Abstract
Genome-wide, DNA mutation analysis in single cells is prone to artifacts associated with cell lysis and whole genome amplification. Here we addressed these issues by developing Single-Cell Multiple Displacement Amplification (SCMDA) and the single-cell variant caller, SCcaller. Validated by comparing SCMDA-amplified single cells with unamplified clones from the same population, the procedure provides a firm foundation for standardizing somatic mutation analysis in single-cell genomics.
Analyzing genetic variants in single cells suffers from artifacts associated with whole genome amplification (WGA). Common artifacts result from cytosine deamination due to single cell lysis and DNA denaturation at elevated temperatures, resulting in artificial CG->TA transitions1. This could explain, for example, the great excess of such mutations found in single neurons2 as compared to similar studies of unamplified neuronal clones3. Amplification errors per se can be enriched through allelic amplification bias, a characteristic of multiple displacement amplification (MDA) - the most frequently used method for WGA from a single cell4 - resulting in artifactual mutation calls (Supplementary Fig. 1a,b). Here we report Single-Cell Multiple Displacement Amplification (SCMDA) and a single-cell variant caller (SCcaller), a validated protocol to accurately identify SNVs across the genome from a single cell after whole genome amplification.
To address cytosine deamination artifacts, we single cell lysis and DNA denaturation is performed on ice using alkaline lysis. To compensate for the much lower effectiveness of cell lysis and DNA denaturation at low temperature we reconfigured MDA, significantly improving the annealing procedure for the hexamer primers (Methods). We then developed SCcaller, which corrects for local allelic amplification bias in SNV calling.
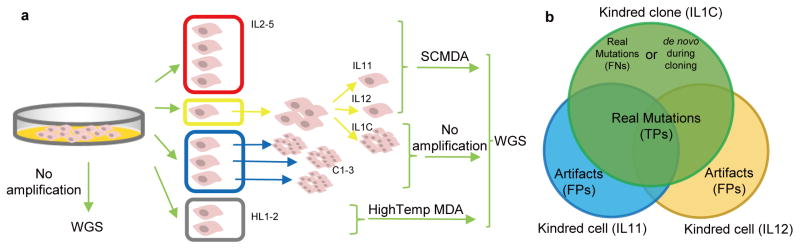
We validated SCMDA and SCcaller by directly comparing SNVs between amplified single cells and unamplified clones derived from cells in the same population of early passage, human primary fibroblasts. We also sequenced SCMDA-amplified single cells and non-amplified clones derived from the same, early growing clone (~5 divisions; 20~30 single cells), reasoning that there should be significant overlap between the single cells and their kindred clone (Fig 1a,b). Finally, we also included single cells after high-temperature lysis and DNA denaturation using a commercially available system (Methods) to confirm artifactual mutations induced through cytosine deamination at high temperature.
Figure 1. Experimental design for validating SNV identification in SCMDA-amplified single cells.
(a) To allow validation for accurate single cell amplification and variant calling, whole genome sequencing (WGS) was performed on (1) four single cells amplified using SCMDA (red); (2) two cells amplified using SCMDA and their non-amplified kindred clone (yellow); (3) three additional, unamplified clones (blue); and (4) two single cells amplified after high temperature lysis (grey). Cell / clone IDs are included in the Figure. (b) The kindred cells and clone are expected to have identical genotypes, including both germline and somatic SNVs. Candidate SNVs identified in both clone and single cells are true positives (TPs). Those found in neither of the cells but only in the clone are false negatives (FNs). Variants found only in one cell are considered false positives (FPs). See Supplementary Note for details. These are conservative assumptions and do not take into account possible de novo mutations in the kindred clone or single cells arising after their divergence. Of note, such events would increase sensitivity and specificity.
Single cells, isolated using the CellRaft system (Methods, Supplementary Fig. 2, 3) were subjected to SCMDA, library preparation and sequencing5 (Methods, Supplementary Note, Supplementary Table 1, 2). As a pre-screen to test for the relative uniformity of amplification we used real-time PCR at 8 specific loci and 66% of 44 cells passed our criteria (Supplementary Note, Supplementary Table 1). Only cells from this group were sequenced. Supplementary Table 3 provides the sequencing statistics, showing that in the single cells on average 85% of the genome was sequenced with a depth of at least 5 reads, as compared to about 90% in the clones and bulk cell population. The genome-wide coverage uniformity of amplification after whole genome sequencing was evaluated using Lorenz plots (Supplementary Fig. 4). The results indicated that, as expected, the unamplified bulk DNA shows the least amount of bias; in addition, amplicon samples produced by SCMDA exhibited less bias than those produced by the commercial, high-temperature lysis system (Supplementary Fig. 4) or by other amplification protocols6,7.
For calling SNVs from the sequencing data, we first predicted the degree of local allelic amplification bias using heterozygous germline SNPs (hSNPs) (Supplementary Fig. 5a–c). Because MDA starts at random positions and elongates to several kilobases, it is possible to predict the degree of allelic bias at a particular locus by considering the degree of bias in neighboring hSNPs using kernel smoothing (Methods, Supplementary Fig. 6 a–d & Supplementary Table 4). We designed SCcaller to adjust allelic amplification bias when estimating the likelihoods of three possibilities, i.e., artifact, heterozygous SNV, and homozygous SNV, for every candidate SNV locus (Methods, Supplementary Fig. 7 & Supplementary Fig. 8 a,b). A likelihood ratio test was used to distinguish real SNVs from artifacts under a certain significance level (α). Its null distribution, which corresponds to the amplification errors, was sampled from the input data. This input-specific null accounts for sample-specific amplification bias, sequencing depth and quality.
We evaluated the accuracy of SCMDA and SCcaller using our kindred single cells and clone combination, because in this system real mutations should be present in both the single cells and the kindred clone (Fig. 1b). For germline SNP calling, SCcaller reached 90.1% sensitivity at a cost of 0.12 false positives (FPs) per million bp (Fig. 2a,b). In comparison, Haplotypecaller from GATK8, a commonly used SNP caller for bulk sequencing data, suffered from more than 7 times as many FPs, i.e., 0.87 FPs per million bp. SCcaller performed similarly on a published MDA-amplified single-cell data set from another laboratory2 and significantly outperformed Haplotypecaller and Monovar9 (Fig. 2c; Supplementary Note).
Figure 2. Accuracy of SCcaller in single cell SNV calling.
Sensitivity and FP rate of germline SNP calling in cell IL11 (a) and IL12 (b). Sensitivity (y-axis) was defined as the ratio of TPs to FNs plus TPs. FP rate per MB on the x-axis is the number of FPs per million bp. Default cutoff (x) refers to α=0.01 by SCcaller. (c) Germline SNP calling using SCcaller was compared with Monovar and Haplotypecaller in a dataset from Lodato et al.2. On the x-axis, the number of artifacts was approximated as the number of SNVs unique to one cell (Supplementary Note). Since these unique SNVs also include real somatic SNVs, this approximation is the upper-bound of the number of artifacts. SCcaller suffered from the smallest number of false positives (<1,000 per cell), as compared to Haplotypecaller (>30,000 per cell) and Monovar (>5,000 per cell). For somatic SNV calling, SCcaller was compared with MuTect, VarScan and Monovar in cell IL11 (d) and IL12 (e). FDR was defined as the ratio of FPs to TPs plus FPs. TPs and FPs were derived from the kindred clone experiment (Fig. 1b, Supplementary Note). (f) Fraction of overlapping somatic SNVs called from MALBAC kindred single cells (Supplementary Note)7. SNPs and SNVs were called in regions with ≥20 sequencing depth. The error bars indicate standard deviations.
We then tested if SCcaller displayed the same unique feature of high specificity, i.e., a low rate of false positive calls, in identifying somatic SNVs using our kindred single cells and clone. We found that SCcaller has a much higher specificity (FDR: 0.308–0.393) than other callers, i.e., MuTect10, VarScan11 and Monovar9 (FDR: 0.745–0.860), most likely by accounting for amplification errors (Fig. 2d,e). Its sensitivity is either higher than that of the other callers (Monovar) or similar (MuTect and VarScan). Sixteen randomly picked SNVs called by SCcaller, were confirmed using Sanger sequencing of the kindred group versus bulk (Supplementary Table 5). Using another published single-cell data set obtained by MALBAC (Multiple Annealing and Looping-Based Amplification Cycles)7, SCcaller reported more than two times higher overlapping somatic SNV calls in all kindred cells than Monovar, MuTect or VarScan (Fig. 2f; Supplementary Note). Of note, the fraction of overlapping calls in this data set was very low (0.3%) as compared to what we obtained with SCMDA (65% on average), likely due to our highly optimized experimental protocol for somatic SNV detection. Hence, applied to different, independently published data sets SCcaller proved superior to other variant callers, including those specifically designed for single cell applications. Of note, SCcaller’s application is limited to genome regions of SNP heterozygosity (Methods).
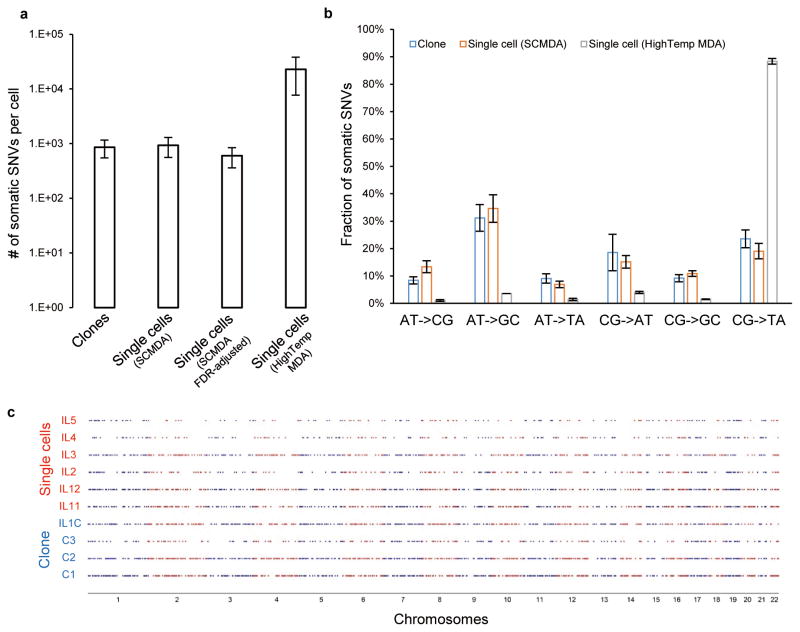
We then extended the somatic SNV detection by SCMDA and SCcaller from the kindred group to all our single cells and clones, comparing the results to those obtained using the commercial procedure with single cell lysis and denaturation at elevated temperature. As expected, in the latter case many more candidate somatic SNVs were detected than in those amplified using SCMDA: an average of 22,929 and 927 per cell, respectively, after adjusting for sequencing depth and coverage (Fig. 3a). Almost all the somatic SNVs detected after high temperature denaturation were CG->TA transitions (Fig. 3b), which is expected when they stem from cytosine deamination. The number of somatic SNVs identified using SCMDA, on average 927±371 (s.d.) per cell, was in the same range as the unamplified clones, on average 856±306 (s.d.) per cell. Of note, these results are also in the same range as previously reported for non-amplified single cell clones obtained through organoid formation (609 per organoid genome)12. The somatic SNVs identified using SCMDA had a very similar spectrum as the clones, with CG->TA mutations making up only 21.3% of all somatic SNVs (Fig. 3b). Of note, the same spectra were obtained when using the other mutation callers (Supplementary Fig. 9). Hence, other callers detected artifacts rejected by SCcaller, but not specifically artifacts resulting from cytosine deamination. Hence, these combined results underscore the validity of SCMDA and SCcaller in providing both reliable mutation identification and reliable mutation spectra for whole genome-amplified single cells.
Figure 3. Frequency, spectrum and distribution of somatic SNVs.
(a) Number of somatic SNVs per cell after correction for sequence depth and coverage (Supplementary Note). (b) Spectrum of somatic SNVs. In a) and b), the error bars indicate standard deviations, and n=4, 6 and 2 for the clones, SCMDA and HighTemp MDA respectively. (c) Genome distribution of somatic SNVs identified. Each row indicates one single cell or clone and each dot represents a somatic SNV. Smaller numbers of somatic SNVs were identified in IL4 and IL5 due to lower sequencing depth (Supplementary Table 3).
The somatic SNVs identified in the human fibroblasts were distributed uniformly across the genome (Fig. 3c), with a moderate depletion of mutations from functional sequence features, such as exons, DNase I hypersensitive sites and 3′ UTRs, similar to what was found for germline polymorphisms in the 1000 Genomes Project (Supplementary Fig. 10a). This is consistent with the presence, in actively proliferating fibroblasts, of considerable selection against deleterious mutations. Somatic SNVs were also depleted from genes expressed in human fibroblasts (P=0.016 one-tailed permutation test, Supplementary Fig. 10b), due to either selection or transcription coupled repair13.
In summary, the validated single-cell whole genome sequencing procedure and variant caller developed allow for the first time to accurately determine the full complement of somatic mutations unique for a cell. This will greatly expand our capability to explore the landscapes of somatic mutagenesis in humans and other organisms, which hitherto was only possible by using clonal amplification, such as organoid technology. Clonal amplification requires extensive cell culture and essentially limits analysis to stem or progenitor cells. Single cell technology allows direct analysis of all types of cells, including postmitotic cells, such as neurons and muscle fibers. Of note, single cell clones are also likely to harbor mutations arising de novo. Indeed, each of the single cells in our kindred groups was found to harbor a unique set of mutations that are not necessarily artifacts. Hence, in contrast to organoid technology SCMDA provides a comprehensive assay to study the landscape of somatic mutagenesis in metazoa, including sub-clonal heterogeneity in both normal and tumor tissue. This will provide increased insight into the pathogenic role of somatic mutations in human aging and disease14.
Online Methods
Cell culture
Primary human dermal fibroblasts from a 6-year-old male were provided by Haeri Choi at Seoul National University, South Korea. Human dermal fibroblasts were grown at 37°C, 3% O2, 10% CO2, in low glucose DMEM medium containing 10% FBS, 100 IU ml−1 penicillin, 100μg ml−1 streptomycin, 2mM L-Glutamine and 1% MEM non-essential amino acids (Gibco).
Preparing single cells
We prepared single cells and clones using the CellRaft array (Cell Microsystems). Following the manufacturer’s instructions, we wetted the CellRaft array with medium before plating the cells. The CellRaft array was covered with a proprietary water-soluble biocompatible coating to prevent air bubble formation when adding liquid to the array. To wet the array, we added 2 ml of the cell culture medium to the array and waited for 3 minutes, then removed the medium; this process was repeated a total of three times.
When cells grew to confluence in a 10-cm plate, we removed the medium, washed the cells with PBS, added 1 ml trypsin (Gibco) and incubated at 37°C for 5 minutes. After confirming that all cells were detached, we added 10 ml complete medium to stop trypsinization and transferred the cell solution to a 10-ml tube. Cells were centrifuged for 5 minutes at 1500 rpm. The supernatant was removed by aspiration and the cell pellet was resuspended in 10 ml fresh medium. This process was performed a total of two times to ensure the removal of cell debris prior to single cell isolation.
The CellRaft array contains 12,000 individual sites, called “rafts”. To pick single cells, we counted cells with the Auto T4 Cellometer (Nexcelom Bioscience) and prepared about 5000 cells in 3 ml of medium. The cell solution was plated onto the CellRaft array and incubated at 37°C, 3% O2, 10% CO2. After 2–4 hours, the fibroblasts were elongated and firmly attached to the array. To remove floating cells (which were probably dead) and avoid contamination from potential cell debris and cell-free DNA in the medium, we removed the medium from the CellRaft array, washed the cells on the CellRaft array twice using 1 ml PBS and added 3 ml fresh complete media. Single rafts containing one cell (as observed by microscopy using a 10x objective, Supplementary Fig. 2) were transferred, using the magnetic wand supplied with the CellRaft system, into 0.2-ml PCR tubes containing 2.5 μl PBS. By observing the PCR tubes under a magnifying glass and the CellRaft array under a microscope before and after the transfer, we validated that there was only one cell in a tube (Supplementary Fig. 3). Tubes containing single rafts were frozen immediately on crushed dry ice and kept at −80°C until use. To validate that there was no contamination from cell-free DNA, we also picked empty rafts as negative controls in whole genome amplification (described in the section on SCMDA).
Single cell clone and kindred cells
To generate single cell clones, we used a modified version of the cloning protocol described previously15. Cells were first plated on the CellRaft array as described above. We kept track of rafts containing single cells using the coordinate markers on the CellRaft. Culturing conditions were the same as described in the cell culture section above, except that the FBS content of the medium was increased from 10% to 20%. Cells were checked for growth daily and medium changed every three days. When the single cell clones reached confluence on the raft, i.e. approximately 10–16 cells per raft, each single raft containing a clone was transferred to a well in a 96-well plate. Upon reaching confluence, the clones were transferred to 24-well plates, then 12-well plates, 6-well plates and 10-cm plates; medium containing 20% FBS was used until the cells were transferred to 12-well plates, at which point FBS supplementation was reduced to 10%.
To generate kindred single cells and clone, we first grew cells to small clones, consisting of approximately 20–30 cells, after transfer to a 96-well plate. We then selected a clone and transferred all of its cells to another CellRaft array. Fifteen kindred cells were isolated from the small clone using the CellRaft system. The remaining cells from the clone were transferred into a well of 96-well plate and grown into a large clone as described above. We sequenced the unamplified DNA from the large clone and the amplified DNA of two of the cells isolated from the 20–30-cell stage clone.
Single cell whole genome amplification by multiple displacement amplification, (SCMDA)
Frozen single cells in PCR tubes were removed from storage at −80°C, quickly spun down by nano-centrifuge, and placed on ice. One μl Exo-Resistant Random Primer (final conc. 12 μM, Thermo Scientific) was then added, followed by 3 μl lysis buffer containing 400mM KOH, 100mM DTT and 10mM EDTA solution. Cell lysis and DNA denaturation was performed on ice for 10 minutes. Then, 3 μl of stop buffer, consisting of 400mM HCl and 600mM Tris-HCl solution (1M, pH7.5), was added to neutralize the lysis buffer, followed by 32 μl of master mix containing 30 μl of MDA reaction buffer and 2 μl of Phi29 polymerase (REPLI-g UltraFast Mini Kit, Qiagen). MDA was carried out in a total volume of 41.5 μl for 1.5 hours at 30°C, followed by an increase in the temperature to 65°C for 3 minutes to inactivate DNA polymerase. After amplification, samples were held at 4°C until purification. Amplified DNA was purified using AMPureXP-beads (Beckman Coulter) and the concentration was measured with the Qubit High Sensitivity dsDNA Kit (Invitrogen Life Science).
As a positive control for the amplification, 1 ng of human genomic DNA in 2.5 μl PBS was also amplified and 2.5 μl of PBS without any template as a negative control. Meanwhile, empty rafts isolated from the CellRaft array were amplified and compared to the controls and single cells to exclude the possibility of contamination by cell-free DNA.
High temperature single cell MDA (HighTemp MDA) was done using the REPLI-g Single Cell Kit (Qiagen). The key difference in the single cell lysis between this procedure and SCMDA is the temperature, i.e., 65°C for the HighTemp method and on ice for SCMDA. The low temperature used by the SCMDA method avoids heat-induced errors in DNA denaturation, mainly cytosine deamination (CG->TA) (Fig. 3b).
Unamplified DNA was extracted from clones using the Quick-gDNA Blood Miniprep Kit (ZYMO Research) and from bulk cell cultures using DNeasy Blood & Tissue Kit (Qiagen).
Library construction and whole genome sequencing
PCR-free libraries were prepared using the Accel-NGS 2S DNA Library Kit (Swift Biosciences). Briefly, 2 μg input DNA was fragmented using Covaris, size-selected and purified using the MinElute Gel Extraction Kit (Qiagen). PCR-free libraries of the DNA from bulk cell populations was prepared using the KAPA LTP Library Preparation Kit (KAPA Biosystems) with Trueseq adapters (Illumina) by the Epigenomics Core at the Albert Einstein College of Medicine. For 3 of the 4 single cell clones (C1, C2 and C3), libraries were prepared by the New York Genome Center (New York, NY). The libraries were sequenced on the Illumina HiSeq 2500, with 2×100 bp or 2×250 bp paired-end reads by the Epigenomics Core at Albert Einstein College of Medicine or the Illumina HiSeq X Ten with 2×150 bp paired-end reads by the New York Genome Center (New York, NY) (Supplementary Note).
Sequence alignment
Raw sequence reads were adapter and quality trimmed using Trim Galore (version 0.3.7), and aligned to human reference genome build 37 using BWA MEM (version 0.7.10)16. After deduplication, the initial mapped reads were indel realigned based on known indels from the 1000 Genomes Project (Phase I)17, and base quality score recalibrated based on known indels from the 1000 Genomes Project (Phase I) and SNVs from dbSNP (build 144) using GATK (version 3.4.46)8.
SCcaller: estimating allelic bias
We used the Nadaraya-Watson kernel-weighted average to estimate the fraction of reads carrying the genotype with more supporting reads (i.e. the major allele fraction, denoted as θ) as a result of allelic bias at genome position x0,
| (1) |
where i denotes heterozygote germline SNPs (hSNPs) from the single cell. We implemented a module in SCcaller to query these hSNPs from a single cell comparing to either a database, e.g. dbSNP, or, preferably, hSNPs called from the bulk cell population. Of note, this list of hSNPs does not have to be precise because bias estimation is a robust procedure and hSNPs are re-called and confirmed together with all the other SNVs in the variant calling step. λ denotes half of the window width for smoothing. We recommend λ=10,000 for MDA-based protocols, which balances prediction accuracy and coverage. The kernel K is defined using the Epanechnikov quadratic kernel,
| (2) |
in which function D is defined as,
| (3) |
in D(t), t refers to the argument of the function D.
To test the accuracy of the prediction, we performed leave-100-out cross-validations and 10-fold cross-validations on hSNPs from a randomly selected region chr1:100,000,000–110,000,000. In the leave-100-out cross-validations we randomly partitioned the hSNPs into subgroups, each with 100 hSNPs. For each subgroup, we predicted the bias of the 100 hSNPs using hSNPs from the other subgroups. In the 10-fold cross-validations, we randomly partitioned the hSNPs into 10 subgroups with equal numbers of hSNPs. For each subgroup, we predicted the bias of the hSNPs using hSNPs from the other 9 subgroups. The strong correlation between prediction and observation (Supplementary Fig. 6 and Supplementary Table 4) indicates high accuracy of the bias estimation.
SCcaller: variant calling
Let “ref” denote the reference allele and “alt” denote the alternative allele. To call the genotype for a single cell, we evaluated the data for a given candidate SNV using three models: (i) model H0: genotype ref/ref - no variant is present at the site and all alternative bases are a result of amplification errors; (ii) model H1: genotype ref/alt - a heterozygous SNV; (iii) model H2: genotype alt/alt - a homozygous SNV. We also took sequencing errors (Phred quality score) into account. Assuming that sequencing errors are independent across reads, we calculated the likelihood of each model using
| (4) |
where j denotes a read covering the site, b denotes the called bases, e denotes error rate transformed from the Phred quality score, r ∈ {A,C,G,T} denotes the reference genotype, and m ∈ {A,C,G,T} denotes the alternative genotype in the data. And Fk is defined as
| (5) |
For H1 and H2, let the estimated variant allele fraction f be,
| (6) |
Following Cibulskis et al10, we assumed each type of substitution error in sequencing has an equal probability of occurring, e/3:
| (7) |
In H0, to account for amplification errors, we set,
| (8) |
This is because with a diploid genome, there are a total of 4 DNA strands in a cell. Errors that occur at the first round of amplification, which are the artifacts most difficult to filter out, will ideally be found in 1/8 of the reads (Supplementary Fig. 1).
We further estimated a null distribution of artifacts using depth and quality from single cell sequencing data itself, and selected criteria η, corresponding to certain α values (0.01 and 0.05 are both reported). This procedure is based on a likelihood ratio test that rejects H0 in favor of H1 when,
| (9) |
where,
| (10) |
In addition, we also designed the SCcaller to calculate the likelihoods of sequencing errors following Cibulskis et al10.
SCcaller: application
SCcaller (v1.0) was used for calling SNVs as follows. The bam file of a single cell was mpileuped for each autosome separately using samtools (v1.3, options -C50 -r chr -Osf referencegenome.fa). To estimate allelic bias in the amplification, hSNPs in the single cell were identified by querying dbSNP (build 144) using option -a hsnp of SCcaller. Next, using option -a varcall in SCcaller, likelihoods of the three models described in the previous section were calculated for all positions in the genome covered by ≥1 reads that differ from the reference genome. Then, using option -a cutoff in SCcaller, η corresponding to α=0.01 was estimated. This cutoff was used to separate real SNVs from amplification artifacts. In addition, SNVs overlapping with indel reads were filtered out. Finally, we queried all SNVs identified in the whole genome sequence of the bulk cell population. SNVs without variant supporting reads, do not overlap with indel reads and have ≥20 wild type reads in the bulk, without overlap with dbSNP, are considered as somatic SNVs.
Potential caveats in using SCcaller
Of note, there are areas of the genome in which SCcaller cannot overcome amplification bias. Those regions include all areas that are haploid, such as the Y chromosome and the X chromosome in males, and other haploid regions, as well as amplified regions. Of note, the amplification products of MDA are generally long, i.e., typically over 10 kb, and not many artifacts will be missed. In the SCcaller, we implemented parameters for lengths of neighboring regions, which can also be specified by the user. With a default setting of 10 kb on both sides of the SNV, 90.5% of total candidate variants (approximately 90% of genomic regions) is covered. Bias of uncovered regions is by default set to the genome wide average (θ=0.75) and can also be specified by the user.
Statistical analysis
Statistic tests, such as permutations and t tests, were performed using R (version 3.3.2) and Microsoft Office Excel (2013) respectively. Statistics of SCcaller are described above and coded in Python (version 2.7).
Public single cell data
Single cell sequencing data from MDA-amplified (previous MDA method) neurons (Brain A)2 and MALBAC-amplified cancer cells7 were downloaded from SRA databases under accession numbers, SRR2141560 to SRR2141570 (previous MDA, 10 single neurons and 1 bulk), SRX204116, SRX202787, SRX202978 and SRX204744 (MALBAC, 3 kindred single cells and 1 bulk). Analysis and variant calling were as described for our SCMDA and HighTemp MDA datasets.
Code availability
SCcaller is freely available at https://github.com/biosinodx/SCcaller.
Supplementary Material
Acknowledgments
This research was supported by NIH (grants AG017242, AG047200, and AG038072 to J.V.) and the Glenn Foundation for Medical Research (J.V.).
Footnotes
Accession Codes
Primary accessions: Sequence Read Archive SRP067062.
Data Availability Statement
All sequencing data are available from the NCBI Sequence Read Archive under accession number SRP067062.
Author contributions
J.V., L.Z., and X.D. conceived this study and designed the experiments. L.Z., B.M. and M.L. performed the experiments. X.D. developed the software. X.D. and T.W. analyzed the data. J.V., X.D., L.Z., B.M. T.W. and A.M. wrote the manuscript.
Competing financial interests
X.D., L.Z., M.L., A.M. and J.V. are co-founders of SingulOmics Corp.
References
- 1.Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Molecular biology and evolution. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- 2.Lodato MA, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science (New York, NY) 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen JL, et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron. 2016;89:1223–1236. doi: 10.1016/j.neuron.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasken RS. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochemical Society transactions. 2009;37:450–453. doi: 10.1042/bst0370450. [DOI] [PubMed] [Google Scholar]
- 5.Gundry M, Li W, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic acids research. 2012;40:2032–2040. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, et al. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11923–11928. doi: 10.1073/pnas.1513988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science (New York, NY) 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zafar H, Wang Y, Nakhleh L, Navin N, Chen K. Monovar: single-nucleotide variant detection in single cells. Nature methods. 2016;13:505–507. doi: 10.1038/nmeth.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature reviews. Molecular cell biology. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 14.Gundry M, Vijg J. Direct mutation analysis by high-throughput sequencing: from germline to low-abundant, somatic variants. Mutation research. 2012;729:1–15. doi: 10.1016/mrfmmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falanga V, et al. Human dermal fibroblast clones derived from single cells are heterogeneous in the production of mRNAs for alpha 1(I) procollagen and transforming growth factor-beta 1. The Journal of investigative dermatology. 1995;105:27–31. doi: 10.1111/1523-1747.ep12312456. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.