Abstract
Background
Common variants in the TCF4 gene are among the most robustly supported genetic risk factors for schizophrenia. Rare TCF4 deletions and loss-of-function point mutations cause Pitt–Hopkins syndrome, a developmental disorder associated with severe intellectual disability.
Methods
To explore molecular and cellular mechanisms by which TCF4 perturbation could interfere with human cortical development, we experimentally reduced the endogenous expression of TCF4 in a neural progenitor cell line derived from the developing human cerebral cortex using RNA interference. Effects on genome-wide gene expression were assessed by microarray, followed by Gene Ontology and pathway analysis of differentially expressed genes. We tested for genetic association between the set of differentially expressed genes and schizophrenia using genome-wide association study data from the Psychiatric Genomics Consortium and competitive gene set analysis (MAGMA). Effects on cell proliferation were assessed using high content imaging.
Results
Genes that were differentially expressed following TCF4 knockdown were highly enriched for involvement in the cell cycle. There was a nonsignificant trend for genetic association between the differentially expressed gene set and schizophrenia. Consistent with the gene expression data, TCF4 knockdown was associated with reduced proliferation of cortical progenitor cells in vitro.
Limitations
A detailed mechanistic explanation of how TCF4 knockdown alters human neural progenitor cell proliferation is not provided by this study.
Conclusion
Our data indicate effects of TCF4 perturbation on human cortical progenitor cell proliferation, a process that could contribute to cognitive deficits in individuals with Pitt–Hopkins syndrome and risk for schizophrenia.
Introduction
Transcription factor 4 (TCF4) is an E-protein basic helix–loop–helix (bHLH) transcription factor that binds to the Ephrussi-box (E-Box) DNA motif.1,2 Common variants in the TCF4 gene are among the most robustly supported genetic risk factors for schizophrenia.3–6 Rare TCF4 deletions and loss-of-function point mutations cause Pitt–Hopkins syndrome,7–11 a developmental disorder associated with severe intellectual disability.
E-proteins show widespread expression and act as transcriptional activators or repressors by forming heterodimers with other bHLH proteins.1 TCF4 is highly expressed in the fetal as well as adult human brain12,13 and is known to dimerize with several bHLH factors that are important for neural development.14–16 Knockout of the TCF4 gene has been reported to affect the differentiation of specific neuronal populations in the mouse hindbrain.15 However, data pertaining to the role of TCF4 in human neural development are currently lacking.
Experimental knockdown of TCF4 expression in human neuroblastoma-derived cells (SH-SY5Y) has been found to alter the expression of genes involved in transforming growth factor (TGF)-β signalling, epithelial to mesenchymal transition and apoptosis.17 Stable knockdown of TCF4 in neural progenitor cells from the human fetal midbrain has been reported to result in gene expression changes more characteristic of differentiating than proliferating cells, suggesting effects on the timing of neural differentiation.18 However, to date, effects of TCF4 manipulation in cells from the developing human cerebral cortex have not been explored. In the present study, we experimentally reduced the endogenous expression of TCF4 in a neural progenitor cell line derived from human fetal neo-cortex in order to explore molecular and cellular mechanisms through which TCF4 perturbation could interfere with early cortical development.
Methods
Cell culture
Experiments were performed using a neural progenitor cell line (CTX0E03) derived from the cortical neuroepithelium of a 12-week human fetus obtained from ReNeuron Ltd (www.reneuron.com) under a material transfer agreement. This cell line has been conditionally immortalized by genomic incorporation of the c-MycERTAM transgene, to stimulate proliferation in the presence of the synthetic drug 4-hydroxy-tamoxifen (4-OHT). The derivation and characteristics of the CTX0E03 cell line are described in detail by Pollock and colleagues.19 Cells were cultured on laminin-coated T75 flasks using a modified DMEM:F12 media, as described previously.20 For the RNA interference experiments, 4-OHT was excluded from the media so that proliferation was not artificially stimulated through c-Myc overexpression.
RNA interference in cultured cells
Two nonoverlapping small interfering RNA (siRNA) targeting all TCF4 messenger RNA (mRNA) transcripts defined by Sepp and colleagues13 were used as 2 separate TCF4 siRNA conditions. The first condition (Cat #s13863) has the sense sequence 5′-GCUCUGAGAUCAAAUCCGAtt-3′ and targets exon 18 of full-length TCF4, as denoted by Sepp and colleagues.13 The second condition (Cat #s13864) has the sense sequence 5′-GAAGGACCCUUACACUCUUtt-3′ and targets exon 15. We used a negative control siRNA (Silencer Negative Control #1, Life Technologies, Cat #AM4611) with no sequence similarity with any human transcript for the control comparison condition. For the microarray experiment, each siRNA was combined with N-TER transfection reagent (Sigma-Aldrich) and added to 4 T75 flasks of separately seeded cells at a final siRNA concentration of 10 nM. For the cell proliferation experiments, each siRNA was combined with N-TER transfection reagent and added to 12 wells (0.33 cm2) of separately seeded cells at a final siRNA concentration of 10 nM. In order to estimate transfection efficiency, an additional T75 flask of seeded cells was transfected with 10 nM BLOCK-iT™ Alexa Fluor Red Fluorescent Control oligonucleotide (Life Technologies) using the same N-TER reagent and visualized after 24 h using fluorescence microscopy. Media (minus both 4-OHT and siRNA) was replaced 48 h after transfection.
RNA and protein preparation
Cells from each T75 flask were harvested for RNA and protein 96 hours after siRNA transfection. Total RNA was extracted from half of the cells in each flask using Tri-Reagent (Life Technologies) according to manufacturer’s instructions. We assessed the integrity of total RNA using the Agilent 2100 Bioanalyzer (Agilent Technologies). All samples had an RNA integrity number (RIN) above 9.5. Protein was extracted from the remaining half of the cells in each flask using radioimmunoprecipitation assay (RIPA) buffer.
Assessment of TCF4 RNA knockdown
Quantitative polymerase chain reaction (qPCR) was used to assess the level of TCF4 RNA knockdown in each TCF4 siRNA condition (relative to the control siRNA condition) before genome-wide gene expression profiling. Total RNA was treated with Turbo DNA-free (Life Technologies) and converted to complementary DNA (cDNA) using random decamers and SuperScript III (Life Technologies). The qPCR primers were designed to amplify exonic sequence included in all known TCF4 transcripts: F: 5′-GAAAGCTGCGTGTCTGAAAA-3′ and 5′-CATCTGTCCCATGTGATTCG-3′. We measured expression of GAPDH, HPRT1 and RPL13A simultaneously as internal control genes. The expression of these 3 housekeeping genes was subsequently found not to differ between siRNA and control conditions in either cell line in the microarray data (all p > 0.05). Reactions were performed using FIREPol EvaGreen qPCR Mix (Solis Biodyne), an MJ Research Chromo 4 (Bio-Rad) and MJ Opticon Monitor analytic software (Bio-Rad). We performed duplicate qPCR reactions to measure expression of each gene in each cDNA sample. Expression of each gene was measured against a standard curve constructed by serial dilution of pooled cDNA. Mean measures of TCF4 expression were divided by the geometric average of the mean measures for the 3 internal control genes to yield a normalized TCF4 expression value for each sample. We compared normalized TCF4 expression values between each TCF4 siRNA condition and the negative control siRNA condition using t tests (2-tailed).
Assessment of TCF4 protein knockdown
Supernatant from whole-cell lysates was mixed with an equal volume of 2× Laemmli buffer and heated to 100°C for 5 min. Equal volumes of 3 samples from each condition were then loaded and separated on a 10% polyacrylamide gel by standard SDS–PAGE. The gel was blotted and probed for TCF4 protein using a C-terminal antibody produced in rabbit (Sigma-Aldrich #SAB4502928) and for β-actin protein using a monoclonal antibody produced in mouse (Abcam #ab8226), with fluorescent secondary antibodies to each. The blot was imaged and densitometry performed using a near-infrared Odyssey scanner. TCF4 immunoreactivity values were normalized to β-actin values within blot and compared between TCF4 and control siRNA conditions using t tests (2-tailed).
Genome-wide gene expression profiling and pathway analysis
We performed genome-wide gene expression profiling of the 12 total RNA samples (4 samples for each siRNA targeting TCF4 and 4 samples for the control siRNA) using the Illumina TotalPrep RNA amplification kit and Illumina HT-12 v4 Bead-Chip arrays. Data were extracted from GenomeStudio software (Illumina), and we applied variance stabilizing transformation followed by robust spline normalization using the lumi Bioconductor package.21 All microarray data from this study have been submitted to the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), with the accession number GSE62085.
Microarray probes showing nominally significant (p < 0.05, uncorrected) differences between each TCF4 siRNA condition and the control siRNA condition were first identified using individual t tests (2-tailed) on normalized microarray data. To limit spurious results arising from low expression genes, we excluded probes that were not detected in all 12 samples with a detection p < 0.05. To refine the data set to those changes most likely to reflect TCF4 knockdown rather than off-target effects of individual TCF4 siRNA, we identified gene expression changes that were significant (p < 0.05) in both TCF4 siRNA conditions and occurred in the same direction (i.e., up- or downregulation) relative to the control siRNA condition. We additionally used Significance Analysis of Microarrays (SAM) software22 to identify high-confidence gene expression changes between each TCF4 siRNA condition and the control siRNA condition, selecting those with a false discovery rate (FDR) < 0.05 and again identifying those changes shared by both siRNA conditions in the same direction relative to the control siRNA. We used the hypergeometric probability test, as used by Rosen and colleagues,23 to assess the probability of the differentially expressed gene lists shared by the 2 siRNA conditions occurring by chance, based on the number of differentially expressed gene probes associated with each siRNA and the number of probes that had a detection p < 0.05 in all samples. The gene set shared by the 2 TCF4 siRNA conditions was subject to Gene Ontology (GO) and KEGG pathway analysis through the DAVID Bioinformatics Resource 6.7,24 which uses a modified Fisher exact test to assess enrichment within predefined gene sets. We tested for enrichment of differentially expressed genes within all KEGG pathways and biological process terms under the comprehensive GOTERM_BP_FAT category, using all gene probes that had a detection p < 0.05 in all samples as the background comparison. We tested for genetic association between schizophrenia and the set of differentially expressed genes shared by the 2 TCF4 siRNA conditions (at p < 0.05) using the latest genome-wide association study (GWAS) data from the Psychiatric Genomics Consortium6 and MAGMA25 for competitive gene set analysis. Summary schizophrenia GWAS statistics were downloaded from www.med.unc.edu/pgc/results-and-downloads. MAGMA was run with default settings and no gene extension window. We excluded TCF4 from these analyses in order to avoid artificial inflation of the genetic association with schizophrenia.
Confirmation of selected gene expression changes
We used qPCR to confirm altered RNA expression of selected genes involved in the cell cycle. Genes were selected on the basis of exhibiting at least a 30% mean difference in expression between each TCF4 siRNA condition and the control siRNA condition in the microarray data. DNAse-treated RNA used for genome-wide expression profiling was converted to cDNA using random decamers and SuperScript III (Life Technologies). The qPCR primers were designed to target the same exons as the microarray probes showing differential expression between siRNA conditions. The microarray data from all samples were used to select an appropriate internal control gene for these assays, as performed previously.20,26 ATP5B was hereby chosen as a suitable internal control because it had the smallest coefficient of variation across samples, it had no significant differences between siRNA conditions, the probe mapped to a single genomic site and it was possible to design primers targeting the same exon as the probe. All primer sequences are shown in Appendix 1, Table S1, available at jpn.ca. Reactions were carried out in triplicate for each sample using FIREPol EvaGreen qPCR Mix (Solis Biodyne) and measured against a standard curve for each gene, as described previously.
Cell proliferation assay
Ninety-six hours after siRNA transfection, 10 uM bromodeoxyuridine (BrdU; Sigma-Aldrich) was added to each of the 12 wells of seeded cells for the TCF4 siRNA#1, TCF4 siRNAi#2 and negative control siRNA conditions and incubated for 6 h. Cells were then fixed in 2% paraformaldehyde, permeabilized and incubated overnight with a rabbit antibody against Ki67 (Abcam # ab15580; 1:500 dilution) and a rat antibody against BrdU (AbD Serotech # MCA2060; 1:500 dilution). The secondary antibodies were Alexa Fluor 594 Donkey Anti-Rabbit IgG and Alexa Fluor 488 Donkey Anti-Rat IgG (Life Technologies #A21207 and #A21208). Cell nuclei were stained by incubation with DAPI (1:1000 dilution).
Image acquisition was performed using the CellInsight NXT High Content Screening Platform (Thermo Scientific) with a 10× objective lens. Image processing and downstream analysis was conducted using the iDEV software package. We used DAPI-stained nuclei to determine valid objects (cells), with objects being excluded based on size and proximity to the edge of the field. We used segmentation to resolve clumped nuclei. Images were acquired for a total of 6 fields per well. Control wells lacking primary antibody were used to set background fluorescence levels. We identified BrdU- and ki67-positive nuclei using the Target Activation bioapplication included in the iDEV software. The average percentage of BrdU- and ki67-positive cells was calculated from the 6 acquired images for each well. The percentages of BrdU- and ki67-positive cells in the 12 wells of each TCF4 siRNA condition were then compared with those in the 12 wells of the negative control siRNA condition using t tests (2-tailed).
Results
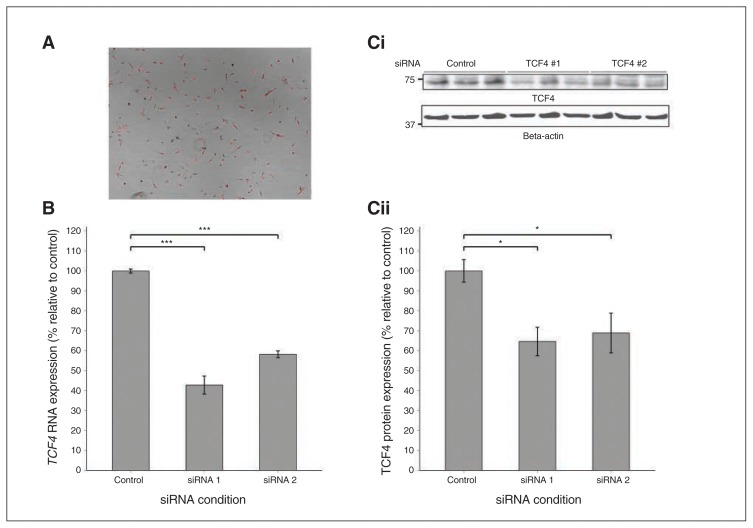
We used 2 nonoverlapping siRNA conditions (TCF4 siRNA #1 and siRNA #2) to transiently knock down TCF4 in cultured neural progenitor cells derived from human fetal neocortex. Transfection efficiency, as indexed by uptake of BLOCK-iT Alexa Fluor Red Fluorescent Control oligonucleotide, was estimated to be above 80% (Fig. 1A). At harvest, mean TCF4 RNA expression (as indexed by qPCR) was reduced by 57% (p < 0.001) in the TCF4 siRNA #1 condition and 42% (p < 0.001) in the TCF4 siRNA #2 condition relative to the control siRNA condition (Fig. 1B). Mean TCF4 protein expression, as indexed by Western blot, was reduced by 35% (p = 0.018) in the TCF4 siRNA #1 condition and 31% (p = 0.05) in the TCF4 siRNA #2 condition relative to the control siRNA condition (Fig. 1C).
Fig. 1.
Knockdown of TCF4 by RNA interference in the CTX0E03 neural progenitor cell line. (A) Uptake of Red Fluorescent oligonucleotide into CTX0E03 neural progenitor cells as an index of small interfering RNA (siRNA) transfection efficiency. Picture taken 24 h after transfection using an Olympus I×70 inverted microscope and AxioVision 4.8 Imaging software (Zeiss). Transfection efficiency was thus estimated to be above 80%. (B) TCF4 RNA expression, as indexed by quantitative polymerase chain reaction (qPCR) at cell harvest (n = 4 per condition). Mean TCF4 RNA expression was reduced by 57% (p < 0.001) in the TCF4 siRNA #1 condition and 42% (p < 0.001) in the TCF4 siRNA #2 condition relative to the control siRNA condition. C) TCF4 protein expression, as indexed by Western blot, at cell harvest in 3 samples from each condition. TCF4 protein, indicated by a band of ~71kDa (i), was reduced by 35% (p = 0.018) in the 3 samples of the TCF4 siRNA #1 condition and 31% (p = 0.05) in 3 samples of the TCF4 siRNA #2 condition relative to those of the control siRNA condition (ii). Error bars represent standard errors of the mean.
Gene expression differed significantly (p < 0.05) between the TCF4 siRNA #1 condition and the control siRNA condition at 1753 microarray probes and between the TCF4 siRNA #2 condition and the control siRNA condition at 1480 microarray probes. Nominally significant gene expression differences were shared by the 2 TCF4 siRNA conditions in the same direction relative to the control condition at 628 probes. This overlap of gene expression changes was highly significant (p < 1 × 10−200). The full list of nominally significant gene expression changes shared by both TCF4 siRNA conditions is provided in Appendix 1, Table S2. A smaller number of directional gene expression changes were shared by the 2 TCF4 siRNA conditions relative to the control siRNA condition at FDR < 0.05. This more stringent analysis identified high-confidence gene expression changes at 39 gene probes that were shared by both TCF4 siRNA conditions (Table 1), an overlap that was highly significant (p = 2.2 × 10−58).
Table 1.
High-confidence gene expression changes observed at a false discovery rate < 0.05 in both TCF4 siRNA conditions compared with the control siRNA condition
| Gene | Microarray probe | Fold-change siRNA#1 | q-value siRNA#1 | Fold-change siRNA#2 | q-value siRNA#2 |
|---|---|---|---|---|---|
| ALDOC | ILMN_1755974 | 0.67 | 0.00 | 0.67 | 0.00 |
| CLIP3 | ILMN_1789733 | 0.64 | 0.00 | 0.67 | 0.00 |
| CRYAB | ILMN_1729216 | 0.50 | 0.00 | 0.56 | 0.00 |
| LOC100008588 | ILMN_3243593 | 0.52 | 0.00 | 0.56 | 0.00 |
| LOC100008589 | ILMN_3251587 | 0.38 | 0.00 | 0.37 | 0.00 |
| RNF19A | ILMN_1812327 | 0.65 | 0.00 | 0.62 | 0.00 |
| TCF4 | ILMN_1814194 | 0.53 | 0.00 | 0.61 | 0.00 |
| IGDCC3 | ILMN_1744635 | 0.70 | 1.97 | 0.68 | 0.00 |
| PNCK | ILMN_1697189 | 0.75 | 1.97 | 0.69 | 0.00 |
| LOC100132394 | ILMN_3249578 | 0.70 | 3.03 | 0.59 | 0.00 |
| APOD | ILMN_1780170 | 0.70 | 0.00 | 0.70 | 3.95 |
| BNIP3 | ILMN_1724658 | 0.69 | 0.00 | 0.68 | 3.95 |
| LOC100134364 | ILMN_3246805 | 0.64 | 0.00 | 0.62 | 3.95 |
| DDX10 | ILMN_1753249 | 1.67 | 0.00 | 1.40 | 4.96 |
| EDNRB | ILMN_1751904 | 0.62 | 0.00 | 0.71 | 4.96 |
| FREM2 | ILMN_1703174 | 0.67 | 0.00 | 0.72 | 4.96 |
| KLF9 | ILMN_1778523 | 0.73 | 1.97 | 0.72 | 3.95 |
| TTYH1 | ILMN_1758497 | 0.73 | 1.97 | 0.70 | 3.95 |
| NTS | ILMN_1764690 | 1.58 | 1.97 | 1.43 | 4.96 |
| SLC22A18 | ILMN_2382505 | 0.73 | 3.03 | 0.69 | 3.95 |
| BIRC5 | ILMN_2349459 | 1.41 | 3.03 | 1.44 | 4.96 |
| HMMR | ILMN_2409220 | 1.37 | 3.03 | 1.39 | 4.96 |
| LOC100131609 | ILMN_3292224 | 1.44 | 3.03 | 1.44 | 4.96 |
| MCM4 | ILMN_1737205 | 1.46 | 3.03 | 1.54 | 4.96 |
| PCDH20 | ILMN_1703572 | 1.43 | 3.03 | 1.45 | 4.96 |
| HMGB2 | ILMN_1654268 | 1.37 | 3.76 | 1.54 | 4.96 |
| KDELC2 | ILMN_1651557 | 1.39 | 3.76 | 1.42 | 4.96 |
| LOC651816 | ILMN_1729115 | 1.53 | 3.76 | 1.53 | 4.96 |
| SPC24 | ILMN_2181432 | 1.39 | 3.76 | 1.50 | 4.96 |
| UBE2C | ILMN_2301083 | 1.35 | 3.76 | 1.38 | 4.96 |
| ADAM19 | ILMN_1713751 | 0.75 | 4.62 | 0.70 | 3.95 |
| CDC45 | ILMN_1670238 | 1.40 | 4.62 | 1.41 | 4.96 |
| HSPH1 | ILMN_1712888 | 1.38 | 4.62 | 1.46 | 4.96 |
| LOC148915 | ILMN_1776052 | 1.34 | 4.62 | 1.38 | 4.96 |
| LOC402112 | ILMN_3213568 | 1.38 | 4.62 | 1.38 | 4.96 |
| MAD2L1 | ILMN_1777564 | 1.38 | 4.62 | 1.42 | 4.96 |
| MCM5 | ILMN_1815169 | 1.30 | 4.62 | 1.39 | 4.96 |
| NUSAP1 | ILMN_2409298 | 1.31 | 4.62 | 1.47 | 4.96 |
| PCNA | ILMN_1694177 | 1.36 | 4.62 | 1.39 | 4.96 |
siRNA = small interfering RNA.
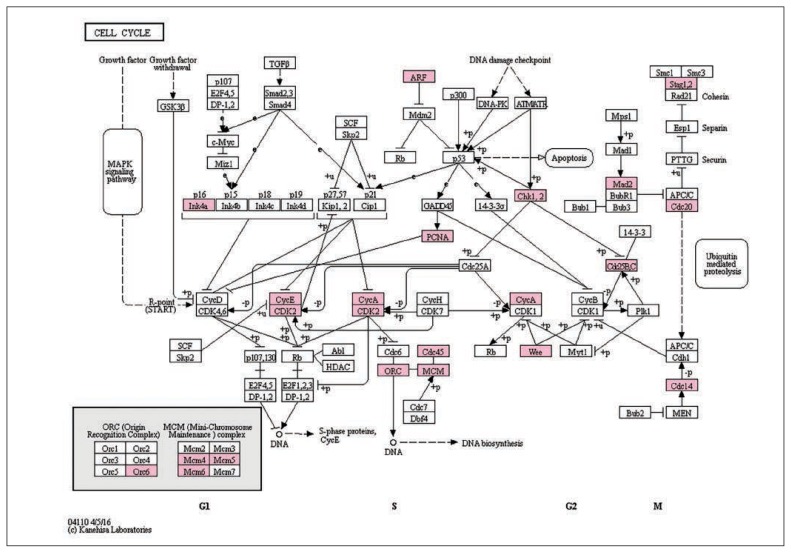
The GO analysis of the shared set of 628 differentially expressed gene probes (p < 0.05) showed a highly significant enrichment of genes involved in the cell cycle. The most significant GO term was “M-Phase” (p = 6.3 × 10−10, Bonferroni-corrected), with the majority of other significant GO terms also relating to the cell cycle (Appendix 1, Table S3). Consistent with the GO analysis, the only significant KEGG term to survive Bonferroni correction was “cell cycle” (p = 0.006, Bonferroni-corrected; Fig. 2). A similar enrichment of the KEGG cell cycle pathway (p = 0.004, Bonferroni-corrected) was observed in the smaller set of 39 genes that were differentially expressed between the control siRNA and each TCF4 siRNA condition at an FDR < 0.05. We confirmed altered expression of selected cell cycle genes showing expression changes following TCF4 knockdown (CDCA3, MAD2L1, MCM5 and PCNA) by qPCR (Appendix 1, Fig. S1).
Fig. 2.
Enrichment of gene expression changes (p < 0.05) shared by the TCF4 small interfering RNA (siRNA)#1 and TCF4 siRNA#2 conditions in the “cell cycle” KEGG pathway (p = 0.006, Bonferroni-corrected). Genes exhibiting altered expression in association with both TCF4 siRNA conditions are indicated in pink. Image generated through the DAVID Bioinformatics Resource 6.7.24
Given that variation in the TCF4 gene confers susceptibility to schizophrenia, we tested for genetic association between the set of genes that were differentially expressed following TCF4 knockdown (at p < 0.05) and schizophrenia using large-scale GWAS data.6 Competitive gene set analysis using MAGMA indicated a nonsignificant trend (p = 0.06) for genetic association between this gene set and the disorder. Notably, 5 of the 9 differentially expressed genes showing the most significant association with schizophrenia (gene-based p < 1 × 10−5) in the MAGMA analysis (GNL3, STAG1, CENPM, NCAPD3 and CDC20) have known roles in the cell cycle. MAGMA-generated schizophrenia association p values for all differentially expressed genes are provided in Appendix 1, Table S4.
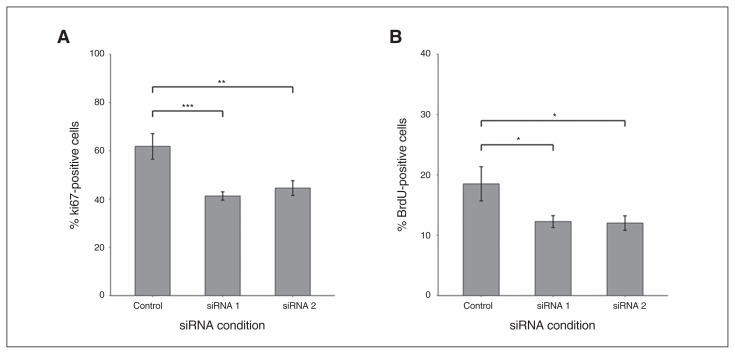
We investigated whether TCF4 siRNA conditions are associated with effects on the proliferation of human cortical progenitor cells in vitro in further experiments using high-content screening. Results are shown in Figure 3. In the TCF4 siRNA#1 condition, we observed a mean 33% decrease in Ki67-positive cells (p < 0.001) and a mean 34% decrease in BrdU-positive cells (p = 0.017) compared with the negative control siRNA. In the TCF4 siRNA#2 condition, we observed a mean 28% decrease in Ki67-positive cells (p = 0.010) and a mean 35% decrease in BrdU-positive cells (p = 0.027) compared with the negative control siRNA condition.
Fig. 3.
Decrease in cell proliferation markers in the CTX0E03 neural progenitor cell line in association with TCF4 small interfering RNA (siRNA) #1 and #2 conditions, as measured by high-content imaging. (A) Compared with the negative control siRNA condition, Ki67-positive cells were reduced by a mean of 33% in association with the TCF4 siRNA#1 condition (p < 0.001) and by a mean of 28% in association with the TCF4 siRNA#2 condition (p = 0.010). (B) Compared with the negative control siRNA condition, BrdU-positive cells were reduced by a mean of 34% in association with the TCF4 siRNA#1 condition (p = 0.017) and by a mean of 35% in association with the TCF4 siRNA#2 condition (p = 0.027); n = 12 per condition. Error bars represent standard errors of the mean. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
In order to explore early neurodevelopmental processes through which genetic perturbation of TCF4 could increase the risk for schizophrenia and Pitt–Hopkins syndrome, we have performed genome-wide expression profiling of a human cortical progenitor cell line following TCF4 knockdown. We found that TCF4 knockdown resulted in altered expression of genes involved in cell cycling. Consistent with the gene expression data, TCF4 siRNA conditions were associated with decreased cortical progenitor cell proliferation in vitro. These data implicate TCF4 in the regulation of human cortical progenitor cell proliferation, a mechanism that could contribute to neurodevelopmental features of schizophrenia and Pitt–Hopkins syndrome.
TCF4 has been previously implicated in neural progenitor cell proliferation in the mouse, where it has been shown to be a genomic target of the zinc finger protein Zac1.27 TCF4 has also been found to participate in the induction of cell cycle arrest in human colorectal cancer cell lines.28 In a study by Chen and colleagues,18 stable knockdown of TCF4 in a neural progenitor cell line derived from the human fetal ventral midbrain brought about changes in gene expression characteristic of differentiating rather than proliferating cells. Although no effect on cell proliferation was observed, the authors speculated that reduced dosage of TCF4 could result in precocious differentiation of some neural progenitors that had prematurely exited the cell cycle.18
The neural progenitor cell line we used for this study was derived from the cortical neuroepithelium of a 12-week human fetus.19 This cell line has been shown to differentiate into β-III tubilin-positive neurons and GFAP-positive astrocytes following removal of 4-OHT and the growth factors bFGF and EGF.19 Although we performed our experiments while these cells were in a proliferative state with growth factors (but not 4-OHT) present, our previous work using the same cell line under similar conditions has shown that gene perturbations do not necessarily affect the expression of cell cycle genes. Thus, our knockdown of the schizophrenia susceptibility gene ZNF804A was found to result in gene expression changes that were enriched for the GO term “cell adhesion,”20 while manipulation of miR-137 caused gene expression changes that were most significantly enriched for the GO term “neuronal differentiation.”26 Although the gene expression changes we observed following TCF4 knockdown were highly enriched for involvement in the cell cycle, differentially expressed genes shared by the 2 TCF4 siRNA conditions (Table 1 and Appendix 1, Table S2) include many related to various other functions in both the developing and mature brain.
At present, our knockdown of TCF4 expression in human neural progenitor cells is most clearly relevant to Pitt– Hopkins syndrome. Although recent studies indicate diverse mechanisms by which Pitt–Hopkins syndrome–associated missense mutations in the TCF4 gene impair TCF4 function,10,11 haploinsufficiency of TCF4 resulting from large heterozygous deletions encompassing the gene is a well-established cause of the disorder.7–9 Importantly, the effects of these deletions are likely to become apparent as soon as TCF4 is expressed. Effects on TCF4 expression are also likely to mediate associations between schizophrenia and common noncoding variants at the TCF4 locus.3–6 However, it is currently unknown when these variants exert their effects and which TCF4 transcripts are affected. Immature neurons derived from induced pluripotent stem cells from individuals with schizophrenia have been found to exhibit increased TCF4 RNA expression compared with those derived from healthy controls,29 suggesting that early developmental disturbances in TCF4 expression are relevant to the disorder. Increased TCF4 RNA expression in fibroblasts and blood from adults with schizophrenia30 and psychosis31 has also been reported.
Although alternative cellular functions of TCF4 could mediate its genetic association with schizophrenia,32 documented effects of other high-confidence schizophrenia susceptibility genes on cell proliferation33,34 support this as a neurobiological risk mechanism for the disorder. Several of the differentially expressed genes showing most significant association with schizophrenia in the present MAGMA analysis have known roles in cell cycling, suggesting that TCF4 could operate through a network of other susceptibility genes involved in neural cell proliferation. Cell cycling disturbances in individuals with schizophrenia are also suggested by observations of altered proliferation of fibroblasts35 and olfactory biopsy cultures36 from patients with the disorder as well as reports of differential expression of cell cycle genes in postmortem brain samples from such patients.37,38 Although, to our knowledge, there are no published data implicating cell cycle dysregulation in individuals with Pitt–Hopkins syndrome, changes in neural progenitor proliferation in the developing neocortex could contribute to the severe intellectual deficits observed in people with this condition.
Limitations
We do not provide a detailed mechanistic explanation of how TCF4 knockdown alters human neural progenitor cell proliferation. As a transcription factor, TCF4 will directly affect the expression of multiple genes. However, our gene expression data do not distinguish between those changes that arise from reduced binding of TCF4 at the gene locus or secondary consequences. Methods based on chromatin immunoprecipitation (e.g., ChIP-Seq) will be crucial in determining the primary targets of TCF4, some of which might mediate the effects on cell proliferation we observe.
Conclusion
We have shown that knockdown of TCF4 causes downstream changes in gene expression and proliferation of human cortical progenitor cells. Early disturbances in TCF4 expression could therefore impact upon the development of the human neocortex through effects on the number and/or subsequent differentiation of certain populations of cortical progenitors, suggesting a plausible neurobiological mechanism contributing to cognitive deficits in individuals with Pitt–Hopkins syndrome and risk for schizophrenia.
Acknowledgements
This work was supported by the Medical Research Council, UK [Grant ID: G0802166]. The authors are grateful to ReNeuron Ltd for providing the human neural cell line used in this study and to The Genome Centre, Barts and The London School of Medicine and Dentistry for microarray analysis.
Footnotes
Competing interests: None declared.
Contributors: M. Hill, B. Williams and N. Bray designed the study. M. Hill, R. Killick, K. Navarrete, A. Maruszak and G. McLaughlin acquired and analyzed the data, which N. Bray also analyzed. N. Bray wrote the article, which all authors reviewed and approved for publication.
References
- 1.Blake DJ, Forrest M, Chapman RM, et al. TCF4, schizophrenia, and Pitt-Hopkins syndrome. Schizophr Bull. 2010;36:443–7. doi: 10.1093/schbul/sbq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarrete K, Pedroso I, De Jong S, et al. TCF4 (e2-2; ITF2): a schizophrenia-associated gene with pleiotropic effects on human disease. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:1–16. doi: 10.1002/ajmg.b.32109. [DOI] [PubMed] [Google Scholar]
- 3.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–7. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg S, de Jong S Irish Schizophrenia Genomics Consortium, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076–81. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amiel J, Rio M, de Pontual L, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–93. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockschmidt A, Todt U, Ryu S, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet. 2007;16:1488–94. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 9.Zweier C, Peippo MM, Hoyer J, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forrest M, Chapman RM, Doyle AM, et al. Functional analysis of TCF4 missense mutations that cause Pitt-Hopkins syndrome. Hum Mutat. 2012;33:1676–86. doi: 10.1002/humu.22160. [DOI] [PubMed] [Google Scholar]
- 11.Sepp M, Pruunsild P, Timmusk T. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum Mol Genet. 2012;21:2873–88. doi: 10.1093/hmg/dds112. [DOI] [PubMed] [Google Scholar]
- 12.de Pontual L, Mathieu Y, Golzio C, et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum Mutat. 2009;30:669–76. doi: 10.1002/humu.20935. [DOI] [PubMed] [Google Scholar]
- 13.Sepp M, Kannike K, Eesmaa A, et al. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5¢ exon usage and splicing. PLoS One. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson P, Jogi A, Grynfeld A, et al. HASH-1 and E2–2 are expressed in human neuroblastoma cells and form a functional complex. Biochem Biophys Res Commun. 2000;274:22–31. doi: 10.1006/bbrc.2000.3090. [DOI] [PubMed] [Google Scholar]
- 15.Flora A, Garcia JJ, Thaller C, et al. The E-protein TCF4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci U S A. 2007;104:15382–7. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzózka MM, Radyushkin K, Wichert SP, et al. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene TCF4 in the brain. Biol Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Forrest MP, Waite AJ, Martin-Rendon E, et al. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8:e73169. doi: 10.1371/journal.pone.0073169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ES, Gigek CO, Rosenfeld JA, et al. Molecular convergence of neurodevelopmental disorders. Am J Hum Genet. 2014;95:490–508. doi: 10.1016/j.ajhg.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollock K, Stroemer P, Patel S, et al. A conditionally immortal clonal stem cell line from human cortical neuroepithelium for the treatment of ischemic stroke. Exp Neurol. 2006;199:143–55. doi: 10.1016/j.expneurol.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Hill MJ, Jeffries AR, Dobson RJ, et al. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21:1018–24. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- 21.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen EY, Wexler EM, Versano R, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–42. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.de Leeuw CA, Mooij JM, Heskes T, et al. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill MJ, Donocik JG, Nuamah RA, et al. Transcriptional consequences of schizophrenia candidate miR-137 manipulation in human neural progenitor cells. Schizophr Res. 2014;153:225–30. doi: 10.1016/j.schres.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt-Edelkraut U, Daniel G, Hoffmann A, et al. Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Mol Cell Biol. 2014;34:1020–30. doi: 10.1128/MCB.01195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst A, Helferich S, Behrens A, et al. The transcription factor ITF-2A induces cell cycle arrest via p21(Cip1) Biochem Biophys Res Commun. 2009;387:736–40. doi: 10.1016/j.bbrc.2009.07.102. [DOI] [PubMed] [Google Scholar]
- 29.Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–5. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattane N, Minelli A, Milanesi E, et al. Altered gene expression in schizophrenia: findings from transcriptional signatures in fibroblasts and blood. PLoS One. 2015;10:e0116686. doi: 10.1371/journal.pone.0116686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirgenes KV, Sonderby IE, Haukvik UK, et al. TCF4 sequence variants and mRNA levels are associated with neurodevelopmental characteristics in psychotic disorders. Transl Psychiatry. 2012;2:e112. doi: 10.1038/tp.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rannals MD, Hamersky GR, Page SC, et al. Psychiatric risk gene transcription factor 4 regulates intrinsic excitability of prefrontal neurons via repression of SCN10a and KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roussel MF, Ashmun RA, Sherr CJ, et al. Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol Cell Biol. 1996;16:2796–801. doi: 10.1128/mcb.16.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun G, Ye P, Murai K, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Lockstone HE, Guest PC, et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J Proteome Res. 2010;9:521–7. doi: 10.1021/pr900867x. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y, Abrahamsen G, McGrath JJ, et al. Altered cell cycle dynamics in schizophrenia. Biol Psychiatry. 2012;71:129–35. doi: 10.1016/j.biopsych.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Katsel P, Davis KL, Li C, et al. Abnormal indices of cell cycle activity in schizophrenia and their potential association with oligodendrocytes. Neuropsychopharmacology. 2008;33:2993–3009. doi: 10.1038/npp.2008.19. [DOI] [PubMed] [Google Scholar]
- 38.Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc Natl Acad Sci U S A. 2009;106:11731–6. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]