ABSTRACT
qnrE1, found in a clinical Klebsiella pneumoniae isolate, was undetectable by PCR assays used for the six qnr families. qnrE1 was located on a conjugative plasmid (ca. 185 kb) and differed from qnrB alleles by 25%. Phylogenetic reconstructions of qnr genes and proteins and analysis of the qnrE1 surroundings showed that this gene belongs to a new qnr family and was likely mobilized by ISEcp1 from the chromosome of Enterobacter spp. to plasmids of K. pneumoniae.
KEYWORDS: PMQR, quinolone, resistance, qnr detection, qnrB
TEXT
The epidemiology of quinolone resistance has dramatically changed since the discovery of the plasmid-mediated quinolone resistance (PMQR) genes in 1998 (1). Among them, the qnr genes encode pentapeptide repeat proteins that protect the quinolone targets, DNA gyrase and topoisomerase IV, from the action of these drugs (2). The qnr genes are grouped into six families: qnrA, qnrB, qnrC, qnrD, and qnrS, all of which have been mainly found in Enterobacteriaceae, and qnrVC, which has been mostly found in Vibrionaceae (2–4). The six gene families differ in sequence by 30% or more from each other, and several alleles have been described for all families except qnrC (http://www.lahey.org/qnrStudies). Inside a given family, the allelic sequences differ on average by 9% or less (qnrA, qnrB, qnrD, and qnrS) or by 16% (qnrVC) (our unpublished data).
Recently, we reported a nationwide survey on PMQR genes based on a collection of 1,058 clinical enterobacteria from Argentina (5). In that study, Klebsiella pneumoniae Q1130, which was isolated in 2007 from the urine specimen of a 78-year-old female ambulatory patient, had a wild-type quinolone resistance-determining region of gyrA and showed low-level quinolone resistance. However, this isolate was negative in the PCR assays for qnrA, qnrB, qnrC, qnrD, and qnrS as well as for other kinds of PMQR genes, such as aac(6′)-Ib-cr and qepA (5). Herein, we investigated the possible presence of an unknown PMQR gene in this isolate.
Horizontal transference of low-level quinolone resistance.
To know whether the low-level quinolone resistance observed in K. pneumoniae Q1130 may be horizontally transferred, a biparental conjugation assay was performed as described previously (6). Susceptibility to nalidixic acid (NAL), ciprofloxacin (CIP), and levofloxacin (LVX) was determined by agar dilution and disc diffusion according to the Clinical and Laboratory Standards Institute (7). The obtained transconjugant, Escherichia coli TC1130, showed MICs of NAL, CIP, and LVX that were 4, 16, and 16 times higher than those of the recipient strain E. coli J53-Azr (Azr indicates resistance to sodium azide), respectively (Table 1). By S1 nuclease assay (8), a unique plasmid band of the same size (ca. 185 kb) was observed for K. pneumoniae Q1130 and its transconjugant (data not shown). These results indicated that K. pneumoniae Q1130 harbored a quinolone resistance mechanism encoded in a conjugative plasmid, which was named pKp1130.
TABLE 1.
Quinolone susceptibility profiles of the clinical isolate K. pneumoniae Q1130, the recipient strains E. coli J53-Azr and TOP10, and their laboratory-derived strains
| Strain | MIC (μg/ml) |
Disc diffusion inhibition zone (mm) |
||||
|---|---|---|---|---|---|---|
| NAL | CIP | LVX | NAL | CIP | LVX | |
| K. pneumoniae Q1130 | 16 | 0.5 | 1 | 14 | 22 | 22 |
| E. coli TC1130a | 16 | 0.25 | 0.5 | 15 | 26 | 25 |
| E. coli J53-Azr | 4 | 0.015 | 0.03 | 23 | 36 | 33 |
| E. coli TOP10(pJET1.2-1130)b | 8 | 0.125 | 0.125 | 22 | 32 | 36 |
| E. coli TOP10(pJET1.2-C)c | 1 | 0.002 | 0.004 | 36 | 48 | 43 |
| E. coli TOP10 | 1 | 0.002 | 0.004 | 35 | 46 | 43 |
Transconjugant selection was done with sodium azide (100 μg/ml) plus CIP (0.12 μg/ml).
This is a qnrE1-harboring transformant selected with ampicillin (50 μg/ml).
This is a qnrE1-negative transformant selected with ampicillin (50 μg/ml).
The qnrVC genes had not been reported in clinical enterobacteria (2–4); therefore, they were the unique PMQR genes that were not previously analyzed in K. pneumoniae Q1130 (5). We confirmed this presumption by testing the presence of qnrVC genes in this clinical isolate and its transconjugant: the PCR assays used (see Table S1 in the supplemental material) rendered negative results in both cases.
Identification of a new qnr gene in pKp1130.
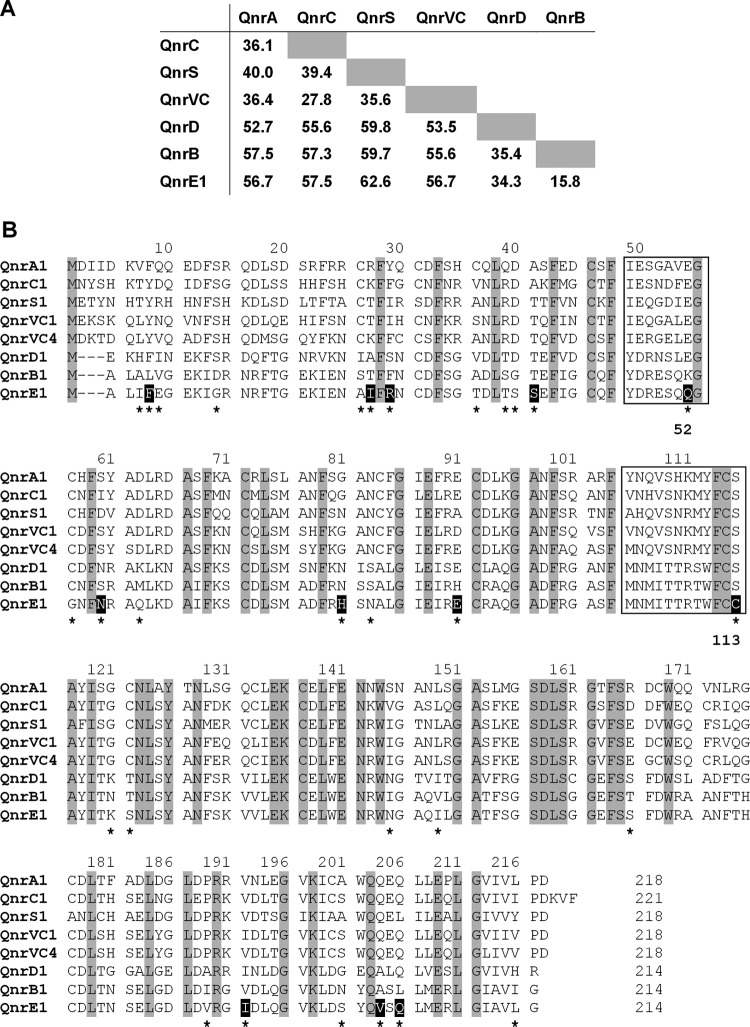
pKp1130 was extracted from E. coli TC1130 with the Qiagen large-construct kit (Qiagen, Hilden, Germany) and sequenced using the MiSeq sequencer (Illumina, San Diego, CA) and CLC Genomics Workbench software v.5.5.1 (CLC bio, Qiagen) for read assembling. Open reading frames (ORFs) were annotated using the RAST Server (rast.nmpdr.org) (9) followed by manual comparative curation and determination of sequence similarity with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Differences between nucleotide or amino acid sequences were calculated as p-distances with MEGA6 (http://www.megasoftware.net/) (10) and expressed as percentages. The sequencing of pKp1130 resulted in 14 contigs, giving a total length of 187,745 nucleotides, which is in agreement with the size estimated by the S1 nuclease assay. A BLAST analysis showed that the best hits (lowest BLAST E values) for the 14 contigs corresponded to sequences of enterobacterial plasmids (data not shown). Nine out of 14 contigs (total length of 154,020 nucleotides) shared 99.8% identity (query cover of 92%) with the plasmid p1 (199,497 bp) of K. pneumoniae NY9 (GenBank accession number CP015386) (see Fig. S1 in the supplemental material). The analysis of the longest contig of pKp1130 (56,313 nucleotides; Contig_5 in Fig. S1) revealed the presence of a 645-bp ORF that showed the highest identity with the qnrB family (average of 75% identity with all of the qnrB alleles previously described and a range of 73.2% [qnrB31 and qnrB53] to 76.4% [qnrB42]) (see Fig. S2A in the supplemental material). This ORF showed 34 nucleotide changes in positions fully conserved among all of the qnrB alleles (Fig. S2B). The protein inferred from this ORF differed from all of the previously known QnrB proteins by an average of 15.8% (34 amino acids), with a range of 14.0% (QnrB1, QnrB17, QnrB66, and QnrB75; 30 amino acids) to 17.8% (QnrB73; 38 amino acids) (Fig. 1A). In addition, the sequence of this putative new protein displayed the typical pentapeptide repeat structure of the Qnr proteins and contained the loops A and B, which have proven to be essential for quinolone-protective activity (11–13) (Fig. 1B). Interestingly, an amino acid change was observed in each loop of the putative new Qnr protein regarding all of the previously described QnrB proteins: K52Q and S113C (QnrB amino acid numbering) for loops A and B, respectively. Only two reports analyzed the effect on quinolone susceptibility driven by substitutions in these positions of QnrB1: the CIP MIC was reduced 16 times in the presence of S113D or S113A (11, 13) while no effect was observed for K52A (13).
FIG 1.
Comparison of Qnr proteins. (A) Paired comparisons of Qnr families. The averages of the percentage of amino acid differences between the indicated families are shown. (B) Pentapeptide repeat structures of Qnr proteins. Only representative variants for the six families are shown. Gaps introduced to maximize the alignment are indicated by hyphens. The amino acid numbering indicated above each block of sequences is based on the longest protein (QnrC1). Residues fully conserved among all of the Qnr variants shown are depicted with a gray-shaded background. The loops A and B (8 and 12 amino acids, respectively), essential for the quinolone-protective activity of the Qnr proteins, are indicated by boxes. The amino acid changes between QnrE1 and QnrB1 are marked with asterisks, and those located in positions for which no changes were found between QnrB1 and any of the remaining QnrB proteins previously described are highlighted with a black-shaded background. The locations in the loops A and B of the changes between QnrE1 and QnrB1 are indicated by bold numbers (QnrB amino acid numbering) below the block of sequences.
These results indicated that the ORF found in pKp1130 may correspond to a new qnr gene, named qnrE1, which is in agreement with the public database repository of qnr sequences (http://www.lahey.org/qnrStudies) (G. A. Jacoby, personal communication).
The genetic context analysis of qnrE1 showed that the insertion sequence ISEcp1 was located 22 bp upstream of this gene while araJ, which encoded a transporter of the major facilitator superfamily, was located 121 bp downstream (see Fig. S3 in the supplemental material). Given that no putative promoter sequences were found in the short stretch that separated the inverted repeat right (IRR) of ISEcp1 from qnrE1, it is very likely that the strong promoter located downstream of the transposase gene of ISEcp1 can drive the expression of qnrE1 (14).
qnrE1 was completely amplified from K. pneumoniae Q1130 by PCR (see primers in Table S1). The sequence of the obtained amplicon (965 bp), based on MiSeq sequencing, was confirmed by direct Sanger sequencing (5). By using the CloneJet PCR cloning kit (Thermo Scientific/Thermo Fisher Scientific, Waltham, MA), this amplicon and an unspecific DNA fragment (976 bp), provided as a control in the cloning kit, were cloned into the vector pJET1.2/blunt, rendering plasmids pJET1.2-1130 and pJET1.2-C, respectively (Fig. S3). Both plasmids were introduced in E. coli TOP10 (Invitrogen/Thermo Fisher Scientific) by electroporation. The identity of the cloned fragment from K. pneumoniae Q1130 was also corroborated by Sanger sequencing (5). The MICs of NAL, CIP, and LVX of E. coli TOP10(pJET1.2-1130) were 8, 64, and 32 times higher, respectively, than those of the qnrE1-negative isogenic strain E. coli TOP10(pJET1.2-C) (Table 1). The levels of resistance conferred by qnrE1 agree well with those observed for other qnr genes (2). These results strongly suggest that the substitutions K52Q and S113C found in loops A and B of QnrE1 do not impair its quinolone-protective activity.
qnrE1 is a member of a new qnr family.
According to the current nomenclature for qnr genes, qnrE1 should be considered to belong to the qnrB family because the genetic differences with the qnrB alleles were lower than 30%, the cutoff proposed for differentiating qnr families (15). However, the previously known qnrB alleles differed in sequence by an average of 8.6% (range, 0.2% to 19.8%) while qnrE1 differed from the former by an average of 25.0% (range, 23.6% to 26.8%). In addition, the forward and reverse primers previously used for the screening of qnrB genes (5) showed a large proportion of nucleotide mismatches (8/19 and 7/22, respectively) regarding the qnrE1 sequence. This fact can explain the lack of amplification observed for K. pneumoniae Q1130 in the PCR for qnrB genes (5). Similarly, we found relevant proportions of mismatches when the sequences of other primers used for qnrB screening in several surveys on PMQR genes (16–18), including degenerate primers (19), were compared with qnrE1.
Therefore, to further analyze the relationship between qnrE1 and the qnrB alleles, we performed phylogenetic reconstructions using two different methods, neighbor-joining (NJ) and maximum likelihood (ML). Phylogenetic trees for DNA or amino acid sequences were constructed with MEGA6 using the general time reversible model, with gamma distribution of the substitution rate heterogeneity over sites (G) and a proportion of invariant sites, or the Jones-Taylor-Thornton model, with G, as the DNA and amino acid substitution models, respectively, that best fitted the data. The reliability of the tree topology was assessed by bootstrapping (20) using 1,000 replicates.
The topologies of the inferred NJ and ML trees for DNA sequences were very similar, and both showed, with high percentages of bootstrap support, that qnrE1 was separated by the same evolutionary distance (0.4 nucleotide substitutions per site) from a cluster that comprised all of the qnrB alleles previously described (Fig. 2; see also Fig. S4 in the supplemental material). The analysis of the NJ and ML trees for amino acid sequences showed equivalent results, with the exception that, as expected, a lower distance (0.2 amino acid substitutions per site in both trees) was observed between QnrE1 and the cluster that comprised all of the QnrB proteins previously described (data not shown).
FIG 2.
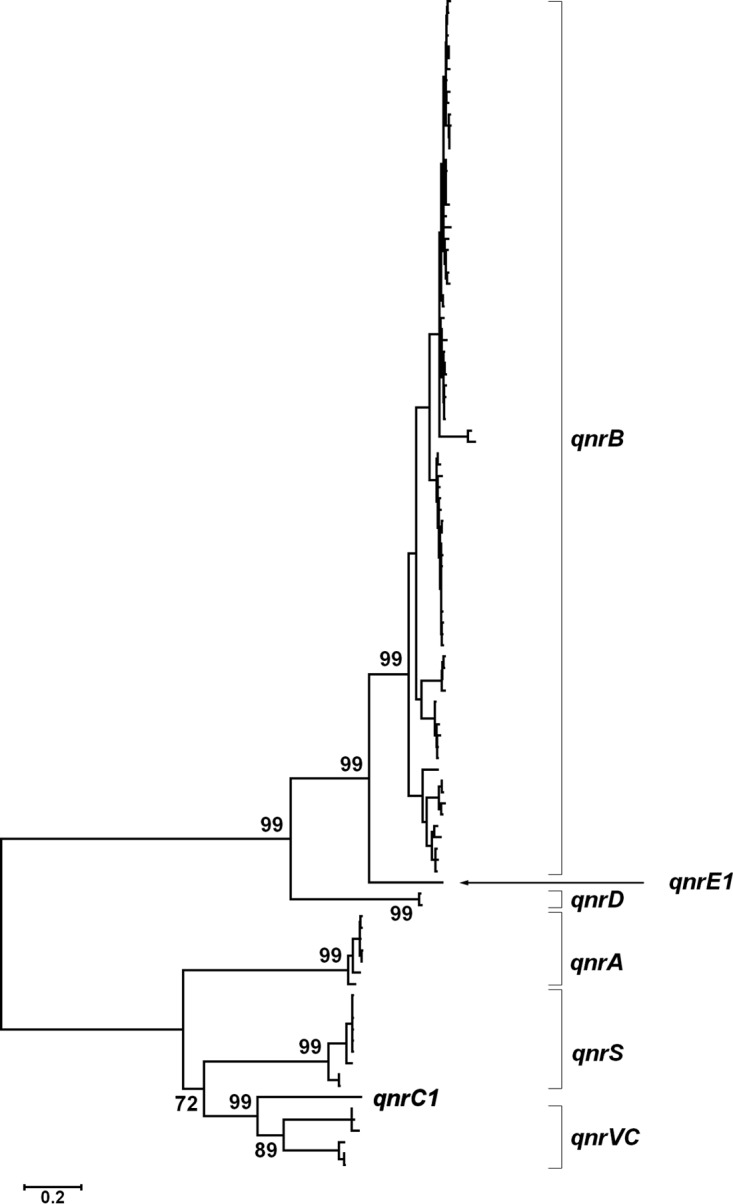
Phylogenetic relationships among qnr genes. The ML tree was generated from an alignment of qnrE1 and all of the qnr genes currently known (accession numbers are indicated in Table S2 in the supplemental material). For simplicity, the taxon names of qnr alleles are excluded (see Table S3 in the supplemental material) and only the bootstrap percentages (over 1,000 replicates) for relevant nodes are shown. The clustering of the alleles for each qnr family is indicated with square brackets at the right. The branch lengths were drawn to the scale shown, which indicates the number of substitutions/site.
Source and mobilization of qnrE1.
To see if there were sequences in the public databases more closely related to qnrE1 than the qnrB alleles, we performed a BLAST search using the complete qnrE1 sequence as the query. Six genes showing more than 90% identity were found.
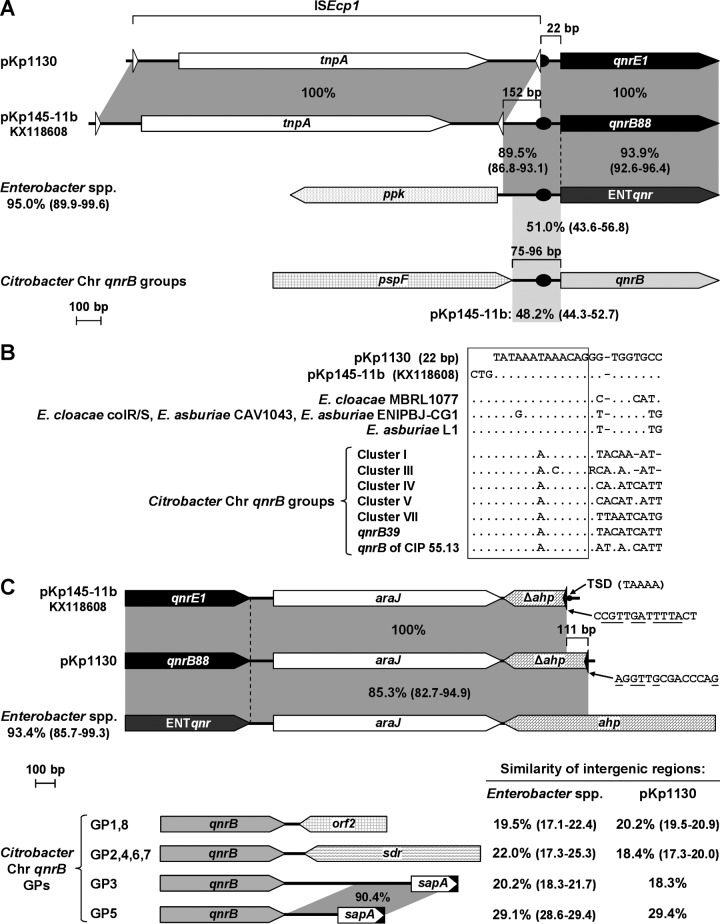
First, the same qnrE1 gene (named qnrB88 in the GenBank report) was described in the plasmid pKp145-11b of a K. pneumoniae isolate from Brazil (GenBank accession number KX118608), which indicates that the distribution of this gene is not limited to Argentina. Second, 5 fluoroquinolone resistance genes (named here as ENTqnr; 96.4% to 92.6% identity with qnrE1) were found in the chromosomes of Enterobacter cloacae MBRL1077, E. cloacae colR/S (21), Enterobacter asburiae CAV1043, E. asburiae ENIPBJ-CG1 (22), and E. asburiae L1 (GenBank accession numbers CP014280, CP010512, CP011591, CP014993, and CP007546, respectively). The 100% identity between pKp1130 and pKp145-11b extended beyond qnrE1 along a 3,951-bp fragment from ISEcp1 to a truncated alkyl hydroperoxidase-encoding gene (Δahp; Fig. 3A and C). The sequence identity in the vicinities of the ahp interruption point found in pKp1130 was confirmed by PCR and Sanger sequencing (5) using the primers araJ-F and R2 (Table S1) and both K. pneumoniae Q1130 and its transconjugant. To gain insights into the possible source of qnrE1 and its occurrence in the plasmids of K. pneumoniae pKp1130 and pKp145-11b, the genetic surroundings of this gene were compared with those of the 5 ENTqnr genes. Both the upstream (excluding ISEcp1) and the downstream regions of qnrE1 showed high identity to those of ENTqnr (Fig. 3A and C), with E. cloacae MBRL1077 showing maximal identities (93.1% and 94.9%, respectively). The results of these comparisons were also consistent with the notion that ISEcp1 was responsible for the mobilization of qnrE1 from the Enterobacter spp. chromosome to pKp1130 and pKp145-11b, truncating the ahp gene. Indeed, we found possible alternative IRRs (23, 24) at the 5′ edges of Δahp in pKp1130 and pKp145-11b, and a 5-bp duplication of the target site, which flanked ISEcp1-qnrE1-araJ-Δahp in pKp145-11b (Fig. 3C). The qnrE1 acquisitions in pKp1130 and pKp145-11b should have occurred through different ISEcp1 insertions in the chromosome of Enterobacter spp. because the region mobilized to pKp1130 begins 152 bp closer to and ends 111 bp farther from qnrE1 than the region mobilized to pKp145-11b (Fig. 3A and C).
FIG 3.
Comparison of genetic surroundings of qnrE1, chromosomal qnr genes of Enterobacter spp. (ENTqnr), and the qnrB alleles. Only relevant regions are shown. A 4,062-bp fragment of the Contig_5 of pKp1130 (see Fig. S1 in the supplemental material) is depicted. In pKp145-11b, qnrE1 was named as qnrB88 according to GenBank report KX118608. The ENTqnr-containing chromosomal regions of E. cloacae MBRL1077, E. cloacae colR/S, E. asburiae CAV1043, E. asburiae ENIPBJ-CG1, and E. asburiae L1 (GenBank accession numbers CP014280, CP010512, CP011591, CP014993, and CP007546, respectively) are represented altogether and indicated as “Enterobacter spp.” (the identity among them is shown below this name); ppk and ahp encode polyphosphate kinase and alkyl hydroperoxidase, respectively. Genes/ORFs are represented by arrow-shaped boxes. Short horizontal square brackets are used to indicate the length of relevant DNA fragments. Percentages show the percentage of identity between sequences and, when multiple paired comparisons were done, the average percentages of identity and range (between brackets) are given. Dark and light gray-shaded areas indicate regions of high and low identity, respectively. (A) qnrE1 upstream region. The qnrB-containing chromosomal regions of Citrobacter spp., which have been associated with seven phylogenetic groups of qnrB genes (26), are represented altogether and indicated as “Citrobacter Chr qnrB groups” (pspF encodes a transcriptional activator of the phage shock protein operon; the YheO-like-encoding gene only found upstream of qnrB in Citrobacter pasteurii CIP 55.13 [26] was omitted for simplicity). Empty triangles indicate inverted repeats of ISEcp1, and black ovals indicate the LexA binding site (truncated in pKp1130). The fragments upstream of all of the qnr genes that contain the LexA boxes (see details in B) are not drawn to scale. (B) Sequence comparison of the 24- to 26-bp fragments upstream of the qnr genes (22 bp in pKp1130) that contain the LexA binding site (boxed). The sequences of the seven Citrobacter Chr qnrB groups (26) are indicated (qnrB of CIP 55.13 and new chromosomal qnrB allele of C. pasteurii CIP 55.13 [26]). Dots indicate nucleotide identity to pKp1130 or pKp145-11b sequences. (C) qnrE1 downstream region. The black triangles indicate the possible alternative IRRs of ISEcp1, and their corresponding sequences are shown (nucleotides identical to IRR are underlined); TSD, target site duplication. The eight genetic platforms (GP1 to GP8) found for the chromosomal qnrB genes of Citrobacter spp. (26) are grouped by sequence similarity of the region depicted in the figure (orf2, putative gene of unknown function; sdr and sapA [truncated at the 3′ end] encode a short-chain dehydrogenase/reductase and a protein involved in antimicrobial peptide resistance, respectively). The identity among the intergenic regions of the GPs in each group and those of Enterobacter spp. or pKp1130 is shown at the right.
The chromosome of Citrobacter spp. has been proposed as the source of qnrB alleles (25, 26). Moreover, the genetic platforms of all of the chromosomal qnrB alleles of Citrobacter spp. available in public databases have been deeply analyzed and classified into eight groups (GP1 to GP8), which are strongly associated with seven phylogenetic groups of qnrB alleles (26). By comparative analyses, we found that the genetic surroundings of the chromosomal qnrB alleles of Citrobacter spp. were quite different from those of qnrE1 or the ENTqnr genes, in both gene content and similarity of intergenic regions (Fig. 3A and C). Interestingly, as observed for the qnrB alleles and qnrD1 (26–29), a LexA binding site was found a few nucleotides upstream of both qnrE1 and ENTqnr genes, although, in pKp1130, it was truncated at its 5′ end by the ISEcp1 insertion (Fig. 3B).
The fact that the source of qnrE1 is different from that of the qnrB alleles is in agreement with the results of the phylogenetic analysis, further supporting the notion that qnrE1 belongs to a new qnr family.
Conclusions.
The new plasmid-located qnrE1 gene described here is undetectable by the PCR detection assays commonly used for the six qnr families currently known. This fact is highly relevant from the point of view of both the epidemiology of PMQR mechanisms and its detection in the clinical setting. Unlike the qnrB alleles, our results strongly suggest that the origin of qnrE1 is the chromosome of Enterobacter spp. and that this gene would have been mobilized from there to plasmids of K. pneumoniae by ISEcp1. These notions may imply that a new source of qnr genes is supplying the reservoir of plasmid-located qnr genes in clinical settings.
Accession number(s).
The qnrE1 sequence depicted in Fig. 3 has been assigned GenBank accession number KY073238.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from ANPCYT (PICT 2015-01728) Buenos Aires, Argentina, to A.P.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02555-16.
REFERENCES
- 1.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol Spectr 2(5):PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pons MJ, Gomes C, Ruiz J. 2013. QnrVC, a new transferable Qnr-like family. Enferm Infecc Microbiol Clin 31:191–192. doi: 10.1016/j.eimc.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca EL, Vicente AC. 2013. Epidemiology of qnrVC alleles and emergence out of the Vibrionaceae family. J Med Microbiol 62:1628–1630. doi: 10.1099/jmm.0.062661-0. [DOI] [PubMed] [Google Scholar]
- 5.Albornoz E, Lucero C, Romero G, Quiroga MP, Rapoport M, Guerriero L, Andres P, Rodriguez C, Galas M, Centrón D, Corso A, Petroni A. 2017. Prevalence of plasmid-mediated quinolone resistance genes in clinical enterobacteria from Argentina. Microb Drug Resist 23:177–187. doi: 10.1089/mdr.2016.0033. [DOI] [PubMed] [Google Scholar]
- 6.Melano R, Corso A, Petroni A, Centrón D, Orman B, Pereyra A, Moreno N, Galas M. 2003. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J Antimicrob Chemother 52:36–42. doi: 10.1093/jac/dkg281. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; approved standard—26th ed. CLSI document M100S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 9.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Martínez JM, Briales A, Velasco C, Conejo MC, Martínez-Martínez L, Pascual A. 2009. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB, and QnrS. J Antimicrob Chemother 63:1128–1134. doi: 10.1093/jac/dkp111. [DOI] [PubMed] [Google Scholar]
- 12.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS. 2011. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J Biol Chem 286:25265–25273. doi: 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacoby GA, Corcoran MA, Mills DM, Griffin CM, Hooper DC. 2013. Mutational analysis of quinolone resistance protein QnrB1. Antimicrob Agents Chemother 57:5733–5736. doi: 10.1128/AAC.01533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano R, Okamoto R, Nagano N, Inoue M. 2007. Resistance to gram-negative organisms due to high-level expression of plasmid-encoded ampC β-lactamase blaCMY-4 promoted by insertion sequence ISEcp1. J Infect Chemother 13:18–23. doi: 10.1007/s10156-006-0483-6. [DOI] [PubMed] [Google Scholar]
- 15.Jacoby G, Cattoir V, Hooper D, Martínez-Martínez L, Nordmann P, Pascual A, Poirel L, Wang M. 2008. qnr gene nomenclature. Antimicrob Agents Chemother 52:2297–2299. doi: 10.1128/AAC.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. 2009. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother 53:639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham DW. 2013. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Med Microbiol 62:1823–1827. doi: 10.1099/jmm.0.064428-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Zhang J, Zheng B, Wei Z, Shen P, Li S, Li L, Xiao Y. 2015. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol 53:766–770. doi: 10.1128/JCM.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briales A, Rodríguez-Martínez JM, Velasco C, de Alba PD, Rodríguez-Baño J, Martínez-Martínez L, Pascual A. 2012. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Int J Antimicrob Agents 39:431–434. doi: 10.1016/j.ijantimicag.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 21.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Yang J, Xiao Y, Li L, Yang F, Jin Q. 2016. Complete genome sequence of a clinical isolate of Enterobacter asburiae. Genome Announc 4:e00523-16. doi: 10.1128/genomeA.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zong Z, Partridge SR, Iredell JR. 2010. ISEcp1-mediated transposition and homologous recombination can explain the context of blaCTX-M-62 linked to qnrB2. Antimicrob Agents Chemother 54:3039–3042. doi: 10.1128/AAC.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge SR. 2007. Genetic environment of ISEcp1 and blaACC-1. Antimicrob Agents Chemother 51:2658–2659. doi: 10.1128/AAC.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacoby GA, Griffin CM, Hooper C. 2011. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother 55:4979–4984. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeiro TG, Novais Â, Branquinho R, Machado E, Peixe L. 2015. Phylogeny and comparative genomics unveil independent diversification trajectories of qnrB and genetic platforms within particular Citrobacter species. Antimicrob Agents Chemother 59:5951–5958. doi: 10.1128/AAC.00027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Jacoby GA, Mills DM, Hooper DC. 2009. SOS regulation of qnrB expression. Antimicrob Agents Chemother 53:821–823. doi: 10.1128/AAC.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briales A, Rodriguez-Martinez JM, Velasco C, Machuca J, Díaz de Alba P, Blazquez J, Pascual A. 2012. Exposure to diverse antimicrobials induces the expression of qnrB1, qnrD, and smaqnr genes by SOS-dependent regulation. J Antimicrob Chemother 67:2854–2859. doi: 10.1093/jac/dks326. [DOI] [PubMed] [Google Scholar]
- 29.Albornoz E, Lucero C, Romero G, Rapoport M, Guerriero L, Andres P, WHONET-Argentina Group, Galas M, Corso A, Petroni A. 2014. Analysis of plasmid-mediated quinolone resistance genes in clinical isolates of the tribe Proteeae from Argentina: first report of qnrD in the Americas. J Glob Antimicrob Resist 2:322–326. doi: 10.1016/j.jgar.2014.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.