Abstract
Extracellular vesicles (EVs) released during cell stress, or demise, can contain a barcode of the cell origin including specific microRNAs (miRNAs). Here we tested the hypothesis that during early alcoholic steatohepatitis (ASH) development, hepatocytes (HCs) release EVs with a miRNA signature that can be measured in circulation. A time course experiment showed that after two weeks of intragastric infusion, a time-point that results in isolated steatosis, there was no increase of blood EVs. After four weeks of infusion mice developed features of early ASH accompanied by a marked increase in the level of EVs in blood (p<0.05), as well as and in culture media of isolated HCs (p<0.001) and hepatic macrophages (p<0.001) with HCs being the predominant source of EVs. The transcriptome analysis of HC-EVs from ASH mice detected differentially expressed miRNAs, including nine significantly up-regulated and four significantly down-regulated miRNAs. Target prediction and pathway analyses of the up-regulated miRNAs identified 121 potential target genes involved in inflammatory and cancer pathways such as NF-κB, EGF, Wnt, and Bcl2. Three miRNAs, let7f, miR-29a, and miR-340, were increased in blood EVs from ASH mice (p<0.05), but not in blood EVs from three other models of chronic liver injury including bile duct ligation, non-alcoholic steatohepatitis, and obese mice, as well as EVs released from hepatocytes exposed to ethanol. The blood EV level (p<0.01) and three miRNAs (p<0.05) were significantly increased in patients with ambulatory mild ALD as compared to non-alcoholics.
Conclusions
Our results reveal that damaged hepatocytes from ASH mice are a key EV source with a specific miRNA cargo, which are specific for ASH-related liver injury. These findings uncover EVs as a potentially novel diagnostic for ASH.
Keywords: ASH, hepatocyte-derived EVs, miRNAs
Introduction
Alcoholic liver disease (ALD) is one of the most common forms of chronic liver disease in the United States and many other countries (1). ALD represents a wide spectrum of liver damage ranging from alcoholic steatosis, alcoholic steatohepatitis (ASH), to cirrhosis, resulting in liver failure and hepatocellular carcinoma (2, 3). Accumulating evidence indicates that hepatocyte stress, and eventually hepatocyte death, plays a central role in inflammation and hepatic injury during the progression of ALD (4–6). Hepatic macrophages (HMs) are involved in the progression of ethanol-induced liver damage (7–9). However, the molecular mechanisms and signaling pathways involved in the crosstalk between stressed, lipid overloaded hepatocytes (HCs) and HMs in ASH remain poorly understood. Moreover, the identification of a specific signature, or “barcode”, from damaged hepatocytes in ASH may help establish mechanistic-based liver-specific and disease-specific biomarkers that may revolutionize the way we diagnose and monitor patients with this condition.
EVs are small membrane vesicles released in a highly regulated manner from damaged or activated cells. EVs are categorized as exosomes or microparticles based on size, less than 100 nm or 100–1000 nm, respectively. Various cells release EVs, which circulate in the plasma of healthy humans and act as a natural delivery system under homeostasis (10). EVs are effective communicators that are generated by a cell of origin or parenteral cell and can act on a number of target cells in the vicinity of the origin in a juxtacrine manner, or at distant sites acting as long range signals (11). EV cargo may reflect the cell of origin as well as the specific stress that induces their formation and release. They transport a variety of bioactive molecules, including mRNA, miRNAs, proteins and lipids that can be transferred among cells, regulating various cellular responses (11, 12). Here we tested the hypothesis that during ASH development hepatocytes release EVs with a miRNA signature that can be detected in blood. To address this hypothesis, we have taken an approach to isolate and culture HCs or HMs from C57BL6 mice after two and four weeks of intragastric continuous ethanol infusion, or pair-fed control diet, and EVs released by the cells were isolated and characterized via flow cytometry, electron microscopy and dynamic light scattering. miRNA encapsulated in HC-EVs was assessed via transcriptome analysis and bioinformatics. miRNA specificity for ASH was investigated using other models of chronic liver injury, cultured hepatocytes exposed to ethanol (EtOH), and ambulatory patients with alcoholic liver disease.
Materials and Methods
Human samples
Patients fulfilling the DSM IV criteria (13) for alcohol dependence and with active alcohol consumption and alcoholic liver disease were compared to individuals without alcohol dependency (weekly alcohol intake of less than 140 g) (non-alcoholic controls). Written informed consent was obtained from all patients and controls. Patient characteristics are shown in Table 1. The study protocol was approved by the Ethics Committee of the Université Catholique de Louvain, in Brussels, Belgium, by the Human Research Protections Program of the University of California San Diego and of the VA San Diego Healthcare System.
Table 1.
Human subject characteristics
| Subjects | Sex | Age | Race* | BMI | |
|---|---|---|---|---|---|
| Non-alcoholics (Controls) | M | 58 | C | 34 | |
| M | 26 | C | 25 | ||
| M | 70 | C | 25 | ||
| M | 69 | C | 36 | ||
| M | 74 | C | 23 | ||
| M | 76 | C | 20 | ||
| M | 34 | C | 27 | ||
| M | 58 | NH | 28 | ||
|
| |||||
| Active alcoholics | M | 51 | C | 19 | ALT=46, AST=59 |
| M | 57 | AI | 36 | ALT=5, AST=74 | |
| M | 58 | C | 18 | ALT=43, AST=95 | |
| M | 46 | C | 30 | ALT=52, AST=141 | |
| M | 42 | C | 21 | ALT=71, AST=50 | |
| M | 44 | C | 37 | ALT=45 | |
| M | 39 | C | 32 | ALT=129, AST=121 | |
| M | 63 | C | 22 | ALT=150, AST=132 | |
| M | 27 | C | 28 | ALT=76, AST=70 | |
| M | 48 | C | 24 | ALT=132, AST=85 | |
| M | 58 | C | 27 | ALT=19, AST=42 | |
| M | 40 | C | 24 | ALT=103, AST=88 | |
| M | 19 | C | 29 | ALT=27, AST=28 | |
| M | 65 | C | 24 | ALT=30, AST=85 | |
| M | 60 | C | 22 | ALT=100, AST=458 | |
| M | 52 | C | 24 | ALT=58, AST=164 | |
| F | 49 | C | 33 | ALT=233, AST=182 | |
| M | 57 | C | 23 | ALT=30, AST=36 | |
| M | 52 | C | 24 | ALT=139, AST=209 | |
| F | 43 | C | 23 | ALT=80, AST=454 | |
| M | 49 | C | 27 | ALT=149, AST=170 | |
C=Caucasian, AA=African American, AI=American Indian, NH=Native Hawaiian
Animal Studies
The use and care of the animals was reviewed and approved by the Institutional Animal Care and Use Committee at the Southern California University. The early ASH mouse model using intragastric infusion was generated as previously described and produced by the Animal Core of the Southern California Research Center for ALPD and Cirrhosis (14). After a one-week acclimatization period with infusion of a control high fat diet, ethanol infusion was initiated at a dose of 22.7g/kg/day which was incrementally increased every two days until it reached 29.2g/kg/day from days 9 to 14 and 30.9g/kg/day from days 15 to 21. At the initial ethanol dose, total caloric intake was set at 533 Cal/kg/day and the caloric percentages of ethanol, dietary carbohydrate (dextrose), protein (lactalbumin hydrolysate) and fat (corn oil) were 29%, 13%, 23%, and 35%, respectively. Vitamin, salt, and trace mineral mix are included at the recommended amounts by the Committee on Animal Nutrition of the National Research Council (AIN-76A, 4.42 g/L and 15.4 g/L, respectively, Dyets Inc, PA). Pair-fed control mice received dextrose solution isocaloric to ethanol plus the basal diet. Following four weeks of intragastric alcohol feeding, mice will be sacrificed for analysis (n=5 pairs).
Liver and blood sample preparation
A part of liver was collected from the right robe before liver perfusion for HC or HM isolation performed by the Integrative Liver Cell Core of the University of Southern California as described below. A piece of liver tissue was fixed in 10% formalin for 24h and embedded in paraffin, incubated in RNAlater Solution (Lifetechnologies, Carlsbad, CA, USA) for RNA isolation, or frozen in liquid nitrogen and stored in −80C. Blood was collected with or without anticoagulant following centrifugation at 12,000 rpm for 15min. The serum or plasma was stored in −80.
Liver histology and Immunostaining
Tissue sections (5μm) were prepared and routinely stained for hematoxylin and eosin. Immunohistochemistry straining for myeloperoxidase (Myeloperoxidase Ab-1, Thermo Scientific, Waltham, MA, USA) was performed in formalin-fixed, paraffin-embedded livers according to the manufacturer’s instruction.
Real-time PCR
Total RNA was isolated from liver tissue using RNeasy Tissue Mini kit (Qiagen, Valencia, CA, USA). The reverse transcripts (the cDNA) were synthesized from 1μg of total RNA using the iScript cDNA Synthesis kit (Bio-Rad). Real-time PCR quantification was performed using SYBR-Green and CFX96 Thermal Cycler from BioRad. Briefly, 10μl of reaction mix contained: cDNA, KAPA SYBR® FAST qPCR master mix, and primers at final concentrations of 200nmol. The sequences of the primers used for quantitative PCR were as follows: tumor necrosis factor-alpha (TNF-α) 5′-CCC TCA CAC TCA GAT CAT CTT CT-3′ and 5′-GCT ACG ACG TGG GCT ACA G-3′; pro interleukin 1-alpha (IL-1α) 5′-GCA CCT TAC ACC TAC CAG AGT-3′ and 5′-AAA CTT CTG CCT GAC GAG CTT-3′; inducible nitric oxide synthase (iNOS) 5′-GTT CTC AGC CCA ACA ATA CAA GA-3′ and 5′-GTG GAC GGG TCG ATG TCA C-3′; Arginase 1 5′-CTC CAA GCC AAA GTC CTT AGA G-3′ and 5′-AGG AGC TGT CAT TAG GGA CAT C-3′; NOD-like receptor family pyrin domain containing 3 (NLRP3) 5′-ATT ACC CGC CCG AGA AAG G-3′ and 5′-TCG CAG CAA AGA TCC ACA CAG-3′; anti-apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) 5′-CTT GTC AGG GGATGA ACT CAA AA-3′ and 5′-GCC ATA CGA CTC CAG ATA GTA GC-3′; pro Caspase 1 5′-ACA AGG CAC GGG ACC TAT G-3′ and 5′-TCC CAG TCA GTC CTG GAA ATG-3′; pro interleukin 1-beta (IL-1β) 5′-GAA ATG CCA CCT TTT GAC AGT G-3′ and 5′-CTG GAT GCT CTC ATC AGG ACA-3′; β_ microglobulin 5′-CCC CAC TGA GAC TGA TAC ATA CG-3′ and 5′-CGA TCC CAG TAG ACG GTC TTG-3′
Cell isolation and cell culture
Liver cells were collected as previously described (15). Briefly, C57BL/6 mouse liver was digested with collagenase perfusion through portal vein and isolated hepatocytes with centrifugation at 50 g for 1 min. Hepatic macrophages were isolated from ASH mice with OptiPrep (Axis-Shield PoC AS, Oslo, Norway) as described before (16) for EV production. HepG2 cells were maintained in Dulbecco’s Modified Eagle Medium (Gibco, Camarillo, CA) supplemented with 10% fetal bovine serum (Cellgro, Manassas, VA), penicillin and streptomycin (Gibco), and sodium pyruvate (Gibco) at 37°C in a 10% CO2 incubator. AML12 cells were maintained in DMEM-12 (Gibco) supplemented with 10% fetal bovine serum (Cellgro, Manassas), penicillin and streptomycin, Insulin-transferrine-selenium (Gibco), and dexamethasone (Roche, Indianapolis, IN) at 37°C in a 10% CO2 incubator.
Immunoblot Analysis
For immunoblot analysis 50μg of isolated hepatocytes lysate was resolved by a 4–20% gradient gel, transferred to a nitrocellulose membrane, and blotted with the appropriate primary antibodies. Membranes were incubated with peroxidase-conjugated secondary antibody (Cell signaling, Danvers, MA, USA). Protein bands were visualized using an enhanced chemiluminescence reagent and digitized using a CCD camera (ChemiDoc®, BioRad, Hercules, CA, USA). A rabbit anti-cleaved caspase 3, anti-caspase 3, anti-phospho-MYPT1, or anti-MYPT1 was purchased from Cell Signaling and anti-beta actin was purchased from GeneTex (Irvine, CA, USA). Protein load was verified using GAPDH (GeneTex), or PORIN (GeneTex) antibody.
Cell-derived or blood EVs isolation
Isolated HCs or HMs from ASH or pair-fed control mice were cultured in medium-2% EVs depleted FBS (SBI, San Diego, CA, USA). Conditioned media of isolated HCs or HMs were collected after 3 h or 24 h of culture (new medium was added after 3 h collection), and cleared from cell debris by centrifugation at 2000×g for 10 minutes at 10°C. Conditioned media of HepG2 or AML12 were collected after 24 h of culture with 250 mM EtOH in serum-free growth medium, and cleared from cell debris by centrifugation at 2000×g for 10 min at 10°C. Cell-derived EVs from conditioned medium or blood EVs from plasma of four weeks of pair-fed control and ASH mice, as well as ten days of bile duct ligation mice, twenty weeks of NASH mice, and twelve weeks of obese mice, were isolated via ultracentrifugation at 100,000×g for 60 minutes at 10°C (Beckman L7-65, Beckman-Coulter, Palo Alto, CA). Pellet (EVs) was resuspended in PBS and stored at −80°C.
Measurement of EVs
Conditioned medium or plasma was incubated with Calcein-AM (Life Technologies) for 30 minutes at room temperature. The number of EVs was determined using 2.5-μm Alignflow alignment beads (Life Technologies) as the size standards for flow cytometry, BD LSRII Flow Cytometer System, (BD Biosciences, San Jose, CA). The data were analyzed using FlowJo software (TreeStar Inc., Ashland, OR).
EV size determination
For dynamic light scattering analysis, entire size was measured by Zetasizer nano ZS90 (Malvern). For transmission electron microscopy, EVs were adhered to 100 mesh Formvar and carbon coated grids for 5 minutes at room temperature. Grids were washed once with water, stained with 1% uranyl acetate (Ladd Research Industries, Williston VT) for 1 minute, dried and viewed using a JEOL 1200 EXII transmission electron microscope. Images were captured using a Gatan Orius 600 digital camera (Gatan, Pleasanton CA).
Quantification of encapsulated miRNAs levels
Encapsulated miRNAs were extracted using miRNase (Quiagen) according to the manufacturer’s instruction. From making template to quantification of miRNAs were performed as previously described (15). Briefly, the templates were made from 10 ng of total RNA using microRNA reverse transcription kit (Life Technologies) with specific primers, which are provided with TaqMan microRNA probes (Life technologies). Real-time PCR quantification for miRNA expression was performed using a TaqMan microRNA expression assay from Life Technologies. Cq value was converted to relative number using power formulation.
miRNA sequencing
Quality and quantity of purified small RNA was assessed using an Agilent Tapestation and a NanoDrop ND-1000, respectively. Libraries were generated from 100 ng of total RNA, using the TruSeq SmallRNA Sample Prep Kit (Illumina, San Diego, CA) following manufacturer’s instructions. Library quality was assessed using a High Sensitivity DNA kit (Agilent, Santa Clara, CA). Libraries were multiplexed and sequenced at 13 pM to an average depth of 7.5 million reads with 50 basepair (bp) single end (SE) reads on an Illumina HiSeq2500 using V4 chemistry.
Sequencing data analysis
The raw sequencing reads in FASTQ format went through preprocessing first to trim 3′ adapter and next to keep only reads whose length is between 17 and 27 bases. Using bowtie, the survived reads were aligned to reference resources such as mouse genome, miRNA precursor sequence, and non-coding RNAs, allowing only up to two mismatches. Only the reads mapped to both genome and precursor, but not to small RNAs other than miRNA were selected for expression level quantification. Then miRNA expression counts were quantified, normalized, calculated for fold-change between the treated and the control groups, and tested for significance of differential expression using negative-binomial model (17). Benjamini-Hochberg method (18) was applied to adjust for multiple testing on the raw p-values of differential expression test. MAGI, a web service for fast miRNA-seq analysis in a Graphics Processing Unit (GPU) infrastructure were used for a pipelined analysis of sequencing data (19).
Pathway analysis
The top 13 differentially expressed miRNAs with raw p-value less than 0.05 were chosen for pathway enrichment analysis. DIANA-miRPath (20) and in-house developed Python scripts were used for pathway analysis. For each miRNA, a set of target genes was created with a threshold of prediction score 0.7 against micro CDS target prediction database. The union set comprising all 13 sets of target gene was mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to derive significantly enriched pathways using Fisher’s exact test. Two heatmaps, miRNA-by-sample and miRNA-by-pathway, were drawn with DIANA-miRPath. Among these, <Pathways In Cancer>, which had 121 genes targeted by 8 miRNAs, was selected for visualization. Cytoscape (21) was used to draw miRNA-gene interaction plot. GeneMANIA (22) was used to infer gene-gene interaction on top of miRNA-gene interaction. Two interaction plots were merged into a single graph.
Statistical analyses
All data are expressed as mean ± SEM unless otherwise noted. Data were analyzed using t-tests in two groups, ASH mice and pair-fed control mice, and One-way Anova in EV functional assay using Graph Pad (Graph Pad Software Inc., CA, USA) for comparison of continuous variables. Data were analyzed using Mann-Whitney in two groups, ALD and non-alcoholic (control) human using Graph Pad. Differences were considered to be significant at p 0.05.
Results
ASH is associated with a marked increase in blood EVs
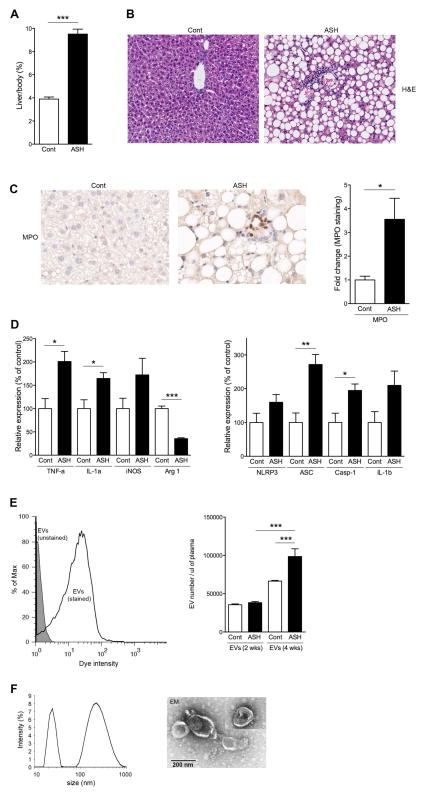
In order to assess the link between changes in blood EV levels with ASH, we used the 4-wk intragastric feeding model which leads to the development of mild ASH. The ratio of liver to body weight was significantly higher (p<0.001) in ASH mice compared to pair-fed control mice (Fig. 1A). As expected, these mice developed the features of ASH including significant steatosis and focal inflammation (Fig. 1B). Focal neutrophil infiltration, assessed via myeloperoxidase (MPO) immunohistochemistry of liver sections, was significantly increased (3.5-fold; p<0.05) in ASH mice compared to pair-fed control mice (Fig. 1C). Markers of pro-inflammatory M1 polarization were significantly increased in the ASH mice including TNF-α (p<0.05) and IL-1 α (p<0.05), and iNOS (M1 macrophage marker), while Arginase 1 (M2 macrophage marker) mRNA level was significantly decreased (p<0.001) in ASH mice compared to pair-fed control mice (Fig. 1D). The inflammasome is known to be a key factor in the progression of alcoholic liver disease (23, 24), thus the mRNA expression levels of inflammasome-related components were assessed via qPCR. NLRP3, ASC (p<0.01), pro-caspase-1 (p<0.05), and pro-IL-1β were up-regulated in ASH mice compared to pair-fed control mice (Fig. 1D), although NLRP3 and pro-IL-1β did not reach statistical significance. Having documented the various histological and molecular features of early ASH, we then assessed whether these features were associated with changes in the levels of EVs in circulation. The stained blood EVs had a distinct peak with 10-fold intensity compared to un-stained blood EVs via flow cytometry analysis (Fig. 1E). The quantification showed that the number of blood EVs was no difference after two weeks between ethanol and pair-fed control mice, whereas it was significantly increased (p<0.05) in ASH mice at 4 weeks (Fig. 1E). The characterization of blood EVs via dynamic light scattering (Fig. 1F) and electron microscopy (Fig. 1F) identified two sub populations of EVs: exosomes and microparticles (MPs).
Fig. 1. Blood EVs are increased in ASH mice.
(A–D) characterization of ASH liver at 4 weeks. (A) Ratio of liver weight to body weight. (B) Haematoxylin-eosin staining or (C) Immunohistochemical staining specific for MPO (neutrophils) of liver sections in pair-fed control or ASH mice. Scale bar, 100 μm. (C) Bar graph shows quantification of MPO positive cells. * p<0.05. (D) Gene expression of inflammatory genes or inflammasome genes as measured by qPCR. All gene expression levels were normalized to housekeeping control, β2 microglobulin, and shown relative to the expression levels of pair-fed control mice. ***p<0.001, **p<0.01, *p<0.05. (E,F) Characterization of blood EVs. (E) Flow cytometry analysis of blood EVs with dye from ASH mice at 4 weeks. The black line represents labeled blood EVs, whereas the gray area represents un-labeled blood EVs (background control). (E) Quantification of stained blood EVs in pair-fed control or ASH mice at 2 or 4 weeks. (F) Dynamic light scattering analysis and (F) Transmission Electron Microscopy of isolated blood EVs at 4 weeks. Values are mean ± SEM.
Hepatocytes are the major source of EV release in the ASH liver
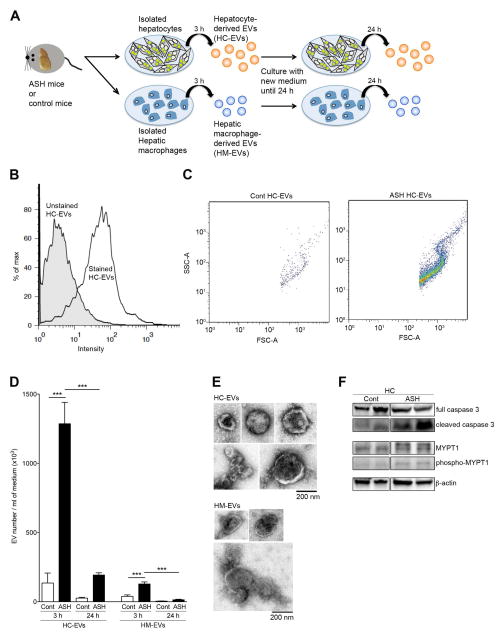
We expected that injured HCs are the major site of EV release from two lines of evidence; 1) cultured HCs exposed to various stressors, such as free fatty acid, cytokines, or alcohol, release EVs (25, 26), and 2) encapsulated miR-122/192 or asialoglycoprotein-receptor (ASGPR1), which is abundant in the HCs, is detected in the blood EVs or miRNAs (27, 28). However, there is no direct evidence that damaged HCs release EVs in liver disease. To address this question, we isolated primary HCs from ASH or pair-fed control mice with cell viability of 90.5% or 94.5%, respectively (Supporting Figure S1), cultured the HCs and assessed the HC-derived EVs (HC-EVs). We also investigated HM-derived EVs (HM-EVs) to determine the quantitative comparison for EV release by these two cell types in the liver from ASH mice. Conditioned medium containing HC-EVs or HM-EVs was collected after 3 h of culture, replenished with fresh medium and collected at 24 h, without any stimulus (Fig. 2A). After removing cell debris carefully with low speed centrifugation, HC-EVs or HM-EVs were stained with calcein and analyzed using flow cytometry. The intensity of stained HC-EVs was 10-fold higher than un-stained HC-EVs (Fig. 2B). The pattern of the calcein positive population was the same between HC-EVs from ASH or control samples (Fig. 2C). The number of HC-EVs and HM-EVs from ASH were significantly increased compared to HC-EVs and HM-EVs from pair-fed control animals (p<0.001) (Fig. 2D). Furthermore, the number of HC-EVs was 10 times higher than HM-EVs in ASH mice, demonstrating that HCs are the key EV producer (Fig. 2D). HC-EV or HM-EV morphology appeared the same when assessed via electron microscopy (Fig. 2E). As previously shown for EVs release during lipotoxicity and ethanol exposure (25, 26, 29), we found that the release of EVs by hepatocytes from ASH mice was associated with increases in, cleaved caspase 3 and phospholated MYPT1 in HCs suggesting that activation of caspases and pho-kinase pathways are also playing a role in their release (Fig. 2F, Supporting Figure S2).
Fig. 2. Hepatocytes are the major source of EV release in ASH mice.
(A) A schematic model: hepatocytes (HC) or hepatic macrophages (HM) were isolated from pair-fed control or ASH mice at 4 weeks and hepatocyte-derived EVs (HC-EVs) or hepatic macrophage-derived EVs (HM-EVs) were isolated from the condition medium. (B) Flow cytometry analysis of HC-EVs with dye from ASH mice. The black line represents labeled HC-EVs, whereas the gray area represents un-labeled HC-EVs (background control). (C) Dot plot of scat fold of HC-EVs from pair-fed control or ASH mice. (D) Quantification of stained HC-EVs or HM-EVs in pair-fed control or ASH mice. ***p<0.001 (E) Transmission Electron Microscopy of isolated HC-EVs or HM-EVs. Values are mean ± SEM. (F) Protein expression of cleaved caspase 3, full caspase 3, phospho-MYPT1, MYPT1, and beta actin in isolated hepatocytes from pair-fed control or ASH mice using immunoblotting.
Differentially expressed miRNAs
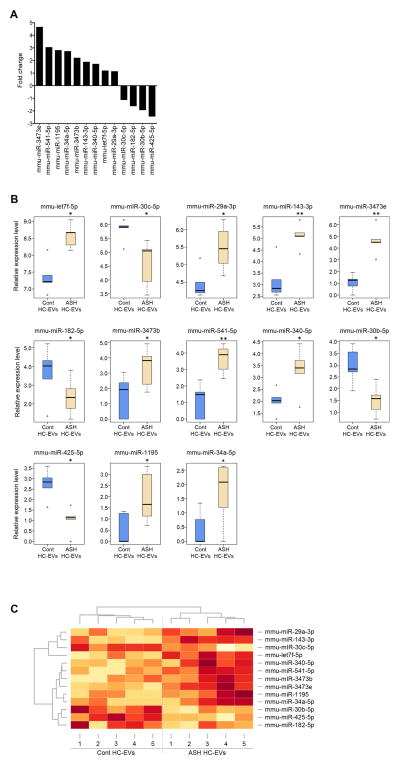
Circulating miRNAs encapsulated in EVs are known to be potentially novel biomarkers in alcoholic hepatitis (30). Therefore, we further investigated whether encapsulated miRNAs in HC-EVs could provide a “barcode” to distinguish ASH from control animals using the miRNA transcriptome. A total of 147 known miRNAs were quantified by alignment with at least 1 read in either group and we found 70 up-regulated miRNAs and 73 down-regulated miRNAs in ASH HC-EVs compared to miRNAs in control HC-EVs. Four miRNAs (miR-690, -673-5p, -540-5p and -540-3p) were detected only in ASH HC-EVs (Supplementary Table 1). Alternatively, 13 miRNAs were significantly up- or down-regulated in ASH HC-EVs compared to control HC-EVs (p<0.05) (Fig. 3A). While miR-122 was the most abundantly expressed, commensurate with previous findings (31), there was no significant difference between HC-EVs from ASH mice and the control group (data not shown). Let-7f showed the highest expression level among the 13 top differentially expressed miRNAs (Fig. 3B). miR-34a, which is known to play a role in hepatic metabolic disease, showed significantly higher expression in ASH samples (32). The heatmap of miRNAs expression (Fig. 3C) showed high within-group similarity and low between-group similarity among 13 different miRNA expression profiles.
Fig. 3. miRNA profile in HC-EVs.
(A) A fold change of 13 significantly up- or down-regulated miRNAs in ASH HC-EVs compared to miRNAs in pair-fed control HC-EVs. (B) Relative miRNA expression level in 13 significantly up- or down regulated miRNA. (C) Heatmap clusters of 13 significantly up- or down-regulated miRNAs by both rows and columns.
Target interactome of significantly up-regulated miRNAs in HC-EVs from ASH
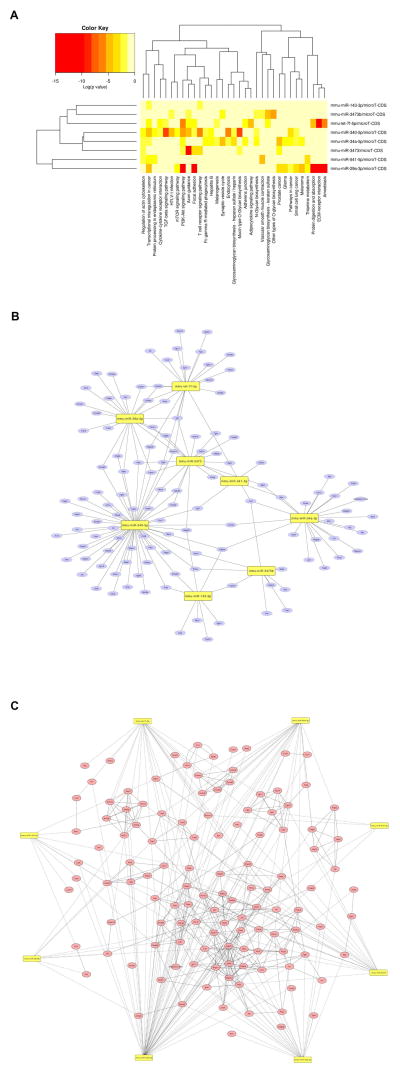
A heatmap of miR by pathway is drawn in Fig. 4A. The row enlists eight top differentially expressed miRNAs and the column contains 33 enriched KEGG pathways. Cancer, transcriptional misregulation in cancer, and PI3K-Akt signaling pathway are highly regulated by eight miRNAs. Eight of the nine significantly up-regulated miRNAs in ASH HC-EVs were involved in inflammatory and cancer pathways with 121 potential target genes, including NF-κB, Jun, IL-6, Pten, smad, EGF, Wnt, and Bcl2 (Fig. 4B) (Supplementary Table 2). While miR-340-5p had the most target genes (n=54), miR-541-5p had the least target genes (n=4). Interestingly, Runx1t1 was targeted by four miRNAs; miR-29a-3p, miR-340-5p, miR-541-5p, and let-7f-5p. In addition, six genes (Pten, Pdgfra, Mapk9, Lamc1, Igf1, and Arf) were targeted by three miRNAs, specifically, let7f-5p, miR-29a-3p, -340-5p, -34a-5p, and -3473 shared the majority of target genes (Fig. 4B). Fig. 4C extends miRNA-gene interaction (dotted line) shown in Fig. 4B by adding gene-gene interaction represented by the solid line. All eight miRNAs were connected through targets, resulting in a larger network influenced by significantly up-regulated miRNA in ASH HC-EVs. While miR-541-5p directly interacts with only 4 genes in Fig. 4B, its network was extended to 19 genes by adding the first neighborhood of target genes. For example, Ikbkb, one of the genes directly targeted by miR-541-5p, has six interacting neighbor genes; Chuk, Dapk1, Ikbkg, Mapk8, Mapk9, and Tgfbr1. Further analysis and validation could delineate their level of involvement in the pathogenesis of ASH.
Fig. 4. Target interactome from significantly up-regulated miRNAs in ASH HC-EVs.
(A) Heatmap clusters of KEGG pathways with nine significantly up- or down-regulated miRNAs by both rows and columns. (B) Target genes in KEGG cancer pathway in each eight miRNAs. (C) Gene-gene interaction with 80 genes from pathways.
miRNA “barcode” derived from HC-EVs can be used to identify ASH using blood EVs
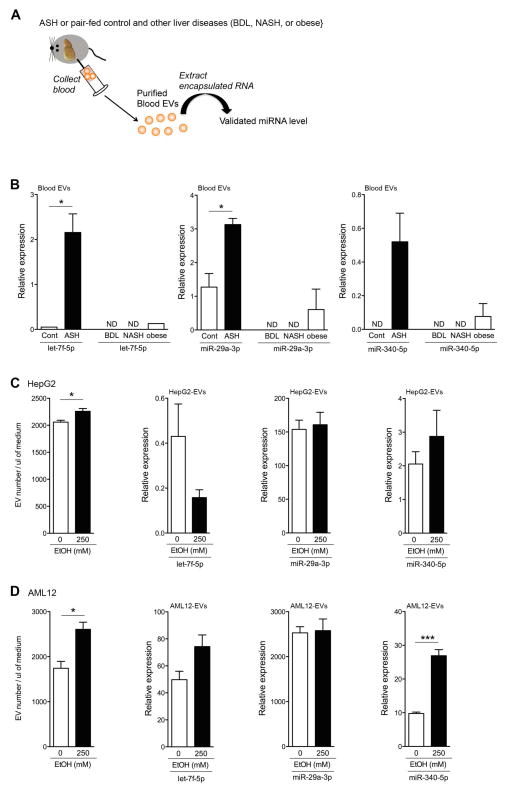
The demonstration that HC-EVs from ASH mice carry a specific miRNA barcode led us to the hypothesis that the miRNA profile present in EVs released by damaged hepatocytes during ASH may be used in blood EVs to non-invasively identify the presence of ASH. To address this question, we purified blood EVs from ASH or pair-fed control mice, extracted the RNA, and performed a qPCR analysis to determine the expression level of seven miRNAs derived from ASH HC-EVs (Fig. 5A). From the seven miRNA tested, three allowed for efficient discrimination between ASH and pair-fed control mice including Let-7f-5p and miR-29a-3p the levels of which were significantly up-regulated in the ASH blood EVs compare to control blood EVs (p<0.05) (Fig. 5B), as well as miR-340 that was only detected in ASH blood EVs (Fig. 5B). miR-3473b levels were similar in blood EVs from both ASH and pair-fed mice while the other miRNAs, including miR-34a, -541, -143- and -1195 levels were below the detectable range in blood EVs by qPCR (data not shown).
Fig. 5. Blood EVs from ASH have miRNA barcode.
(A) A schematic model: blood EVs from ASH or pair-fed control, as well as bile duct ligation (BDL), non-alcoholic steatohepatitis (NASH), and obese, mice were purified from the blood and checked the miRNA level in blood EVs. (B) Relative Cq value of let-7f-5p, miR-29a-3p, and miR-340-5p via quantitative RT-PCR analysis. (C–D) Quantification of stained hepatocyte-derived EVs from HepG2 (C) or AML12 (D) without or with EtOH 250 mM. Relative Cq value of let-7f-5p, miR-29a-3p, and miR-340-5p in HepG2-derived EVs (C) or AML12-derived EVs (D) via quantitative RT-PCR. *p<0.05. Values are mean ± SEM.
miRNA barcode, is specific for alcoholic liver injury and present in human alcoholic patients
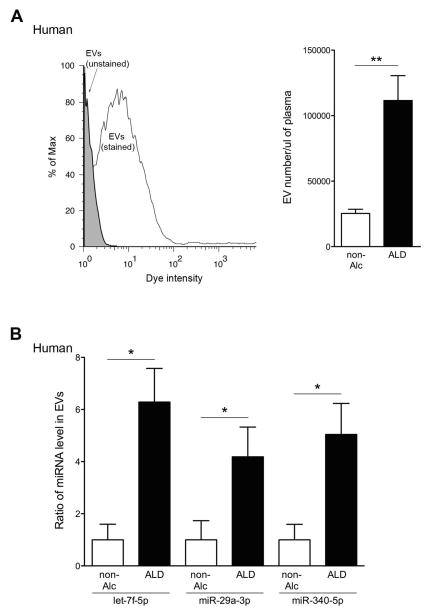
To assess the specificity of the identified miRNA barcode in ASH mice, we assessed three miRNAs (let-7f, miR-29a, and miR340) in blood EVs from different liver disease mouse models, including BDL, NASH, and obese mice (Fig. 5A). Three miRNAs were below the detectable range in blood EVs from BDL and NASH mice and were expressed at low levels in blood EVs from obese mice by qPCR (Fig. 5B). In order to further explore whether the miRNA barcode identified reflects alcohol exposure, we treated hepatocytes with EtOH and assessed miRNA level in hepatocyte-derived EVs in vitro. As previously shown (25), the release of hepatocyte-derived EVs were significantly increased in HepG2 and AML12 treated with 250 mM EtOH (p<0.05) (Fig. 5C and 5D), however, three miRNAs level in HepG2- or AML12-derived EVs was not increased, except miR-340 in AML12-derived EVs (Fig. 5C and 5D). These results strongly suggest that the identified miRNA profile is a specific marker of ALD-related liver injury. Finally, in order to address the potential translational significance of our findings for patients with ALD, we conducted an early phase pilot study were we quantified three miRNAs level in blood EVs from a small group of ambulatory patients with ALD versus individuals without alcohol dependency (non-alcoholics). Blood EVs were stained with calcein and analyzed using flow cytometry. The intensity of stained blood EVs was higher than un-stained blood EVs (Fig. 6A). Blood EVs were significantly increased in patients with ALD compared to non-alcoholics (p<0.01) (Fig. 6A). Similar findings were observed for the three miRNAs in blood EVs (let-7f: p<0.05, miR-29a: p<0.05, and miR-340: p<0.05) (Fig. 6B).
Fig. 6. Blood EVs from patients with ALD have miRNA barcode.
(A) Flow cytometry analysis of blood EVs with dye from non-alcoholics (non-Alc) or patients with alcoholic liver disease (ALD). The black line represents labeled blood EVs, whereas the gray area represents un-labeled blood EVs (background control). (A) Quantification of stained blood EVs in non-Alc or ALD. **p<0.01. (B) Relative Cq value of let-7f-5p, miR-29a-3p, and miR-340-5p via quantitative RT-PCR analysis. **p<0.05. Values are mean ± SEM.
Discussion
The key finding of this study is the demonstration that in experimental alcoholic steatohepatitis (ASH) mice there is an increased production and release of extracellular vesicles (EVs), predominantly by hepatocytes, that contain a miRNA signature detectable in blood EVs as a “barcode” to identify ASH. Alcoholic liver disease (ALD) is a wide spectrum disease that includes steatosis, steatohepatitis or ASH, and in severe cases fibrosis and/or cirrhosis. Steatosis, characterized by fat accumulation in hepatocytes, is by far the most common form of ALD and tends to follow a benign clinical course that resolves upon cessation of alcohol consumption (33, 34). A subgroup of patients with ALD progress to ASH, which is characterized by the persistence of fatty liver accompanied by a robust inflammatory response. In later stages of ALD collagen deposition and regenerative nodules can result in the development of fibrosis and cirrhosis, respectively. Although much progress has been made in the understanding of the mechanisms by which alcohol damages the liver, there is still currently a paucity of pharmacologic strategies to stop, or reverse disease progression. One of the key factors limiting the development of effective therapies is the lack of reliable, mechanism-based non-invasive biomarkers that can help identify patients at risk for disease progression. In the current study, we set out to test the hypothesis that during ASH development fat-laden hepatocytes release EVs with a miRNA signature that can be measured in circulation. In order to do this, we developed various novel experimental approaches that allowed us to directly test our hypothesis. Recent studies from our lab, as well as others, have demonstrated that the number of blood EVs increases in murine models of nonalcoholic and alcoholic liver disease (28, 30), while in vitro exposure of primary rat hepatocytes to saturated fatty acids (26), or Huh7.5 or HepG2 cells to ethanol (25, 29), results in the release of EVs into the media. The current study builds on these findings and demonstrates that both exosomes and microparticles are increased in the circulation of mice with ASH induced by continuous intragastric ethanol feeding. Moreover, we were able to demonstrate for the first time that both HCs and HMs isolated from this mouse model produce large quantities of EVs and that HCs are the major source for EV release in the ASH liver. Data from our lab, as well as others, have previously showed that EVs are efficiently internalized into target cells and the transferring of their composition, such as proteins, lipids, or miRNAs, is a key mechanism by which EVs modulate cell signaling in target cells (12, 13)(35). miRNAs have an especially crucial role in gene regulation with the majority of miRNAs binding to the 3′UTR region of the target mRNA leading to translational suppression, and some miRNAs bind to their target mRNA with a perfect match resulting in gene suppression through mRNA cleavage (14). Indeed, we, and others have recently shown an inverse correlation between expression levels of certain miRNAs in the liver and those in blood EVs, or plasma, suggesting the potential for using specific miRNAs as sensitive and informative biomarkers of liver damage (15–20). There are mainly two forms of circulating miRNAs: encapsulated miRNAs in EVs (EV-miRNAs), and complex miRNA with non-coding RNA binding proteins known as Argonaute proteins (Ago-miRNA) (21). Ago-miRNA cannot integrate into recipient cells due to its size and lack of bioactivity for penetration (22), whereas EV-miRNAs can efficiently penetrate recipient cells with their miRNAs (23). Our analysis identified 13 significantly up- or down-regulated miRNAs in ASH HC-EVs compared to control HC-EVs. Eight miRNAs from the nine significantly up-regulated in ASH HC-EVs (miR-541, miR-3473, miR-143, miR-29a, let-7f, miR-340, miR-34a, and miR-3473b) were involved in inflammatory and cancer pathways and potentially regulate 121 target genes, including Bcl2, Jun, IL-6, Pten, smad, and Wnt families (35, 36). Furthermore, miR34a has been found to play an important role in liver damage and remodeling during ALD with both Caspase-2 and Sirtuin 1 being direct targets of this miRNA (38).
Development of non-invasive reliable biomarkers that allow an assessment of the severity of ALD in patients is greatly needed. Our finding that EVs derived from hepatocytes from ASH mice elucidated a profile of differentially expressed miRNAs, in conjunction with the increased level of EVs in blood, led us to explore the possibility that a miRNA signature can be measured in circulation and exploited for diagnostic purposes. Indeed, we found that three out of seven miRNAs tested allowed for the efficient discrimination between ASH and pair-fed control mice and between ASH and other causes of liver injury including cholestatic and metabolic demonstrating the specificity of the miRNA profile identified. In addition, a short-term 2 week course of gastric infusion, a time point that is associated with only minimal changes in the liver did not result in an increase in circulating EVs. While, consistent with previous studies (25, 29). In vitro, exposure of hepatocytes to ethanol did enhance the EV release. However, when we explored the miRNA profile we found that this treatment did not mimic the one found in hepatocytes from ASH mice and in blood from these mice. Taken together, these results suggest that quantitation of EV release and the miRNA barcode are markers for ALD related liver injury that allows for distinction between ASH and fatty liver and is not a mere consequence of ethanol exposure. We further tested the translational significance of our findings in a pilot small human study consistent of a group of patients with ambulatory ALD and non-alcoholics (controls). This cohort was selected as our intragastric model is a model of early chronic ASH.. Previous reports suggested that blood exosomes or EVs are increased in hospitalized patients with acute alcoholic hepatitis (29, 30). We observed a significant increase in blood EVs in patients with ambulatory ALD. We further found that our identified profile of three miRNAs (let-7f, miR-29, and miR-340) was significantly increased in these patients compared to non-alcoholics. These data hold the promise that this approach may allow for the creation of a customized miRNA array that would serve as a “barcode” to identify ASH and future larger prospective studies in humans are warranted.
In summary, the current study demonstrates in an established murine model of mild ASH that EVs are increased in circulation, with hepatocytes being the key source of these EVs, and carry a specific miRNA cargo. These findings uncover EVs as potential novel biomarkers and therapeutic targets for ASH.
Supplementary Material
Acknowledgments
Funding: The work was supported by NIH grants P50AA011999 (Pilot Project Program), R21 AA023574, and Gilead research scholars program in liver disease to AE; NIH grants U54HL108460 to JK and LOM; NIH grants U01AA021908 to SBH; NIH grants U01AA021856 to BS; NIH grants P50AA011999 (Pilot Project Program and Animal Core), R24AA12885 (Integrative Liver Cell Core), U01AA018663, and 5I01BX001991 (Department of Veterans Affairs Merit Review) to HT; NIH grants U01 AA022489 and DK082451 to AEF;.
We thank the UCSD IGM Genomics Center, especially Kristen Jepsen for siRNA sequencing advice and assistance. The authors would like to thank Dr. Marilyn Farquhar for the use of the electron microscopy facility, and Timo Meerloo for electron microscopy sample preparation.
Abbreviations
- ALD
alcoholic liver disease
- ASH
alcoholic steatohepatitis
- HCC
hepatocellular carcinoma
- EV
extracellular vesicle
- HC
hepatocyte
- HM
hepatic macrophage
- HC-EV
hepatocyte-derived EV
- HM-EV
hepatic macrophages-derived EV
- miRNA
microRNA
Footnotes
Conflict of Interest: The authors state no conflict of interest.
References
- 1.Mathurin P, Bataller R. Trends in the management and burden of alcoholic liver disease. J Hepatol. 2015;62:S38–46. doi: 10.1016/j.jhep.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol. 2015;12:231–242. doi: 10.1038/nrgastro.2015.35. [DOI] [PubMed] [Google Scholar]
- 3.Bataller R, Gao B. Liver fibrosis in alcoholic liver disease. Semin Liver Dis. 2015;35:146–156. doi: 10.1055/s-0035-1550054. [DOI] [PubMed] [Google Scholar]
- 4.Feldstein AE, Gores GJ. Apoptosis in alcoholic and nonalcoholic steatohepatitis. Front Biosci. 2005;10:3093–3099. doi: 10.2741/1765. [DOI] [PubMed] [Google Scholar]
- 5.Barnes MA, Roychowdhury S, Nagy LE. Innate immunity and cell death in alcoholic liver disease: role of cytochrome P4502E1. Redox Biol. 2014;2:929–935. doi: 10.1016/j.redox.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner C, Galluzzi L, Kepp O, Kroemer G. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Park PH, Thakur V, Pritchard MT, McMullen MR, Nagy LE. Regulation of Kupffer cell activity during chronic ethanol exposure: role of adiponectin. J Gastroenterol Hepatol. 2006;21(Suppl 3):S30–33. doi: 10.1111/j.1440-1746.2006.04580.x. [DOI] [PubMed] [Google Scholar]
- 8.Tsukamoto H. Redox regulation of cytokine expression in Kupffer cells. Antioxid Redox Signal. 2002;4:741–748. doi: 10.1089/152308602760598882. [DOI] [PubMed] [Google Scholar]
- 9.Miller AM, Horiguchi N, Jeong WI, Radaeva S, Gao B. Molecular mechanisms of alcoholic liver disease: innate immunity and cytokines. Alcohol Clin Exp Res. 2011;35:787–793. doi: 10.1111/j.1530-0277.2010.01399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eguchi A, Mulya A, Lazic M, Radhakrishnan D, Berk MP, Povero D, Gornicka A, et al. Microparticles release by adipocytes act as “find-me” signals to promote macrophage migration. PLoS One. 2015;10:e0123110. doi: 10.1371/journal.pone.0123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ball SA, Tennen H, Poling JC, Kranzler HR, Rounsaville BJ. Personality, temperament, and character dimensions and the DSM-IV personality disorders in substance abusers. J Abnorm Psychol. 1997;106:545–553. doi: 10.1037//0021-843x.106.4.545. [DOI] [PubMed] [Google Scholar]
- 14.Ueno A, Lazaro R, Wang PY, Higashiyama R, Machida K, Tsukamoto H. Mouse intragastric infusion (iG) model. Nat Protoc. 2012;7:771–781. doi: 10.1038/nprot.2012.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Chi F, Guo T, Punj V, Lee WN, French SW, Tsukamoto H. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest. 2015;125:1579–1590. doi: 10.1172/JCI76468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 19.Kim J, Levy E, Ferbrache A, Stepanowsky P, Farcas C, Wang S, Brunner S, et al. MAGI: a Node. js web service for fast microRNA-Seq analysis in a GPU infrastructure. Bioinformatics. 2014;30:2826–2827. doi: 10.1093/bioinformatics/btu377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negash AA, Gale M., Jr Hepatitis regulation by the inflammasome signaling pathway. Immunol Rev. 2015;265:143–155. doi: 10.1111/imr.12279. [DOI] [PubMed] [Google Scholar]
- 24.Petrasek J, Bala S, Csak T, Lippai D, Kodys K, Menashy V, Barrieau M, et al. IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J Clin Invest. 2012;122:3476–3489. doi: 10.1172/JCI60777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Povero D, Eguchi A, Niesman IR, Andronikou N, de Mollerat du Jeu X, Mulya A, Berk M, et al. Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Sci Signal. 2013;6:ra88. doi: 10.1126/scisignal.2004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arrese M, Eguchi A, Feldstein AE. Circulating microRNAs: emerging biomarkers of liver disease. Semin Liver Dis. 2015;35:43–54. doi: 10.1055/s-0034-1397348. [DOI] [PubMed] [Google Scholar]
- 28.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, et al. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma VK, Li H, Wang R, Hirsova P, Mushref M, Liu Y, Cao S, et al. Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J Hepatol. 2016;64:651–660. doi: 10.1016/j.jhep.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabb DW. Pathogenesis of alcoholic liver disease: newer mechanisms of injury. Keio J Med. 1999;48:184–188. doi: 10.2302/kjm.48.184. [DOI] [PubMed] [Google Scholar]
- 34.Bergheim I, McClain CJ, Arteel GE. Treatment of alcoholic liver disease. Dig Dis. 2005;23:275–284. doi: 10.1159/000090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Povero D, Panera N, Eguchi A, Johnson CD, Papouchado BG, de Araujo Horcel L, Pinatel EM, et al. Lipid-Induced Hepatocyte-Derived Extracellular Vesicles Regulate Hepatic Stellate Cells via MicroRNA Targeting Peroxisome Proliferator-Activated Receptor-γ. Cell Mol Gastroenterol Hepatol. 2015;1:646–663. doi: 10.1016/j.jcmgh.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.