Abstract
Urine as a true non-invasive sampling source holds great potential for biomarker discovery. While approximately 2000 proteins can be detected by mass spectrometry in urine from healthy people, the amount of these proteins vary considerably. A systematic evaluation of a large number of samples is needed to determine the range of the variations. Current biomarker studies often measure limited number of urine samples in the discovery phase, which makes it difficult to determine whether proteins differentially expressed between control and disease groups represent actual difference, or are just physiological variations among the individuals, leads to failures in the validation phase with the increased sample numbers. Here, we report a streamlined workflow with capacity of measuring 8 urine proteomes per day at the coverage of > 1500 proteins. With this workflow, we evaluated variations in 497 urine proteomes from 167 healthy donors, establishing reference intervals (RIs) that covered urine protein variations. We demonstrated that RIs could be used to monitor physiological changes by detecting transient outlier proteins. Furthermore, we provided a RIs-based algorithm for biomarker discovery and validation to screen for diseases such as cancer. This study provided a proof-of-principle workflow for the use of urine proteome for health monitoring and disease screening.
Keywords: Reference intervals, Urine proteome, Cancer, Biomarker, Mass spectrometry
Highlights
-
•
Proteomics revealed that human urine proteomes were highly variable yet within a defined range.
-
•
Reference intervals of the human urine proteome were established from 500 samples in an international two-center setting.
-
•
Reference intervals were used to facilitate monitoring physiological and pathological changes, including cancer screening.
Urine was found to contain about 2000 proteins with great variability in abundance. The levels of urine proteins hold great potential for biomarker discovery in disease diagnosis, early detection and health monitoring. We developed a workflow to establish the reference interval (RI) of variation for each urine protein for healthy people, allowing us to find proteins that are out of the range and their association with changes in physiology and diseases including cancer. Our study paved a way to use urine proteins for health monitoring and disease screening in the future.
1. Introduction
Urine is a commonly used biological fluid for discovery of disease markers, diagnostics, and health status monitoring. Urine presents several distinct advantages over blood. For example, its sampling is truly non-invasive, therefore can be repeated frequently; the urine proteome is also simpler than the plasma proteome and more amenable to proteomic analysis (An and Gao, 2015, Shao et al., 2011b). Proteins in urine originate from glomerular filtration of plasma and secretion of urogenital system (Pisitkun et al., 2004, Pisitkun et al., 2006, Sun et al., 2005, Wang et al., 2006) and changes in urinary protein composition can reflect physiological and pathological status of the human body (Decramer et al., 2008, Wu and Gao, 2015).
Much effort has been made to characterize protein composition of urine using mass spectrometry (MS) during the last decade (Adachi et al., 2006, Kentsis et al., 2009, Khristenko et al., 2016, Marimuthu et al., 2011, Nagaraj and Mann, 2011, Sun et al., 2009, Thongboonkerd et al., 2002). Databases, such as Max-Planck Unified Proteome database (http://mapuproteome.com/) (Zhang et al., 2007), the Human Kidney and Urine Proteome Project (http://www.hkupp.org/) (Yamamoto et al., 2008), the Human Urinary Proteome Database (http://mosaiques-diagnostics.de/diapatpcms/mosaiquescms/front_content.php?idcat=257) (Coon et al., 2008), Urinary Protein Biomarker (UPB) database (http://www.mybiosoftware.com/upb-20130710-urine-protein-biomarker-database.html) (Shao et al., 2011a), and Urine Proteomics.org (http://urineproteomics.org/databases.html) (Kentsis et al., 2009), documented lists of urinary proteins, providing convenient resources for keeping track of published urine proteomes. However, none of these databases provided quantitative information about the urine proteins.
A number of clinical proteomics studies have generated a long list of candidate urine protein biomarkers of various diseases (Beretov et al., 2015, Goodison et al., 2009, Rodriguez-Suarez et al., 2014, Shao, 2015); however, no biomarkers derived from ‘discovery’ studies were successfully translated into clinical practice to influence patient care and management (Fuzery et al., 2013, Mischak et al., 2010). One reason for these translational failures was the small sample size included in the studies, which did not have enough statistical power to distinguish between the difference resulting from pathological changes and the physiological variations among different individuals (Rifai et al., 2006, Rodriguez-Suarez et al., 2014).
At present, proteomics studies have revealed that intra-personal variation of urine proteomes is substantial (Nagaraj and Mann, 2011); the variation is further compounded by normal geno-proteomic differences among individuals. However, due to the lack of systematic evaluation of variations in human urine proteomes based on large population or long time span of sampling, it is still not clear whether these variations reflect fluctuation within a certain range in healthy persons and further, whether it is feasible to establish a protein reference range for the human urine proteome.
In clinical diagnosis, a normal range for a laboratory test is customarily established by a reference interval (RI) for its distributions in the population. Measurements of hundreds to thousands of people are required to make reliable empirical estimates (CLSI, 2010, Thompson et al., 2004). Typically, 2.5th and 97.5th percentiles of RI, which cover 95% of reference population are used as lower and upper limits, where outlier values are thought to signal potential problems for the test subject (CLSI, 2010). This approach is effective and widely used in diagnostics. But the concept has not been adopted in proteomics, as it requires the measurement of large number of samples.
Here we report a streamlined workflow to measure urine proteome from high-speed sediment of urine at the level of > 1500 proteins within 3 h of MS running time. We measured 497 samples from 167 healthy donors, enabling us to evaluate day-to-day and inter-personal variations of the human urine proteome in a two-center setting. This dataset allowed us to establish intra-personal and pan-human RIs that covered variations of the human urine proteome. We presented examples using these RIs to identify outlier proteins that associated with physiological or pathological states, which might be used for health monitoring. Our study paved a way for biomarker discovery and validation for disease diagnosis and health monitoring by using urine proteome.
2. Materials and Methods
2.1. Sample Collection and Preparation
Midstream of the first-morning urine was obtained and stored at − 80 C. Informed consents were signed by all test subjects and the study was approved by the Institutional Review Boards, Baylor College of Medicine (BCM) and Beijing Proteome Research Center (BPRC), respectively. Research adhered to the standards indicated by the Declaration of Helsinki. We used 10–20 ml of urine samples to establish the method. After establishing the standard operation procedure (SOP), 20 ml was used in the remaining experiments. Twenty milliliters of urine samples were centrifuged at 200,000 g for 70 min to save the pellets. We used a previously described method (Pisitkun et al., 2004) with modifications to remove uromodulin (UMOD; GeneID 7369). Briefly, 400 μl of resuspension buffer (50 mM Tris, 250 mM sucrose, pH 8.5) and dithiotheitol (DTT) was added to the pellets to a final concentration of 50 mM and the suspension was then heated at 65 °C for 30 min. Then wash buffer (10 mM TEA, 100 mM NaCl, pH 7.4) was added and a second ultracentrifugation was carried out for 30 min. The sediments were dissolved in sodium dodecyl sulfate (SDS) buffer (1% SDS, 50 mM Tris, pH 8.5) and half of the samples were used for SDS-PAGE. Resolved proteins were visualized with Coomassie Brilliant Blue and 6 gel pieces were subjected to in-gel digestion by trypsin as previously described (Malovannaya et al., 2010). Sample metadata were summarized in Supplementary Table 1.
2.2. NanoHPLC-MS Analysis
The extracted peptides were re-suspended in 20 μl of loading solution (5 % methanol containing 0.1 % formic acid) and 5 μl was analyzed. Thermo Fisher Q Exactive and LTQ Orbitrap VelosPro coupled to nLC-1000 were used. A homemade trap column (2 cm × 75 μm) and an analytical column (10 cm × 75 μm), both packed with Reprosil-Pur Basic C18 (3 μm, Dr. Maisch GmbH, Germany) were used. A 75 min gradient of 5–28 % acetonitrile at a flow rate of 400 nl/min was used for on-line HPLC-MS.
For Q Exactive, the full MS scan range was set to 375–1300 m/z and trap size for MS1 and MS2 were 3 × 106 and 2 × 105, respectively. The mass resolution for MS1 and MS2 were 140,000 and 17,500 respectively. The top 25 ions were selected for higher energy collision dissociation (HCD) with collision energy set at 27. For LTQ Orbitrap VelosPro, the full MS scan range was set to 375–1300 m/z and trap size for MS1 and MS2 were 3 × 106 and 3 × 104, respectively. The mass resolution for MS1 was 100,000. The top 20 ions were selected for collision induced dissociation (CID) with collision energy set at 29. Dynamic exclusion was used after 1st identification with 10 s repeat duration and 30 s exclusion duration.
2.3. Protein Identification and Label-free Quantification
Proteome Discoverer (PD, V1.4, ThermoFisher) with Mascot (Mascot V2.3, Matrix Science) was used to search raw data against Human RefSeq database (the 2013.07.04). Mass tolerance for precursor ions was set to 20 ppm; mass tolerances of fragment ions were 0.02 and 0.5 Da for Q Exactive and LTQ Orbitrap VelosPro, respectively. Carbamidomethylation of cysteine, oxidation of methionine, acetylation of protein N-terminal were included as variable modifications. A maximum of one missed cleavages was allowed. All assigned peptides were filtered with 1 % false discovery rate (FDR) at peptide level. We only kept identifications with ≥ 2 unique peptides (1 % FDR and ion score > 20), which was stricter than 1 % FDR at the protein level.
All identified peptides were quantified with peak areas derived from their MS1 intensity. The process was as followed: 1) MS raw data were converted to the MS-platform independent mzXML format; 2) the spectral assignments from PD1.4 were then channeled through an in-housed pipeline to construct Extracted Ion Chromatogram (XIC) peaks with their corresponding intensity values included in mzXML data. For protein quantification, intensity based absolute quantification (iBAQ) algorithm (Schwanhausser et al., 2011) was used. To normalize the differences in loading amounts among samples, we then converted iBAQ value to FOT (fraction of total) - iBAQ value of each protein divided by the sum of all iBAQ values of all proteins in the sample. Thus, FOT number is a relative concentration for the protein in the total measurable proteome. FOTs of most proteins in a sample were very small and more than five decimal values were common. These small numbers would be visually difficult for human eyes. Therefore, we multiplied the FOT number with 105 to obtain iFOT5 to make easier visualization of values. All missing values were substituted with zero.
2.4. An algorithm for Screening Cancer
We first only kept proteins with ≥ 2 strict peptides (1 % FDR and ion score > 20). In the rest 450 normal samples, we randomly selected 350 samples and calculate their RIs based on the iFOT5 values. We used nonparametric 99.5th percentile values as the upper limits for selecting outlier proteins. We then randomly selected 45 cancer samples as the training data set to find outlier proteins that are outside of the RI upper limits, resulting in ~ 500 proteins. We then applied the same scheme on the validation dataset and to obtain the outlier pools, we then calculated the p-value for an overlap between the cancer outlier pool and the background proteins (total 15,447) based on the hypergeometric test (equivalent to the Fisher exact test) (Draghici et al., 2003). We repeated this procedure 20 times to remove potential random drawing errors and for each time the receiver operating characteristic curve (ROC) curve based on the test is plotted. We obtained final cancer outlier pool with 509 proteins from the ROC curve of the best run with the largest area under the curve (AUC) (0.957), and the hypergeometric test's p-value cut-off 1.78 × 10− 8 was obtained by setting the specificity at 0.95.
2.5. Statistical Analyses
Personal and pan-human RIs were calculated with non-parametric percentile method which is not dependent on distribution of data. In cancer screening, the p-value for an overlap between cancer outlier pool and sample-specific outlier proteins was calculated with hypergeometric test.
3. Results
3.1. Development of a Streamlined Workflow for Profiling Urine Proteome
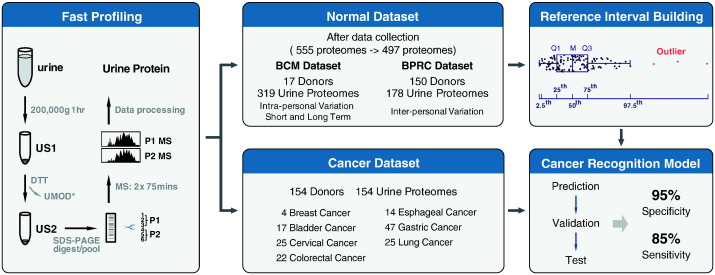
We first established a procedure to measure urine proteomes with a throughput of up to 8 samples per day per MS (Fig. 1). We used a single-step ultracentrifugation to collect high-speed sediment of the urine. After ultracentrifugation, majority of the most abundant protein, UMOD (Pisitkun et al., 2006, Raimondo et al., 2013), were removed by reduction with DTT (Supplementary Fig. 1). Samples were then separated by SDS-PAGE, cut into 6 slices and in-gel digested with trypsin. Extracted peptides were pooled and subjected to 2 MS runs, which took 3 h of machine time and typically resulted in the identification of ~ 1500 proteins. In the present study, we acquired 555 urine proteomes from 180 healthy donors. Of these data, 57 (10 % of 555) urine proteomes from 13 (7 % of 180) test subjects were collected during the method establishment phase, during which centrifugation at low speed before ultracentrifugation or different temperatures in DTT reduction step for removing protein UMOD were tested to optimize the procedure. To keep technical consistency, these 57 urine proteomes were removed and the remaining 497 urine proteomes from 167 healthy donors were included in further analysis (Fig. 1).
Fig. 1.
A schematic description of the workflow for fast profiling of urine proteome, establishing a reference interval (RI) of urine proteins for healthy human being and a cancer recognition algorithm. High-speed sediment of urine was collected by ultracentrifugation and uromodulin was removed by reduction with dithiotheitol. Then protein pellet was subjected to 1D SDS-PAGE for separation. After in-gel digestion, samples were pooled and measured by 2 MS runs. Normal dataset was acquired in an international two-center mode. Totally, 555 urine proteomes from 180 healthy donors were acquired. Of these data, 57 urine proteomes from 13 test subjects were collected during the method establishment phase in which centrifugation at low speed before ultracentrifugation or different temperatures in DTT reduction step for removing protein UMOD were tested to optimize our procedure. To keep technical consistency, these 57 urine proteomes were removed and only remaining 497 urine proteomes from 167 healthy donors were included in further analysis, with a BCM dataset (17 donors, 319 urine proteomes) and a BPRC dataset (150 donors, 178 urine proteomes). Personal and pan-human RI of urine proteomes were established with data in normal dataset. Cancer dataset (154 donors, 154 urine proteomes) included 7 types of solid tumors. Utilizing pan-human RI and data in cancer dataset, a cancer recognition algorithm was established to discriminate normal and cancer samples.
To assess technical variation in the workflow, we divided 16 urine samples collected from 5 individuals over 5 days into 2 MS samples each, and processed them in parallel. The Venn diagrams (Supplementary Fig. 2a) showed an approximately 90 % (1389/1695–2094/2190) overlap between the same-sample splits at the level of ~ 1200–1600 protein identifications; the squares of Pearson correlation coefficients for quantification (in iFOT5, Supplementary Fig. 2b) were between 0.93 and 0.96. These results indicated that combined variation in both sample processing and MS running was small.
To demonstrate the stability of LC-MS systems, we presented the quality control (QC) data of LC-MS (Velos and Q-Exactive, ThermoFisher) used for acquiring all data in this study. QC data were acquired every three days to monitor the performance of each machine in Baylor College of Medicine (BCM) and Beijing Proteome Research Center (BPRC). Two hundred nanograms of commercial HeLa cell lysates (ThermoFisher) or 500 ng of home-made 293 T cell lysates were used for this purpose at BCM and BPRC, respectively. Twenty sets of QC data from each machine that span two months were used to demonstrate the stability of LC-MS. Analysis of Pearson correlation were performed to evaluate not only inter-day variations from each machine but also inter-machine variations in either center, as shown in Supplementary Fig. 2c and d. Coefficients of correlation ranged from 0.83 to 0.97 for all QC data in both centers. Besides technical variation from machines, these comparisons also included variations from different batch of QC samples. Results from QC data clearly suggested that the measurements from LC-MS platforms in both BCM and BPRC were stable and repeatable.
3.2. Intra-individual Variation and Personal Reference Interval of the Urine Proteome
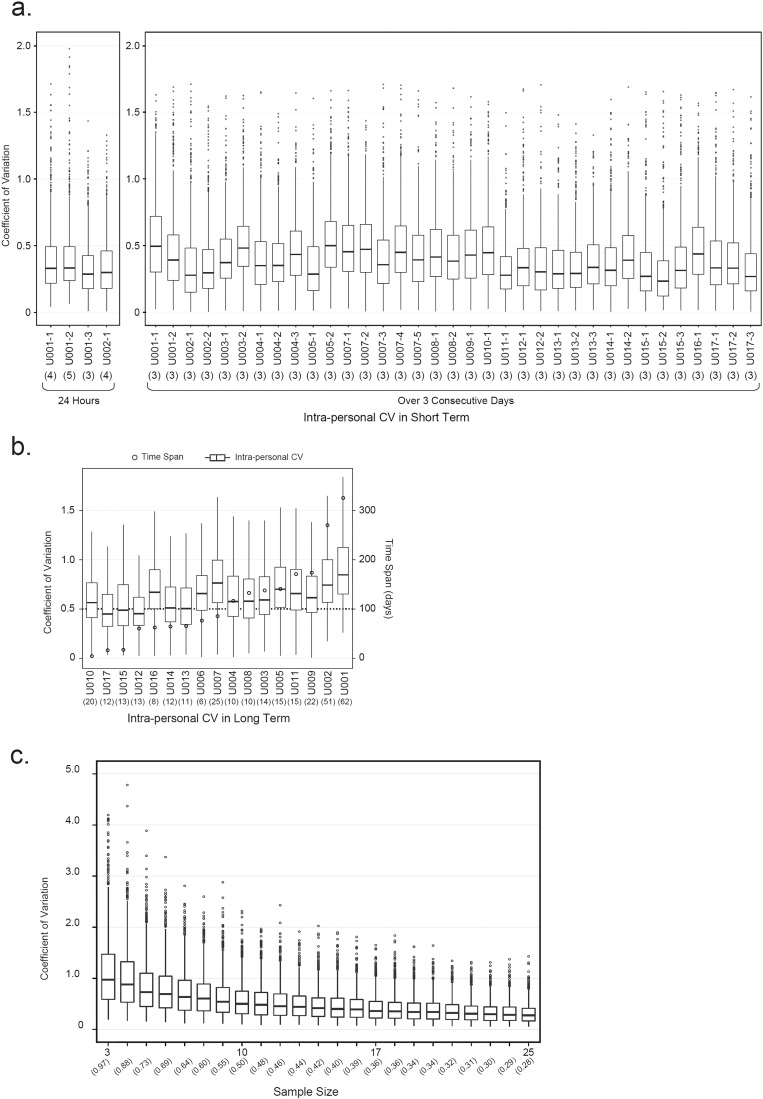
We recruited 17 apparently healthy individuals from Baylor College of Medicine (BCM) to investigate short-term (within 24 h and over three consecutive days) and long-term (months) intra-individual variations in the urine proteome (Supplementary Table 2). To record 24-hour change in the urine proteome, urine samples from a test subject were separately collected in every ~ 6 h and measured. In this way, we obtained a total of four datasets from test subject U001 (three datasets) and U002 (one dataset) containing 3 to 5 urine proteomes in each dataset. Intra-personal coefficient of variations (CV) were calculated for all datasets using only proteins with non-zero quantification values. Variations of urine proteomes within 24 h were represented with the spread of intra-personal CVs from each dataset in a box-plot format (Fig. 2a). While the median CVs of four 24-hour datasets ranged from 0.29 to 0.33, the maximum CV was as high as 2.0 for proteins that varied substantially. Furthermore, we investigated intra-personal variations during a few day period. One urine sample was collected in each morning from a test subject over 3 consecutive days. In this way, we acquired 35 datasets from 16 donors, resulting in at least 1 dataset for each person and as many as 5 datasets for one person. Intra-personal CVs were calculated from proteins with non-zero quantification values in all three samples in the 3-day dataset. As shown in Fig. 2a, the median CVs within 3 days ranged from 0.23 to 0.5, slightly higher than intra-personal variations within 24 h.
Fig. 2.
The effects of sampling time span and sample size on intra-personal variations of the urine proteome. (a) Intra-personal variations of urine proteomes in short term (24 h and 3 consecutive days) of urine sampling. Coefficients of variation (CVs) were calculated from proteins quantified in all samples in each dataset. Number in parenthesis under X-axis represented sample number in each dataset. For variations within 24 h (median CVs 0.29–0.33), 4 datasets were acquired from two individuals (U001 and U002); For variations over 3 consecutive days (median CVs 0.23–0.5), 35 datasets were acquired from 16 donors (U001–U005 and U007–U017). (b) Intra-personal variations of human urine proteomes in long term (> 60 days) of urine sampling. Number in parenthesis under X-axis represented sample number in each dataset. Of 17 donors in BCM dataset, intra-personal CVs for long term (> 60 days) could be calculated from 14 donors (U001–U009, U011–U014, and U016). For convenience of comparison, intra-personal CVs from U010, U015, and U017 (time span for urine sampling are 5, 17, and 18 days, respectively) were also plotted in the same graph (the first 3 boxes). Median CVs for 17 donors range from 0.45–0.87. (c) The effect of sample size on intra-personal variation of human urine proteome. The graph was plotted according to the dataset of U001. Number in parenthesis under X-axis represented median CV in each dataset. CVs in Y-axis were calculated with 100 iterations of mean iFOT5 for each protein in each subset size.
Intra-personal variations in long term (> 60 days) were evaluated with datasets from 14 donors that resulted in 6 to 62 proteomes in each dataset, with the longest time span of 314 days (Supplementary Table 2). The spreads of intra-personal CVs were displayed in Fig. 2b with median CVs ranging from 0.45 to 0.87. It was clear that intra-personal variation in long term was larger than that in short term (Fig. 2a and b). As shown in Fig. 2b, intra-personal CVs did not change linearly with the time span. Thus, intra-personal variations were relatively stable over a period, making it possible to obtain personal RIs of the urine proteome.
Next, we investigated how many urine samples were sufficient to establish stable personal ranges. For the top 2 largest personal datasets (containing 62 and 51 samples from U001 and U002, respectively), we tested a sub-sampling method to determine the effect of sample size on variation. We used proteins that were detected and quantified in at least 10% of the samples (7/62 for U001 and 6/51 for U002) to focus on proteins that were frequently detected in the urine. We sub-sampled sets of 3 to 25 samples, and made100 iterations for each subset. We calculated CVs of mean iFOT5 for each protein in each subset and plotted overall spreads of CVs for all proteins against the sample size (Fig. 2c and Supplementary Fig. 3). It was clear that overall spreads of CVs improved with sample size and become stable with 15 to 20 samples. This suggested that approximately 15 to 20 samples were sufficient to establish reliable personal RIs for frequently detected proteins in the urine.
3.3. Comparison of BCM with BPRC Dataset, Inter-personal Variations, and Pan-human Reference Intervals
The BCM dataset focused on measuring diverse samples from a small population (17 subjects, 319 urine proteomes). We also acquired a BPRC dataset at Beijing Proteome Research Center (BPRC) in Beijing, China, in which samples from a relative large population (150 subjects, 178 urine proteomes) were collected. For both datasets, collection, storage and preparation of samples as well running of the LC-MS systems followed the same standard operation procedure and were performed at each site independently, resulting in 497 urine proteomes from 167 test subjects combined (Fig. 1).
We first evaluated the compatibility of the two datasets using proteins detected in > 10% of the samples in each dataset (32/319 for BCM and 18/178 for BPRC). We named these proteins as frequently identified proteins. The BCM and BPRC datasets contained 1950 and 2071 frequently identified proteins, respectively, with 1770 proteins in common. The totally identified urine proteins in both datasets were 5405 at 1 % protein FDR level. The frequently identified 2000 proteins constituted the common human urine proteome, while the other 3000 proteins reflected the normal geno-proteomic differences among individuals under different physiological conditions. As keratins (KRTs) were often found as contaminations from the sample operators, we did not include KRTs in the protein list for urine protein RIs.
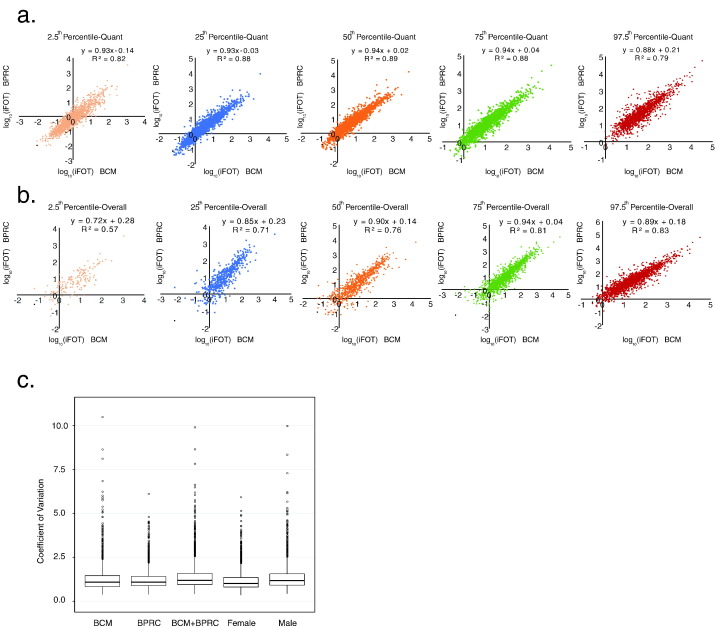
We calculated the 2.5th, 25th, median (50th), 75th, and 97.5th percentile values from common proteins with their non-zero iFOT5 values from both datasets. The Pearson correlations were shown in Fig. 3a. The reference intervals (2.5th and 97.5th percentile) in both datasets were highly correlated. These tight correlations from two independent datasets suggested that abundance of proteins in urine fluctuated within a stable range.
Fig. 3.
A comparison between the BCM and BPRC datasets and inter-individual variations of the urine proteome. (a) Correlations of 2.5th, 25th, Median (50th), 75th, and 95th percentile values calculated from only non-zero quantification values (iFOT5) of proteins (1770) commonly identified in at least 10% of samples in both BCM and BPRC datasets. (b) Correlations of 2.5th, 25th, Median (50th), 75th, and 95th percentile values calculated from both zero and non-zero quantification values (iFOT5) of proteins (1770) commonly identified in at least 10% of samples in both BCM and BPRC datasets. (c) Inter-individual variations of urine proteomes from 5 datasets including BCM (17 donors, 319 urine proteomes), BPRC (150 donors, 178 urine proteomes), BCM plus BPRC (167 donors, 497 urine proteomes), female (69 donors, 154 urine proteomes), and male (98 donors, 343 urine proteomes) datasets.
In order to see the impact of zero values (used for imputing missing values in both datasets) on percentile values, we also included both zero and non-zero values to calculate percentile values and correlation between the two datasets (Fig. 3b). The correlation was not high for the 2.5th percentile values then improved considerably for higher percentile values, and for above the 75th percentile values, they were as good as those calculated from the non-zero values only. Therefore, it might not matter that much including or not including zero values to calculate and use 75th percentile values and above. For the later analyses to identify outliers, we always used the high percentile and did not use the lower percentile values for picking outliers.
Next, we calculated inter-personal CVs of frequently identified proteins from the 5 datasets, namely the BCM, BPRC, BCM plus BPRC, female, and male datasets, containing 1950, 2071, 2025, 2011 and 1883 proteins, respectively. The CV spreads of the 5 datasets were similar (Fig. 3c), with median CVs ranging from 1.01 to 1.19. The CV for the female dataset was slightly lower than that for the male dataset (median CVs 1.01 vs 1.17). It was clear that the inter-personal variation was significantly larger than intra-personal variation (Figs. 3c and 2b). These datasets allowed us to calculate 5 RIs for frequently detected urine proteins. The RIs derived from combined BCM and BPRC dataset was designated as the tentative pan-human RIs (Supplementary Table 3).
Among the 109 blood protein biomarkers approved by Food and Drug Administration (FDA) (Anderson, 2010), forty-six of them were found in the urine proteome with a RI (Supplementary Table 4). Additionally, six hundred and sixty-nine proteins in the common urine proteome were associated with 656 Online Mendelian Inheritance in Man (OMIM) (Hamosh et al., 2005) phenotypes. Of them, forty proteins were linked with autosomal dominant or recessive diseases and 13 proteins were associated with X-linked diseases (Supplementary Table 4). Together it suggests that measuring human urine proteome have the potential to monitor ~ 50 % (46/109) of the blood protein biomarkers approved by FDA and offer the possibility to screen genetic diseases.
3.4. Potential Applications of Personal and Pan-human Reference Intervals in Monitoring Health Status
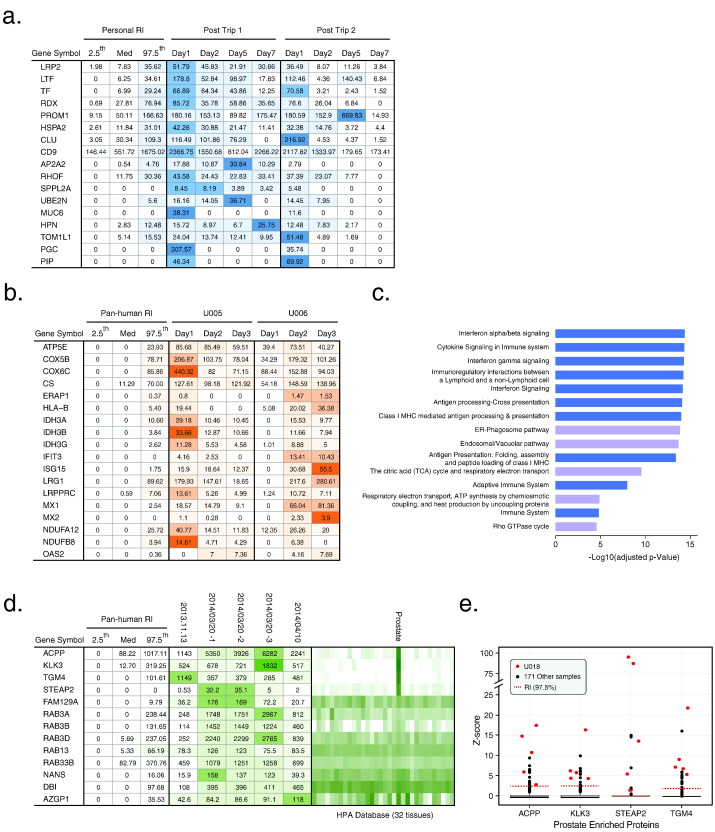
To test whether the urine proteome can be used to monitor physiological and pathological states, we first analyzed urine samples from one individual (U002) after trans-continental travels from Beijing, China to Houston, Texas, USA, which took over 18 h of flight time and might confer physiological stress on the traveler. Two post-flight studies (3 months apart) were performed with urine samples collected on sequential days after the travel for up to a week. From the first trip, we detected 377, 209, 223, and 110 outlier proteins that were above 97.5th percentile of personal RIs (Supplementary Table 5) on day 1, 2, 5 and 7, respectively; we were able to repeatedly detect 44 outlier proteins on day 1 from the second trip (Supplementary Table 5), indicating that some physiological changes may be reproducible. As exemplified in Fig. 4a, levels of some of the outliers demonstrated downward trend of change post travel and gradually returned to their normal levels. We concluded that our workflow is capable of detecting changes in urine in response to physiological stresses.
Fig. 4.
Outlier proteins in urine proteome register physiological changes. (a) Outlier proteins and their changes up to a week from two transcontinental trips (3 months apart). The empirical 2.5th, Median (Med) and the 97.5th percentile values are shown as the “normal” range. (b) Consistent outlier proteins and their changes in three days from two people who caught flu. (c) Top 15 pathways enriched by Reactome analysis of the common outlier proteins in (b). (d) Persistent tissue-specific outlier proteins detected from an individual. Outlier proteins of three different samples from test subject U018 (the 2014.03.20 sample was measured three times). The expression levels (mRNA in 32 tissues from the Human Protein Atlas) are shown as the green-scale heat map, with darker green representing higher expression levels. (e) Abundance distributions of the 4 prostate-enriched outlier proteins from the U018 test subject (measured 5 times) and the other 171 donors in the combined BCM and BPRC dataset. Occasionally, these 4 proteins were also detected as outlier proteins in the other male subjects but they were not persistent, nor were they outliers with the 4 prostate enriched proteins at the same time for any other test subjects.
We next analyzed urine samples collected from two test subjects (U005 and U006) after they experienced flu-like symptoms such as fever, runny nose and sneeze. U005 and U006 lived in the same household and presumably caught the same flu strain. With the pan-human RIs, we found 103 outliers above the 97.5th percentile of RIs (Supplementary Table 5). Pathway analysis using Reactome Pathway Database (http://www.reactome.org) (Croft et al., 2011) revealed that many outliers were enriched in pathways associated with immune response (Fig. 4c, Supplementary Table 5). For example, upregulation of IFIT1, IFIT3, OAS2, ISG15, PML, MX1, and MX2 were indicative of activation of antivirus response of the adaptive immunity (Fig. 4b) (Haller et al., 2006, Stetson and Medzhitov, 2006). Notably, ISG15, MX1 and OAS2 are known interferon-stimulated genes (Diamond and Farzan, 2013, Haller et al., 2006), and have been validated as antiviral proteins in gene knockout mouse models (Diamond and Farzan, 2013, Haller et al., 2006, Sadler and Williams, 2008). These results suggested that the anti-virus response left a trace in the urine in test subjects who caught flu.
We also encountered an interesting test subject (U018) whose three urine samples were collected and measured on November 13th, 2013, March 20th, 2014, and April 10th, 2014 respectively. We found 13 common outliers (above 97.5th percentile of pan-human RIs) in all samples and, according to Human Protein Atlas (Uhlen et al., 2015), four outliers (KLK3, TGM4, ACPP, and STEAP2) were prostate enriched proteins (Fig. 4d). Four other prostate enriched proteins (RDH11, KLK2, NEFH, and SLC30A4) were also detected as outliers in one of the 3 measurements. While these proteins were occasionally detected as outliers in other male subjects, their levels went back to normal range in the follow-up measurement, and never were 4 of them detected as outliers at the same time from any single test subject (Fig. 4e). Thus, U018 seemed to possess a set of unique prostate enriched outlier proteins that may or may not indicate any physiological or pathological conditions at present, but may benefit from continuous monitoring.
3.5. Application in Cancer Screening
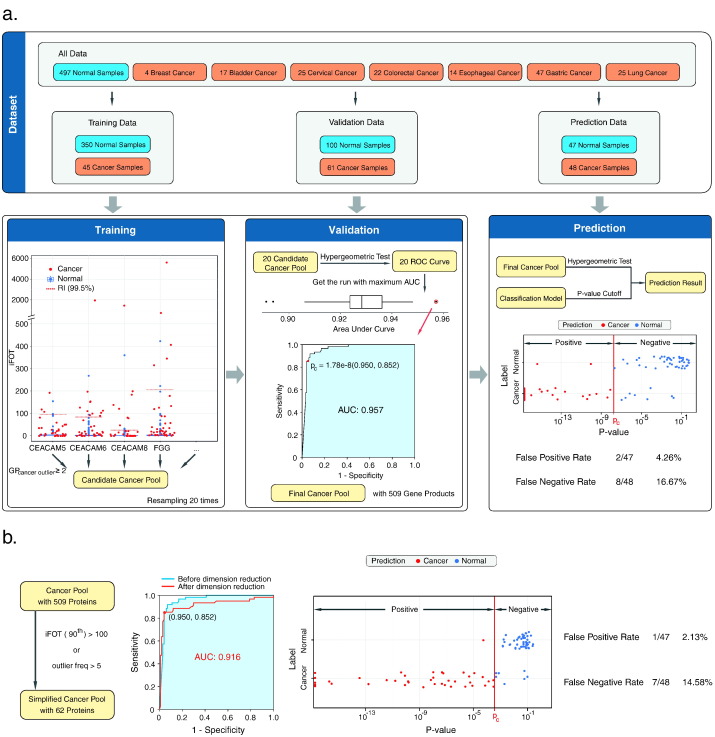
To investigate whether the RIs can be used for disease screening, we measured 154 samples from patients with bladder (17 cases), breast (4), cervical (25), colorectal (22), esophageal (14), gastric (47), and lung cancers (25), and used the data and the pan-human RIs to establish a cancer recognition algorithm (Fig. 5a).
Fig. 5.
A workflow that combines marker discovery and validation in one and a cancer recognition algorithm using pan-human reference interval to distinguish samples from healthy people and cancer patients. (a) A schematic description of the workflow for establishing a cancer recognition algorithm. Normal and cancer urine proteomes were split into 3 parts for training, validation and prediction. The candidate cancer outlier pool was first generated by filtering out outlier proteins in at least 2 samples from the training dataset. Then its performance for classifying cancer and normal samples were tested with the validation dataset and the hypergeometric distribution test. Then a receiver operating characteristic curve (ROC) and area under curve (AUC) were generated. The process was iterated 20 times to obtain the final candidate cancer outlier pool (509 proteins) – “cancer markers”. At last, the prediction was done according to the cancer markers and the hypergeometric test. (b) Reduction of cancer marker pool and its performance in classifying cancer and normal samples. Reduction of dimension according to outlier frequency or its abundance reduced the cancer markers from 509 to 62 proteins with slightly improved power in classifying cancer and normal samples.
The 497 normal samples were randomly divided into 3 groups. Group I consisted of 350 samples for establishing the pan-human RIs; Group II contained 100 samples for validation; and the remaining 47 samples as Group III were used for further prediction. The 154 cancer samples were similarly divided into 3 groups with 45, 61, and 48 samples for training, validation and prediction, respectively. While Group I and II samples were permutated in each iteration, the prediction sets (Group III) never participated in training or validation.
To establish a cancer outlier pool, we randomly picked 350 normal and 45 cancer samples as training dataset to find outlier proteins above 99.5th percentile of the pan-human RIs. We kept the outlier protein only if it was found in at least 2 samples in the cancer training dataset. The validation dataset (61 cancer and 100 normal samples) were individually compared with the cancer outlier pool and the significance of overlap between them was calculated with a hypergeometric distribution test (Draghici et al., 2003) to obtain a p-value. When all p-values from cancer and normal samples were combined, a ROC curve was obtained and the AUC was calculated to assess the performance and to determine the cutoff p-value (Pc) for classifying cancer. To minimize sampling bias, this process was iterated 20 times to obtain the final cancer outlier pool that contained 509 proteins, with the best AUC (0.957) of ROC (Supplementary Table 6, Fig. 5a). We then took the outlier proteins from the prediction set (48 cancer and 47 normal samples) to calculate p-values with the hypergeometric distribution test. This algorithm misclassified 2 samples as cancer out of the 47 normal samples (false positive rate 4.26 %), and 8 as normal out of the 48 cancer samples (false negative rate 16.67 %).
To further reduce the number of proteins in the cancer outliner pool, we applied the following constraints: 1) keep the outliers if it appeared in > 5 cancer samples; or 2) if its iFOT5 at 90th percentile of the RIs in the cancer training set was > 100. With these restrictions, the cancer outlier pool was reduced from 509 to 62 proteins (Supplementary Table 6), resulting in a ROC with AUC of 0.916. Using the Pc with 95 % specificity, this algorithm misclassified 1 sample from the 47 normal samples (false positive rate 2.13 %) and 7 from the 48 cancer samples (false negative rate 14.58 %) (Fig. 5b). These results suggested that reduction of cancer outlier pool by removing less common or low abundant proteins could actually improve the performance of the cancer recognition algorithm.
4. Discussion
Here, we presented a streamlined workflow to measure several hundred urine proteomes in an international, two-center setting. We aimed to address several common and critical issues that might be relevant to clinical urine proteomics, including 1) how variable is the human urine proteome? 2) Can we establish reference intervals and how many samples are sufficient? 3) Can we find a way to discover urine biomarkers for disease diagnosis and health status monitoring?
We were able to establish RIs for 2000 commonly detected urine proteins. As we measured 497 urine proteomes and only included proteins that were detected in > 10 % of the samples, every protein was detected in > 49 times with protein FDR < 1 %, thus the identification was very stringent and error in protein identification was negligible. There were 902 proteins with median value (50th percentile value) greater than zero, these were relatively abundant urine proteins. The rest of the 1098 proteins were not detected in half of the samples but were detected in > 10 % of the samples. Their abundance may not be high, but their RIs (up to 97.5th percentile) were accurate as they were derived from at least 12 detectable measurements. The 99.5th percentile value may not be accurate for a small portion of the proteins as they may be calculated from fewer than 12 detectable measurements.
The current disease marker discovery and validation programs often utilized two cohorts of samples - disease and control (Fuzery et al., 2013, Pavlou et al., 2013, Rodriguez-Suarez et al., 2014) and unfortunately, many markers from these studies did not pass the validation phase. This was due in part to the “noise markers” resulting from physiological fluctuation or individual differences (Gao, 2013). With limited sample size in the discovery phase, it was understandably difficult to cover the physiological variations of the control group. A case in point was the protein TMEM256. As the second most abundant protein in our dataset, its normal RI for the 2.5th percentile value was 7.1 while the 97.5th percentile value was 9305 (see Supplementary Table 3), spanning a factor of 1329 folds within the normal range! TMEM256 was identified as a candidate prostate cancer marker with ratio of 140 for the cancer and control group (Overbye et al., 2015), later it did not validate from another cohort where the difference between cancer and control was not significant when measured with immune assays (Wang et al., 2017). By establishing a normal RI as presented here, most of the “noise markers” or highly variable markers could be filtered out in the first step. We had demonstrated the feasibility of obtaining comparable data in an international, two-center (BCM in USA and BPRC in China) setting. These datasets allowed us to evaluate intra- and inter-personal variations in urine proteome and to establish personal and pan-human RIs, which could be used to identify associations between the urine proteins and physiological or pathological conditions. We showed that subtle, but consistent changes could be detected as outliers of the urine proteome after trans-continental travel or catching flu, demonstrating a “proof-of-principle” for health state monitoring. While outlier proteins detected in trans-continental travel were also detected under normal conditions at lower abundances, many outliers detected with flu symptom were rarely detected under normal conditions. This suggests that trans-continental travel was a milder physiological perturbation, whereas catching flu led to more profound changes. One interesting observation was the consistent increase of proteins involved in gastric function (PGC, MUC6) after inter-continental travel, indicating that digestive system may experience major stress. This was consistent with a common complaint of stomach discomfort suffered by many travelers adjusting to day and night jetlag. We believe that this is the instance where MS-based urine proteomics was used to measure, in an unbiased manner, significant deviations from normal protein levels in urine due to minor physiological and pathological changes. These results are suggestive that pathological conditions are likely to leave discernable footprints in the urine proteome. While these 2 examples are intriguing, systematic monitoring of urine proteome for more cases were required to implicate any biological significance. Nevertheless, our approach could be adopted for high risk population screening before the disease symptoms became apparent, and might have the potential to discover early disease markers.
We also found that if the underlying cause were physiological variation, outlier proteins often went back to the normal range in the follow up measurement, as exemplified in the intercontinental travel case. Persistent detection of outlier proteins may be indicative of non-physiological conditions. This is an intriguing hypothesis now and more experiments are needed to test it. The outlier proteins enriched in prostate as persistently detected in U018 may be one example. These proteins were also sporadically detected as outliers in other test subjects, but they went back to levels within RIs in the follow up measurements, demonstrating a transient nature of the normal physiological response. Periodical measurements of a person's urine proteome could establish a personal health archive that would be valuable for detecting future health issues.
Our study pointed to a way for biomarker analysis or discovery and the possibility for cancer screening using the urine proteome. The algorithm for cancer screening illustrated a way to detect cancer with reduced false positive rate while keeping false negative rate in a reasonable range. Importantly, our algorithm demonstrated that with pan-human RIs, it was possible to use relatively small number of samples (106 samples covering 7 cancers) to obtain a cancer outlier pool that could distinguish samples from cancer patients and healthy individuals. Many proteins in cancer outlier pool have clear relationship with cancer. For example, CEACAM1, CEACAM6, and CEACAM8 (see Supplementary Table 6) are carcinoembryonic antigens that are utilized as tumor maker in clinic (Hammarstrom, 1999). IDH2, MTOR and SF3B1 (see Supplementary Table 6) are encoded by cancer driver genes (Rubio-Perez et al., 2015). IDH2 (isocitrate dehydrogenase 2) catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) while reducing NADP + to NADPH (DeBerardinis et al., 2007). The dysfunction of IDH through mutation or alteration in expression level has been observed in numerous types of cancers (Lv et al., 2012). MTOR, mammalian target of rapamycin, has emerged as a critical effector in cell-signaling pathways commonly deregulated in human cancers (Guertin and Sabatini, 2007). Together, these examples suggested that proteins closely related to cancer could be picked up by our algorithm.
We noted several limitations to the current study that warranted further development. First, the current RIs were defined in iFOT and it was only a relative concentration in the total measurable urine proteome. The total urine proteome was used for normalization and therefore, RIs had no unit. We could not define the RIs in a clinical lab more friendly fashion, i.e. in the unit of g or mol per mol creatinine (to normalize for differences in urinary output). Second, we measured the high-speed sediment of urine, not the total urine, to get rid of the most abundant soluble urine proteins to favor MS detection, thus, our RIs must be used with the same operational procedures – high-speed urine sediment, SDS-PAGE then followed by LC-MS measurements. Third, although the pan-human RIs established in this study were based on the large dataset of urine proteomes from healthy persons, the coverage of population was still limited when considering factors of genders and age. Most of test subjects were in the age group of 25 to 55, while older people and children were not included. They were expected to have unique and different proteomes and must be dealt with separately. Furthermore, as the dataset only include data from people of Chinese ethnicity, cautions should be taken when comparing them to data from people of other ethnicities. Diversities of urine samples in gender, age, and ethnicities should be considered in future studies. Additionally, while our workflow is a good compromise between throughput and the depth of the urine proteome, it would be more useful to automate the system, decrease sample preparation time, adopt a gel-free method and further increase sample throughput (Court et al., 2015, Santucci et al., 2015, Yu et al., 2014).
In summary, we showed that the human urine proteome contained rich information related to health and disease. We envisioned that urine proteome analysis, as a true noninvasive analytical tool, would find widespread applications in health monitoring, disease detection and even genetic disease screening in the near future.
The following are the supplementary data related to this article.
Supplementary Figs. 1–3 and Supplementary Table 2.
Metadata of urine samples in this study.
Pan-human reference intervals.
Plasma protein markers and OMIM phenotype related proteins that can be detected in common urine proteome.
Urine proteomes from U002 acquired after two trans-continental travels (3 months apart).
Cancer outlier pool.
Conflicts of Interests
The authors declare no competing financial interests.
Author Contributions
J.Q., G.W., B.Z., and F.H. conceived the idea; J.Q., G.W., B. Z, Y. W. and W.L. designed the experiments; C.S. and X.M. collected clinical urine samples and provided clinical interpretation of the data; W.L., T.L., S.Y.J., M.V.H., C.D., and W.S. performed experiments, X.N., A.M., Y.H., T.S., W.Z., Y.H., Y.Q., and G.S. analyzed data; W.L., X.N., A.M., Y.W. and J.Q. wrote the paper.
Acknowledgments and Funding Sources
This project was supported by an internal research grant from the Joint Center for Translational Medicine (BPRC-Baodi-001) between Beijing Proteome Research Center and Tianjin Baodi Hospital, and an International Collaboration Grant 2014DFB30010 from the Ministry of Science and Technology of China.
Contributor Information
Bei Zhen, Email: zp1963@sina.com.
Guangshun Wang, Email: wgsTMUBH@163.com.
Jun Qin, Email: jqin@bcm.edu.
References
- Adachi J., Kumar C., Zhang Y., Olsen J.V., Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M., Gao Y. Urinary biomarkers of brain diseases. Genomics Proteomics Bioinformatics. 2015;13:345–354. doi: 10.1016/j.gpb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N.L. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin. Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- Beretov J., Wasinger V.C., Millar E.K., Schwartz P., Graham P.H., Li, Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . EP28A3CE-Defining, Establishing, and Verifying Reference Intervals in the Clinical Laboratory. third ed. 2010. A guideline for global application developed through the Clinical and Laboratory Standards Institute consensus process. Approved Guideline. [Google Scholar]
- Coon J.J., Zurbig P., Dakna M., Dominiczak A.F., Decramer S., Fliser D., Frommberger M., Golovko I., Good D.M., Herget-Rosenthal S., Jankowski J., Julian B.A., Kellmann M., Kolch W., Massy Z., Novak J., Rossing K., Schanstra J.P., Schiffer E., Theodorescu D., Vanholder R., Weissinger E.M., Mischak H., Schmitt-Kopplin P. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin. Appl. 2008;2:964. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court M., Garin J., Masselon C.D. Urine sample preparation and fractionation for global proteome profiling by LC-MS. Methods Mol. Biol. 2015;1243:175–186. doi: 10.1007/978-1-4939-1872-0_10. [DOI] [PubMed] [Google Scholar]
- Croft D., O'kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., Jupe S., Kalatskaya I., Mahajan S., May B., Ndegwa N., Schmidt E., Shamovsky V., Yung C., Birney E., Hermjakob H., D'eustachio P., Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C.B. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decramer S., Gonzalez De Peredo A., Breuil B., Mischak H., Monsarrat B., Bascands J.L., Schanstra J.P. Urine in clinical proteomics. Mol. Cell. Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S., Khatri P., Martins R.P., Ostermeier G.C., Krawetz S.A. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Fuzery A.K., Levin J., Chan M.M., Chan D.W. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin. Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Urine-an untapped goldmine for biomarker discovery? Sci. China Life Sci. 2013;56:1145–1146. doi: 10.1007/s11427-013-4574-1. [DOI] [PubMed] [Google Scholar]
- Goodison S., Rosser C.J., Urquidi V. Urinary proteomic profiling for diagnostic bladder cancer biomarkers. Expert Rev Proteomics. 2009;6:507–514. doi: 10.1586/epr.09.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Haller O., Kochs G., Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., Mckusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A., Monigatti F., Dorff K., Campagne F., Bachur R., Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin. Appl. 2009;3:1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristenko N.A., Larina I.M., Domon B. Longitudinal urinary protein variability in participants of the space flight simulation program. J. Proteome Res. 2016;15:114–124. doi: 10.1021/acs.jproteome.5b00594. [DOI] [PubMed] [Google Scholar]
- Lv Q., Xing S., Li Z., Li J., Gong P., Xu X., Chang L., Jin X., Gao F., Li W., Zhang G., Yang J., Zhang X. Altered expression levels of IDH2 are involved in the development of colon cancer. Exp. Ther. Med. 2012;4:801–806. doi: 10.3892/etm.2012.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A., Li Y., Bulynko Y., Jung S.Y., Wang Y., Lanz R.B., O'malley B.W., Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimuthu A., O'meally R.N., Chaerkady R., Subbannayya Y., Nanjappa V., Kumar P., Kelkar D.S., Pinto S.M., Sharma R., Renuse S., Goel R., Christopher R., Delanghe B., Cole R.N., Harsha H.C., Pandey A. A comprehensive map of the human urinary proteome. J. Proteome Res. 2011;10:2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischak H., Allmaier G., Apweiler R., Attwood T., Baumann M., Benigni A., Bennett S.E., Bischoff R., Bongcam-Rudloff E., Capasso G., Coon J.J., D'haese P., Dominiczak A.F., Dakna M., Dihazi H., Ehrich J.H., Fernandez-Llama P., Fliser D., Frokiaer J., Garin J., Girolami M., Hancock W.S., Haubitz M., Hochstrasser D., Holman R.R., Ioannidis J.P., Jankowski J., Julian B.A., Klein J.B., Kolch W., Luider T., Massy Z., Mattes W.B., Molina F., Monsarrat B., Novak J., Peter K., Rossing P., Sanchez-Carbayo M., Schanstra J.P., Semmes O.J., Spasovski G., Theodorescu D., Thongboonkerd V., Vanholder R., Veenstra T.D., Weissinger E., Yamamoto T., Vlahou A. Recommendations for biomarker identification and qualification in clinical proteomics. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- Nagaraj N., Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J. Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- Overbye A., Skotland T., Koehler C.J., Thiede B., Seierstad T., Berge V., Sandvig K., Llorente A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 2015;6:30357–30376. doi: 10.18632/oncotarget.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou M.P., Diamandis E.P., Blasutig I.M. The long journey of cancer biomarkers from the bench to the clinic. Clin. Chem. 2013;59:147–157. doi: 10.1373/clinchem.2012.184614. [DOI] [PubMed] [Google Scholar]
- Pisitkun T., Johnstone R., Knepper M.A. Discovery of urinary biomarkers. Mol. Cell. Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo F., Morosi L., Corbetta S., Chinello C., Brambilla P., Della Mina P., Villa A., Albo G., Battaglia C., Bosari S., Magni F., Pitto M. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. BioSyst. 2013;9:1220–1233. doi: 10.1039/c3mb25582d. [DOI] [PubMed] [Google Scholar]
- Rifai N., Gillette M.A., Carr S.A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Suarez E., Siwy J., Zurbig P., Mischak H. Urine as a source for clinical proteome analysis: from discovery to clinical application. Biochim. Biophys. Acta. 2014;1844:884–898. doi: 10.1016/j.bbapap.2013.06.016. [DOI] [PubMed] [Google Scholar]
- Rubio-Perez C., Tamborero D., Schroeder M.P., Antolin A.A., Deu-Pons J., Perez-Llamas C., Mestres J., Gonzalez-Perez A., Lopez-Bigas N. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27:382–396. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci L., Candiano G., Petretto A., Bruschi M., Lavarello C., Inglese E., Righetti P.G., Ghiggeri G.M. From hundreds to thousands: widening the normal human Urinome (1) J. Proteome. 2015;112:53–62. doi: 10.1016/j.jprot.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shao C. Urinary protein biomarker database: a useful tool for biomarker discovery. Adv. Exp. Med. Biol. 2015;845:195–203. doi: 10.1007/978-94-017-9523-4_19. [DOI] [PubMed] [Google Scholar]
- Shao C., Li M., Li X., Wei L., Zhu L., Yang F., Jia L., Mu Y., Wang J., Guo Z., Zhang D., Yin J., Wang Z., Sun W., Zhang Z., Gao Y. A tool for biomarker discovery in the urinary proteome: a manually curated human and animal urine protein biomarker database. Mol. Cell. Proteomics. 2011;10(M111):010975. doi: 10.1074/mcp.M111.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C., Wang Y., Gao Y. Applications of urinary proteomics in biomarker discovery. Sci. China Life Sci. 2011;54:409–417. doi: 10.1007/s11427-011-4162-1. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Sun W., Chen Y., Li F., Zhang L., Yang R., Zhang Z., Zheng D., Gao Y. Dynamic urinary proteomic analysis reveals stable proteins to be potential biomarkers. Proteomics Clin. Appl. 2009;3:370–382. doi: 10.1002/prca.200800061. [DOI] [PubMed] [Google Scholar]
- Sun W., Li F., Wu S., Wang X., Zheng D., Wang J., Gao Y. Human urine proteome analysis by three separation approaches. Proteomics. 2005;5:4994–5001. doi: 10.1002/pmic.200401334. [DOI] [PubMed] [Google Scholar]
- Thompson I.M., Pauler D.K., Goodman P.J., Tangen C.M., Lucia M.S., Parnes H.L., Minasian L.M., Ford L.G., Lippman S.M., Crawford E.D., Crowley J.J., Coltman C.A., Jr. Prevalence of prostate cancer among men with a prostate-specific antigen level < or = 4.0 ng per milliliter. N. Engl. J. Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- Thongboonkerd V., Mcleish K.R., Arthur J.M., Klein J.B. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., Von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., Von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wang L., Li F., Sun W., Wu S., Wang X., Zhang L., Zheng D., Wang J., Gao Y. Concanavalin A-captured glycoproteins in healthy human urine. Mol. Cell. Proteomics. 2006;5:560–562. doi: 10.1074/mcp.D500013-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang L., Skotland T., Berge V., Sandvig K., Llorente A. Exosomal proteins as prostate cancer biomarkers in urine: from mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharm. Sci. 2017;98:80–85. doi: 10.1016/j.ejps.2016.09.023. [DOI] [PubMed] [Google Scholar]
- Wu J., Gao Y. Physiological conditions can be reflected in human urine proteome and metabolome. Expert. Rev. Proteom. 2015;12:623–636. doi: 10.1586/14789450.2015.1094380. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Langham R.G., Ronco P., Knepper M.A., Thongboonkerd V. Towards standard protocols and guidelines for urine proteomics: a report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop, 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics. 2008;8:2156–2159. doi: 10.1002/pmic.200800138. [DOI] [PubMed] [Google Scholar]
- Yu Y., Suh M.J., Sikorski P., Kwon K., Nelson K.E., Pieper R. Urine sample preparation in 96-well filter plates for quantitative clinical proteomics. Anal. Chem. 2014;86:5470–5477. doi: 10.1021/ac5008317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Y., Adachi J., Olsen J.V., Shi R., De Souza G., Pasini E., Foster L.J., Macek B., Zougman A., Kumar C., Wisniewski J.R., Jun W., Mann M. MAPU: Max-Planck Unified database of organellar, cellular, tissue and body fluid proteomes. Nucleic Acids Res. 2007;35:D771–D779. doi: 10.1093/nar/gkl784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–3 and Supplementary Table 2.
Metadata of urine samples in this study.
Pan-human reference intervals.
Plasma protein markers and OMIM phenotype related proteins that can be detected in common urine proteome.
Urine proteomes from U002 acquired after two trans-continental travels (3 months apart).
Cancer outlier pool.