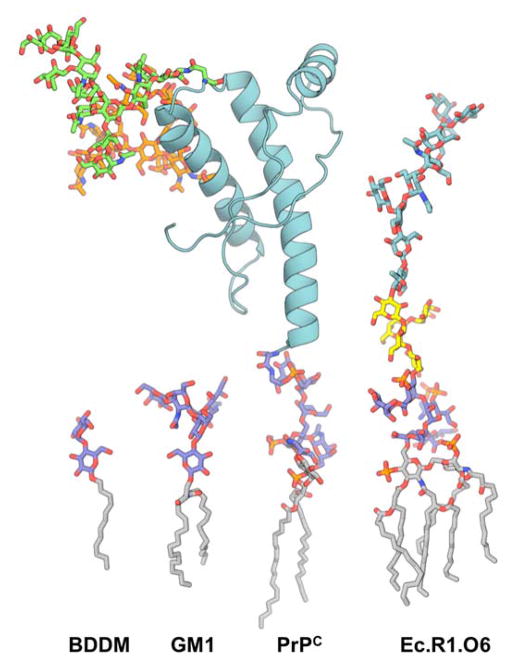
Figure 5.
Structure of glycoconjugates: BDDM for dodecyl-beta-D-maltoside, GM1 for monosialotetrahexosyl ganglioside, PrPC for a glycosylphospatidylinositol (GPI)-anchored human prion protein (cyan) with two N-glycans (green) [94], and Ec.R1.O6 for E. coli lipopolysaccharide (LPS) with R1 core (blue for the inner core and yellow for the outer core) and two repeating units of O6 antigen (cyan) [95].
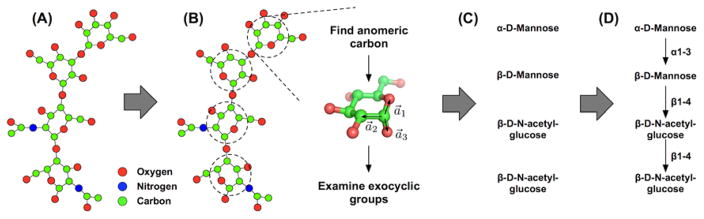
Figure 6.
Illustration of carbohydrate annotation procedure in Glycan Reader. (A) Molecular topology is built using HETATM and CONECT records in a PDB file. (B) Potential carbohydrate molecules are examined for anomeric carbon, stereochemistry of each ring carbon atoms, and exocyclic groups. (C) Carbohydrate type is annotated. (D) Glycosidic linkages are assigned between monosaccharides. Figure reproduced with permission from Journal of Computational Chemistry [93].
Glycolipids have been widely studied because of their importance in a range of biological functions. However, computational studies of glycolipids are still challenging due to their structural complexity arising from glycosidic linkages between sugars. Glycolipid Modeler facilitates generation of an appropriate glycolipid structure. The module provides two options to generate a glycolipid structure and the corresponding PSF: (1) pre-defined glycolipid sequences and (2) glycolipid sequences specified by users. Currently, Glycolipid Modeler supports 171 pre-defined glycolipids with 51 lipid types and 11 carbohydrate types. In addition, it allows users to introduce various chemical modifications to carbohydrates.
LPS Modeler is designed to simplify generation of structural models of complex lipopolysaccharide (LPS) molecules. LPS Modeler provides the options to select bacterial type, lipid A type, core type, the number of O-antigen repeating units, and O-antigen type. Currently, LPS Modeler supports LPS structures of 14 bacteria species with 32 lipid A types, 49 core types, and 222 O-antigen types. The selected LPS sequence is displayed, so that a user can check and modify the sugar sequence.
2.8 Experimentally guided simulation
Double electron-electron resonance (DEER) is a powerful technique to obtain distance information in the range of 18 to 80 Å by measuring the dipolar coupling between two unpaired electron spins. The distance distributions obtained from DEER experiments provide valuable protein structural information in a specific functional state. DEER Spin-Pair Distributor provides an interface to calculate the spin-pair distance distribution of labeled sites in a protein using MD simulations. Specially parameterized dummy spin-labels that mimic the methanethiosulfonate (MTS) spin labels are attached to user-specified labeling sites [96, 97]. The simulation is performed for 10 ns using CHARMM on the server, which takes from 30 min (for 60-residue protein) to 10 hours (for 5,000-residue protein). Following the link in the email sent to users after the job is done, the distributions between any labeled pair could be displayed on the web browser. The calculated distribution can be used to guide the selection of the label sites for further experiments as well as validate different protein structure models.
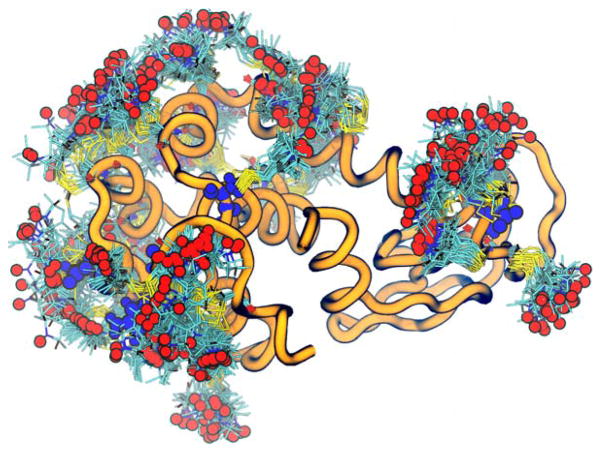
The distance distributions from DEER experiments can also be used with computational modeling and simulation to model/refine the protein structure that best match the DEER data. Restrained-ensemble MD (reMD) has been recently developed to simultaneously incorporate multiple distance distribution constraints for MD simulation [96, 98, 99]. In this method, each spin label is represented using multiple molecular fragments in all-atom detail (Figure 7), and the distance distribution between two selected labels is restrained to the corresponding experimental distance distribution. Taking advantage of the spin label function in PDB Reader & Manipulator [100], DEER reMD Prepper provides a quick and easy way to setup an reMD simulation.
Figure 7.
reMD simulation of lysozyme in solution. Multiple spin labels are added to one protein residue with the nitroxide groups shown as red spheres. The protein is shown as orange ribbon. The backbone atoms of the restrained residues are shown in blue spheres. Water molecules are removed for clarity.
MDFF and xMDFF are two structural refinement methods that incorporate experimental data from cryo-electron microscopy and X-ray crystallography into MD simulations [101, 102]. The EM and X-ray data are added to the molecular mechanics FF as an external potential and the simulation is performed in real space. An important feature of these methods is that the macromolecule remains flexible and stereochemically correct during the simulation. MDFF/xMDFF Utilizer helps users to prepare MDFF and xMDFF simulation systems using the CHARMM all-atom or PACE CG FF in various environments including vacuum, implicit solvent, explicit solvent, and bilayer.
2.9 Implicit solvent based simulation
PBEQ Solver [103] allows the characterization of the electrostatic potential on a macromolecular surface by solving the Poisson-Boltzmann (PB) equation using the PBEQ module [104, 105] in CHARMM. In addition, one can use this module to calculate (1) solvation free energy of biomolecules, (2) protein-protein (DNA or RNA) electrostatic interaction energy, and (3) pKa of a selected titratable residue in proteins.
Implicit solvent methods are a popular approximate description of solvent or membrane environments and CHARMM offers various implicit solvent models that users can apply to their systems. Implicit Solvent Modeler helps users to setup various implicit solvent models in CHARMM, and run molecular simulations with Langevin dynamics. The available implicit solvent models are: ACE (Analytical Continuum Electrostatics Potential) [106], EEF1/IMM1 (Effective Energy Function) [107, 108], GBMV (Generalized Born using Molecular Volume) [109, 110], GBSW (Generalized Born with a simple Switching) [111, 112], SASA (Solvent Accessible Surface Area implicit solvation model) [113], and SCPISM (Screened Coulomb Potentials Implicit Solvent Model) [114]. In addition, users can also set up molecular simulations of membrane(-bound) proteins in membrane environments using EEF1/IMM1 [108], GBMV [110], and GBSW [111].
Brownian dynamics (BD) using a realistic, accurate potential of mean force (based on implicit solvent approximation) is an effective approach for simulating ion transport through wide aqueous molecular pores [115–117]. GCMC/BD Ion Simulator [118] prepares input files for grand canonical Monte Carlo (GCMC) BD simulations of channel proteins [119, 120]. The webserver is designed to help users to avoid most of the technical difficulties and issues encountered in setting up and simulating complex pore systems.
2.10 Supported force fields and MD engines
Depending on the purpose of simulation and the characteristics of simulation system, different simulation methods and FFs can be applied. For example, to investigate atomistic structure and dynamic properties, an all-atom simulation system and a specific all-atom FF are necessary, while a large biological system requires CG methods. CHARMM-GUI supports various FFs: the CHARMM FF for all-atom additive simulations [36–40], the Martini [68] and PACE CG FFs [69] for CG simulations, and the Drude FF [121, 122] for polarizable all-atom simulations. Note that a Drude FF-based simulation system can be conveniently set up through Drude Prepper by simply uploading a PSF file and an equilibrated coordinate file obtained from MD simulation using the non-polarizable all-atom CHARMM FF.
CHARMM-GUI has been supporting various MD engines such as CHARMM [22], NAMD [23], GROMACS [24], AMBER [25], OpenMM [29], and CHARMM/OpenMM [30], and was recently extended to support GENESIS [26], LAMMPS [27], and Desmond [28]. While CHARMM, NAMD, OpenMM, CHARMM/OpenMM, and GENESIS support the standard CHARMM topology and parameter file formats (e.g., psf, prm, rtf, and str), other MD engines use different topology and parameter formats, requiring format conversion. We have developed several Python programs to generate the corresponding topology and parameter files (top and itp for GROMACS, dms for Desmond, and data file for LAMMPS) using CHARMM topology and parameter files, so that users can readily use the simulation input and data files for any supported MD engine.
3. CONCLUDING DISCUSSION
We have briefly reviewed the current functional modules in CHARMM-GUI, demonstrating diverse biomolecular simulation systems that CHARMM-GUI supports. It should be stressed that CHARMM-GUI also improves the reproducibility and quality of biomolecular simulations by providing simulation input files with generally accepted standards and well-validated protocols for all major simulation programs. The CHARMM-GUI development project is ongoing. More modules will be available in Input Generator for advanced simulations techniques, such as QM and QM/MM calculations [123–126], NMR structure calculation and refinement [127–130], multi-site λ-dynamics [131–133], constant pH simulations [134–145], grid-based energy correction map [146, 147], and various replica-exchange enhanced sampling methods [148]. In terms of system building, our effort will be placed toward making Glycolipid Modeler and LPS Modeler available in Membrane Builder and other modules, enabling crowding and cellular-scale system modeling, and extending CHARMM-GUI for nanoscale material sciences. The current and future developments with more tutorials and illustrations are expected to open a door allowing CHARMM-GUI users to perform even more advanced biomolecular and nanomaterial simulations of a diverse range of chemical system.
Acknowledgments
The CHARMM-GUI development has been support in part by grants from NSF MCB-0918374, MCB-1157677, MCB-1516154, DBI-1145987, DBI-1561372, IIA-1359530 (to WI), MCB-1149187, DBI-1145652 (to JBK) and from NIH GM092950 (to WI), GM087519,(to WI and BR), GM072558 (to ADM and BR), GM070855, GM051501 (to ADM). Parts of simulations were performed using the Extreme Science and Engineering Discovery Environment (XSEDE) allocations: MCB070009 (to WI) and MCB-100139 (to JBK), which are supported by NSF ACI-1053575.
References
- 1.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux B, MacKinnon R. The cavity and pore helices in the KcsA K+ channel: Electrostatic stabilization of monovalent cations. Science (New York, NY) 1999;285(5424):100–102. doi: 10.1126/science.285.5424.100. [DOI] [PubMed] [Google Scholar]
- 3.Im W, Roux B. Ions and counterions in a biological channel: A molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. Journal of Molecular Biology. 2002;319(5):1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 4.Im W, et al. Novel free energy calculations to explore mechanisms and energetics of membrane protein structure and function. Journal of Computational Chemistry. 2009;30(11):1622–1633. doi: 10.1002/jcc.21320. [DOI] [PubMed] [Google Scholar]
- 5.Lindorff-Larsen K, et al. How fast-folding proteins fold. Science (New York, NY) 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 6.Mori T, et al. Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2016;1858(7 Part B):1635–1651. doi: 10.1016/j.bbamem.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernèche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414(6859):73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 8.Huang N, Banavali NK, MacKerell AD. Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase. Proceedings of the National Academy of Sciences. 2003;100(1):68–73. doi: 10.1073/pnas.0135427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dror RO, et al. Biomolecular simulation: a computational microscope for molecular biology. Annual review of biophysics. 2012;41(1):429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 10.Knapp B, Schreiner W. Graphical User Interfaces for Molecular Dynamics—Quo Vadis? Bioinformatics and biology insights. 2009;3:103. [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Q, Nussinov R. Making biomolecular simulations accessible in the post-nobel prize era. PLOS Comput Biol. 2014;10(8):e1003786. doi: 10.1371/journal.pcbi.1003786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller BT, et al. CHARMMing: A New, Flexible Web Portal for CHARMM. Journal of Chemical Information and Modeling. 2008;48(9):1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glycam Biomolecule Builder. Available from: http://www.glycam.com.
- 14.Hospital A, et al. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012;28(9):1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 15.Vanommeslaeghe K, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. Journal of chemical information and modeling. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. Journal of chemical information and modeling. 2012;52(12):3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yesselman JD, et al. MATCH: An atom - typing toolset for molecular mechanics force fields. Journal of computational chemistry. 2012;33(2):189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoete V, et al. SwissParam: a fast force field generation tool for small organic molecules. Journal of computational chemistry. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Roux B. Automated force field parameterization for nonpolarizable and polarizable atomic models based on ab initio target data. Journal of chemical theory and computation. 2013;9(8):3543–3556. doi: 10.1021/ct4003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanquelef E, et al. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Research. 2011;39(suppl 2):W511–W517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo S, et al. CHARMM-GUI: A web-based graphical user interface for CHARMM. Journal of Computational Chemistry. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 22.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 25.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung J, et al. GENESIS: a hybrid-parallel and multi-scale molecular dynamics simulator with enhanced sampling algorithms for biomolecular and cellular simulations. Wiley Interdiscip Rev Comput Mol Sci. 2015;5(4):310–323. doi: 10.1002/wcms.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plimpton S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. Journal of Computational Physics. 1995;117(1):1–19. [Google Scholar]
- 28.Bowers KJ, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. SC 2006 Conference, Proceedings of the ACM/IEEE. 2006 [Google Scholar]
- 29.Eastman P, et al. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. Journal of Chemical Theory and Computation. 2013;9(1):461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arthur EJ, Brooks CL. Parallelization and improvements of the generalized born model with a simple sWitching function for modern graphics processors. J Comput Chem. 2016;37(10):927–939. doi: 10.1002/jcc.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo S, Kim T, Im W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE. 2007;2(9):e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jo S, et al. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophysical Journal. 2009;97(1):50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu EL, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. Journal of Computational Chemistry. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vriend G. WHAT IF: A molecular modeling and drug design program. Journal of Molecular Graphics. 1990;8(1):52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, et al. The Protein Data Bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. The journal of physical chemistry B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denning EJ, et al. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. Journal of computational chemistry. 2011;32(9):1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart K, et al. Optimization of the CHARMM additive force field for DNA: Improved treatment of the BI/BII conformational equilibrium. Journal of chemical theory and computation. 2011;8(1):348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guvench O, et al. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate–protein modeling. Journal of chemical theory and computation. 2011;7(10):3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. Journal of chemical theory and computation. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutsias EA, et al. A kinematic view of loop closure. Journal of Computational Chemistry. 2004;25(4):510–528. doi: 10.1002/jcc.10416. [DOI] [PubMed] [Google Scholar]
- 42.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W, et al. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. Journal of Computational Chemistry. 2012;33(31):2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Case DA, et al. AMBER 14. University of California; San Francisco: 2014. [Google Scholar]
- 45.Jmol: an open-source Java viewer for chemical structures in 3D. Available from: http://www.jmol.org.
- 46.Darden T, York DM, Pedersen LG. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. Journal of Chemical Physics. 1993;98(12):10089–10092. [Google Scholar]
- 47.Lee J, et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. Journal of Chemical Theory and Computation. 2016;12(1):405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomize MA, et al. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22(5):623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 50.Woolf TB, Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proceedings of the National Academy of Sciences. 1994;91(24):11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woolf TB, Roux B. Structure, energetics, and dynamics of lipid–protein interactions: a molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins: Structure, Function, and Bioinformatics. 1996;24(1):92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 52.Denisov IG, et al. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. Journal of the American Chemical Society. 2004;126(11):3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie TK, et al. Chapter Eleven - Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. In: Nejat D, editor. Methods in Enzymology. Academic Press; 2009. pp. 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klon AE, et al. Molecular Belt Models for the Apolipoprotein A-I Paris and Milano Mutations. Biophysical Journal. 2000;79(3):1679–1685. doi: 10.1016/S0006-3495(00)76417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi Y, et al. CHARMM-GUI PACE CG Builder for Solution, Micelle, and Bilayer Coarse-Grained Simulations. Journal of Chemical Information and Modeling. 2014;54(3):1003–1009. doi: 10.1021/ci500007n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi Y, et al. CHARMM-GUI Martini Maker for Coarse-Grained Simulations with the Martini Force Field. Journal of Chemical Theory and Computation. 2015;11(9):4486–4494. doi: 10.1021/acs.jctc.5b00513. [DOI] [PubMed] [Google Scholar]
- 57.Cheng X, et al. CHARMM-GUI Micelle Builder for Pure/Mixed Micelle and Protein/Micelle Complex Systems. Journal of Chemical Information and Modeling. 2013;53(8):2171–2180. doi: 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- 58.Cheng X, et al. Molecular dynamics simulation strategies for protein–micelle complexes. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2016;1858(7):1566–1572. doi: 10.1016/j.bbamem.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gruner SM, et al. Lipid polymorphism: the molecular basis of nonbilayer phases. Annual review of biophysics and biophysical chemistry. 1985;14(1):211–238. doi: 10.1146/annurev.bb.14.060185.001235. [DOI] [PubMed] [Google Scholar]
- 60.Rand R, et al. Membrane curvature, lipid segregation, and structural transitions for phospholipids under dual-solvent stress. Biochemistry. 1990;29(1):76–87. doi: 10.1021/bi00453a010. [DOI] [PubMed] [Google Scholar]
- 61.Leikin S, et al. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes. Biophysical journal. 1996;71(5):2623. doi: 10.1016/S0006-3495(96)79454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z, Rand R. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophysical journal. 1997;73(1):267. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuller N, Rand R. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophysical journal. 2001;81(1):243–254. doi: 10.1016/S0006-3495(01)75695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marrink S-J, Mark AE. Molecular view of hexagonal phase formation in phospholipid membranes. Biophysical journal. 2004;87(6):3894–3900. doi: 10.1529/biophysj.104.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sodt AJ, Pastor RW. Bending free energy from simulation: correspondence of planar and inverse hexagonal lipid phases. Biophysical journal. 2013;104(10):2202–2211. doi: 10.1016/j.bpj.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi Y, et al. CHARMM-GUI HMMM Builder for Membrane Simulations with the Highly Mobile Membrane-Mimetic Model. Biophysical Journal. 2015;109(10):2012–2022. doi: 10.1016/j.bpj.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohkubo YZ, et al. Accelerating Membrane Insertion of Peripheral Proteins with a Novel Membrane Mimetic Model. Biophysical Journal. 2012;102(9):2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monticelli L, et al. The MARTINI Coarse-Grained Force Field: Extension to Proteins. Journal of Chemical Theory and Computation. 2008;4(5):819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 69.Han W, Schulten K. Further Optimization of a Hybrid United-Atom and Coarse-Grained Force Field for Folding Simulations: Improved Backbone Hydration and Interactions between Charged Side Chains. Journal of Chemical Theory and Computation. 2012;8(11):4413–4424. doi: 10.1021/ct300696c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marrink SJ, de Vries AH, Mark AE. Coarse Grained Model for Semiquantitative Lipid Simulations. The Journal of Physical Chemistry B. 2004;108(2):750–760. [Google Scholar]
- 71.Marrink SJ, et al. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. The Journal of Physical Chemistry B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 72.Yesylevskyy SO, et al. Polarizable Water Model for the Coarse-Grained MARTINI Force Field. PLoS Comput Biol. 2010;6(6):e1000810. doi: 10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Jong DH, et al. Improved Parameters for the Martini Coarse-Grained Protein Force Field. Journal of Chemical Theory and Computation. 2013;9(1):687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 74.Arnarez C, et al. Dry Martini, a Coarse-Grained Force Field for Lipid Membrane Simulations with Implicit Solvent. Journal of Chemical Theory and Computation. 2015;11(1):260–275. doi: 10.1021/ct500477k. [DOI] [PubMed] [Google Scholar]
- 75.Periole X, et al. Combining an Elastic Network With a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. Journal of Chemical Theory and Computation. 2009;5(9):2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 76.Wassenaar TA, et al. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. Journal of Chemical Theory and Computation. 2014;10(2):676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 77.Rzepiela AJ, et al. Reconstruction of atomistic details from coarse-grained structures. Journal of Computational Chemistry. 2010;31(6):1333–1343. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- 78.Stansfeld PJ, Sansom MS. From coarse grained to atomistic: a serial multiscale approach to membrane protein simulations. Journal of Chemical Theory and Computation. 2011;7(4):1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 79.Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. Journal of Computer-Aided Molecular Design. 2012;26(1):15–26. doi: 10.1007/s10822-011-9517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woo H-J, Roux B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(19):6825–6830. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deng Y, Roux B. Calculation of Standard Binding Free Energies: Aromatic Molecules in the T4 Lysozyme L99A Mutant. Journal of Chemical Theory and Computation. 2006;2(5):1255–1273. doi: 10.1021/ct060037v. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, Deng Y, Roux B. Absolute binding free energy calculations using molecular dynamics simulations with restraining potentials. Biophysical Journal. 2006;91(8):2798–2814. doi: 10.1529/biophysj.106.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. The Journal of Physical Chemistry B. 2009;113(8):2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mobley DL, Dill KA. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure. 2009;17(4):489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee HS, Seok C, Im W. Potential Application of Alchemical Free Energy Simulations to Discriminate GPCR Ligand Efficacy. Journal of Chemical Theory and Computation. 2015;11(3):1255–1266. doi: 10.1021/ct5008907. [DOI] [PubMed] [Google Scholar]
- 86.Jo S, et al. CHARMM-GUI Ligand Binder for absolute binding free energy calculations and its application. Journal of Chemical Information and Modeling. 2013;53(1):267–277. doi: 10.1021/ci300505n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beglov D, Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. The Journal of chemical physics. 1994;100(12):9050–9063. [Google Scholar]
- 88.Im W, Berneche S, Roux Bt. Generalized solvent boundary potential for computer simulations. The Journal of Chemical Physics. 2001;114(7):2924–2937. [Google Scholar]
- 89.Weeks JD, Chandler D, Andersen HC. Role of repulsive forces in determining the equilibrium structure of simple liquids. The Journal of chemical physics. 1971;54(12):5237–5247. [Google Scholar]
- 90.Deng Y, Roux B. Hydration of amino acid side chains: nonpolar and electrostatic contributions calculated from staged molecular dynamics free energy simulations with explicit water molecules. The Journal of Physical Chemistry B. 2004;108(42):16567–16576. [Google Scholar]
- 91.Jiang W, Hodoscek M, Roux B. Computation of absolute hydration and binding free energy with free energy perturbation distributed replica-exchange molecular dynamics. Journal of chemical theory and computation. 2009;5(10):2583–2588. doi: 10.1021/ct900223z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang W, Roux B. Free energy perturbation Hamiltonian replica-exchange molecular dynamics (FEP/H-REMD) for absolute ligand binding free energy calculations. Journal of chemical theory and computation. 2010;6(9):2559–2565. doi: 10.1021/ct1001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jo S, et al. Glycan Reader: automated sugar identification and simulation preparation for carbohydrates and glycoproteins. Journal of Computational Chemistry. 2011;32(14):3135–3141. doi: 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu EL, et al. Insight into early-stage unfolding of GPI-anchored human prion protein. Biophysical journal. 2015;109(10):2090–2100. doi: 10.1016/j.bpj.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu EL, et al. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophysical journal. 2013;105(6):1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Islam SM, et al. Structural Refinement from Restrained-Ensemble Simulations Based on EPR/DEER Data: Application to T4 Lysozyme. The Journal of Physical Chemistry B. 2013;117(17):4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Islam SM, Roux B. Simulating the Distance Distribution between Spin-Labels Attached to Proteins. The Journal of Physical Chemistry B. 2015;119(10):3901–3911. doi: 10.1021/jp510745d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roux B, Islam SM. Restrained-Ensemble Molecular Dynamics Simulations Based on Distance Histograms from Double Electron–Electron Resonance Spectroscopy. The Journal of Physical Chemistry B. 2013;117(17):4733–4739. doi: 10.1021/jp3110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen R, et al. Structural Refinement of Proteins by Restrained Molecular Dynamics Simulations with Non-interacting Molecular Fragments. PLoS Comput Biol. 2015;11(10):e1004368. doi: 10.1371/journal.pcbi.1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jo S, et al. Chapter Eight-CHARMM-GUI PDB Manipulator for Advanced Modeling and Simulations of Proteins Containing Nonstandard Residues. Advances in protein chemistry and structural biology. 2014;96:235–265. doi: 10.1016/bs.apcsb.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trabuco LG, et al. Flexible Fitting of Atomic Structures into Electron Microscopy Maps Using Molecular Dynamics. Structure. 2008;16(5):673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McGreevy R, et al. xMDFF: molecular dynamics flexible fitting of low-resolution X-ray structures. Acta Crystallographica Section D. 2014;70(9):2344–2355. doi: 10.1107/S1399004714013856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jo S, et al. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acid Research. 2008;36(Web Server issue):W270–5. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Im W, Beglov D, Roux B. Continuum solvation model: computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Computer Physics Communications. 1998;111(1):59–75. [Google Scholar]
- 105.Nina M, Im W, Roux Bt. Optimized atomic radii for protein continuum electrostatics solvation forces. Biophysical chemistry. 1999;78(1):89–96. doi: 10.1016/s0301-4622(98)00236-1. [DOI] [PubMed] [Google Scholar]
- 106.Schaefer M, Karplus M. A Comprehensive Analytical Treatment of Continuum Electrostatics. J Phys Chem. 1996;100:1578–1599. [Google Scholar]
- 107.Lazaridis T, Karplus M. Effective energy functions for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 108.Lazaridis T. Effective energy functions for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 109.Lee MS, Salsbury FR, Jr, Brooks CL., III Novel generalized Born methods. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 110.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes. J Chem Phys. 2005;122:124706. doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 111.Im W, Feig M, Brooks CL., III An Implicit Membrane Generalized Born Theory for the Study of Structures, Stability, and Interactions of Membrane Proteins. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Im W, Lee MS, Brooks CL., III Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 113.Ferrara P, Apostolakis J, Caflisch A. Evaluation of a fast implicit solvent model for molecular dynamics simulations. Proteins-Structure Function and Genetics. 2002;46(1):24–33. doi: 10.1002/prot.10001. [DOI] [PubMed] [Google Scholar]
- 114.Hassan SA, Guarnieri F, Mehler EL. A General Treatment of Solvent Effects Based on Screened Coulomb Potentials. J Phys Chem B. 2000;104:6478–6489. [Google Scholar]
- 115.Eisenberg B, Hyon Y, Liu C. Energy variational analysis of ions in water and channels: Field theory for primitive models of complex ionic fluids. The Journal of Chemical Physics. 2010;133(10):104104. doi: 10.1063/1.3476262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu B, et al. Electrodiffusion: A continuum modeling framework for biomolecular systems with realistic spatiotemporal resolution. The Journal of chemical physics. 2007;127(13):135102. doi: 10.1063/1.2775933. [DOI] [PubMed] [Google Scholar]
- 117.Lee KI, et al. Brownian dynamics simulations of ion transport through the VDAC. Biophysical journal. 2011;100(3):611–619. doi: 10.1016/j.bpj.2010.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee KI, et al. Web interface for brownian dynamics simulation of ion transport and its applications to beta-barrel pores. Journal of Computational Chemistry. 2012;33(3):331–339. doi: 10.1002/jcc.21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Im W, Seefeld S, Roux B. A grand canonical Monte Carlo–Brownian dynamics algorithm for simulating ion channels. Biophysical Journal. 2000;79(2):788–801. doi: 10.1016/S0006-3495(00)76336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Im W, Roux Bt. Brownian dynamics simulations of ions channels: a general treatment of electrostatic reaction fields for molecular pores of arbitrary geometry. The Journal of Chemical Physics. 2001;115(10):4850–4861. [Google Scholar]
- 121.Lopes PE, et al. Polarizable force field for peptides and proteins based on the classical drude oscillator. Journal of chemical theory and computation. 2013;9(12):5430–5449. doi: 10.1021/ct400781b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Savelyev A, MacKerell AD. All-atom polarizable force field for DNA based on the classical drude oscillator model. Journal of computational chemistry. 2014;35(16):1219–1239. doi: 10.1002/jcc.23611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Field MJ, Bash PA, Karplus M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. Journal of Computational Chemistry. 1990;11(6):700–733. [Google Scholar]
- 124.Gregersen BA, Lopez X, York DM. Hybrid QM/MM study of thio effects in transphosphorylation reactions. Journal of the American Chemical Society. 2003;125(24):7178–7179. doi: 10.1021/ja035167h. [DOI] [PubMed] [Google Scholar]
- 125.Cui Q, Karplus M. Molecular properties from combined QM/MM methods. I. Analytical second derivative and vibrational calculations. The Journal of Chemical Physics. 2000;112(3):1133–1149. [Google Scholar]
- 126.Knott BC, et al. Carbohydrate–Protein Interactions That Drive Processive Polysaccharide Translocation in Enzymes Revealed from a Computational Study of Cellobiohydrolase Processivity. Journal of the American Chemical Society. 2014;136(24):8810–8819. doi: 10.1021/ja504074g. [DOI] [PubMed] [Google Scholar]
- 127.Cheng X, et al. Solid-state NMR-restrained ensemble dynamics of a membrane protein in explicit membranes. Biophysical journal. 2015;108(8):1954–1962. doi: 10.1016/j.bpj.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee J, et al. Application of solid-state NMR restraint potentials in membrane protein modeling. Journal of Magnetic Resonance. 2008;193(1):68–76. doi: 10.1016/j.jmr.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chakravorty DK, et al. Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods. Journal of biomolecular NMR. 2013;56(2):125–137. doi: 10.1007/s10858-013-9729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen J, Im W, Brooks CL. Refinement of NMR structures using implicit solvent and advanced sampling techniques. Journal of the American Chemical Society. 2004;126(49):16038–16047. doi: 10.1021/ja047624f. [DOI] [PubMed] [Google Scholar]
- 131.Knight JL, Brooks CL., III Multi-Site lambda-dynamics for simulated Structure-Activity Relationship studies. J Chem Theory Comput. 2011;7(9):2728–2739. doi: 10.1021/ct200444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Armacost KA, Goh GB, Brooks CL., III Biasing Potential Replica Exchange Multi-Site λ-Dynamics for Efficient Free Energy Calculations. J Chem Theor Comp. 2015;11(3):1267–77. doi: 10.1021/ct500894k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Armacost KA, Goh GB, Brooks CL., III Predicting Binding Free Energies in a Combinatorial Chemical Space Using Multisite Lambda Dynamics. J Am Chem Soc. 2015 doi: 10.1021/acs.jpclett.8b01284. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee MS, Salsbury FR, Jr, Brooks CL., III Constant-pH molecular dynamics using continuous titration coordinates. Proteins. 2004;56(4):738–52. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 135.Khandogin J, Brooks CL., III Constant pH Molecular Dynamics with Proton Tautomerism. Biophys J. 2005;89(1):141–57. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Khandogin J, Brooks CL., III Linking folding with aggregation in Alzheimer s beta-amyloid peptides. Proc Natl Acad Sci U S A. 2007;104(43):16880–5. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang BW, Brunetti L, Brooks CL., III Probing pH-dependent dissociation of HdeA dimers. J Am Chem Soc. 2011;133(48):19393–8. doi: 10.1021/ja2060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goh GB, Knight JL, Brooks CL., III Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent. J Chem Theory Comput. 2012;8(1):36–46. doi: 10.1021/ct2006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Foit L, et al. Chaperone activation by unfolding. Proc Natl Acad Sci U S A. 2013;110(14):E1254–62. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goh GB, Knight JL, Brooks CL., III pH-dependent dynamics of complex RNA macromolecules. J Chem Theory Comput. 2013;9(2):935–943. doi: 10.1021/ct300942z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Law SM, Zhang BW, Brooks CL., III pH-sensitive residues in the p19 RNA silencing suppressor protein from carnation Italian ringspot virus affect siRNA binding stability. Protein Sci. 2013;22(5):595–604. doi: 10.1002/pro.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goh GB, et al. Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism. Proteins. 2014;82(7):1319–31. doi: 10.1002/prot.24499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Goh GB, Laricheva EN, Brooks CL., III Uncovering pH-dependent transient states of proteins with buried ionizable residues. J Am Chem Soc. 2014;136(24):8496–9. doi: 10.1021/ja5012564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Panahi A, Brooks CL., III Membrane Environment Modulates the pKa Values of Transmembrane Helices. J Phys Chem B. 2015;119(13):4601–7. doi: 10.1021/acs.jpcb.5b00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zeng X, Mukhopadhyay S, Brooks CL., III Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion. Proc Natl Acad Sci U S A. 2015;112(7):2034–9. doi: 10.1073/pnas.1414190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang M, MacKerell AD., Jr Conformational Sampling of Oligosaccharides Using Hamiltonian Replica Exchange with Two-Dimensional Dihedral Biasing Potentials and the Weighted Histogram Analysis Method (WHAM) Journal of chemical theory and computation. 2015;11(2):788–799. doi: 10.1021/ct500993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang M, Huang J, MacKerell AD., Jr Enhanced conformational sampling using replica exchange with concurrent solute scaling and Hamiltonian biasing realized in one dimension. Journal of chemical theory and computation. 2015;11(6):2855–2867. doi: 10.1021/acs.jctc.5b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mori T, et al. Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2016;1858(7):1635–1651. doi: 10.1016/j.bbamem.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]