Abstract
Microglia are the resident inflammatory cells of the central nervous system (CNS) and have important roles in development, homeostasis and a variety of neurologic and psychiatric diseases. Difficulties in procuring human microglia have limited their study and hampered the clinical translation of microglia-based treatments shown to be effective in animal disease models. Here, we report the differentiation of human induced pluripotent stem cells (iPSC) into microglia-like cells by exposure to defined factors and co-culture with astrocytes. These iPSC-derived microglia (iPS-MG) have the phenotype, gene expression profile and functional properties of brain-isolated microglia. Murine iPS-MG generated using a similar protocol have equivalent efficacy to primary brain-isolated microglia in the treatment of murine syngeneic intracranial malignant gliomas. The ability to generate human microglia facilitates the further study of this important CNS cell type and raises the possibility of their use in personalized medicine applications.
INTRODUCTION
Microglia reside in the healthy CNS in a resting but surveillant state1,2, and promote homeostasis through reciprocal signaling interactions with neurons. In response to CNS injury, microglia can migrate to sites of damage, secrete inflammatory cytokines, phagocytose foreign matter and debris, and generate reactive oxygen species3–5. Beneficial properties of microglia include activation of innate and adaptive immune responses during infections and stimulation of neuronal plasticity, neurite outgrowth and synaptogenesis following ischemic strokes. Microglia can secrete factors capable of destroying glioma cells in vitro6,7 and in vivo8 and the intratumoral injection of LPS stimulates microglia and macrophages to diminish tumor growth in mice9. Recently, microglia derived from non-glioma human subjects have been shown to induce the expression of genes that control cell cycle arrest and differentiation, and markedly mitigate the sphere-forming capacity of glioma patient-derived brain tumor initiating cells in culture10. Microglia may also contribute to the progression of diseases such multiple sclerosis, Parkinson’s disease, HIV dementia, amyotrophic lateral sclerosis, Huntington’s disease, Pick’s disease, brain tumors and prion disease4,11. In disorders such as Alzheimer’s disease, microglia can have either positive or negative effects depending on the disease stage, the local microenvironment and the presence of disease-associated gene variants12,13
The therapeutic use of microglia has been demonstrated in experimental animal models of human diseases. Myeloablative conditioning with lethal irradiation or busulfan followed by bone marrow transplantation results in the brain engraftment and microglial differentiation of myeloid progenitor cells14. In genetically engineered mice with obsessive-compulsive disorder, or CNS lysosomal storage, application of this conditioning-transplantation paradigm using wildtype bone marrow cells has been shown to cure or improve symptoms15. A similar treatment strategy using gene-modified bone marrow cells has been shown to restore declines in general activity, rearing behavior, and food intake in an experimental model of induced Parkinson’s disease16,17. Collectively, these studies demonstrate the therapeutic potential of normal or gene-modified microglia, but the clinical translation of these results requires a source of autologous cells that can readily engraft in the diseased or injured brain, preferably without the need for lethal irradiation or busulfan mediated myeloablation.
We report here the sequential differentiation of human iPSC into myeloid progenitor-like intermediate cells and then into cells with the phenotypic, transcriptional and in vitro functional characteristics of brain-derived microglia. To demonstrate the potential in vivo use of such cells, murine iPS-MG generated using an analogous method were used to treat syngeneic intracranial malignant glioma bearing animals. The ability to generate human iPS-MG in particular may facilitate the study of the role of microglia in health and disease.
RESULTS
Human iPSCs differentiate into microglia-like cells via a hematopoietic progenitor-like intermediate cell
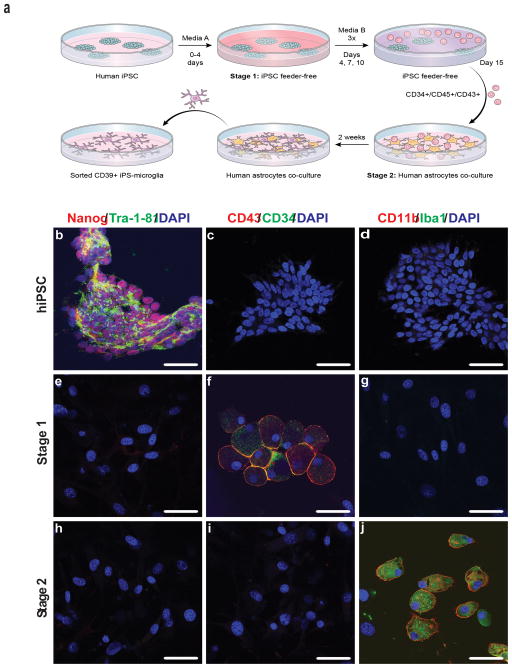
The well characterized human iPSC line NCRM-5 was obtained from the NIH Center for Regenerative Medicine (NIH CRM). iNC-01 transgene-free human iPSC were generated from peripheral blood CD34+ hematopoietic stem/progenitor cells. Given the myeloid lineage of microglia, a two-stage protocol in which human iPSC are first differentiated into hematopoietic progenitor-like cells (iPS-HPC) and then into hiPS-MG was devised (Fig. 1a). NCRM-5 hiPSC were differentiated on OP9 feeder layers, whereas for differentiation of iNC-01 hiPSC, a feeder-free differentiation protocol was developed. Prior to differentiation to iPS-HPC, iPSC express the stem cell markers Nanog and Tra-1-81, but not the hematopoietic progenitor cell markers CD3418,19 and CD4320 or the microglial markers CD11b and Iba1 (Fig. 1b–d). Differentiation of iPSCs to iPS-HPC (stage 1) results in the loss of Nanog and Tra-1-81 expression and gain of the hematopoietic markers CD34 and CD43 (Fig. 1e–g). Subsequent culture of iPS-HPC on astrocyte monolayers (stage 2) supplemented with GM-CSF, M-CSF, and IL-3 results in the loss of CD34 and CD43 expression and the gain of CD11b and Iba1 expression in as early as 7 days (Fig. 1h–j). These CD34-, CD43-, CD11b+ and Iba1+ microglia-like cells continue to increase in number over the ensuing one week. At the end of stage 2 differentiation, ~9% of the mixed astrocyte-microglial cultures are positive for ENTPD1 (CD39), a plasma membrane protein, specific for microglia (Supplementary Fig. 1). On average, 1 × 106 NCRM-5 and iNC-01 iPSC result in 2 × 106 and 3 × 106 microglia-like cells, respectively. A fibroblast derived iPSC (ND.1) results in a slightly lower yield of 8 × 105 microglia-like cells per 1 × 106 iPSC, and Inc-07s-1 (an additional feeder-free iPSC) results in a yield of 2 × 106 microglia-like cells per 1 × 106 iPSC.
Figure 1. Human iPSCs are differentiated into microglia-like cells.
(a) Schematic representation of the process used to differentiate human iPSCs into microglia-like cells. (b–j) Phenotypic staining at various steps of the differentiation process. Human iPSCs, Stage 1 hematopoietic progenitor-like cells, and Stage 2 hiPS-microglia immunostained with antibodies to Nanog and Tra-1-81 (b,e,h), CD34 and CD43 (c,f,i), CD11b and iba 1 (d,g,j), respectively. DAPI was used to visualize nuclei. The figure is representative of three independent experiments done in replicates (n=6). Scale bars, 50 μm.
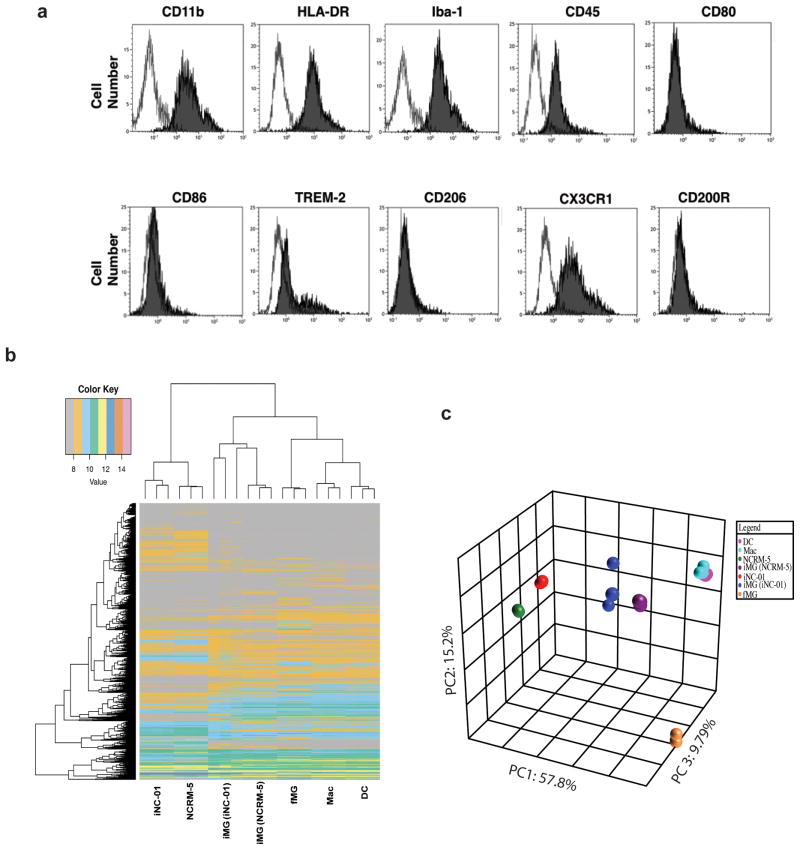
Following stage 2 differentiation, the resulting human iPS-MG cells were harvested and phenotypically characterized using immunocytometry staining. In addition to CD11b and Iba-1, the human iPS-MG express HLA-DR, CD45, TREM-2 and CX3CR1 but are negative for CD86, CD206, CD200R and CD80 expression (Fig. 2a). Because the phenotypes of other immune system phagocytic cells such as dendritic cells (DC) and macrophages (Mac) can resemble that of microglia, the gene expression signatures of human iPS-MG, DC and Mac were compared. Also included in the analyses were the human iPSC lines iNC-01 and NCRM-5 from which the human iPS-MG were derived, and commercially obtained primary human fetal brain-isolated microglia (fMG). 53,617 gene fragment expression values were generated per biological triplicate sample and the 60% of data found to be noise-biased was discarded. After subjecting the data to one-factor ANOVA under Benjamini and Hochberg false discovery rate multiple comparison correction conditions, we restricted our analysis to 18,302 gene fragments with a corrected p-value <0.05. Using Tukey’s Post-Hoc analysis, we then quantified the number of these differentially expressed genes having both a p-value < 0.05 and an absolute fold-change in expression ≥ 1.5 (14,475 genes) in pairwise comparisons of all the experimental groups. Based on correlation-based clustering analysis, the gene expression patterns of iPS-MG resemble that of fMG as well as DC and Mac, consistent with differentiation toward the myeloid lineage. The cells that most differ from iPS-MG in terms of gene expression are iPSCs. These gene expression relationships can be visualized using correlation-based clustering (Fig. 2b) and covariance-based principal component analysis (Fig. 2c) for Ingenuity microglia annotated genes. Recently, a unique microglial genes signature has been identified in both fetal and adult human microglia21. This human microglial signature consists of six genes: P2RY12 , GPR34, MERTK, C1QA, PROS1, and GAS6. We extracted individual gene expression data for these six microglia genes from the human microarray dataset described above and find that all six of these genes are highly expressed in the iPS-MG, but not in the iPSC, Mac and DC (Supplementary Fig. 2). RT-PCR analyses confirm the expression of these genes in the iPS-MG but not the iPSC, with the exception of P2RY12 which is expressed in both, but is greater in iPS-MG (Table 1). Expression of this distinct microglial gene signature supports the microglia-like identity of the iPS-MG and distinguishes them from other myeloid cell types such as DC and Mac.
Figure 2. Phenotypic and Genotypic analysis indicate human microglia-like cells.
(a) Cells from stage 2 (hiPSC-MG) were harvested using CD11b+ beads; immunocytochemical staining for microglial markers was carried out and analyzed by flow-cytometry. iPS-MG express CD11b, HLA-DR, Iba-1, CD45, TREM-2, CX3CR1, but do not express the macrophage marker CD206, T-cell stimulatory molecule like CD86, or the myeloid lineage marker CD200R. Histogram, filled-primary antibody and blank is the istotype control. The figure is representative of three independent experiments (n=3). (b) Heat map, and (c) Three-dimensional principal-component analysis examining expression levels of microglia genes between iPS-MG derived from iNC-01 and NCRM-5, fetal microglia (fMG), dendritic cells (DC), macrophages (Mac), and human induced pluripotent stem cells (iNC-01 and NCRM-5). Expression levels for each class were averaged from three independent cell culture or differentiation experiment samples (n=3).
Table 1. RT-PCR validation for expression of the six microglial signature genes in iPS-MG.
Expression levels were normalized to ACTB. The table is representative of three independent experiments with at least two technical replicates for each experiment (n=7).
| Genes | iPS | iPS-MG |
|---|---|---|
| P2RY12 | 1.00000 +/− 0.15935 SEM | 3.88033 +/− 0.51938 SEM |
| GPR34 | no expression | 4.35370 +/− 0.32043 SEM |
| MERTK | 1.00000 +/− 0.15251 SEM | 2.44567 +/− 0.16386 SEM |
| C1QA | 0.00009 +/− 0.00002 SEM | 4.35370 +/− 0.34634 SEM |
| PROS1 | 0.02689 +/− 0.03585 SEM | 4.35370 +/− 0.35904 SEM |
| GAS6 | 1.00000 +/− 0.12571 SEM | 4.90290 +/− 0.86260 SEM |
Human iPS-MG demonstrate functional properties of microglia
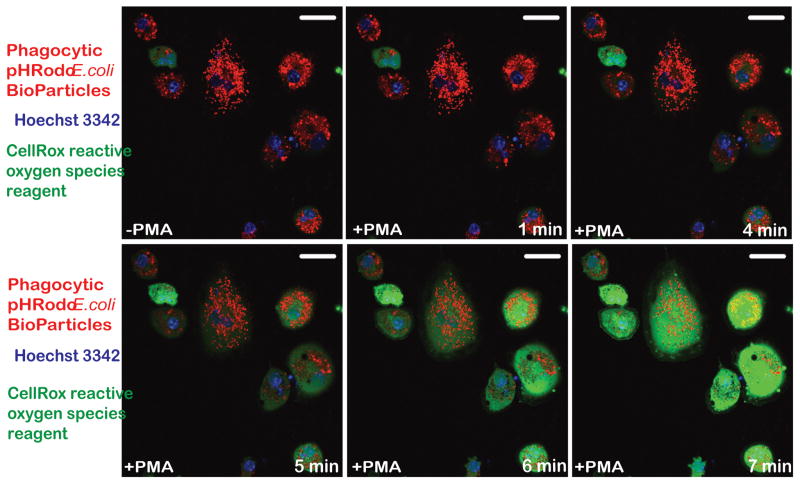
In addition to expressing the unique microglial gene signature shown above, microglia-like cells should also demonstrate the characteristic functional properties of microglia such as the phagocytosis of foreign particles, the production of reactive oxygen species (ROS) and the secretion of inflammatory cytokines. Under baseline culture conditions, iPS-MG actively phagocytose pHrodo E. Coli BioParticles conjugates as evidenced by the acidification and resulting visualization of these red fluorescent particles within circular intracellular structures (-PMA) (Fig. 3). Following stimulation with PMA, iPS-MG produce reactive oxygen species as evidenced by the oxidation and resulting green fluorescence of the CellROX® Green Reagent indicator dye over time (Fig. 3). The mean corrected fluorescence intensities of control cells in the phagocytosis and ROS experiments were 2,383,034.40 and 4,363,224.28 relative units, respectively. After the addition of the pHrodo E. Coli BioParticles or the CellRox reagents, the mean corrected fluorescence intensities of the cells at 7 mins were 18,489,482.66 and 5,104,373.38 relative units, respectively. Also, following 16 hours of stimulation with lipopolysaccharide (LPS), the concentration of TNFα in iPS-MG culture media was found to be 60 ± 7.6 pg/mL. The concentration of TNFα in iPS-MG culture media prior to LPS stimulation, and in fibroblast culture media before and after LPS stimulation was undetectable (detection limits: TNFα > 15.6 pg/ml).
Figure 3. hiPSC-microglia demonstrate functional abilities consistent with human microglia.
Confocal imaging of hiPSC-MG reveals a noticeable increase in phagocytosis of pHrodo™ Red E. coli BioParticles (red fluorescence) without treatment with phorbol myristrate acetate (-PMA), however upon stimulation with PMA, they show production of reactive oxygen species (green fluorescence) starting at t=5 min. Hoechst 3342 (blue fluorescence) was used to visualize nuclei. The figure is representative of three independent experiments carried out in replicates (n=6). Scale bars, 50 μm.
Murine iPSCs differentiate into microglia-like cells via a hematopoietic progenitor-like intermediate
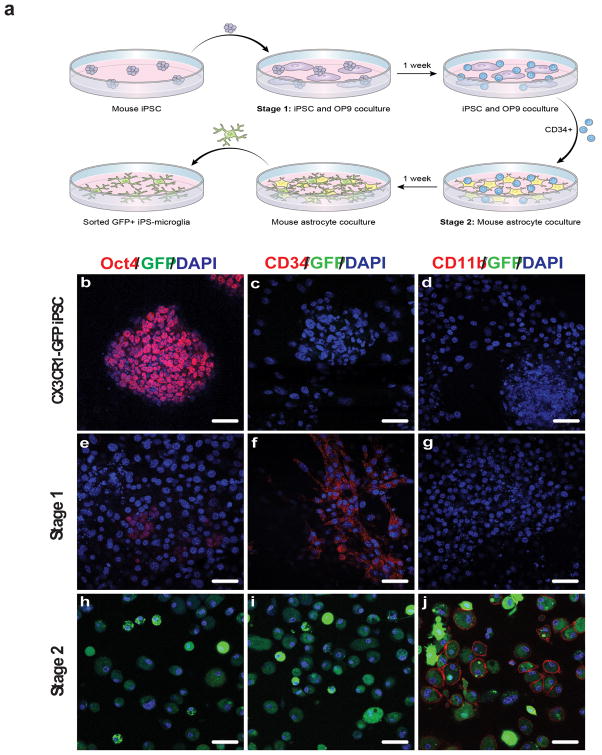
To demonstrate the potential therapeutic value of iPS-MG, murine iPS-MG were differentiated from murine iPSC using a two stage method analogous to that for human iPS-MG (Fig. 4a). Murine iPSCs generated by the lentiviral reprogramming of murine embryonic fibroblasts isolated from Cx3cr1gfp/+ knockin mice were used as a starting cell population. Expression of the fractalkine receptor CX3CR1 is a necessary, but not sufficient, criterion for defining murine microglia, and the microglia of Cx3cr1gfp/+ knockin mice express green fluorescent protein (GFP) under control of the Cx3cr1 promoter22. GFP expression by iPS-MG was used to facilitate their in vitro and in vivo identification.
Figure 4. Mouse iPSCs are differentiated into microglia using a two-stage process.
(a) Schematic of the murine iPSC differentiation process to microglia-like cells. (b–j) Cx3cr1gfp/+ miPSCs, Stage 1 hematopoietic progenitor-like cells, and Stage 2 miPS-MG immunostained with antibodies to Oct4 (b,e,h), CD34 (c,f,i), CD11b (d,g,j), and endogeneous CX3CR1-GFP expression (b–j). DAPI was used to visualize nuclei. Cells expressing the CX3CR1-GFP reporter (h,i,j) only became evident at the completion of Stage 2 culture. The figure is representative of three independent experiments done in replicates (n=6). Scale bars, 50 μm.
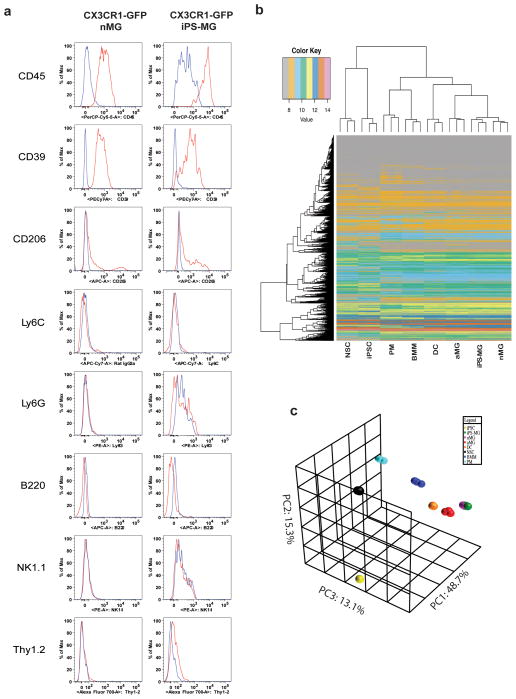
Cx3cr1gfp/+ iPSC express Oct4, but not CD34, CD11b or GFP (Fig. 4b–d). Following co-culture on OP9 murine stromal cell layers, cells are directed to an intermediate iPS-HPC (stage 1) state with loss of Oct4 expression and gain of CD34 expression (Fig. 4e,f). At stage 1, none of the co-cultured cells express either CD11b or GFP (Fig. 4g). Subsequent culture of iPS-HPC on astrocyte monolayers supplemented with GM-CSF, M-CSF, and IL-3 (stage 2) results in the appearance of GFP+ cells in as early as three days (Fig. 4h–j). These GFP+ cells continue to increase in number over the following 7 to 14 days and require only bi-weekly media changes for maintenance. GFP+ cells constitute ~4% of the mixed astrocyte-microglia cell culture (Supplementary Fig. 3). Loss of CD34 expression and gain of CD11b expression occur over this time period as well, consistent with the generation of murine iPS-MG (Fig. 4i,j). On average, for every 1 × 106 murine iPSC, the yield of microglia-like cells is 3 × 106. The iPS-MG also express CD39 and CD45, but not Ly6C, Ly6G, B220, Thy1.2 and NK1.1, a phenotypic profile that closely resembles that of neonatal brain-isolated microglia (nMG) (Fig. 5a)18,22. The yields following the differentiation of adult fibroblast derived iPSC are equivalent to that of embryonic fibroblast derived iPSC.
Figure 5. miPS-microglia display phenotypic markers and gene expression consistent with primary neonatal microglia.
(a) Flow cytometric analysis of neonatal microglia (nMG) and iPS-MG demonstrates comparable levels of cell-surface markers. The panels for each cell type were gated on live GFP+ cells. Red, primary antibody. Blue, Isotype. The figure is representative of two independent experiments (n=2). (b) Heat map examining expression levels and relationships of all genes post-noise analysis between neonatal primary microglia (nMG), iPS-MG, adult primary microglia (aMG), dendritic cells (DC), bone marrow macrophages (BMM), peritoneal macrophaes (PM), mouse Cx3cr1gfp+/−-iPSC (iPSC), and neural stem cell (NSC). (c) Three-Dimensional Principal Component Analysis (PCA) of the microarray data. iPS-MG total RNA was compared with that of other cells types as listed above. Expression levels for each class were averaged from three independent cell culture or differentiation experiments (n=3).
The gene expression signatures of murine iPSC, iPS-MG, nMG, adult brain-isolated microglia (aMG), bone-marrow derived macrophages (BMM), peritoneal macrophages (PM) and dendritic cells (DC) were compared. Also included in the analyses were nestin+ neural stem cells (NSC) given previous reports of the differentiation of ES cells into microglia using a neuronal differentiation protocol23. 35,556 gene fragment expression values were generated per biological triplicate sample and the 20% of data found to be noise-biased was discarded. After subjecting the data to one-factor ANOVA under Benjamini and Hochberg false discovery rate multiple comparison correction conditions, we restricted our analysis to 26,842 gene fragments with a corrected p-value <0.05. Using Tukey’s Post-Hoc analysis, we then quantified the number of these genes having both a p-value < 0.05 and an absolute fold-change in expression ≥ 1.5 in pairwise comparisons of all the experimental groups resulting in 20,232 genes. The results of this a posteriori analysis indicate that the gene expression patterns of iPS-MG most closely resemble that of nMG. The next-closest resemblance is to that of aMG. The cells which most differ from iPS-MG in terms of gene expression are iPSCs and NSC. These gene expression relationships can be visualized using correlation-based clustering (Fig. 5b) and covariance-based principal component analyses (Fig. 5c).
Murine iPS-MG demonstrate in vitro and in vivo functional properties of native microglia
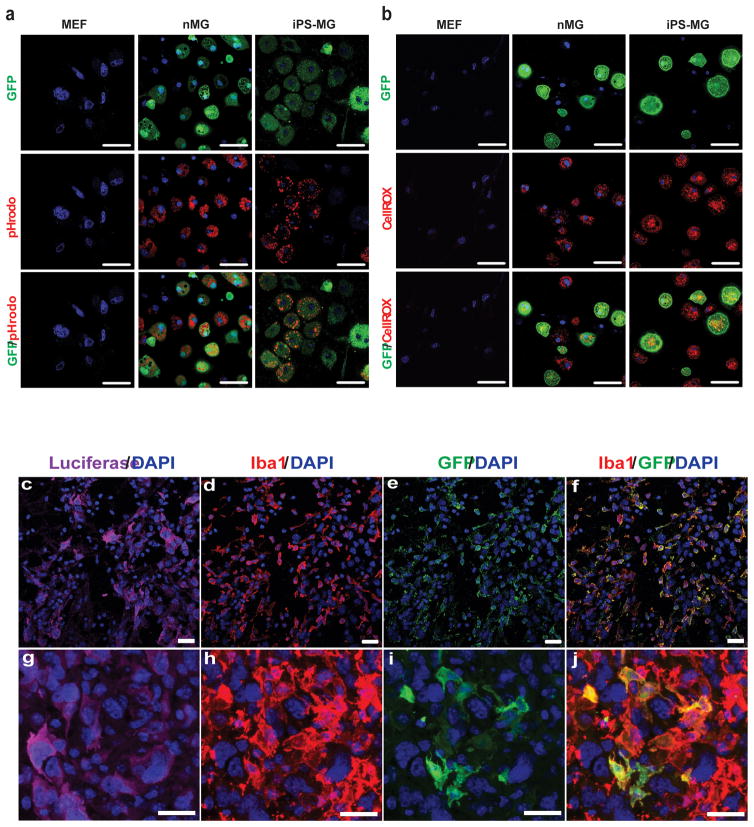
In addition to their phenotypic and gene expression resemblance to nMG, murine iPS-MG also demonstrate the in vitro functional properties of nMG. Following stimulation with PMA, both cell types actively phagocytose pHrodo E. Coli BioParticles conjugates as evidenced by the acidification and resulting visualization of red fluorescent particles within circular intracellular structures (Fig. 6a). The mean cell fluorescence intensities of nMG and iPS-MG after the addition of the pHrodo E. Coli BioParticles were qualitatively similar at 140,166.72 and 152,096.58 relative units, respectively. Under baseline culture conditions, both nMG and iPS-MG generate ROS, as evidenced by the oxidation and resulting red fluorescence of the CellROX® Deep Red Reagent indicator dye within cells (Fig. 6b). The mean cell fluorescence intensities after the addition of the CellRox reagent were 338,876.45 relative units for nMG and 235,213.10 relative units for iPS-MG. Control fibroblasts neither phagocytose foreign particles nor produce ROS under the same conditions (Fig. 6a, b).
Figure 6. miPSC-microglia demonstrate functional abilities of neonatal microglia.
(a) Confocal imaging of GFP+ iPS-MG, nMG and control mouse embryonic fibroblasts (MEF) treated with the phagocytosis reagent pHrodo™ Red E. coli BioParticles after stimulation with phorbol myristrate acetate (PMA). Hoescht 33342 was used to visualize nuclei. The figure is representative of three independent experiments each performed in replicates (n = 6). Scale bars, 50 μm. (b) Confocal imaging of GFP+ iPS-MG and nMG, as well as mouse embryonic fibroblasts treated with reactive oxygen species reagent CellROX® Deep Red Reagent. Hoescht 33342 was used to visualize nuclei. The figure is representative of three independent experiments performed in replicates (n=6). Scale bars, 50 μm. (c–j) miPS-microglia engraft in brain tissue and migrate to sites of pathology. Brain sections from C57BL/6 mice injected in the right hemisphere with luciferase-expressing GL261 tumor cells, followed by injection of iPS-MG into the contralateral cortex. The sections were stained for luciferase (c,g), Iba1 (d,h), endogenous CX3CR1-GFP production (e,i), or Iba1 and endogenous GFP (f,j). DAPI was used to visualize nuclei. Sections viewed under 40x (c–f) and 63x (g–j) magnification show iPS-MG to be evident at the periphery of the tumor. The figure is representative of two independent experiments performed in replicate (n=4). Scale bars, 20 μm.
Migration to sites of CNS pathology such as brain tumors is an in vivo functional hallmark of microglia24. The ability of murine iPS-MG to migrate to and infiltrate experimental gliomas in mice was therefore assessed. One week following the intracranial implantation of syngeneic, luciferase-expressing malignant glioma cells, iPS-MG were injected into the cerebral hemisphere contralateral to the tumor site. Brains were immunohistochemically analyzed one week later, and luciferase-expressing tumor cells were found in areas of high nuclear density (Fig. 6c, g). There was an expected abundance of infiltrating Iba-1+ microglia in these areas (Fig. 6d, h) and while many of them were GFP− and represent native microglia (Fig. 6e, i), many were also Iba-1+/GFP+ and represent the iPS-MG cells previously injected into the contralateral hemisphere (Fig. 6f, j). These results demonstrate the ability of iPS-MG to engraft within an unconditioned brain and migrate from one hemisphere to the other in response to a tumor.
Murine iPS-MG increase the survival of intracranial malignant glioma-bearing mice
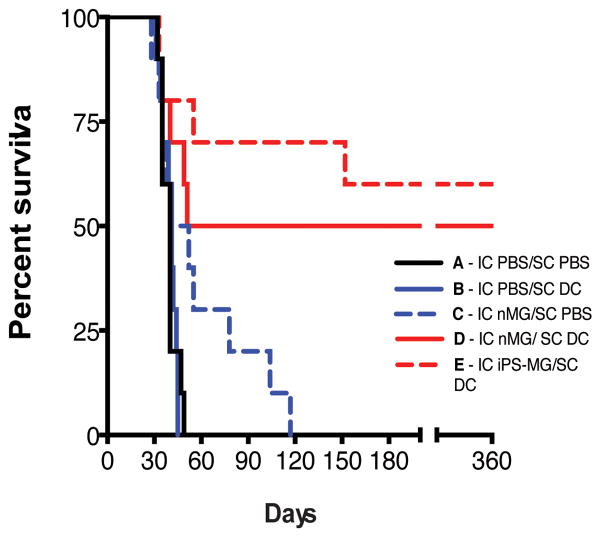
The ability of native microglia to phagocytose tumor cell debris, load antigens onto major histocompatibility class II (MHC II) molecules, and display co-stimulatory molecules such as B7-1 and B7-2 suggest their ability to present tumor antigens in vivo25,26. Additionally, microglia are capable of directly killing tumor cells by producing nitric oxide, reactive oxygen species, and TNF-α4. As a potential clinical application of iPS-MG, we assessed the ability of intracranially injected nMG or iPS-MG, in combination with subcutaneously injected dendritic cells (DC)27, to prolong the survival of malignant glioma-bearing mice. C57BL/6 mice were intracranially implanted with syngeneic GL261 malignant glioma tumor cells expressing luciferase to facilitate the in vivo imaging of tumor size and growth. One week following implantation, animals were imaged to confirm tumor engraftment and randomized into five groups. Each control or treatment group was then injected intracranially (IC) and subcutaneously (SC) with different combinations of vehicle control (phosphate buffered saline - PBS) or cells (DC, nMG or iPS-MG) pulsed in vitro with tumor cell lysates as shown in Supplementary Table 1.
The median survival of the untreated control group (Group A - IC and SC injection of PBS) was 40 days while that of the SC DC treated group (Group B) and IC nMG treated group (Group C) were 41 days (P = 0.859, log-rank test) and 46 days (P = 0.086, log-rank test), respectively. The combination IC nMG/SC DC treated group (Group D) had a median survival of 205.5 days (P = 0.008, log-rank test, df=1, χ2= 6.983) while the median survival of the IC iPS-MG /SC DC (Group E) group was undefined as more than 50% of the animals were alive at the end of the one year observation period (P = 0.001, log-rank test, df=1, χ2= 10.22). P-values are in comparison to Group A (Supplementary Table 1 and Fig. 7). Treatment of animals with cells pulsed in vitro with murine embryonic fibroblast lysates prior to injection did not prolong survival and demonstrates the antigen specificity of the anti-tumor treatments (data not shown).
Figure 7. Murine nMG and iPS-MG prolong the survival of tumor bearing animals.
Kaplan-Meier survival plot of intracranial tumor bearing mice treated with intracranial (IC) injections of PBS, neonatal microglia (nMG) or iPSC derived microglia (iPS-MG) and subcutaneous (SC) injections of PBS or dendritic cells (DC) as indicated in Supplementary Table 1. Statistical analysis for the survival curves was carried out using the log-rank test. A p value of < 0.05 was considered statistically significant. P-values are in comparison to Group A. For the combination IC nMG/SC DC treated group (Group D): P = 0.008, log-rank test, df=1, χ2= 6.983; while for the IC iPS-MG /SC DC (Group E): P = 0.001, log-rank test, df=1, χ2= 10.22. (n=10 animals for each group).
DISCUSSION
Experimental mouse models of human diseases in which microglia have been genetically modified or ablated have been especially useful in demonstrating the beneficial and detrimental effects of microglia on the pathogenesis of a variety of brain and spinal cord disorders. A major obstacle to the further laboratory study and clinical translation of these findings has been the lack of an abundant source of normal and disease-specific human microglia. One partial solution to overcoming this obstacle has been the differentiation of ES cells into microglia-like cells using a modified neuronal differentiation protocol. ES cells can be readily expanded to large numbers prior to differentiation and mouse ES cell-derived microglia are capable of engrafting in the unconditioned CNS23,28–30. While these protocols represent a significant advance in the study of microglia, they require several weeks to complete and do not mirror the normal microglial developmental process. In contrast to neurons and macroglia, microglia are of mesodermal rather than neuroectodermal origin and arise from embryonic yolk sac myeloid precursors that travel to the brain and spinal cord before differentiating into mature, functional cells within the CNS31–33. An analogous protocol for the differentiation of human ES cells has not been reported and even if one were to be developed, the translation of ES cell-derived microglia to the clinical therapeutic setting would likely be constrained by the unavailability of patient-specific ES cells as a starting population, histocompatibility issues and ethical concerns. To address these shortcomings, a protocol for the differentiation of human iPSC into microglia-like cells was developed. Human iPSC can be readily generated from normal and disease-specific individuals and also expanded to large numbers in vitro. Additionally, a protocol utilizing a myeloid rather than neural cell intermediate state was purposely designed to be more consistent with the in vivo developmental lineage of microglia.
Our results above demonstrate the differentiation of both human and mouse iPSC into cells with not only the phenotype and consensus gene expression signature, but also the in vitro and in vivo functional properties, of primary brain-isolated microglia. In the correlation-based clustering analysis, the gene expression patterns of the murine iPS-MG most closely resemble that of neonatal microglia and next closely that of adult microglia. In contrast, in the human analysis, the human iPS-MG do not cluster as tightly with the commercially obtained human fetal microglia. Extraction of the individual gene expression data for the six consensus human microglia genes (P2RY12, GPR34, MERTK, C1QA, PROS1, and GAS6) from the microarray dataset does however demonstrate the high expression of all six genes in the iPS-MG, but not in the iPSC, Mac, DC or the commercially obtained human fetal microglia. While there are robust protocols for the isolation of murine microglia, similar protocols and source tissue for the isolation of authentic human microglia are currently lacking. This underscores the need for protocols such as ours.
Very recently, another protocol for the differentiation of human iPSC and ES cells into microglia-like cells was reported34. Similar to ours, cells are differentiated through a myeloid rather than neural intermediate and the final yields of microglia-like cells from a given number of starting iPSC are similar. The major advantage of the other method appears to be the use of defined factors rather than co-culture with astrocytes to induce final differentiation. The use of astrocytes derived from the same starting iPSC as that used to make the microglia-like cells, would, however avoid the introduction of any allogeneic cells into the differentiation process, an important consideration for potential clinical uses. An apparent advantage of our astrocyte co-culture protocol compared to the defined factor protocol is the use of starting iPSC propagated under serum and feeder free conditions rather than those grown on a feeder layer of murine embryonic fibroblasts. The avoidance of any xenogeneic cells in the differentiation process will facilitate the future clinical use of the human microglia-like cells. An additional difference in the two protocols is the time required for the differentiation of the iPSC into microglia-like cells. Whereas our mouse and human protocols require 2 and 4 weeks, respectively, the defined factor protocol requires 8 weeks for both species. Finally, the microglia-like behavior of the cells generated using the defined factor protocol was demonstrated using an elegant in vitro 3D organotypic neuroglial environment. In contrast, we demonstrate the engraftment, migration and potential clinical use of our microglia-like cells in an in vivo model that does not require myeloablative conditioning of the host animal with lethal irradiation.
The potential applications of normal and disease-specific human iPS-MG include their use in disease modeling, drug discovery studies and toxicity screening assays35. Genetic mutations of microglia-expressed genes have been associated with the pathogenesis and prognosis of a variety of human diseases. Microglia derived from the iPSC of patients with such diseases may allow for the direct study of the roles of those gene products in disease development and progression. One such example is superoxide dismutase-1 (SOD1), a mutation of which is implicated in amyotrophic lateral sclerosis (ALS), a neurodegenerative disorder affecting adult motor neurons. Over 100 different SOD1 mutations have been identified and the role of these mutations in human motor neuron and microglial function could be modelled using normal and disease-specific iPS-MG36. iPS-MG could also be used in high throughput screens of SOD1 active drugs and microglia-specific toxicity assays. Another potential application of human iPS-MG is their use in personalized medicine applications. The murine iPS-MG generated here are able to engraft within an unconditioned brain and migrate to an area of pathology. Furthermore, the murine iPS-MG, in combination with DC, are an effective cell therapy for intracranial malignant glioma tumors. iPS-MG could also potentially be used for the treatment of diseases such as ALS, or as a delivery vector for gene based therapies of diseases such as multiple sclerosis or Parkinson’s disease16,17,28,36.
ONLINE METHODS
Mouse Strains
C57BL/6NCr mice were obtained from NCI-Frederick. Cx3cr1gfp/gfp knockin mice were obtained from the Jackson Laboratory. Heterozygous Cx3cr1gfp/+ knockin mice were generated by breeding Cx3cr1gfp/gfp mice to C57BL/6NCr mice.
Creation of Cx3cr1gfp/+ miPSC and human iPSC cell lines
Fibroblasts isolated from embryonic day 13 Cx3cr1gfp/+ knockin mice were reprogrammed to the iPSC state using the Dox-inducible polycistronic m4F2A lentivirus kit (ST000043; Stemgent) according to the manufacturer’s instructions. Confirmation of the iPSC state was obtained using alkaline phosphatase staining, immunocytochemistry staining for Oct4, Sox2, and SSEA-1, teratoma formation, and chimera formation following blastocyst injection (Supplementary Figs. 4a–e). Karyotype analysis revealed the cells to be 40,XY (Supplementary Fig. 4f). All animal usage was approved by the NINDS/NIDCD Animal Care and Use Committee. Human iPSC NCRM-5 was established from human cord blood-derived CD34+ cells using episomal plasmids (Lonza contract) made available to NIH Center for Regenerative Medicine and currently available through RUCDR Infinite Biologics. The iNC-01 cell line was obtained from Dr. Harry L. Malech’s laboratory of Host Defenses and Research Technologies Branch, NIAID, NIH. The iNC-01 cells were reprogrammed from human peripheral blood hematopoietic stem/progenitor cells (CD34+) cells that had been mobilized into the donor’s peripheral blood by G-CSF treatment. The reprogramming method involved transduction with integrating STEMCCA-loxP lentivirus expressing human Oct4/Klf4/Sox2/c-myc from a single transcript, followed by Cre-mediated excision of the vector37. Authentication and characterization of the cell line as iPSC arising from the healthy donor included38, TRA-1-60 and alkaline phosphatase staining for pluripotency markers, 3-germ layer characterization by teratoma formation in immunodeficient NSG mice followed by H&E staining and histology characterization, 3-germ layer characterization by embryoid body differentiation and immunofluorescence microscopy after staining for smooth muscle actin (mesoderm), α-fetoprotein (endoderm), and class III b-tubulin (ectoderm). Additional characterization included DNA fingerprinting to confirm that the origin of the cells matched the donor blood sample by DNA microsatellite analysis at 7 loci in comparison with PBMCs derived from that patient using the AmpFLSTR COfiler kit (Applied Biosystems), and genome-wide DNA methylation analysis at >485,000 CpG methylation sites in comparison with embryonic stem cells, other iPSC lines, and primary hematopoietic cells using Infinium HumanMethylation450 BeadChip (Illumina). The iNC-01 iPSC line was further characterized molecularly for additional pluripotency markers by global proteome analysis using electrospray ionization mass spectrometry (ESI-MS) and MALDI-TOF/TOF mass spectrometry in comparison with embryonic stem cells, other iPSC lines, and somatic cells38 and by expression analysis of 754 microRNAs using the TaqMan Human MicroRNA Array A+B v3.0.
Cell culture
Cx3cr1gfp/+ iPSC were maintained on a mitotically inactivated C57BL/6 background MEF feeder layer (Globalstem) using Knockout DMEM with 10% ES-qualified fetal bovine serum, non-essential amino acids, sodium pyruvate, 2-mercaptoethanol, penicillin/streptomycin (all from Gibco), and leukemia inhibitory factor (LIF) (Millipore). iPSCs were passaged onto fresh MEF layers every three days. Primary mouse astrocytes were isolated from postnatal day 5 C57BL/6 mice using an Astrocyte Isolation Starter Kit (Miltenyi) as per the manufacturer’s instructions and maintained in animal astrocyte media (Lonza), supplemented with astrocyte growth supplement (Lonza), fetal bovine serum, and penicillin/streptomycin as per the manufacturer’s instructions. Media was changed every other day and astrocytes were passaged at a 1:5 ratio upon reaching confluency. OP9 stromal cells (CRL-2749, ATCC) were maintained in Alpha-MEM media (Gibco) supplemented with 20% fetal bovine serum and penicillin/streptomycin on 0.1% gelatin-coated tissue culture treated dishes (Sigma). Media was changed twice weekly, and OP9 cells were passaged upon reaching confluency. Normal nMG were isolated from postnatal day 2 brains using the Neural Tissue Dissociation Kit and CD11b MicroBeads (Miltenyi) according to manufacturer’s directions and cultured in DMEM (Life Technologies) with 10% fetal bovine serum, Glutamax, non-essential amino acids, sodium pyruvate, 2-mercaptoethanol (Gibco), and sodium bicarbonate (Sigma). Microglia were plated on six-well temperature-responsive tissue culture plates (Upcell) at 2e6 cells per well in 2 mL of media. Adult murine microglia were purified from mixed glial cultures. Dendritic cells and bone marrow-derived macrophages were isolated following culture of whole bone marrow with recombinant murine IL-4 (RND Systems) and GM-CSF (Peprotech), or M-CSF (Peprotech) containing media, respectively. Peritoneal macrophages were harvested by saline lavage. Neural stem cells were purchased from Millipore. Firefly luciferase-expressing GL261 cells (National Cancer Institute) were cultured in Neurobasal media (Gibco), supplemented with N2 and B27 additives (Gibco), Glutamax, and penicillin-streptomycin. NCRM-5 and iNC-01 hiPSCs were maintained as feeder-free on matrigel (BD)–coated 6-well plates and split every 3 days. The cells were split 1:6 or 1:12 using EDTA (0.5mM in Ca2+ and Mg2+ free PBS with 1.8mg/ml NaCl). Essential 8 (Invitrogen) medium was utilized for growth and maintenance of NCRM-5 hiPSC, and maintained in humidified 37°C incubator with 5% CO2 and 21% O239. mTESR (StemCell Technologies) media was used for iNC-01 cells. The cell lines were regularly tested for mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (Lonza).
Microglial Differentiation
Human iPSC myeloid differentiation on OP9 feeder layers
NCRM-5 human iPSCs were harvested from a well of a 6 well plate using EDTA according to Chen et al.39 and resuspended in OP9 differentiation medium (ODM - MEM α with 20% FBS and 100 μM MTG). All steps were cultured in a humidified 37°C incubator with 5% CO2 and 21% O2. On day 0, iPSC aggregates were added to a confluent OP9 (ATCC) plate. On day 1, medium was removed, and 20 ml of fresh ODM was added to the plate. On day 4 and 6, half of the medium was replaced with fresh medium. On day 9, cells were harvested using 5 ml of collagenase solution (1 mg/ml) by incubating it for 30 min at 37°C (Life technologies). Next, collagenase was aspirated, and 5 ml of trypsin/EDTA solution (0.05%/0.5 mM) was added, and incubated for 15 min at 37°C. Cell suspension was collected, and MACS sort of CD34+/CD43+ cells was carried out according to Miltenyi manufacturer’s directions. Subsequently, pellet from the previous step was resuspended using myeloid progenitor expansion medium [ODM with 200 ng/ml GM-CSF (Berlex)] at a concentration of ~5.0 × 104 cells/ml. pHEMA-coated T75 flasks were washed with 20 ml PBS and 10–20 ml of CD34+ cell suspension was added, and incubated for 2 days. On day 11, expanded CD34+ cells were centrifuged and resuspended in astrocyte differentiation medium [ADM - IMDM with 10% FBS, 20 ng/ml GM-CSF, 20 ng/ml M-CSF (Peprotech) and 20 ng/ml IL3 (Peprotech)]. Expanded CD34+ cells were plated onto confluent 10 cm plates of human astrocytes at a density of 1–2 × 105 cells/10 ml/plate. On day 15, 10 ml of ADM was added. On days 18–25, media was aspirated and plates were harvested using 5 ml of trypsin/EDTA and incubated for 5–10 min. Subsequently, cell suspension from plate were washed, and MACS sorted for CD11b+ cells according to Miltenyi manufacturer’s directions. The CD11b+ cells were collected and resuspended using ADM, and used for further characterization, expansion or used in experiments as desired.
Feeder-free differentiation of human iPSC-microglia on astrocytes
iNC-01 hiPSC were cultured feeder-free in mTESR1 media (Stemcell technologies). hiPSCs were passaged when 70% confluent using PBS/EDTA/NaCl to loosen colonies from the plate, and replated onto a matrigel-coated plate at the ratio of 1:5 to 1:8. The cultures were further incubated for 2 days at 37°C, 5% CO2, 5% 02. Medium was changed to fresh mTeSR1 on day -1. On Day 0, STEMdiff APEL medium (Stemcell technologies) was pre-warmed to RT (3 ml per well of a 6 well plate). Media A was prepared by adding the desired growth factors just before use. Media A consisted of Stemdiff APEL medium (Stemcell technologies), 30 ng/ml hVEGF (Peprotech), 30 ng/ml hBMP4 (Peprotech), 40 ng/ml hSCF (Peprotech), and 50 ng/ml hActivin A (Invitrogen). mTESR1 medium was aspirated from iPSC culture wells and replaced with freshly prepared Media A. The cultures were further incubated for 4 days at 37°C, 5% CO2, 5% 02. No further media changes were carried out during this time. On Days 4, 7 and 10, media A was removed and replaced with media B, and incubated for 3 days at 37°C, 5% CO2, 5% 02. Media B was prepared by adding the growth factors to Stemdiff APEL medium (3 ml per well), 300 ng/ml hSCF (Peprotech), 300 ng/ml hFlt3L (Peprotech), 10 ng/ml hIL-3 (Peprotech), 10 ng/ml hIL-6 (Peprotech), 50 ng/ml hG-CSF (Peprotech), 25 ng/ml hBMP4 (Peprotech). Cells were harvested on Day 15 to analyze for hematopoietic differentiation using 1 ml. of accutase (Innovative Cell Technologies) per well. Cells from 3-wells of a 6-well plate culture were analyzed for CD34, CD45 and CD43 expression by FACs, and other half were plated onto one 10-cm dish of human astrocyte in 10 ml. of media C [500 ml. IMDM (Gibco), 10% defined FBS (Gibco), 5 ml. Penicillin/Streptomycin (Gibco)], 20 ng/ml. of hIL3 (Peprotech), 20 ng/ml of hGMCSF (Peprotech) and 20 ng/ml of hM-CSF (Peprotech). The astrocyte-hematopoietic progenitor co-culture plates were further incubated at 37°C, 5% CO2, for 1 to 2 weeks for differentiation to microglia. CD39+ microglia was isolated using MACS (Miltenyi).
Mouse iPSC-microglia differentiation on OP9 feeder layers
Cx3cr1gfp/+ iPSC were differentiated into microglia using a two-stage protocol. In the first stage, a modified protocol for the hematopoietic differentiation of iPSC40, Cx3cr1gfp/+ iPSC colonies were dissociated into a single-cell suspension using 1 mg/mL collagenase (Invitrogen) and Accutase (Stemcell Technologies) and plated at a density of 1.5e6 cells per 15-cm tissue culture dish of confluent OP9 stromal cells. This Stage 1 culture was maintained in alpha-MEM with 10% defined FBS (Hyclone), 2-mercaptoethanol, and penicillin/streptomycin for seven days. On day four, the media was fully replaced. After seven days of Stage 1 culture, cells were harvested by dissociation with trypsin (Gibco) and transferred onto a confluent astrocyte monolayer for Stage 2 culture. Stage 2 cultures were maintained in DMEM/F12 media (Gibco) with 10% defined FBS, 2 mM 2-mercaptoethanol, 2 mM Glutamax (Gibco), non-essential amino acids (Gibco), 20 ng/mL recombinant murine GM-CSF (Peprotech), 20 ng/mL recombinant murine M-CSF (Peprotech), 20 ng/mL recombinant murine IL-3 (Peprotech), and penicillin/streptomycin. Media in Stage 2 cultures was replaced twice weekly. Purification of iPS-MG from Stage 2 cultures was carried out using anti-CD11b or anti-CD39 magnetic beads (Miltenyi) according to manufacturer’s instructions followed by optional FACS sorting of iPS-MG by gating on GFP+ cells.
Immunofluorescence
iPSC cultures, Stage 1 and Stage 2 cultures were established in 8-well permanox-coated chamber slides (Thermo Fisher). Cells were fixed with 4% paraformaldehyde, blocked with Background Buster (Innovex Biosciences) and incubated with primary antibodies overnight at 4°C. Cells were washed with PBS and incubated with the appropriate secondary antibodies for 1 hour at RT. For the hiPSC, rabbit anti-human Nanog (Stemgent, Cat # 09-0020) and mouse anti-human Tra-1-81 (1:100) (Stemgent, Cat # 09-0011) was used as primary antibodies and anti-rabbit alexa fluor 546 and anti-mouse alexa fluor 488 were used as the secondary antibodies (Molecular Probes). For the stage 1, differentiated cells were stained with rabbit anti-human CD34 (abcam, Cat # ab81289) and APC-conjugated-mouse anti-human CD43 (BD, Cat # ab81289, Clone # 560198), and the signal for CD34 was detected using anti-rabbit alexa fluor 488 (Invitrogen, Cat # A-11008). For stage 2, microglia-like cells were detected using the mouse anti-human CD11b (abcam, Cat # ab91145) and rabbit anti-human Iba1 (Wako, Cat # 019-19741). Secondary antibodies used in this case were anti-mouse alexa fluor 546 (Invitrogen, Cat # A-11003) and anti-rabbit alexa fluor 488. For mouse cells, the following primary/secondary antibody combinations were used: rabbit anti-mouse Oct4 polyclonal (Stemgent, Cat # 09-0023)/anti-rabbit AlexaFluor 546 antibody (Invitrogen, Cat # A10040); rat mAb CD34 mAb [MEC 14.7] (abcam, Cat # ab8158)/ anti-rat AlexaFluor 647 (Invitrogen, Cat # A-21247) antibody; anti-mouse CD11b mAb directly conjugated to AlexaFluor 647 (BD Biosciences, Cat # 557686, clone # M1/70)/secondary not needed. All primary antibodies were used at a 1:100 dilution in PBS with 0.1% BSA, while secondary antibodies were used at a 1:200 dilution in the same buffer. Following staining, the slides were mounted using the Vectashield mounting medium with DAPI (Vector labs) and imaged using the Zeiss LSM 510 confocal microscope. All isotype control primary mAb stained cells lacked fluorescence labeling. All the antibodies used are commercially validated antibodies.
FACS analyses
CD11b+-sorted cells were collected from the astrocyte feeder cells using MACs (Miltenyi). DAPI was used to determine LIVE/DEAD cells. Next, the cells were washed and stained with primary antibodies such as Ms anti-human anti-CD11b (1:200) (abcam, Cat # ab91145), and HLA-DR (1:200) (abcam, Cat # [CR3/43] ab7856), rabbit pAB anti-CD45 (1:200) (abcam, ab10558), mouse mAB anti-CD86 (1:200) (abcam, Cat # [BU63] ab23556), rabbit anti-human Iba1 (1:100) (Wako, Cat # 019-19741), CD80 (1:200) (abcam), rabbit pAB CX3CR1 (1:200) (abcam, Cat # ab8021), goat pAb TREM-2 (1:200) (abcam, Cat # ab85851) antibodies, and their corresponding isotype controls for 1 hr at 4°C. After incubation, the cells were washed and stained with corresponding secondary antibodies either tagged with corresponding Alexa fluor 488 or Alexa fluor 546 for 45 min on ice, and subsequently the signals were analyzed using Moflo Astrios Cell Sorter (Beckman Coulter, Inc). For mouse iPS-MG, CD11b+-sorted cells were collected from the astrocyte feeder cells using MACs (Miltenyi). Subsequently, the cells were incubated with Fc receptor block consisting of Rat anti-mouse CD16/32 (1:100) (BD Pharmingen, Cat # 553142) and Mouse IgG (1:50) (Jackson Immunoresearch) for 15 min at 4°C. Staining was carried out for the dead cells using LIVE/DEAD® Fixable Blue Dead Cell Stain Kit for UV excitation (Molecular Probes) for 30 min at 4°C. Next, the cells were washed and stained with fluorochrome-conjugated Rat anti-mouse Ly6C-APC-Cy7 (1:400) (Biolegend, Cat # 128026, clone # HK1.4), Rat anti-mouse Ly6G-PE (1:200) (BD Pharmingen, Cat # 551461, clone # 1A8), Rat anti-mouse CD45-PerCP-Cy5.5 (1:50) (BD, Cat # 550994, clone # 30-F11 ), Rat anti-mouse CD39-PE-Cy7 (1:200) (eBioscience, Cat # 25-0391-80, clone # 24DMS1), Rat anti-mouse Thy1.2-Alexa fluor 700 (1:1200) (Biolegend, Cat # 105320, clone # 30-H12), Anti-mouse NK1.1-PE (1:200) mAb (eBioscience, Cat # 12-5941-83, clone # PK136), Rat anti-mouse B220-APC (1:1000) (BD Pharmingen, Cat # 553092, clone # RA3-6B2) antibodies, and their corresponding isotype controls for 1 hr at 4°C. All antibodies used are commercially validated. After incubation, the cells were washed twice and fixed with 1% paraformaldehyde. The signals were analyzed using a LSR II flow-cytometer (BD).
Microarray hybridization
For human iPS-MG, total RNA was isolated from NCRM-5 and iNC-01 hiPSC lines, iPS-MG and human fetal microglia (Clonexpress) using miRNeasy kit (Qiagen). RNA from human dendritic and bone marrow macrophages (AllCells) was also used. Isolated total RNA was amplified with the WT Expression Kit (Ambion), and labeled with the GeneChip WT Terminal Labeling and Controls Kit (Affymetrix). Labeled cRNA were hybridized to the Affymetrix GeneChip Human Gene 2.0 ST Array (Affymetrix, Inc) in blinded interleaved fashion. The Affymetrix Gene Chip Scanner 3000 was used to scan each array and gene probe intensities were generated using the Affymetrix AGCC software (Affymetrix, Inc).
RNA was obtained from murine nMG, adult microglia, Cx3cr1gfp/+ iPSC, iPS-MG, peritoneal macrophages, bone marrow-derived macrophages, bone marrow-derived dendritic cells, and neural stem cells. RNA was extracted from cells using the miRNeasy mini kit (Qiagen), amplified with the WT Expression Kit (Ambion), and labeled with the GeneChip WT Terminal Labeling and Controls Kit (Affymetrix). Labeled cRNA were hybridized to the Affymetrix GeneChip Mouse Gene 1.0 ST Array (Affymetrix, Inc) in blinded interleaved fashion. The Affymetrix Gene Chip Scanner 3000 was used to scan each array and gene probe intensities were generated using the Affymetrix AGCC software (Affymetrix, Inc).
Microarray data analysis
Gene fragment data summarization and normalization was performed separately for mouse and human samples using the Partek Genomic Suite v2 (http://www.partek.com) with the “RMA” option selected under default settings. Subsequent analysis of these data were also performed separately for mouse and human while the analyses performed were the same. Data quality was assured via Tukey box plot, PCA scatter plot and correlation-based heat map using functions in “R” (http://cran.r-project.org/). Lowess modeling of the data (CV~mean expression) was used to characterize noise for the system and discard noise-biased data. Differential expression was tested for via ANOVA under BH correction condition followed by TukeyHSD post-hoc test. Gene fragments found to have a corrected p-value < 0.05 by ANOVA and a post-hoc p-value < 0.05 were deemed to have significant differential expression between the corresponding cell types if the absolute difference of means was >= 1.5X. Visualization of cell type relationships post testing was accomplished using the heat.map.2() functions supported in R and the 3D-PCA option supported in Partek.
Real-time PCR
Total RNA was isolated from human iPSC and human iPS-MG using miRNeasy kit (Qiagen). One μg of RNA was subsequently treated with amplification grade DNAse I (Life Technologies) to remove any contaminating genomic DNA. 250 ng of total RNA was used for cDNA synthesis using iScriptTM cDNA Synthesis Kit (Bio-Rad). All Q-PCRs were performed in 20 μl volume using SYBR Green PCR master mix (Applied Biosystems) in a 96-well optic tray using CFX96 Real-Time PCR machine (Bio-Rad). The reactions were conducted in duplicate, and samples without reverse transcriptase were used as a non-template control. Primers specific for the six microglial genes were obtained from Integrated DNA Technologies. Q-PCR reactions were run with an initial denaturation temperature of 95°C for 10 min, which was followed by 40 cycles of three-step amplification [denatuartion at 95°C for 30 s, ~60°C (dependent on the annealing temperature of the primer) annealing for 20s, and extension at 72°C for 20s]. The optimal annealing temperature for gene specific primers was determined prior to the experimental runs. The housekeeping gene beta-actin was used as a reference.
Phagocytosis assay
5e4 miPS-MG, nMG or MEFs were cultured for 48 hours on 4-well CC2 treated glass chamber slides (Thermo Fisher). Cells were stimulated for 15 minutes with 100 nM phorbol myristate acetate (PMA) diluted in dye-free MEM media (GIBCO) prior to the incubation of cells with pHrodo™ Red E. coli BioParticles® Conjugate (Life Technologies) for 1.5 hours at 37°C. Hoechst 33342 (Thermo Scientific) was used to stain the nuclei. Human-iPS-microglia were treated similarly however, they were not activated with PMA prior to incubation with the bioparticles. Live cell imaging was carried out using an LSM 510 confocal microscope using excitation/emission wavelengths of 560/585 nm (Zeiss). Nuclei were visualized with 20 μM Hoescht 33342. Corrected total cell fluorescence intensity for the confocal images was calculated using Image J.
Reactive oxygen species assay
5e4 miPS-MG, nMG or MEFs were cultured as above for the phagocytosis assay. Cells were then treated with 20 μM CellROX® Deep Red Reagent (Life Technologies) diluted in dye-free MEM media for 30 min at 37°C. Human iPS-MG were treated with PMA and treated with CellROX® Green Reagent (Life Technologies) dye. Nuclei were stained with Hoecst 33342 dye. Live cell imaging was performed at 640/645 nm using the Zeiss LSM 510 confocal microscope. Analysis of the confocal images for corrected total fluorescence intensity was carried out using Image J.
miPSC-MG engraftment studies
Animals were anesthetized and placed in a stereotaxic frame (Stoelting). Coordinates used for stereotactic injection were 3 mm right lateral and 1 mm posterior to bregma, with an injection depth of 3.5 mm. Male C57BL/6NCr 6-week old mice were injected with 3μL of a 5e4 cells/μL suspension of luciferase-expressing GL261 cells in dPBS, resulting in the implantation of 1.5e5 cells/animal. One week later, animals were injected 3 mm left lateral and 1 mm posterior to bregma, with an injection depth of 3.5 mm, with 3 μL of an 8.33e4 cells/μL suspension of miPS-MG in dPBS, resulting in the implantation of 2.5e5 cells/animal. One day later, animals were sacrificed and perfused with 4% paraformaldehyde. Brains were removed, post-fixed in 4% paraformaldehyde at 4°C for four hours, cryoprotected with 15% and 30% sucrose solutions, and cut into 8μm-thick tissue sections using a cryostat. The sections were stained with goat anti-luciferase (1:1000) (Fitzgerald, Cat # 70C-CR2029GAP), chicken anti-GFP (1:200) (abcam, Cat # ab13970), and rabbit anti-Iba1 (1:200) (Wako, Cat # 019-19741) primary antibodies, followed by donkey anti-goat AlexaFluor 647 (Invitrogen, 1:100), donkey anti-chicken AlexaFluor 488 (Jackson, 1:100), and donkey anti-rabbit AlexaFluor 555 (Invitrogen, 1:100) secondary antibodies, respectively. Images were taken using a Zeiss LSM 510 confocal microscope. Mice were housed 4 animals/cage and monitored regularly for signs of distress and clinical deterioration. The animals were on a 12 hour light and dark cycle. All animal studies were approved by the NINDS/NIDCD Institutional Animal Care and Use Committee.
Antigen loading and cell preparation
One day pre-injection, floating DCs were harvested and plated in six-well tissue culture plates at 2e6 cells per well in 2 mL of media. Both DCs and microglia were incubated with the antigen at a concentration of 100 μg/mL overnight. On the day of injection, cells were washed with PBS twice to remove the antigen, then detached from tissue culture plates, washed once more in PBS, and resuspended at a concentration of 8.33e7 cells/mL (IC microglia) or 1.25e6 cells/mL (SC dendritic cells).
Tumor implantation and treatment delivery of mice with mouse neonatal brain-isolated or mouse iPSC microglia
Tumors were implanted into 6 weeks old, male C57BL/6NCr mice using either a stereotaxic frame or a syringe-pump (Harvard Apparatus) with custom cannulated nylon bolt (Plastics One). Coordinates used for the injection were 3 mm right lateral and 1 mm posterior to bregma, with an injection depth of 3.5 mm. C57Bl/6 mice were anesthetized and injected with 3μL of a 5e4 cells/μL suspension of luciferase-expressing GL261 cells in dPBS, resulting in the implantation of 1.5e5 cells/animal. Five days after tumor implantation, animals underwent high-resolution bioluminescent imaging using the Xenogen IVIS imaging system to confirm tumor engraftment. Animals were block randomized by tumor size into experimental or control groups. Previous pilot experiments demonstrated a 20% three month survival rate for control animals and an 80% three month survival rate for treated animals. A power analysis anticipating similar results, equal numbers of animals per group, a two-tailed type I error rate of 0.05 and a type II error rate of 0.2 indicated the need for 10 animals per group. One week post tumor implantation, animals were treated with 1.5e5 cells in IC injections and 2.5e5 cells in SC injections or 5 μL of a 2e5 cells/μL suspension of iPS-MG (sorted for GFP+ cells) in dPBS, resulting in the implantation of 1e6 cells/animal. No repeat injections were administered and there was no immediate post-treatment mortality. Therefore, no animals were excluded from the analyses once randomized. Mice were housed 4 animals/cage in coded cages and were on a 12 hour light and 12 hour dark cycle. Animals were monitored daily in a blinded fashion regarding treatment and those showing signs of distress or clinical deterioration as defined in the experimental protocol or found dead were deemed expired. The experimental or control treatment received by the expired animal was subsequently identified. All animal studies were approved by the NINDS/NIDCD Institutional Animal Care and Use Committee.
Statistical analysis
Differential expression for the microarray experiments was tested for via ANOVA under BH correction condition followed by TukeyHSD post-hoc test. Gene fragments found to have a corrected P < 0.05 by ANOVA and a post-hoc P < 0.05 were deemed to have significant differential expression between the corresponding cell types if the absolute difference of means was >= 1.5X. Statistical analysis for the survival curves for animal treatment study was carried out using the log-rank test (Mantel-Cox test). A P value of < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health.
Footnotes
ACCESSION CODES
Data for both the mouse and human samples are available for download from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/); see SuperSeries GSE78116.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
DATA AVAILABILITY
Data for both the mouse and human samples are available for download from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/); see SuperSeries GSE78116. Also, additional data that support the findings of this study are available from the corresponding authors upon request.
AUTHOR CONTRIBUTIONS
J.K.P. conceived the project; H.P., M.J.S., D.M.I., Y.C. , D.B.M., and J.K.P. designed experiments; H.P., M.J.S., D.M.I., Y.C., A.B.S., G.K. and M.A.B. performed experiments; H.P., M.J.S., D.B.M., and J.K.P. analyzed the data; K.R.J. performed microarray data analysis; A.G.E., C.L.S. and S.G. provided technical support; H.L.M. provided conceptual advice; H.P., M.J.S., D.B.M. and J.K.P. wrote the manuscript.
References
- 1.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 2.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 3.Cardona AE, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang SY, et al. Induction of glioma apoptosis by microglia-secreted molecules: The role of nitric oxide and cathepsin B. Biochim Biophys Acta. 2009;1793:1656–1668. doi: 10.1016/j.bbamcr.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Mora R, et al. TNF-alpha- and TRAIL-resistant glioma cells undergo autophagy-dependent cell death induced by activated microglia. Glia. 2009;57:561–581. doi: 10.1002/glia.20785. [DOI] [PubMed] [Google Scholar]
- 8.Brantley EC, et al. Nitric oxide-mediated tumoricidal activity of murine microglial cells. Transl Oncol. 2010;3:380–388. doi: 10.1593/tlo.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chicoine MR, et al. The in vivo antitumoral effects of lipopolysaccharide against glioblastoma multiforme are mediated in part by Toll-like receptor 4. Neurosurgery. 2007;60:372–380. doi: 10.1227/01.NEU.0000249280.61761.2E. discussion 381. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar S, et al. Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat Neurosci. 2014;17:46–55. doi: 10.1038/nn.3597. [DOI] [PubMed] [Google Scholar]
- 11.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 12.Gandy S, Heppner FL. Microglia as dynamic and essential components of the amyloid hypothesis. Neuron. 2013;78:575–577. doi: 10.1016/j.neuron.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 14.Capotondo A, et al. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc Natl Acad Sci U S A. 2012;109:15018–15023. doi: 10.1073/pnas.1205858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen SK, et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–785. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biju K, et al. Macrophage-mediated GDNF delivery protects against dopaminergic neurodegeneration: a therapeutic strategy for Parkinson’s disease. Mol Ther. 2010;18:1536–1544. doi: 10.1038/mt.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biju KC, et al. Bone marrow-derived microglia-based neurturin delivery protects against dopaminergic neurodegeneration in a mouse model of Parkinson’s disease. Neurosci Lett. 2013;535:24–29. doi: 10.1016/j.neulet.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butovsky O, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122:3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sedgwick JD, Ford AL, Foulcher E, Airriess R. Central nervous system microglial cell activation and proliferation follows direct interaction with tissue-infiltrating T cell blasts. J Immunol. 1998;160:5320–5330. [PubMed] [Google Scholar]
- 20.Vodyanik MA, Thomson JA, Slukvin Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napoli I, Kierdorf K, Neumann H. Microglial precursors derived from mouse embryonic stem cells. Glia. 2009;57:1660–1671. doi: 10.1002/glia.20878. [DOI] [PubMed] [Google Scholar]
- 24.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2015;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang I, Han SJ, Kaur G, Crane C, Parsa AT. The role of microglia in central nervous system immunity and glioma immunology. J Clin Neurosci. 2010;17:6–10. doi: 10.1016/j.jocn.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain SF, et al. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8:261–279. doi: 10.1215/15228517-2006-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Gool S, et al. Dendritic cell therapy of high-grade gliomas. Brain Pathol. 2009;19:694–712. doi: 10.1111/j.1750-3639.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beutner C, et al. Engineered stem cell-derived microglia as therapeutic vehicle for experimental autoimmune encephalomyelitis. Gene Ther. 2013;20:797–806. doi: 10.1038/gt.2012.100. [DOI] [PubMed] [Google Scholar]
- 29.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. Generation of microglial cells from mouse embryonic stem cells. Nat Protoc. 2010;5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchiya T, et al. Characterization of microglia induced from mouse embryonic stem cells and their migration into the brain parenchyma. J Neuroimmunol. 2005;160:210–218. doi: 10.1016/j.jneuroim.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 31.Eglitis MA, Mezey E. Hematopoietic cells differentiate into both microglia and macroglia in the brains of adult mice. Proc Natl Acad Sci U S A. 1997;94:4080–4085. doi: 10.1073/pnas.94.8.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 34.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 36.Clement AM, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 37.Merling RK, et al. Transgene-free iPSCs generated from small volume peripheral blood nonmobilized CD34+ cells. Blood. 2013;121:e98–107. doi: 10.1182/blood-2012-03-420273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pripuzova NS, et al. Development of a protein marker panel for characterization of human induced pluripotent stem cells (hiPSCs) using global quantitative proteome analysis. Stem Cell Res. 2015;14:323–338. doi: 10.1016/j.scr.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senju S, et al. Characterization of dendritic cells and macrophages generated by directed differentiation from mouse induced pluripotent stem cells. Stem Cells. 2009;27:1021–1031. doi: 10.1002/stem.33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.