Abstract
Overexpression of epithelial cell adhesion molecule (EpCAM) has been implicated in advanced endometrial cancer, but its roles in this progression remain to be elucidated. In addition to its structural role in modulating cell-surface adhesion, here we demonstrate that EpCAM is a regulatory molecule in which its internalization into the nucleus turns on a transcription program. Activation of EGF/EGFR signal transduction triggered cell-surface cleavage of EpCAM, leading to nuclear internalization of its cytoplasmic domain EpICD. ChIP-seq analysis identified target genes that are co-regulated by EpICD and its transcription partner, LEF-1. Network enrichment analysis further uncovered a group of 105 genes encoding functions for tight junction, adherent and cell migration. Furthermore, nanomechanical analysis by atomic force microscope (AFM) revealed increased softness and decreased adhesiveness of EGF-stimulated cancer cells, implicating acquisition of an epithelial-mesenchymal transition (EMT) phenotype. Thus, genome editing of EpCAM could be associated with altering these nanomechanical properties towards a less aggressive phenotype. Using this integrative genomic-biophysical approach, we demonstrate for the first time an intricate relationship between EpCAM-regulated transcription and altered biophysical properties of cells that promote EMT in advanced endometrial cancer.
Introduction
Epithelial cell adhesion molecule (EpCAM) is a cell-surface protein known to mediate cell-cell and cell-matrix interactions (1,2). The extracellular domain of EpCAM (or EpEX) contains an N-terminal sequence, a thyroglobulin-like domain, and a C-terminal domain followed by a transmembrane domain and an intracellular domain (or EpICD) (3-5). The EpEX on the surface of one cell can bind to another EpEX on neighboring cells thereby holding these cells together (6). This EpCAM-mediated homophilic adhesion is further supported through inner interactions between EpICD and cytoplasmic fibers via α-actinin that serves as an intracellular bridge to stabilize the entire adhesion unit (1,2,7).
While EpCAM supports normal adhesion functions for epithelial cells, its transient down-regulation may promote epithelial-to-mesenchymal transition (EMT) for cancer cell migration and invasion (8). Also, circulating tumor cells (CTCs) bound to seed metastases in cancers of epithelial origin display very diverse levels of EpCAM expression, possibly related to their stage of EMT and invasiveness (9). In endometrial cancer, malignant cells must undergo EMT to facilitate myometrial invasion (10). However, upregulated EpCAM is frequently observed in endometrial tumors and is known to promote invasion by preventing cell-cell adhesion (11). Conditional knockout of EpCAM in a murine model attenuates the motility and migration of epidermis-resident Langerhans cells, further suggesting the role of EpCAM as a negative regulator for cell adhesion (12).
Therefore, these earlier studies indicate a paradoxical role of EpCAM for both cell adhesion and migration. On one hand, EpCAM mediates cell-cell contacts and thus prevents cell migration, but on the other hand the molecule can be switched to promote cell invasion. Recent studies reveal that regulated intra-membrane proteolysis (RIP) of EpCAM with γ-secretase results in shedding of the EpEX from the cell surface and release of EpICD into the cytoplasm (13). While the cleavage of EpEX may lead to a decrease in cell-cell adhesion and thereby promote cell movement, the event alone is insufficient to explain multifaceted influences of EpCAM on advanced cancer invasion and metastasis. It has been suggested that internalized EpICD subsequently forms a complex with β-catenin in the nucleus that regulate an oncogenic transcription program (13-15). Nevertheless, the molecular mechanisms underpinning this pleiotropic effect of EpCAM on advanced endometrial cancer development remain to be elucidated.
Here we report that activation of epidermal growth factor receptor (EGFR) signaling by a ligand triggers EpCAM cleavage leading to nuclear internalization of EpICD in endometrial cancer cells. The internalized EpICD interacts with LEF1 in β-catenin-mediated complexes that regulate gene transcription responsible for cell motility and migration. Atomic force microscopy (AFM) detected changes in nanomechanical properties of ligand-stimulated endometrial cancer cells, supporting the acquisition of an EMT phenotype. We also determined whether nanomechanical properties are reversed in cells carrying genome-edited EpCAM. In addition to removal of cell-surface EpCAM that lessens cell adhesiveness, we demonstrate how these nanomechanical changes are notably influenced through a transcription program regulated by its cleaved fragment, EpICD, for cancer cell progression.
Materials and Methods
Cell culture and treatment
Endometrial cancer cell lines (RL95-2, Ishikawa, and AN3CA) were obtained from the American Type Culture Collection (ATCC) and passaged in our laboratory for fewer than 6 months after resuscitation. All cell lines were regularly authenticated according to the guidelines provided by the ATCC based on morphology. For treatment, cells were serum starved in 10% heat-inactivated charcoal stripped FBS for overnight before adding EGF (10 ng/mL, PEPROTECH), Irresa (100 nM/mL, Tocris Bioscience) and/or DAPT (10 μM/mL, Selleckchem).
Immunofluorescence staining and Western blotting
Cells seeded onto cover glass slides were incubated with antibody against the extracellular-surface domain (EpEX) of EpCAM (AbD Serotec). Images were visualized by Nikon Eclipse microscope (Nikon Instruments). Western blot analysis was performed as described previously (16). Antibodies against α-tubulin (membrane/cytoplasmic fraction) and Lamin B1 (nuclear fraction, Abcam) were used as loading controls. Details of image quantification are described in Supplementary Material and Methods.
Proliferation and migration assays
The assays were performed in the Incucyte Zoom System (Essen Bioscience). Details of proliferation and migration are described in Supplementary Material and Methods.
ChIP-seq and network-based pathway analysis
Cells at different time points of EGF treatment were performed as previously described (17). Immunoprecipitation was carried out using antibodies against EpCAM (Santa Cruz Biotechnology) and LEF1 (Santa Cruz Biotechnology) on SX-8G-Star Compact Automated System following the manufacturer's instructions (Diagenode). Pull-down products were used to prepare libraries for high-throughput sequencing. Network-based pathway analysis was performed using algorithm NetPEA (18). Details are described in the Supplementary Material and Methods.
CRISPR/Cas9 genome-editing of EpCAM
LentiCRISPR-Cas9 pX330 plasmid was purchased from Addgene (Cambridge) and the oligo pairs targeting the first exon of EpCAM were designed according to the instructions (19). These cells diluted into single cells were seeded into 96-well plates. Stable clones were cultured for two months under Puromycin selection. Selected clones were verified by sequencing to ensure the success of EpCAM-editing, and the absence of EpCAM in these clones was assessed by Western blotting. Details are described in the Supplementary Material and Methods and all the primers used are listed in the Supplementary Table S1.
Reverse transcription PCR (RT-PCR) and BioMark system
RNA was isolated from EGF-treated and control cells and subjected to RT-PCR using SuperScript® III RT in the StepOnePlus™ Real-Time PCR Systems (Life Technologies). The BioMark system (Fluidigm) was used to examine the effects of EGF on the expression of EMT-related genes. The ΔΔCt was calculated for each gene using those of GAPDH and UBB for normalization. Primer sequences for RT-PCR and BioMark system are listed in Supplementary Table S1.
Nanomechanical imaging of cells with Atomic Force Microscopy (AFM)
Cells cultured to keep confluence below 50% were imaged in Petri dishes using a Nanoscope Catalyst (Bruker) atomic force microscope mounted on a Nikon Ti inverted epi-fluorescent microscope. For scanning, individual cells without forming colonies and physically contacting other cells were selected. To achieve the highest consistency of the data under the applied conditions, cells from a single dish were imaged only for up to 90 minutes. To determine mechanical properties of the cells, the Peak Force Quantitative Nanomechanical Mapping (PF-QNM) mode was performed through the software controlling the AFM (Bruker). SCANASYST-AIR (Bruker) probes with a spring constant of the nominal value 0.02 N/m was used and an exact value of the constant for each probe with the thermal tuning was determined. Details are described in the Supplementary Material and Methods.
Probing of cell surface with molecular recognition AFM
Adhesion forces between an extracellular domain of EpCAM molecules and an AFM probe functionalized with anti-EpCAM antibodies were measured to detect presence, evaluate adhesive properties, and determine distribution of EpCAM molecules on a cell surface. For probe functionalization, the procedure presented by Gruber (http://www.jku.at/biophysics/content) was followed. The modified SCANASYST-AIR (Bruker) probes were chemically modified in three steps: 1) probe activation with amine groups, 2) coupling of cross-linkers equipped long flexible hydrophilic chain to the amine groups, and 3) attaching monoclonal mouse (R&D Systems, MAB960), polyclonal goat anti-human EpCAM (AF960), or anti-Rpt5/S6a subunit of the human 26S proteasome (Enzo Life Sciences) antibodies to the free end of the cross-linker chain. Details are described in the Supplementary Material and Methods.
Accession numbers
ChIP-seq data are available at the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE72948.
Results
EGF stimulation promotes membranous proteolysis of EpCAM and nuclear internalization of EpICD
Overexpression of EpCAM is frequently observed in endometrial tumors (Supplementary Fig. S1) (20). However, it remains to be determined how this aberrant upregulation of EpCAM affects its cell-surface functions. Using an antibody recognizing EpEX (the extracellular-surface domain), we conducted immunofluorescence analysis of two endometrial cancer cell lines - RL95-2 with moderate expression of EpCAM and Ishikawa with high EpCAM expression (Fig. 1A). RL95-2 cells exhibited marked reduction of EpEX staining 24 hours after EGF stimulation, compared to that of untreated cells (Fig. 1A, upper-left panels and Supplementary Fig. S2A). This reduction was not noticed in Ishikawa cells with low expression of EGFR (Fig. 1A lower-left panels and Supplementary Fig. S2A and B). AN3CA cells, displaying no EpEX staining, were used as negative controls (Supplementary Fig. S2C).
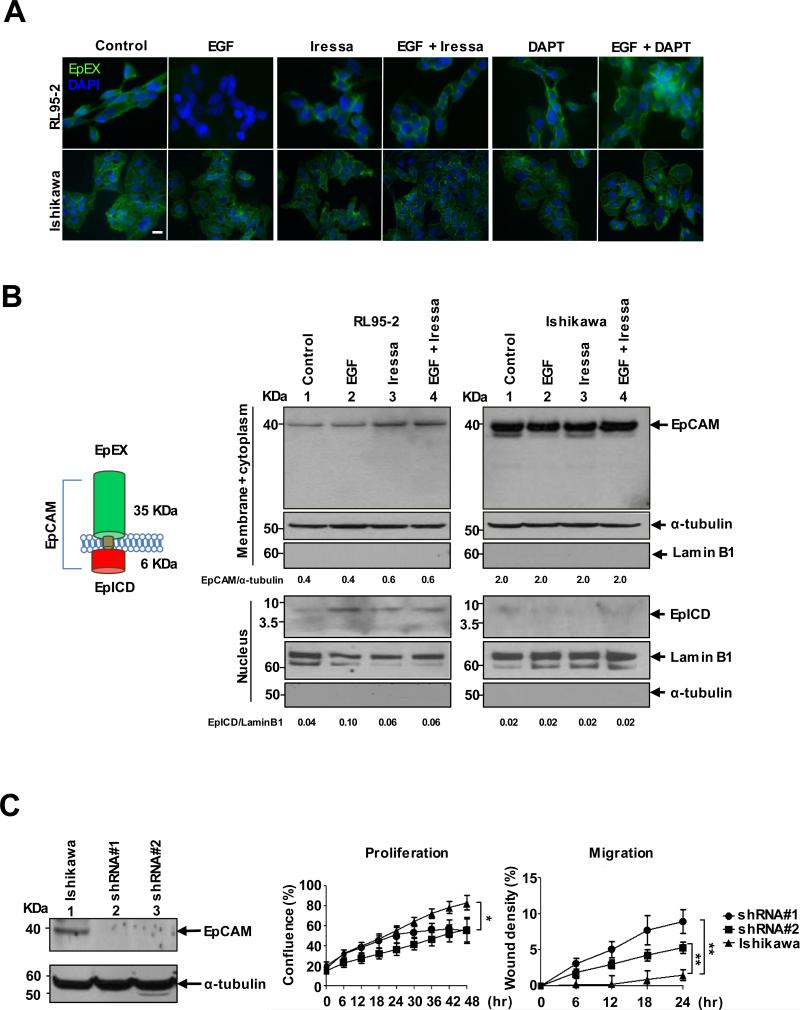
Figure 1.
Membranous proteolysis and nuclear translocation of EpCAM in EGF-stimulated cells. A, Immunofluorescence staining of EpEX in EGF- and/or Iressa and/or DAPT-treated RL95-2 and Ishikawa cells; scale bar, 10 μm. B, Western blot of EpCAM/EpICD of membrane/cytoplasmic and nuclear fractions in RL95-2 and Ishikawa cells. The relative band intensity is provided under the panels. C, shRNA knockdown of EpCAM enhanced migration and reduced proliferation in in Ishikawa cells determined as means ± SD from 3 independent experiments. *p< 0.05, **p<0.01.
We then determined whether this reduction of EpEX could be mediated through the canonical EGFR pathway. RL95-2 and Ishikawa cells were treated with the EGFR tyrosine phosphorylation inhibitor Iressa (100 ng/mL) in the presence or absence of EGF stimulation. This treatment prevented the loss of EpEX in RL95-2 cells, but had no effect on EGFR-low Ishikawa cells (Fig. 1A, middle panels and Supplementary Fig. S2A). To further investigate whether the reduction of EpEX could be attributed to an EGFR-mediated cleavage of EpCAM, we treated EGF-stimulated cells with DAPT (10 mg/mL). DAPT is an inhibitor of proteolytic activity of γ-secretase, involved in the release of EpICD (12). Apparently, the EpEX was retained upon stabilization of EpCAM by the inhibition of EpICD cleavage (Fig. 1A, upper-right panels). Again, the treatment had no effect on low-EGFR expressing Ishikawa cells (Fig. 1A lower-right panels and Supplementary Fig. S2A).
The cell-surface cleavage of EpCAM may cause the release of its intracellular domain EpICD from the cytoplasm to the nucleus (13). Alternatively, this proteolysis leads to degradation of the entire EpCAM protein. To differentiate these events, we conducted Western blotting of membrane/cytoplasmic and nuclear fractions in RL95-2 and Ishikawa cells treated with EGF and/or Iressa (Fig. 1B). The EGF stimulation led to an increase in nuclear accumulation of EpICD (6-kDa) in RL95-2 cells (left panel-lanes 1 and 2). However, this EGFR-mediated proteolysis of EpCAM was partially blocked by Iressa, leading to the accumulation of EpCAM in the membrane/cytoplasmic fraction (left panel-lanes 3 and 4). The EGF-dependent EpCAM cleavage event was not noticed in EGFR-low Ishikawa cells (Fig. 1B, right panel). Taken together, these results suggest that the activation of EGF/EFGR signaling not only causes an increase in EpCAM expression, but also renders a cleavage event leading to membranous dissociation of EpEX and nuclear accumulation of EpICD. Interestingly, while the knockdown of EpCAM by shRNA led to increased migration but decreased proliferation of Ishikawa cells (Fig. 1C), this downregulation had an opposite effect in RL95-2 cells in a subsequent study (see the migration assay in Supplementary Fig. S2D). Whereas the former event attenuates EpCAM-mediated cell surface adhesion, the latter may cause an EpICD-mediated transcription program for EMT.
EpICD-LEF1-coregulated targets are involved in tight junction and adherent function
To globally survey this EGF-mediated transcription program, we performed a time-course ChIP-seq in RL95-2 cells stimulated with the ligand for 0, 12, 24, and 48 hours. An in-house peak-calling algorithm, BELT, was developed to map EpICD binding sites in the genome (21). Overall, we identified a range from 11,813 to 12,291 peaks across the four time points of treatment with 5% of false discovery rate (Supplementary Fig. S3A). While the majority of binding peaks were located on intergenic regions, we found increased EpICD-binding events (up to 2016 loci) frequently located at transcription start sites (TSSs) and/or proximal regions, especially in 12- and 24-hour EGF treatments (Fig. 2A and Supplementary Fig. S3B). Although initial pathway analysis revealed that a great number of target loci are involved in housekeeping functions, we also identified a subgroup of loci linked to cell migration and oncogenic pathways (Supplementary Fig. S3C).
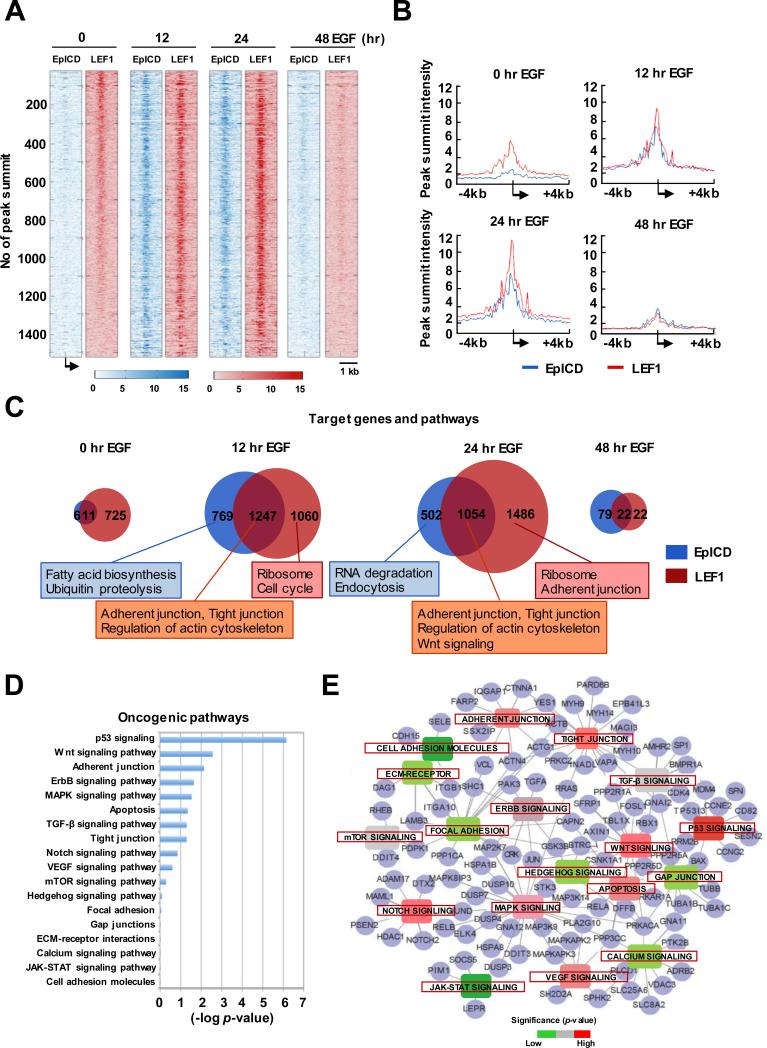
Figure 2.
Co-occupancies of EpICD and LEF1 in target loci involved in cell mobility functions. A, ChIP-seq peak summit alignments of EpICD and LEF1 in TSS regions in RL95-2 cells treated with EGF at different time points. Each row represents the same genes and the intensities of peaks are presented in colored scale bar. B, Peak summit intensity of target loci at different time points of EGF treatment. C, Venn diagrams of target loci and their biological functions at different time points of EGF stimulation. D, Oncogenic pathway analysis of 1247 EpICD-LEF1-regulated target loci. E, Interconnected signaling network of a subset of EpICD-LEF1-regulated target loci (n=105) involved in cell mobility functions.
Because EpICD has been shown to form β-catenin-mediated transcription complexes with LEF1 that is also overexpressed in endometrial tumors (Supplementary Fig. S1), we examined LEF1 binding profiles in RL95-2 cells by ChIP-seq (Supplementary Fig. S4). When integrating LEF1 and EpICD datasets, we further identified 1247 co-binding events that were mapped to TSSs of genes (i.e., based on peak summit intensities at 12 hours of EGF stimulation; Fig. 2B and Supplementary Fig. S5). The co-binding event means both peaks are located in +/− 1Kb from TSS of the same gene. Prior to EGF stimulation, LEF1 alone was found to bind the majority (>70%) of these regions, possibly attributed to activated β-catenin signaling in RL95-2 cells. After the stimulation, the number of co-binding events was increased and peaked around 12 to 24 hours, but drastically decreased at 48 hours of the treatment.
Next, we conducted KEGG pathway analysis to compare biological functions of 1247 EpICD-LEF1 co-bound loci relative to those of singular loci (22-24). While the majority of singular loci were related to biosynthesis and housekeeping functions, these co-bound loci were found to encode oncogenic functions associated with 18 signal transduction pathways (Fig. 2C-D). In an interconnected signaling network, we specifically identified 105 of these co-bound loci that are highly involved in tight junction, adherent, and focal adhesion (Fig. 2E). In addition, aberrant expression patterns of these candidate genes could be linked to advanced development of endometrial cancers in a Cancer Genome Atlas (TCGA) endometrial cancer cohort (Supplementary Fig. S6). Since these genes may play a role in endometrial cancer cell migration and invasion, we wanted to further dissect the mechanisms underlying this advanced phenotype.
Genome editing of EpCAM alters expression profiles of target genes linked to EMT
From the ChIP-seq data, we identified a group of genes co-bound by EpICD and LEF1 that are highly involved in cell mobility functions (i.e., adherent and tight junctions). We therefore determined whether disruption of EpCAM expression altered migration behaviors in RL95-2 cells. Toward this end, we used the CRISPR/Cas9 genome-editing system for which specific primers targeting the start codon ATG and the first exon of EpCAM were designed to disrupt its open reading frame. Two EpCAM-edited clones were selected; Clone 1 harbored a frameshift mutation whereas Clone 2 had a deletion of two codons from the region encoding the signal sequence (Fig. 3A, left panel). Western blot analysis confirmed the absence of EpCAM protein in Clone 1 and minute traces of the protein in Clone 2 (right panel). We then explored if these EpCAM-edited cells become less aggressive than wildtype cells. In a wound-healing assay, we observed that these EpCAM-edited cells showed a reduced ability to migrate during the course of EGF treatment, as compared to wildtype cells (p<0.005 for Clone 1 and p<0.05 for Clone 2; Supplementary Fig. S2D). This EpCAM gene disruption, however, did not influence the growth of these clonal cells.
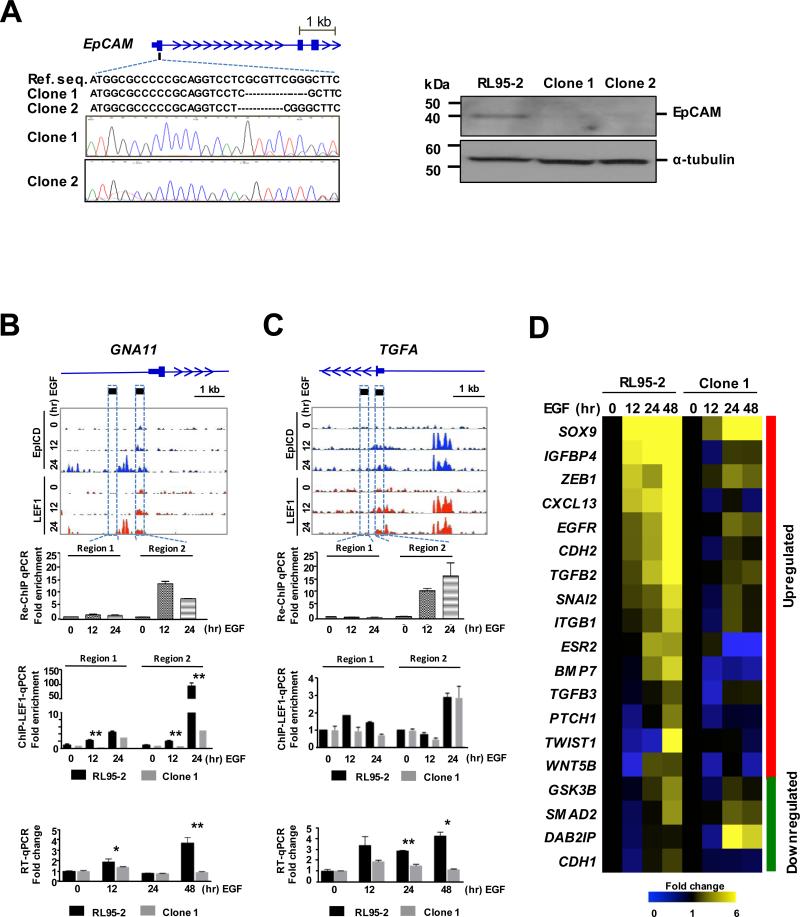
Figure 3.
Altered expression of EpICD-LEF1-regulated loci in EpCAM-edited cells. A, CRISPR/Cas9 genome editing of EpCAM. Left panel: Altered nucleotide sequences of the first exon of EPCAM in two edited clones. Right panel: Western blot of EpCAM in RL95-2 cells and two EpCAM-edited clones. B-C, Upper panels: Genomic binding landscape of EpICD and LEFT in two target loci, GNA11 and TGFA at three time points of EGF stimulation. Middle panels: re-ChIP-qPCR of EpICD and LEF1 co-binding and ChIP-qPCR of LEF1 binding, respectively. Fold enrichment is the fold change compared to control, the value is calculated as % of Input. Fold enrichment = % input of EGF-treated sample / % input of 0-hr EGF sample. Lower panels: RT-qPCR of mRNA expression in RL95-2 wildtype and EpCAM-edited cells in response to EGF stimulation. The results were determined as means ± SD from 3 independent experiments. *p<0.05, **p<0.01. D, Expression heat map of 19 genes involved in epithelial-mesenchymal transition (EMT) in RL95-2 and EpCAM-edited cells stimulated with EGF.
We then investigated whether this genome editing could alter the EpICD-LEF1 co-regulated transcription program in RL95-2 cells. A sequential (LEF1 then EpICD) chromatin immunoprecipitation (or re-ChIP)-qPCR assay confirmed this co-binding event at the 5’-ends of one target locus, GNA11 (see Region 2 at the 12- and 24-hour time points after EGF stimulation in Fig. 3B, upper panel). Then, we conducted a separate ChIP-qPCR study to determine whether the disruption of EpCAM has an effect on LEF1 binding. As shown in Fig. 3B (middle panel), a marked reduction (~20-fold) of LEF1 binding occurred at the GNA11 locus in EpCAM-edited cells relative to wildtype cells (p<0.01). As a result, a biphasic increase in the expression of GNA11 was no longer present in EpCAM-edited cells after the EGF stimulation (lower panel). However, a second co-regulated locus, TGFA, responded somewhat differently to the EpCAM disruption (Fig. 3C). Although the binding of LEF1 on TGFA was not overtly affected in EpCAM-edited cells, the EGF-mediated increase in the expression of this gene was significantly attenuated (p<0.01) likely attributed to the depletion of EpICD binding. Similar patterns of gene expressions were observed in EpCAM-edited clones (Supplementary Fig. S7A and 7B).
Since increased cell motility is a hallmark of EMT, a process linked to cancer progression and metastasis (25,26), we additionally determined whether the genome-edited EpCAM could influence the transcription programming of EMT. Microfluidic RT-qPCR was conducted to simultaneously analyze expression profiles of 19 known EMT-related genes in EpCAM-edited cells (Clone 1) and wildtype cells stimulated with EGF for 48 hours (Fig. 3D). As expected, EGF-stimulated wildtype cells showed up-regulation of mesenchymal gene markers and down-regulation of epithelial gene markers. However, these expression changes were less apparent in EpCAM-edited cells. Together, the finding suggests that this EpICD-LEF1 co-regulated gene transcription is directly linked to EMT for promoting endometrial cancer cell migration.
Genome editing of EpCAM affects nanomechanical properties of EGF-responsive cells
To gain insight into the mechanism of EpCAM-dependent changes in the mobility of EGF-stimulated RL95-2 cells, we employed single-cell imaging by atomic force microscopy (AFM). Beyond imaging of cell morphology and surface topography, the technique allows for high resolution and non-destructive mapping of cell elasticity and adhesion by recording a mechanical response of the micro-sized probe to its interaction with a cell (Fig. 4A, left panel) (27,28). For AFM, elasticity and adhesion constitute major components of a cellular nanomechanical phenotype (Fig. 4A, right panel). Elasticity is defined as the ability of a cell to return to its original shape after the removal of distorting force. The Young modulus is a measure of elasticity and is expressed in units of pressure (i.e., kilopascal or kPa). The higher value of the Young modulus, the more rigid and less soft is the object. Adhesion, recorded in the units of force (i.e., piconewton or pN), quantifies forces needed to separate dissimilar objects, here an AFM tip from a cell. The higher value of adhesion, stickier is the object. Based on collected images, the Young modulus and adhesion for each cell was calculated. For consistency, only single cells, not in contact with neighboring cells, were analyzed. Untreated RL95-2 cells were relatively rigid with the Young modulus broadly extending from 2.3 to 34 kPa (Fig. 4B-C). This diversity was severely decreased when the cells were stimulated with EGF. Moreover, after 12- and 24-hour treatments the cells were about 10 times softer than control cells (Fig. 4B-C). The effect of EGF treatment was much less pronounced after 48 hours, showing again a higher variance of increased cell rigidity. This pattern of EGF-induced changes was followed by cell adhesiveness. Again, untreated cells and cells treated for 48 hours were very variable and the most adhesive. After 12 hours, EGF-exposed cells were 6 times less adhesive with much smaller variance (Fig. 4C). This nanomechanical change appeared not to be associated with apoptosis in EGF-treated or control cells (Supplementary Fig. S8A).
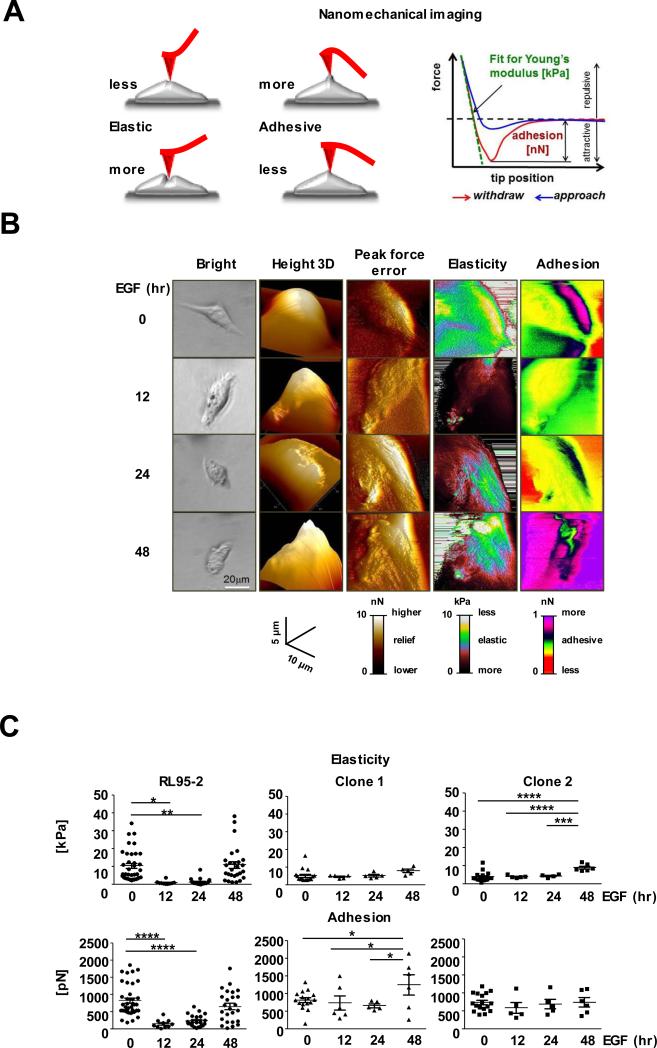
Figure 4.
Altered nanomechanical features of EGF-responsive RL95-2 cells detected by Atomic Force Microscopy (AFM). A, Left panel: a scheme of interactions of an AFM probe (red) with a cell (grey) illustrating cell mechanical responses to the tip indentation. Right panel: a force plot depicts dependence of the force challenging the tip on tip distance (position) from a cell surface. B, Examples of nanomechanical features of individual RL95-2 cells responding to the EGF treatment. Components of the mechanical phenotype of the same cell are arranged in columns. Images of cells at the specified time points of EGF treatment are in rows. C, Elasticity and adhesion of individual RL95-2 cells and two EpCAM-edited clones. EGF stimulation of RL95-2 cells increased cell elasticity but decreased their surface adhesion. The nanomechanical phenotype of the EpCAM-edited clones was refractory to the EGF treatment. Vertical line represents mean ± SD. *p< 0.05, **p<0.01, ***p<0.005, ****p<0.001.
EpCAM-edited cells became refractory to the EGF treatment and exhibited relatively little cell-to-cell variations in their mechanical properties. Interestingly, cells from both clones were about two times softer than wildtype RL95-2 cells, although their adhesiveness remained unchanged when compared to wildtype cells. Apparently, the absence of EpCAM did not uniformly affect the nanomechanical properties in the edited clones.
To further investigate whether EGF stimulation and an increase of cell softness are accompanied by re-organization of cytoskeleton (29), we performed immunofluorescence analysis of two related structural proteins, α-tubulin and actin in wildtype and EpCAM-edited cells (Supplementary Fig. S8B). The differences in content and organization of actin cytoskeleton between control and EGF-treated cells or between wildtype and EpCAM-edited cells are evident in single cell images.
Molecular recognition AFM confirms gradual removal of EpCAM from the surface of EGF-responsive cells
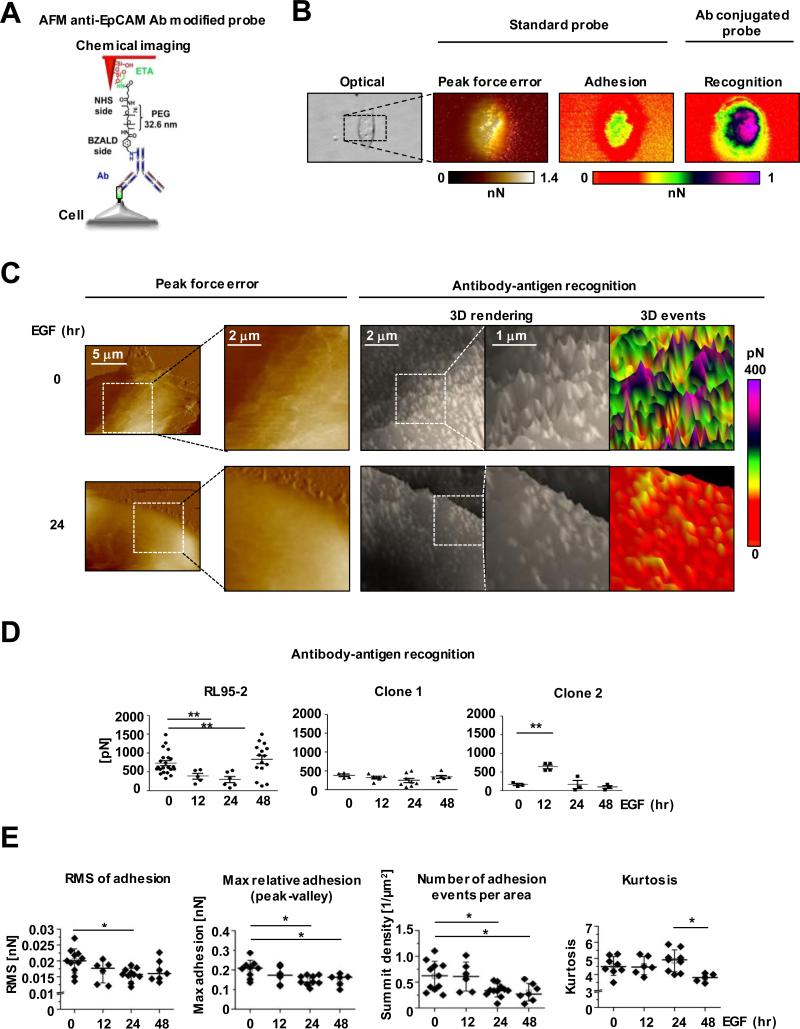
To gain understanding of the cell-surface distribution of EpCAM, we used EpCAM-specific recognition AFM. In this technique, AFM measures forces between a target molecule (receptor) and an AFM tip chemically functionalized with a “bait” molecule (ligand). In particular, forces of separation of the ligand and receptor are measured. Additionally, this AFM mode enables to correlate recognition maps, indicative of a specific distribution of the target molecule with topographical and mechanical features of a cell assessed with a standard, not modified tip. Here we functionalized ScanAsyst-Air probes with antibodies recognizing the EpEX domain (Fig. 5A). We examined the same RL95-2 cells first with the standard, and then with the modified probe (Fig. 5B). Under the applied imaging conditions, the standard tip produced a smooth surface relief for all the tested cells. As expected, adhesion between the standard tip and cell membrane was relatively weak. In contrast, the modified probe was adhering much more strongly to the cell membrane, suggesting the rich presence of EpEX molecules recognized by specific antibodies attached to the probe (Fig. 5B and Supplementary Fig. S9A). It was clear that the map of recognition events did not spatially correlate with the adhesion image collected with the unmodified probe. As a negative control, we also determined that a probe modified with antibody recognizing intracellular, non-membrane Rpt5 protein, which is a part of the 26S proteasome, failed to detect any strong recognition events on RL95-2 cells (Supplementary Fig. S9B). Also, only very weak, presumably nonspecific events remained when the anti-EpCAM-Abs-modified tip was used to probe cells that were pre-incubated with the same Abs (Supplementary Fig. S9C). To the contrary, EpCAM molecules were detected as dense uniformly distributed sharp “warps” on a cell surface of positive control cells with strong EpCAM expression (Supplementary Fig. S9D-E). Closer examination of the recognition maps of RL95-2 cells revealed that EpCAM molecules were presented as regular small adhesion “hills” relatively uniformly dispersed on a cell surface (Fig. 5C). Upon stimulation with EGF, the number of recognition events and their apparent strength (i.e., height of the hills) decreased in a time-dependent manner, from about 780 pN for untreated cells to 390 and 300 pN for the 12- and 24-hour treatments (Fig. 5D). On the other hand, the EpCAM-edited clones showed very low strength of recognition and no time dependent changes upon EGF exposure. These results strongly indicate that EpCAM molecules are removed from the membrane surface of EGF-treated RL95-2 cells and are missing from the EpCAM-edited cells. One surprising effect was an increase of the strength of recognition events in RL95-2 cells 48 hours after EGF treatment (Fig. 5D). We speculate that at the last step of EpCAM removal from the cell surface, the antibody-decorated probe interacts with small isolated islands of EpCAM molecules. This way, many EpCAM molecules that were obscured from interactions with ligands by crowding, became exposed and receptive to antibodies. The notion of the EGF-dependent loss of EpCAM molecules from the RL95-2 cell surface was further strengthened by the analysis of surface roughness of the recognition maps (Fig. 5E and Supplementary Fig. S9E). Indeed, the root mean square (RMS), a commonly used parameter describing surface roughness, as well as average roughness and maximal relative adhesion, all decreased with the EGF exposure time. Taken together, this novel approach provides a visualization tool to directly monitor the EGF-dependent removal of surface EpCAM to the point of only isolated islands remaining in endometrial cancer cells, further confirming the aforementioned EpCAM cleavage event by immunofluorescence analysis and Western blotting.
Figure 5.
Gradual removal of EpCAM molecules from the surface of RL95-2 cells detected by chemical recognition atomic force microscopy (AFM). A, A scheme of the anti-EpCAM-antibody-conjugated AFM tip. ETA - ethanolamine, PEG76 - polyethylene glycol 76, NHS - N-hydroxysuccinimide, and BZALD - benzaldehyde. B, Images of the same single control RL95-2 cell acquired with bright field microscopy and AFM. Peak Force Error and adhesion images were rendered with the non-modified tip, the recognition image was captured with the functionalized tip. C, AFM imaging with the functionalized tip detected loss of EpCAM molecules after the 24-hour treatment of RL95-2 cells with EGF. The 3D Events panels were plane corrected. D, Decrease of recognition forces measured with the functionalized probe reveals gradual removal of EpCAM molecules from the cell surface. The EpCAM-edited cells did not register recognition events. E, Roughness analysis of recognition events confirms gradual disappearance of EpCAM molecules from a RL95-2 cell surface and detects formation of EpCAM islands. Root mean square (RMS) of adhesion and the maximal relative adhesion systematically decreased with the EGF exposure time. Events density dropped abruptly after the 12-hour exposure but kurtosis increased indicating a more pointed surface.
Discussion
In addition to its structural role for cell-surface adhesion, EpCAM has a functional role that promotes a transcription program of EMT within the nucleus. This unique process is initiated through activation of the EGF/EGFR signaling, leading to membranous cleavage of EpEX and nuclear accumulation of EpICD. While the EpCAM cleavage was previously reported in advanced cancer types (30-33), the mechanistic cause underlying this process has not been elucidated. Using a combined genomic-biophysical approach, we found that this cleavage event triggered a concerted effort resulting in promotion of an EMT phenotype in endometrial cancer cells. Whereas our ChIP-seq analysis identified an EpICD-LEF1 co-regulated transcription module associated with tight junction and adherent junction, AFM analyses additionally detected nanomechanical changes in cell-surface properties of EGF-stimulated cells. Our integrative approach notably demonstrates that a nanomechanical phenotype can be directed through a transcription program for advanced cancer progression (see the model in Fig. 6).
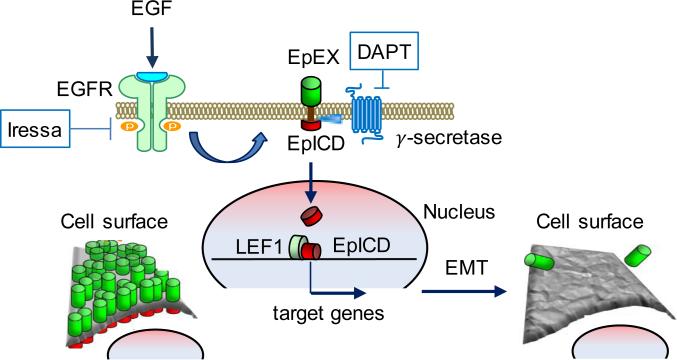
Figure 6.
A proposed model of cell response to the EGF treatment. EGF binding to an EGFR triggers cleavage of membrane EpCAM to the intracellular EpICD part that after nuclear translocation together with LEF1 targets expression of genes responsible for cell mobility. Simultaneous removal of the extracellular EpEX part reduces cell adhesion and increases cell elasticity. Therefore, both parts of the EpCAM molecule using distinct molecular mechanisms collaborate toward formation of the more invasive phenotype supporting tumor progression.
This nanomechanical phenotype is a complex result of the unique properties of cell membrane, underlying cytoskeleton and cell turgor, which together reflect an epithelial-mesenchymal transition for promoting cell mobility (34). Nanomechanical parameters, most notably elasticity and adhesion, have been proposed to provide a rich resource for robust characterization of human cells, including distinguishing between cancer and normal cells, and also for fine stratification of cancer cells according to their malignancy (35,36). The increase in softness and decrease in adhesiveness could be traced to EMT. The switch from the apical-basal epithelial phenotype into the spindle-shaped mesenchymal counterpart signals readiness of the cell for invasion (37). This EMT process is accompanied by profound changes in cytoskeleton architecture and in membrane characteristics, including distribution and type of surface proteins and lipid rafts, detectable by AFM (38). In our case, the low rigidity (low Young modulus; soft cells) of EGF-stimulated RL95-2 cells accompanies acquisition of an aggressive phenotype attributed to the described EpCAM-mediated transcription program. Interestingly, the rigidity of EpCAM-edited cells was relatively higher than EGF-treated wildtype cells with the surface EpCAM cleaved-off. This observation suggests that the EpCAM editing directly affects properties of the cytoskeleton, independent of the EpICD/LEF1-regulated transcription program in wildtype cells.
Contrary to the EpCAM-edited cells, the adhesiveness of RL95-2 cells decreased upon EGF treatment. Such decrease is consistent with increased mobility and increased invasiveness of these cells. The phenomenon has been noted in biophysical studies of cancer cells before and is not at odds with EpCAM as a surface adhesion molecule (27). First, EpCAM is not the sole adhesion molecule on the cell surface, with other cadherins likely playing the most important role and responding to EMT signaling (39,40). Second, the standard AFM probe detects adhesion between the cell and any surface, not the protein-specific cell–to-cell adhesion moderated by EpCAM. Therefore, the loss of EpCAM alone does not need to translate into changes in general cell adhesion when the EMT signaling is disrupted. The result is consistent with a recent report by Tsaktanis and colleagues (41) that a cleavage or knockout of EpCAM did not affect adhesion of cells population to the matrix. Our finding stresses possible distinct roles played by EpEX and EpICD. It also suggests that EpCAM is not only responsible for cell adhesion but influences elasticity as well.
Supporting our previous observation that EGF treatment led to changes in an epithelial morphology to a more fibroblastic mesenchymal appearance in RL95-2 cells (16), the AFM results further provide quantitative proof of their elevated softness and reduced adhesiveness, leading to an increase in cell movement. This enhanced cell movement follows EGF stimulation in a time-dependent manner. Again, the EpCAM-edited cells showed a different response from wildtype RL95-2 cells, suggesting that the presence of EpCAM is essential for EGF-stimulated advanced phenotype of endometrial cancer cells.
Molecular recognition AFM enabled us to bring the single cell surface mapping to the single molecule level. With the help of AFM probes functionalized with EpEX specific antibodies, we were able to precisely follow the abundance and distribution of EpCAM molecules by monitoring the receptor (EpEX) - ligand (antibodies) interactions on the cell surface without the limitation of resolution of optical methods. The molecular recognition AFM was successfully applied before in single cell studies (42-45). In our studies we successfully monitored events of EpEX-antibody binding and separation. The expected force of single molecule antibody-protein antigen separation is in the range of 100 pN - 200 pN (46). We routinely observed events of at least twice as high forces. Such outcome was expected taken into account the likely dense cover of EpEX receptors and polyvalent recognition events with simultaneous binding of multiple ligands attached to the probe to multiple receptors on the cell surface. Furthermore, recognition events on the EpCAM-edited cells were only registered by very weak forces similar to cells treated with EGF for 12 and 24 hours. Therefore, we conclude that likely EpCAM molecules were absent from a surface of the EpCAM-edited cells. This is in agreement with our immunofluorescence and Western blotting data. In addition to recording the strength of ligand-receptor interactions, the molecular recognition mapping enables exploration of distribution pattern of recognition events by analyzing the roughness of the maps. Roughness is a morphology-derived parameter independent of the overall geometric shape of a cell (47). In our case, decreasing roughness indicated that the EpCAM molecules gradually disappear from the surface of EGF-treated endometrial cancer cells.
In summary, our studies show that EGF/EGFR-mediated cleavage of EpCAM initiates dual actions. First, the cleavage triggers EpICD-LEF1-mediated transcription program promoting EMT. Second, the loss of EpEX directly affects cell membrane and changes nanomechanical properties toward a more aggressive phenotype. Since many of the EpICDLEF1 target genes are involved in cell adhesion functions, the transcription program supplements the direct actions in cytoskeleton remodeling and further modulation of the nanomechanical phenotype. Thus, the two processes act in concert to support and enhance invasiveness of endometrial cancer cells.
Supplementary Material
Acknowledgements
The authors acknowledge the assistance of the Next-Generation Sequencing Core for process ChIP-seq samples and the BioAnalytics and Single-Cell Core for atomic force microscopy studies at the University of Health Science Center-San Antonio.
Grant Support
This work was supported by NIH grants R01CA172279, U54CA113001, and P30CA054174; the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP150600; the University of Texas System STAR award; gift of the Cancer Therapy and Research Center Foundation; and the Max and Minnie Tomerlin Voelcker Fund (all authors received). Y-T. Hsu. is a recipient of predoctoral fellowship from the CPRIT grant RP140105.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol. 1994;125:437–46. doi: 10.1083/jcb.125.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzar M, Bakker HA, Briaire-de-Bruijn IH, Fleuren GJ, Warnaar SO, Litvinov SV. Cytoplasmic tail regulates the intercellular adhesion function of the epithelial cell adhesion molecule. Mol Cell Biol. 1998;18:4833–43. doi: 10.1128/mcb.18.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavsic M, Guncar G, Djinovic-Carugo K, Lenarcic B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat Commun. 2014;5:4764. doi: 10.1038/ncomms5764. [DOI] [PubMed] [Google Scholar]
- 4.Pavsic M, Lenarcic B. Expression, crystallization and preliminary X-ray characterization of the human epithelial cell-adhesion molecule ectodomain. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1363–66. doi: 10.1107/S1744309111031897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baeuerle P, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–23. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17-1A antigen (Ep-CAM). J Mol Med. 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- 7.Balzar M, Briaire-de Bruijn IH, Rees-Bakker HA, Prins FA, Helfrich W, de Leij L, et al. Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol Cell Biol. 2001;21:2570–80. doi: 10.1128/MCB.21.7.2570-2580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CW, Liao MY, Lin WW, Wang YP, Lu TY, Wu HC. Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial-mesenchymal transition gene expression in colon cancer. J Biol Chem. 2012;287:39449–59. doi: 10.1074/jbc.M112.386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CL, Mahalingam D, Osmulski P, Jadhav RR, Wang CM, Leach RJ, et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate. 2013;73:813–26. doi: 10.1002/pros.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colas E, Pedrola N, Devis L, Ertekin T, Campoy I, Martinez E, et al. The EMT signaling pathways in endometrial carcinoma. Clin Transl Oncol. 2012;14:715–20. doi: 10.1007/s12094-012-0866-3. [DOI] [PubMed] [Google Scholar]
- 11.Nubel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, et al. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res. 2009;7:285–99. doi: 10.1158/1541-7786.MCR-08-0200. [DOI] [PubMed] [Google Scholar]
- 12.Gaiser M, Lämmermann T, Feng X, Igyarto B, Kaplan D, Tessarollo L, et al. Cancer-associated epithelial cell adhesion molecule (EpCAM; CD326) enables epidermal Langerhans cell motility and migration in vivo. Proc Natl Acad Sci USA. 2012;109:97. doi: 10.1073/pnas.1117674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 14.Chaves-Pérez A, Mack B, Maetzel D, Kremling H, Eggert C, Harréus U, et al. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene. 2013;32:641–50. doi: 10.1038/onc.2012.75. [DOI] [PubMed] [Google Scholar]
- 15.Jachin S, Bae JS, Sung JJ, Park HS, Jang KY, Chung MJ, et al. The role of nuclear EpICD in extrahepatic cholangiocarcinoma: association with beta-catenin. Int J Oncol. 2014;45:691–8. doi: 10.3892/ijo.2014.2472. [DOI] [PubMed] [Google Scholar]
- 16.Hsu YT, Gu F, Huang YW, Liu J, Ruan J, Huang RL, et al. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clin Cancer Res. 2013;19:6272–85. doi: 10.1158/1078-0432.CCR-13-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu P-YY, Hsu H-KK, Singer GA, Yan PS, Rodriguez BA, Liu JC, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–44. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Ruan J. Network-based Pathway Enrichment Analysis. Proceedings IEEE International Conference on Bioinformatics and Biomedicine. 2013:218–21. doi: 10.1109/BIBM.2013.6732493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Sahwi K, Bellone S, Cocco E, Casagrande F, Bellone M, Abu-Khalaf M, et al. Overexpression of EpCAM in uterine serous papillary carcinoma: implications for EpCAM-specific immunotherapy with human monoclonal antibody adecatumumab (MT201). Mol Cancer Ther. 2010;9:57–66. doi: 10.1158/1535-7163.MCT-09-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan X, Bonneville R, Apostolos J, Wu W, Jin VX. W-ChIPeaks: a comprehensive web application tool for processing ChIP-chip and ChIP-seq data. Bioinformatics. 2011;27:428–30. doi: 10.1093/bioinformatics/btq669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–28. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 27.Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK. AFM-based analysis of human metastatic cancer cells. Nanotechnology. 2008;19:384003. doi: 10.1088/0957-4484/19/38/384003. [DOI] [PubMed] [Google Scholar]
- 28.Osmulski P, Mahalingam D, Gaczynska ME, Liu J, Huang S, Horning AM, et al. Nanomechanical biomarkers of single circulating tumor cells for detection of castration resistant prostate cancer. Prostate. 2014;74:1297–307. doi: 10.1002/pros.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gradya ME, Compostoa RJ, Eckmannb DM. Cell elasticity with altered cytoskeletal architectures across multiple cell types. J Mech Behav Biomed. 2016;61:197–207. doi: 10.1016/j.jmbbm.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Ralhan R, He HC, So AK, Tripathi SC, Kumar M, Hasan MR, et al. Nuclear and cytoplasmic accumulation of Ep-ICD is frequently detected in human epithelial cancers. PLoS One. 2010;5:e14130. doi: 10.1371/journal.pone.0014130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maghzal N, Vogt E, Reintsch W, Fraser JS, Fagotto F. The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J Cell Biol. 2010;191:645–59. doi: 10.1083/jcb.201004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong D, Moser P, Kasal A, Seeber A, Gastl G, Martowicz A, et al. Loss of membranous expression of the intracellular domain of EpCAM is a frequent event and predicts poor survival in patients with pancreatic cancer. Histopathology. 2014;64:683–92. doi: 10.1111/his.12307. [DOI] [PubMed] [Google Scholar]
- 33.Ralhan R, Cao J, Lim T, Macmillan C, Freeman JL, Walfish PG. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC Cancer. 2010;10:331. doi: 10.1186/1471-2407-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tartibi M, Liu YX, Liu GY, Komvopoulos K. Single-cell mechanics - an experimental-computational method for quantifying the membrane-cytoskeleton elasticity of cells. Acta Biomaterialia. 2015;27:224–235. doi: 10.1016/j.actbio.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 35.Lekka M, Laidler P, Gil D, Lekki J, Stachura Z, Hrynkiewicz AZ. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur Biophys J. 1999;28:312–16. doi: 10.1007/s002490050213. [DOI] [PubMed] [Google Scholar]
- 36.Lekka M, Pogoda K, Gostek J, Klymenko O, Prauzner-Bechcicki S, Wiltowska-Zuber J, et al. Cancer cell recognition--mechanical phenotype. Micron. 2012;43:1259–66. doi: 10.1016/j.micron.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21:89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orsini F, Cremona A, Arosio P, Corsetto PA, Montorfano G, Lascialfari A, et al. Atomic force microscopy imaging of lipid rafts of human breast cancer cells. Biochim Biophys Acta. 2012;1818:2943–49. doi: 10.1016/j.bbamem.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Camand E, Peglion F, Osmani N, Sanson M, Etienne-Manneville S. N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J Cell Sci. 2012;125:844–57. doi: 10.1242/jcs.087668. [DOI] [PubMed] [Google Scholar]
- 40.Chen GT, Waterman ML. Cancer: leaping the E-cadherin hurdle. EMBO J. 2015;34:2307–09. doi: 10.15252/embj.201592757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsaktanis T, Kremling H, Pavšič M, von Stackelberg R, Mack B, Fukumori A, et al. Cleavage and Cell Adhesion Properties of Human Epithelial Cell Adhesion Molecule hEpCAM. J Biol Chem. 2015;290:24574–91. doi: 10.1074/jbc.M115.662700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chtcheglova LA, Waschke J, Wildling L, Drenckhahn D, Hinterdorfer P. Nano-scale dynamic recognition imaging on vascular endothelial cells. Biophys J. 2007;93:11–3. doi: 10.1529/biophysj.107.109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laurent VM, Duperray A, Sundar Rajan V, Verdier C. Atomic force microscopy reveals a role for endothelial cell ICAM-1 expression in bladder cancer cell adherence. PLoS One. 2014;9:e98034. doi: 10.1371/journal.pone.0098034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat Cell Biol. 2000;2:313–17. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Chtcheglova LA, Zhu R, Hinterdorfer P, Zhang B, Tang J. Nanoscale organization of human GnRH-R on human bladder cancer cells. Anal Chem. 2014;86:2458–64. doi: 10.1021/ac403304g. [DOI] [PubMed] [Google Scholar]
- 46.Schwesinger F, Ros R, Strunz T, Anselmetti D, Guntherodt HJ, Honegger A, et al. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc Natl Acad Sci USA. 2000;97:9972–77. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis LW, Lewis PD, Wright CJ, Conlan RS. Atomic force microscopy comes of age. Biol Cell. 2010;102:133–43. doi: 10.1042/BC20090127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.