ABSTRACT
In this study, an ertapenem-nonsusceptible Escherichia coli isolate was investigated to determine the genetic basis for its carbapenem resistance phenotype. This clinical strain was recovered from a patient that received, 1 year previously, ertapenem to treat a cholangitis due to a carbapenem-susceptible extended-spectrum-β-lactamase (ESBL)-producing E. coli isolate. Whole-genome sequencing of these strains was performed using Illumina and single-molecule real-time sequencing technologies. It revealed that they belonged to the ST131 clonal group, had the predicted O25b:H4 serotype, and produced the CTX-M-15 and TEM-1 β-lactamases. One nucleotide substitution was identified between these strains. It affected the ompR gene, which codes for a regulatory protein involved in the control of OmpC/OmpF porin expression, creating a Gly-63-Val substitution. The role of OmpR alteration was confirmed by a complementation experiment that fully restored the susceptibility to ertapenem of the clinical isolate. A modeling study showed that the Gly-63-Val change displaced the histidine-kinase phosphorylation site. SDS-PAGE analysis revealed that the ertapenem-nonsusceptible E. coli strain had a decreased expression of OmpC/OmpF porins. No significant defect in the growth rate or in the resistance to Dictyostelium discoideum amoeba phagocytosis was found in the ertapenem-nonsusceptible E. coli isolate compared to its susceptible parental strain. Our report demonstrates for the first time that ertapenem resistance may emerge clinically from ESBL-producing E. coli due to mutations that modulate the OmpR activity.
KEYWORDS: OmpR, ST131, avibactam, carbapenem, envZ, ESBL, temocillin
INTRODUCTION
Carbapenems are broad-spectrum antibiotics usually reserved for severe life-threatening infections. Some enterobacterial isolates have developed carbapenem resistance, which resulted in limited options for the treatment of infections caused by these organisms. Except for the members of the Proteeae tribe, carbapenem resistance in Enterobacteriaceae is almost always attributable to the production of β-lactamases, which can be distinguished according to their carbapenemase activity. True carbapenemases (e.g., KPC, OXA-48, and metallo-β-lactamases [MBLs]) confer per se resistance to carbapenems, whereas extended-spectrum β-lactamases (ESBLs) and AmpC-type enzymes require an additional mechanism of resistance, such as decrease in the uptake of antibiotics by porin deficiency (1, 2) or efflux system overexpression (3), to be responsible for carbapenem resistance.
Decreased bacterial cell permeability due to loss or alteration of the outer membrane porins F (OmpF) and/or C (OmpC) constitutes one of the chief mechanisms of carbapenem resistance in Escherichia coli clinical isolates in combination with ESBL or AmpC production (4–9). By reducing the antibiotic concentration inside the periplasm, porin change can amplify the β-lactamase effects of ESBLs and AmpC-type enzymes toward weakly hydrolyzed substrates, such as carbapenems (1, 2). The susceptibility to ertapenem, which is generally less stable against β-lactamases than other carbapenems (10), is more affected.
The OmpC and OmpF coding genes are transcriptionally regulated by a two-component signal transduction regulatory system (TCS) consisting of the OmpR and EnvZ proteins (11). EnvZ is an inner membrane protein that is phosphorylated by intracellular ATP at histidine 243 in response to signal related to the environmental osmolarity. It transfers this phosphoryl group to aspartic acid 55, which is located in the N-terminal receiver domain of OmpR, the response regulator, thus permitting the transmission of the signal to the C-terminal effector domain of OmpR, which contains a winged helix-turn-helix DNA binding motif (12). By binding to their promoter regions, the activated OmpR induces the transcriptional activation of ompC and ompF genes.
The aim of this study was to identify potential gene(s) associated with the reduced ertapenem susceptibility exhibited by an E. coli clinical isolate. Comparative genome analysis of this isolate and its parental ertapenem-susceptible strain allowed detection of a mutation within the ompR gene.
RESULTS
Antibiotic susceptibility profiles.
The MIC of ertapenem against E. coli ErtR (1 μg/ml) was 30-fold higher than that against the corresponding parental E. coli ErtS strain (0.032 μg/ml). The E. coli ErtR derivative also appeared to be significantly less susceptible to other β-lactams, such as ceftazidime and meropenem, than the parental E. coli ErtS strain (Table 1). No changes in the MICs for any other antimicrobial agents tested, including gentamicin, amikacin, and tigecycline, were observed (Table 1). Both clinical isolates were resistant to sulfamethoxazole-trimethoprim (>32 μg/ml) and ciprofloxacin (>32 μg/ml) but remained susceptible to gentamicin (0.5 μg/ml), amikacin (4 μg/ml), tigecycline (0.5 μg/ml), colistin (0.5 μg/ml), and fosfomycin (2 μg/ml). Moreover, no decrease of ertapenem MIC was observed in the presence of phenylalanine-arginine β-naphthylamide dihydrochloride (PAβN), revealing that the MIC increase probably was not associated with an efflux mechanism. In contrast, addition of clavulanate to the Mueller-Hinton (MH) agar plates resulted in a significant decrease of ertapenem MIC for the E. coli ErtR mutant isolate, suggesting the contribution of an Ambler class A β-lactamase, such as CTX-M-15, to carbapenem resistance (Table 1).
TABLE 1.
MIC values for the E. coli clinical isolates and the E. coli ErtR (pOmpR-WT) recombinant clone
| β-Lactama | MIC (μg/ml) for: |
|||
|---|---|---|---|---|
| E. coli ErtS | E. coli ErtR | E. coli ErtR MH-CLAVb | E. coli ErtR (pOmpR-WT) | |
| Amoxicillin | >32 | >32 | 32 | >32 |
| Amoxicillin-CLAV | 8 | 16 | ND | 8 |
| Piperacillin | >32 | >32 | 4 | >32 |
| Piperacillin-TZB | 4 | 16 | ND | 4 |
| Temocillin | 2 | 4 | 2 | 2 |
| Cefoxitin | 4 | 16 | 4 | 4 |
| Ceftazidime | 16 | 64 | 0.5 | 16 |
| Ceftazidime-avibactam | 0.125 | 0.25 | ND | 0.125 |
| Cefepime | >32 | >32 | 1 | >32 |
| Aztreonam | >32 | >32 | 0.5 | >32 |
| Ertapenem | 0.032 | 1 | 0.032 | 0.032 |
| Imipenem | 0.125 | 0.25 | 0.125 | 0.125 |
| Meropenem | 0.032 | 0.25 | 0.032 | 0.032 |
CLAV, clavulanic acid at 2 μg/ml; TZB, tazobactam at 4 μg/ml.
MH-CLAV, MICs obtained using clavulanate (2 μg/ml)-containing Mueller-Hinton agar plates. ND, not determined.
Genotyping analysis.
Comparative genome sequencing pointed to one mutation in the genome of isolate ErtR compared to that of isolate ErtS (see below), confirming that the isolates were closely related. The genome of E. coli ErtS includes a chromosome of 5,188,809 bp with a GC content of 54.92% and 3 plasmids: an IncF plasmid of 135,180 bp containing the IncFII, FIA, and FIB replicons, an IncI1 plasmid of 88,514 bp, and an IncN plasmid of 55,001 bp. The functional annotation of the chromosome indicates a total of 5,202 predicted coding sequences (CDSs). The strain belonged to the B2 phylogroup and the global epidemic clone ST131. The genome sequence-inferred serotype was O25b:H4, and the fimH allele was H30.
Resistance determinants.
blaCTX-M-15 was detected in the two E. coli clinical isolates, which was consistent with the ESBL phenotype, and blaTEM-1 was detected as well. The β-lactamase genes were carried on the IncF plasmid. In addition, the genes for tetracycline resistance (tetA), macrolide resistance [mph(A)], sulfonamide resistance (sul1), and trimethoprim resistance (dfrA25) were detected. The latter genes were carried by the IncN plasmid, except the mph(A) gene, which was harbored by the IncF plasmid. Point mutations associated with quinolone resistance were detected in gyrA (S83L and D87N) and parC (E84V).
Virulence factors.
Comparison with the virulence factor database (https://cge.cbs.dtu.dk/services/VirulenceFinder/) and with the study of Lefort et al. (13) showed that sat, iha, iss, sfa, irp2, fyuA, and iucC genes were harbored by both isolates. Sat is a secreted autotransporter toxin that alters the structural and functional components of intercellular junctions (14), iha is an iron-regulated gene homologue adhesin that augments adherence to uroepithelial cells (15), sfa codes for the S fimbrial adhesin (13), iss is a serum survival gene associated with a 20-fold increase in complement resistance (16), and irp2, fyuA, and iucC genes belong to the iron capture systems (13).
Genomic analysis of reduced ertapenem susceptibility.
The quality of the assembled reads obtained by high-throughput sequencing (HTS) is disclosed in Table S1 in the supplemental material. Comparative genomic analysis of E. coli ErtR with E. coli ErtS, after subtracting the sequence errors detected during the reference analysis by mapping the MiSeq E. coli ErtS reads against the PacBio RS II E. coli ErtS reference sequence, revealed one mutation in the ompR gene, which was confirmed after Sanger sequencing.
The ompR G188→T transversion created a Gly-63-Val substitution in the amino-terminal phosphorylation domain of OmpR. Because the ompR gene codes for a protein that regulates the expression of the OmpC and OmpF outer membrane porins, it was tempting to speculate that it plays a role in the acquired ertapenem resistance. In order to determine the role of the Gly-63-Val replacement in ertapenem resistance, a complementation experiment, which consisted of transferring the wild-type ompR gene into the E. coli ErtR derivative, was carried out. The wild-type allele of the ompR gene was amplified from the DNA extract of E. coli ErtS and subsequently cloned into pCR-BluntII-Topo, giving rise to the pOmpR-WT recombinant plasmid. Sequence analysis revealed that the orientation of the cloned insert was under the transcriptional control of the lacZ promoter flanking the cloning site. The E. coli ErtR (pOmpR-WT) recombinant clone was fully susceptible to ertapenem (0.032 μg/ml) (Table 1). This strongly suggests that alteration of OmpR accounted for ertapenem resistance in the E. coli ErtR derivative.
OMP profile.
SDS-PAGE analysis of outer membrane proteins (OMPs) revealed a strong decrease in the amount of both major porins OmpC and OmpF for the E. coli ErtR isolate compared to its parental E. coli ErtS strain (Fig. 1). The presence of a band of the same intensity for the porin OmpA for both strains indicated that the decrease of OmpC/OmpF was not due to a technical problem.
FIG 1.
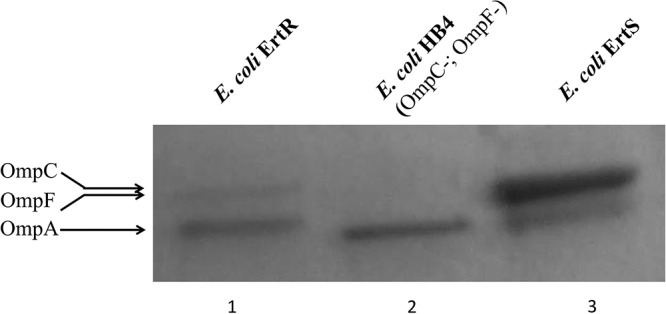
OMP profiles of E. coli ErtR compared to its parental strain, E. coli ErtS. OMPs were profiled by SDS-PAGE. Lane 1, E. coli ErtR; lane 2, E. coli HB4 ΔompC-ompF strain (control strain); lane 3, E. coli ErtS. The horizontal arrows on the left indicate the positions of OMPs OmpC-OmpF and OmpA. Note the same amount of OmpA protein in the three lanes.
Structure-activity relationship.
The Gly-63 residue is well conserved among various OmpR proteins in the Enterobacteriaceae family and among other response regulators, such as DrrB (17) (Fig. 2), suggesting that this amino acid plays a crucial role in the structure-activity relationship. A virtual model of the amino-terminal phosphorylation domain of the OmpR-G63V variant was built using the X-ray crystal structures of receiver domains of a homologous response regulator, Thermotoga maritima DrrB. It showed that the Gly-63-Val replacement, which is located in the N-terminal extremity of the α-3 helix, was located in the vicinity of the active site, in front of the β-3 strand that harbors the histidine-kinase phosphorylation site Asp-55 (Fig. 3) (12). Moreover, the molecular modeling assay revealed that the Gly-63-Val substitution induced a displacement of 0.4 Å of the α-carbon of Asp-55, whereas the α-carbon of residue 63 moved from its initial position by 0.3 Å (data not shown). Replacement of Gly-63 by an asparagine or arginine residue, which contains a bulky side chain, displaced the α-carbon of Asp-55 by 0.6 Å or 1.5 Å, respectively (data not shown). This result suggested that the replacement of a hydrogen atom by a more bulky lateral-side site at position 63 would result in a steric hindrance that displaced the histidine-kinase phosphorylation site.
FIG 2.

Multiple alignment of the amino acid sequences of various homologues of the OmpR-PhoB subfamily. Proteins studied include OmpR of E. coli ErtS (this study), OmpR of M. morganii (GenBank accession no. KJY03066), OmpR of S. marcescens (GenBank accession no. KFL01930), OmpR of C. freundii (GenBank accession no. AKL57621), OmpR of P. mirabilis (GenBank accession no. KGA91828), OmpR of K. pneumoniae (GenBank accession no. CDO12110), OmpR of E. cloacae (GenBank accession no. AHE72147), OmpR of P. aeruginosa (GenBank accession no. BAP53865), CheY of E. coli (GenBank accession no. BAA15698), NtrC of E. coli (GenBank accession no. ACT31142), NarL of E. coli (GenBank accession no. CAA48935), PhoP of E. faecalis (GenBank accession no. KGJ35834), CheY of E. faecalis (GenBank accession no. KOA04247), PhoB of E. coli (GenBank accession no. ACJ50526), DrrB of T. marina (PDB entry 1P2F), and CpxR of E. coli (GenBank accession no. BAE77397). Numbering of the amino acid residues is according to that of OmpR of E. coli (12). Shaded amino acids at positions 55 and 63 represent highly conserved residues among response regulators.
FIG 3.
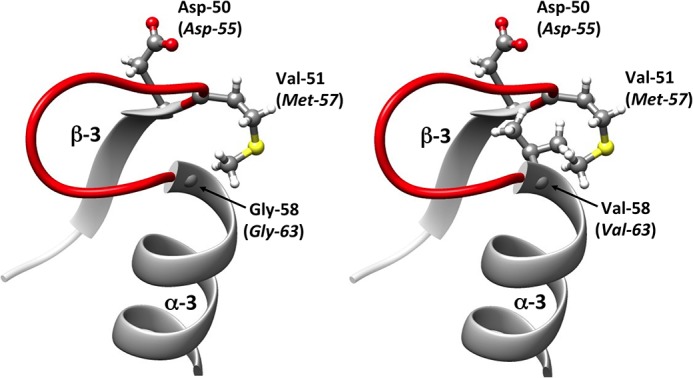
Representation of the secondary structures surrounding the aspartate residue (Asp) at position 50/55. The protein variant was modeled from the crystallographic structure of the DrrB response regulator of Thermotoga maritima (PDB entry 1P2F) by the introduction of an amino acid substitution followed by energy minimization. The amino acids belong to DrrB, whereas those italicized in parentheses are their corresponding residues in the homolog E. coli OmpR protein. The secondary structure in red refers to the loop lying between the α-3 helix and the β-3 strand. (Left) Regulator response DrrB of Thermotoga maritima with glycine residue at position 58 (corresponding to the Gly-63 residue in the native OmpR). (Right) Variant of DrrB with a Gly-58-Val substitution, corresponding to the Gly-63-Val replacement in the OmpR-G63V variant.
As depicted in Fig. 2, OmpR and DrrB differed by several amino acids that were in the loop between the Gly-63 and Asp-55 residues. Structural analysis of DrrB was performed by changing these residues, followed by energy minimization. No significant displacement of the α-carbon of Asp-55 was detected (<1 Å), suggesting that the structural differences between OmpR and DrrB did not affect the position of the histidine-kinase phosphorylation site (data not shown).
Bacterial growth analysis.
To assess the fitness cost potentially related to the OmpR alteration, the bacterial growth of the E. coli ErtS and E. coli ErtR clinical isolates was determined (Fig. 4). This procedure revealed that the maximum growth rates (MGRs) of the E. coli ErtS and E. coli ErtR isolates were not significantly different. Moreover, the relative fitness of the E. coli ErtR mutant compared to the parental E. coli ErtS isolate was not significantly different (confidence interval, 1.004 ± 0.002).
FIG 4.
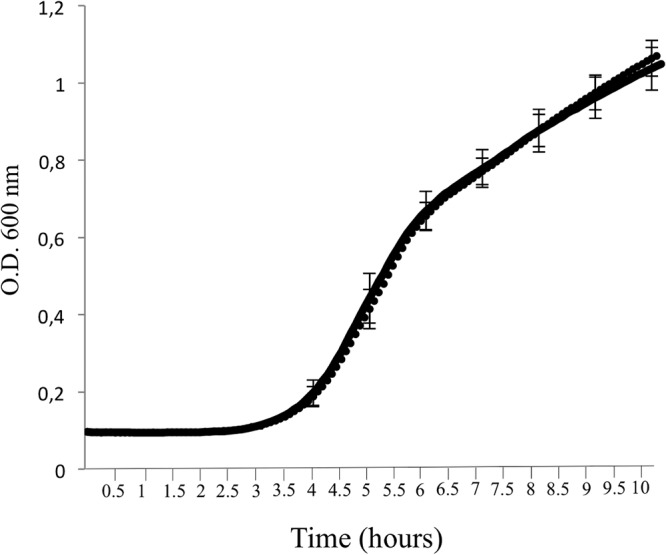
Growth curves of E. coli strains in LB. Growth of E. coli ErtS (in dotted line) and E. coli ErtR (in solid line). The lines are surperimposed. The error bars indicate the standard deviations of the means for five experiments. Values of the optical density (O.D.) at 600 nm were collected every 5 min. The maximum growth rate (MGR) of E. coli ErtS was 1.45 ± 0.12 h−1, whereas the MGR of E. coli ErtR was 1.46 ± 0.14 h−1.
Experimental virulence assay using the social amoeba Dictyostelium discoideum.
As B2 phylogroup strains possess extraintestinal virulence potential (18), the resistance to phagocytosis of ErtS parental and ErtR mutant strains was studied in a D. discoideum model. The virulent phylogroup B2 E. coli 536 strain, as well as the nonvirulent phylogroup A E. coli B REL606 strain, were also included in the experiment as controls. Three days after plating, with 102, 103, 104, and 105 amoeba cells, plaques were observed only for E. coli B REL606. Six days after plating, no plaque was observed for E. coli ErtS, E. coli ErtR, and E. coli 536, in agreement with a retained virulence (Fig. 5).
FIG 5.
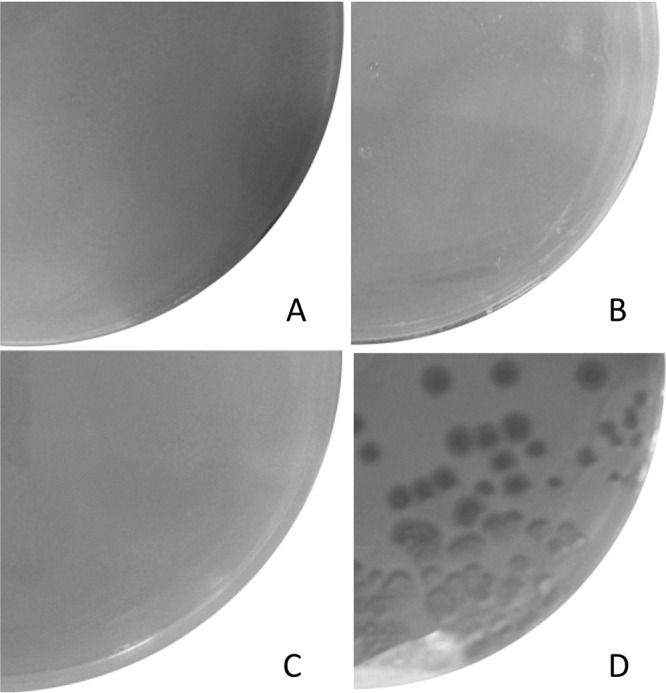
Examples of virulence phenotypes observed in the D. discoideum amoeba model. Bacteria (108 CFU) were plated on petri dishes containing HL5 agar with D. discoideum at variable concentrations and examined at day 6. (A) E. coli 536 with 103 amoebae. Amoebae did not form lysis plaque, indicating a grazing-resistant phenotype. (B) E. coli ErtS with 103 amoebae. Amoebae did not form lysis plaque. (C) E. coli ErtR with 103 amoebae. Amoebae did not form lysis plaque. (D) E. coli B REL 606 with 102 amoebae. Amoebae fed on the lawn of bacteria, forming large lysis plaques, in agreement with a grazing-sensitive phenotype.
DISCUSSION
One of the chief mechanisms of carbapenem resistance in E. coli and other Enterobacteriaceae is decreased bacterial cell permeability due to loss or alteration of the outer membrane porins F and/or C (19, 20). So far, molecular studies revealed that reduced permeation of carbapenems in E. coli clinical isolates was ascribed to direct alteration in the porin structure or deletion of the ompC and ompF genes (4–8). It stresses for the first time that carbapenem resistance in E. coli clinical isolates can also result from functional change in a chromosomally encoded regulatory factor. Contribution of the regulatory system to ertapenem resistance has been raised previously by Warner et al., who described an ertapenem-resistant E. coli clinical isolate with a mutated marR gene (21). Physiologically, the marR gene codes for a repressor that downregulates the expression of the multiple antibiotic regulon marAB, which controls the expression of efflux pump systems and is also involved in the control of porin expression. However, the ompF gene of this strain was also inactivated by a partial deletion, revealing that the marR mutation alone did not account for the lack of permeability (21).
The regulation of the porins is implemented in E. coli by a complex regulatory network whose EnvZ-OmpR TCS is a central element, since phosphorylated OmpR is absolutely required for OmpC and OmpF expression (22). Our study showed that the Gly-63-Val substitution in OmpR renders the expression of ompC and ompF inactive. These results are in agreement with the in vitro studies of Tängdén et al. and Adler et al., which demonstrated that a single-step EnvZ- or OmpR-inactivating mutation was enough to significantly increase the MIC for ertapenem due to a subsequent reduction of OmpC and/or OmpF of up to 90% (23, 24). Our report provides further evidence for the clinical relevance of OmpR change in the emergence of ertapenem resistance among E. coli clinical isolates.
As described previously, lack of permeability can magnify the hydrolytic activity of β-lactamases against substrates that are poorly hydrolyzed (1, 2). By changing the porin profile, OmpR-inactivating mutation highlighted the weak carbapenemase activity of an Ambler class A β-lactamase that was produced by the E. coli ErtR isolate. The MIC of E. coli ErtR for meropenem was increased compared to that of E. coli ErtS (0.125 μg/ml and 0.032 μg/ml, respectively), yet the MIC still was within the range of susceptibility according to the CLSI definition of clinical breakpoints (25). On the other hand, the MIC of E. coli ErtR for imipenem remained unchanged. This discrepancy, which was also evidenced in the study of Tängdén et al. (23), could be attributable to the intrinsic hydrolysis spectrum of CTX-M β-lactamases. The full restoration of the MIC value of E. coli ErtR for meropenem by clavulanic acid supported this hypothesis. These findings suggest that exposure to ertapenem or meropenem, at therapeutic or suboptimal concentrations, respectively, favors the selection of carbapenem-nonsusceptible (CNS) ESBL-producing E. coli mutants. In addition, the E. coli ErtR isolate remained susceptible to temocillin (4 μg/ml) and the ceftazidime-avibactam combination (0.25 μg/ml), suggesting that the alteration of the ompR allele did not affect the in vitro activity of these compounds in E. coli. Additional studies are needed to establish what the roles of ceftazidime-avibactam and temocillin might be as substitutes for carbapenems to reduce the emergence of CNS E. coli.
Molecular modeling suggested that the Gly-63-Val replacement impacted the structure of the amino-terminal receiver domain of OmpR. To the best of our knowledge, this is the first time that an OmpR variant presents a structural alteration at position 63, at the extremity of the α-3 helix in the vicinity of the β-3 strand that harbors the histidine-kinase phosphorylation site Asp-55. The OmpR variants that were previously described had structural changes at different positions, such as R45S, L112R, D100E, R209C, and I83T replacements (12, 23). The glycine at position 63 is well conserved among various response regulators, suggesting that it plays a crucial role in the structure-activity relationship of this protein family. According to our modeling study, the Gly-63-Val substitution may induce a steric hindrance between the α-3 helix and the β-3 strand, which might consecutively displace the Asp-55 residue from its most competent position for accepting the phosphoryl group from the histidine residue of the cognate sensor EnvZ.
OmpR not only controls the expression of the OmpC and OmpF porins but also is involved in the regulation of other metabolic pathways, such as curli fimbria expression (26). An OmpR-inactivating mutation therefore might impact pathogen physiology and reduce bacterial fitness. Adler et al. showed that OmpR inactivation caused a 20% reduction in growth rate (24). These results were in agreement with those obtained by Tsai et al., who showed that OmpK35/ompK36 porin deficiency in K. pneumoniae could decrease fitness and virulence (27). Interestingly, the OmpR-G63V change did not alter the fitness and the experimental virulence in an amoeba model of the E. coli ErtR clinical isolate, suggesting that this structural modification modulated the functional properties of OmpR. The resulting porin expression was reduced enough to confer decreased ertapenem susceptibility without impairing virulence and fitness. It appears that the OmpR-G63V change confers an adaptive benefit for survival by resisting ertapenem without reducing phagocytosis resistance and fitness.
The global increase in ESBL-producing E. coli is closely tied to the pandemic clonal group ST131. Postulated explanations for the success of ST131 include higher transmissibility, a greater ability to colonize in the intestine or urinary tract, enhanced virulence, and multidrug resistance (MDR) status (28). The genetic background of the E. coli ErtR isolate, which belonged to the ST131 complex, further emphasizes the clinical relevance of our report, since it highlights the potential risk of emergence of ertapenem resistance among the CTX-M-producing E. coli pandemic clones without decreasing fitness.
Concluding remarks.
Our report demonstrates, for the first time, that ertapenem resistance may emerge clinically from ESBL-producing E. coli due to structural changes in OmpR. It provides more insights into the structure-activity relationship of this response regulator.
MATERIALS AND METHODS
Bacterial strains.
The clinical isolate E. coli ErtS, which was identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), was recovered in a blood culture from a 49-year-old man admitted at the Teaching Hospital of Amiens (Amiens, France) and diagnosed with cholangitis in August 2011. The patient immediately underwent an endoscopic retrograde cholangio-pancreatography, and a probabilistic antibiotic treatment with amoxicillin-clavulanate was started. The antibiotic treatment was modified once the microbiological documentation identified the carbapenem-susceptible ESBL-producing E. coli isolate ErtS in the blood cultures. Imipenem was then substituted for the previous antibiotic for 2 days. The patient was discharged home with a switch from imipenem to ertapenem, which was monitored during 1 month. In July 2012, the patient was admitted to an intensive care unit (ICU) to support insufficient breathing. He was diagnosed with bronchial intraepithelial neoplasia. The ertapenem-nonsusceptible ESBL-producing E. coli isolate ErtR was recovered in his bronchial aspirate. The patient died 2 weeks later from complications of malignancy.
The wild-type E. coli TOP10 strain (Invitrogen, Cergy Pontoise, France) was used as the recipient for the transformation experiment. The virulent phylogroup B2 E. coli 536 strain and the nonvirulent phylogroup A E. coli B REL606 strain were used as a control in the assay of resistance to Dictyostelium discoideum amoeba phagocytosis (18).
Antimicrobial susceptibility testing.
MICs were determined by an agar dilution technique on Mueller-Hinton (MH) agar (Sanofi-Diagnostics Pasteur, Paris, France) with an inoculum of 104 CFU per spot and were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (25). MICs of β-lactams in combination with clavulanic acid (Sigma-Aldrich, Saint Quentin Fallavier, France) (2 μg/ml) for the E. coli ErtR isolate were performed to determine the contribution of the ESBL to carbapenem resistance. The MIC of ertapenem in combination with phenylalanine-arginine β-naphthylamide dihydrochloride (PAβN; 26.3 μg/ml; Sigma-Aldrich), which is an inhibitor of resistance-nodulation-cell division (RND) pumps of enterobacteria (29), was also determined. A 2-fold decrease in MIC after addition of PAβN was considered significant (29).
Whole-genome sequencing and comparative genomic analysis.
The complete genome sequence of the ertapenem-susceptible E. coli isolate ErtS was obtained by single-molecule real-time sequencing using a Pacific Bioscience (PacBio) RS II sequencer (GATC Biotech, Constance, Germany) to generate a reference sequence. The genome annotation of the E. coli isolate ErtS was carried out using the MicroScope platform (http://www.genoscope.cns.fr/agc/microscope) (30). Thereafter, the genomes of E. coli ErtS and E. coli ErtR were sequenced twice using the same technology. DNA libraries were prepared using the Nextera library construction protocol, according to the manufacturer's instructions, after genomic DNA purification with a NucleoSpin tissue kit (Macherey-Nagel, Hoerdt, France) and were sequenced on a MiSeq sequencer (Illumina, Paris, France). High-throughput sequencing (HTS) data were analyzed using the PALOMA bioinformatic pipeline implemented in the MicroScope platform (30). In a first step, the HTS data were preprocessed to assess their quality. This step includes options such as read trimming, merging, or splitting paired-end reads. In a second step, MiSeq E. coli ErtR reads were mapped onto the PacBio RS II ErtS reference sequence using the SSAHA2 package (31). Only unique matches having an alignment score equal to at least half of their length were retained as seeds for full Smith-Waterman realignment (32) with a region extended on both sides by five nucleotides of the reference genome. All computed alignments then were screened for discrepancies between read and reference sequences, and in fine, a score based on coverage, allele frequency, quality of bases, and strand bias was computed for each detected event to assess its relevance. To filter sequencing errors, read-mapping errors, and assembly errors, we performed, in a third step, an analysis consisting of mapping the MiSeq E. coli ErtS reads against the PacBio RS II E. coli ErtS reference sequence (same analysis as that described above). More than 90% of the small-variation events that were detected in this reference analysis had less than 8 supporting reads and a score under 0.5. These criteria were then applied to filter out false-positive events from the first comparative genomic analysis (second step, described above).
Typing methods.
The genome sequence-inferred serotype and the sequence type (ST) were identified with software available from the Center for Genomic Epidemiology (SerotypeFinder 1.1, with threshold for 85% identity and a minimum length of 60%, and MSLT 1.8, respectively; www.genomicepidemiology.org/) by uploading the genome of E. coli ErtS. Virulence genes were identified with software from the Center for Genomic Epidemiology (VirulenceFinder 1.5, with a threshold of 90% identity and a minimum length of 60%) and by direct comparison with the sequences described in the study of Lefort et al. (13). The fimH-based subtyping was determined by BLAST comparison against public fimH gene sequences. Isolates with a fimH allele that differed by only one nucleotide (i.e., one single-nucleotide polymorphism) compared with an established fimH allele reference sequence were considered to belong to the corresponding fimH allele group (33).
PCR, Sanger sequencing, and complementation experiments.
Genomic DNA of E. coli isolates was extracted with the QIAmp kit (Qiagen, Courtaboeuf, France). PCR amplification of the ompR gene was performed with primers OmpR-F (5′-CACGCTTACAAATTGTTGCG-3′) and OmpR-R (5′-GTGGCGAGAAGCGCAATCG-3′), yielding a 745-bp amplification product that contained the entire coding sequence of the ompR gene. The PCR product was purified using a QIAquick PCR purification kit (Qiagen, Courtaboeuf, France) and Sanger sequenced (GATC Biotech) for confirmation of the whole-genome sequencing results. The PCR product was polished using Pfu DNA polymerase (Fermentas, St Rémy Les Chevreuses, France) and subsequently cloned into pCR-BluntII-Topo (Life Technologies SAS, Saint Aubin, France). The recombinant plasmid was purified using the QIAquick plasmid midi kit (Qiagen) and transformed into E. coli strain ErtR, as described previously (34).
Examination of porin expression.
OMPs were isolated according to the rapid procedure of Carlone et al. (35) and separated by SDS-PAGE, as previously described (19).
Protein structure analysis.
A multiple-sequence alignment of OmpR analogues was built using CLUSTAL Omega, which is available at the European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/msa/clustalo/). The variant model OmpR-Gly-63-Val was generated by the SWISS MODEL server (http://swissmodel.expasy.org) using the deposited X-ray crystal structure of the response regulator DrrB of Thermotoga maritima (PDB entry 1P2F) (17). The amino acid substitution Gly-58-Val in DrrB, which corresponds to the Gly-53-Val replacement in the homologous response regulator OmpR, was introduced. The model then was optimized by energy minimization. Structures of the model and the native protein were aligned to determine the displacement of the substituted residue and the surrounding amino acids. A representative figure of the model and the native regulator was generated using the UCSF Chimera software package, version 1.5.3 (36, 37).
Bacterial growth.
Cells grown overnight in lysogeny broth (LB) were washed and transferred into fresh LB at a dilution of 1:250. The growth rates were then measured for E. coli ErtS and E. coli ErtR isolates every 5 min at 37°C by monitoring the optical density at 600 nm (Tecan microplate reader) in LB medium. Experiments were repeated 5 times. The maximum growth rate (MGR) was calculated as the time point at which the maximum value of the derivative of the smoothed function was observed. MGRs were compared by strain using a Welch test as described previously (38).
The fitness of the E. coli ErtR mutant relative to the parental E. coli ErtS isolate was also estimated by a competition experiment, as described previously (39), in LB after 24 h of incubation. Briefly, the relative proportion of each strain was evaluated by plating dilutions on selective and nonselective LB agar plates. The competitive index (CI) was calculated as the logarithm of the number of antibiotic-resistant CFU divided by the inferred number of sensitive CFU and divided again by the same ratio in the inoculum. The experiment was repeated three times.
Experimental virulence assay using the social amoeba D. discoideum.
Bacterium-amoeba interaction was assessed in triplicate by the coculture of D. discoideum (AX3) and each E. coli strain in HL5 medium (5 g · liter−1 proteose peptone, 5 g · liter−1 thiotone E peptone, 10 g · liter−1 glucose, 5 g · liter−1 yeast extract, 0.35 g · liter−1 Na2HPO4 7H2O, 0.35 g · liter−1 KH2PO4, pH 6.5) incubated for 6 days at 24°C as described previously (18). A volume (300 μl) of an E. coli isolate culture grown overnight was plated on HL5 agar petri dishes and allowed to dry for 20 min under a sterile airflow. The same volume of amoeba culture (D. discoideum AX3) was added on these plates and allowed to dry for 20 min under a sterile airflow. Plates were covered with Parafilm and incubated for 6 days at 24°C. Three replicates were performed for each experiment. Plates were screened at day three and day six to assess the occurrence of bacterial lysis plaques corresponding to phagocytosis of the bacteria by the amoeba (grazing-sensitive phenotype). The presence of a lawn of bacteria indicated an absence of phagocytosis (grazing-resistant phenotype).
Accession number(s).
The genomes of E. coli ErtS and E. coli ErtR have been deposited in the European Nucleotide Archive (ENA) under accession numbers PRJEB12281 and PRJEB11796, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank AstraZeneca Pharmaceuticals (Waltham, MA, USA) for providing avibactam. We also thank the LABGeM team and the National Infrastructure France Genomique for technical support.
This study was supported by a Programme Hospitalier de Recherche (PHRC) of the Centre Hospitalier Universitaire d'Amiens.
This work was partially supported by a grant from the Fondation pour la Recherche Médicale (équipe FRM 2016, DEQ20161136698).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00014-17.
REFERENCES
- 1.Mammeri H, Guillon H, Eb F, Nordmann P. 2010. Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC β-lactamases. Antimicrob Agents Chemother 54:4556–4560. doi: 10.1128/AAC.01762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3203–3206. doi: 10.1128/AAC.48.8.3203-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez A, Canle D, Latasa C, Poza M, Beceiro A, del Mar Tomás M, Fernández A, Mallo S, Pérez S, Molina F, Villanueva R, Lasa I, Bou G. 2007. Cloning, nucleotide sequencing, and analysis of the AcrAB-TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob Agents Chemother 51:3247–3253. doi: 10.1128/AAC.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia JH, Siu LK, Su LH, Lin HS, Kuo AJ, Lee MH, Wu TL. 2009. Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J Chemother 21:621–626. doi: 10.1179/joc.2009.21.6.621. [DOI] [PubMed] [Google Scholar]
- 5.Goessens WH, van der Bij AK, van Boxtel R, Pitout JD, van Ulsen P, Melles DC, Tommassen J. 2013. Antibiotic trapping by plasmid-encoded CMY-2 β-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother 57:3941–3949. doi: 10.1128/AAC.02459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Héritier C, Spicq C, Nordmann P. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 42:3831–3833. doi: 10.1128/JCM.42.8.3831-3833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillon H, Tande D, Mammeri H. 2011. Emergence of ertapenem resistance in an Escherichia coli clinical isolate producing extended-spectrum β-lactamase AmpC. Antimicrob Agents Chemother 55:4443–4446. doi: 10.1128/AAC.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oteo J, Delgado-Iribarren A, Vega D, Bautista V, Rodriguez MC, Velasco M, Saavedra JM, Perez-Vazquez M, Garcia-Cobos S, Martinez-Martinez L, Campos J. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents 32:534–537. doi: 10.1016/j.ijantimicag.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Hawser SP, Bouchillon SK, Lascols C, Hackel M, Hoban DJ, Badal RE, Canton R. 2012. Susceptibility of European Escherichia coli clinical isolates from intra-abdominal infections, extended-spectrum β-lactamase occurrence, resistance distribution, and molecular characterization of ertapenem-resistant isolates (SMART 2008-2009). Clin Microbiol Infect 18:253–259. doi: 10.1111/j.1469-0691.2011.03550.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones RN, Sader HS, Fritsche TR. 2005. Comparative activity of doripenem and three other carbapenems tested against Gram-negative bacilli with various β-lactamase resistance mechanisms. Diagn Microbiol Infect Dis 52:71–74. doi: 10.1016/j.diagmicrobio.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Hall MN, Silhavy TJ. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol 146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 12.Mattison K, Oropeza R, Byers N, Kenney LJ. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol 315:497–511. doi: 10.1006/jmbi.2001.5222. [DOI] [PubMed] [Google Scholar]
- 13.Lefort A, Panhard X, Clermont O, Woerther PL, Branger C, Mentré F, Fantin B, Wolff M, Denamur E, COLIBAFI Group. 2011. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J Clin Microbiol 49:777–783. doi: 10.1128/JCM.01902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guignot J, Chaplais C, Coconnier-Polter MH, Servin AL. 2007. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell Microbiol 9:204–221. doi: 10.1111/j.1462-5822.2006.00782.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HL, Tarr PI. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect Immun 73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuba PJ, Leon MA, Banerjee A, Palchaudhuri S. 1989. Cloning and DNA sequence of plasmid determinant iss, coding for increased serum survival and surface exclusion, which has homology with lambda DNA. Mol Gen Genet 216(2-3):287–292. [DOI] [PubMed] [Google Scholar]
- 17.Robinson VL, Wu T, Stock AM. 2003. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol 185:4186–4194. doi: 10.1128/JB.185.14.4186-4194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adiba S, Nizak C, Van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. doi: 10.1371/journal.pone.0011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 20.Dupont H, Gaillot O, Goetgheluck AS, Plassart C, Emond JP, Lecuru M, Gaillard N, Derdouri S, Lemaire B, Girard de Courtilles M, Cattoir V, Mammeri H. 2015. Molecular characterization of carbapenem-non-susceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother 60:215–221. doi: 10.1128/AAC.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner DM, Yang Q, Duval V, Chen M, Xu Y, Levy SB. 2013. Involvement of MarR and YedS in carbapenem resistance in a clinical isolate of Escherichia coli from China. Antimicrob Agents Chemother 57:1935–1937. doi: 10.1128/AAC.02445-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batchelor E, Walthers D, Kenney LJ, Goulian M. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol 187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tängdén T, Adler M, Cars O, Sandegren L, Löwdin E. 2013. Frequent emergence of porin-deficient subpopulations with reduced carbapenem susceptibility in ESBL-producing Escherichia coli during exposure to ertapenem in an in vitro pharmacokinetic model. J Antimicrob Chemother 68:1319–1326. doi: 10.1093/jac/dkt044. [DOI] [PubMed] [Google Scholar]
- 24.Adler M, Anjum M, Andersson DI, Sandegren L. 2013. Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J Antimicrob Chemother 68:51–59. doi: 10.1093/jac/dks368. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol 180:2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai Y-K, Fung C-P, Lin J-C, Chen J-H, Chang F-Y, Chen T-L, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 55:1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. [DOI] [PubMed] [Google Scholar]
- 29.Chollet R, Chevalier J, Bollet C, Pages J-M, Davin-Regli A. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob Agents Chemother 48:2518–2523. doi: 10.1128/AAC.48.7.2518-2523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, Mercier J, Renaux A, Rollin J, Rouy Z, Roche D, Scarpelli C, Médigue C. 2017. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 45:D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning Z, Cox AJ, Mullikin JC. 2001. SSAHA: a fast search method for large DNA databases. Genome Res 11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith TF, Waterman MS. 1981. Identification of common molecular subsequences. J Mol Biol 147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 33.Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J Clin Microbiol 51:2991–2999. doi: 10.1128/JCM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mammeri H, Poirel L, Fortineau N, Nordmann P. 2006. Naturally occurring extended-spectrum cephalosporinases in Escherichia coli. Antimicrob Agents Chemother 50:2573–2576. doi: 10.1128/AAC.01633-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol 24:330–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. Swiss-model: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GC, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon J-M, Garry L, Clermont O, Denamur E, Arlet G, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b:H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. doi: 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol 192:4885–4893. doi: 10.1128/JB.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


