Abstract
Motivation
Optical mapping is a technique for capturing fluorescent signal patterns of long DNA molecules (in the range of 0.1–1 Mbp). Recently, it has been complementing the widely used short-read sequencing technology by assisting with scaffolding and detecting large and complex structural variations (SVs). Here, we introduce a fast, robust and accurate tool called OMBlast for aligning optical maps, the set of signal locations on the molecules generated from optical mapping. Our method is based on the seed-and-extend approach from sequence alignment, with modifications specific to optical mapping.
Results
Experiments with both synthetic and our real data demonstrate that OMBlast has higher accuracy and faster mapping speed than existing alignment methods. Our tool also shows significant improvement when aligning data with SVs.
Availability and Implementation
OMBlast is implemented for Java 1.7 and is released under a GPL license. OMBlast can be downloaded from https://github.com/aldenleung/OMBlast and run directly on machines equipped with a Java virtual machine.
Supplementary information
Supplementary data are available at Bioinformatics online
1 Introduction
Despite recent advances in sequencing technology, read lengths of typical next generation sequencing instruments and the latest PacBio sequencer are still on the order of 100 bp and 10 kbp, respectively (Van Dijk et al., 2014). Optical mapping, however, can produce molecules of lengths on the order of 0.1 Mbp to even 1 Mbp (Lam et al., 2012). Optical mapping complements sequencing by resolving complex regions and has been used in several areas including guided sequence assembly (Lin et al., 2012), guided sequence scaffolding (Dong et al., 2013), structural variation (SV) detection (Cao et al., 2014) and pathogen identification (Miller, 2013). In fact, optical mapping was used by the Assemblathon 2 organizers to validate the sequence assemblies provided by various teams (Bradnam et al., 2013). With an increasing number of optical maps and multiple applications, bioinformatics tools specific for optical mapping are needed (see Mendelowitz and Pop, 2014 for a recent survey).
Aligning optical maps is very different from aligning short reads from sequencing due to the vast discrepancy in data type. Various types of error in optical maps pose challenges to error tolerant alignment. Notwithstanding the problems, fast, robust and accurate alignment is fundamental to any downstream analysis of optical maps. In this article, we introduce OMBlast for aligning optical maps, and compare its performance with other alignment methods on synthetic data based on different prokaryotic and eukaryotic organisms of varying genome sizes at different error rates. We also studied the effects of SVs on alignment performance. Finally we tested our alignment method on optical maps generated from an actual experiment. Our results show that OMBlast is both fast and accurate, and its performance is superior to other methods when SVs are present.
This paper is organized as follows. Section 2 gives an overview of optical mapping and outlines the terms used for the remainder of this paper. The methods behind OMBlast are explained in Section 3. In Section 4, we evaluate our method against several existing tools using both synthetic and real data sets. Finally, Section 5 summarizes our findings and offers directions for future work.
2 Background
2.1 Overview
In recent years, two commercialized methods have renewed interest in optical mapping so that it could complement current short-read sequencing methods. Both OpGen Inc. and Bionano Genomics Inc. have independently devised ways of generating high-throughput data of optical maps. OpGen Inc.'s method relies on the static imaging of restriction maps (Dimalanta et al., 2004) while Bionano Genomics Inc.'s is based on continuous imaging of nicked DNA molecules attached with fluorescent labels passing through nanotubes (Das et al., 2010). These two technologies produce optical maps with similar data types. An optical map contains a series of smaller DNA segments, separated by fluorescent labels in Bionano Genomics Inc.mi system, or by digestion cut in OpGen Inc.'s system (Supplementary Figure S1). As data generation becomes easier, a good alignment method for handling large amounts of data becomes important for further downstream analysis.
2.2 Terminology
Since the two methods were independently developed, the terminology used to describe errors differs slightly; we summarize them in Supplementary Figure S1. Irrespective of the technology, an optical map consists of two pieces of information: (i) the size of each segment situated between two consecutive cuts/nicks and (ii) the order of the segments. As a result, assuming a single optical map i consists of ni segments, these segments are separated by fluorescent signals [alternatively, cuts in OpGen data (From this point onward, we will always use the term ‘signals’)]. We can represent such data as an -tuple such that each element is the absolute location of the signal (i.e. number of base pairs) relative to the beginning of the molecule.
In this article, we consider the problem of aligning one optical map against another. This problem setting is analogous to the mapping of a query DNA sequence against another DNA sequence. Instead of processing sequences made up of the four bases ({A, C, G, T}), an optical map consists of signal positions which are positive integers. We denote one of these optical maps as the reference and the other as the query. Usually the experimentally generated optical maps are aligned to a longer optical map obtained from either in silico digestion of the reference sequence, or an optical map assembly, but this is not a restriction on how the OMBlast software can be used. The tuples of signals are denoted as R and Q. Let the xth signal on the reference and the yth signal on the query be represented as Rx and Qy, respectively. We define the matching signal pair to be the pairing of a reference signal with a query signal.
Our aim is to output the best alignment of each query against the reference, in terms of the similarity between the patterns of fluorescent labels on the reference and query. As illustrated in Supplementary Figure S1, this is non-trivial since the optical map itself can contain various types of error. For example, an optical map can have missing and extra signals. Furthermore, unlike short-read sequencing which provides single base resolution, optical mapping is accurate to only 0.7–1.5 kbp for OpGen (Nagarajan et al., 2008) and Bionano data (Lam et al., 2012). Measurement error (i.e. error in measuring the length of the DNA segments) and scaling factor [i.e. an indicator of the extent that a DNA molecule is stretched, which remains constant within a DNA molecule but varies between different DNA molecules (Reinhart et al., 2015)] also exist in optical mapping data.
Other than the experimental and instrumental errors, the mismatch between reference and query due to genetic variation also poses challenges to alignment. Single nucleotide polymorphism (SNP) could toggle ‘on’ a new enzyme site or ‘off’ an existing enzyme site. Segment size matching is also affected by the presence of large insertions or deletions. In more complicated cases, part of the query could be reversely and separately aligned to the reference due to SVs. Despite the challenges, genetic variations carry important biological insights and need to be accurately identified. To overcome these obstacles, an alignment method has to perform both local alignment and joining of the separate local alignments of different parts of a query map in order to capture various types of genetic variations.
2.3 Related work
Some publicly available alignment software for optical mapping includes RefAligner (Shelton et al., 2015), the one described by Valouev et al. (2006) (whose software we will call Valouev hereafter), Scaffolding using Optical Map Alignment (SOMA) (Nagarajan et al., 2008), and TWIN (Muggli et al., 2014). Both Valouev and SOMA are based on dynamic programming (DP) coupled with scoring functions which quantify how well a query matches the reference. Valouev employs probability distributions to model the types of errors found in optical maps while SOMA uses scoring functions that model the distribution of segment lengths as well as to what extent the lengths agree between the query and the reference. Meanwhile, TWIN takes a different approach by constructing an FM-Index on the segment lengths of the reference and then performing a near-exact match of the query by using the metric used by SOMA. Another program for aligning optical maps is RefAligner, a tool developed by Bionano Genomics Inc. for performing alignment using dynamic programming.
In this article, we introduce OMBlast for the alignment of optical maps. Although dynamic programming is also used in certain modules within OMBlast, our method is mainly based on a seed-and-extend approach (Altschul et al., 1990), as implied by the name of our software. By using a seed-and-extend approach, our framework is superior in speed, and provides better alignments in complex regions by joining local alignments, in comparison to the dynamic programming approaches for global alignments.
3 Methods
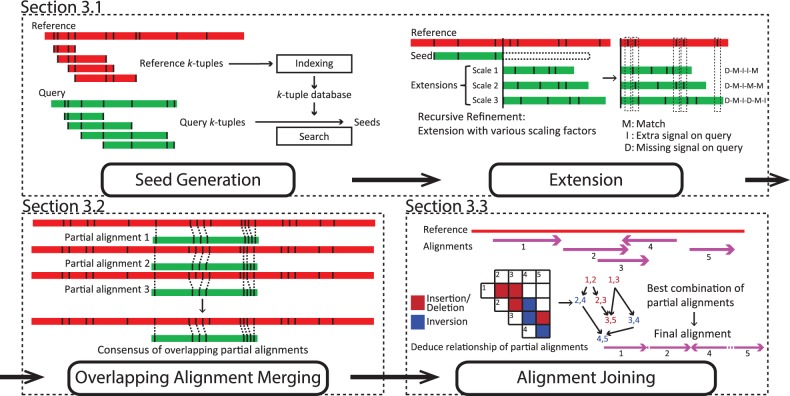
Our objective is to find the best way to locate regions on the reference optical map that is most similar to the query optical map. BLAST has been proven to be a fast and accurate seed-and-extend method for handling sequencing data (Altschul et al., 1990). We introduced some novel elements to adapt the key ideas of seed-and-extend to handle optical maps. As illustrated in Figure 1, the entire algorithm is broken down into three main modules. The first module aligns queries against the reference using a seed-and-extend approach and yields partial alignments as local matches. Next, the second module merges overlapping partial alignments into consensus partial alignments. Finally, the last module joins the consensus partial alignments into a global match of the query. In this module, merged partial alignments will be trimmed and joined for the final alignment. We explain each of the three modules below.
Fig. 1.
Overview of the OMBlast algorithm, which comprises of three modules: seed-and-extend (Details in Supplementary Figures S2-S6), overlapping alignment merging (Details in Supplementary Figures S7-S8) and alignment joining (Details in Supplementary Figures S9-S11)
3.1 Alignment
3.1.1 Seed generation phase
A basic assumption behind the seed-and-extend approach is that good alignments usually involve some aligned regions on the reference and the query that are almost identical. Therefore, the seeding module of OMBlast aims at efficiently identifying all regions on the reference that are almost identical to each region on the query, such that these matched regions can be further processed by the other modules to form complete alignments.
Specifically, efficient seeding is performed by matching k-tuples (the ‘seeds’). Consider a reference with n segments and a query with m segments. For convenience, here we represent the reference by the lengths of its segments, , following their order of occurrence on the reference. Similarly, the query is represented by its segment lengths . A k-tuple on the reference is the list of segment lengths for k consecutive segments, ri, ,…, , for any i between 1 and . The k-tuples on the query are defined in the same way. The goal of seeding is to find, for each k-tuple on the query, all matching k-tuples on the reference. A query k-tuple is said to match a reference k-tuple if all pairs of the corresponding segment lengths (qj, ri),…, match, where two segment lengths qx and ry match if they are considered sufficiently close subject to the level of errors tolerated:
| (1) |
where Ts and Tm represent the maximum scaling and measuring errors tolerated, respectively.
To find out all matching k-tuples efficiently, two searching strategies are implemented in OMBlast, namely (A) Sorted list merging and (B) Binning. Method A begins with an indexing phase whereby every reference k-tuple has its k segment lengths and segment IDs placed into k corresponding lists ((ri, i) to the first list, (, i + 1) to the second list etc.). Each list is independently sorted according to the segment lengths. With this index, to find all matches for a query k-tuple , qj is first used to perform two binary searches from the first sorted list, to find out the smallest and largest reference segment lengths that satisfy Inequalities (1). The corresponding IDs of these segments are recorded. The length of the second query segment is then used to perform a similar search from the second sorted list. Among the matching segments, only those segments (say, x) with their previous segment (x − 1) also retrieved from the previous search are recorded. This process continues until either no segments are reported in an iteration, in which case the query has no matches in the reference, or searching to all k lists has been performed, in which case all k-tuples on the reference that match the query k-tuple are obtained.
In Method B, k-tuples are converted to strings and matched k-tuple pairs are identified by looking for identical strings. Specifically, segment sizes are discretized into bins of 5 kbp, and each bin is represented by a single character. For each reference k-tuple, the segment lengths are converted to the corresponding bin characters and the resulting string is stored in a hash table. When searching for the matches of a query k-tuple, each segment length of it is first used to determine the minimum and maximum matching segment lengths based on Inequalities 1, and the characters of all the bins that overlap this range are recorded. The query k-tuple is then converted to a set of strings by considering all combinations of the recorded characters of each segment, and each string is used to search the reference hash table for identical matches. Finally, a post-filtering is performed to remove false hits, which can happen because bin characters rather than exact segment lengths were used in the searches.
Methods A and B produce identical results, and their running time is compared in Supplementary Material Section S4. Based on the results, Method B with k =3 usually achieves the best performance, and is thus set as the default option of OMBlast and used in our experiments.
For both methods, in order to reduce the amount of time spent on regions of low complexity, a query k-tuple is removed if it matches over 10 other query k-tuples.
3.1.2 Extension phase
The seeding phase produces matching query and reference k-tuples that are almost identical. The extension phase takes each matching pair and attempts to extend it to include more adjacent segments on the query and reference that are also similar. In order to perform these extensions efficiently, we assume that the factor involved in scaling errors remains constant within a single DNA molecule, as suggested previously (Reinhart et al., 2015). Based on this assumption, for a given scaling factor, we can compute the error-corrected segment length of each query segment by dividing it by the scaling factor. These corrected segment lengths are then used to extend the matching k-tuples from the seeding phase, by checking whether the accumulated segment lengths from the reference and query also match according to Inequalities (2).
| (2) |
where Tm represents the maximum measuring errors tolerated, and represent the accumulated query segment length (scaling error-corrected) and accumulated reference segment length, respectively. Since the actual scaling factor is unknown, we use a process called ‘recursive refinement’ (Supplementary Figure S4) to search for it based on the extension results. Briefly, a set of reasonable values of the scaling factor are used at the beginning to start the extension of the first extra segment. Scaling factor values that lead to poor extensions are dropped, and the remaining ones are used in the next iteration for extending another extra segment. This process is repeated until no more extensions can be performed without introducing five consecutive mismatches. The consequence of such early termination is minimal (Supplementary Figure S6) since local alignments involving different parts of a query can be further joined together, to be described in Section 3.3.
As shown later in Section 4.3, a notable benefit of using a constant scaling factor over dynamic programming (which allows the scaling factor to change flexibly from segment to segment) is the drastic reduction of running time, while alignment accuracy is still preserved.
3.2 Overlapping alignment merging
The seed-and-extend module produces local matches as partial alignments. Some partial alignments are overlapping as they are extended from different seeds in close proximity on the reference. This module aims at merging these overlapping partial alignments into consensus partial alignments. We define two partial alignments as overlapping if they are on the same orientation and share at least one matching signal pair (See Supplementary Material Section S2.2 for further details).
In order to effectively construct a consensus partial alignment that favors fewer extra and missing signals from a set of overlapping partial alignments, we convert the overlapping partial alignments into a weighted directed acyclic graph. Specifically, each matching signal pair is represented as a node in the graph. From each partial alignment with a list of m matching signal pairs , we build a directed edge between every consecutive matching signal pairs . The weight of an edge is inversely proportional to the number of missing signals , and extra signals (see Supplementary Material Section S2.2 for further details). Next, we use a dynamic programming approach that marks the cumulative weight to the nodes in a bottom-up manner. Each node’s cumulative weight is assigned the highest weight among each sum of weight of incoming edge and node, or zero if the node has no incoming edge or the highest weight is negative. After traversing all nodes in the graph, we select the node with highest weight and use backtracking to obtain the path leading to the highest weight. Collectively, matching signal pairs from the best path are taken as the consensus partial alignment.
3.3 Alignment joining
Previous modules produce consensus partial alignments as local matches for a query. Here, we aim at picking and joining these partial alignments into a complete, final alignment as the global match for the query. This module constructs a link for every two partial alignments that could be connected, and from them finds the best series of links to obtain the chain of partial alignments as the final alignment.
First, the module checks if a pair of partial alignments, px and py, could be connected based solely on their reference position, with details described in Supplementary Material Section S2.3. For every link, we assign them a connection relationship , which is taken as the penalty for connection later on. Next, a weighted directed acyclic graph is constructed with these links as nodes. Edges are automatically created for every two nodes with relationship and , as the second partial alignment of the former node is the first partial alignment of the latter node. The weight is dependent on the score of the second partial alignment and the relationship penalty. We use a dynamic programming approach to update the weight of nodes in a bottom-up manner, similar to Section 3.2. Finally, we backtrack from the node with the highest weight to obtain the path which indicates the best combinations of , where the partial alignments are selected as the components of the final alignment.
Using this approach, local partial alignments separated by high errors in the data or SVs could be joined into the global, final alignment. As demonstrated in Section 4.3, OMBlast could handle queries with SVs better, and even capture potential sites of inversion.
3.4 Score calculations
OMBlast employs a scoring system that assigns the final alignments with a quality score based on the extent of the tolerated errors. This score reflects the alignment quality that could be ranked and compared with qualities of other alignments. At first, the score for each partial alignment is calculated, followed by a combined score for the overall alignment. The score for the ith partial alignment p is calculated as:
| (3) |
where tm, tes and tms represent the score for a match, extra signal and missing signal, respectively, as provided by the user. Meanwhile, the variables um, ues and ums represent the number of matches, extra signals and missing signals, and s is the scaling factor representing the ratio of the aligned query length to the aligned reference length. Note that in the score calculation, we assume the reference is a true correct reference. When one reverses the role of reference and query, the score for partial alignment is slightly different. The score o for the overall alignment with q partial alignments is calculated as:
| (4) |
where denotes the relationship penalty between partial alignment pi and as indicated in Supplementary Figure S9. Briefly, this is composed of a constant penalty from either one of the relationships (Insertion/Deletion or Inversion), and the number of extra and missing signals between the two partial alignments. OMBlast also employs an alternative scoring scheme that takes into account uniqueness in alignments (see Supplementary Material Section S2.3.4). More information on parameter selections could be found in the software manual of OMBlast.
4 Experiments
The performance of OMBlast was evaluated against RefAligner, Valouev, SOMA and TWIN. The software versions and where these tools were obtained are summarized in Supplementary Table S2. In our experiments, we evaluated OMBlast using both simulated and real data sets.
4.1 Data sets
4.1.1 Simulated data
We downloaded the genomes of four prokaryotic and eukaryotic organisms from GenBank, namely Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans and Homo sapiens (see Supplementary Table S4 for the corresponding accession numbers). We then digested them in-silico with the nicking enzyme BspQI, which recognizes the sequence GCTCTTC. The result is the four reference optical maps summarized in Supplementary Table S5.
From each of these four reference optical maps, we extracted 1000 optical maps at random to act as queries. This procedure was performed three times, so that error bars in the graphs below represent SDs. Variations on these data sets were produced through the introduction of four different error rates (i.e. none, low, medium and high). In general, as the error rate increases, more extra and missing signals are induced and with a larger variation in scaling error. Supplementary Material Section S5.3 summarizes the details of the simulated data generation.
The SVs considered included insertions and deletions of various sizes as well as sequence inversions. Since optical mapping aims at resolving large SVs, we incorporated 50 and 100 kbp insertions and deletions, and inversions equal to half the length of the optical map (See Supplementary Material Section S5.3.2 for details). All simulation scripts and data sets are available in the Git repository.
4.1.2 Real data
Although synthetic data allow us to measure the performance of OMBlast using various error models against a known gold standard, it is important to verify our method using a real data set generated from an actual experiment.
To test how our alignment method performs on real data sets, we generated two sets of optical maps by using DNA from a sample of Acinetobacter baumannii 718 532 (Yim et al., 2015) and a sample of E. coli K-12 MG 1655. The DNA was loaded onto the Bionano Iryschip for optical mapping data generation (Supplementary Material Section S8.1). The test results of A. baumannii and E. coli are included in Section 4.3.5 and Supplementary Material Section S8.5 respectively. We also tested the alignment methods on a human data set from the individual YH produced previously (Cao et al., 2014) and the results are included in Section 4.3.6.
4.2 Analytic method of simulation results
Performance of the various alignment methods is compared using running time and curves that plot precision versus recall. All of these values are averaged across the three replicates.
We performed experiments on an Intel Xeon CPU E5-4640 (2.40 GHz) with 512 GB of main memory running the Ubuntu 14.04.2 operating system. Running time includes only the amount of time used by the alignment method (also called the ‘user time’).
As for the accuracy of alignment methods, each method returned an alignment score for each query molecule. Although the meaning and even the range of these scores varied between alignment methods, most systems returned a single score for each alignment, which we used to rank the results. Valouev provided two sets of scores—we used the one that gave the better results. The list of alignments was sorted by score such that the best alignments appeared at the top of the list.
Then, we calculated precision and recall levels at regular intervals of 10% of the entire result list to yield points on the curves. Precision was defined as the percentage of query molecules that were aligned correctly to the reference while recall was the percentage of correctly aligned results out of the 1000 molecules. More details on precision and recall analysis are described in Supplementary Material Section S3.3.
4.3 Experimental results
We tuned the default parameters for OMBlast empirically. We took into account the trade-offs between the results portrayed in precision-recall graphs and running times. The default values of OMBlast perform well, albeit not the best, for data sets with different error rates from various species. Users need not set them unless they think these values are not suitable for their purpose. Details of the preliminary experiments are described in Supplementary Material Section S4 with default values highlighted in the graphs. The default values in OMBlast were used throughout the following analysis unless otherwise mentioned. Note that the default parameters were tuned against Bionano data. However, OMBlast could also be applied on OpGen data, as depicted in Supplementary Material Section S8.6.
4.3.1 Running time
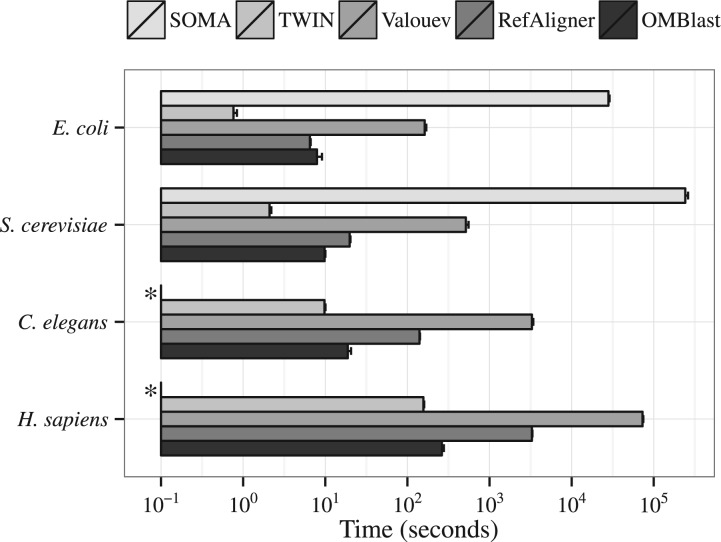
Figure 2 shows the alignment speed of the alignment methods across the four species, with the error rate fixed at medium and SVs being absent. RefAligner was about 10 times faster than Valouev across all organisms. The speed of OMBlast was similar to that of RefAligner for small organisms like E. coli but OMBlast ran faster than RefAligner as the genome size increased. For H. sapiens, OMBlast was about 10 times faster than RefAligner. SOMA was the slowest alignment method, and was 1000 times slower than OMBlast for E. coli and S. cerevisiae. Because of this, our experiments with SOMA on the C. elegans and H. sapiens data sets could not complete within a reasonable time limit. TWIN was the fastest among all alignment methods, but was only slightly faster than OMBlast for H. sapiens. TWIN achieves the higher speed by sacrificing error tolerance, which leads to much lower accuracy, as explained later in Section 4.3.3. The presence of SVs has little impact on the running time of each of the alignment methods (Results not shown).
Fig. 2.
Alignment speed (user time) of different alignment methods at medium error across four species, without any SVs. Along the horizontal axis is the user time in seconds on a logarithmic scale. (*) Results of SOMA for the C. elegans and H. sapiens data sets are missing from this graph
4.3.2 Memory
Except TWIN, memory usage of the other alignment methods was inversely correlated with the mapping speed (Supplementary Figure S23). TWIN used the least memory despite its shortest running time. OMBlast used the most memory in the alignment process.
The reported number does not reflect the true memory usage of OMBlast because OMBlast is written in Java, which performs garbage collection automatically and reclaims memory only when needed. To check the minimum memory required by OMBlast, a range of thresholds was applied to limit Java’s memory usage. The minimum memory requirement for OMBlast was 250 MB, but higher running times were observed, which could be attributed to the system being pre-occupied with garbage collection. When the memory limit was set to 5 GB, which was similar to the memory usage of RefAligner, OMBlast attained its shortest running time (Supplementary Figure S26).
4.3.3 Alignment accuracy without SVs
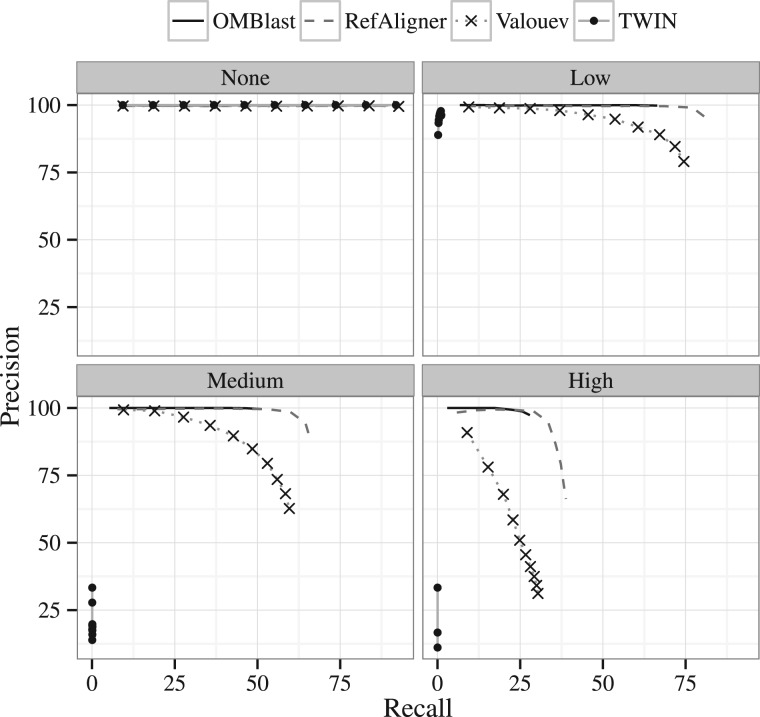
Next, we examine the performance of the various alignment methods in the absence of SVs. Due to the exceptionally long running time as described in Section 4.3.1, SOMA was only run on the E. coli and S. cerevisiae data sets. Figure 3 and Supplementary Material Section S6.2 show the precision-recall graphs on H. sapiens and other data sets respectively, across all four error rates. Intuitively, as the error rate increases, more molecules are not aligned, or are aligned to the incorrect location, lowering both precision and recall.
Fig. 3.
Precision-recall graphs for the H. sapiens data set for the four error rates. Results for SOMA are unavailable because the executions for this data set could not complete
There is only tiny difference among the five programs in the absence of errors. However, the difference in performance becomes more noticeable as error rate increases. TWIN, as indicated by Muggli et al. (2014), is not suitable for data with extra or missing signals. Its performance drops much more significantly than other alignment methods upon introduction of any error even with only measurement and resolution errors introduced. (For more details please refer to Supplementary Material Section S6.2). Valouev performs better than SOMA and TWIN in terms of both precision and recall. OMBlast and RefAligner perform the best among all alignment methods and their precision-recall graphs are very similar. The only difference appears at the end of the precision-recall graphs, where RefAligner tends to report more alignments. In this way, RefAligner attains higher recall than OMBlast, but sacrifices precision in the process.
4.3.4 Alignment accuracy with SVs
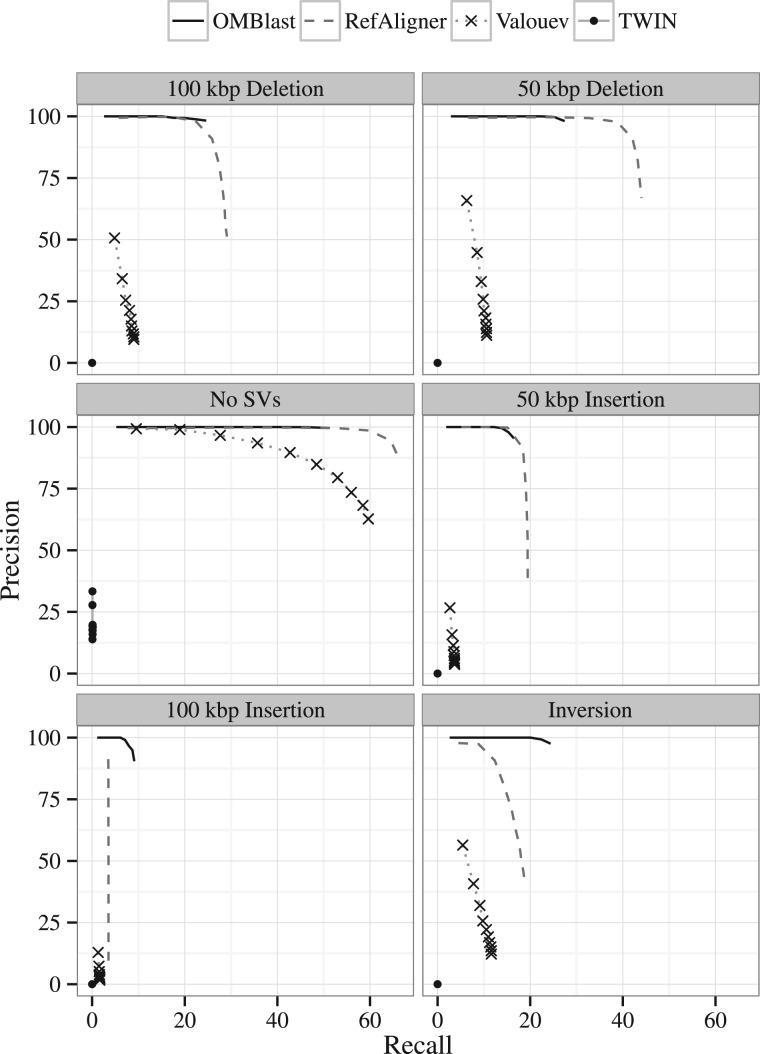
Next, we considered the performance of the alignment methods in the presence of artificially created SVs. The trade-offs between precision and recall are summarised in the precision-recall graphs of Figure 4 for the H. sapiens data set with a medium error rate and various SVs incorporated as different sub-panels. TWIN, as mentioned before, does not perform well due to the lack of error tolerance. The performance of Valouev dropped significantly upon any SVs being introduced. OMBlast and RefAligner performed the best. Like the results without SVs, RefAligner sacrifices precision for higher recall for 50 kbp insertions, 50 kbp deletions and 100 kbp deletions. Among these cases, precision of RefAligner drops even more significantly to beyond 75%. OMBlast outperforms RefAligner on both precision and recall for the 100 kbp insertion and inversion data sets.
Fig. 4.
Precision-recall graphs of the alignment methods (without SOMA) in the absence (left, center panel) and presence (all other panels) of different SVs. The other panels from left to right, top to down are: deletion of 100 kbp, deletion of 50 kbp, insertion of 50 kbp, insertion of 100 kbp and inversion. The synthetic H. sapiens data set is used, with a medium error rate
It is generally harder to achieve high recall in the presence of large insertions as compared with the presence of large deletions. This can be attributed to (i) extra signals appearing in the inserted part as noise and (ii) reduced number of signals on the molecules that come from the reference due to the inserted part. OMBlast can handle inversions with similar performance as 50 kbp deletions.
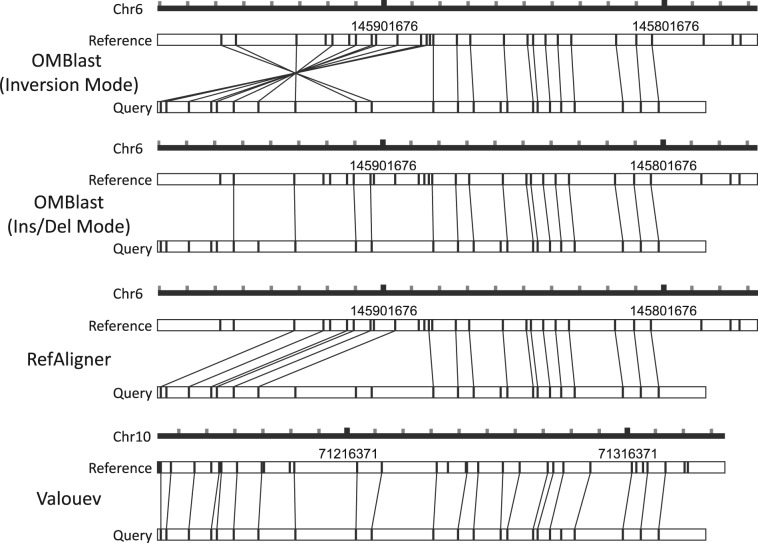
Figure 5 depicts an example of an alignment of an inversion-containing query. Valouev, RefAligner and OMBlast with Insertion/Deletion mode ‘on’ have failed to align the query correctly. Valouev aligns the query to an incorrect genomic position. Alignments of RefAligner and OMBlast with Insertion/Deletion mode ‘on’ are located at the correct genomic region, but they could not reflect the presence of an inversion in the molecule. OMBlast with Inversion mode ‘on’ provides the correct alignment with the indicated inversion.
Fig. 5.
An example of aligning simulated data in the presence of inversion. The scale represents the genomic position of the alignment and each tick mark on the scale represents 10 kbp. Only OMBlast with Inversion mode ‘on’ provides the correct alignment. Valouev aligns the query at a completely wrong genomic region. RefAligner and OMBlast with Insertion/Deletion mode ‘on’ align the query at the correct place, but the alignment is in fact incorrect and could not reflect the presence of inversion
4.3.5 A. baumannii data
Since no complete genome was available, the data were filtered (Supplementary Material Section S8.3) and aligned against the concatenated scaffolds assembled from A. baumannii 718532 (Yim et al., 2015) and the representative genome ATCC_17978 (NC_009085.1).
In the absence of an artificially created set of correct answers, we chose to compare the alignment methods with each other in a pair-wise fashion. We define consistency as the ratio of the number of consistent alignments between two alignment methods to the number of molecules with alignments that are reported by both alignment methods. Two alignments from two alignment methods are consistent if their corresponding regions on the reference and query overlap. Limitations in the comparison include false alignments not being captured, and the value is less accurate when an alignment method produces very few results. Our findings show that OMBlast and RefAligner produced highly consistent results, while SOMA had low consistency with other alignment methods (Table 1). Valouev has reported the highest number of alignments (Table 2), because it attempted to align almost all optical maps. Similar consistency values were obtained when using ATCC_17978 as the representative genome (Supplementary Material Section S8.5).
Table 1.
Consistency of pairs of optical mapping alignment methods on the concatenated scaffold of A. baumannii 718 532
| OMBlast | RefAligner | Valouev | SOMA | |
|---|---|---|---|---|
| OMBlast | 99.4% | 84.1% | 51.2% | |
| RefAligner | 87.2% | 48.5% | ||
| Valouev | 34.5% |
Percentages represent the ratio of consistent alignments to the total number of alignments reported by both alignment methods.
Table 2.
Number of alignments reported by each alignment methods on the A. baumannii data set with 4531 filtered optical maps
| No. of Alignments | 718 532 | ATCC_17978 |
|---|---|---|
| OMBlast | 1821 | 1937 |
| RefAligner | 1944 | 1745 |
| Valouev | 4495 | 4492 |
| SOMA | 333 | 281 |
| TWIN | 0 | 0 |
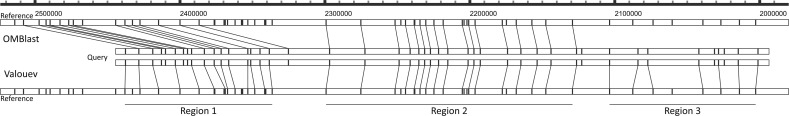
Aligning molecules with SVs with actual experimental data is exemplified by the alignment shown in Figure 6. Only OMBlast and Valouev have output alignments for the target molecule on the representative genome ATCC_17978. The alignments of OMBlast and Valouev are coherent in region 2. However, Valouev aligns region 1 by matching the query to the reference without considering the potential deletion event. OMBlast, instead, aligns region 1 to a region nearby and explicitly indicates a deletion event in the reference. Valouev attempts to align region 3, but such an alignment is of lower confidence than region 2.
Fig. 6.
The alignment of molecule 28925 (query) from the A. baumannii data set on to the ATCC_17978 in silico genome map (reference). Although RefAligner forgoes the alignment of this molecule, OMBlast and Valouev align the molecule to similar genomic regions. Three sub-regions are highlighted in the alignments. One could distinguish the alignment confidence by looking at the number of extra or missing signals, and the deviation in difference in size of segments between the query and the reference. Region 2 is consistently aligned by OMBlast and Valouev. In contrast, the alignment of Valouev in region 1 appears less correct than the alignment of OMBlast. Valouev attempts to align region 3 but such an alignment is of lower confidence
4.3.6 Human data
To investigate the performance of alignment methods on high-throughput human data, a real data set from the individual YH (Cao et al., 2014) was downloaded for alignment against the human reference genome hg38. Here we only compared the performance of OMBlast and RefAligner, because Valouev alignments could not be completed within a month. Duplicated optical maps were removed (Supplementary Material Section S8.2) and further filtering by the number of signals and length was performed (Supplementary Material Section S8.3).
To report on the accuracy, we developed a precision estimation scheme that we call precision metrics. The precision metrics estimate the precision of real results based on the relationship between alignment scores and the alignment accuracy observed from the alignment results of the simulated data in Section 4.3.3. First, alignments from the simulated data sets were sorted by descending order of alignment score and separated into 10 equally-sized bins. The accuracy of each bin can be calculated based on the number of correct and wrong simulated alignments inside. After constructing these bins, each alignment from the real data set was categorized into a bin according to their alignment score so that the accuracy of the bin could be assigned to that alignment.
Precision metrics were then calculated by summing up the assigned accuracy across the alignments. Our findings show that both alignment methods yield largely consistent results. Although OMBlast aligns fewer optical maps than RefAligner (Table 3, Total alignments) due to the more stringent default options, it attains higher estimated overall precision (Table 3, Estimated Pecision). To compare the estimated precision under the same recall level as OMBlast, we pruned the RefAligner results so that only the top 382 429 alignments with the highest score were retained. From this pruned result set, the estimated precision is very similar to that of the observed OMBlast results. The pruning of the RefAligner results illustrates the trade-off between precision and recall of the two systems.
Table 3.
Alignment statistics of OMBlast and RefAligner on the YH data set
| OMBlast | RefAligner |
||
|---|---|---|---|
| Unpruned | Pruned | ||
| Total optical maps | 1 457 446 | 1 457 446 | 1 457 446 |
| Total alignments | 382 429 | 843 768 | 382 429 |
| Consistency | 98.4% | N/A | N/A |
| Depth of coverage | 21.2% | 45.3% | 27.0% |
| Fraction of | 91.6% | 97.9% | 96.3% |
| genome aligneda | |||
| Estimated precision | 99.5 ± 0.5% | 82.0 ± 1.2% | 99.7 ± 0.2% |
| Precise alignments | 380 466 ± 1893 | 691 990 ± 9841 | 381 113 ± 775 |
| Unique alignments | 44 432 | 505771 | N/A |
| Estimated precision | 99.5 ± 0.5% | 71.5 ± 1.7% | N/A |
Precision metrics were estimated from the simulated data set.
Portion of the genome covered by at least one alignment. The percentage approaches 100% with increasing data coverage. A low percentage implies that some regions are always missed by the alignment method. Also, if the reference used in the alignment is not perfect and contains errors, a very high percentage suggests non-specific alignment at misaligned regions.
5 Discussion
We have described OMBlast, a fast alignment method aimed at aligning high-throughput optical maps while preserving overall alignment precision and recall. The underlying algorithm of OMBlast is a seed-and-extend method, where seeds are initially formed from consecutive segment lengths. Using this approach, OMBlast can retrieve local alignments from various portions of the molecules. After extension, overlapping partial alignments are merged and then joined. These two steps further link the local alignments and give a more comprehensive alignment result, which is extremely useful when handling data with SVs.
Experiments with synthetic data without SVs have shown that alignment with OMBlast gives recall levels that are just below RefAligner and Valouev. However, precision is high and running time is equal to or less than the current state-of-the-art. RefAligner and OMBlast achieve the best trade-off between precision and recall. More importantly, OMBlast has superior performance with data with insertions, deletions or inversions due to the seed-and-extend approach coupled with alignment joining.
Properly dealing with SVs for larger eukaryotic genomes is paramount—it is one of the main applications of optical mapping (Cao et al., 2014). Furthermore, assisted scaffolding of NGS contigs requires accurate alignment of contigs on optical mapping scaffolds. This underlying technique of OMBlast is crucial in handling a wide range of variations and mis-assemblies in the contigs during scaffolding. Since optical mapping assembly relies on pairwise alignment, a high mapping speed would help accelerate the process. Instead, RefAligner and Valouev do not support combination of partial alignments and thus could not handle the inverted alignment of a sub-region.
We note that our experiments included both SOMA and TWIN. Since SOMA had noticeably longer running times, it is not suitable for larger eukaryotic genomes. Also, Muggli et al. (2014) indicated that TWIN is not applicable to data with SVs. Both SOMA and TWIN are designed for sequence assembly finishing and not alignment of high-throughput optical maps, the focus of OMBlast. Of course, SOMA and TWIN have other advantages, which may influence the future direction for OMBlast. We will further demonstrate the use of OMBlast in scaffolding, especially in polyzygous genome. In addition, a more complicated relationship system will be built for alignment in complex regions to include other cases of genome arrangements. In this manuscript, we demonstrated the ability of OMBlast to align optical maps correctly and the alignment could reflect the presence of SV. Yet we did not present any comprehensive SV results. An automated, statistically-based method is still required for proper SV detection and verification of aligned optical maps as one direction for our future work. Our experiments have shown that all alignment methods have difficulties in dealing with SVs. OMBlast performs well, but the low recall clearly indicates that there remains room for improvement.
Supplementary Material
Acknowledgements
The authors would like to thank the anonymous reviewers for their insightful comments that greatly improved the article.
Funding
This work is partially supported by the Health and Medical Research Fund 12110542, RGC Collaborative Research Fund (CUHK3/CRF/11G and C4042-14G), Theme-based research schemes T12-403/11, T12-401/13R and T12-402/13-N of the Hong Kong Government, and the Lo Kwee-Seong Biomedical Research Fund and Lee Hysan Foundation.
Conflict of Interest: none declared.
References
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bradnam K.R. et al. (2013) Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H. et al. (2014) Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. GigaScience, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.K. et al. (2010) Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res., 38, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimalanta E.T. et al. (2004) A microfluidic system for large DNA molecule arrays. Anal. Chem., 76, 5293–5301. [DOI] [PubMed] [Google Scholar]
- Dong Y. et al. (2013) Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol., 31, 135–141. [DOI] [PubMed] [Google Scholar]
- Lam E.T. et al. (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol., 30, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.C. et al. (2012) AGORA: assembly guided by optical restriction alignment. BMC Bioinformatics, 13, 189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelowitz L., Pop M. (2014) Computational methods for optical mapping. GigaScience, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.M. (2013) Whole-genome mapping: a new paradigm in strain-typing technology. J. Clin. Microbiol., 51, 1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggli M.D. et al. (2014). Efficient indexed alignment of contigs to optical maps In Brown D., Morgenstern B., editors, Proceedings of the 14th International Workshop on Algorithms in Bioinformatics (WABI 2014), Volume 8701 of Lecture Notes in Computer Science, pp. 68–81. Springer Berlin Heidelberg. [Google Scholar]
- Nagarajan N. et al. (2008) Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics, 24, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart W.F. et al. (2015) Distribution of distances between DNA barcode labels in nanochannels close to the persistence length. J. Chem. Phys., 142, 064902.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J.M. et al. (2015) Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics, 16, 734.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A. et al. (2006) Alignment of optical maps. J. Comput. Biol., 13, 442–462. [DOI] [PubMed] [Google Scholar]
- Van Dijk E.L. et al. (2014) Ten years of next-generation sequencing technology. Trends Genet., 30. [DOI] [PubMed] [Google Scholar]
- Yim A.K.Y. et al. (2015) Draft genome sequence of extensively drug-resistant Acinetobacter baumannii strain CUAB1 from a patient in Hong Kong, China. Genome Announce., 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.