Abstract
Although originally discovered as neuronal growth cone-collapsing factors, repulsive guidance molecules (RGMs) are now known as key players in many fundamental processes, such as cell migration, differentiation, iron homeostasis, and apoptosis, during the development and homeostasis of many tissues and organs, including the nervous, skeletal, and immune systems. Furthermore, three RGMs (RGMa, RGMb/DRAGON, and RGMc/hemojuvelin) have been linked to the pathogenesis of various disorders ranging from multiple sclerosis (MS) to cancer and juvenile hemochromatosis (JHH). While the molecular details of these (patho) biological effects and signaling modes have long remained unknown, recent studies unveil several exciting and novel aspects of RGM processing, ligand–receptor interactions, and downstream signaling. In this review, we highlight recent advances in the mechanisms-of-action and function of RGM proteins.
RGMs: A Small Gene Family with Widespread Effects
Guidance molecules, initially observed to direct growing axons during embryogenesis [1], also have crucial roles in the morphogenesis and homeostasis of non-neuronal tissues by controlling a plethora of cellular processes, ranging from cell division and migration to differentiation and death. The discovery of RGMa in 2002 revealed a new family of guidance molecules (Box 1). Since then, four RGMs have been found in vertebrates: RGMa, RGMb (or DRAGON), RGMc (or hemojuvelin), and RGMd (only present in fish). Invertebrates, such as Caenorhabditis elegans, have one RGM gene.
Box 1. A Brief History of the Repulsive Guidance Molecule.
During nervous system development, axons travel long distances to reach their synaptic partner cells. Axons are guided to their targets by many different attractive and repulsive guidance molecules present in their environment [1]. A neuronal system in which this process of axon guidance has been studied extensively is the retinotectal system (i.e., axonal projections from retinal neurons in the eye to the tectum in the brain). Gradients of topographic guidance cues had been postulated decades ago to drive the process of retinotectal map formation. However, identification of these cues remained elusive for nearly 50 years. In 1990, Bonhoeffer and colleagues discovered a tectum-derived lipid-anchored RGM with a molecular weight of 33/35 kDa [63], later named ‘RGM’ [64]. RGM was the first graded topographic guidance molecule and its amino acid sequence was published in 2002 [29]. RGM is part of a small gene family that contains four members: RGMa (or RGM), RGMb (DRAGON), RGMc (hemojuvelin), and RGMd (only present in fish). Work from different groups has shown that RGMa and RGMb are expressed in a largely nonoverlapping pattern in the CNS and other tissues, whereas RGMc is mostly absent from the CNS and expressed in liver and skeletal muscle. In addition to axon guidance, RGMs are now known to subserve a multitude of physiological functions ranging from immune system function to the regulation of iron homeostasis. Furthermore, in adult humans with traumatic brain injury, cerebral stroke, MS, Parkinson's disease and Alzheimer's disease, RGM proteins (mostly RGMa, but also in some indications RGMb) are re-expressed and accumulate at sites of damage or injury. This suggests that targeting RGMs may be a promising therapeutic strategy for different brain diseases. Consequently, several studies have successfully explored the effect of neutralizing RGMs in experimental disease models. These showed, for example, that neutralization of RGMa, a known inhibitor of axon regeneration, following spinal cord injury results in enhanced functional recovery [13]. The first clinical trials targeting RGM with a highly selective RGMa-specific antibody (ABT-555) will tell whether such regeneration promotion of damaged axons is also observed in humans with MS or suffering from spinal cord injury.
RGMs are membrane-associated glycosylphosphatidylinositol (GPI)-linked proteins that harbor an N-terminal signal peptide, an RGD motif (RGMa and RGMc), and a partial von Willebrand type D (vWFD) structural domain (Figure 1A). Each RGM displays tissue-specific expression, is subjected to distinct biosynthetic and processing steps, and has not only unique, but also shared biological functions [2]. RGMs bind the type 1 transmembrane protein Neogenin (Figure 1A) and many of the reported biological effects of RGMs rely on Neogenin receptor functions, such as axon guidance or neuronal survival [3,4]. RGMs also serve as co-receptors for bone morphogenetic proteins (BMPs) (Figure 1A) to regulate iron metabolism, skeletal development [5–10] and axon regeneration [11]. In addition to these physiological roles, and as discussed below, RGMs have been implicated in various diseases and are considered to be promising targets in the treatment of MS, spinal cord injury, stroke, anemia, and inflammation [5,11–15].
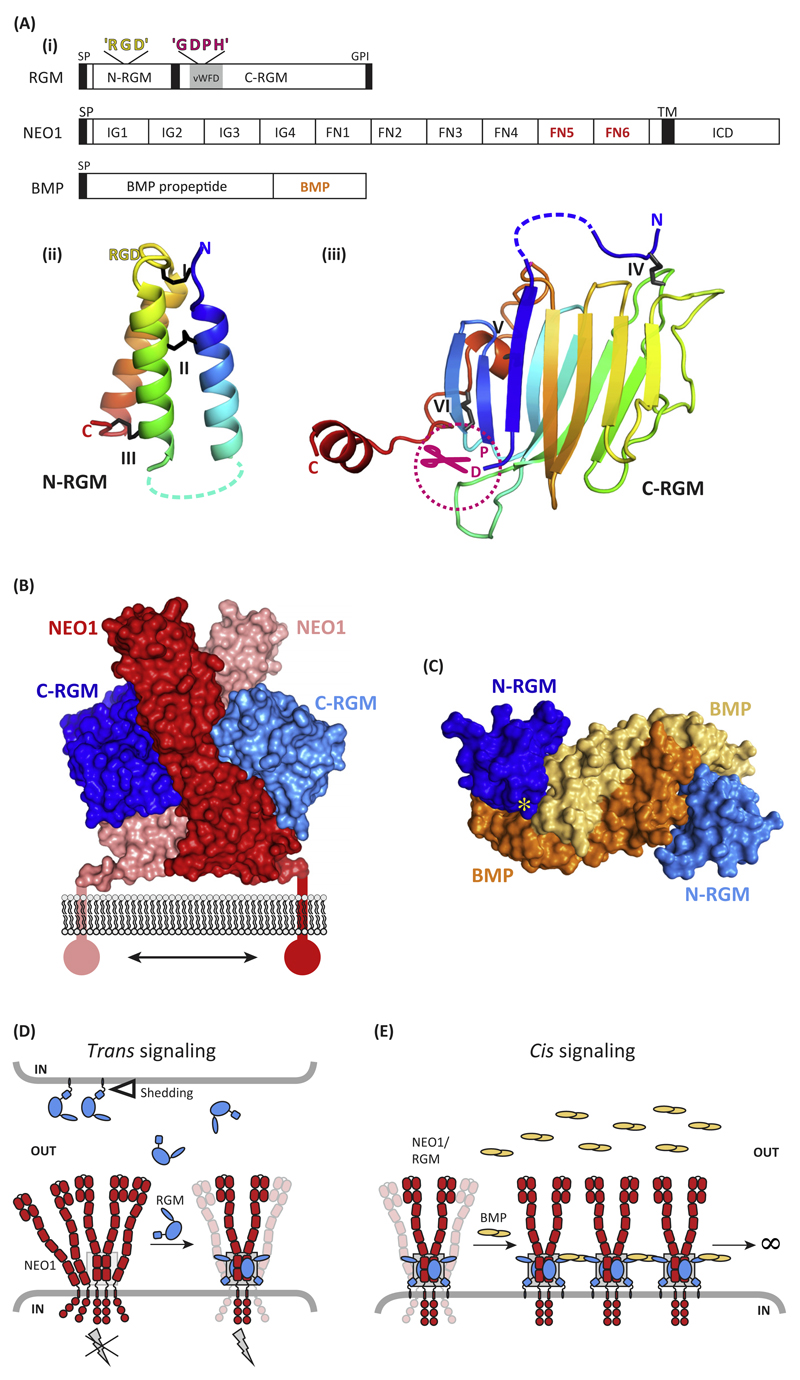
Figure 1. Molecular Determinants of Repulsive Guidance Molecules (RGMs) and Their Interactions with Neogenin (NEO1) and Bone Morphogenetic Protein (BMP) Ligands.
(A) (i) Schematic representation and domain organization of RGMs, Neogenin, and BMPs. Both the N-terminal domain (ii) [N-RGM; Protein Data Bank (PDB) ID 4UI1] and C-terminal domain (iii) (C-RGM; PDB ID 4BQ6) form distinct domains stabilized by several intramolecular disulfide bonds. Structures are shown in cartoon in rainbow coloring (blue, N terminus; red, C terminus). Note that RGD motifs are potential integrin-binding sites, but that no binding of RGMs to integrins has been reported so far. (B) The RGM–Neogenin complex (PDB ID 4BQ6). Two C-RGM molecules (blue) act as a molecular staple bringing together two Neogenin receptors (red). (C) The N-RGM–BMP complex (PDB ID 4UI1). The disulfide-linked BMP2 dimer binds two molecules of N-RGM in its wing region. The yellow asterisk indicates the position of the ‘RGD’ motif. (D) Model for RGM-mediated signaling in trans. The RGM ectodomain can be shed by proteolytic or phospholipase activity (open triangle). RGM binding to preclustered Neogenin results in stabilization and dimerization of the Neogenin ectodomain, subsequently activating downstream signaling (gray lightning bolt). The gray box highlights the RGM–Neogenin signaling hub observed in the crystal structure. (E) Model of RGM-mediated signaling in cis. RGMs can act as a physical protein bridge bringing together Neogenin and the BMP ligand, resulting in clustering. Abbreviations: FN, fibronectin; GDPH, autoproteolysis cleavage motif; GPI, glycosylphosphatidylinositol anchor; ICD, intracellular domain; IG, immunoglobulin; L, flexible linker; RGD, Arg-Gly-Asp; SP, signal peptide; TM, transmembrane; vWFD, von Willebrand Factor type D.
Although the molecular mechanisms underlying the biological effects and signaling modes of RGMs have long remained unknown, recent work has unveiled several exciting and novel aspects of RGM processing, ligand–receptor interactions, and downstream signaling. These insights include high-resolution structural data of binary or tertiary protein complexes, the unique processing of RGMs into protein fragments with distinct functions, and the identification of a novel molecular mechanism to control ligand-induced ectodomain shedding of Neogenin. In this review, we discuss recent highlights in RGM research, from novel structural data to previously unexplored signaling mechanisms and cellular functions.
Structural Insight into Ligand–Receptor Interactions
For many years, RGMs posed a molecular puzzle because of a general lack of structural homologies to any known protein fold. Recent studies have shed light on their 3D structure and identified two ordered and disulfide-stabilized domains that are connected by a flexible linker [16,17] (Figure 1A–C). The RGM N-terminal domain (N-RGM) comprises a three-helix bundle that harbors the ‘RGD’ motif. RGD motifs are traditionally known to be important in integrin-mediated adhesion, but no binding of RGMs to, or signaling through, integrins has been reported. The C-terminal domain (C-RGM) forms a tight β-sandwich structure and harbors a ‘GDPH’ cleavage site, which mediates autoproteolysis (Figure 1A).
C-RGM is the major high-affinity interaction site for Neogenin [16], with an additional Neogenin-binding site positioned in N-RGM, as suggested by antibody-blocking experiments [11]. In the C-RGM-Neogenin (NEO1) complex structure, two RGM molecules act as a molecular staple bringing together the juxtamembrane fibronectin type III (FNIII) domains of two Neogenin receptors (Figure 1B) [16]. This Neogenin region is necessary and sufficient for high-affinity RGM–Neogenin interactions, because single-point mutants of interface residues (either in RGM or Neogenin) abolish binding [16,18]. The RGM–Neogenin complex architecture is proposed to induce the dimerization of Neogenin and to position its C termini in close proximity to each other. These data hint at intracellular dimerization as a mechanism of signal transduction through the plasma membrane.
RGMs are crucial activators of BMP signaling and bind to BMP ligands with high affinity [17–19]. Crystal structures of all human RGMs in complex with the BMP ligand BMP2 revealed a common mode of binding and identified N-RGM as the high-affinity interaction site for BMP ligands [17] (Figure 1C). This analysis also informed a potential molecular mechanism for JHH (see Glossary)-linked mutations in RGMc that occur at the binding interface with the BMP ligand, because these mutations disrupt the RGMc–BMP interaction. RGMc controls levels of hepcidin through BMP signaling and reduced hepcidin expression leads to the iron overload in the liver, heart, and pancreas observed in JHH [8–10].
Glossary.
A disintegrin and metalloproteinase domain-containing protein 17 (ADAM17): also known as tumor necrosis factor-∝ converting enzyme (TACE); a protease that induces ectodomain shedding of Neogenin and other cell surface proteins.
Adherens junction (AJs): a protein complex located at the junction between epithelial cells. These proteins mediate cell–cell adhesion and are linked to the actin cytoskeleton of the cell.
Experimental autoimmune encephalomyelitis (EAE): animal model for studying brain inflammation. Administration of different antigens (e.g., myelin or MOG) induces demyelinating disease of the CNS.
Guanine nucleotide exchange factor (GEF): proteins that stimulate monomeric GTPases by triggering the exchange of GDP for GTP.
Hepcidin: a small peptide secreted predominantly by hepatocytes that is essential for iron metabolism. It functions to degrade the iron exporter ferroportin.
Juvenile hemochromatosis (JHH): a rare genetic disorder characterized by the accumulation of iron in various organs of the body. Mutations in RGMc can cause JHH and a reduction in hepcidin.
WAVE regulatory complex (WRC): group of proteins that regulate actin dynamics by stimulating the actin-nucleating activity of the Arp2/3 complex at the plasma membrane.
WRC-interacting receptor sequence (WIRS): conserved peptide motif that mediates binding to WRC.
A comparison between the structures of the RGM–BMP and BMP–BMP type I receptor ectodomain complexes showed that RGM and the BMP type I receptor ectodomain of BMP-R1A share the same binding site on the BMP ligand. This was an unexpected discovery, because simultaneous binding of the BMP type I and type II receptors to the BMP ligand is an essential requirement for canonical, SMAD-dependent downstream signaling. So, how can RGM activate canonical BMP signaling despite it competing with BMP-receptor binding? Since the RGM–BMP interaction is pH dependent, unlike the BMP–BMP receptor complex [17], an endocytosis-linked mechanism for RGM-activated BMP signaling has been proposed. In this model, the RGM–BMP complex (potentially together with the BMP type II receptor) might be targeted into endosomes, which are enriched with BMP type I receptors [20]. The lower pH of the endosomal environment might then lead to dissociation of the RGM–BMP complex and replacement by the BMP type I receptor, leading to potentiation of SMAD signaling provided by the endosomal environment compared with the cell surface [20,21]. However, further work will be required to test this model, for example demonstrating the involvement of RGMs in BMP ligand endocytosis and showing that the endosome functions as a platform for BMP signaling. It will be interesting to see whether all RGM family members act via the same mechanism or whether the biological context, including available interaction partners, has a role in directing how RGMs affect the BMP pathway. However, localization of the signaling machinery in close vicinity to the nucleus is a favorable way of activating target gene transcription. Such a mechanism has been suggested for other extracellular signaling systems, including the epidermal growth factor and transforming growth factor signaling pathways [21–23].
Much evidence suggests that the Neogenin and BMP signaling pathways are functionally linked, most likely through the actions of RGMs [9,24–27]. The most compelling evidence is the iron overload observed in the livers of mice in which the gene encoding Neogenin (NEO) has been knocked out [28]. Structural experiments showing a direct physical interaction with RGMs acting as a link between these two signaling pathways led to the crystal structure of a ternary complex comprising the BMP ligand BMP2, the full-length extracellular domain of RGMb, and the two membrane proximal FNIII domains of Neogenin [17]. These results, together with X-ray solution scattering and super-resolution microscopy [17], suggest a model by which RGMs mediate clustering of BMP and Neogenin dimers on the cell membrane. These data show that RGMs are a structural bridge between Neogenin and BMP signaling, and inform that a clustering mechanism may be important in the activation of these signaling pathways.
Cell Biology, Molecular Regulation, and Downstream Signaling of RGMs
RGMs signal through both trans (intercellular) and cis (same cell) interactions. Trans signaling is relevant for cell functions such as axon growth and guidance [29,30], early stages of neurulation [31], CD4+ T cell adhesion and activation [32], and leukocyte migration [33]. This can be either contact dependent (adhesive) or mediated by gradients established by cleaved extracellular RGM isoforms. The location of the RGM-binding site on the Neogenin-FNIII domains, close to the plasma membrane, and the RGM-Neogenin complex architecture [16] indicate a binding mode for which the release of the RGM ectodomain expressed by neighboring cells might be a prerequisite (Figure 1D). By contrast, RGM ectodomain shedding may not be required in situations where both Neogenin and RGM are expressed on the same cell surface (cis signaling), such as in hepatocytes [25] and chondrocytes [9] (Figure 1E).
Work over the past decade has led to the identification of several (co)receptors for RGMs as well as components of downstream signaling pathways that mediate their biological effects. Furthermore, RGMs have been implicated in a plethora of cell biological effects (e.g., axon growth and iron homeostasis), in different organs (e.g., brain, skeleton, and immune system), and in relation to diverse human disorders (e.g., spinal cord injury and cancer). In addition, recent data indicate that both RGMs and Neogenin are proteolytically processed. This processing serves to release RGMs from the cell surface in certain situations as well as to diversify the effects of the RGM signaling pathway and to control signaling duration following ligand binding. Here, we highlight some of the most recent insights into RGM processing and signaling in the context of specific biological functions, and refer readers to other reviews for a detailed discussion of other signaling mechanisms and biological effects [2,8,10,34,35].
Autocatalytic and Proteolytic Cleavage of RGMc in the Control of Body Iron Levels
All RGMs contain an autocatalytic Gly-Asp-Pro-His (GDPH) cleavage site (Figure 1A), which is known to be unstable under mildly acidic conditions due to a specific conformation and/or general acid catalysis [24]. Incubation of purified recombinant RGMc at pH 5.5 increases autocatalytic cleavage, supporting the idea that RGMc undergoes partial autocatalytic cleavage within the GDPH sequence [24]. The GDPH site is found in the loop connecting two β sheets (Figures 1A and 3A) and is highly conserved across species. Autocatalytic cleavage does not lead to a secreted fragment of RGM, since the resulting two polypeptides are joined covalently by disulfide bonds to form a stable structural unit. Autocatalytic cleavage appears to be important for the correct folding of the protein [16]. A clue about the functional importance of RGM autocatalytic cleavage comes from genetic studies in patients with JHH. Several JHH-associated mutations are located in the vicinity of the autocatalytic cleavage site for RGMc [16,36,37]. These mutants are often retained in the ER and have low signaling activities, suggesting that autocatalytic processing is necessary for RGMc plasma membrane expression and for its iron regulatory function. Furthermore, experiments using noncleavable RGMa mutants reveal that RGMa autocatalytic processing is also required for its growth inhibitory effects on axons [38]. Thus, autocatalytic processing appears to be a general and important feature of RGMs.
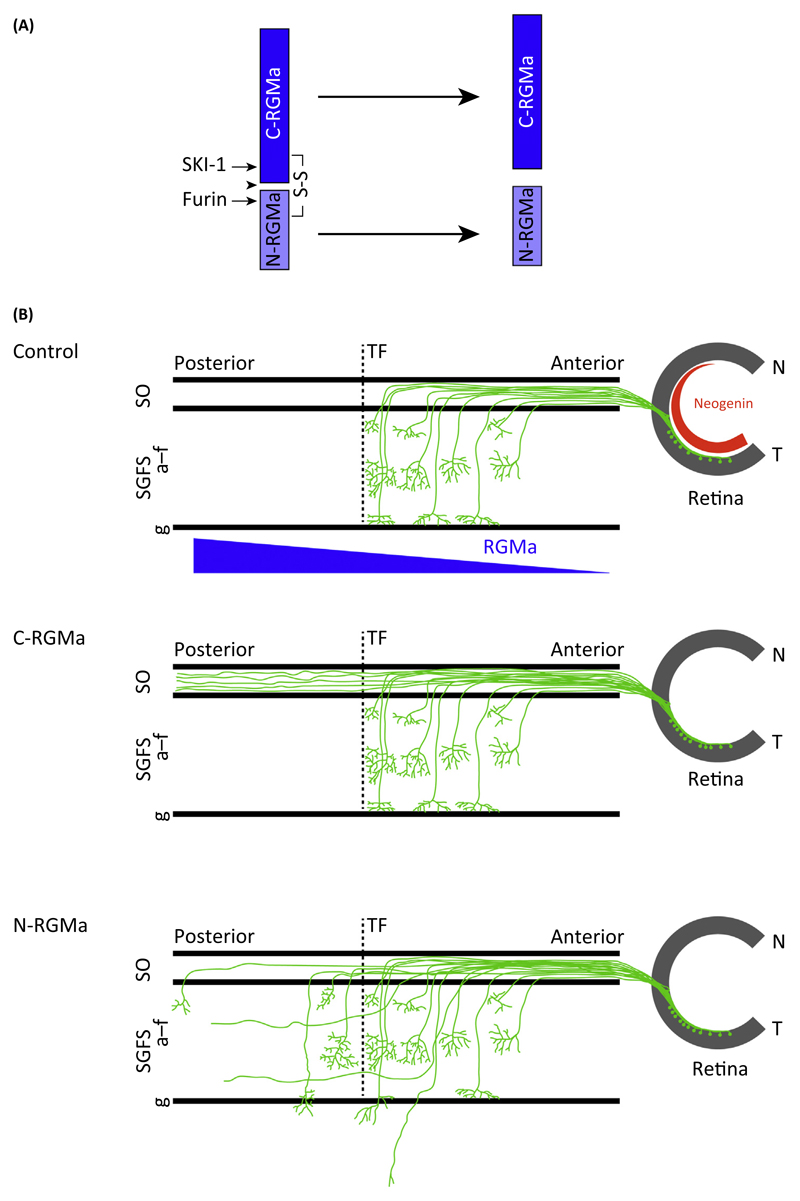
Figure 3. Proteolytic Processing of Repulsive Guidance Molecules (RGMa) Generates N- and C-RGMa Fragments that Regulate Distinct Aspects of Retinotectal Path Finding in vivo.
(A) Autocatalytic processing (arrowhead) and proteolysis by subtilisin kexin isozyme-1 (SKI-1) and furin generates C- and N-RGMa fragments. (B) Ectopic expression of C- and N-RGMa peptides in the chick optic tectum results in distinct axon-targeting defects. Normally, two gradients of Neogenin (blue) in the eye and RGMa (in the tectum) allow correct anterior–posterior targeting of retinal axons. In control experiments, all axons from retinal ganglion cells in the eye terminate before the terminal front (TF) in the tectum and arborize in layers a–f of the stratum griseum et fibrosum superficiale (SGFS). Ectopic expression of C-RGMa results in axonal overshooting beyond the TF, with aberrant retinal axons remaining restricted to the superficial stratum opticum (SO) layer. By contrast, ectopic expression of N-RGMa induces overshooting beyond the TF and into deeper layers (beyond SGFS layer g). Abbreviations: N, nasal; T, temporal.
In addition to autocatalytic cleavage, proteolytic processing of RGMc by furin and serine protease matriptase-2 (TMPRSS6) has a role in the regulation of body iron levels. Membrane-bound RGMc acts as a co-receptor for BMPs to regulate hepcidin expression resulting in increased iron absorption. RGMc cleavage by furin at a specific C-terminal cleavage site, not present in RGMa or RGMb, releases a 42-kDa soluble protein, which acts as a decoy receptor that competes with membrane-bound RGMc for binding to BMP ligands, thereby suppressing hepcidin expression [39]. In addition, the serine protease matriptase-2 (TMPRSS6) binds and cleaves cell surface RGMc. However, the RGMc fragment shed by matriptase-2 has reduced ability to bind BMPs and fails to repress BMP-induced hepcidin expression in vitro [40,41]. Unlike furin, it is thought that matriptase-2 impacts iron homeostasis mainly by reducing levels of membrane-bound RGMc. However, other work suggests that the interaction between matriptase-2 and RGMc is more complex, (e.g., independent of protease activity) [42]. Further work is needed to assess the requirement of RGMc cleavage by matriptase-2 in vivo and the precise link between matriptase-2, RGMc, BMP signaling, and hepcidin [10]. Nevertheless, these studies reveal important roles for proteolytic processing of RGMc in the regulation of hepcidin expression and iron levels.
Ectodomain Shedding of Neogenin
Over the past few years, many groups have confirmed the ability of RGMs to induce axon repulsion, neurite growth inhibition, and growth cone collapse using different types of neuron [11,13,29,43–46]. The role of RGMs in axon guidance and neurite growth inhibition during development has been shown in frog and chick embryos, while in vivo evidence for the neurite growth inhibitory effects of RGMs in mammals is provided by the ability of RGM blockage to promote central nervous system (CNS) axon regeneration. Other important effects of RGM on cell death, migration, and differentiation have also been confirmed (Boxes 1 and 2) [11,13,47–49]. The neurite growth inhibitory effects of RGMs have been studied extensively and rely on signaling by Rho-GTPases downstream of RGMa. Binding of RGMa to Neogenin activates RhoA through the guanine nucleotide exchange factor (GEF) LARG in an Unc5B-dependent manner [50]. In parallel, RGM–Neogenin interactions trigger a reduction in Ras activity through focal adhesion kinase (FAK) and p120GAP [51] (Figure 2, Key Figure). However, these signaling events provide a simplified view of RGM signaling and, as discussed below, precise regulation of the sensitivity of neurons to RGM of processing of RGMs into distinct polypeptides appears to be a prerequisite for the formation of appropriate neuronal networks.
Box 2. Mutations and Knockouts of RGMs.
With the exception of RGMc, not much is known about the effects of in vivo loss of function of RGMs. RGMb maps to chromosome 5q15 and RGMb gene knockout in mice results in death at 2–3 weeks after birth [65]. The olfactory epithelium of RGMb-knockout mice displays an increase in dividing progenitor cells in addition to supernumerary sustentacular support cells [49]. A role for RGMb in proliferation is also suggested in different cancers. Reduced RGMb expression, probably by RGMb promoter hypermethylation, is associated with poor prognosis in patients with non-small lung cancer, and overexpression of RGMb in a highly metastatic mouse model has a suppressive effect on cancer progression [66]. This suggests that RGMb acts as a tumor suppressor, possibly by inhibiting SMAD activation. This idea is also supported by the observation that the RGMb gene is inactivated by frame-shift mutations in a subtype of colorectal cancer [67].
A potential tumor suppressor role for RGMa is suggested in human colon cancer where (epi)genetic inactivation of RGMa results in strongly decreased levels of RGMa not only in colorectal cancer (CRC) tissue, but also in CRC cell lines and adenomas [68]. The RGMa gene maps to chromosome 15q26.1 and RGMa gene knockout in mice can result in early embryonic lethality, due to failure of neural tube closure (only 50% of the expected homozygous mice are born). Surviving RGMa-knockout mice show no defects in retinotectal map formation [47]. This is not an unexpected finding since RGMb may compensate for RGMa loss of function. Unfortunately, double RGMa/RGMb-knockout mice are subject to early embryonic or postnatal lethality, while conditional RGMa/RGMb double-knockout mice are not yet available.
RGMc maps to chromosome 1q21.1 and was identified as the second gene mutation that results in JHH [69]. HAMP, encoding hepcidin, was the first gene linked to JHH. Many mutations have been identified in RGMc resulting in premature stop codons or missense substitutions of highly conserved residues. Two studies analyzing RGMc-knockout mice showed massively increased serum iron levels and very low hepcidin expression [47,70]. RGMc-knockout mice also display an interesting retinal phenotype, with abnormal vasculogenesis and angiogenesis, and reactive gliosis of microglial and Müller-type glial cells [71].
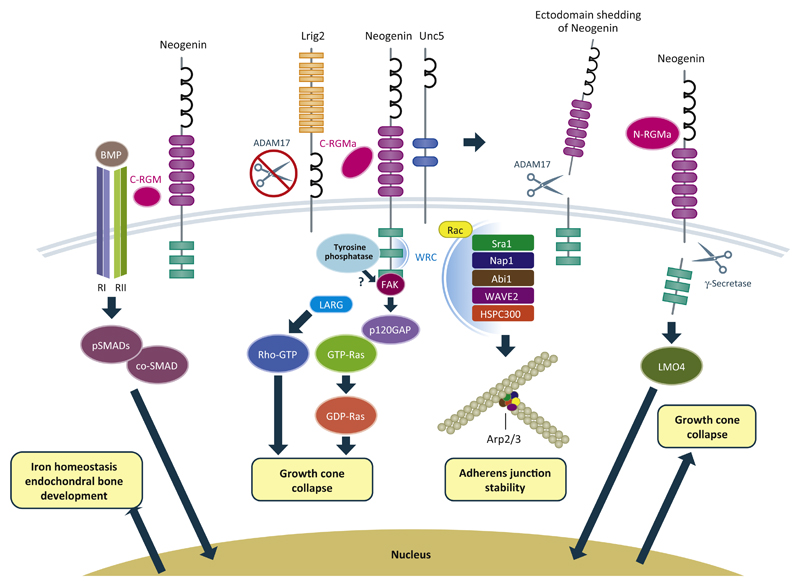
Figure 2.
RGMs act as co-receptors for bone morphogenetic proteins (BMPs) and have been proposed to act as a structural bridge between BMPs and Neogenin. A recently proposed model suggests that RGMs induce endocytosis of the BMP receptor complex, thereby activating canonical SMAD signaling. Interactions between RGMs and BMP signaling have been implicated in iron homeostasis and endochondral bone development. Binding of RGM to Neogenin inhibits interactions between Lrig2 and Neogenin, allowing ectodomain shedding by A disintegrin and metalloproteinase domain-containing protein 17 (ADAM17) leading to signal termination. In general, RGM–Neogenin binding leads to the activation of RhoA through Unc5 and LARG, and inactivation of Ras through focal adhesion kinase (FAK) and p120 RasGAP to induce growth cone collapse. However, signaling is dependent on the proteolytic processing of RGMs, since C-RGM triggers RhoA-dependent signaling, while the effects of N-RGM rely on shedding of the Neogenin intracellular domain by γ-secretase and LMO4. The Neogenin intracellular domain has been proposed to move into the nucleus, possible together with LMO4, and regulate gene transcription. In epithelial cells, Neogenin binds and localizes the WAVE regulatory complex (WRC), leading to actin nucleation by Arp2/3, which also requires activation by Rac1 and adherence junction stability. The extent to which these signaling pathways are specific to select cell types or cellular functions remains to be determined.
Key Figure
Signaling Mechanisms Downstream of Repulsive Guidance Molecules (RGMs)
The finding that cell surface shedding of Neogenin by A disintegrin and metalloproteinase domain-containing protein 17 (ADAM17) desensitizes axons to RGMa suggested a role for proteolysis in Neogenin signaling [52]. The extracellular part of Neogenin binds to, and is cleaved by, ADAM17. This cleavage event induces ectodomain shedding and thereby reduces Neogenin cell surface expression. However, how cleavage of Neogenin by ADAM17 is initially prevented to allow cleavage only after ligand binding remained unknown. Recent findings showed that the transmembrane leucine-rich repeat protein Lrig2 negatively regulates ADAM17-mediated guidance receptor proteolysis in neurons [48]. Lrig2 binds Neogenin and prevents premature Neogenin shedding by ADAM17 (Figure 2). RGMa reduces Lrig2–Neogenin interactions, providing ADAM17 access to Neogenin and allowing this protease to induce ectodomain shedding. This study identified a unique ligand-gated mechanism that controls receptor shedding by ADAMs and shows that the functions of Lrig2 are required for the effects of RGM on neurite growth, cortical neuron migration, and regenerative failure.
Proteolysis of RGMs Into Different Functional Protein Fragments
Proprotein convertases (PCs) form a family of nine proteinases. Two of these, furin and subtilisin kexin isozyme-1 (SKI-1), process RGMa into C-terminal membrane-bound (C-RGM) and N-terminal soluble (N-RGM) fragments (Figure 3A) [38,46]. Interestingly, these cleavage events are dependent on RGM autoproteolysis. D149A and H151A mutations in the autocatalytic cleavage sequence do not alter RGMa processing toward the cell surface, but abolish processing by both SKI-1 and furin [38]. The functional significance of C-RGMa and N-RGMa fragments in vivo has been studied using the chick retinotectal system. Retinal ganglion cell (RGC) neurons reside in the eye and send their axons in a topographic manner through the optic nerve to the tectum in the brain (Figure 3B). RGMa is expressed in an anterior low to posterior high gradient in the embryonic tectum, whereas Neogenin is expressed in the retina in a temporal high to nasal low gradient (Figure 3B). The RGMa gradient in the tectum restricts temporal RGC axons to the anterior part of the tectum, whereas nasal axons can target the posterior part. When retinal axons reach the tectum, they first extend within the most superficial layer of the optic tectum, the stratum opticum (SO). Once axons reach the appropriate anterior–posterior coordinates in the tectum, they turn into deeper layers to establish terminal arbors within layers a–f of the stratum griseum et fibrosum superficiale (SGFS) (Figure 3B) [27]. Ectopic expression of C-RGMa throughout the tectum leads to axon overshooting mainly in the superficial layer, while ectopic N-RGMa induces overshooting of RGC axons into deeper layers of the tectum (Figure 3B). In agreement with a role for C-RGMa and N-RGMa in retinotectal path finding, in vivo inhibition of C-RGMa or N-RGMa with recombinant antibodies in chick leads to defects in the targeting of RGC axons [46].
It is unknown why ectopic tectal expression of C-RGMa and N-RGMa differently affects retinal axon targeting in vivo. A possible explanation is that the peptides activate distinct downstream signaling pathways that exert quantitatively or quantitatively distinct effects on embryonic axons. Indeed, while both C-RGMa and N-RGMa require Neogenin to regulate axon development, N-RGMa influences axon growth and guidance in vitro, while C-RGMa only affects axon growth. Furthermore, these effects are induced through the activation of distinct downstream pathways. C-RGMa leads to stimulation of the signaling cascade involving activation of LARG, RhoA, and ROCK, whereas signaling downstream of N-RGMa is thought to rely on γ-secretase cleavage of the intracellular domain of Neogenin [53] (Figure 2). Many cell surface receptors undergo cleavage by γ-secretase, triggering the subsequent release of their intracellular domain (ICD). Often this intracellular cleavage event is preceded by proteolytic release of the receptor ectodomain followed by shuttling of the ICD into the nucleus. NeICD harbors NLS and NES sequences, binds various nuclear proteins, and acts as a transactivator of gene transcription [54]. γ-Secretase cleavage is required for RGMa-mediated axon repulsion in vitro and ectopic NeICD expression induces RGC axon targeting effects in vivo [53]. One of the binding partners of NeICD is LIM-only protein 4 (LMO4), a transcriptional co-activator [55]. LMO4 is required for the axon-repulsive activity of RGMa and for targeting of RGC axons in the tectum by N-RGMa. Recent evidence indicates that DCC/Frazzled (Fra), a close homolog of Neogenin, is also processed by γ-secretase to release its intracellular domain (Fra-ICD) [56]. Fra-ICD shuttles between the cytoplasm and the nucleus, where it works as a transcriptional factor that regulates Commissureless expression to control axon midline crossing. NeICD may serve a similar function and act as a transcription factor. It is interesting to note that LMO4 functions as a novel co-factor of Neurogenin 2 (NGN2) in the developing cortex [57]. LMO4 binds NGN2 to form a multiprotein transcription complex. This complex is recruited to the E-box containing enhancers of NGN2 target genes, which regulate various aspects of cortical development and activate NGN2-mediated transcription [57]. It will be interesting to determine whether LMO4 and NeICD (and perhaps NGN2) form a transcription complex that regulates genes involved in axon growth. While in vivo work suggests that the axon-repulsive effects of N-RGMa are independent of LARG, in vitro studies have shown that LMO4 knockdown inhibits RhoA activation by RGMa [55]. Therefore, further studies are needed to probe the role of RhoA downstream of N-RGMa and to address other questions, such as whether γ-secretase is required in vivo for the effects of N-RGMa.
Together, these studies reveal that RGMa processing by PCs generates distinct RGMa fragments that signal through different signaling cascades (LARG–RhoA–ROCK versus NeICD– LMO4) to exert specific biological effects. This suggests that proteolytic processing of RGMs, together with the ability of RGMs to signal in trans and cis and their link to different signaling systems (Neogenin and BMP), functions to diversify the effects of these proteins. This helps to explain how a small family of proteins can regulate a disproportionally large number of biological events in different tissues and organ systems. Finally, it is also interesting to note that pre-incubation of Neogenin with C-RGMa abolishes Neogenin–N-RGMa binding, while vice versa N-RGMa reduces interactions between C-RGMa and Neogenin [38]. Therefore, it is possible that the local concentration of C-RGMa or N-RGMa determines which of the two peptides will prevalently interact with Neogenin to influence axons.
Lipid Raft Localization of Neogenin
RGMs have a crucial role in BMP signaling. For an extensive description of the signaling pathways involved in these effects and the proposed role of Neogenin, we refer readers to other reviews [8,10,58]. However, it should be noted that, whereas early studies failed to implicate BMPs in RGM-mediated effects on developing neurons, more recent work suggests that RGMs function through BMPs to affect neurons [27,45,59]. A compelling example of the functional interplay between RGMs, Neogenin, and BMPs is the role of these proteins in endochondral bone development during skeleton formation. During this process, Neogenin controls chondrocyte maturation by promoting BMP-induced receptor association with lipid rafts, thus enhancing effective BMP receptor concentration or BMP-binding affinity and increasing SMAD phosphorylation and downstream gene transcription [9]. How does Neogenin localize BMP receptors to lipid rafts? RGMs were found to form a protein bridge between Neogenin and BMP receptors, thereby inducing the formation of a multimeric receptor complex. Since RGMs contain GPI domains that localize these proteins to lipid rafts [29], the authors proposed that RGMs are responsible for moving the Neogenin–RGM–BMP receptor complex into lipid rafts. The presence of Neogenin in lipid rafts is required not only during endochondral bone development, but also for its neurite growth inhibitory and neuron death-inducing effects in the nervous system [15,27]. Interference with RGM–Neogenin binding using specific protein fragments or anti-RGMa antibodies causes Neogenin to move out of lipid rafts and prevents proapoptotic and neurite growth inhibitory effects. Application of these tools in models for brain or axonal injury (e.g., middle cerebral artery occlusion or optic nerve crush) promotes regeneration and functional recovery [11,15,27]. This suggests that interfering with the lipid raft localization of Neogenin represents a powerful means of neutralizing the detrimental effects of RGM–Neogenin following injury or disease.
Actin Regulation During Epithelial Cell Adhesion
Epithelial morphogenesis is fundamental to organogenesis in the embryo. Epithelial sheets undergo choreographed movements to generate complex structures, such as the neural tube. Early depletion of Neogenin or RGMa in the neuroepithelium leads to loss of adhesion and apicobasal polarity and, as a result, a failure in neural tube closure [31,47,60,61]. E-cadherin-mediated cell–cell adhesion found at adherens junctions (AJs) has a key role in maintaining the fidelity of the epithelium. Junctional stability requires reciprocal interactions between the cadherins and the circumferential actin ring running parallel to the AJ. The actin ring undergoes continuous turnover and failure to rebuild the ring causes loss of adhesion. Interestingly, recent work identified Neogenin as a key component of the actin nucleation machinery governing AJ stability [62]. Neogenin promotes the formation of stable actin rings at AJs by spatially coupling Arp2/3-mediated actin nucleation to the AJ via recruitment of the wave regulatory complex (WRC) (Figure 2). A direct interaction between the Neogenin WRC-interacting receptor sequence (WIRS) domain and the WRC is crucial for the restricted localization of the WRC and Arp2/3 to the junction. Neogenin is not sufficient to activate Arp2/3, which requires activation of Rac and Sra1 binding. Neogenin also affects E-cadherin recycling, but whether this effect depends on its ability to control actin dynamics is unknown. Knockdown of RGMa induces defects in AJ stability and WRC localization similar to those observed following Neogenin depletion [62]. This suggests that RGMa and Neogenin act together at the AJ. However, the precise mode-of-action of RGMa at the AJ, (e.g., whether it functions in cis or trans with Neogenin) remains unknown. Given that growth cone steering is highly dependent on actin regulation, it will be interesting to determine whether the induction of actin nucleation by Neogenin has a role at neuronal growth cones as well.
Immune Cell Signaling and Multiple Sclerosis
A new role of RGMa in the immune system has become recently apparent [32,33]. Bone marrow-derived dendritic cells express RGMa and Neogenin is expressed by CD4+ T lymphocytes. Binding of RGMa to Neogenin-positive CD4+ T cells induces activation of the small GTPase Rap1, thereby increasing adhesion to intracellular adhesion molecule-1 (ICAM-1). Thus, in contrast to Neogenin-induced growth cone collapse, which results in rapid loss of adhesion [45], binding between RGMa and Neogenin on immune cells can trigger enhanced adhesion. This increase in ICAM-1-adhesion may facilitate invasion of immune cells into the MS brain, making RGMa a novel therapeutic target for disease. An antibody blocking RGMa was able to improve disease scores in commonly used MS mouse models. Treatment of mice with an anti-RGMa antibody reduced invasion of inflammatory cells into the CNS. The antibody also affected T cell proliferation and cytokine production in a mouse model and in isolated peripheral blood mononuclear cells (PBMCs) from patients with MS [32], indicating a T cell immune suppressive effect. These experiments suggest that blocking RGMa may reduce inflammatory disease. However, another study reported that RGMa inhibits migration of RGMa-expressing leukocytes (T and B lymphocytes, monocytes, and granulocytes) via chemo- and contact repulsion and RGMa suppressed inflammation in a zymosan-induced peritonitis model [33]. These seemingly contrasting results might be explained by the different types of models used and by differences in underlying signaling complexes. Whereas the focus of both studies was the adaptive immune system, a third study focused on the innate immune system (microglia cells and macrophages) and its role in MS-associated neurodegeneration [11]. Highly inflammatory microglial cells and macrophages have been postulated to have an important role in progressive MS and are the target of several new therapeutic drug approaches. Systemic treatment of experimental autoimmune encephalomyelitis (EAE) rats with RGMa-specific antibodies resulted in significant and highly reproducible functional improvement, reduction of the size of the microglial lesion, enhanced axon regeneration into the inflammatory lesion, and signs of remyelination [11]. One of the first symptoms of MS in 20–30% of patients is an optic neuritis, an inflammatory attack of the optic nerve. Such an attack can have a strong impact on RGCs and their axons forming the retinal nerve fiber layer (RNFL). In a tEAE optic neuritis model, systemic application of RGMa-specific antibodies dramatically reduced degeneration of the RNFL, suggesting that RGMa is involved not only in inhibition of axon regeneration, but also in cell death regulation [11]. How does RGMa contribute to neurodegeneration in MS? Recent work shows that IL-17-expressing CD4+ T cells (Th17 cells) strongly express RGMa and that Th17 cells induce neuronal cell death probably via RGMa–Neogenin-induced dephosphorylation of Akt [14]. Together, these studies implicate RGMa in immune regulation and disease. However, future work is needed to unravel the precise molecular details of these effects.
Concluding Remarks
It is an exciting time to study RGMs. During the first RGM symposium (Awaji, Japan, April 2–3, 2016), leading RGM experts from all over the world presented new and promising data that showed that, since their original discovery as tectum-derived axon repellents, RGM proteins have emerged as pleiotropic regulators of a multitude of cell biological processes in many different tissues. Often, these novel cellular functions have only been probed in one specific tissue and, therefore, an important future goal is to assess whether some of the newly discovered functions of RGMs also contribute to the development or homeostasis of other tissues (see Outstanding Questions). For example, given the important role of regulation of the actin cytoskeleton in growth cone collapse, it is tempting to speculate that Neogenin may also control WRC-Arp2/3 signaling in growth cones, as has been shown recently at AJs. Furthermore, while the ability of RGMs to control BMP signaling in regulating iron homeostasis and endochondral bone formation have been firmly established, the contribution of BMPs to other RGM-mediated effects remains largely unexplored. It has become clear that RGMs have diverse binding partners (e.g., BMPs and Neogenin), while these binding partners can also interact with proteins unrelated to RGMs, such as Netrin-1 or BMP receptors. An important challenge is to understand how these interactions are regulated. Do different binding partners compete for binding on RGMs or Neogenin? For example, since RGMa and Netrin-1 both bind the Neogenin FNIII region, do these proteins compete for binding to Neogenin? Do ligands, such as RGMs and Netrins, activate similar or distinct signaling cascades downstream of Neogenin? How can RGMs signal axon repulsion and cell adhesion through the same receptor? Thus far, many studies have used vertebrate models to dissect the functions and signaling pathways of RGMs. However, many invertebrate species have a RGM gene and, therefore, represent excellent models to address outstanding questions about RGM biology.
Outstanding Questions.
Do recently identified signaling and proteolytic processing mechanisms have a general role in RGM-mediated functions in different organs?
How are complex ligand–receptor interactions between BMPs, RGMs, Neogenin, BMP receptors, and Netrin-1 regulated? Do different Neogenin ligands compete for binding and do they trigger similar signaling cascades?
How can RGMs signal cell collapse and adhesion through the same receptor?
How do changes in RGMs lead to disease in- and outside the nervous system?
RGMs are being implicated in an ever-increasing number of diseases, ranging from cancer and MS to JHH. An interesting observation in the CNS is the consistent upregulation of RGMs following a variety of insults (e.g., immune-mediated, neurodegeneration, or trauma). The first results of blocking RGMs in experimental models are promising and a first clinical trial of anti-RGMa blocking antibodies in patients with MS is currently underway. In addition to blocking RGM function, it will be important to better understand how RGMs normally function, because this will undoubtedly contribute to our ability to define the pathogenic mechanisms underlying specific disorders and to eventually design novel and more effective therapeutic strategies.
Trends.
Structural data hint at intracellular dimerization of Neogenin as a mechanism of signal transduction through the plasma membrane, suggest endocytosis-linked activation of BMP signaling by RGM, and identify RGMs as a structural bridge between BMP and Neogenin signaling.
RGM and Neogenin proteins control actin dynamics by direct interactions with the WAVE regulatory complex and may achieve part of their biological effects through transcriptional regulation mediated by nuclear translocation of the Neogenin intracellular domain.
RGMs serve as crucial BMP co-receptors in the control of iron homeostasis and endochondral bone formation.
Proteolytic processing regulates and diversifies the biological effects of RGMs.
Changes in the function or expression of RGMs underlie various disorders and blocking these cues or influencing their downstream signaling pathways has great therapeutic potential.
Acknowledgments
We thank Hidekiyo Harada for help with preparing Figure 3. Work on RGMs in the laboratories of the authors was supported in part by a VICI grant from the Netherlands Organization for Scientific Research (ALW) and by a grant from the UU strategic theme Dynamics of Youth to R.J.P. C.S. is supported by a Cancer Research UK Senior Research Fellowship (C20724/A14414) and a European Research Council (ERC) Consolidator award (647278).
References
- 1.Kolodkin AL, Pasterkamp RJ. SnapShot: axon guidance II. Cell. 2013;153:722.e1. doi: 10.1016/j.cell.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Severyn CJ, et al. Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem J. 2009;422:393–403. doi: 10.1042/BJ20090978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajagopalan S, et al. Neogenin mediates the action of repulsive guidance molecule. Nat Cell Biol. 2004;6:756–762. doi: 10.1038/ncb1156. [DOI] [PubMed] [Google Scholar]
- 4.Matsunaga E, et al. RGM and its receptor neogenin regulate neuronal survival. Nat Cell Biol. 2004;6:749–755. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- 5.Babitt JL, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 6.Babitt JL, et al. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- 7.Samad TA, et al. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- 8.Corradini E, et al. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 20:389–398. doi: 10.1016/j.cytogfr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, et al. Neogenin regulation of BMP-induced canonical Smad signaling and endochondral bone formation. Dev Cell. 2010;19:90–102. doi: 10.1016/j.devcel.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian C, Liu J. Repulsive guidance molecules (RGMs) and neogenin in bone morphogenetic protein (BMP) signaling. Mol Reprod Dev. 2013;80:700–717. doi: 10.1002/mrd.22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demicheva E, et al. Targeting repulsive guidance molecule A to promote regeneration and neuroprotection in multiple sclerosis. Cell Rep. 2015;10:1887–1898. doi: 10.1016/j.celrep.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, et al. RNA interference against repulsive guidance molecule A improves axon sprout and neural function recovery of rats after MCAO/reperfusion. Exp Neurol. 2012;238:235–242. doi: 10.1016/j.expneurol.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Hata K, et al. RGMa inhibition promotes axonal growth and recovery after spinal cord injury. J Cell Biol. 2006;173:47–58. doi: 10.1083/jcb.200508143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanabe S, Yamashita T. Repulsive guidance molecule-a is involved in Th17-cell-induced neurodegeneration in autoimmune encephalomyelitis. Cell Rep. 2014;9:1459–1470. doi: 10.1016/j.celrep.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Shabanzadeh AP, et al. Uncoupling Neogenin association with lipid rafts promotes neuronal survival and functional recovery after stroke. Cell Death Dis. 2015;6:e1744. doi: 10.1038/cddis.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell CH, et al. Structure of the repulsive guidance molecule (RGM)-neogenin signaling hub. Science. 2013;341:77–80. doi: 10.1126/science.1232322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Healey EG, et al. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat Struct Mol Biol. 2015;22:458–465. doi: 10.1038/nsmb.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, et al. Neogenin interacts with hemojuvelin through its two membrane-proximal fibronectin type III domains. Biochemistry. 2008;47:4237–4245. doi: 10.1021/bi800036h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Q, et al. Repulsive guidance molecule (RGM) family proteins exhibit differential binding kinetics for bone morphogenetic proteins (BMPs) PLoS One. 2012;7:e46307. doi: 10.1371/journal.pone.0046307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartung A, et al. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol Cell Biol. 2006;26:7791–7805. doi: 10.1128/MCB.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Guglielmo GM, et al. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 22.Vieira AV, et al. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 23.Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 24.Zhang A-S, et al. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J Biol Chem. 2005;280:33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang A-S, et al. Hemojuvelin-neogenin interaction is required for bone morphogenic protein-4-induced hepcidin expression. J Biol Chem. 2009;284:22580–22589. doi: 10.1074/jbc.M109.027318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C, et al. The neogenin/DCC homolog UNC-40 promotes BMP signaling via the RGM protein DRAG-1 in C. elegans. Development. 2013;140:4070–4080. doi: 10.1242/dev.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tassew NG, et al. Modifying lipid rafts promotes regeneration and functional recovery. Cell Rep. 2014;8:1146–1159. doi: 10.1016/j.celrep.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Lee D-H, et al. Neogenin inhibits HJV secretion and regulates BMP-induced hepcidin expression and iron homeostasis. Blood. 2010;115:3136–3145. doi: 10.1182/blood-2009-11-251199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monnier PP, et al. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- 30.Wilson NH, Key B. Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev Biol. 2006;296:485–498. doi: 10.1016/j.ydbio.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Kee N, et al. Neogenin and RGMa control neural tube closure and neuroepithelial morphology by regulating cell polarity. J Neurosci. 2008;28:12643–12653. doi: 10.1523/JNEUROSCI.4265-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muramatsu R, et al. RGMa modulates T cell responses and is involved in autoimmune encephalomyelitis. Nat Med. 2011;17:488–494. doi: 10.1038/nm.2321. [DOI] [PubMed] [Google Scholar]
- 33.Mirakaj V, et al. Repulsive guidance molecule-A (RGM-A) inhibits leukocyte migration and mitigates inflammation. Proc Natl Acad Sci USA. 2011;108:6555–6560. doi: 10.1073/pnas.1015605108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita T, et al. Neogenin and repulsive guidance molecule signaling in the central nervous system. Curr Opin Neurobiol. 2007;17:29–34. doi: 10.1016/j.conb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 35.De Vries M, Cooper HM. Emerging roles for neogenin and its ligands in CNS development. J Neurochem. 2008;106:1483–1492. doi: 10.1111/j.1471-4159.2008.05485.x. [DOI] [PubMed] [Google Scholar]
- 36.Lanzara C, et al. Spectrum of hemojuvelin gene mutations in 1q–linked juvenile hemochromatosis. Blood. 2004;103:4317–4321. doi: 10.1182/blood-2004-01-0192. [DOI] [PubMed] [Google Scholar]
- 37.Lee PL, et al. Hemojuvelin (HJV) mutations in persons of European, African-American and Asian ancestry with adult onset haemochromatosis. Br J Haematol. 2004;127:224–339. doi: 10.1111/j.1365-2141.2004.05165.x. [DOI] [PubMed] [Google Scholar]
- 38.Tassew NG, et al. SKI-1 and Furin generate multiple RGMa fragments that regulate axonal growth. Dev Cell. 2012;22:391–402. doi: 10.1016/j.devcel.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Silvestri L, et al. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007;109:4503–4510. doi: 10.1182/blood-2006-08-041004. [DOI] [PubMed] [Google Scholar]
- 40.Silvestri L, et al. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111:924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- 41.Maxson JE, et al. Matriptase-2- and proprotein convertase-cleaved forms of hemojuvelin have different roles in the down-regulation of hepcidin expression. J Biol Chem. 2010;285:39021–39028. doi: 10.1074/jbc.M110.183160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guillem F, et al. Inactive matriptase-2 mutants found in IRIDA patients still repress hepcidin in a transfection assay despite having lost their serine protease activity. Hum Mutat. 2012;33:1388–1396. doi: 10.1002/humu.22116. [DOI] [PubMed] [Google Scholar]
- 43.Brinks H, et al. The repulsive guidance molecule RGMa is involved in the formation of afferent connections in the dentate gyrus. J Neurosci. 2004;24:3862–3869. doi: 10.1523/JNEUROSCI.5296-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunaga E, et al. Repulsive guidance molecule plays multiple roles in neuronal differentiation and axon guidance. J Neurosci. 2006;26:6082–6088. doi: 10.1523/JNEUROSCI.4556-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conrad S, et al. Neogenin-RGMa signaling at the growth cone is bone morphogenetic protein-independent and involves RhoA, ROCK, and PKC. J Biol Chem. 2007;282:16423–16433. doi: 10.1074/jbc.M610901200. [DOI] [PubMed] [Google Scholar]
- 46.Tassew NG, et al. Sustained in vivo inhibition of protein domains using single-chain Fv recombinant antibodies and its application to dissect RGMa activity on axonal outgrowth. J Neurosci. 2009;29:1126–1131. doi: 10.1523/JNEUROSCI.5385-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niederkofler V, et al. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci. 2004;24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Erp S, et al. Lrig2 negatively regulates ectodomain shedding of axon guidance receptors by ADAM proteases. Dev Cell. 2015;35:537–552. doi: 10.1016/j.devcel.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Kam JWK, et al. RGMB and neogenin control cell differentiation in the developing olfactory epithelium. Development. 2016;143:1534–1546. doi: 10.1242/dev.118638. [DOI] [PubMed] [Google Scholar]
- 50.Hata K, et al. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J Cell Biol. 2009;184:737–750. doi: 10.1083/jcb.200807029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Endo M, Yamashita T. Inactivation of Ras by p120GAP via focal adhesion kinase dephosphorylation mediates RGMa-induced growth cone collapse. J Neurosci. 2009;29:6649–6662. doi: 10.1523/JNEUROSCI.0927-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okamura Y, et al. TACE cleaves neogenin to desensitize cortical neurons to the repulsive guidance molecule. Neurosci Res. 2011;71:63–70. doi: 10.1016/j.neures.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee P, et al. γ-secretase and LARG mediate distinct RGMa activities to control appropriate layer targeting within the optic tectum. Cell Death Differ. 2016;23:442–453. doi: 10.1038/cdd.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldschneider D, et al. The neogenin intracellular domain regulates gene transcription via nuclear translocation. Mol Cell Biol. 2008;28:4068–4079. doi: 10.1128/MCB.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaffar G, et al. LIM-only protein 4 interacts directly with the repulsive guidance molecule A receptor Neogenin. J Neurochem. 2008;107:418–431. doi: 10.1111/j.1471-4159.2008.05621.x. [DOI] [PubMed] [Google Scholar]
- 56.Neuhaus-Follini A, Bashaw GJ. The intracellular domain of the Frazzled/DCC receptor is a transcription factor required for commissural axon guidance. Neuron. 2015;87:751–763. doi: 10.1016/j.neuron.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asprer JST, et al. LMO4 functions as a co-activator of neurogenin 2 in the developing cortex. Development. 2011;138:2823–2832. doi: 10.1242/dev.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mueller TD. RGM co-receptors add complexity to BMP signaling. Nat Struct Mol Biol. 2015;22:439–440. doi: 10.1038/nsmb.3037. [DOI] [PubMed] [Google Scholar]
- 59.Ma CHE, et al. The BMP coreceptor RGMb promotes while the endogenous BMP antagonist noggin reduces neurite outgrowth and peripheral nerve regeneration by modulating BMP signaling. J Neurosci. 2011;31:18391–18400. doi: 10.1523/JNEUROSCI.4550-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mawdsley DJ, et al. The Netrin receptor Neogenin is required for neural tube formation and somitogenesis in zebrafish. Dev Biol. 2004;269:302–315. doi: 10.1016/j.ydbio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Kee N, et al. Netrin-1 is required for efficient neural tube closure. Dev Neurobiol. 2013;73:176–187. doi: 10.1002/dneu.22051. [DOI] [PubMed] [Google Scholar]
- 62.Lee NK, et al. Neogenin recruitment of the WAVE regulatory complex maintains adherens junction stability and tension. Nat Commun. 2016;7:11082. doi: 10.1038/ncomms11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stahl B, et al. Biochemical characterization of a putative axonal guidance molecule of the chick visual system. Neuron. 1990;5:735–743. doi: 10.1016/0896-6273(90)90227-7. [DOI] [PubMed] [Google Scholar]
- 64.Müller BK, et al. Chromophore-assisted laser inactivation of a repulsive axonal guidance molecule. Curr Biol. 1996;6:1497–1502. doi: 10.1016/s0960-9822(96)00754-3. [DOI] [PubMed] [Google Scholar]
- 65.Xia Y, et al. Dragon (Repulsive Guidance Molecule b) Inhibits IL-6 Expression in macrophages. J Immunol. 2011;186:1369–1376. doi: 10.4049/jimmunol.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, et al. Repulsive guidance molecule B inhibits metastasis and is associated with decreased mortality in non-small cell lung cancer. Oncotarget. 2016;7:15678–15689. doi: 10.18632/oncotarget.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hesson LB, et al. Integrated genetic, epigenetic, and transcriptional profiling identifies molecular pathways in the development of laterally spreading tumors. Mol Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-16-0175. [DOI] [PubMed] [Google Scholar]
- 68.Li VSW, et al. Frequent inactivation of axon guidance molecule RGMA in human colon cancer through genetic and epigenetic mechanismS. Gastroenterology. 2009;137:176–187. doi: 10.1053/j.gastro.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 69.Papanikolaou G, et al. Mutations in HFE2 cause iron overload in chromosome 1q–linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 70.Huang FW. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tawfik A, et al. Deletion of hemojuvelin, an iron-regulatory protein, in mice results in abnormal angiogenesis and vasculogenesis in retina along with reactive gliosis. Investig Opthalmology Vis Sci. 2014;55:3616. doi: 10.1167/iovs.13-13677. [DOI] [PMC free article] [PubMed] [Google Scholar]