Abstract
Background & Aims
Nonalcoholic fatty liver disease (NAFLD) is a consequence of defects in diverse metabolic pathways that involve hepatic accumulation of triglycerides. Features of these aberrations might determine whether NAFLD progresses to nonalcoholic steatohepatitis (NASH). We investigated whether the diverse defects observed in patients with NAFLD are due to different NAFLD subtypes with specific serum metabolomic profiles, and whether these can distinguish patients with NASH from patients with simple steatosis.
Methods
We collected liver and serum from methionine adenosyltransferase 1a knockout (MAT1A-KO) mice, which have chronically low level of hepatic S-adenosylmethionine (SAMe) and spontaneously develop steatohepatitis, as well as C57Bl/6 mice (controls); the metabolomes of all samples were determined. We also analyzed serum metabolomes of 535 patients with biopsy-proven NAFLD (353 with simple steatosis and 182 with NASH) and compared them with serum metabolomes of mice. MAT1A-KO mice were also given SAMe (30 mg/kg/day for 8 weeks); liver samples were collected and analyzed histologically for steatohepatitis.
Results
Livers of MAT1A-KO mice were characterized by high levels of triglycerides, diglycerides, fatty acids, ceramides, and oxidized fatty acids, as well as low levels of SAMe and downstream metabolites. There was a correlation between liver and serum metabolomes. We identified a serum metabolomic signature associated with MAT1A-KO mice that was also present in 49% of the patients; based on this signature, we identified 2 NAFLD subtypes. We identified specific panels of markers that could distinguish patients with NASH from patients with simple steatosis for each subtype of NAFLD. Administration of SAMe reduced features of steatohepatitis in MAT1A-KO mice.
Conclusions
In an analysis of serum metabolomes of patients with NAFLD and MAT1A-KO mice with steatohepatitis, we identified 2 major subtypes of NAFLD and markers that differentiate steatosis from NASH in each subtype. These might be used to monitor disease progression and identify therapeutic targets for patients.
Keywords: mouse model, lipid metabolism, one-carbon metabolism, prognostic
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD), the leading cause of chronic liver disease in Western countries, starts with the excessive accumulation of hepatic triglycerides (TG), which in some cases progresses to nonalcoholic steatohepatitis (NASH), a condition characterized by the appearance of inflammation, cellular injury with or without fibrosis together with steatosis1. Although patients with simple steatosis are thought to have better prognosis, the overall morbidity and mortality is increased in NASH patients2. Hepatic steatosis arises when de novo lipogenesis (DNL) and the uptake of fatty acids (FA) from circulation saturate the capacity of the liver to oxidize FA and their elimination as TG in the form of very-low density lipoproteins (VLDL)1. As there are different causes that can lead to steatosis, we hypothesized that different NAFLD subtypes may exist reflecting the variety of mechanisms causing liver fat accumulation, and that each subtype would be characterized by a unique serum metabolomic signature. This is relevant because a better understanding of NAFLD biology will facilitate the development of personalized treatment.
We assessed this hypothesis by first analyzing the liver metabolome of MAT1A-KO mice3 to determine the mechanism causing liver fat accumulation in this model of steatohepatitis. This model is relevant to humans, as NASH patients frequently show reduced MAT1A mRNA levels4. Then, we examined if the liver metabolome is reflected in serum in mice. This is an important consideration, since serum and not liver will be the basis for future classification of human NAFLD into subtypes. After that, we investigated whether a large cohort of NAFLD patients could be classified into different subtypes based on their similarity with the serum metabolome of MAT1A-KO mice, and searched for the existence of serum metabolic biomarkers that distinguished NASH from simple steatosis in each subtype. Finally, we assessed if SAMe administration improved steatohepatitis in MAT1A-KO mice.
MATERIALS AND METHODS
Animal experiments
Eight-month old MAT1A-KO5 male mice (in a C57Bl/6 background) with increases in levels of liver enzymes (ALT and AST) and hepatic lipid accumulation (determined by ultrasound) were given orally by gavage with vehicle (water) (n=12) or SAMe (Abbott, Chicago, IL; 30 mg/kg/day, n=12) for 8 weeks before sacrificing. Age-matched wild-type male sibling littermates showing normal liver serum enzymes and ultrasound were also treated with vehicle for the same duration (n=11). Animals were bred and housed in the CIC bioGUNE animal unit, accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The mice were housed in groups using high quality wood pellet hygienic litter bedding (Lignocel HBK 1500–3000, Rettenmaier & Sönne, Germany) and in the presence of enrichment materials. Animals were fed with standard commercial chow animal diet (Ref. 2914, Envigo, Barcelona, Spain). Submandibular and retro orbital blood samples were collected at the beginning and at the end of the experiment. Blood samples were deposited in serum separator gel tubes (Microtainer, Becton-Dickinson, Franklin Park, NJ) and centrifuged (6000 rpm, 15 min, 4°C) for serum separation. Livers were remo ved and snap frozen in liquid nitrogen, OCT cryo-compound embedded or formalin fixed. All procedures were performed during the light cycle and were approved by the Diputación de Bizkaia upon a favorable assessment by the Institutional Animal Care and Use Committee at CIC bioGUNE.
Patients
Our study included total of 535 patients who underwent liver biopsy analysis (353 with diagnosis of simple steatosis and 182 of NASH), seen at 11 participating hospitals. Among them, 377 patients were previously described by Barr et al6, and 158 additional patients were recruited since 2013 for this study by three hospitals that also participated in the first study (Hospital Universitario Marqués de Valdecilla, Hospital Universitario Príncipe de Asturias, and Hospital Virgen de Valme). Principal component analysis (PCA) of the metabolomics data showed that patients cluster together independently of the hospital of origin (supplementary Figure 1). All patients were recruited using the following inclusion criteria: (1) age 18–75 years; (2) no known acute or chronic disease except for obesity or type 2 diabetes based on medical history, physical examination, and standard laboratory tests; (3) alcohol consumption was less than 20 g/day for women and 30 g/day for men. Exclusion criteria included viral-, autoimmune-, hemochromatosis- and drug-induced causes of liver disease. All of the subjects were of Caucasian origin. Additional details have been published6. The institutional review board at each of the participating hospitals approved the study and written informed consent was obtained from all patients. For all subjects, blood was drawn under fasting conditions on the morning the diagnostic liver biopsy was performed. Serum was separated and stored at −80°C until analysis. Clinical data (Table 1) were collected retrospectively using patient records and laboratory values obtained at the time of biopsy.
Table 1. Clinicopathological characteristics of the NAFLD patients included in the study.
All diagnoses were established histologically in liver biopsy specimens. Additional classification as M-subtype, non-M-subtype or indeterminate is based on results detailed in section “Discovery of two human NAFLD subtypes”. Values are given as percentage or mean±standard deviation of the mean. Results that were significantly different (P<.05) among the M-subtype, non-M-subtype or indeterminate groups are indicated.
| Total | M-subtype | Non-M-subtype | Indeterminate | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Simple steatosis | NASH | Simple steatosis | NASH | Simple steatosis | NASH | Simple steatosis | NASH | |
| N (%) | 535 | 353 (66%) | 182 (34%) | 174 (66%) a | 88 (34%) d,e | 103 (61%) c | 68 (39%) | 76 (75%) | 26 (25%) |
| % (female) | 69% | 70% | 68% | 72% a | 77% d,e | 62% c | 57% | 77% | 65% |
| Age (years) | 44.77 ± 11.35 | 44.04 ± 11.54 | 46.07 ± 10.94 | 40.31 ± 10.77 a,b | 43.64 ± 9.68 d | 48.35 ± 11.29 | 49.30 ± 11.44 | 46.77 ± 10.89 | 45.36 ± 11.81 |
| BMI (kg/m2) | 44.76 ± 10.78 | 44.50 ± 10.65 | 45.21 ± 11.02 | 48.19 ± 9.71 a,b | 49.72 ± 8.67 d,e | 40.54 ± 10.34 | 40.66 ± 11.18 f | 41.12 ± 10.30 | 42.01 ± 12.02 |
| Fasting glucose (mg/dL) |
113.59 ± 41.26 | 111.39 ± 35.86 | 117.31 ± 48.96 | 112.21 ± 35.97 | 119.21 ± 51.93 e | 106.32 ± 27.47 | 119.89 ± 50.82 f | 117.10 ± 45.01 | 101.05 ± 21.03 |
| Total fasting cholesterol (mg/dL) |
188.71 ± 41.08 | 187.84 ± 41.88 | 190.66 ± 39.45 | 176.88 ± 35.29 a | 175.01 ± 27.60 e | 196.59 ± 47.31 | 192.11 ± 43.89 | 191.69 ± 39.20 | 213.67 ± 23.56 |
| Fasting HDL- cholesterol (mg/dL) |
47.02 ± 13.70 | 48.90 ± 14.26 | 41.83 ± 10.51 | 45.67 ± 10.96 | 42.85 ± 11.80 | 50.06 ± 14.77 | 40.66 ± 10.07 | 52.50 ± 17.37 | 48.50 ± 9.47 |
| Fasting LDL- cholesterol (mg/dL) |
109.81 ± 33.14 | 109.14 ± 33.39 | 111.35 ± 32.87 | 109.17 ± 32.89 | 104.17 ± 23.98 | 107.78 ± 34.34 | 113.13 ± 37.22 | 112.29 ± 34.36 | 119.50 ± 19.33 |
| Fasting triglycerides (mg/dL) |
150.33 ± 100.19 | 150.57 ± 106.95 | 149.78 ± 83.33 | 124.88 ± 47.39 a | 134.38 ± 96.15 | 164.48 ± 92.57 | 156.71 ± 79.30 | 170.99 ± 177.11 | 145.62 ± 83.08 |
| Alanine transaminase (ALT) (U/L) |
39.70 ± 33.09 | 35.38 ± 28.39 | 44.62 ± 37.23 | 29.93 ± 20.54 a | 34.01 ± 19.45 d | 39.11 ± 32.68 | 55.56 ± 45.07 | 38.22 ± 30.97 | 52.76 ± 51.28 |
Significant at P<.05, between M-subtype and non-M-subtype patients with simple steatosis.
Significant at P<.05, between M-subtype and indeterminate patients with simple steatosis.
Significant at P<.05, between non-M-subtype and indeterminate patients with simple steatosis.
Significant at P<.05, between M-subtype and non-M-subtype patients with NASH.
Significant at P<.05, between M-subtype and indeterminate patients with NASH.
Significant at P<.05, between non-M-subtype and indeterminate patients with NASH.
Diagnoses were established histologically in liver biopsy specimens. The histological diagnosis of NAFLD was established by a single liver pathologist in each participating hospital using the scoring system defined by Kleiner et al.7 Following assessment, patients were classified by the pathologists into two histological groups: (1) Simple steatosis (hepatic steatosis alone), and (2) NASH (presence as determined by the pathologist). None of the patients had cirrhosis.
Metabolomic analysis
To measure metabolites measurements in serum, we combined a liquid chromatography (LC)-single quadrupole-mass spectrometry (MS) amino acid analysis system was with 2 separate LC-time of flight-MS based platforms that analyzed methanol and chloroform or methanol extracts for lipid analysis. For liver samples, the previous 3 LC-MS platforms were completed with a methanol/water extract analysis, covering polar metabolites. For details see Supplementary Methods. Absolute concentration of TG, DG, FA, phosphatidylethanolamine (PE), phosphatidylcholine (PC), methionine, SAMe and methylthioadenosine (MTA) was determined as described in Supplementary Methods.
Proteomics
For proteomics measurements, liver samples were prepared and analyzed by LC/MS/MS as described in Supplementary Methods. DGAT2 and SCD1 were determined by immunoblotting.
Global DNA methylation profiles
Global DNA methylation was analyzed by Reduced Representation Bisulfite Sequencing, digesting DNA with TaqI and MspI and preparing libraries with NEXTflex™ Bisulfite-Seq Kit (Bioo-Scientific) (see details in Supplementary Methods). Libraries were sequenced in a HiScanSQ (Illumina Inc.).
Hepatocyte mitochondrial membrane potential
The JC-1 dye (5,5,6,6-tetrachloro-1,1,3,3-tetraethylbenzimidazolyl-carbocyanineiodide) was used to determine mitochondrial membrane potential in hepatocytes isolated from MAT1A–KO and WT mice as described in Supplementary Methods.
Histology
Histological staining, hematoxylin and eosin, Sudan III red, Sirius Red and F4/80 immunostaining are described in Supplementary Methods.
Statistical analysis
Data are represented as means ± standard deviation of the mean. Differences between groups were tested using Student’s t-test. Significance was defined as P<0.05. The ‘volcano plot’ analysis was performed as an effective and easy-to-interpret graph that summarizes both fold-change and t-test criteria. It is a scatter-plot of the negative log10-transformed P-values from the t-test against the log2 fold change. All calculations were performed using statistical software package R v.3.1.1 (R Development Core Team, 2011; http://cran.r-project.org). Multivariate PCA modelling was performed with the software SIMCA 14.1 (Umetrics, Sweden).
A hierarchical clustering algorithm based on metabolites ion intensity was used to visualize the differences in metabolite signatures between samples, as well as the ward’s minimum variance method as agglomeration method. Biochemically related compounds were generally found to cluster together. The maximum of the average of the individual silhouette widths was calculated for the clusters8. The cluster analysis was calculated with the cluster R package9. The heatmap was realized with the pheatmap R package (Kolde R. Pheatmap: Pretty Heatmaps. R package version 1.0.8. http://CRAN.R-project.org/package=pheatmap).
RESULTS
SAMe depletion alters one-carbon metabolism
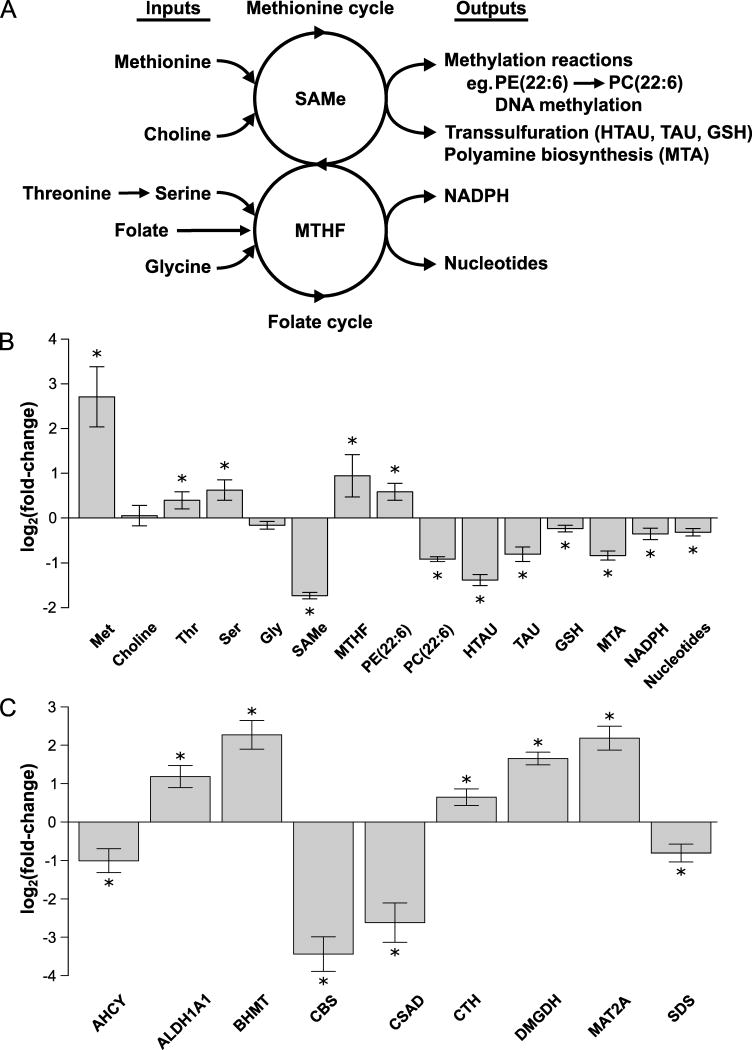
SAMe and folate metabolism are connected—they contribute to the 1-carbon metabolism that circulates 1-carbon units from different amino acids (methionine, threonine, serine and glycine) and nutrients (choline, folate) to generate a large variety of outputs, including methylation of DNA and PE rich in polyunsaturated FA (PUFA) to form PC rich in PUFA and syntheses of glutathione (GSH), polyamines, NADPH and nucleotides (Figure 1a)3. MAT1A deletion resulted in a reduction in hepatic SAMe, as reported in a previous study5, and in the content of downstream metabolites such as PC(22:6), methylthioadenosine (MTA, a biomarker of polyamine synthesis), hypotaurine, taurine and GSH (biomarkers of the transsulfuration pathway), NADPH (Figure 1b) and DNA methylation (supplementary Figure 2).
Figure 1. SAMe depletion alters one-carbon metabolism.
A) Schematic representation of one-carbon metabolism. One-carbon metabolism circulates one-carbon units from different inputs (methionine, choline, serine, threonine, glycine), via SAMe and methyltetrahydrofolate (MTHF), into a large variety of outputs, such as DNA and phospholipid methylation, glutathione (GSH), polyamines, NADPH and nucleotide synthesis. B) Relative fold-change (log2) in the hepatic content of the main metabolites involved in one-carbon metabolism in MAT1A-KO as compared to WT mice. MAT1A deletion induced a reduction in hepatic SAMe content and downstream metabolites, such as phosphatidylcholine with docosahexaenoic acid PC(22:6), methylthioadenosine (MTA, a biomarker of polyamine biosynthesis), GSH, hypotaurine (HTAU) and taurine (TAU) (three key metabolites of the transsulfuration pathway), NADPH and nucleotides. MAT1A ablation resulted also in the accumulation of methionine (Met) and upstream metabolites, such as serine (Ser), threonine (Thr), MTHF and phosphatidylethanolamine with docosahexaenoic acid PE(22:6). C) Relative fold-change (log2) in the protein content of enzymes involved in hepatic one-carbon metabolism in MAT1A-KO as compared to WT mice. MAT1A deletion led to abnormal protein content of numerous enzymes involved in one-carbon metabolism. AHCY, adenosylhomocysteinase; ALDH1A1, aldehyde dehydrogenase 1a1; BHMT, betaine-homocysteine S-methyltransferase; CBS, cystathionine β-synthase; CSAD, cysteine sulfinic acid decarboxylase; CTH, cystathionine γ-lyase; DMGDH, dimethylglycine dehydrogenase; MAT2A, methionine adenosyltransferase 2a; and SDS, serine dehydratase. *P<.05.
MAT1A deletion led also to the accumulation of methionine and upstream metabolites, such as threonine, serine, PE(22:6) and methyltetrahydrofolate (MTHF) in liver, and to abnormal protein content of numerous enzymes involved in one-carbon metabolism (AHCY, ALDH1A1, BHMT, CBS, CSAD, CTH, DMGDH, MAT2A and SDS) (Figure 1b,c).
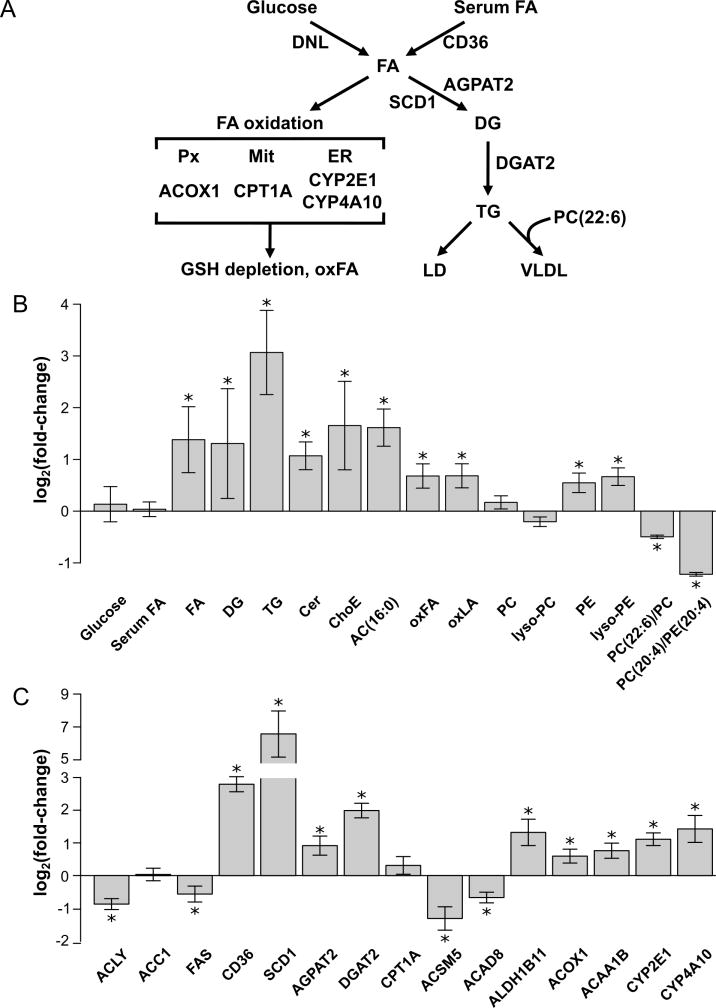
SAMe depletion activates FA uptake, desaturation and esterification impairing FA oxidation and VLDL secretion
SAMe depletion was associated with hepatic accumulation of FA, DG and TG (Figure 2a,b). Analysis of proteomics data, searching for proteins differentially expressed in MAT1A-KO that are involved in lipid metabolism, revealed that the content of CD36, a FA transporter whose overexpression correlates with TG accumulation in human NAFLD10, was significantly augmented, as well as the protein content of SCD1, which is the rate-limiting enzyme in the synthesis of monounsaturated FA, the major FA of TG and membrane phospholipids (Figure 2c). The protein content of AGPAT2 and DGAT2, two additional key enzymes in TG biosynthesis, was also increased in MAT1A-KO liver (Figure 2c). The content of the main enzymes involved in DNL was either normal (ACC1) or significantly reduced (ACLY and FAS) (Figure 2c). Our results also showed an accumulation of palmitoylcarnitine (AC16:0), the rate-limiting substrate in mitochondrial FA oxidation, despite having normal protein content of CPT1A, which suggested an impaired oxidation of FA in MAT1A-KO mice (Figure 2b,c). Indeed, hepatocytes isolated from MAT1A-KO mice showed a loss of mitochondrial membrane polarization (supplementary Figure 3a,b). Notably, mitochondrial polarization was restored in MAT1A-KO hepatocytes upon incubation with SAMe (supplementary Figure 3a,b). Moreover, the content of oxidized FA was augmented in MAT1A-KO, which agrees with the increased protein content of ALDH1B1, a critical mitochondrial enzyme involved in lipid peroxidation (Figure 2b,c). Accordingly, MAT1A-KO livers showed decreased content of several enzymes that catalyze the mitochondrial oxidation of FA (ACSM5 and ACAD8), and augmented content of key enzymes involved in peroxisomal (ACOX1 and ACAA1B) and endoplasmic reticulum (CYP2E1 and CYP4A10) FA oxidation (Figure 2c). Finally, SAMe depletion caused a reduction in the ratio PC(22:6)/PC and PC(20:4)/PE(20:4), both indicators of reduced PEMT activity (Figure 2b). PEMT activity is needed for VLDL assembly and export, and its deletion makes mice more prone to develop fatty liver11. Notably, MAT1A-KO mice have reduced VLDL secretion12. Collectively, the aforementioned results indicate that the increased flow of peripheral fat stored in the adipose tissue to the liver by way of the serum FA pool, together with an impaired export of TG into VLDL and decreased oxidation of FA in the mitochondria, is the main source of lipids contributing to fatty liver in MAT1A-KO mice.
Figure 2. SAMe depletion activates FA uptake and esterification, while FA oxidation and VLDL secretion are impaired.
A) Schematic representation of hepatic lipid metabolism. Hepatic fatty acids (FA) originate from serum and through de novo lipogenesis (DNL). FA can either be oxidized in the mitochondria (Mit) or esterified to form triglycerides (TG), which are stored in lipid droplets (LD), used to form other lipids, such as phospholipids, ceramides (Cer) and cholesteryl esters (ChoE) (not shown), or exported into blood as very low density lipoproteins (VLDL). The formation of VLDL particles requires phosphatidylcholine (PC) molecules rich in polyunsaturated FA (PUFA), such as PC(22:6). The rate-limiting step in mitochondrial β-oxidation is carnitine palmitoyltransferase 1a (CPT1A), which forms palmitoylcarnitine (AC16:0). The accumulation of FA in the cytoplasm increases their oxidation in peroxisomes (Px) and endoplasmic reticulum (ER). The first step in Px β-oxidation is acyl-CoA oxidase 1 (ACOX1), which generates reactive oxygen species (ROS). ER ω-oxidation, which is catalyzed by cytochrome P450 (CYP) enzymes, such as CYP2E1 and CYP4A10, also generates ROS. In its turn, ROS induces glutathione (GSH) depletion and produces oxidized FA (oxFA), such as linoleic acid (LA) derived oxidized FA (oxLA), which can lead to fibrosis and cell death. B) Relative fold-change (log2) in the hepatic content of the main metabolites involved in lipid metabolism in MAT1A-KO as compared to WT mice. AC(16:0), palmitoylcarnitine; oxFA, oxidized FA; oxLA, linoleic acid (18:2)-derived oxidized FA; PC, phosphatidylcholine; lyso-PC, lyso-phosphatidylcholine; PE, phosphatidylethanolamine; lyso-PE, lyso-phosphatidylethanolamine; PC(22:6)/PC, ratio PC with docosahexaenoic acid/total PC; PC(20:4)/PE(20:4) and ratio PC/PE with arachidonic acid. C) Relative fold-change (log2) in the content of proteins involved in liver lipid metabolism in MAT1A– KO as compared to WT mice. DNL enzymes: ACLY, citrate lyase; ACC1, acetyl-CoA carboxylase 1; FAS, and fatty acid synthase. FA transport: CD36, fatty acid translocase. FA esterification: SCD1, stearoyl-CoA desaturase; AGPAT2, 1-acylglycerol-3-phosphate O-acyltransferase 2; and DGAT2, diacylglycerol acyltransferase 2. Mitochondrial FA β-oxidation: CPT1A, carnitine palmitoyltransferase 1a; ACSM5, acyl-CoA synthetase medium chain family member 5; ACAD8; acyl-CoA dehydrogenase family member 8; and ALDH1B1; aldehyde dehydrogenase 1 family member B1. Peroxisomal FA β-oxidation: ACOX1, acyl-CoA oxidase 1; and ACAA1B, acyl-CoA acetyltransferase. Endoplasmic reticulum FA ω-oxidation: CYP2E1 and CYP4A10. *P<.05.
These findings prompted us to speculate that excess FA may be rerouted towards other pathways, such as ceramide synthesis. Consistent with this hypothesis, livers from MAT1A-KO mice showed an accumulation of ceramides (Figure 2b). Moreover, MAT1A-KO mice displayed high levels of cholesteryl esters, PE and lyso-PE, which agrees with the work of others showing the importance of lyso-phospholipids in human NASH13,14, and a decrease in the PC/PE ratio (Figure 2b,c), which would indicate that a deficiency in liver SAMe has far reaching effects upon lipid metabolism.
Discovery of two human NAFLD subtypes
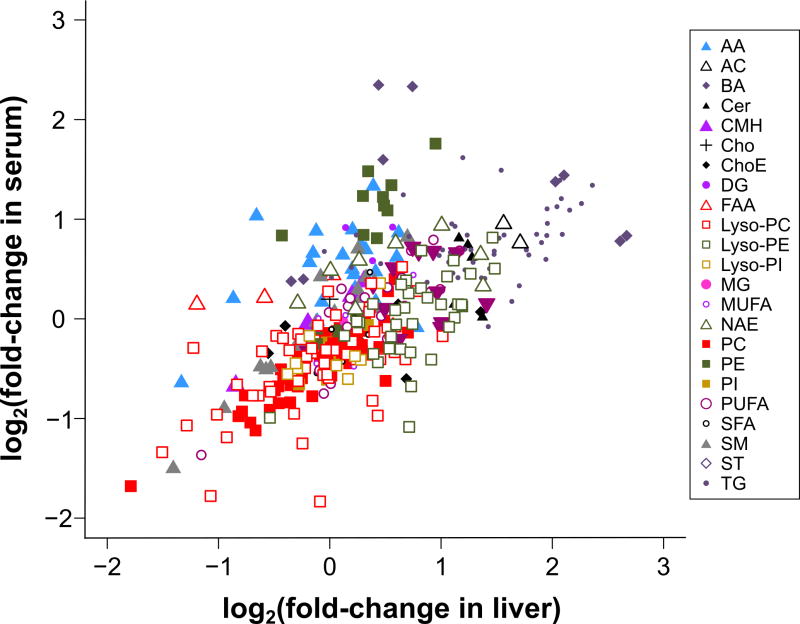
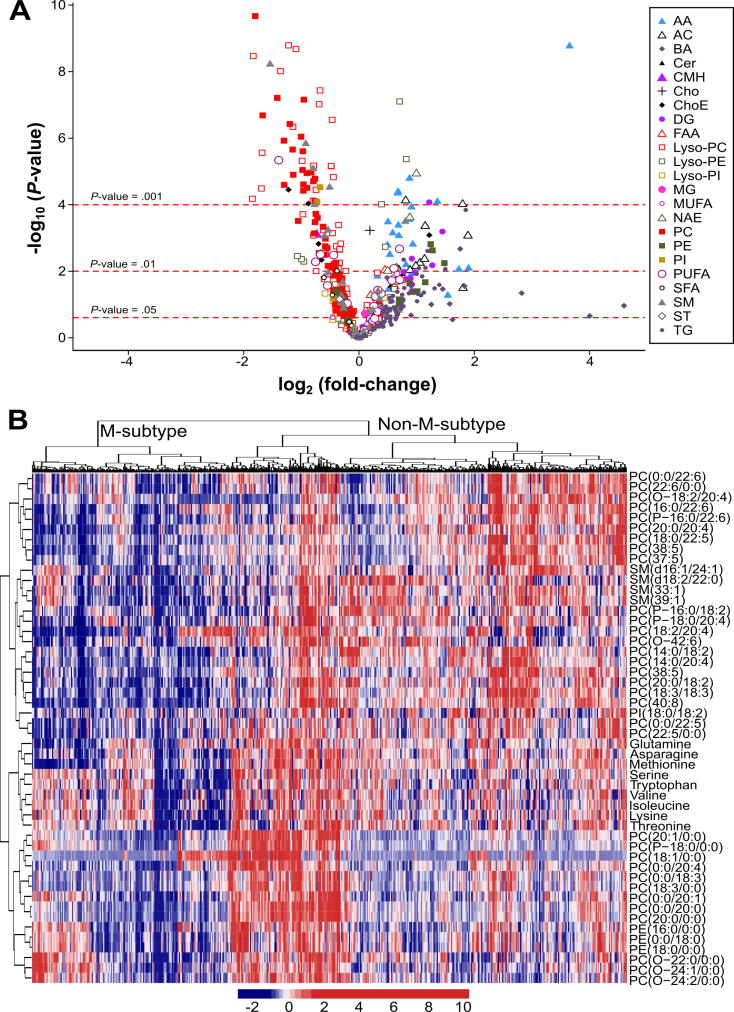
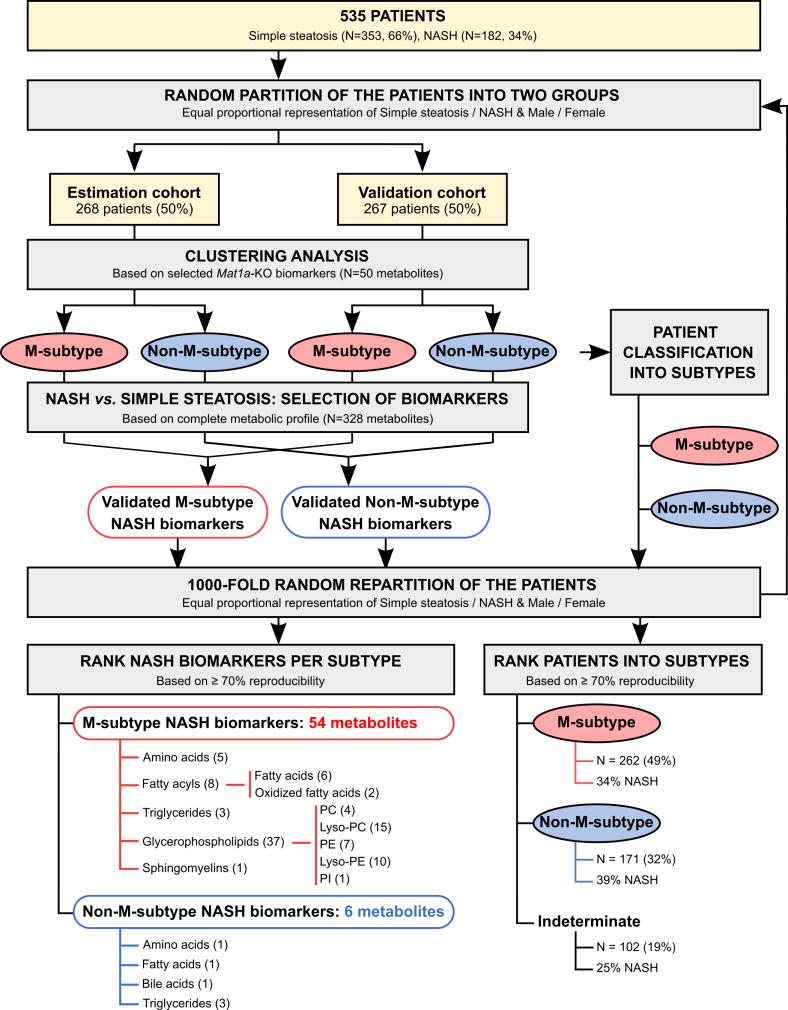
To investigate if NAFLD patients display alterations in hepatic metabolism similar to those observed in MAT1A-KO mice, we first analyzed if the serum metabolomic profile of MAT1A-KO mice reflected the liver metabolomic profile. Accordingly, we compared the liver metabolomic profile of MAT1A-KO and WT mice and generated a list with the fold-change and P-values for each metabolite (supplementary Table 1). Then we proceeded similarly for the serum metabolomic profiles of MAT1A-KO and WT mice and generated a second list showing the fold-change and P-values for each metabolite (supplementary Table 1). Finally, we analyzed if the serum metabolomic profile reflected hepatic metabolism by comparing the common metabolites in both sets of data. As shown in Figure 3, there is a statistically significant correlation (R2=0.45, P<1E-04) between both metabolomic profiles. Subsequently, we selected the top 50 serum metabolites that more significantly differentiated between MAT1A-KO and WT mice (Figure 4a). Silhouette cluster analysis8 revealed that this signature sub-classified a cohort of 535 patients with biopsy proven NAFLD (353 diagnosed with simple steatosis and 182 with NASH) into two main clusters, a first cluster showing a serum metabolic profile similar to that observed in the MAT1A-KO mice (M-subtype) and a second cluster showing a different profile (non-M-subtype) (Figure 4b). For validation, we followed the procedure described in Figure 5. First, samples were randomly partitioned (50/50) into two cohorts (estimation and validation) with equal proportional representation of steatosis/NASH and male/female. Then, clustering analysis based on the selected top 50 MAT1A-KO biomarkers generated two main clusters and patients were classified into M and non-M-subtypes. Based on the complete metabolic profile (N=328 metabolites) of the human serum samples, biomarkers that significantly differentiated between NASH and steatosis were selected and validated by comparison between the results in estimation and validation cohorts. After 1,000-fold repetition of this random partition, each time with equal proportional representation of steatosis/NASH and male/female, the frequency distribution of the metabolites that significantly differentiated between NASH and steatosis in the M and non-M-subtypes was determined, and those showing a reproducibility of at least 700 times in 1,000 repetitions selected. A rank NASH biomarkers list per subtype, showing the reproducibility and p-value, was generated (supplementary Table 2). The M-subtype NASH biomarker list contained 54 metabolites: 5 amino acids, 8 fatty acyls (6 FA and 2 oxidized FA), 3 TG, 37 phospholipids (4 PC, 15 lyso-PC, 7 PE, 10 lyso-PE and 1 phosphatidylinositol) and 1 sphingomyelin (Figure 5 and supplementary Table 2a). Twenty-nine of these metabolites had a reproducibility ≥90%. Interestingly, 25 of the 54 biomarkers were lyso-phospholipids. The non-M-subtype NASH biomarkers list consisted of 6 metabolites: 1 amino acid, 1 FA, 1 bile acid and 3 TG (Figure 5 and supplementary Table 2b).
Figure 3. The serum metabolomic profile reflects hepatic metabolism.
Comparison of liver and serum metabolomics profiles of MAT1A-KO mice. Each point represents the log2(fold-change) of individual metabolic ion features of MAT1A-KO compared to WT mice in serum and liver. A list with the log2(fold-change) and P-value for each individual metabolite in serum and liver is given in supplementary Table 1. R2=0.45, P=1E-04. AA, amino acids; AC, acyl carnitines; BA, bile acids; Cer, ceramides; CMH, monohexosylceramides; Cho, cholesterol; ChoE, cholesteryl esters; DG, diglycerides; FAA, fatty acyl amides; PC, phosphatidylcholines; Lyso-PC, lyso- phosphatidylcholines; PE, phosphatidylethanolamines; Lyso-PE, lyso-phosphatidylethanolamines; PI, phosphatidylinositols; Lyso-PI, lyso-phosphatidylinositols; MG, monoglycerides; SFA, MUFA and PUFA, saturated, monounsaturated and polyunsaturated fatty acids, respectively; NAE, N-acylethanolamines; SM, sphingomyelins; ST, steroids; TG, triglycerides.
Figure 4. Identification of a subset of NAFLD patients showing a Mat1a-KO serum metabolomic profile.
A) Volcano plot representation indicating the -log10(P-value) and log2(fold-change) of individual serum metabolic ion features of MAT1A-KO compared to WT mice. AA, amino acids; AC, acyl carnitines; BA, bile acids; Cer, ceramides; CMH, monohexosylceramides; Cho, cholesterol; ChoE, cholesteryl esters; DG, diglycerides; FAA, fatty acyl amides; PC, phosphatidylcholines; Lyso-PC, lyso-phosphatidylcholines; PE, phosphatidylethanolamines; Lyso-PE, lyso-phosphatidylethanolamines; PI, phosphatidylinositols; Lyso-PI, lyso-phosphatidylinositols; MG, monoglycerides; SFA, MUFA and PUFA, saturated, monounsaturated and polyunsaturated fatty acids, respectively; NAE, N-acylethanolamines; SM, sphingomyelins; ST, steroids; TG, triglycerides. B) Heatmap representation of the serum metabolomic profile from 535 patients with biopsy-confirmed NAFLD. Each data point corresponds to the relative ion abundance of a given metabolite (vertical axis) in an individual patient’s serum. Metabolite selection is based on the top 50 serum metabolites that more significantly differentiated between MAT1A-KO and WT mice. The hierarchical clustering is based on optimum average silhouette width, obtaining the classification of the samples into two groups: first cluster resembles the serum metabolomic profile observed in the MAT1A-KO mice (M-subtype), while second cluster shows a different metabolomic profile (non-M-subtype).
Figure 5. Scheme for the identification and validation of NAFLD subtypes and NASH biomarkers.
Serum samples from 535 patients with biopsy proven NAFLD (353 simple steatosis and 182 NASH) were randomly partitioned (50/50) into two cohorts (estimation and validation cohort) with equal proportion of steatosis/NASH and male/female. Clustering analysis, based in the 50 serum metabolites that more significantly differentiated between MAT1A-KO and WT mice (see Figure 4), generated two main clusters and patients were classified into M and non-M-subtypes. Based on the complete metabolic profile (N=328 metabolites) of the human serum samples, biomarkers that significantly differentiated between NASH and simple steatosis were selected and validated by comparison between the results in estimation and validation cohorts. After 1,000-fold repetition of this random partition, each time with equal proportional representation of simple steatosis/NASH and male/female, the frequency distribution of the metabolites that significantly differentiated between NASH and simple steatosis in the M- and non-M-subtypes was determined, and those showing a reproducibility of at least 700 times in 1,000 repetitions selected. The frequency distribution of the NAFLD patients into the M- and non-M-subtypes was also calculated. Following the criteria based on ≥70% reproducibility, 262 patients (49%) were classified as M-subtype and 171 (32%) as non-M-subtype. The remaining 102 patients (19%) showed a reproducibility of less than 70% and could not be classified as either M- or non-M-subtype (indeterminate group). PC, phosphatidylcholine; Lyso-PC, lyso- phosphatidylcholine; PE, phosphatidylethanolamine; Lyso-PE, lyso- phosphatidylethanolamine; PI, phosphatidylinositol.
The frequency distribution of the NAFLD patients into the M- and non-M-subtypes was also calculated (Figure 5). Following the criteria based on ≥70% reproducibility, 262 patients (49%) were classified as M-subtype and 171 (32%) as non-M-subtype. The remaining 102 patients (19%) showed a reproducibility of less than 70% and could not be classified as either M- or non-M-subtype (indeterminate group). Supplementary Table 3 summarizes the serum metabolites associated with the MAT1A-KO serum metabolomics profile in the human subtypes.
Although there was a significant excess of females in the M-subtype steatosis and NASH groups compared to the non-M-subtype (Table 1), PCA of serum metabolome showed no differences between females and males (Supplementary Figure 4). In this study, female mice were not tested. The BMI in the M group was significantly higher while the age and ALT was lower than in the non-M-subtype in both, simple steatosis and NASH (Table 1). Among the M-subtype group, the percent of NASH patients (34%) was the same as that in the total cohort of NAFLD patients (34%) and slightly, but significantly, lower than that observed in patients classified as non-M-subtype (39%) and higher than in the indeterminate group of patients (25%) (Table 1).
Administration of SAMe improves steatohepatitis
Bioavailability of orally administered SAMe is poor due to a significant first-pass effect and rapid hepatic metabolism15. SAMe half-life is about 5 min16. Consistent with this, we previously observed that following SAMe intra peritoneal injection liver SAMe content rose rapidly, reaching a peak in 15 minutes, recovering basal levels 4 hours after injection17. Here we found that the concentration of serum and liver SAMe in MAT1A -KO mice that received SAMe was not significantly different from KO mice that received the vehicle (not shown), which agrees with the short half-life of SAMe since mice were sacrificed 24 hours after the last administration of SAMe. Analysis of the differentially methylated DNA regions (DMRs) in SAMe-given MAT1A-KO mice compared to vehicle-given MAT1A-KO animals showed that SAMe administration increased DNA methylation (supplementary Figure 2), which confirms the intracellular utilization of orally administered SAMe. DMRs identified in MAT1A-KO mice that received SAMe were distributed across all chromosomes and the majority of these regions were hypomethylated in mutant mice that received the vehicle. The relevance of these findings is confirmed by the observation that human NASH associates with hypomethylation of liver DNA18.
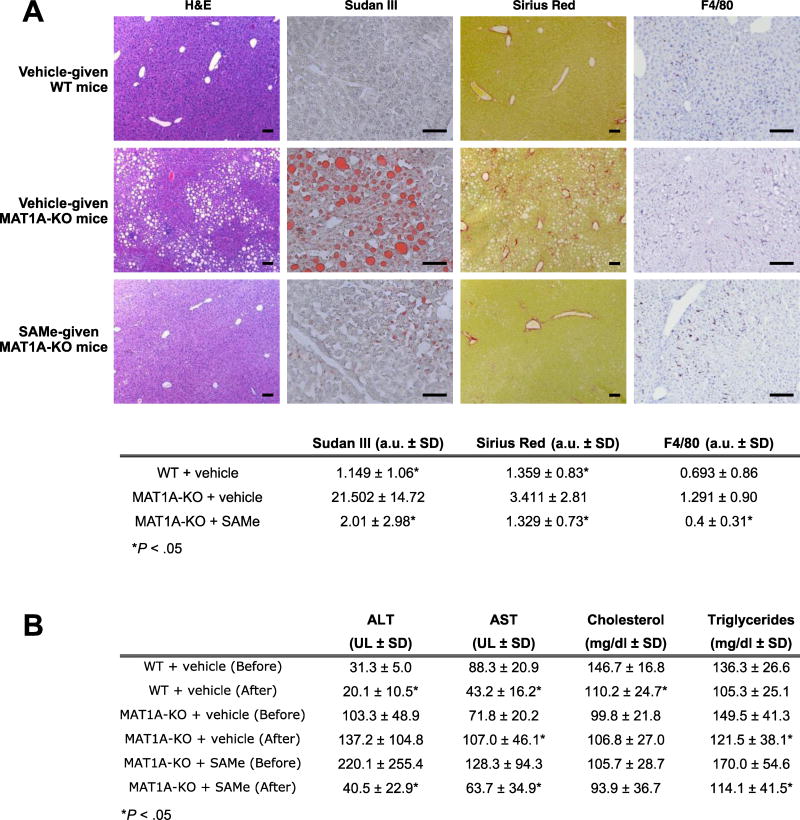
Finally, we found that SAMe administration improved liver function, as indicated by a reduction in serum transaminases and liver histology (Figure 6). Mice that received SAMe displayed a reduction of lipid accumulation, as quantified by morphometry of Sudan red stained area (11-fold as compared to MAT1A-KO mice that received vehicle, P=2.0E-04) (Figure 6a). Mice given SAMe showed also decreased liver fibrosis, as quantified by morphometry of Sirius red staining (3-fold as compared to MAT1A-KO mice given the vehicle, P=2.6E-02) (Figure 6a). Furthermore, livers from MAT1A-KO mice given SAMe exhibited a reduction of inflammation as quantified by morphometry of F4/80 (3-fold as compared to MAT1A-KO mice given the vehicle, P=3.0E-03) (Figure 6a).
Figure 6. Effect of SAMe administration on histology and serology in MAT1A KO mice.
A) Representative images of hematoxylin and eosin (H&E), Sudan III red, Sirius red and F4/80 immunofluorescence staining of liver tissues after eight weeks SAMe (30 mg/kg/day) or vehicle administration are shown. Sizing bars correspond to 100 µm for H&E and Sirius Red, and 50 µm for Sudan III and F4/80. Quantitative analyses are shown in the table. Results that were significantly different (P<.05) to vehicle-given MAT1A-KO mice are indicated. Data shown represent mean of twelve vehicle-given MAT1A-KO, twelve SAMe-treated MAT1A-KO and eleven vehicle-given wild type (WT) animals. B) Effect of SAMe administration on serum parameters. For each group of animals (WT + vehicle, MAT1A-KO + vehicle, and MAT1A-KO + SAMe), results that were significantly different (P<.05) before and after administration are indicated. Twelve SAMe-given MAT1A-KO, twelve vehicle-treated MAT1A-KO and eleven vehicle-given WT mice were analyzed.
DISCUSSION
Our results provide evidence that MAT1A protein functions as an integrator of the cellular metabolic status and that its deletion not only affects hepatic metabolism downstream of SAMe (DNA and phospholipid methylation, polyamine and GSH synthesis) but also leads to an imbalance in the circulation of one-carbon units from specific amino acids (methionine, threonine, serine and glycine) to folates, altering an unparalleled diversity of cellular processes ranging from the biosynthesis of lipids, proteins and amino acids, to the regulation of mitochondrial polarization and function. Genetic and functional evidence support the importance of the activity of the serine-glycine-folate-methionine pathway in tumorigenesis19. We propose that these changes, although many are likely to be part of a compensatory response, will create a metabolic environment that favors steatohepatitis development and, perhaps, its progression to hepatocellular carcinoma in MAT1A-KO mice20. Our results show that SAMe depletion induced hypomethylation of DNA regions in MAT1A-KO as compared to WT mice. In human NAFLD, hepatic DNA hypomethylation has been found to be associated with more advanced NAFLD18. Although much remains to be learned in this area, this is probably an important driver of NASH in MAT1A-KO mice as SAMe administration significantly decreased the number of DMRs and improved liver function and histology.
Our results unveiled the existence of three NAFLD metabolic phenotypes, M-, non-M- and indeterminate. All three metabolomic phenotypes were found in both simple steatosis and NASH in approximately the same proportions, which suggests that patients of the M-subtype are not at higher risk to develop NASH than those with a non-M-subtype. Whether the natural course of the different subtypes differ cannot be addressed. This, however, does not preclude the diagnostic value of identifying patients of the M-subtype, as around 50% of all NAFLD patients show this phenotype and those are likely to benefit from SAMe or other specific treatments to be developed in the future. The M phenotype occurred significantly more frequently in female as compared to the non-M-subtype. The BMI in the M-subtype was also significantly higher, while the age and ALT were significantly lower than in the non-M-subtype. We obtained a second metabolic signature that could separate NAFLD patients of the M-subtype into NASH and simple steatosis. This signature included methionine, several PE, lyso-PE, PC and lyso-PC. As these metabolites are biomarkers of impaired one-carbon metabolism, these results support the concept that altered SAMe level is a determinant that switches from benign steatosis to NASH. This agrees with the finding that NASH patients with more advanced disease often show decreased MAT1A expression4.
The finding that the serum metabolomic profile of MAT1A-KO mice reflects the liver metabolomics profile supports the hypothesis that patients with an M metabolic phenotype have lower MAT1A activity. As the M metabolic phenotype was found in both simple steatosis and NASH, this would suggest that impaired SAMe synthesis occurs early in the development of NAFLD in this subgroup of patients. Human liver MAT1A protein levels show a Gaussian frequency distribution with a large variation between individuals21. Whether individuals with lower MAT1A protein have lower hepatic SAMe content and are at higher risk to develop NAFLD, needs to be determined. Variations in hepatic MAT1A protein among individuals do not correlate with age or any common MAT1A genetic polymorphism21, and there is evidence indicating that MAT1A expression is not regulated by the circadian rhythm22. Whether microRNAs, which have been found to regulate MAT1A expression23, are involved in determining MAT1A protein content is not known. Alternatively, the situation may be more complicated and may be that the M-subtype was related to alterations in MAT1A enzymatic activity and other enzyme heterogeneities.
Since there is considerable interest in the use of SAMe to treat NASH24, these findings suggest that NASH patients showing an M-subtype serum metabolomic profile may be best suited to benefit from SAMe treatment. The present results showing that SAMe treatment improved NASH in mice deficient in SAMe synthesis supports this concept. The non-M-subtype may be a heterogeneous group where NAFLD and its progression to NASH may result from alterations in different biochemical pathways. Comparison of other mouse models of NAFLD with the non-M-subtype may clarify this point.
Supplementary Material
Acknowledgments
Grant Support:
This work was supported by NIH grant R01AT001576 (SCL and JMM), Plan Nacional of I+D SAF 2014-52097R (JMM) and SAF 2014-54658R (MLM-C), MINECO-ISCiii PIE14/00031 (MLM-C and JMM), CIBERehd-ISCiii (JMM, MLM-C and AMA), Basque Government Health Department 2013111114 and Spanish-AECC (MLM-C). Mice studies were partially supported by Abbott Laboratories GmbH, Hannover, Germany.
Author contribution
CA, acquisition of data, analysis and interpretation of data, statistical and informatics analysis, drafting of the manuscript, study supervision
DF-R, animal experimentation, acquisition of data, analysis and interpretation of data, study supervision
MV-R, animal experimentation, acquisition of data, analysis and interpretation of data, study supervision
IM-A, statistical and informatics analysis
NN, acquisition of data, analysis and interpretation of data
SMVL, acquisition of data, analysis and interpretation of data
JLL, acquisition of data, analysis and interpretation of data, informatics analysis
RM, acquisition of data
CPI, acquisition of data
VGJ, technical
MI-L, analysis and interpretation of data
LC-V, interpretation of data
IM, statistical and informatics analysis
AR, acquisition and interpretation of data
JC, human samples
AM-D, human samples
MR-G, human samples
HS, study concept and design
JP, study concept and design
JVE, acquisition and interpretation of data
PA, acquisition of data
MN, critical revision of the manuscript for important intellectual content
JMF-P, acquisition of data, analysis and interpretation of data
JA, acquisition of data, analysis and interpretation of data, study supervision
AMA, acquisition of data, analysis and interpretation of data, informatics analysis
MLM-C, study concept and design, critical revision of the manuscript for important intellectual content, study supervision
SCL, study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, obtained funding JMM, study concept and design, analysis and interpretation of data, drafting of the manuscript, obtained funding, study supervision
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations:
Complete list of abbreviations included in Supplementary materials: “Supplemental abbreviations”.
Disclosures:
JMM is Abbott, Galmed and OWL Metabolomics’ consultant and/or speaker.
MN is Abbott’s consultant and has been on the scientific advisory board or speaker for OWL and Echosens.
HS and JP, Abbott’s employees.
CA, IM-A, RM, MI-L and IM, OWL Metabolomics’ employees.
DF-R, MV-R, NN, SMVL, JLL, CPI, VGJ, LC-V, AR, JC, AM-D, MR-G, JVE, PA, JMF-P, JA, AMA, MLM-C and SCL, nothing to declare.
REFERENCES
- 1.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo P. Long-term mortality in nonalcoholic fatty liver disease: is liver histology of any prognostic significance? Hepatology. 2010;51:373–375. doi: 10.1002/hep.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moylan CA, Pang H, Dellinger A, et al. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59:471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu SC, Alvarez L, Huang ZZ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr J, Caballeria J, Martínez-Arranz I, et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;11:2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 8.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- 9.Maechler M, Rousseuw PJ, Struyf A, et al. Cluster: cluster analysis basics and extensions. R package version 2.0.4 [Google Scholar]
- 10.Greco D, Kotronen A, Westerbacka J, et al. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1281–G1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- 11.Vance DE. Physiological roles of phosphatidylethanolamine N-methyltransferase. Biochim Biophys Acta. 2013;1831:626–632. doi: 10.1016/j.bbalip.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Cano A, Buque X, Martinez-Una M, et al. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology. 2011;54:1975–1986. doi: 10.1002/hep.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han MS, Park SY, Shinzawa K, et al. Lysophosphatidylcholine as a death effector in the lipoapoptosis of hepatocytes. J Lipid Res. 2008;49:84–97. doi: 10.1194/jlr.M700184-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Kakisaka K, Cazanave SC, Fingas CD, et al. Mechanisms of lysophosphatidylcholine-induced hepatocyte lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G77–G84. doi: 10.1152/ajpgi.00301.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loehrer FM, Schwab R, Angst CP, et al. Influence of oral S-adenosylmethionine on plasma 5-methyltetrahydrofolate, S-adenosylhomocysteine, homocysteine and methionine in healthy humans. J Pharmacol Exp Ther. 1997;282:845–850. [PubMed] [Google Scholar]
- 16.Mudd SH, Poole JR. Labile methyl balances for normal humans on various dietary regimens. Metabolism. 1975;24:721–735. doi: 10.1016/0026-0495(75)90040-2. [DOI] [PubMed] [Google Scholar]
- 17.Lu SC, Ramani K, Ou X, et al. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50:462–471. doi: 10.1002/hep.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy SK, Yang H, Moylan CA, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Chantar ML, Corrales FJ, Martinez-Cruz LA, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 21.Ji Y, Nordgren KK, Chai Y, et al. Human liver methionine cycle: MAT1A and GNMT gene resequencing, functional genomics, and hepatic genotype-phenotype correlation. Drug Metab Dispos. 2012;40:1984–1992. doi: 10.1124/dmd.112.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J-S, Coon SL, Blackshaw S, et al. Methionine adenosyltransferase:adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J Biol Chem. 2005;280:677–684. doi: 10.1074/jbc.M408438200. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Cho ME, Li TW, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285–298. doi: 10.1172/JCI63861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anstee QM, Day CP. S-adenosylmethionine (SAMe) therapy in liver disease: a review of current evidence and clinical utility. J Hepatol. 2012;57:1097–1109. doi: 10.1016/j.jhep.2012.04.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.