Abstract
Innate lymphoid cells (ILCs) are important regulators in various immune responses. Current paradigm states that all newly-made ILCs originate from common lymphoid progenitors (CLP) in the bone marrow. Id2, an inhibitor of E protein transcription factors, is indispensable for ILC differentiation. Unexpectedly, we found that ectopically expressing Id1 or deleting two E protein genes in the thymus drastically increased ILC2 counts in the thymus and other organs where ILC2 normally reside. Further evidence suggests a thymic origin of these mutant ILC2s. The mutant mice exhibit augmented spontaneous infiltration of eosinophils and heightened responses to papain in the lung and increased ability to expulse the helminth parasite, Nippostrongylus brasiliensis. These results prompt the question whether the thymus naturally has the capacity to produce ILC2s and E proteins restrain such a potential. The abundance of ILC2s in Id1 transgenic mice also offers a unique opportunity for testing the biological functions of ILC2s.
INTRODUCTION
Innate lymphoid cells (ILCs) play pivotal roles in mounting innate immunity against pathogens as well as in contributing to the pathogenesis of immunological illnesses (1,2). Therefore, understanding the regulation of their ontology will provide insights into therapeutic strategies to modulate the function of these ILCs. Rapid progress has led to the notion that ILCs originate from lymphoid-primed multipotent progenitors (LMPP) or common lymphoid progenitors (CLP), the same progenitors that give rise to adaptive lymphoid cells (1–3). Several populations of ILC progenitors have been identified and are thought to develop from CLP in the bone marrow. They cannot differentiate into adaptive lymphoid cells while retaining the ability to generate all ILC subsets (2,4–6). However, it is unclear if ILC differentiation follows this single linear path or if multipotent progenitors distinct from LMPP or CLP or those residing outside of the bone marrow also possess the ability to produce ILCs. For example, progenitors thought to lie upstream of CLP in the hierarchy of hematopoiesis have been shown to travel to the thymus (7,8), which then propagate into the earliest T cell progenitors called ETP (9). Whether progenitors seeding the thymus can generate all ILC subsets has not been fully investigated but thymus can be a fertile ground for ILC2s, because it provides ample IL-7 and Notch signals, essential for ILC2 differentiation (10–12). T cell progenitors also possess transcription factors necessary for ILC2 differentiation, which include Bcl11b, GATA3 and TCF1 (10,13–16). Although the thymus is not essential for ILC2 production, CD4 and CD8 double-negative thymocytes (DN1 and DN2) were shown to give rise to ILC2 cells when cultured extensively with IL-7 and IL-33 (17,18). However, their capacity to produce ILC2 in vivo has not been demonstrated.
Id2 is a member of the family of helix-loop-helix Id proteins, consisting of Id1-4, which all inhibit the DNA-binding activity of a group of E protein transcription factors encoded by the E2A, HEB and E2-2 genes (19,20). Id2-deficient mice lack not only NK cells but also all ILC subsets (21). Id2 is highly expressed in both ILCs and ILC progenitors (6,22,23). In the thymus, E2A and HEB are both expressed and play redundant roles in controlling T cell differentiation (24–30). Ablation of both E2A and HEB blocks T cell development at the DN3 stage whereas loss of either of these genes leads to partial impairment (31). Id2 and Id3 are known to be involved in later stages of T and NKT cell development in the thymus and the Id3 gene is known to be activated by pre-TCR and TCR signaling (32–36).
Although the Id1 gene is normally not expressed in lymphoid cells, we have previously used it as a tool to inhibit all E proteins in thymocytes by ectopically expressing Id1 off the lck proximal promoter (plck). Homozygous Id1 transgenic mice were found to have T cell development blocked at the DN stage in the thymus, lacking any T lineage committed cells (37–39). Re-examination of Id1 transgenic mice with ILC2 parameters revealed a large number of ILC2s in the thymus and in peripheral organs such as the lung, mesenteric lymph nodes (MLN), and spleen. Augmented ILC2 production in the thymus is found to be cell autonomous and likely responsible for the accumulation of ILC2 in the periphery. Consistent with the role of Id proteins as inhibitors of E proteins, deletion of both E2A and HEB genes in the thymus (31) using the plck-Cre transgene also led to similar phenotypes as Id1 transgenic mice. Id1 transgenic ILC2s display overlapping but distinct transcriptional profiles compared to wild type ILC2s. Furthermore, Id1 transgenic mice exhibit augmented recruitment of eosinophils to the lung before and after challenges with an allergen, papain. These mice are more adept at expulsing the helminth parasite, Nippostrongylus brasiliensis (N. brasiliensis). Together, our findings suggest that specific down-regulation of E proteins in the thymus highlights a previously unappreciated potential of the thymus as an organ for ILC2 production.
MATERIALS AND METHODS
Mice and in vivo treatments
C57BL/6 (CD45.2), TCRβ−/−, and B6-CD45.1 mice were purchased from the Jackson Laboratory and Charles River Laboratory. pLck-Id1 transgenic mice and LckCre;E2Af/f;HEBf/f mice are as described (38,40). All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Oklahoma Medical Research Foundation (OMRF).
For protease challenge, 10 μg papain was intra-nasally administrated into mice every 24 h on day 0, day 1, and day 2. Mice were sacrificed 12 h after the last challenge. Bronchoalveolar lavage fluid (BALF) and lungs were analyzed.
For infections with N. brasiliensis, mice were subcutaneously inoculated with 500 third-stage infective larvae (L3) and sacrificed 6 days later for analysis of ILC2 responses. BALF, lungs, mediastinal and mesenteric lymph nodes were analyzed, and adult worm burden was determined by enumerating worms present in the proximal half of the small intestine.
Isolation of Lung Hematopoietic Cells
Perfused lungs were cut into small fragments and digested in Hank’s balanced salt solution containing 2.5 mg/ml collagenase (Sigma) and 160 U/ml DNase I (Sigma) for 30 m. Cells were filtered by using a 70μm cell strainer.
Flow Cytometry and Cell Sorting
All antibodies (Abs) used were purchased from BioLegend unless specified otherwise. Abs in the lineage (Lin) cocktail included anti-FcεR (MAR-1; eBioscience), anti-B220 (RA3-6B2), anti-CD19 (ID3; BD Biosciences), anti-Mac-1 (M1/70; BD Biosciences), anti-Gr-1 (R86-8C5; BD Biosciences), anti-CD11c (HL3; BD Biosciences), anti-NK1.1 (PK 136), anti-Ter-119 (Ter-119), anti-CD3 (145-2C11), anti-CD8α (53-6.7; BD Biosciences), anti-CD5 (53-7.3), anti-TCRβ (H57-597), and anti-γδTCR (GL-3; BD Biosciences). Additional Abs used included anti-CD45.2 (104), anti-CD45.1 (A20), anti-CD4 (RM4-5; BD Biosciences), anti-cKit (2B8), anti-Sca-1(D7), anti-Thy1.2 (53-2.1), anti-IL-5 (TRFK5), anti-IL-13 (eBio13A; eBioscience), anti-Siglec F (E50-2440; BD Biosciences), anti-IL7Rα (A019D5), anti-α4β7 (DATK32; eBioscience), anti-CD25 (PC61), anti-PLZF (Mags.21F7; eBioscience), and anti-ST2 (DIH9).
Cell sorting was performed on a FACSAria II (BD Biosciences), and flow cytometric analysis was performed on a LSR-II (BD Biosciences). Intracellular staining of transcription factors was done using Foxp3 Staining Buffer kit (eBioscience). Staining of cytokines was carried out using Cytofix/cytoperm kit (BD Bioscience).
Bone Marrow Transplantation
To generate mixed bone marrow chimeras, 105 sorted Lin−Thy1− bone marrow cells from CD45.2+ C57BL/6 or Id1 transgenic mice, were mixed with 105 wild type (WT) CD45.1+Lin−Thy1− cells and intravenously transferred into lethally irradiated WT hosts (CD45.1). Recipients were analyzed 7 weeks after transplantation.
RNA-Seq analysis
Total RNA was purified from ILC2 cells of Id1 transgenic thymus and MLN as well as from MLN ILC2s cultured as described (16). Approximately 300,000 cells were used in the preparation of each sample. RNA-Seq was performed in duplicates by the Genomics Core Facility at OMRF using Illumina Truseq Stranded mRNA library prep kits followed by sequencing on an Illumina Hiseq 2500 in Rapid mode with 50bp, paired-end reads (Illumina, San Diego, CA). In addition, transcriptomes of WT ILC2 cells from small intestine (GSE47851) (16) and WT DN3 thymocytes (GSE31235) (41) were included in our analyses. Sequencing reads were aligned to the Mus musculus genome reference (mm9) using Star ver. 2.4.0h. Gene-level read counts were determined using HTSeq ver. 0.5.3p9 with the Ensembl Mus musculus NCBIM37 (release 67) annotation. Non- and low-expressed genes were filtered out and 20,471 genes remained in the analysis. The read counts were normalized using DESeq ver. 1.18.0. Principal component analysis (PCA) was performed using the Fluidigm SINGuLAR Analysis Toolset 2.0 in R (42). The top-ranked PCA genes were determined based on the maximum absolute value of each gene loading score in the first three principal components. Hierarchical clustering using complete linkage and Euclidean distance as the similarity metric and heat map analyses were performed using Cluster 3.0 and Java TreeView respectively (43,44).
Statistics
Pairwise comparison was performed using Student’s t test or one-way ANOVA.
RESULTS
Down-regulation of E proteins promotes ILC2 production
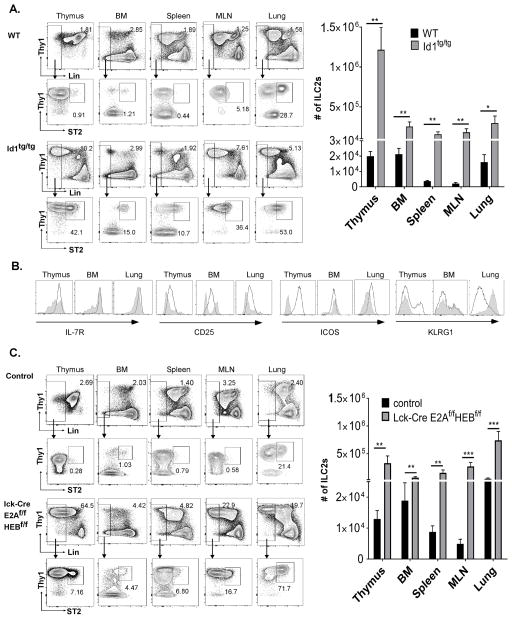
To study the collective role of E proteins in T cell development, we previously created Id1 transgenic mice by using the proximal promoter of the lck gene, known to direct gene expression specifically in the thymus at an early stage. In homozygotes (Id1tg/tg), T cell differentiation is completely blocked (37–39). Within the CD4 and CD8 double negative compartment depleted of B, myeloid, or NK cells, we detected subsets of cells with atypical CD44 and CD25 profiles (Fig. S1A). Interestingly, these cells could produce IL-5 and IL-13 but very little IFNγ and IL-17 upon stimulation with PMA and ionomycin (Fig. S1B). Further analyses of the lineage negative fraction (Lin−) for the expression of ILC2 surface markers such as Thy1 and ST2 revealed an approximate 60 fold increase in ILC2 counts when comparing Id1 transgenic to WT thymuses (Fig. 1A). These cells were also found to be able to produce IL-5 and IL-13 (Fig. S1C). Moreover, the ILC2 levels in other organs such as the lung, spleen, bone marrow and MLN were also 10–100 fold higher (Fig. 1A). We also detected increased numbers of ILC2s in the white adipose tissue, blood and small intestine of the transgenic mice (data not shown). In heterozygous transgenic mice (Id1tg), the numbers of ILC2 in different organs were also dramatically elevated even though T cell development was not completely blocked (data not shown). In contrast, the numbers of ILC1 in the liver and ILC3 in the small intestine of Id1 transgenic mice were not dramatically altered (data not shown).
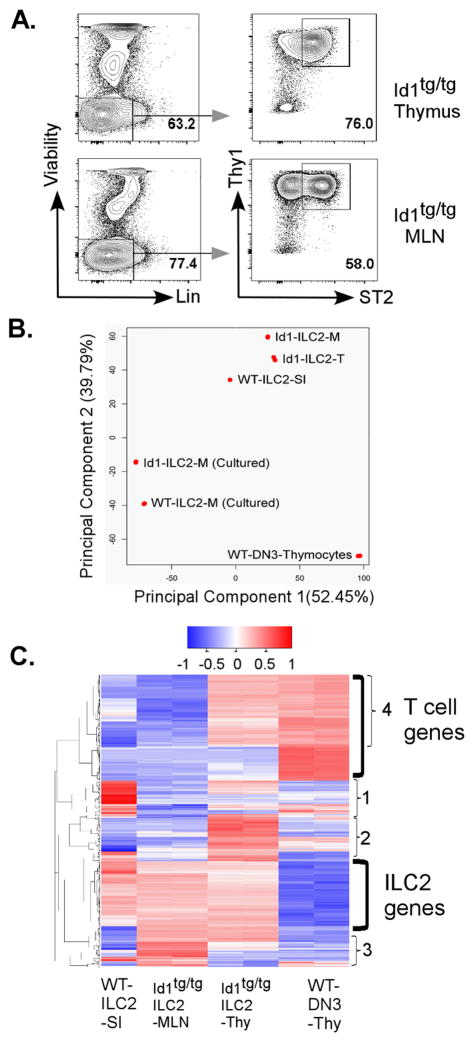
Figure 1. Down-regulation of E protein function augments ILC2 production.
(A) Analyses of Lin− cells from C57BL/6J (WT) and Id1tg/tg mice. Cells from indicated tissues of mice of indicated genotypes were stained for lineage markers (FcεR, B220, CD19, Mac-1, Gr-1, CD11c, NK1.1, Ter-119, CD3, CD8α, TCRβ and γδTCR) together with antibodies against Thy1 and ST2. The lineage negative gates and expression profiles of Thy1 and ST2 on Lin− cells are presented. The percentage of ILC2s defined as Lin−Thy1+ST2+ is shown. Bar graphs show average cell numbers in each tissue (n=4–9). (B) Additional analyses for the indicated markers on ILC2s defined as in (A). Shaded peaks represent the profiles of WT ILC2s in indicated tissues and the solid lines depict those of Id1tg/tg ILC2s. (C) Analyses of Lin− cells from plck-Cre/E2Af/f;HEBf/f and littermate controls without plck-Cre as described for (A) (n=6–8). Bar graphs show data pooled from five (A) or three (C) independent experiments. Error bars are SEM. Student’s t test was used for statistical analyses. * p< 0.05; ** p< 0.01 and *** p < 0.001.
Further characterization of ILC2s (Lin−Thy1+ST2+) was carried out with additional markers associated with ILC2 phenotypes, which include IL-7Ra, CD25, ICOS and KLRG1. While the expression of these markers on wild type ILC2s in different organs are variable, Id1 transgenic ILC2s in the thymus, BM and lung displayed a similar pattern, namely Lin−Thy1+ST2+IL-7Ra+ICOS+CD25intKLRG1± (Fig. 1B). This result suggests that the putative ILC2s found in Id1 transgenic mice carry multiple characteristics of well-recognized ILC2s. Interestingly, the fact that Id1 transgenic ILC2s from different organs exhibit a similar phenotype raised the possibility that these cells come from the same source such as the thymus.
To test if Id1 acted by inhibiting E protein transcription factors (encoded by the E2A, HEB and E2-2 genes), we generated plck-Cre/E2Af/f /HEBf/f mice to specifically delete both the E2A and HEB genes with the plck-Cre transgene, whose expression is known to be restricted to the thymus. These mice were reported to have impaired T cell development (31) but we found them to possess large numbers of ILC2s in the thymus and other organs (Fig. 1C), suggesting that E protein ablation in the thymus enhances the production of ILC2 cells, a phenotype similar to that found in Id1 transgenic mice.
Augmented ILC2 differentiation in the thymus is cell autonomous
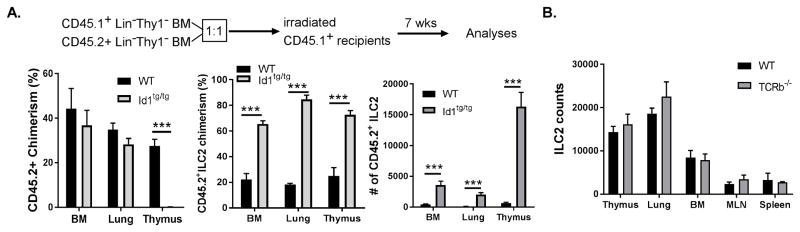
To determine if the Id1 transgene promoted ILC2 differentiation in a cell autonomous manner, we created mixed bone marrow chimeras where Lin−Thy1.2− bone marrow cells from CD45.2+ WT or Id1tg/tg mice were mixed with equal numbers of CD45.1+Lin−Thy1.1− WT cells and injected into lethally irradiated CD45.1+C57BL/6 hosts. Seven weeks later, the recipients were analyzed for chimerism and ILC2 production (Fig. 2A). Bone marrow reconstitution with CD45.2+ WT and Id1tg/tg cells achieved 44% and 37% chimerism, respectively. Likewise, CD45.2+ cells also represented substantial fractions of hematopoietic cells in the lung. While WT CD45.2+ thymocytes were abundant, Id1 transgenic cells in the thymus were barely detectable due to a complete block in T cell development. In contrast, ILC2s of Id1tg/tg origin in the bone marrow, lung and thymus vastly outnumbered CD45.1+ILC2s derived from the CD45.1+ donors or hosts even though the chimerism of WT CD45.2+ILC2s was only about 20% in each of these locations (Fig. 2A). These results suggest that the promotion of ILC2 production by the Id1 transgene is likely through cell autonomous mechanisms rather than alterations in secreted factors which would have stimulated ILC2 differentiation from both transgenic and CD45.1+ progenitors. Admittedly, the number of ILC2 produced from Id1 donors in the thymus of these recipients (Fig. 2A) was not as high as that found in Id1tg/tg mice themselves (Fig. 1A). ILC2 differentiation of Id1tg/tg progenitors was probably hindered by robust T cell development from CD45.1+ progenitors, which competes for space and cytokines. However, the massive production of ILC2 in Id1tg/tg mice is not caused by a blockage of T cell development itself because TCRβ−/− mice, despite the block in T cell development and lack of regulatory T cells, do not have dramatically augmented ILC2 counts (Fig. 2B). We have also ruled out the possibility that Id1 expression stimulates the proliferation of ILC2s since no increase in cycling cells was detected in various tissues of Id1tg/tg mice as measured using Ki67 and DAPI staining (Fig. S2A). In fact, WT thymic ILC2s had a larger fraction of cells in S-G2 phases than Id1tg/tg counterparts, possibly due to more robust homeostatic proliferation. Likewise, we obtained no evidence of increased survival of Id1 transgenic ILC2s in the thymus or periphery. Instead, Id1 transgenic ILC2s appeared to exhibit increased apoptosis in the bone marrow (Fig. S2B).
Figure 2. Augmented ILC2 production in Id1 transgenic mice is cell intrinsic.
(A) Mixed bone marrow chimera. Lin−Thy1− BM cells of C57BL/6 (CD45.2+) or Id1 transgenic (CD45.2+) mice were mixed with the same cells from CD45.1+ B6 mice at 1:1 ratio and transplanted into lethally irradiated CD45.1+ B6 mice (5 per cohort). Total CD45+ and ILC2 cells in indicated organs were analyzed 7 weeks after transplantation. Chimerism was defined as percent of CD45.2+ fractions of CD45.1+ plus CD45.2+ cells. The numbers of CD45.2+ILC2s are shown on the right. Error bars are SEM. Data shown is a representative of three independent experiments. (B) Comparison of ILC2 counts in WT and TCRβ−/− mice. ILC2s were defined as described in the legend for Fig. 1 except that anti-CD5 was also included in the lineage cocktail. Student’s t test was used for statistical analyses.
Data from the transplantation studies also showed that ILC2s generated from donor bone marrow readily repopulated adult lymphoid and non-lymphoid organs in irradiated recipients (Fig. 2A). Although the representation of WT CD45.2+ILC2s was somewhat lower than the overall chimerism, possibly due to the radio-resistance of host ILC2s, Id1 transgenic donors supplied a large number of ILC2s to all of the organs examined. These results suggest that transplanted bone marrow progenitors are capable of giving rise to ILC2s either in the bone marrow or thymus and these ILC2s can home to peripheral tissues such as the lung of irradiated hosts. These results are in contrast to the observations made with parabiotic mice (45), which showed little exchange of lung ILC2s between the two mice under physiological conditions.
ILC2 differentiation in the bone marrow is not perturbed in Id1 transgenic mice
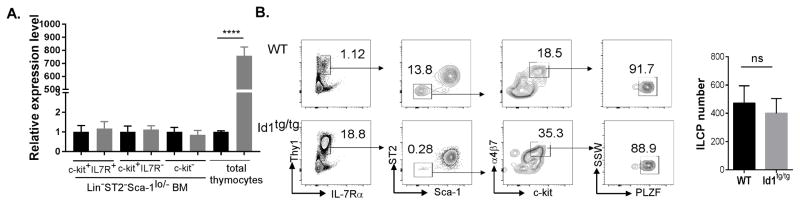
To verify whether increased production of ILC2s is due to the specific expression of the Id1 transgene in the thymus, we analyzed Id1 mRNA in total thymocytes of Id1tg mice and different fractions of lineage negative BM of Id1tg/tg mice. Because ILC2 cells generated in the thymus can migrate to the BM may express the transgene, ILC2 cells were excluded from the BM preparations (Fig. 3A). The c-kit+/loIL-7R+ fraction likely contains common lymphoid and ILC progenitors whereas the c-kit+/loIL-7R− fraction includes myeloid progenitors. The c-kit− fraction consists of the remainder of the Lin-Sca-1lo/−ST2− BM. While Id1 levels were 760 fold higher in Id1tg thymus than WT controls, the levels in the different populations of bone marrow were comparable, suggesting that the transgene was not expressed in the bone marrow though the possibility of the transgene expression in rare unknown subsets have not been ruled out (Fig. 3A).
Figure 3. The frequency of ILC progenitors was not affected by thymus-specific expression of the Id1 transgene.
(A) The Id1 transgene was specifically expressed in transgenic thymocytes. Total Id1 mRNA levels in thymocytes and bone marrow cells of WT and Id1tg mice were determined using qRT-PCR and normalized against the levels of β-actin. Bone marrow cells of WT and Id1tg/tg mice were fractionated by first enriching for Lin−Sca-1lo/−ST2− cells and then dividing the resulting population into c-kit+IL-7Rα+, c-kit+IL-7Rα− and c-kit− subsets. The c-kit+ fractions include both c-kithi and c-kitint cells. The normalized Id1 levels relative to the WT fractions are shown. (B) Similar numbers of bone marrow ILC progenitors (Lin−Thy1+IL7Rα+ ST2−Sca-1−α4β7+PLZF+) were found in WT and Id1tg/tg mice. Gating strategy was shown on the left and the average ILCP number was shown on the right (n=3). Data shown are representative of at least two independent experiments. Student’s t test was used for statistical analyses.
To further verify if any undetectable levels of Id1 expression in rare progenitors promoted ILC2 differentiation in the bone marrow, we determined the numbers of PLZF-positive ILC progenitors (ILCP) in the bone marrow (6). We found that WT and Id1tg/tg mice have similar numbers, suggesting early stages of ILC differentiation in the bone marrow was not altered (Fig. 3B). The large number of Lin−Thy1+ST2+ cells in the transgenic bone marrow, which can also be characterized as Lin−Sca−1+ CD25int (data not shown), could be attributed to increased thymus output.
Lineage tracing data support the thymic origin of excess ILC2s in Id1 transgenic mice
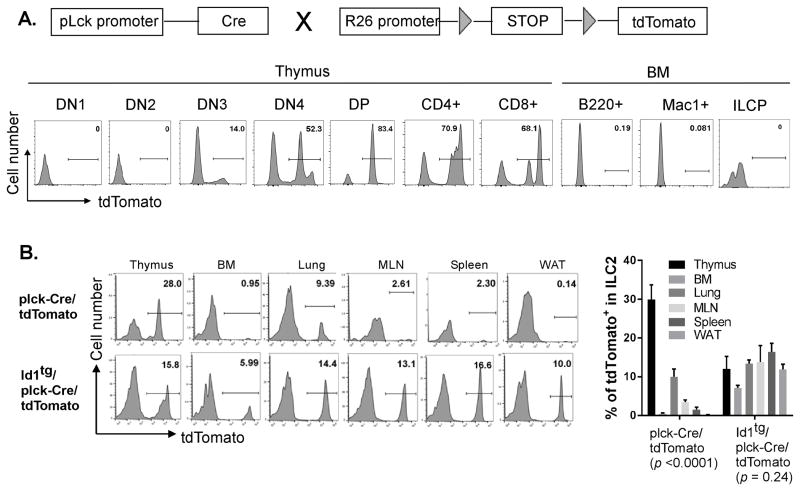
In an attempt to mark ILC2s generated in the thymus, we took advantage of the thymus-specific expression driven by the proximal promoter of the lck gene (plck). We created plck-Cre/ROSA26-Stop-tdTomato mice with or without the Id1 transgene and first confirmed that Cre expression labeled most cells in the thymus, but not non-T cells including ILCPs in the bone marrow (Fig. 4A). Within the CD4 and CD8 double negative compartment of thymocytes, an appreciable proportion of DN3 cells were tdTomato+ but DN1 and DN2 cells had very low levels of reporter expression (Fig. 4A). In the absence of the Id1 transgene, a significant fraction of ILC2s in the thymus were tdTomato+ but BM ILC2s were not (Fig. 4B). Interestingly, a remarkable percentage of lung ILC2s expressed tdTomato, but other peripheral organs such as white adipose tissue (WAT) had much lower fractions of tdTomato+ ILC2s (Fig. 4B). This result suggests that thymus-derived ILC2s preferentially home to the lung under natural conditions. Whether thymus-derived ILC2s play an important role in lung physiology or pathophysiology however remains to be investigated in the future.
Figure 4. Lineage tracing data suggest the thymic origin of Id1 transgenic ILC2s.
(A) The Cre transgene is driven by the proximal promoter of the lck gene in plck-Cre mice, which were crossed with ROSA26-STOP-tdTomato reporter mice. Percentages of tdTomato+ cells in subsets of thymocytes and B220+ or Mac-1+ bone marrow cells as well as ILCP are shown. CD4, CD8, CD19, B220, Mac-1, Gr1, FcγR, Ter119, NK1.1, and TCRγδ negative thymocytes were further defined by c-kit and CD25 expression in DN populations: DN1 (C-kit+CD25−), DN2 (C-kit+CD25+), DN3 (C-kit−CD25+) and DN4 (C-kit−CD25−). (B) Analyses of tdTomato expression in ILC2s in mice of indicated genotypes. To ensure the elimination of any potential contamination of T lineage cells, ILC2s were defined as in Fig. 2B except that the Lin− population was further gated for TCRβ−TCRγδ− based on staining with antibodies against these TCRs conjugated with different fluorophores from that used for lineage staining, and analyzed for Thy1 and ST2. It is noted that the fluorescence intensity in WT thymic ILC2s was reproducibly lower than their Id1tg counterparts. Bar graph shows the average percentage of tdTomato+ cells in the ILC2 population of pooled data from several experiments (n= 3–10). One–way ANOVA was used to determine the statistical significance of the variations of tdTomato levels in different tissues in each strain.
Id1tg/plck-Cre/ROSA26-Stop-tdTomato mice had about 12% of tdTomato+ILC2s in the thymus. This percentage was lower than that found in mice on the wild type background, probably because Id1 expression beginning at the DN1 or DN2 stages (39) diverted the cells at these stages to ILC2 before tdTomato was turned on. Nevertheless, the percentages of tdTomato+ ILC2s were found to be around 10% in all organs of Id1 transgenic mice (Fig. 4B). These results are in contrast to the observations in wild type mice, where ILC2s in the BM, MLN, spleen and white adipose tissue showed little tdTomato expression (Fig. 4B). One-way ANONA analyses showed that the difference in tdTomato percentages in different organs on the wild type background was highly significant (p <0.0001) whereas that on the Id1 transgenic background was not (p=0.24). Therefore, these findings, combined with data shown in Figs 1–3, suggest that ILC2s overproduced in the Id1 transgenic thymus, supply these cells to all peripheral organs, leading to the dramatic increases of ILC2s with similar characteristics of the thymic ILC2s in these organs.
Id1 transgenic ILC2s display ILC2 signature gene expression
As a means of further phenotyping Id1 transgenic ILC2s, we performed RNA-seq analyses of ILC2s (Lin−Thy1+ST2+) freshly isolated from Id1tg/tg thymus and MLN (Fig. 5A) and compared data reported by Yagi et al. for WT ILC2s from small intestine (16). We also cultured Id1tg/tg ILC2s from MLN in the presence of IL-7, IL-25 and IL-33 in the same manner as Yagi et al (16). In addition, we included gene expression profiles of WT DN3 thymocytes in our analyses (41). Principal component analysis (PCA) showed that Id1tg/tg ILC2s from the thymus and MLN closely resemble each other and share substantial similarities with WT ILC2s from the small intestine (Fig. 5B). In contrast, these ILC2s are distinct from DN3 thymocytes. After culturing for a week, WT and Id1tg/tg ILC2s remain closely related, though the cultured cells have very different transcriptomes from freshly isolated cells. These results help validate the identity of the Id1 transgenic ILC2s.
Figure 5. Id1 transgenic ILC2s have overlapping and distinct trancriptomes from WT ILC2s.
(A) Representative sorting data for ILC2s from the thymus and MLN of Id1tg/tg mice. For each sort, single cell preparations from three mice were depleted with antibodies against CD3, B220, Mac1, Ter119 and NK1.1. The unbound cells were stained and gated as indicated. Approximately 300,000 ILC2s were obtained from each sort. (B) Principal component analyses. RNA-seq analyses of Id1tg/tg ILC2s from the thymus (T) and mesenteric lymph nodes (M) compared to WT ILC2 from small intestine (SI) and CD4−CD8−CD44−CD25+ thymocytes (DN3). Data of MLN ILC2 cells cultured in vitro for 7 days were also included for both WT and Td1tg/tg mice. (C) Hierarchical clustering and heat map of top 500 PCA genes. The log2 of gene-level read counts were centered on the mean and normalized.
Indeed, hierarchical clustering and heat map analyses of the top 500 PCA genes in different preparations of ILC2s and DN3 thymocytes revealed large blocks of genes specifically expressed in DN3 or ILC2 cells (Fig. 5C). Id1tg/tg ILC2s share a group of over 100 ILC2 signature genes with WT ILC2s, which include Rora, Rarg, Icos, Il1rl1, Klrg1, Arg1, IL2rb, IL13 and IL6 (Supplemental Table 1). Noticeably, there are also small clusters of genes that are different among WT ILC2s from the small intestine, and Id1tg/tg ILC2s from the MLN and thymus (clusters 1–3, Fig. 5C and supplemental Table 1). These differences may be due to the influences of tissue environments, Id1 overexpression, or the maturity of the ILC2 cells even though the possibility of technical variations among data generated in different laboratories has not been ruled out. Interestingly, ILC2s from the thymus also transcribe some genes found in DN3 developing T lineage cells (cluster 4). This is unlikely due to T cell contamination in the preparation, as Id1tg/tg mice do not have T cells. Although a few of the genes in cluster 4 are related to cell cycle control, the similarities also raise a question whether both DN3 and ILC2 cells in the thymus are derived from common progenitors. Taken together, the transcriptome analyses provide further evidence to suggest that the phenotypic ILC2s found in Id1 transgenic mice exhibit characteristics of wild type ILC2s.
Incidentally, comparison of the transcript levels of E and Id proteins between Id1tg/tg thymic ILC2 and WT DN3 cells revealed a slight decrease (around 2.5 fold) in E2A and HEB levels in ILC2s. In contrast, Id1 expression was 725 fold higher as a result of transgene expression. Id2 levels in Id1tg/tg cells were 59 fold higher, which is consistent with their commitment to the ILC2 lineage. However, Id3 expression in transgenic ILC2s decreased by 55-fold probably due to a lack of pre-TCR signaling, which drives Id3 expression (34). The levels of E2A, HEB, Id2 and Id3 were similar in all ILC2 preparations analyzed. Therefore, these results reflect the expression patterns of E and Id proteins in their respective cell type.
Id1 transgenic mice exhibit spontaneous inflammation and hypersensitivity to allergen stimulation
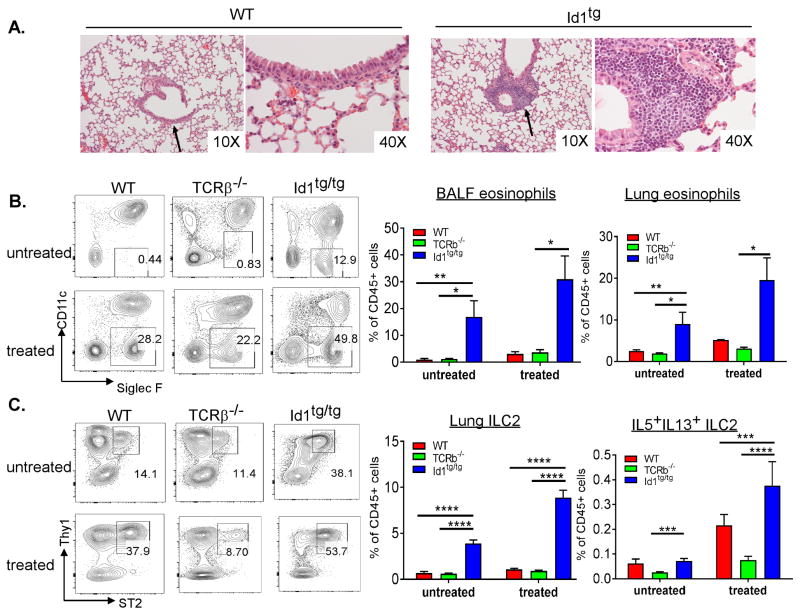
To test the functionality of Id1 transgenic ILC2s, we examined the type 2 immune responses in the lung. First of all, histological examination revealed that naive Id1 transgenic lungs showed more pronounced infiltration of lymphoid-looking cells around small bronchioles, a location where ILC2s are thought to congregate (Fig. 6A). This is consistent with the dramatic increases in the numbers of ILC2s but not B or T cells in the lung in Id1 transgenic mice. Moreover, marked elevation in steady-state eosinophil counts were observed in the BALF and lung of untreated Id1tg/tg mice, which suggests increased ILC2 function (Fig. 6B). Since Id1tg/tg mice lack any T cells, we used TCRβ−/− mice to control for the lack of T helper 2 and regulatory T cell functions and they did not show elevated eosinophil recruitment.
Figure 6. Id1 transgenic mice exhibit spontaneous inflammation and hyper response to allergen.
(A) H&E staining of lung sections of 2-month old WT and Id1tg/tg lungs. Areas with higher magnification are pointed by arrows. Representative graphs from two mice of each genotype are shown. (B) Eosinophil infiltration in mice of indicated genotypes following intra-nasal exposure to papain or in untreated controls. Eosinophil gates in total BALF were shown on the left, percent of eosinophils in total CD45+ cells of BALF and lungs of untreated and treated mice were shown on the right (n=3–6). (C) Lung ILC2s in the same mice as (B). CD45+Lin− cells were analyzed for Thy1 and ST2 expression and ILC2s are gated as shown. Percent of lung ILC2s and IL5+IL13+ILC2s of total CD45+ cells were shown on the right. Error bars are SEM. Student’s t test was used for statistical analyses. * p< 0.05; and *** p < 0.001. Data shown are representative of 2–3 experiments.
When the mice were treated with papain, an allergen delivered intra-nasally, substantial increases in eosinophil recruitment were seen in mice of all three genotypes. In addition, marked increases of IL-5 and IL-13 producing ILC2 cells were detected (Fig. 6C). These results suggest that Id1 transgenic ILC2s can mount type 2 immune responses and the elevated ILC2 counts in the lung of Id1tg/tg mice leads to greater allergic reactions in steady state and upon allergen exposure.
Id1 transgenic mice exhibit enhanced ability to expulse helminth parasites
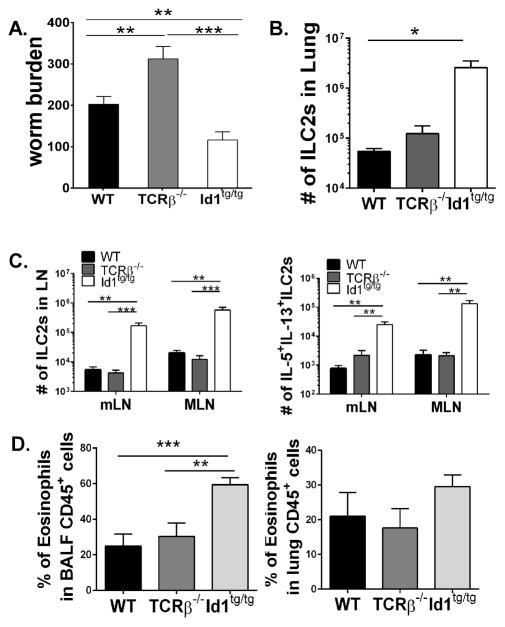
One of the major functions of ILC2s is to help the hosts clear parasite infection. Infection with helminth parasite such as N. brasiliensis has been widely used as a test for ILC2 functions. We inoculated WT, TCRβ−/− and Id1tg/tg transgenic mice with 500 third-stage infective larvae (L3) subcutaneously and measured adult worm burden in the small intestine 6 days later (Fig. 7A). At this time point, WT mice retained about 200 adult worms, while T cell deficient TCRβ−/− mice had significantly greater numbers of worms. In contrast, Id1tg/tg mice, despite their T cell defect, exhibited enhanced worm clearance compared to both WT and TCRβ−/− mice, suggesting enhanced worm expulsion in the presence of excessive ILC2s (Fig 7B). Consistent with the data on worm burden, Id1tg/tg mice harbored many more ILC2s capable of producing IL-5 and IL-13 (Fig. 7C) and recruited significantly larger numbers of eosinophils in the BALF and lung (Fig. D). These results suggest that the large numbers of ILC2 cells in Id1 transgenic mice lead to augmented type 2 immune responses.
Figure 7. Augmented helminth expulsion by Id1 transgenic mice.
Mice of indicated genotypes were inoculated subcutaneously with 500 third-stage N. brasiliensis larvae. Six days later mice were analyzed. (A) Worm counts in the small intestine (n=5–9). (B). ILC2 cells in the lung. (C) ILC2 cells in mediastinal (mLN) and mesenteric lymph nodes (MLN). (D) Percent of Eosinophils in the CD45+ fraction in the BALF and lung. * < 0.05; **, p < 0.01 and *** p < 0.001. Error bars are SEM. Data shown are representative of at least 3 experiments.
DISCUSSION
The unexpected findings in our Id1 transgenic mice taught us several important lessons: (1) Multipotent progenitors that normally seed the thymus or T cell precursors are capable of generating functional ILC2s; (2) E proteins suppress the innate lymphoid fate while driving T cell differentiation; (3) Elevated ILC2 counts lead to heightened, spontaneous and induced type 2 immune responses. These findings also prompt the question as to whether down-regulation of E protein activities simply highlight a natural capacity of the thymus as a second site for ILC2 production. Indeed, previous studies have shown that DN thymocytes are able to give rise to ILC2 cells in vitro (17,18). The phenotypes of Id1 transgenic mice are the first examples that illustrate the extraordinary capacity of the thymus to generate ILC2s in vivo. This is likely due to the ideal environment the thymus provides and the intrinsic properties of thymic T cell progenitors poised for ILC2 differentiation, namely the combination of Notch and IL-7 signaling and the presence of critical transcription factors such as TCF1, GATA3 and Bcl11b (10,13,15,16). It would be extremely interesting to determine how E proteins restrain such a potential.
It would also be informative to determine the developmental stages at which T cell progenitors can be hijacked to become ILC2s by down-regulating E protein function. Reporter expression induced by plck-Cre is helpful in addressing this question. Although ILCs have been shown to express some levels of the lck gene (46), patterns of gene expression driven by the proximal promoter of the lck gene in transgenic constructs are unlikely the same as that of the native lck gene. Indeed, we have shown plck-Cre only marks T lineage cells and ILC2s in the thymus but not ILC2s in the bone marrow, MLN, spleen, white adipose tissue and skin (Fig. 4B and Bajana and Sun, data not shown). The tdTomato+ fraction in thymic ILC2s is generally higher than that in DN3 thymocytes (Fig. 4A and B), which would mean that ILC2s arise after the DN3 stage, or that tdTomato is turned on independently in ILC2s in the thymus. Although the Id1 transgene is also driven by the proximal promoter of the lck gene, its expression is known to occur at the DN1 to DN2 stage (39). As a result, a significant portion of ILC2s were probably derived from T cell progenitors before plck-Cre is expressed, leading to a lower percentage of tdTomato+ ILC2s in the Id1 transgenic thymus.
The fact that about 10% of lung ILC2s were labeled with tdTomato suggests that thymus-derived ILC2s preferentially home to the lung though the possibility that tdTomato is expressed independently in the lung remains to be ruled out. Considering that only 30% of the thymic ILC2s were tdTomato+, the actual fraction of lung ILC2s that come from the thymus could be as high as 30%. If thymic ILC2s indeed contribute to the pool of lung ILC2s, it would be interesting to determine if these cells have distinct properties and serve different functions, which could be of great significance in understanding certain illnesses such as childhood asthma.
Finally, the heightened type 2 immune responses seen in Id1 transgenic mice illustrate the impact of augmented ILC2 production. This observation complements findings obtained using animals stimulated with IL-33, which might have effects beyond expanding ILC2. It is also interesting to point out that the surface phenotypes of Id1 transgenic ILC2s in the lung are different from their wild type counterparts, e.g. KLRG1 expression (Fig. 1B), even though they both can mount type 2 immune responses, exemplifying the heterogeneity of ILC subsets (47,48). Thus, this Id1 transgenic mouse model will be useful for investigating different aspects of ILC2 function implicated in respiratory disorders, dermatitis, obesity and glucose intolerance.
Supplementary Material
Acknowledgments
We thank Dr. Yuan Zhuang for providing E2Af/f and HEBf/f mice, Dr’s. Fotini Gounari, Samantha Webb and Sandra Bajana for valuable advice.
This work was supported by funds from the NIH (R01 AI126851-01 and R21 AI117895), and Presbyterian Health Foundation to XHS as well as the COBRE grant (P30GM110766) to CGM. XHS holds the Lew and Myra Chair in Biomedical Research.
Footnotes
Accession number:
[geo] GSE94597 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94597)
Reference List
- 1.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 2.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo M, I, Weissman L, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014:3. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, Wang H, Wang X, Yu S, Zhou X, Cam M, Xue HH, Bhandoola A. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 8.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc Natl Acad Sci U S A. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, Huang J, Haczku A, Pear WS, Artis D, Bhandoola A. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentek R, Munneke JM, Helbig C, Blom B, Hazenberg MD, Spits H, Amsen D. Modulation of Signal Strength Switches Notch from an Inducer of T Cells to an Inducer of ILC2. Front Immunol. 2013;4:334. doi: 10.3389/fimmu.2013.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roediger B, Kyle R, Yip KH, Sumaria N, Guy TV, Kim BS, Mitchell AJ, Tay SS, Jain R, Forbes-Blom E, Chen X, Tong PL, Bolton HA, Artis D, Paul WE, Fazekas de St GB, Grimbaldeston MA, Le GG, Weninger W. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong C, Zhu J. Bcl11b drives the birth of ILC2 innate lymphocytes. J Exp Med. 2015;212:828. doi: 10.1084/jem.2126insight1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Wang C, Clare S, Wang J, Lee SC, Brandt C, Burke S, Lu L, He D, Jenkins NA, Copeland NG, Dougan G, Liu P. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med. 2015;212:865–874. doi: 10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, Belz G, Liu P, Fallon PG, McKenzie AN. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med. 2015;212:875–882. doi: 10.1084/jem.20142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, Belkaid Y, Zhao K, Zhu J. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, Hardman CS, Hwang YY, Fallon PG, McKenzie AN. Transcription factor RORalpha is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentek R, Munneke JM, Helbig C, Blom B, Hazenberg MD, Spits H, Amsen D. Modulation of Signal Strength Switches Notch from an Inducer of T Cells to an Inducer of ILC2. Front Immunol. 2013;4:334. doi: 10.3389/fimmu.2013.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr Top Dev Biol. 2014;110:189–216. doi: 10.1016/B978-0-12-405943-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 20.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 21.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 22.Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, Shi W, Ritchie ME, Naik SH, Huntington ND, Carotta S, Belz GT. Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Rep. 2016;17:436–447. doi: 10.1016/j.celrep.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang Y, Barndt RJ, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi A, Yamasaki S, Takase K, Nakatsu F, Arase H, Onodera M, Saito T. E2A and HEB activate the pre-TCR alpha promoter during immature T cell development. J Immunol. 2001;167:2157–2163. doi: 10.4049/jimmunol.167.4.2157. [DOI] [PubMed] [Google Scholar]
- 28.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- 31.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Wu D, Jiang N, Zhuang Y. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol. 2013;191:5052–5064. doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 34.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras- ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 35.Verykokakis M, Krishnamoorthy V, Iavarone A, Lasorella A, Sigvardsson M, Kee BL. Essential Functions for ID Proteins at Multiple Checkpoints in Invariant NKT Cell Development. J Immunol. 2013;191:5973–5983. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu T, Wang H, Simmons A, Bajana S, Zhao Y, Kovats S, Sun XH, Alberola-Ila J. Increased level of E protein activity during invariant NKT development promotes differentiation of invariant NKT2 and invariant NKT17 subsets. J Immunol. 2013;191:5065–5073. doi: 10.4049/jimmunol.1301546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, Xu M, Nie L, Peng XC, Jimi E, Voll RE, Nguyen T, Ghosh S, Sun XH. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-kappaB activities. Immunity. 2002;16:9–21. doi: 10.1016/s1074-7613(02)00264-9. [DOI] [PubMed] [Google Scholar]
- 38.Kim D, Peng XC, Sun XH. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang HC, Perry SS, Sun XH. Id1 attenuates Notch signaling and impairs T-cell commitment by elevating Deltex1 expression. Mol Cell Biol. 2009;29:4640–4652. doi: 10.1128/MCB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Hoon MJ, Imoto S, Miyano S. Statistical analysis of a small set of time-ordered gene expression data using linear splines. Bioinformatics. 2002;18:1477–1485. doi: 10.1093/bioinformatics/18.11.1477. [DOI] [PubMed] [Google Scholar]
- 43.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 44.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 45.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, Brandt C, Kane L, Campos LS, Lu L, Belz GT, McKenzie AN, Teichmann SA, Dougan G, Liu P. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature. 2016;539:102–106. doi: 10.1038/nature20105. [DOI] [PubMed] [Google Scholar]
- 48.Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, Lim S, Kurioka A, Fergusson JR, Tang CL, Kam MH, Dennis K, Lim TK, Fui AC, Hoong CW, Chan JK, Curotto de LM, Narayanan S, Baig S, Shabeer M, Toh SE, Tan HK, Anicete R, Tan EH, Takano A, Klenerman P, Leslie A, Tan DS, Tan IB, Ginhoux F, Newell EW. Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.