Abstract
Summary
Ancient DNA has emerged as a remarkable tool to infer the history of extinct species and past populations. However, many of its characteristics, such as extensive fragmentation, damage and contamination, can influence downstream analyses. To help investigators measure how these could impact their analyses in silico, we have developed gargammel, a package that simulates ancient DNA fragments given a set of known reference genomes. Our package simulates the entire molecular process from post-mortem DNA fragmentation and DNA damage to experimental sequencing errors, and reproduces most common bias observed in ancient DNA datasets.
Availability and Implementation
The package is publicly available on github: https://grenaud.github.io/gargammel/ and released under the GPL.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
DNA retrieved from subfossils, also called ancient DNA (aDNA), is increasingly used to reconstruct population histories (Leonardi et al., 2016). The analysis of aDNA data remains, however, challenging due to a number of factors that can affect downstream inferences. First, DNA tends to degrade over time, leading to fragments of limited sizes (30–80 bp) showing substantial nucleotide misincorporations (Briggs et al., 2007). Second, environmental microbes tend to colonize the organism postmortem (Green et al., 2009). As a result, the endogenous DNA fraction can sometimes be extremely reduced, making shotgun sequencing approaches uneconomical. Third, such exogenous sequences can impact the reconstruction of ancient genomes if not properly identified during read alignment. In the case of aDNA retrieved from hominin species, the DNA from present-day humans, which can be introduced at any stage including during the excavation and in the laboratory, is particularly problematic as it mixes unrelated population histories within a single sample.
Ancient DNA researchers often use simulations to test the robustness of summary statistics aimed at inferring population parameters. While some packages have simulated platform-specific errors due to sequencing, no packages are currently available to properly simulate aDNA sequence datasets, including their most prominent characteristics, such as such as damage, fragmentation, human and microbial contamination.
Here, we present gargammel, a package that simulates aDNA sequence datasets from a set of genome references representing the microbial fraction, the endogenous fraction and the present-day human contamination. The package can simulate most common features of aDNA sequences, including post-mortem DNA damage and base misincorporations. In addition, it simulates base compositional bias due to the molecular tools used in library preparation, sequencing bias against GC-rich fragments and errors introduced by the sequencing platform.
2 Methods
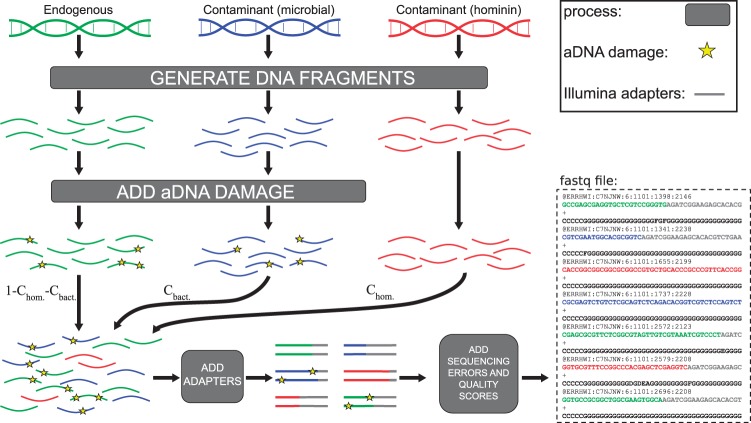
Our algorithm reflects the entire molecular and experimental process leading to the retrieval of aDNA fragments (Fig. 1). In its simplest mode, the user first provides three sets of references in fasta format: (i) the microbial contaminant, (ii) endogenous genome and (iii) the present-day human contamination. The user can also provide full microbial profiles, including taxonomic abundances, to represent more complex sources of microbial contamination. In this case, corresponding (or closely related) microbial genomes will be automatically downloaded from NCBI. The user either provides the desired endogenous coverage or a fixed number of fragments to simulate. The endogenous genome can contain 1 sequence for haploid organisms or 2 sequences as to simulate a diploid organism where fragments are sampled from each with equal probability.
Fig. 1.
Gargammel flowchart. A precise number of fragments are selected from the endogenous, present-day human contaminants and microbial genomes. Damage characteristic of aDNA is added and the various types of fragments, contaminants and endogenous ones, are combined, sequencing adapters are added in silico and sequencing errors with corresponding quality scores are produced. Microbial contamination occurs at a rate of Cbact. whereas present-day human contamination occurs at a rate of Chom. Molecular damage can be added using different models for all three sources via command-line options
Fragments are selected from all three sets depending on the desired composition of the final set (e.g. 70% microbial, 20% endogenous and 10% present-day human contamination). The size of the fragments can be selected from a user-specified distribution.
As aDNA base composition can be different from modern DNA (Jónsson et al., 2013) and vary together with the molecular tools used during library preparation (Seguin-Orlando et al., 2013), the base composition can also be modeled. Subsequently, post-mortem deamination is added according to the parameters of standard aDNA damage models (Briggs et al., 2007) or a user-specified matrix of position-specific misincorporation rates.
Fragmented aDNA templates can be shorter than the read length. Gargammel proceeds by adding the necessary length of the sequencing adapter. Finally, the ART sequencing simulator (Huang et al., 2012) is used on the resulting sequences to produce Illumina reads with sequencing errors and quality scores. The various subprograms of the pipeline are called by a wrapper script and are detailed in the Supplementary Methods. Finally, the wrapper script combines reads from the three sources to create the final sequence set.
3 Features
We tested gargammel for its ability to reproduce empirical features found in six previously released aDNA datasets (see Supplementary Results). These include: (i) size distribution, (ii) base composition, (iii) GC-bias due to the DNA polymerase used for library amplification and; (iv) DNA misincorporation. The results presented in the Supplementary Results show a high consistency between observed and simulated distributions to show the applicability of gargammel as a sequence simulator for aDNA.
Gargammel provides researchers with the opportunity to perform various inquiries to evaluate the robustness of various analyses to aDNA properties. In the Suppl. Results, we present two such types of analyses. First, we evaluated the potential impact of present-day contamination on admixture tests based on the D-statistics (Durand et al., 2011). Simulated sequences were obtained from coalescence models without any admixture. We find that amounts of present-day human contamination in ancient human datasets can create spurious signals of admixture depending on the coalescent model, especially when a distant outgroup is used in the test and when the modern contamination source originates from a population coalescing deeply with the endogenous individual. Second, we evaluated whether microbial fragments of increasing size could impact the false positive alignment rate against the human reference genome (see Schubert et al., 2012 for an evaluation of aDNA mapping). We identified a size threshold (35 bp) that reduces such impact for the microbial community identified in the 12.8 kyr-old Clovis individual (Rasmussen et al., 2014). Such tests can be adapted to any other situation of interest.
Supplementary Material
Acknowledgments
We thank Weichun Huang and all members of the Paleomix group at the Centre for GeoGenetics for fruitful discussions.
Funding
This work was supported by the Danish Council for Independent Research, Natural Sciences (4002-00152B); the Danish National Research Foundation (DNRF94); the Villum Fonden (miGENEPI), and; Initiative d’Excellence Chaires d’attractivité, Université de Toulouse (OURASI).
Conflict of Interest: none declared.
References
- Briggs A.W. et al. (2007) Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U. S. A., 104, 14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E.Y. et al. (2011) Testing for ancient admixture between closely related populations. Mol. Biol. Evol., 28, 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.E. et al. (2009) The Neandertal genome and ancient DNA authenticity. EMBO J., 28, 2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. et al. (2012) ART: a next-generation sequencing read simulator. Bioinformatics, 28, 593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H. et al. (2013) mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics, 29, 1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi M. et al. (2016) Evolutionary patterns and processes: lessons from ancient DNA. Syst. Biol., syw059.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. et al. (2014) The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature, 506, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M. et al. (2012) Improving ancient DNA read mapping against modern reference genomes. BMC Genomics, 13, 178. ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin-Orlando A. et al. (2013) Ligation bias in Illumina next-generation DNA libraries: implications for sequencing ancient genomes. PLOS One, 8, e78575.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.