Abstract
Chloroviruses are large double-stranded DNA (dsDNA) viruses that infect certain isolates of chlorella-like green algae. They contain up to approximately 400 protein-encoding genes and 16 transfer RNA (tRNA) genes. This review summarizes the unexpected finding that many of the chlorovirus genes encode proteins involved in manipulating carbohydrates. These include enzymes involved in making extracellular polysaccharides, such as hyaluronan and chitin, enzymes that make nucleotide sugars, such as GDP-l-fucose and GDP-d-rhamnose and enzymes involved in the synthesis of glycans attached to the virus major capsid proteins. This latter process differs from that of all other glycoprotein containing viruses that traditionally use the host endoplasmic reticulum and Golgi machinery to synthesize and transfer the glycans.
Keywords: chloroviruses, giant viruses, hyaluronan synthesis, chitin synthesis, nucleotide sugar synthesis, glycan synthesis, glycan structures, glycoproteins
1. Introduction
In discussing enzymes involved in manipulating carbohydrates, one usually does not consider viruses to play a role in this important subject. However, as described in this review, chloroviruses (family Phycodnaviridae) that infect certain isolates of single-celled, eukaryotic chlorella-like green algae are an exception to this process because they encode enzymes involved in making extracellular polysaccharides, nucleotide sugars and the synthesis of glycans attached to their major capsid glycoproteins.
The plaque-forming chloroviruses are large icosahedral (190 nm in diameter), double-stranded DNA (dsDNA)-containing viruses (genomes of 290 to 370 kb) with an internal lipid membrane. They exist in inland waters throughout the world with titers occasionally reaching thousands of plaque-forming units (PFU) per mL of indigenous water. Known chlorovirus hosts, which are normally endosymbionts and are often referred to as zoochlorellae [1,2], are associated either with the protozoan Paramecium bursaria, the coelenterate Hydra viridis or the heliozoan Acanthocystis turfacea [3,4,5,6]. Four such zoochlorellae and their viruses are Chlorella variabilis NC64A and its viruses (referred to as NC64A viruses), Chlorella variabilis Syngen 2–3 and its viruses (referred to as Osy viruses), Chlorella heliozoae SAG 3.83 and its viruses (referred to as SAG viruses) and Micractinium conductrix Pbi and its viruses (referred to as Pbi viruses). The zoochlorellae are resistant to virus infection when they are in their symbiotic relationship, because the viruses have no way of reaching their hosts.
The genomes of 43 chloroviruses infecting these four hosts have been sequenced, assembled and annotated [6,7,8,9,10,11]. Collectively, the viruses encode genes from 643 predicted protein families; however, any given chlorovirus only has 330 to 416 protein-encoding genes (PEGs). Thus, the genetic diversity among these viruses is large, and many of the proteins are unexpected for a virus. With the exception of homologs solely in other chlorovirus members, about 50% of their PEGs do not match anything in the databases.
The prototype chlorovirus Paramecium bursaria chlorella virus type 1 (PBCV-1) is an NC64A virus [12]. PBCV-1 is an icosahedron (190 nm in diameter) with a spike-like structure at one vertex and a few external fibers that extend from some of the viral capsomeres [5,13]. The outer capsid layer covers a single lipid bilayered membrane, which is essential for infection. The PBCV-1 major capsid protein (named Vp54) is a glycoprotein, and three Vp54s form a trimeric capsomere, which has pseudo-six-fold symmetry. A proteomic analysis of PBCV-1 virions revealed that the virus contains 148 virus-encoded proteins and at least one host-encoded protein [10]. The PBCV-1 genome is a linear ~331-kb, non-permuted dsDNA molecule with covalently-closed hairpin termini. Identical ~2.2-kb inverted repeats flank each 35-nucleotide-long, incompletely base-paired, covalently closed hairpin loop [14,15]. The remainder of the PBCV-1 genome contains primarily single-copy DNA that encodes ~416 putative proteins and 11 transfer RNAs (tRNAs) [5]. The G + C content of the PBCV-1 genome is 40%; in contrast, its host nuclear genome is 67% G + C. PBCV-1 and other chlorovirus genomes contain methylated bases, which occur in specific DNA sequences. The methylated bases are part of chlorovirus-encoded DNA restriction and modification systems [16].
As the title of this review indicates, many of the chlorovirus genes encode enzymes involved in various aspects of carbohydrate metabolism. We have listed putative chlorovirus genes involved in carbohydrate metabolism, which are encoded by the 43 chloroviruses whose genomes have been sequenced, in Table 1, Table 2 and Table 4. Recombinant proteins have been produced from some of these genes, and the proteins have been characterized (indicated in bold in the tables). When some of the genes were initially cloned and the recombinant proteins characterized, the genes were hybridized to many other chlorovirus genomes by dot blots to determine the distribution of the genes. Because of the large number of viruses, these experimental results are not included in the tables, unless the virus genome was subsequently sequenced.
Table 1.
Chlorovirus encoded enzymes involved in the synthesis of polysaccharides.
| Host | Viruses | HAS 1 | CHS 2 | CBP 3 |
|---|---|---|---|---|
| NC64A | AN69C | 390R | 395L, 438L | |
| AR158 | C418R | C423L, C475L | ||
| CviK1 | 102R | 365R | 370L, 414L | |
| CvsA1 | 375R | 380L, 427L | ||
| IL-3A | 386L, 432L | |||
| IL-5-2s1 | 134L | 503L, 562L | ||
| KS-1B | 314L, 360L | |||
| MA-1D | 485L | 362R | 367L, 472L | |
| MA-1E | 113L | 407L, 451L | ||
| NE-JV-4 | 390L | |||
| NY-2B | 542L, 484L | |||
| NY2A | B139R, B472R | B480L | ||
| NYs-1 | 137R | 360R, 495L, 555L | ||
| PBCV-1 | a A098R | A333L, A348Dl, A383R | ||
| SYN | OSY-NE-5 | 038R | 167L, 184L | |
| Pbi | AP110A | 152L, 175R | 828R | |
| CVA-1 | 150L, 169R | 834R | ||
| CVB-1 | 177L | 791R | ||
| CVG-1 | 146R | 792R | ||
| CVM-1 | 165L, 186R | 832R | ||
| CVR-1 | 838R | |||
| CZ-2 | 798R | |||
| Can18-4 | 163R | 839R | ||
| FR483 | N124R | N690R | ||
| FR5L | 151L | 797R | ||
| MT325 | M128R | M701R | ||
| NE-JV-1 | 278R, 282R | 734L | ||
| NW665 | 133R | 821R | ||
| OR0704.2.2 | 116R | 804R | ||
| SAG | ATCV-1 | Z734R | ||
| Br0604L | 834R | 431R | ||
| Can0610SP | 438R, 442R | |||
| Canal-1 | 746R | 405R | ||
| GM0701 | 852R | 436R | ||
| MN0810 | 087R, 900R | 466R, 468R, 531L | ||
| MO0605SPH | 435R | |||
| NE-JV-2 | 462R | |||
| NE-jv-3 | 431R | |||
| NTS-1 | 893R | 461R, 463R, 529L | ||
| OR0704.3 | 431R | |||
| TN603 | 869R | 425R | ||
| W10606 | 457R |
1 Hyaluronan synthase; 2 chitin synthase; 3 chitin binding proteins, except for the chitinase proteins reported in Table 5; a the recombinant protein has the predicted activity. The numbers refer to the protein names, and the R and L refer to the strand orientation.
Table 2.
Chlorovirus encoded enzymes involved in sugar metabolism.
| Host | Viruses | GFAT 1 | UDP-GlcDH 2 | GMD 3 | GMER 4 | UGD 5 | AT 6 | D-LD 7 | ADP-RGH 8 | FRD 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 109R | 384R, 487R | 129R | 334L | 739L | 055R | |||
| AR158 | C132R | C413R, C729L | C155R | C344L | C767L | |||||
| CviK1 | 105R | 359R, 662L | 122R | 312L | 742L | 055R | ||||
| CvsA1 | 064R | 368R | 083R | 321L | 716L | 131R | ||||
| IL-3A | 104R | 375R, 685L | 126R | 326L | 726L | 051R | ||||
| IL-5-2s1 | 130R | 492R, 858L | 106L | 417L | 896L | |||||
| KS-1B | 066R | 261L | 643L | |||||||
| MA-1D | 481L | 355R, 838L | 456L | 284L | 872L | |||||
| MA-1E | 116R | 396R | 134R | 355L | 806L | 045R | ||||
| NE-JV-4 | 131R | 340L | 740L | 064R | ||||||
| NY-2B | 473R, 836L | 185R | 408L | 881L | ||||||
| NY2A | B143R | B465R | B163R | B395L | B853L | |||||
| NYs-1 | 143R | 483R, 846L | 167R | 404L | 879L | |||||
| PBCV-1 | a A100R | a A609L | a A118R | a A295L | A654L | A053R | ||||
| SYN | OSY-NE-5 | 039R | 045R | 139L | 340L | 015R | 308R | |||
| Pbi | AP110A | 071R | 146L | 893R | 053L | |||||
| CVA-1 | 056R | 144L | 900R | 040L | 205R | |||||
| CVB-1 | 071R | 172L | 856R | 056L | 221R | |||||
| CVG-1 | 050R | 812L | 857R | |||||||
| CVM-1 | 069R | 159L | 893R | 052L | 222R | |||||
| CVR-1 | 062R | 151L | 906R | 046L | 210R | |||||
| CZ-2 | 059R | 718L | 865R | 048L | 917L | |||||
| Can18-4 | 061R | 859L | 908R | 048L | 212R | |||||
| FR483 | N035R | N712L | N747R | N170R | ||||||
| FR5L | 087R | 145L | 863R | 076L | ||||||
| MT325 | M036R, M037R | M719L | M758R | M026L | ||||||
| NE-JV-1 | 081R | 291R | 861R | 810L | ||||||
| NW665 | 046R | 846L | 889R | 189R | ||||||
| OR0704.2.2 | 062R | 722L | 862R | 051L | ||||||
| SAG | ATCV-1 | Z571L | a Z804L | Z282L | Z544R | Z147L | Z295L | |||
| Br0604L | 667L, 839R | 934L | 332L | 631R | 173L | 350L | ||||
| Can0610SP | 687L | 965L | 338L | 658R | 170L | 355L | ||||
| Canal-1 | 605L, 751R | 847L | 329L | 576R | 188L | 343L | 898L | 886R | ||
| GM0701 | 664L, 856R | 954L | 337L | 629R | 180L | 354L | ||||
| MN0810 | 887L | 720L, 904R | 992L | 365L | 689R | 204L | 379L | |||
| MO0605SPH | 656L | 897L | 341L | 625R | 181L | 359L | 943R | |||
| NE-JV-2 | 708L | 981L | 367L | 672R | 186L | 383L | ||||
| NE-JV-3 | 679L | 935L | 332L | 648R | 175L | 347L | 981R | |||
| NTS-1 | 714L, 898R | 1012L | 378L | 681R | 188L | 391L | ||||
| OR0704.3 | 676L | 960L | 335L | 639R | 179L | 354L | ||||
| TN603 | 659L, 873R | 966L | 326L | 625R | 179L | 342L | ||||
| W10606 | 679L | 916L | 360L | 651R | 185L | 375L | 962R |
1 Glutamine-fructose-6-phosphate aminotransferase; 2 UDP-glucose-6-dehydrogenase; 3 GDP-d-mannose dehydratase; 4 GDP-4-keto-6-deoxy-d-mannose epimerase/reductase (=GDP-l-fucose synthase 2); 5 UDP-d-glucose 4,6-dehydratase, 6 acetyltransferase; 7 d-lactate dehydrogenase; 8 ADP-ribosylglycohydrolase; 9 fumarate reductase; a the recombinant proteins have the predicted activities. The numbers refer to the protein names, and the R and L refer to the strand orientation.
2. Chlorovirus Encoded Polysaccharide Synthesizing Enzymes
Three PBCV-1 encoded enzymes are involved in the synthesis of the extracellular matrix polysaccharide hyaluronan (also referred to as hyaluronic acid), including glycosyltransferase Class I hyaluronan synthase (HAS; Table 1) [17,18]. Until the has gene (a098r) was discovered in PBCV-1, hyaluronan was only thought to occur in vertebrates and a few pathogenic bacteria, where it forms an extracellular capsule, presumably to avoid the immune system [19,20]. Hyaluronan is an essential constituent of the extracellular matrix in vertebrates and consists of ~10,000 or more alternating β-1,4-glucuronic acid (GlcA) and β-1,3-N-acetylglucosamine (GlcNAc) residues. Typically, the HAS enzyme is located on the inner surface of the plasma membrane. The newly-synthesized hyaluronan then moves through the membrane and cell wall to the extracellular matrix.
PBCV-1 also encodes two enzymes involved in the biosynthesis of hyaluronan precursors, glutamine:fructose-6-phosphate amidotransferase (GFAT, gene a100r) and UDP-glucose dehydrogenase (UDP-GlcDH, gene a609l; Table 2) [21]. All three PBCV-1 genes involved in hyaluronan synthesis are expressed early during virus infection, and all three transcripts decrease significantly by 60 min post-infection (PI) [18,21]. However, these three genes do not function like an operon, although two of the genes, a98r and a100r, are adjacent to one another and are co-linear in the PBCV-1 genome. In contrast, a609l is located ~240 kb away and is transcribed in the opposite orientation [17]. The identification of these three genes led to the discovery that hyaluronan lyase-sensitive hair-like fibers begin to accumulate on the surface of PBCV-1-infected host cells by 15 min PI. By 4 h PI, the infected cells are covered with a dense fibrous hyaluronan network (Figure 1) [18].
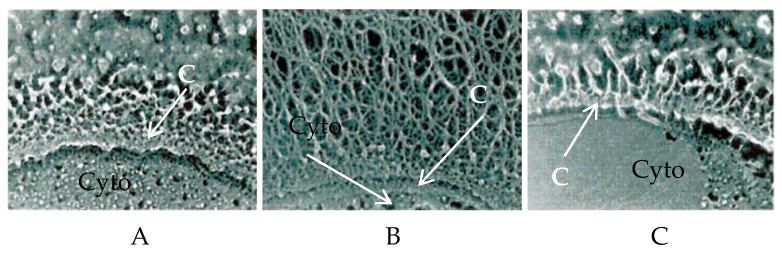
Figure 1.
Location of hyaluronan on the surface of infected Chlorella variabilis NC64A cells and ultrastructural changes in the algal cell wall after chlorovirus Paramecium bursaria chlorella virus type 1 (PBCV-1) infection. The figure shows the cross-sections of (A) the surface of the uninfected cells; (B) cells at 4 h post-infection (PI); and (C) cells at 4 h PI that were treated with hyaluronan lyase. Note that after treatment with hyaluronan lyase, the cell surface resembles the surface of uninfected cells. C is the cell wall, and Cyto is cytoplasm. Micrographs were taken from Graves et al. [18] with permission.
Three additional enzymes are needed to convert glucosamine-6-phosphate (GlcN-6P) to UDP-N-acetylglucosamine (UDP-GlcNAc), and these enzymes (EC2.3.1.4, EC5.4.2.3, EC2.7.7.23) are encoded by the host [22]. This is not surprising because the host NC64A cell wall is predicted to contain chitin, which is a polymer of GlcNAc residues, and so, the alga must encode these enzymes.
The has gene that encodes hyaluronan synthase is present in 12 of the 43 chloroviruses isolated from diverse geographical regions, including 5 NC64A viruses, 6 Pbi viruses and 1 Osy virus (Table 1). In contrast, the udp-glcdh gene is present in 40 of the 43 viruses, 14 of which have two copies of the gene, while the gfat gene is present in 27 chloroviruses, including 11 of the 14 NC64A viruses, all 14 Pbi viruses, one of the 13 SAG viruses and the only Osy virus that has been sequenced (Table 2). Both of these latter two genes are present in all of the 12 viruses that have a has gene, except for the one Osy virus that lacks a udp-glcdh gene.
Surprisingly, 19 of the 31 chloroviruses that lack a has gene have a gene encoding a chitin synthase (CHS). Chitin, an insoluble linear homopolymer of β-1,4-linked-GlcNAc residues, is a common component of insect exoskeletons, shells of crustaceans and fungal cell walls [23]. Chitin is rare in algal cell walls, although it has been reported to exist in some green algae [24]. Like the has gene, the chs gene is expressed as early as 10 min PI and peaks at 20–40 min PI, and the transcript disappears at 120–180 min PI. Furthermore, cells infected with chs-containing viruses produced chitin fibers on the external surface of their hosts [25]. As discussed below, many of the chloroviruses also encode chitinases and chitosanases.
At least one chlorovirus, CVK2, has replaced the PBCV-1 has gene with a 5-kb region containing chs, udp-gdh2 (a gene encoding a second UDP-GlcDH) and two other ORFs [26]. Therefore, at least some chloroviruses have changed from HAS viruses to CHS viruses or vice versa, by swapping genes.
Two NC64A chloroviruses have both has and chs genes, and at least one of them forms both hyaluronan and chitin on the surface of their infected cells [25,27]. Finally, 12 chloroviruses lack both genes, and no extracellular polysaccharides are formed on the surface of cells infected with at least one of these viruses [18]. The fact that many chloroviruses encode enzymes involved in extracellular polysaccharide biosynthesis suggests that the polysaccharides, which require a large expenditure of ATP for their synthesis, are important in the virus life cycles. However, the extracellular hyaluronan does not play an obvious role in the interaction between PBCV-1 and its algal host because neither plaque size nor plaque numbers were altered by including either hyaluronidase or free hyaluronan in the top agar of the PBCV-1 plaque assay [17].
The three genes involved in synthesizing hyaluronan have probably been obtained rather recently in evolutionary terms because the coding portions of the PBCV-1 gfat and udp-glcnc genes are 44% G + C, while the has gene is 46.7% G + C. In contrast, PBCV-1, as well as all the NC64A viruses, have a 40% G + C content [11,21].
Currently, it is not known how or why the chloroviruses acquired these polysaccharide-synthesizing genes. We have considered the following possible evolutionary advantages for acquiring these genes: (1) the polysaccharides prevent infection by a second chlorovirus; (2) they cause the infected cells to clump with uninfected host cells, thus increasing the probability that progeny viruses can infect healthy host cells; (3) they prevent paramecia from taking up infected algal cells, (4) the chloroviruses have another host in nature, and this other host is attracted to or binds to hyaluronan or chitin on virus-infected algae, which would facilitate progeny-virus infections; or (5) polysaccharides increase the functional diameter of the infected cell, which might facilitate consumption by a predator. This could aid virus movement in the water column. In regards to the first possibility, it is known that attachment of other viruses to PBCV-1-infected cells at 4 h PI is inhibited when the external surface of the host is covered with hyaluronan fibers [18]. However, this is unlikely to be the explanation for the presence of hyaluronan because normally the host, C. variabilis NC64A, is only infected by one virus, and this restriction occurs in the first few min PI [28,29]. In regards to the second possibility, host cells often clump shortly after infection, and this phenomenon, which does not always occur, could be due to hyaluronan production. The last three possibilities have not been explored experimentally.
We have experimentally tried to address the question: does the presence of hyaluronan and/or chitin on the exterior surface of the host cell wall confer an evolutionary advantage to a virus that has one or both of these genes? To answer the question, chlorella cells were co-infected with combinations of chloroviruses that: (1) have both genes; (2) only have the has gene; (3) only have the chs gene; and (4) lack both genes. The resulting lysates were then added to fresh cells and allowed to replicate and lyse. After five passages, progeny viruses were plaqued, and 20 plaques were randomly picked to determine if one virus type dominated. However, after repeating these experiments several times, no consistent pattern was obtained [30].
To ideally conduct this experiment, one would like to either add the chs gene to the PBCV-1 genome so that both genes are present, replace the has gene with the chs gene or remove the has gene so that PBCV-1 lacked both genes. Unfortunately, this experimental protocol is currently not possible because procedures are not available for reverse genetic manipulation of chlorovirus genomes. Therefore, in the experiments described above, viruses were selected that had the desired properties and also had similar growth kinetics as PBCV-1.
In addition to not knowing why the chloroviruses acquired the has and chs genes, another question is: how are the newly-forming hyaluronan and/or chitin fibers moved through the membrane and the complex cell wall to the exterior of the algal host from the plasma membrane? This phenomenon would appear to be equivalent to pushing a thread through a furnace filter. One would expect the polysaccharide fibers to bunch up underneath the cell wall. In fact, this happened when the viral has gene was expressed in cultured tobacco cells [31]. Could a pilot protein(s) that is attached to the leading end of the polymer guide the hyaluronan chain through the wall?
3. Chlorovirus Encoded Nucleotide Sugar Metabolism Enzymes
Many chloroviruses also encode enzymes involved in nucleotide sugar metabolism, as well as other sugar metabolic enzymes (Table 2). Two enzymes encoded by all of the NC64A, SAG and Syn chloroviruses, GDP-D-mannose 4,6 dehydratase (GMD) and GDP-4-keto-6-deoxy-d-mannose epimerase reductase (GMER) (Table 2), comprise a highly-conserved pathway in bacteria, plants and animals that converts GDP-d-mannose to GDP-l-fucose (Figure 2) [32]. Fucose is found in glycoconjugates of many organisms, where it often plays a fundamental role in cell-cell adhesion and recognition [33]. The Pbi chloroviruses lack both gmd and gmer genes (Table 2) even though the glycans attached to the major capsid protein from the three evaluated Pbi viruses have fucose [34,35].
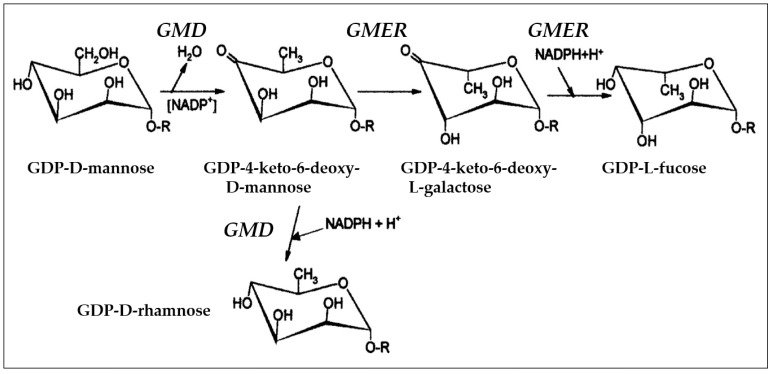
Figure 2.
Scheme of the biosynthesis of GDP-l-fucose and GDP-d-rhamnose. PBCV-1 GDP-d-mannose 4,6 dehydratase (GMD) catalyzes both the dehydration of GDP-d-mannose to the intermediate GDP-4-keto-6-deoxy-d-mannose and the NADPH-dependent reduction of this latter compound to GDP-d-rhamnose. NADP+ serves as the cofactor for GMD during the internal oxidoreduction reaction involved in the dehydration process. The epimerization and the NADPH-dependent reduction of the 4-keto group leading to GDP-l-fucose are carried out by PBCV-1 GDP-4-keto-6-deoxy-D-mannose epimerase reductase (GMER). Figure was taken from Tonetti et al. [32] with permission.
In vitro reconstruction of the pathway using recombinant PBCV-1 GMD and GMER proteins resulted in the synthesis of GDP-l-fucose as expected. Unexpectedly, however, the PBCV-1 GMD also catalyzed the NADPH-dependent reduction of the intermediate GDP-4-keto-6-deoxy-d-mannose, to form GDP-d-rhamnose. That is, the enzyme has two activities, and both sugars are produced in the infected cell [32]. The PBCV-1 recombinant GMD has another property that is unusual. Unlike recombinant GMDs from many other organisms, the viral encoded enzyme is very stable when stored at either 4 °C or −20 °C [32]. The PBCV-1 GMD enzyme was crystalized, and the structure resembles other GMDs [36].
A recombinant GMD protein encoded by another chlorovirus, Acanthocystis turfacea chlorella virus 1 (ATCV-1), which has 53% amino acid identity with the PBCV-1 GMD, was also characterized because the amino acid differences between the two enzymes suggested they might have slightly different properties. In fact, the ATCV-1 GMD does not form GDP-d-rhamnose, and so, it lacks the second enzyme activity [37]. Both GMD enzymes bound NADPH tightly, and this association was essential for the stabilization and function of both enzymes, even though NADP+ is the co-enzyme required to initiate the GMD catalytic cycle. Phylogenetic analyses established that the PBCV-1 GMD is the most evolutionarily diverged of all the GMDs, whereas the ATCV-1 GMD was in a clade of bacterial GMDs [37].
The GMER enzymes from PBCV-1 and ATCV-1 have 63% amino acid identity to each other and phylogenetically are more similar to one another and to other GMERs than are the two GMDs. The possible evolutionary consequences of these differences have been discussed previously [37]. Both fucose and rhamnose are constituents of the glycans attached to the PBCV-1 and ATCV-1 major capsid proteins (see below). However, the PBCV-1 glycan contains three rhamnose residues, with one in the D-configuration, whereas only one with L-configuration is present in the ATCV-1 glycan. Perhaps there was enough natural selection pressure on the PBCV-1 GMD gene to evolve to synthesize GDP-d-rhamnose, whereas the ATCV-1 GMD did not face this pressure.
ATCV-1 and all of the SAG viruses, however, encode another enzyme, UDP-d-glucose 4,6-dehydratase (UGD), that is one of two enzymes involved in the synthesis of L-rhamnose [38], and this enzyme may contribute to rhamnose synthesis. The PBCV-1 host chlorella, which is closely related to the ATCV-1 host chlorella, encodes the second enzyme in the rhamnose pathway [22], and so the host is predicted to be able to synthesize the rhamnose required for ATCV-1 glycan synthesis.
4. Unusual Attachment of Glycans to the Chlorovirus Major Capsid Proteins
Structural proteins of many viruses, such as rhabdoviruses, herpesviruses, poxviruses and paramyxoviruses, are glycosylated. Glycans contribute to the protease resistance and the antigenicity of these viruses. Most virus glycans are linked to Asn in the protein via N-acetylglucosamine, although some viruses also have O-linked glycans attached to either Ser or Thr residues via an amino sugar, usually N-acetylglucosamine or N-acetylgalactosamine. Typically, viruses use host-encoded glycosyltransferases and glycosidases located in the endoplasmic reticulum (ER) and Golgi apparatus to add and remove N-linked sugar residues from virus glycoproteins either co-translationally or shortly after translation of the protein. This post-translational processing aids in protein folding, progression in the secretory pathway and in the regulation of host-virus interactions [39,40,41,42]. After folding the protein, virus glycoproteins are transported by host-sorting and membrane-transport functions to virus-specified regions in host membranes where they displace host glycoproteins. Progeny viruses then bud through these virus-specific target membranes, which is usually the final step in the assembly of infectious viruses. Thus, nascent viruses only become infectious after budding through the membrane, usually the plasma membrane, as they exit the cell. Consequently, the glycan portion of virus glycoproteins is host-specific. The theme that emerges from these viruses is that virus glycoproteins are synthesized and glycosylated by the same processes as host glycoproteins. Therefore, the only way to alter virus protein glycosylation is to either grow the virus in a different host or have a mutation that alters the virus protein glycosylation site.
Unlike the process described above, glycosylation of the chlorovirus major capsid proteins differs from that scenario because the viruses encode most, if not all, of the machinery for the process. In addition, the process occurs in the cytoplasm. The conclusion that the chlorovirus PBCV-1 major capsid protein (Vp54, gene a430l) is glycosylated by a different mechanism than that used by other characterized viruses originally arose from antibody studies [43]. Rabbit polyclonal antiserum prepared against intact PBCV-1 particles inhibited virus plaque formation by agglutinating the virions. However, spontaneously-derived, antiserum-resistant, plaque-forming variants of PBCV-1 occurred at a frequency of 10−5–10−6. At the time of the 1993 publication, these antiserum-resistant variants fell into four serologically-distinct classes; two additional antigenic variants have subsequently been isolated for a total of six variants (Table 3). Polyclonal antisera prepared against members of each of these antigenic classes react predominately with the Vp54 equivalents from the viruses in the class used for the immunization. Each of the Vp54 proteins from the antigenic variants migrated faster on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) than those of the strains from which they were derived, indicating a lower molecular weight. However, all of the de-glycosylated Vp54 proteins migrated at the same rate on SDS-PAGE, indicating that the differences resided in the size of the attached glycans. In addition, the nucleotide sequence of the a430l gene in each of the variants was identical to the wild-type gene, which verified that the polypeptide portion of Vp54 was not altered in the mutants. Western blot analyses of Vp54 proteins isolated from the variants, before and after removing the glycans with trifluoromethane-sulfonic acid or altering the glycan with periodic acid, also supported the notion that the antigenic variants reflected differences in the Vp54 glycans, not the Vp54 polypeptide [43].
Table 3.
PBCV-1 antigenic variants that affect the molecular weight of the major capsid glycoprotein.
| Antisera Classes | Predicted MW (kDa) c | SDS-PAGE Estimates (kDa) d | |
|---|---|---|---|
| Class a | Label b | ||
| + | Wild-type | 54.1 | 54 |
| F | CME6 | 54 | not determined |
| A | P91 | 52.8 | 53 |
| E | EPA-15 | 52.8 | not determined |
| B | EPA-2 | 51.6 | 52 |
| C | E1L3 | 51.1 | 51 |
| D | P1L6 | 50.5 | 50.5 |
a Listed in order of predicted molecular weight based on nuclear magnetic resonance (NMR) analysis (De Castro et al., [44,45]); b representative mutant strain label; c the gene encoding the PBCV-1 major capsid protein (a430l) is wild-type in sequence and does not vary among antisera classes; d Graves et al. [46]. MW, molecular weight.
All of the glycan antigenic variants form plaques on their C. variabilis NC64A host, so one can infer that the glycans are not directly involved in virus infection and virus replication. However, anecdotal evidence suggests that the glycans are important in virus stability because the variants with the smallest glycans do not remain infectious in storage as long as wild-type virus.
Additional observations supported the concept that PBCV-1 Vp54 glycosylation was unusual: (1) unlike viruses that acquire their glycoproteins(s) by budding through a plasma membrane, which results in infectious particles, plaque-forming PBCV-1 particles accumulate inside the host 30–40 min before virus release [47]; (2) all of the antigenic variants were grown in the same host so the glycan differences are not due to the host; (3) polyclonal antibodies to Vp54, the major capsid protein, do not react with host glycoproteins; (4) the Vp54 protein lacks an ER and Golgi signal peptide; (5) unlike most glycoproteins that exhibit size micro-heterogeneity, PBCV-1 Vp54 appears homogeneous on SDS-PAGE; in addition, mass spectrometry analysis only revealed one satellite peak that differed from the main peak by 140 Da, the approximate weight of either one arabinose or xylose residue [46]; and (6) the ability to easily crystallize Vp54 as a homotrimer provided additional evidence that the protein is essentially homogeneous [48,49].
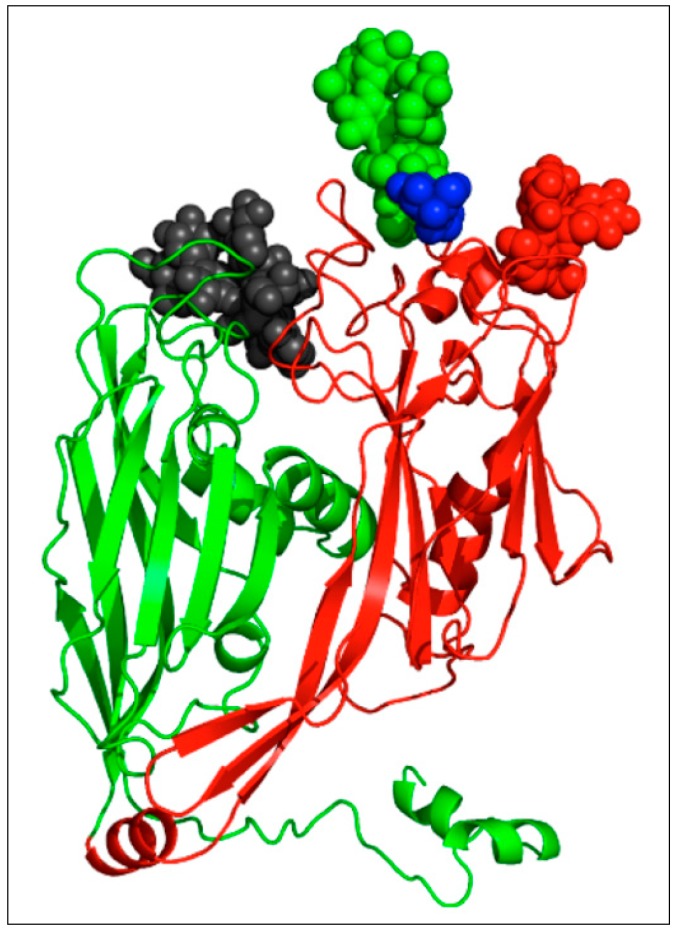
Evidence that the N-linked Vp54 glycans are not attached to the Vp54 protein by a traditional N-linkage was initially obtained from the X-ray crystal structure of the protein. The structure revealed that the protein had four N-linked glycans at Asn positions 280, 302, 399 and 406 [48]. None of these Asn were located in an Asn-X-(Thr/Ser) sequon sequence commonly recognized by ER located glycosyltransferases [50,51,52]. This finding also explained why prior attempts to remove Vp54 glycans with enzymes that cleave traditional N-linked glycans were unsuccessful [53] Nandhagopal et al. [48] also reported that Vp54 contained two O-linked glycans. However, re-examination of the X-ray crystal data (Figure 3) indicates that no O-linked glycans are present in the protein [49], which agrees with our unsuccessful attempts to detect them by chemical procedures.
Figure 3.
Structure of the revised PBCV-1 Vp54 monomer. The two jelly-roll domains are colored in green and red, respectively. The glycans located on the surface are shown as a space-filling representation of their atoms and are colored according to the residue they are attached to (Asn-280: green, Asn-302: black, Asn-399: red, Asn-406: blue). Taken from De Castro et al. [49] with permission.
5. Glycan Structures Attached to Chlorovirus Major Capsid Proteins
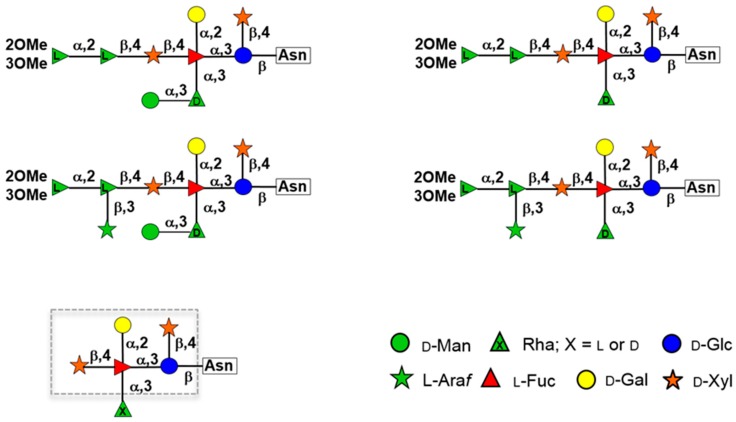
The structures of the PBCV-1 Vp54 N-linked glycans were reported recently, and they consist of 8–10 neutral monosaccharide residues, producing a total of four glycoforms (Figure 4) [54]. These structures do not resemble any structure previously reported in the three Domains of Life. Among their most distinctive features are: (1) the four glycoforms share a common core structure, and the four glycoforms are related to the non-stoichiometric presence of two monosaccharides, L-arabinose and D-mannose; the most abundant glycoform consists of nine neutral monosaccharide residues organized in a highly-branched fashion; (2) the glycans are attached to the protein by a β-glucose linkage, which is rare in nature and has only been reported in glycoproteins from a few organisms [55,56,57,58]; and (3) the glycoform contains a dimethylated rhamnose as the capping residue of the main chain, a hyper-branched fucose residue and two rhamnose residues with opposite absolute configurations.
Figure 4.
Structures of PBCV-1 Vp54 N-glycans. Arabinose and mannose are not stoichiometric substituents and create four different glycoforms. The two on the left are the most abundant, and both have mannose. The structure at the bottom represents the conserved core oligosaccharide that is present in all of the chloroviruses studied to date. Residues within the box are those strictly conserved, while rhamnose (outside the box) is a semi-conserved element because its absolute configuration is virus dependent. The figure was modified from De Castro et al. [34,54] with permission.
Attempts to fit the Vp54 glycan structures into the original Vp54 X-ray crystal structure [48] were unsuccessful and led to a re-examination of the original structure. This re-examination produced a structure that was compatible with the four N-linked glycan structures (Figure 3) [49]. As mentioned above, the revised structure lacks the two O-linked glycans reported originally.
The PBCV-1 Vp54 has a molecular weight of 53,790 Da. The a430l gene encodes Vp54 with a predicted molecular weight of 48,165 Da so the combined sugars have a molecular weight of 5625 Da, which is about the weight of the four glycans. Vp54 was also reported to have a myristic acid attached to the carboxyl portion of the protein [59]. However, myristic acid has not been observed in any of the recent Vp54 structural experiments, and so, its status is currently unknown. The structures of the Vp54 glycans from the PBCV-1 antigenic variants, referred to above, are currently being determined, and as expected, the structures are truncated forms of the wild-type PBCV-1 glycans [44]. PBCV-1 particles were reported to have two additional glycoproteins in addition to Vp54 [59]. Both of these glycoproteins react with the PBCV-1 antibody, and so, the glycan structures are predicted to be similar or identical to the glycans associated with Vp54. The gene encoding one of these proteins (Vp260) was identified (gene a122r). Gene a122r homologs are common in the chloroviruses, and some of the viruses have as many as five copies of the gene [60]. The role that Vp260 plays in the PBCV-1 virion is unknown.
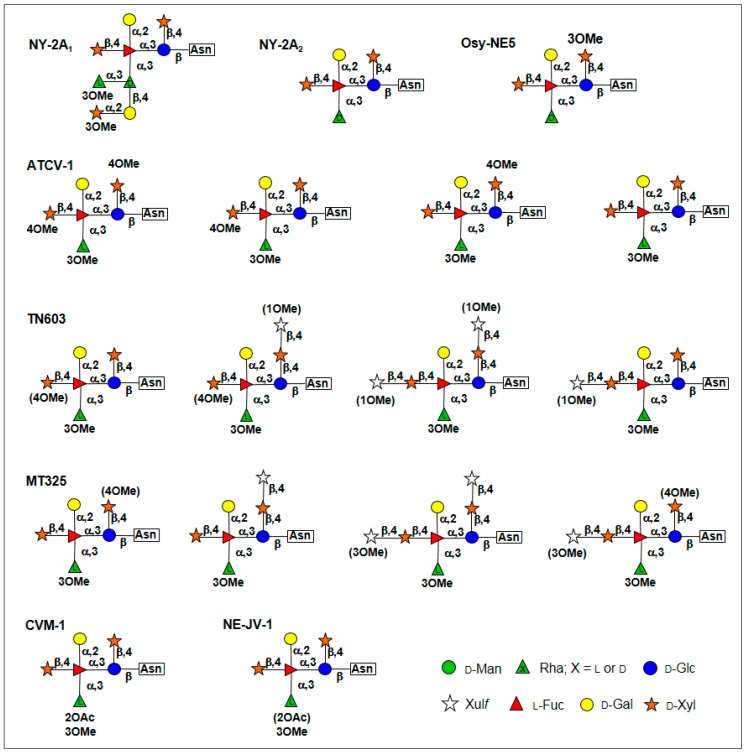
The glycan structures of the major capsid proteins from seven more chloroviruses, which represent all four chlorovirus types, were recently reported (Figure 5) [6,34,35]; collectively, all of the glycans have a common core region (outlined in Figure 4). The common core region consists of a pentasaccharide with a β-glucose linked to an Asn residue, which is not located in the typical sequon Asn-X-(Thr/Ser). The glucose has a terminal xylose unit and a hyperbranched fucose, which is in turn substituted with a terminal galactose and a second xylose residue. The third position of the fucose unit is always linked to a rhamnose, which is a semi-conserved element because its configuration is virus dependent. Additional decorations occur on this core N-glycan and represent a molecular signature for each chlorovirus.
Figure 5.
Structures of N-glycans from seven chloroviruses representing all four chlorovirus types. Substituents in brackets are not stoichiometric. All sugars are in the pyranose form, except where specified. Virus NY-2A is an NC64A virus, virus Osy-NE5 an Osy virus, viruses ATCV-1 and TN603 SAG viruses and MT325, CVM-1 and NE-JV-1 Pbi viruses. This figure was modified from [6,34,35] with permission.
6. Chlorovirus PBCV-1 Encoded Glycosyltransferases
In addition to the two glycosyltransferases, hyaluronan synthase and chitin synthase previously described, the 43 chloroviruses collectively encode eight putative glycosyltransferases (Table 4). Six of these eight glycosyltransferase-encoding genes are in PBCV-1; they are scattered throughout the PBCV-1 genome. None of these six PBCV-1 encoded glycosyltransferases have an identifiable signal peptide that would target them to the ER. Furthermore, with the exception of PBCV-1 glycosyltransferases A473L (six transmembrane domains; CESA CelA-like) and A219/222/226R (nine transmembrane domains; CXCX-2), none of the four remaining PBCV-1 encoded glycosyltransferases are predicted to have transmembrane domains. Therefore, these enzymes are expected to be soluble proteins. The genes for the six PBCV-1 encoded glycosyltransferases are expressed early during PBCV-1 infection [61]. Thus, assuming the enzymes are stable, they would be available for adding sugars to the Vp54 glycans during virus replication.
Table 4.
Chlorovirus encoded enzymes involved in synthesizing glycans attached to virus major capsid proteins.
| Host | Viruses | EXT 1 | GT-A 2 | GT-GT4 3 | GT-GTA 4 | CESA CelA-Like 5 | CSCS-2 6 | GT 7 | GT17 8 |
|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 078L | 123R | 559R | 065R | 104R | 255R | ||
| AR158 | C093L | C150R | C661L | C265R | C559R | ||||
| CviK1 | 080L | 117R | 594L | 518L | 242R | ||||
| CvsA1 | 039L | 077R | 611L | 535L | 247R | ||||
| IL-3A | 071L | 120R | 606L | 060R | 099R | 240R | |||
| IL-5-2s1 | 175R | 109L | 773L | 313R | 649R | ||||
| KS-1B | 024L | 060R | 528L | 009R | 046L | 170R | |||
| MA-1D | 531R | 4549L | 753L | 194R | 637R | ||||
| MA-1E | 533R | 128R | 702L | 626L | 281R, 210R | ||||
| NE-JV-4 | 085L | 124R | 631L | 074R | 108L | 250R | |||
| NY-2B | 116L | 180R | 754L | 160R | 323R | 633R | |||
| NY2A | 107L | B159R | B736L | B618R | |||||
| NYs-1 | 098L | 162R | 760L | 187R | 641R | ||||
| PBCV-1 | A075L | A111/114R | A546L | A064R | A473L | A219/222/226R | |||
| SYN | OSY-NE-5 | 025L | 044R | 283L | 097L | ||||
| Pbi | AP110A | 013L | 548R | 226R | 970R | ||||
| CVA-1 | 016L | 532R | 220R | 971R | |||||
| CVB-1 | 025L | 538R | 232R | 811L | 918R | ||||
| CVG-1 | 019L | 520R | 217R | 815L | 920R | ||||
| CVM-1 | 022L | 550R | 237R | 953R | |||||
| CVR-1 | 020L | 545R | 225R | 977R | |||||
| CZ-2 | 012L | 532R | 380L | 822L | 932R | ||||
| Can18-4 | 020L | 557R | 229R | 862L | 971R | ||||
| FR483 | N012L | N472R | N191R | N715L | N805R | ||||
| FR5L | 046L | 537R | 819L | 926R | |||||
| MT325 | M009L | M467R | M186R | M721L | M813R | ||||
| NE-JV-1 | 079L | 464L | 801R | 930R | |||||
| NW665 | 015L | 532R | 849L | 955R | |||||
| OR0704.2.2 | 017L | 549R | 382L | 823L | 923R | ||||
| SAG | ATCV-1 | Z830R | Z120R | Z667L | Z178L, Z823R, Z417L | Z425R | Z347R | ||
| Br0604L | 959R | 137R | 225L, 952R, 483L | 489R | 399R | ||||
| Can0610SP | 1007R | 140R | 789L | 210L, 1002R, 978L, 487L | 495R | 407R | |||
| Canal-1 | 874R | 141R | 164L | 871R | 447R | 380R | |||
| GM0701 | 977R | 141R | 752L | 228L, 975R, 486L | 493R | 405R | |||
| MN0810 | 1009R | 165R | 244L | 424R | |||||
| MO0605SPH | 932R | 138R | 164L | 230L, 926R, 479L | 488R | 412R | |||
| NE-JV-2 | 1020R | 150R | 804L | 1015R, 992L, 516L | 523R | 438R | |||
| NE-JV-3 | 970R | 145R | 778L | 215L, 210L, 964R, 484L | 493R | 407R | |||
| NTS-1 | 1044R | 156R | 1016L, 516L | 441R | |||||
| OR0704.3 | 1006R | 146R | 944L | 1001R, 972L, 485L, 219L | 493R | 404R | |||
| TN603 | 991R | 141R | 226L, 986R, 475L | 483R | 400R | ||||
| WI0606 | 951R | 141R | 233L, 945R, 506L | 514R | 430R |
1 Exotosin glycosyltransferase; 2 glycosyltransferase family A; 3 glycosyltransferase GT4-type super family; 4 glycosyltransferase GTA-type super family; 5 CESA CelA-like cellulose synthase catalytic subunit (UDP-glucose as substrate); 6 cellulose synthase catalytic subunit; 7 glycosyltransferase; 8 glycosyltransferase family 17.
The PBCV-1 a064r gene encodes a 638-amino acid protein with three predicted domains. The N-terminal 211 amino acid domain resembles a “fringe-class” of glycosyltransferases (GT-GTA) and contains the last four of the five conserved motifs characteristic of this group of glycosyltransferases [62,63], including the proposed catalytic amino acids, the Asp-X-Asp sequence in motif 3 and the first Asp residue in motif 5. However, spacing between some of the four motifs differs from that of the fringe-glycosyltransferases. As mentioned above, the A064R protein, which is only present in five NC64A viruses, lacks both an identifiable signal peptide that would target the protein to the ER and a membrane-spanning motif, in contrast to “fringe” glycosyltransferases.
The 211-amino acid A064R glycosyltransferase domain was cloned, and the recombinant protein was crystallized [64]. The 1.6 Å crystal structure of the peptide has a mixed α/β fold containing a central, six-stranded β sheet flanked by α helices. The overall fold is similar to the catalytic domains in retaining glycosyltransferases in the GT-A group, family 34, although the amino acid similarity between them is low. Zhang et al. [64] suggested that the A064R glycosyltransferase bound to UDP-glucose better than to UDP-galactose or UDP-N-acetyl glucosamine. However, these binding experiments were conducted prior to knowing the Vp54 glycan structures. Now, there is evidence that the glycosyltransferase domain adds L-rhamnose to the distal xylose residue in the core structure [45].
Analysis of the six PBCV-1 antigenic variants revealed mutations in a064r that correlated with a specific antigenic class, B (EPA-1) (Table 3). The a064r gene in all six of these antigenic variants was sequenced to determine if mutations in a064r correlated with the EPA-1 antigenic variation [46]. The a064r sequences from three of the mutants had single nucleotide substitutions, which produced a single amino acid substitution in the glycosyltransferase portion of the A064R protein. Two of the amino acid substitutions occurred in the Asp-X-Asp motif (domain 3), and the other one was in domain 4. A fourth variant had an extra base in the coding sequence, which created a frame shift mutation in the gene. Finally, the entire gene was deleted in the other two antigenic variants.
Dual infection experiments with some of the different antigenic variants established that viruses containing wild-type a064r complemented and recombined with viruses that contained variant a064r to form wild-type virus. Therefore, it was concluded that a064r encodes a glycosyltransferase involved in the synthesis of the Vp54 glycan [46].
As noted above, the protein product of the a064r gene contains three domains with domain 1 being the glycosyltransferase. Domain 2 does not match anything in GenBank, but the C-terminal domain 3 is predicted to be a methyltransferase. We suspect that this C-terminal domain of approximately 200 amino acids is involved in methylating the terminal L-rhamnose in the Vp54 glycan [45].
A homolog of PBCV-1 glycosyltransferase, A546L (GT-GT4), has also been produced and crystallized [65]. The a546l gene homolog was from another NC64A chlorovirus NY-2A (gene b736l), and the 396-amino acid protein resembles members in the GT4 family of glycosyltransferases in the CAZy classification [66,67]. However, its biochemical function remains to be elucidated.
Of the eight glycosyltransferases encoded by the 43 chloroviruses, only two of them, homologs of PBCV-1 A111/114R and A075L, are present in all of the viruses, and so, they are predicted to be involved in the synthesis of the core glycan structure. A111/114R is especially interesting because it is predicted to have at least two glycosyltransferase catalytic domains.
Now that structures of the glycans from the chlorovirus major capsid proteins are becoming available, one can begin to characterize the viral encoded glycosyltransferases biochemically. One question that needs to be addressed is: Are the sugars added sequentially to the Vp54 protein backbone or are the glycans initially synthesized independently of Vp54, possibly on a lipid carrier and then attached to the protein in a single step? A slight variation of these two possibilities is that the core glycan is synthesized independently of the protein and then attached to Vp54. Additional sugars could then be added sequentially to these core glycans [68]. We suspect that this viral encoded glycosylation pathway represents a previously undescribed pathway, possible even a pathway that existed in eukaryotes prior to the ER and Golgi glycosylation pathway [18].
7. Additional Chlorovirus Encoded Sugar Metabolism Enzymes
Besides the chlorovirus-encoded enzymes described above, the viruses have four additional genes predicted to encode enzymes involved in sugar metabolism (Table 2). Recombinant proteins have not been produced from any of these genes, and so, it is unknown if they encode functional enzymes. These putative enzymes include an acetyltransferase (AT) encoded by all 43 chloroviruses, a D-lactate dehydrogenase (D-LD) encoded by 32 chloroviruses, fumarate reductase (FRD) encoded by five chloroviruses and ADP-ribosyl glycohydrolase (ADP-RGH) encoded by nine chloroviruses, all but two of which are Pbi viruses. The roles these putative enzymes play in the viral life cycles are unknown.
8. Chlorovirus-Encoded Polysaccharide Degrading Enzymes
In addition to the polysaccharide synthesizing enzymes described above, the chloroviruses also encode polysaccharide-degrading enzymes (Table 5). The chloroviruses are unique among viruses infecting eukaryotic organisms in that they, like bacteriophages, need to penetrate a rigid algal cell wall to initiate infection. The icosahedral shaped chlorovirus PBCV-1 has a spike-like structure at one vertex [13], which appears to make the initial contact with the cell wall of its host, C. variabilis NC64A [69]. Attachment is immediately followed by host cell wall degradation at the point of contact by a virus-packaged enzyme(s) [70]. After wall degradation, the viral internal membrane fuses with the host membrane to produce a narrow (~5 nm in inner diameter), membrane-lined tunnel, which allows entry of the viral DNA and some viral proteins [71]. This membrane fusion results in immediate host membrane depolarization [72] and potassium ion efflux [73]. This process results in an empty capsid remaining on the host cell surface.
Table 5.
Chlorovirus encoded enzymes involved in degrading polysaccharides.
| Host | Viruses | CHI 1 | CHIS 2 | GUN 3 | BCHIL 4 | Lysin 5 | GH 6 | CD 7 | ALGL 8 |
|---|---|---|---|---|---|---|---|---|---|
| NC64A | AN69C | 297R | 331L | 102L | 204R | 540R | 387L | 250L | |
| AR158 | C342L | C126L | C220R | C681L | C415L | C263L | |||
| CviK1 | 309L | 99L | 194R | 610L | 362L | 237L | |||
| CvsA1 | 318L | 058L | 200R | 627L | 371L, 373L | 243L | |||
| IL-3A | 285R | 323L | 097L | 196R | 624L | 378L | 235L | ||
| IL-5-2s1 | 373R | 415L | 140R | 262R | 796L | 495L | 310L | ||
| KS-1B | 211R | 257L | 048R | 132R | 546L | 166L | |||
| MA-1D | 241R, 242R | 283L | 491R | 147R | 777L | 359L | 191L | ||
| MA-1E | 352L | 114R | 194R, 229R | 717L | 399L | 277L | |||
| NE-JV-4 | 298R | 337L | 110R | 205R | 648L | 246L | |||
| NY-2B | 367R | 406L | 158L | 270R | 777L | 476L | 319L | ||
| NY2A | B393L | B137L | B239R | B756L | B469L | B288L | |||
| NYs-1 | 360R | 403L | 134L | 251R | 487L | ||||
| PBCV-1 | a A260R | a A292L | a A094L | a A181/182R | a A561L | a A215L | |||
| SYN | OSY-NE-5 | 119R | 138L | 037L | 117L | 290L | 099R | ||
| Pbi | AP110A | 106R | 122R | 174L | 942R | 306R | 158L | 338R | |
| CVA-1 | 102R | 114R | 167L | 942R | 297R | 156L | 327R | ||
| CVB-1 | 129R | 143R | 890R | 310R | 183L | 342R | |||
| CVG-1 | 105R | 113R | 142L | 896R | 303R | 329R | |||
| CVM-1 | 122R | 133R | 184L | 927R | 330R | 171L | 360R | ||
| CVR-1 | 109R | 121R | 174L | 948R | 163L | 335R | |||
| CZ-2 | 070R | 079R | 114L | 902R | 263R | 293R | |||
| Can18-4 | 113R | 121R | 160L | 944R | 316R | 349R | |||
| FR483 | N087R | N119L | N779R | N262R | N293R | ||||
| FR5L | 098R | 106R | 897R | 300R | 157L | 336R | |||
| MT325 | M085R | M091R | M124L | M791R | M258R | M289R | |||
| NE-JV-1 | 328L | 218L | 275L | 088R | 592L | 050L | 269L | 472L | |
| NW665 | 088R | 128L | 921R | 281R | 315R | ||||
| OR0704.2.2 | 074R | 082R | 113L | 895R | 257R | 292R | |||
| SAG | ATCV-1 | Z780L | Z204R | Z819L | Z814L | Z511L | Z771L | ||
| Br0604L | 253R, 254R | 950L | 902R, 942L | 518L | 832L | 895L | |||
| Can0610SP | 244R | 985R, 1001L | 936R, 996R | 613L | 919L | ||||
| Canal-1 | 815L | 242R | 866L | 138L, 855L | 546L | 743L | 806L | ||
| GM0701 | 251R | 972L | 917R, 963L | 526L | 846L | 910L | |||
| MN0810 | 271R | 082L | 162L, 1003L | 659L | 896L | 959L | |||
| MO0605SPH | 871L | 257R | 920L | 911L | 588L | 862L | |||
| NE-JV-2 | 950L | 258R | 1000R, 1013L | 1008R | 634L | 932L | |||
| NE-JV-3 | 907L | 244R | 957L | 948L | 603L | 897L | |||
| NTS-1 | 973L | 276R | 1024R | 1039R | 556L | 886L | 961L | ||
| OR0704.3 | 251R | 979R, 999L | 918R, 994R | 604L | 910L | ||||
| TN603 | 253R | 983L | 939R, 977L | 520L | 867L | 931L | |||
| WI0606 | 889L | 262R | 938L | 929L | 612L | 878L |
1 Chitinase; 2 chitosanase; 3 1-3-beta glucanase; 4 bifunctional chitinase/lysozyme; 5 lysin homologs from encoded by PBCV-1 CDS A561L; 6 glycosyl hydrolase; 7 chitin deacetylase; 8 alanine alginate lyase; a the recombinant proteins have the predicted activity.
In addition to virus entry into the host cells, nascent infectious PBCV-1 viruses exit the cells at 6–8 h PI by lysis of the plasma membrane and the cell wall. Therefore, it is not surprising that the chloroviruses encode polysaccharide-degrading enzymes in order to enter and exit the host cell. In fact, PBCV-1 encodes five such enzymes (Table 5), including two chitinases [74,75], a chitosanase [74,76], a β-1,3 glucanase [77] and an alkaline alginate lyase [78] or a polysaccharide lyase, cleaving chains of β- or α-1,4-linked glucuronic acids [79,80]. Recombinant proteins have been produced from each of these genes and shown to have the expected activity. Interestingly, the β-1,3 glucanase gene is expressed very early and disappears by 60 min PI. The protein is also made very early and disappears by 90 min PI [77]. Therefore, this enzyme is unlikely to be involved in either viral entry or viral exit from the cell. One possible function for the enzyme is to degrade host β-1,3 glycans, which might serve as host storage polysaccharides. Gene transcripts from the other four polysaccharide-degrading enzymes are present throughout the viral life cycles [74,81].
Experiments conducted about 30 years ago established that a crude enzyme preparation made from PBCV-1 lysates, named lysin, had good wall degrading activity and could be used to produce C. variabilis NC64A protoplasts [82,83]. Therefore, it was assumed that one or more of the five PBCV-1 encoded enzymes would be packaged in the PBCV-1 virion and be responsible for degrading the host cell wall at the point of infection. In fact, Yamada et al. [76] reported that a chitosanase activity was packaged in a closely-related chlorovirus, CVK2. However, an ensuing report [75] indicated that the CVK2 chitosanase activity was due to incomplete purification of the virion. Subsequently, a PBCV-1 proteome study identified 148 virus-encoded proteins and one host-encoded protein in highly purified virions [10]. Surprisingly, none of the five polysaccharide-degrading enzymes were packaged in the PBCV-1 virions.
Consequently, the 148 virus-encoded proteins packaged in the PBCV-1 particles were re-examined for possible polysaccharide or cell wall degrading activity. This effort revealed that one of the PBCV-1-encoded proteins packaged in the virion, A561L, has a putative glycosyl hydrolase domain. A recombinant protein produced from this domain has cell wall degrading activity, and the protein is under active investigation [84]. Homologs of the A561L domain (named A561L lysin) are present in most of the chloroviruses (Table 5), but not all. For example, viruses NYs-1 and CVR-1 appear to lack an a561l gene homolog encoding this domain, and the similarity between the predicted A561L homolog from viruses NY-2B and WI0606 is not very high. The apparent absence of the protein from these viruses deserves to be investigated further because one would expect the enzyme(s) that degrades the host cell wall during virus infection would be highly conserved.
Twenty-four of the 43 chloroviruses encode a protein that has a polysaccharide deacetylase domain (Table 5). Viruses in three of the four types (Osy being the exception) have the gene, but it is also missing in some viruses in each of the three types, so the gene is clearly not required for the success of the viruses. Its role might be to remove the acetyl group from chitin during host cell wall degradation.
In addition to these glycolytic enzymes, 42 of the 43 chloroviruses, encode a functional glycosylase protein that initiates pyrimidine photodimer excision [85,86]. The enzyme is part of a DNA repair pathway.
9. Conservation of the Chlorovirus Encoded Sugar Enzymes
Only three of the 21 chlorovirus encoded proteins listed in Table 1, Table 2 and Table 4 are present in all 43 chloroviruses, and these would be considered to be core proteins. The three are an acetyltransferase (AT), an exostosin glycosyltransferase (EXT) and a family A glycosyltransferase (GT-A). As noted above, we predict that the two glycosyltransferases are involved in the synthesis of the glycan core attached to the major capsid protein. The predicted function of the acetyltransferase is unknown.
Two of the seven viral encoded proteins involved in polysaccharide degrading activity are conserved in all of the viruses, the chitosanase (CHIS) and a putative bifunctional chitinase/lysozyme (BCHIL) (Table 4). Presumably these two enzymes play a role in the release of the nascent viruses from the cell. As indicated above, they are not packaged in the PBCV-1 virion, and so, they are not involved in the immediate early virus infection process.
The presence or absence of some of the chlorovirus sugar encoding enzymes displays some interesting patterns. For example, the GMD and GMER encoding genes are present in all of the NC64A, SAG and Osy viruses and absent in all of the Pbi viruses. This observation would suggest that the three virus types that have these genes would be more closely related to each other than to the Pbi viruses. However, a phylogenetic tree that shows the evolutionary relationship between the 43 viruses based on 29 concatenated core proteins [6] indicates that SAG and Pbi viruses are in the same branch and that the NC64A and Osy viruses are in a separate branch. Therefore, these results would imply that the SAG and NC64A/Osy viruses either acquired the genes separately after the four virus types had separated from a common ancestor or that the chlorovirus ancestor had both genes and for some reason, they were lost in the Pbi lineage.
Most of the other protein patterns are even more difficult to explain. For example, the GFAT encoding gene is present in all of the Pbi viruses and present in 11 of the 14 NC64A viruses and one SAG virus. Several of the other genes have similar complicated patterns and await explanations.
10. Sugar Enzymes Coded by Other Large DNA Viruses
This review has focused on carbohydrate enzymes encoded by the chloroviruses, primarily because these enzymes have been the most intensively studied. However, as new giant viruses are being discovered and their genomes sequenced, it is clear that some of them encode putative enzymes involved in carbohydrate manipulations. The most extensively studied of these other large DNA viruses is Acanthamoeba polyphaga mimivirus, which has genes encoding both glycosyltransferases and nucleotide sugars (see the recent review by Piacente et al. [87]). Other large DNA viruses encoding putative sugar manipulating enzymes include prasinoviruses (family Phycodnaviridae like the chloroviruses) that infect small marine green algae, including Ostreococcus, Bathycoccus and Micromonas species; these viruses have clusters of putative genes for enzymes involved in nucleotide-sugar metabolism and glycosyltransferases [88]. Similar genes are present in other members of the Mimiviridae family, including Phaeocystis globosa virus [89] and Cafeteria roenbergensis virus [90]. Putative glycosyltransferase-encoding genes have also been reported in the genomes of pandoraviruses [91], Pithovirus sibericus [92] and Mollivirus sibericus [93].
In conclusion, it is becoming clear that virus-encoded sugar-manipulating enzymes and glycosylation systems can no longer be considered a hallmark solely of cellular organisms, but that some viruses also encode unique and complex glycan systems, which are still largely unknown. One encourages young glycobiologists to consider working on some of these systems.
Acknowledgments
The authors thank the many undergraduate students, graduate students, postdocs and collaborators who have worked on various aspects of the chlorovirus encoded sugar enzymes over the past 25 years. This research has been supported by grants from NIH, NSF, DOE and the Stanley Medical Research Institute over the years.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Karakashian S.J., Karakashian M.W. Evolution and symbiosis in the genus Chlorella and related algae. Evolution. 1965;19:368–377. doi: 10.2307/2406447. [DOI] [Google Scholar]
- 2.Kodama Y., Suzuki H., Dohra H., Sugii M., Kitazume T., Yamaguchi K., Shigenobu S., Fujishima M. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics. 2014;15:1. doi: 10.1186/1471-2164-15-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada T., Onimatsu H., Van Etten J.L. Chlorella viruses. In: Maramorosch K., Shatkin A.J., editors. Advances in Virus Research. Vol. 66. Elsevier Inc; San Diego, CA, USA: 2006. pp. 293–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunigan D.D., Fitzgerald L.A., Van Etten J.L. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Van Etten J.L., Dunigan D.D. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 2012;17:1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quispe C.F., Esmael A., Sonderman O., McQuinn M., Agarkova I., Battah M., Duncan G.A., Dunigan D.D., Smith T.P., De Castro C., et al. Characterization of a new chlorovirus type with permissive and non-permissive features on phylogenetically related algal strains. Virology. 2017;500:103–113. doi: 10.1016/j.virol.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald L.A., Graves M.V., Li X., Feldblyum T., Hartigan J., Van Etten J.L. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology. 2007;358:459–471. doi: 10.1016/j.virol.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald L.A., Graves M.V., Li X., Feldblyum T., Nierman W.C., Van Etten J.L. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007;358:472–484. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald L.A., Graves M.V., Li X., Hartigan J., Pfitzner A.J., Hoffart E., Van Etten J.L. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology. 2007;362:350–361. doi: 10.1016/j.virol.2006.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunigan D.D., Cerny R.L., Bauman A.T., Roach J.C., Lane L.C., Agarkova I.V., Wulser K., Yanai-Balser G.M., Gurnon J.R., Vitek J.C., et al. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J. Virol. 2012;86:8821–8834. doi: 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeanniard A., Dunigan D.D., Gurnon J.R., Agarkova I.V., Kang M., Vitek J., Duncan G., McClung O.W., Larsen M., Claverie J.-M., et al. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics. 2013;14:158. doi: 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Etten J.L., Burbank D.E., Kuczmarski D., Meints R.H. Virus infection of culturable chlorella-like algae and development of a plaque assay. Science. 1983;219:994–996. doi: 10.1126/science.219.4587.994. [DOI] [PubMed] [Google Scholar]
- 13.Cherrier M.V., Kostyuchenko V.A., Xiao C., Bowman V.D., Battisti A.J., Yan X., Chipman P.R., Baker T.S., Van Etten J.L., Rossmann M.G. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strasser P., Zhang Y.P., Rohozinski J., Van Etten J.L. The termini of the chlorella virus PBCV-1 genome are identical 2.2-kbp inverted repeats. Virology. 1991;180:763–769. doi: 10.1016/0042-6822(91)90089-T. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Strasser P., Grabherr R., Van Etten J.L. Hairpin loop structure at the termini of the chlorella virus PBCV-1 genome. Virology. 1994;202:1079–1082. doi: 10.1006/viro.1994.1444. [DOI] [PubMed] [Google Scholar]
- 16.Nelson M., Zhang Y., Van Etten J.L. DNA methyltransferases and DNA site-specific endonucleases encoded by chlorella viruses. In: Jost J.P., Saluz H.P., editors. DNA methylation: molecular biology and biological significance. Birkhauser Verlag Publ; Basel, Switzerland: 1993. pp. 186–211. [DOI] [PubMed] [Google Scholar]
- 17.DeAngelis P.L., Jing W., Graves M.V., Burbank D.E., Van Etten J.L. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- 18.Graves M.V., Burbank D.E., Roth R., Heuser J., DeAngelis P.L., Van Etten J.L. Hyaluronan synthesis in virus PBCV-1-infected chlorella-like green algae. Virology. 1999;257:15–23. doi: 10.1006/viro.1999.9628. [DOI] [PubMed] [Google Scholar]
- 19.DeAngelis P.L. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pathogens, and algal viruses. Cell. Mol. Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeAngelis P.L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002;268:317–326. doi: 10.1002/ar.10163. [DOI] [PubMed] [Google Scholar]
- 21.Landstein D., Graves M.V., Burbank D.E., DeAngelis P., Van Etten J.L. Chlorella virus PBCV-1 encodes functional glutamine: fructose-6-phosphate amidotransferase and UDP-glucose dehydrogenase enzymes. Virology. 1998;250:388–396. doi: 10.1006/viro.1998.9388. [DOI] [PubMed] [Google Scholar]
- 22.Blanc G., Duncan G., Agarkova I., Borodovsky M., Gurnon J., Kuo A., Lindquist E., Lucas S., Pangilinan J., Polle J., et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gooday G.W., Humphreys A.M., McIntosh W.H. Roles of chitinases in fungal growth. In: Muzzarelli R., Jeuniaux C., Gooday G.W., editors. Chitin in nature and technology. Springer; Boston, MA, USA: 1986. pp. 83–91. [Google Scholar]
- 24.Kapaun E., Loos E., Reisser W. Cell wall composition of virus-sensitive symbiotic Chlorella species. Phytochemistry. 1992;31:3103–3104. doi: 10.1016/0031-9422(92)83453-6. [DOI] [Google Scholar]
- 25.Kawasaki T., Tanaka M., Fujie M., Usami S., Sakai K., Yamada T. Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology. 2002;302:123–131. doi: 10.1006/viro.2002.1572. [DOI] [PubMed] [Google Scholar]
- 26.Ali M., Kawasaki T., Yamada T. Genetic rearrangements on the chlorovirus genome that switch between hyaluronan synthesis and chitin synthesis. Virology. 2005;342:102–110. doi: 10.1016/j.virol.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 27.Yamada T., Kawasaki T. Microbial synthesis of hyaluronan and chitin: New approaches. J. Biosci. Bioeng. 2005;99:521–528. doi: 10.1263/jbb.99.521. [DOI] [PubMed] [Google Scholar]
- 28.Chase T.E., Nelson J.A., Burbank D.E., Van Etten J.L. Mutual exclusion occurs in a Chlorella-like green alga inoculated with two viruses. J. Gen. Virol. 1989;70:1829–1836. doi: 10.1099/0022-1317-70-7-1829. [DOI] [PubMed] [Google Scholar]
- 29.Greiner T., Frohns F., Kang M., Van Etten J.L., Käsmann A., Moroni A., Hertel B., Thiel G. Chlorella viruses prevent multiple infections by depolarizing the host membranes. J. Gen. Virol. 2009;90:2033–2039. doi: 10.1099/vir.0.010629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis G., Van Etten J.L. Does extracellular hyaluronan and/or chitin confer an evolutionary advantage to the chloroviruses? Unpublished.
- 31.Rakkhumkaew N., Kawasaki T., Fujie M., Yamada T. Prolonged synthesis of hyaluronan by chlorella cells infected with chloroviruses. J. Biosci. Bioeng. 2013;115:527–531. doi: 10.1016/j.jbiosc.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Tonetti M., Zanardi D., Gurnon J.R., Fruscione F., Armirotti A., Damonte G., Sturla L., De Flora A., Van Etten J.L. Paramecium bursaria chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J. Biol. Chem. 2003;278:21559–21565. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- 33.Ma B., Simala-Grant J.L., Taylor D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158r–184r. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 34.De Castro C., Speciale I., Duncan G., Dunigan D.D., Agarkova I., Lanzetta R., Sturiale L., Palmigiano A., Garozzo D., Molinaro A., et al. N-linked glycans of chloroviruses sharing a core architecture without precedent. Angew. Chem. Int. Ed. 2016;55:654–658. doi: 10.1002/anie.201509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speciale I., Agarkova I., Duncan G., Van Etten J.L., De Castro C. Structure of the N-glycan from the chlorovirus NE-JV-1. A. Van. Leeuw. 2017 doi: 10.1007/s10482-017-0861-3. In press. [DOI] [PubMed] [Google Scholar]
- 36.Rosano C., Zuccotti S., Sturla L., Fruscione F., Tonetti M., Bolognesi M. Quaternary assembly and crystal structure of GDP-D-mannose 4,6 dehydratase from Paramecium bursaria chlorella virus. Biochem. Biophys. Res. Commun. 2006;339:191–195. doi: 10.1016/j.bbrc.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Fruscione F., Sturla L., Duncan G., Van Etten J.L., Valbuzzi P., De Flora A., Di Zanni E., Tonetti M. Differential role of NADP+ and NADPH in the activity and structure of GDP-D-mannose 4,6-dehydratase from two chlorella viruses. J. Biol. Chem. 2008;283:184–193. doi: 10.1074/jbc.M706614200. [DOI] [PubMed] [Google Scholar]
- 38.Chothi M.P., Duncan G.A., Armirotti A., Abergel C., Gurnon J.R., Van Etten J.L., Bernardi C., Damonte G., Tonetti M. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J. Virol. 2010;84:8829–8838. doi: 10.1128/JVI.00770-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doms R.W., Lamb R.A., Rose J.K., Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 40.Olofsson S., Hansen J.E. Host cell glycosylation of viral glycoproteins--a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 1998;30:435–440. doi: 10.1080/00365549850161386. [DOI] [PubMed] [Google Scholar]
- 41.Flint S.J., Racaniello V.R., Rail G.F., Skalka A.M., Enquist L.W. Principles of Virology. 4th ed. Vol. 1. ASM Press; Washington, DC, USA: 2015. p. 569. [Google Scholar]
- 42.Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang I.N., Li Y., Que Q., Bhattacharya M., Lane L.C., Chaney W.G., Van Etten J.L. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. USA. 1993;90:3840–3844. doi: 10.1073/pnas.90.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Castro C., Speciale I., Agarkova I., Molinaro A., Tonetti M., Duncan G.A., Van Etten J.L. Glycan structures of chlorovirus PBCV-1 major capsid protein antigenic variants. Manuscript in preparation.
- 45.De Castro C., Speciale I., Agarkova I., Molinaro A., Duncan G., Lowary T., Lee S., Jimenez-Barbero J., Tonetti M., Van Etten J.L. Disclosure of the first viral encoded glycosyl transferase: Domain 1 of A064R gene of Paramecium bursaria chlorella virus encodes for a functional b-L-rhamnosyl transferase. Manuscript in preparation.
- 46.Graves M.V., Bernadt C.T., Cerny R., Van Etten J.L. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology. 2001;285:332–345. doi: 10.1006/viro.2001.0937. [DOI] [PubMed] [Google Scholar]
- 47.Van Etten J.L., Burbank D.E., Xia Y., Meints R.H. Growth cycle of a virus, PBCV- 1, that infects chlorella-like algae. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-X. [DOI] [PubMed] [Google Scholar]
- 48.Nandhagopal N., Simpson A.A., Gurnon J.R., Yan X., Baker T.S., Graves M.V., Van Etten J.L., Rossmann M.G. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Castro C., Speciale I., Molinaro A., Van Etten J.L., Klose T., Rossmann M.G. Major capsid protein of chlorovirus PBCV-1 structure combining X-ray and carbohydrate molecular modeling approaches. Proc. Natl. Acad. Sci. USA. In revision. [DOI] [PMC free article] [PubMed]
- 50.Zielinska D.F., Gnad F., Wisniewski J.R., Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz F., Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Abu-Qarn M., Yurist-Doutsch S., Giordano A., Trauner A., Morris H.R., Hitchen P., Medalia O., Dell A., Eichler J. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 2007;374:1224–1236. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 53.Chaney W.G., Van Etten J.L. Inability to cleave glycans from chlorovirus PBCV-1 major capsid protein by traditional glycosidases. Unpublished.
- 54.De Castro C., Molinaro A., Piacente F., Gurnon J.R., Sturiale L., Palmigiano A., Lanzetta R., Parrilli M., Garozzo D., Tonetti M.G., et al. Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc. Natl. Acad. Sci. USA. 2013;110:13956–13960. doi: 10.1073/pnas.1313005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wieland F., Heitzer R., Schaefer W. Asparaginylglucose: novel type of carbohydrate linkage. Proc. Natl. Acad. Sci. USA. 1983;80:5470–5474. doi: 10.1073/pnas.80.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mengele R., Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 1992;267:8182–8185. [PubMed] [Google Scholar]
- 57.Schreiner R., Schnabel E., Wieland F. Novel N-glycosylation in eukaryotes: laminin contains the linkage unit beta-glucosylasparagine. J. Cell. Biol. 1994;124:1071–1081. doi: 10.1083/jcb.124.6.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gross J., Grass S., Davis A.E., Gilmore-Erdmann P., Townsend R.R., St Geme J.W. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 2008;283:26010–26015. doi: 10.1074/jbc.M801819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Que Q., Li Y., Wang I.N., Lane L.C., Chaney W.G., Van Etten J.L. Protein glycosylation and myristylation in chlorella virus PBCV-1 and its antigenic variants. Virology. 1994;203:320–327. doi: 10.1006/viro.1994.1490. [DOI] [PubMed] [Google Scholar]
- 60.Chuchird N., Nishida K., Kawasaki T., Fujie M., Usami S., Yamada T. A variable region on the chlorovirus CVK2 genome contains five copies of the gene for Vp260, a viral-surface glycoprotein. Virology. 2002;295:289–298. doi: 10.1006/viro.2002.1408. [DOI] [PubMed] [Google Scholar]
- 61.Yanai-Balser G.M., Duncan G.A., Eudy J.D., Wang D., Li X., Agarkova I.V., Dunigan D.D., Van Etten J.L. Microarray analysis of chlorella virus PBCV-1 transcription. J. Virol. 2010;84:532–542. doi: 10.1128/JVI.01698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan Y.P., Schultz J., Mlodzik M., Bork P. Secreted fringe-like signaling molecules may be glycosyltransferases. Cell. 1997:9–11. doi: 10.1016/S0092-8674(00)81852-8. [DOI] [PubMed] [Google Scholar]
- 63.Bruckner K., Perez L., Clausen H., Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y., Xiang Y., Van Etten J.L., Rossmann M.G. Structure and function of a chlorella virus encoded glycosyltransferase. Structure. 2007;15:1031–1039. doi: 10.1016/j.str.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiang Y., Baxa U., Zhang Y., Steven A.C., Lewis G.L., Van Etten J.L., Rossmann M.G. Crystal structure of a virus-encoded putative glycosyltransferase. J. Virol. 2010;84:12265–12273. doi: 10.1128/JVI.01303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coutinho P.M., Deleury E., Davies G.J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 67.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 68.Van Etten J.L., Gurnon J.R., Yanai-Balser G.M., Dunigan D.D., Graves M.V. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta. 2010;1800:152–159. doi: 10.1016/j.bbagen.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X., Xiang Y., Dunigan D.D., Klose T., Chipman P.R., Van Etten J.L., Rossmann M.G. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc. Natl. Acad. Sci. USA. 2011;108:14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meints R.H., Lee K., Burbank D.E., Van Etten J.L. Infection of a chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- 71.Milrot E., Shimoni E., Dadosh T., Unger T., Van Etten J.L., Minsky A. Bacteriophage-like infection of the large eukaryote-infecting Paramecium bursaria chlorella virus-1. PLoS Pathogens. Submitted. [DOI] [PMC free article] [PubMed]
- 72.Frohns F., Käsmann A., Kramer D., Schäfer B., Mehmel M., Kang M., Van Etten J.L., Gazzarrini S., Moroni A., Thiel G. Potassium ion channels of chlorella viruses cause rapid depolarization of host cells during infection. J. Virol. 2006;80:2437–2444. doi: 10.1128/JVI.80.5.2437-2444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neupärtl M., Meyer C., Woll I., Frohns F., Kang M., Van Etten J.L., Kramer D., Hertel B., Moroni A., Thiel G. Chlorella viruses evoke a rapid release of K+ from host cells during early phase of infection. Virology. 2008;372:340–348. doi: 10.1016/j.virol.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 74.Sun L., Adams B., Gurnon J., Ye Y., Van Etten J.L. Characterization of two chitinase genes and one chitosanase gene encoded by chlorella virus PBCV-1. Virology. 1999;263:376–387. doi: 10.1006/viro.1999.9958. [DOI] [PubMed] [Google Scholar]
- 75.Hiramatsu S., Ishihara M., Fujie M., Usami S., Yamada T. Expression of a chitinase gene and lysis of the host cell wall during Chlorella virus CVK2 infection. Virology. 1999;260:308–315. doi: 10.1006/viro.1999.9824. [DOI] [PubMed] [Google Scholar]
- 76.Yamada T., Hiramatsu S., Songsri P., Fujie M. Alternative expression of a chitosanase gene produces two different proteins in cells infected with chlorella virus CVK2. Virology. 1997;230:361–368. doi: 10.1006/viro.1997.8486. [DOI] [PubMed] [Google Scholar]
- 77.Sun L., Gurnon J.R., Adams B.J., Graves M.V., Van Etten J.L. Characterization of a β-1,3-glucanase encoded by chlorella virus PBCV-1. Virology. 2000;276:27–36. doi: 10.1006/viro.2000.0500. [DOI] [PubMed] [Google Scholar]
- 78.Suda K., Tanji Y., Hori K., Unno H. Evidence for a novel chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999;180:45–53. doi: 10.1111/j.1574-6968.1999.tb08776.x. [DOI] [PubMed] [Google Scholar]
- 79.Sugimoto I., Onimatsu H., Fujie M., Usami S., Yamada T. vAL-1, a novel polysaccharide lyase encoded by chlorovirus CVK2. FEBS Lett. 2004;559:51–56. doi: 10.1016/S0014-5793(04)00022-5. [DOI] [PubMed] [Google Scholar]
- 80.Ogura K., Yamasaki M., Yamada T., Mikami B., Hashimoto W., Murata K. Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action. J. Biol. Chem. 2009;284:35572–35579. doi: 10.1074/jbc.M109.068056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sugimoto I., Hiramatsu S., Murakami D., Fujie M., Usami S., Yamada T. Algal-lytic activities encoded by Chlorella virus CVK2. Virology. 2000;277:119–126. doi: 10.1006/viro.2000.0590. [DOI] [PubMed] [Google Scholar]
- 82.Meints R.H., Burbank D.E., Van Etten J.L., Lamport D.T. Properties of the chlorella receptor for the virus PBCV-1. Virology. 1988;164:15–21. doi: 10.1016/0042-6822(88)90614-9. [DOI] [PubMed] [Google Scholar]
- 83.Agarkova I., Hertel B., Zhang X., Lane L., Tchourbanov A., Dunigan D.D., Thiel G., Rossmann M.G., Van Etten J.L. Dynamic attachment of chlorovirus PBCV-1 to Chlorella variabilis. Virology. 2014;466–467:95–102. doi: 10.1016/j.virol.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agarkova I.V., Lane L.C., Dunigan D.D., Quispe C., Duncan G.A., Milrot E., Minsky A., Esmael A., Van Etten J.L. Identification of the chlorovirus PBCV-1 enzyme involved in degrading the host cell wall during virus infection. Manuscript in preparation. [DOI] [PMC free article] [PubMed]
- 85.Furuta M., Schrader J.O., Schrader H.S., Kokjohn T.A., Nyaga S., McCullough A.K., Lloyd R.S., Burbank D.E., Landstein D., Lane L., et al. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl. Environ. Microbiol. 1997;63:1551–1556. doi: 10.1128/aem.63.4.1551-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCullough A.K., Romberg M.T., Nyaga S., Wei Y., Wood T.G., Taylor J.S., Van Etten J.L., Dodson M.L., Lloyd R.S. Characterization of a novel cis-syn and trans-syn-II pyrimidine dimer glycosylase/AP lyase from a eukaryotic algal virus, Paramecium bursaria chlorella virus-1. J. Biol. Chem. 1998;273:13136–13142. doi: 10.1074/jbc.273.21.13136. [DOI] [PubMed] [Google Scholar]
- 87.Piacente F., Gaglianone M., Laugieri M.E., Tonetti M.G. The autonomous glycosylation of large DNA viruses. Int. J. Mol. Sci. 2015;16:29315–29328. doi: 10.3390/ijms161226169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moreau H., Piganeau G., Desdevises Y., Cooke R., Derelle E., Grimsley N. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 2010;84:12555–12563. doi: 10.1128/JVI.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santini S., Jeudy S., Bartoli J., Poirot O., Lescot M., Abergel C., Barbe V., Wommack K.E., Noordeloos A.A., Brussaard C.P., et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fischer M.G., Allen M.J., Wilson W.H., Suttle C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Philippe N., Legendre M., Doutre G., Coute Y., Poirot O., Lescot M., Arslan D., Seltzer V., Bertaux L., Bruley C., et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 92.Legendre M., Bartoli J., Shmakova L., Jeudy S., Labadie K., Adrait A., Lescot M., Poirot O., Bertaux L., Bruley C., et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Legendre M., Lartigue A., Bertaux L., Jeudy S., Bartoli J., Lescot M., Alempic J.M., Ramus C., Bruley C., Labadie K., et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]