Abstract
Intercellular epithelial junctions formed by classical cadherins, β-catenin, and the actin-binding protein α-catenin link the actin cytoskeletons of adjacent cells into a structural continuum. These assemblies transmit forces through the tissue and respond to intracellular and extracellular signals. However, the mechanisms of junctional assembly and regulation are poorly understood. Studies of cadherin-catenin assembly in a number of metazoans have revealed both similarities and unexpected differences in the biochemical properties of the cadherin·catenin complex that likely reflect the developmental and environmental requirements of different tissues and organisms. Here, we report the structural and biochemical characterization of HMP-1, the Caenorhabditis elegans α-catenin homolog, and compare it with mammalian α-catenin. HMP-1 shares overall similarity in structure and actin-binding properties, but displayed differences in conformational flexibility and allosteric regulation from mammalian α-catenin. HMP-1 bound filamentous actin with an affinity in the single micromolar range, even when complexed with the β-catenin homolog HMP-2 or when present in a complex of HMP-2 and the cadherin homolog HMR-1, indicating that HMP-1 binding to F-actin is not allosterically regulated by the HMP-2·HMR-1 complex. The middle (i.e. M) domain of HMP-1 appeared to be less conformationally flexible than mammalian α-catenin, which may underlie the dampened effect of HMP-2 binding on HMP-1 actin-binding activity compared with that of the mammalian homolog. In conclusion, our data indicate that HMP-1 constitutively binds β-catenin and F-actin, and although the overall structure and function of HMP-1 and related α-catenins are similar, the vertebrate proteins appear to be under more complex conformational regulation.
Keywords: actin, α-catenin (a-catenin), Caenorhabditis elegans (C. elegans), cell adhesion, small-angle X-ray scattering (SAXS), X-ray crystallography, HMP-1, HMP-2, four helix bundle
Introduction
Cell-cell adhesion is a defining feature of multicellular organisms, and underlies tissue morphogenesis and homeostasis. In the adherens junction, the extracellular region of classical cadherins mediates the physical interaction between cells. The cytoplasmic domain of cadherins binds to β-catenin, which in turn binds to α-catenin, a filamentous actin (F-actin)-binding protein. The cadherin·β-catenin·α-catenin complex binds stably to actin in a force-dependent manner, thereby physically linking cadherins to the actomyosin cytoskeleton. In addition to β-catenin and F-actin, α-catenin binds to other F-actin-binding proteins including vinculin, which is recruited to nascent junctions upon a force-dependent conformational change in α-catenin, and is thought to further stabilize the linkage to the cytoskeleton (1).
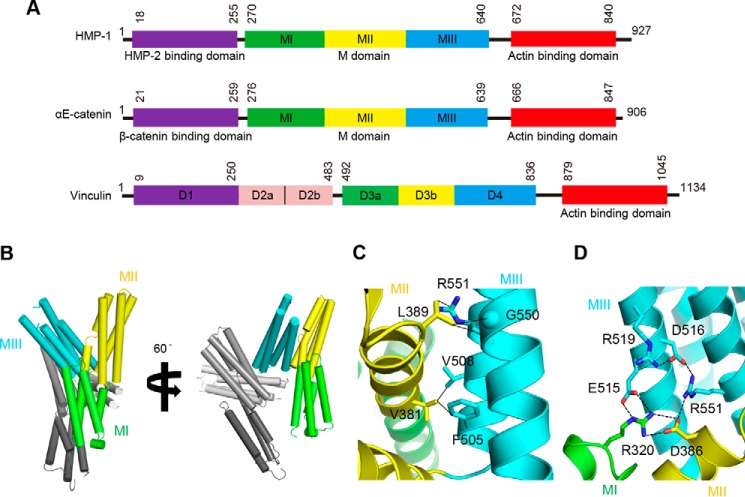
Crystal structures of mammalian αE-catenin revealed that it comprises a series of helical bundle domains (Fig. 1A) (2). The N-terminal (N) domain contains two 4-helix bundles, designated NI and NII, which share a central, long α-helix. The N domain binds to β-catenin and, in the case of mammalian αE-catenin, mediates homodimerization. This is followed by the middle (M) domain, comprised of three tandem 4-helix bundles, MI, MII, and MIII, that contain binding sites for vinculin, α-actinin, and afadin. A flexible loop connects the M domain to the C-terminal actin-binding domain (ABD).3 The structure of vinculin, another junctional protein that is a paralog of αE-catenin, is similar but contains an extra domain inserted between the N and M domains (Fig. 1A).
Figure 1.
Overall domain structure of HMP-1 and crystal structure of the HMP-1 M domain. A, domain structures of HMP-1, mouse αE-catenin, and chicken vinculin. Both HMP-1 and αE-catenin have similar domain structures, including a β-catenin-binding domain, M domain, and ABD. Domain boundaries are labeled with residue numbers. B, crystal structure of the HMP-1 M domain shown in two orientations. There are two molecules in an asymmetric unit. In one molecule, MI is colored green, MII yellow, and MIII cyan, as shown in A. In the other molecule, MI to MIII are colored from light gray to dark gray. C, non-polar interactions between MII and MIII bundles. Interacting residues are labeled and their side chains are shown as sticks. Cα atom of Gly is represented as a sphere. Non-polar interactions within 3.8 Å distance are shown with solid lines. D, polar interaction network among three 4-helix bundles. Interacting atoms within 3.4 Å distance are connected with dashed lines and each residue is labeled and its side chain is shown as sticks.
Vertebrate α-catenin functions as a sensor of mechanical force. Binding of β-catenin to the N domain weakens the affinity of vertebrate α-catenins for actin (3–6), but application of the mechanical force to αE-catenin overcomes this inhibition (7), indicating allosteric regulation of α-catenin actin-binding activity by both its binding partners and force. Tension-dependent conformational changes in αE-catenin recruit vinculin to cell-cell junctions (8, 9). Biochemical and structural data show that vinculin binds to the MI bundle, which exists in a conformational equilibrium in which its two central helices can dissociate from the bundle and bind to vinculin. This equilibrium is shifted toward the “closed,” four-helix bundle state through intramolecular interactions with the MIII domain, also known as the “adhesion modulation domain.” It is thought that mechanical force alters the relative positions of MI and MIII and thereby removes this inhibition (10, 11).
The nematode Caenorhabditis elegans has a homologous junctional complex consisting of HMR-1, a classical cadherin homolog, HMP-2, a β-catenin homolog, and HMP-1, the α-catenin homolog (12). Despite sequence homologies, the C. elegans proteins exhibit different biochemical and biophysical properties than their mammalian counterparts. The cytoplasmic domain of HMR-1 is shorter than that of classical vertebrate cadherins and binds to HMP-2 more weakly; in both cases phosphorylation of the cadherin increases affinity for β-catenin (13, 14). Most metazoans express a single β-catenin that functions in both cadherin-mediated cell adhesion and Wnt signaling, whereas C. elegans expresses several β-catenin homologs, of which only HMP-2 functions in adhesion. HMP-1 is a monomeric α-catenin homolog that binds HMP-2 and the phosphorylated HMR-1·HMP-2 complex, but it was reported that it does not bind actin in vitro, even though its ABD and association with actin are essential for its function in vivo (15). Differences in biochemical properties between mammalian αE-catenin and homologs in zebrafish, fruit fly, and cnidarians have also been described (16). Thus, understanding the biochemical function of the C. elegans proteins promises to provide insights into shared and unique roles of the cadherin·catenin complex in multicellular organisms. Here, we present structural and biochemical data on C. elegans HMP-1 and its interactions with other junctional components.
Results and discussion
Crystal structure of the HMP-1 M domain
The middle (M) domain of mammalian αE-catenin is conformationally flexible, which enables force-dependent binding to vinculin (10, 11), whereas little is known about the conformation and flexibility of the HMP-1 M domain. Therefore, we determined the crystal structure of the HMP-1 M domain to understand its structural and functional characteristics. The HMP-1 M domain (residues 270–646) was overexpressed, purified, and crystallized (18), and the structure determined at 2.4 Å resolution (Table 1).
Table 1.
Data collection and refinement statistics
| HMP-1 M domain | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9795 |
| Space group | P212121 |
| Unit cell parameters a, b, c (Å) | 74.9, 81.5, 151.4 |
| Resolution (Å) (last shell) | 50–2.4 (2.5–2.4) |
| Unique reflections | 36,804 (3,587) |
| Completeness (%) | 99.3 (98.9) |
| Multiplicity | 3.8 (3.8) |
| I/σ(I) | 15.7 (5.6) |
| Rmergea | 0.085 (0.25) |
| CC1/2 | 0.999 (0.978) |
| Refinement | |
| No. of reflections working set (test set) | 35,763 (1793) |
| Rcryst/Rfreeb | 0.20/0.25 |
| Bond length r.m.s. deviation from ideal (Å) | 0.004 |
| Bond angle r.m.s. deviation from ideal (°) | 0.77 |
| Ramachandran analysisc | |
| % Favored regions | 96 |
| % Allowed regions | 4 |
| % Outliers | 0 |
aRmerge = Σh Σi|Ii(h) − 〈I(h)〉|/Σh Σi Ii(h), where Ii(h) is the ith measurement of reflection h, and 〈I(h)〉 is the weighted mean of all measurements of h.
br = Σh|Fobs(h)| − |Fcalc(h)‖/Σh|Fobs(h)|. Rcryst and Rfree were calculated using the working and test reflection sets, respectively.
cAs defined in MolProbity.
The structure of the HMP-1 M domain consists of three 4-helix bundles, and is similar to low resolution structures of the corresponding region of αE-catenin visualized in crystals of nearly full-length αE-catenin (PDB codes 4IGG and 4K1N). The HMP-1 M domain crystals have two crystallographically independent copies in the asymmetric unit (Fig. 1B) that are structurally very similar (r.m.s. deviation = 1.2 Å for 348 Cα positions). Size exclusion chromatography verified that the M domain is a monomer in solution (data not shown). The first two 4-helix bundles, MI and MII, are connected by a continuous α helix, and the last helix of MII and the first helix of MIII are connected by a single amino acid, Thr500, which packs against Val381 and Ile384 in MII, and Phe505 in MIII.
The relative position of MII and MIII is stabilized by extensive polar and nonpolar interactions between them (Fig. 1, C and D). Phe505 and Val508 in MIII interact with Val381 in MII. The aliphatic portion of the Arg551 side chain and Cα of Gly550 in MIII engage in van der Waals interactions with Leu389 in MII. In addition to these nonpolar interactions, polar interactions also stabilize the overall MII-MIII conformation, including a salt bridge between Arg551 and Asp386 and hydrogen bonding of His512 with Asp382 and Ser385. Arg551 and Asp386 form additional polar interactions with charged residues on neighboring bundles, thereby connecting MI, MII, and MIII and stabilizing the overall M domain structure. Overall, 5 linked polar interactions lie at the core of the M domain (Fig. 1D). In addition, two more salt bridges, Asp504 (MIII)-Arg378 (MI) and Arg554 (MIII)-Asp497 (MII), stabilize a compact conformation of the M domain.
Structural and functional comparison of the M domains of HMP-1, αE-catenin, and vinculin
The overall structure of the HMP-1 M domain is more similar to that of the αE-catenin M domain (r.m.s. deviation value of 2.1 Å for 309 Cα positions) than the vinculin M domain (r.m.s. deviation value of 3.9 Å for 268 Cα positions) (Fig. 2, A and B). The D3b and D4 bundles of vinculin superimpose well with MII and MIII of HMP-1 (r.m.s. deviation = 1.3 Å for 206 Cα positions), whereas the first bundle (vinculin D3a, HMP-1 MI) is positioned differently. This appears to be caused by the presence of the vinculin D2 domain, which is absent in HMP-1 and other α-catenins. The D2 bundle makes close contacts with D3a, likely leading to a different subdomain arrangement in vinculin (Fig. 2B).
Figure 2.

Structural comparisons of HMP-1 M with the corresponding regions of αE-catenin and vinculin. Structural alignment of the HMP-1 M domain with the αE-catenin M domain (A) and the corresponding region of vinculin (B). An additional 4-helix bundle domain denoted as D2 in vinculin shown as a surface model. αE-catenin and vinculin are colored orange (A) and pink (B), respectively.
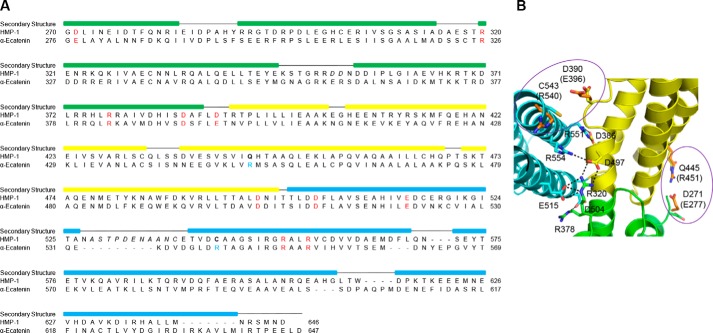
Molecular dynamics simulations and mutagenesis identified six salt bridges that stabilize the relative positions of the three M domain bundles in mammalian αE-catenin (19). Sequence alignment of the αE-catenin and HMP-1 M domains (Fig. 3A) reveals that four of six salt bridges are conserved in HMP-1: Asp497-Arg554, Arg378-Asp504, Arg320-Glu515, and Asp386-Arg551 (Fig. 3B). The other two salt bridges are not conserved, as the positively charged arginine is replaced by an uncharged cysteine in one pair (Asp390-Cys543) and by a neutral glutamine in another (Asp271-Gln445) (Fig. 3B). The four conserved salt bridges in HMP-1, together with the aforementioned polar and nonpolar interactions, appear to be sufficient to maintain the observed M domain crystal structure. This is supported by the observation that crystal structures of the MII-MIII fragment of αE-catenin (PDB codes 1H6G and 1L7C) are very different from both the full M domain structure of αE-catenin (PDB codes 4IGG and 4K1N) and HMP-1. Variation in the relative position of MII and MIII in MII-MIII fragment crystals indicates that all three bundles are required to keep the M domain in a stable conformation. This may also be due in part to the low pH of these crystals, which could disrupt stabilizing salt bridges.
Figure 3.
Sequence alignment between HMP-1 M and αE-catenin M. A, structure-based sequence alignment of the HMP-1 M domain with mouse αE-catenin M domain. Conserved residues involved in forming salt bridges are labeled in red; the italicized residues are not visible in the structure. The colored rectangular box on top of the sequence represents the helical region. B, residues in the HMP-1 M domain structure homologous to those from αE-catenin that form the six salt bridges that stabilize the αE-catenin M domain. Polar interactions are shown as dashed lines in the four conserved salt bridges. Non-conserved residues are circled and corresponding residues of αE-catenin are colored orange and labeled in parentheses.
The dynamic formation and breakage of salt bridges within the M domain are likely important for structural stability and may be related to the function of αE-catenin as a mechanosensor (19). Binding to vinculin is force dependent, and is mediated by the MI bundle, which unfolds such that its two central helices become co-linear and bind to the vinculin D1 domain (20, 21). It has been proposed that αE-catenin adopts two distinct conformations depending on mechanical stress. In the absence of tension, the vinculin-binding site in MI is largely inaccessible, as shown by the relatively weak affinity of αE-catenin for vinculin (Kd = 2 μm) (20). As force is applied to the protein, the salt bridges stabilizing the M domain structure are broken and the MI bundle is freed to interact with vinculin (19). When MIII is deleted, the KD drops to 5 nm, indicating that stabilization of the M domain by the MIII bundle inhibits vinculin binding (20).
Given the structural similarity between the HMP-1 and αE-catenin M domains, it is possible that the HMP-1 M domain undergoes open-closed conformational changes similar to mammalian αE-catenin. Such changes could regulate the accessibility of a binding site for an actin-binding protein such as vinculin, although HMP-1 and the vinculin homolog DEB-1 are not co-expressed in epithelial cells in C. elegans (22). Distinct tissue expression could reflect an evolutionary change in gene expression without an alteration in the intrinsic ability of the two proteins to interact. Alternatively, a loss of affinity between HMP-1 and DEB-1 could have ultimately led to their functional dissociation over evolutionary time. To assess the latter possibility, we tested whether the HMP-1 M domain can bind to vertebrate vinculin and/or DEB-1.
The D1 domain of vinculin, consisting of the N-terminal 259 amino acids, encompasses the full α-catenin-binding site (20). Vinculin D1 and the corresponding region of DEB-1 were expressed and purified with N-terminal GST tags for pulldown assays. Even though the DEB-1 D1 domain is highly homologous to vinculin D1 (53% sequence identity and 77% sequence similarity) (supplemental Fig. S1), DEB-1 D1 was not easily expressed as a soluble protein in Escherichia coli, and the small amount of soluble material collected from cell lysis eluted as a high molecular weight aggregate during gel filtration chromatography. To obtain soluble DEB-1 D1 protein, we introduced the point mutation C206S, which removes a non-conserved, solvent-exposed residue that should not affect overall structural integrity, nor the potential HMP-1-binding site based on a structure-based sequence alignment with the vinculin·αE-catenin complex (PDB code 4E18). DEB-1 C206S D1 was expressed successfully as a soluble protein and used for binding assays.
GST pulldown assays indicated that the HMP-1 M domain binds to GST-vinculin D1, whereas no binding was detected to GST-DEB-1 D1 (supplemental Fig. S2). Isothermal titration calorimetry (ITC) measurements showed that the HMP-1 M domain binds to vinculin D1 with 1:1 stoichiometry and a Kd of 0.60 μm (Table 2, supplemental Fig. S3), an affinity similar to that of the αE-catenin M domain for vinculin D1 (Kd = 2 μm) (20). Like the interaction between αE-catenin and vinculin, the HMP-1-vinculin interaction was endothermic, which strongly suggests that binding to vinculin involves unfurling of the MI bundle as described for αE-catenin (20). Unlike mammalian α-catenin, however, removing the HMP-1 MIII bundle did not increase affinity for vinculin (Kd = 0.57 μm; Table 2). When both MII and MIII were removed, the vinculin binding affinity to MI increased by ∼200-fold (Kd = 3 nm), essentially the same as the affinity of αE-catenin MI-MII for vinculin (Table 2).
Table 2.
ITC measurement of HMP-1 binding to HMP-2, vinculin D1, and DEB-1 D1
| Proteins | KD | ΔH | TΔS | ΔG | |
|---|---|---|---|---|---|
| nm | kcal mol−1 | ||||
| HMP-1 + HMP-2 | |||||
| HMP-1(FL) | HMP-2(13-678) | 1 | −21.5 | −9.2 | −12.3 |
| HMP-1(FL) | HMP-2(13-678)·pHMR-1(cyto80) | 3.9 | −22.9 | −11.3 | −11.6 |
| HMP-1 + vinculin | |||||
| HMP-1 M | Vinculin D1 | 600 | 18.5 | 27.0 | −8.5 |
| HMP-1 MI-MII | Vinculin D1 | 572 | 13.1 | 21.6 | −8.5 |
| HMP-1 MI | Vinculin D1 | 3.1 | 7.6 | 19.2 | −11.6 |
| HMP-1 + DEB-1 | |||||
| HMP-1 M | DEB-1 D1 | NDa | ND | ND | ND |
| HMP-1 MI-MII | DEB-1 D1 | ND | ND | ND | ND |
| HMP-1 MI | DEB-1 D1 | 150 | −21.8 | 12.5 | −9.3 |
a ND, not determined.
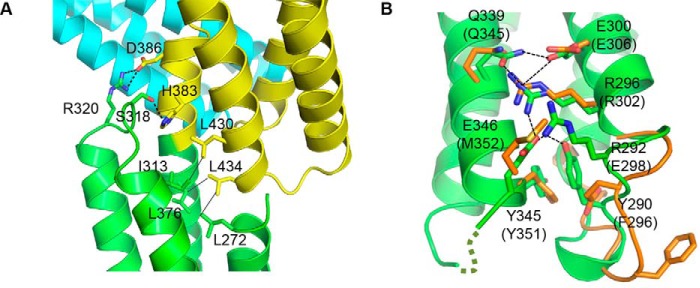
We previously showed that, unlike mammalian αE-catenin, the HMP-1 MI region is partially protected from protease digestion in full-length HMP-1 (15). That observation, together with the vinculin binding data presented here, suggest that although the underlying mechanism of binding is the same in both HMP-1 and αE-catenin, MI-MII forms a more stable structure in HMP-1 than in mammalian αE-catenin. The HMP-1 M domain crystal structure reveals that Ser318 and Arg320, located on a loop connecting the second and third helices of the MI bundle, form polar interactions with His383 and Asp386, respectively, in MII (Fig. 4A). In αE-catenin, Ser318 is replaced with Cys324, which does not contact MII. Moreover, a more extensive polar interaction network is found between the second and third helices in HMP-1 MI relative to αE-catenin. In particular, Glu346 stabilizes polar interactions between two helices through hydrogen bonds with Tyr290, Arg292, and Arg296 (Fig. 4B). In addition, the loop containing Phe296 and Glu298 in αE-catenin appears to be flexible, as it adopts different conformations in crystallographically independent structures, which places these residues in different positions that may facilitate or disrupt the inter-helical interactions, making the MI bundle more dynamic.
Figure 4.
Intramolecular interaction of HMP-1 M domain. A, a detailed view of HMP-1 M domain intramolecular interaction. The MI and MII bundles stabilize one another through polar and hydrophobic interactions. Each interacting residue is labeled and represented as sticks. Polar and non-polar interactions are shown as dashed and solid lines, respectively. B, intramolecular interactions between the second and third helices in the MI domains of HMP-1 and αE-catenin compared. Residues involved in extensive polar interactions in HMP-1 are shown as sticks and interacting atoms are connected with dashed lines. The corresponding residues of αE-catenin are shown and labeled in parentheses. Glu298 and Phe296 of αE-catenin do not structurally align well with Arg292 and Tyr290 of HMP-1, respectively.
To test whether the HMP-1 MI bundle could bind to DEB-1, GST pulldown and ITC experiments were performed using the purified HMP-1 MI bundle (supplemental Figs. S2 and S3). HMP-1 MI binds to DEB-1 D1 with Kd of 150 nm, which is 50 times weaker than its binding to vinculin (Table 2). Assuming the affinity for DEB-1 is similarly weakened by the presence of MII and/or MIII as seen in the binding to vinculin, the HMP-1 M domain would be predicted to interact with DEB-1 with Kd of ∼7.5 μm, which likely explains why no binding was observed by ITC or pulldown.
The weaker binding between HMP-1 and DEB-1 can be attributed to differences in the binding interface as well as the less dynamic structure of the HMP-1 M domain relative to that of αE-catenin. The amino acids engaged in the formation of the αE-catenin·vinculin complex are well conserved in HMP-1 and DEB-1, respectively, except for two regions (supplemental Table S1). Met352 of αE-catenin, which contacts Met26 of vinculin, is replaced by the polar residue Glu346 in HMP-1. In DEB-1, His63 replaces Thr64 of vinculin, and thereby removes the polar interaction with Arg329 present in αE-catenin (Arg323 of HMP-1). We speculate that functional disassociation may have allowed the HMP-1 M domain to evolve a distinct mechanism of regulation and/or function.
Overall architecture of full-length HMP-1
To further investigate differences between α-catenin and HMP-1, we attempted to crystallize full-length HMP-1. These attempts failed, perhaps due to the flexibility of the C-terminal ABD and the preceding linker.
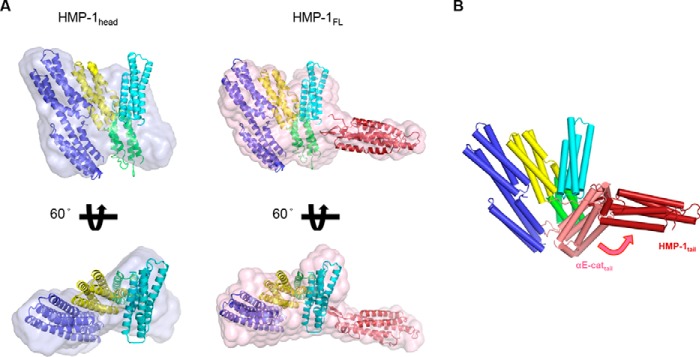
Therefore, we visualized the full-length structure of HMP-1 at low resolution by small angle X-ray scattering (SAXS) (supplemental Fig. S4, Table 3). The values of the radius of gyration (Rg) and the maximum particle size (Dmax) were 42 and 150 Å, respectively, in agreement with an earlier study (23). Comparison of the SAXS-derived envelope of full-length HMP-1 to that of the HMP-1 “head” comprising the N and M domains indicated that the protruding stalk of the envelope likely corresponds to the C-terminal ABD (Fig. 5A). HMP-1 is a monomer in solution regardless of protein concentration or temperature (15). There are no crystal structures of monomeric full-length α-catenins available. Therefore, crystal structures of the N and ABD domains of αN-catenin, as well as our HMP-1 M domain structure were used to model the full-length HMP-1 into the SAXS envelope (PDB codes 4P9T and 4K1O) (21, 24). By comparing the full-length HMP-1 envelope with that of the HMP-1 head, the tail domain was docked into the protruding portion of the envelope such that its N terminus is in reasonable proximity to the C-terminal end of MIII.
Table 3.
SAXS data collection and derived parameters
| HMP-1 head | HMP-1 FL | |
|---|---|---|
| Data collection | ||
| Instrument | SSRL 4–2 | PAL 4C |
| Detector distance (m) | 1.7 | 1, 4 |
| Wavelength(A), energy (KeV) | 1.127, 11.0 | 0.734, 16.9 |
| q-range (Å−1) | 0.007 − 0. 5 | 0.006 − 0.69 |
| Exposure time (s) | 10 × 1 | 10 × 1 |
| Temperature (K) | 288 | 277 |
| Concentration (mg/ml) | 1, 2.5, 5, 9 | 1.5, 3.5, 7.5 |
| Structural parameters | ||
| I(0) (cm−1) from Guinier | 0.11 (at 2.5 mg/ml) | 0.16 (at 1.5 mg/ml) |
| Rg (Å) from Guinier | 36 | 42 |
| Rg (Å) from P(r) | 35.45 | 42.81 |
| Dmax (Å) from P(r) | 127 | 150 |
| Software employed | ||
| Data processing | PRIMUS | PRIMUS |
| Inverse Fourier transform | GNOM | GNOM |
| ab initio modeling | GASBOR, DAMAVER | GASBOR, DAMAVER |
| Three-dimensional graphics representation | PyMOL | PyMOL |
Figure 5.
Model of full-length HMP-1. A, a HMP-1 full-length model constructed within an averaged and filtered envelope from SAXS data analysis is shown together with the SAXS envelope of the HMP-1 head in two different orientations. The HMP-1 M domain and crystal structures of the N and C domains of αN-catenin (PDB codes 4P9T and 4K1O) were fitted within the molecular envelope using the head domain of αE-catenin as a guide (PDB code 4IGG). The N and C domains are colored blue and red, respectively. B, structural alignment of the HMP-1 full-length model with one protomer of the dimeric structure of αE-catenin (PDB code 4IGG). The aligned head domain of αE-catenin is omitted and the differently positioned tail domain of αE-catenin is shown and colored salmon.
Superposition of our HMP-1 SAXS model and a crystal structure of the αE-catenin dimer lacking the first 81 residues reveal very different orientations of their respective tail domains, which pack against the M domain (Fig. 5B) (5). However, another crystal structure of full-length dimeric αE-catenin indicates that the ABD is highly mobile in solution (21). SAXS analysis of full-length αN-catenin, which exists predominantly as a monomer in solution, revealed an elongated shape with Rg = 51 Å and Dmax = 175 Å (21); the authors of that study positioned the ABD next to MIII almost linearly, leaving it freely available for F-actin binding. These overall dimensions are larger than those of HMP-1. Thus, the overall size of HMP-1 determined by SAXS is smaller than that of monomeric αN-catenin, and the C-terminal ABD appears to protrude in a relatively fixed orientation from the head, indicating a more compact structure with relatively limited flexibility of its ABD.
Full-length HMP-1 binds actin constitutively
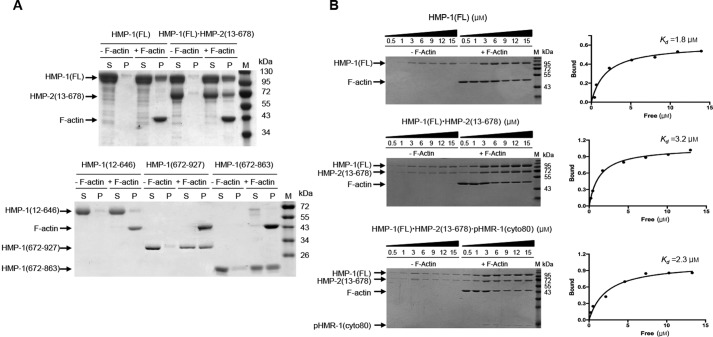
Our SAXS model of HMP-1 shows its ABD exposed to solvent, suggesting it could also bind to F-actin. We have shown that the ABD is essential for HMP-1 function in vivo (15). However, we reported previously that the purified isolated HMP-1 ABD bound actin but full-length HMP-1 did not (15). We therefore re-tested the F-actin-binding activity of several different purified HMP-1 constructs, including the full-length protein, in standard actin pelleting assays. We found that the full-length HMP-1 protein does bind actin (Fig. 6A). Sequence alignment based on the αN-catenin ABD structure indicates that HMP-1 has a 64-residue unstructured region following the ABD helical bundle. Deletion of the C-terminal 23 amino acids of the ABD, which are not conserved in other α-catenin ABDs, does not affect the ability of this domain to bind F-actin (15). We made an even shorter ABD construct in which the unstructured C-terminal tail was removed entirely (residues 672–863). This minimal ABD binds to F-actin comparably to the full ABD + C-terminal tail (residues 672–927), suggesting that the non-conserved tail region is not involved in F-actin binding. All constructs containing the minimal ABD, including full-length HMP-1 and HMP-1-ΔN (270–863; data not shown), bound to F-actin, whereas HMP-1 constructs devoid of the ABD did not (Fig. 6A), indicating that HMP-1 binds to F-actin through its ABD.
Figure 6.
Actin pelleting assays. A, F-actin binding assays were carried out with HMP-1 full-length (HMP-1(FL)), HMP-1·HMP-2 binary complex (HMP-1(FL) + HMP-2(13–678)), tailless HMP-1 (HMP-1(12–646)), and two different constructs of the ABD (HMP-1(672–927) and HMP-1(672–863)) in the presence and absence of F-actin (labeled as +F-actin and −F-actin, respectively). Equal amounts of supernatant (S) and pellet (P) fractions after high speed centrifugation were loaded onto an SDS-PAGE gel and stained with Coomassie Blue. Each protein band and actin are marked with arrows. B, F-actin pelleting assays for HMP-1(FL), a binary complex, HMP-1(FL)·HMP-2(13–678), and a ternary complex, HMP-1·HMP-2(13–678)·pHMR-1(cyto80). of the pellet was loaded for 0.5, 1, 3, and 6 μm samples of HMP-1, whereas of the pellet was loaded for 9, 12, and 15 μm samples. The sample loading amount was the same for both binary and ternary complexes, except for 6 μm, for which only of the pellet was used. Concentration of bound HMP-1 was plotted against free HMP-1 concentration using GraphPad to obtain Kd. For each protein, at least three different experiments were performed and a representative SDS-PAGE gel and saturation curve are shown.
We sought to find the cause of this experimental discrepancy between our previous report (15) and these results. We found that in some of the experiments reported in the original paper, the “full-length” HMP-1 construct had acquired an insertion in the M-ABD linker, apparently due to recombination in the bacteria used for expression of the recombinant protein. The inserted sequence created a frameshift and deleted the ABD. We verified that the constructs used in the present work contained the native sequence, so we conclude that the full-length HMP-1 protein binds F-actin.
HMP-2 binds strongly to HMP-1 and has little effect on the binding affinity of HMP-1 for actin
HMP-1 forms a ternary complex with HMP-2 and the phosphorylated cytoplasmic domain of HMR-1 (pHMR-1(cyto)) (15). The interaction between HMP-2 and non-phosphorylated HMR-1 is much weaker than that of β-catenin with E-cadherin, and only pHMR-1(cyto) forms a stable complex with HMP-2 in vitro and in vivo (13). Mammalian αE- and αN-catenins bind to a cadherin·β-catenin binary complex with Kd of ∼1 nm, whereas they bind an order of magnitude more weakly to β-catenin alone (Kd of 10–20 nm) (25). Using ITC, we tested whether pHMR-1 binding similarly increases the affinity of HMP-2 toward HMP-1. An N-terminal 12-amino acid deletion mutant of HMP-2 (HMP-2(13–678)) was used instead of full-length HMP-2 to improve solubility (13). The Kd value of the interaction between HMP-1 and HMP-2 was measured as ∼1 nm, and the binary complex of pHMR-1(cyto) and HMP-2 bound to HMP-1 with a similar Kd (Table 2, supplemental Fig. S5). Thus, the HMP-2 affinity for HMP-1 is intrinsically higher than in the mammalian system, and HMR-1 binding does not enhance the affinity of HMP-2 for HMP-1. The E-cadherin cytoplasmic domain is longer than that of HMR-1; crystal structures show that its C-terminal region forms two short helices that bind to the N-terminal armadillo repeats of β-catenin (26), and modeling suggests that it forms additional contacts with α-catenin (25). In contrast, sequence and structural data show that HMR-1 lacks this C-terminal helical region, and thus cannot form contacts with HMP-1 in the ternary complex. We speculate that selective pressure to maintain a ∼1 nm affinity complex of cadherin, β-catenin, and α-catenin, combined with the truncated tail of HMR-1, requires a higher affinity of HMP-2 for HMP-1.
We reported previously that HMP-1 forms a ternary complex with HMP-2 and phosphorylated HMR-1 (15), but that the ternary complex did not bind actin. Re-evaluation of this result showed that both the binary HMP-2·HMP-1 complex and the ternary complex of pHMR-1, HMP-2, and HMP-1 bound to F-actin with Kd values in the single micromolar range, similar to HMP-1 alone (Fig. 6B and supplemental Fig. S6). This behavior is different from the effect of β-catenin on the actin-binding activity of vertebrate α-catenins. β-Catenin binding decreases the affinity >10 times in the zebrafish cadherin·catenin complex (3), and about 2–3 times in the αT-catenin·β-catenin complex (6). The underlying structural reason for this allosteric behavior is not fully understood. However, if β-catenin binding alters actin-binding properties by conformational changes transmitted through the M domain, the relative rigidity of the HMP-1 M domain may dampen this allosteric effect. We speculate that differences in the M domain structure alter the detailed force-dependent behavior of cadherin·catenin complex binding to F-actin.
Conclusions
Knowledge of α-catenin function is essential for understanding the regulation of cell-cell adhesion as well as the evolution of multicellularity in metazoans. Our data indicate that the overall structure and function of HMP-1 and other α-catenins are similar, but more complex conformational regulation appears to have arisen in the vertebrate proteins. First, α-catenin and actin-binding proteins vinculin and α-actinin are co-expressed with α-catenin in mammalian epithelial cells, whereas their homologs in C. elegans are not (22). These additional partners may influence the properties of α-catenin through allosteric effects. Second, mechanical force is a key regulator of mammalian α-catenin, because the inhibitory effect of β-catenin on actin binding by αE-catenin is overcome by mechanical force, and force also relieves the autoinhibition of vinculin binding by the MI domain. The effects of force on the C. elegans cadherin·catenin complex remain to be established. More broadly, studies of α-catenins from cnidarians, nematodes, fish, and mammals have revealed significant functional differences despite their overall homology (16). The challenge in the future will be to understand how different properties of α-catenins relate to molecular structure as well as the mechanical environment of their respective tissues.
Experimental procedures
Protein expression and purification
Details of expression constructs for full-length HMP-1 (HMP-1(FL)), HMP-1 actin-binding domain (HMP-1(672–927)), HMP-2(13–678), HMR-1(cyto80), and chicken vinculin D1 have been reported (13, 15, 20). Constructs of HMP-1, including HMP-1 head domain (HMP-1(12–646)), HMP-1 M domain (HMP-1(270–646)), HMP-1 MI–MII (HMP-1(270–504)), HMP-1 MI (HMP-1(270–386)), and HMP-1 minimal actin-binding domain (HMP-1(672–863)) were designed based on the structure of mammalian αE-catenin (2). The DEB-1 D1 domain was subcloned into a pGEX-TEV vector and the C206S mutation was introduced using QuikChange site-directed mutagenesis (Agilent Technologies Inc.). Most constructs of HMP-1 (HMP-1(12–646), HMP-1 M, HMP-1 MI–MII, HMP-1 MI, HMP-1(672–863), and HMP-1(672–927)), HMP-2(13–678), HMR-1(cyto80), vinculin D1, and DEB-1 C206S D1 were expressed as GST fusion proteins in E. coli Rosetta (DE3) cells, and HMP-1(FL) was expressed in E. coli BL21 (DE3) pLysS cells. Cells were grown to an A600 of 0.7 at 37 °C and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 20 °C (HMP-1(FL)), 16 h at 18 °C (HMP-1(672–863) and HMP-1(672–927)), or 4 h at 30 °C (HMP-2(13–678), HMR-1(cyto80), vinculin D1, HMP-1(12–646), HMP-1 MI, HMP-1 MI–MII, and HMP-1 M). The purification method for the HMP-1 M domain was previously reported (18) and HMP-1(FL), HMP-1(12–646), HMP-1 MI, and HMP-1 MI–MII were purified using the same method. Briefly, each protein was purified using GST affinity chromatography, and the GST tag was removed on the glutathione agarose beads by tobacco etch virus (TEV) protease treatment. Each cleaved protein was further purified on a HiTrap Q anion exchange column followed by Superdex 200 size exclusion chromatography in GF buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm DTT). In the case of HMP-1(672–863) and HMP-1(672–927), the HiTrap Q purification step was skipped, and a nickel-nitrilotriacetic acid column was used to remove His6-tagged TEV protease. Each HMP-1 protein was collected from the flow-through fraction and loaded on Superdex 200 size exclusion column in GF buffer.
For GST pulldown assays, vinculin D1 and DEB-1 C206S D1 were purified as GST fusion proteins. Instead of treatment with TEV protease after GST affinity purification, each GST fusion protein was eluted with 20 mm reduced glutathione (GSH), 50 mm Tris-HCl, pH 8.0, and 50 mm NaCl, and further purified by HiTrap Q anion exchange and Superdex 200 size exclusion chromatography.
The binary complex was formed by mixing purified HMP-1(FL) and HMP-2(13–678) in an equimolar ratio. The mixture was incubated for 1 h at room temperature, then injected on to a Superdex 200 gel filtration column. HMP-1(FL) and HMP-2(13–678) coeluted from the column and the coeluted fractions were pooled, concentrated to about 5 mg/ml, and stored at −80 °C before use.
For ternary complex purification, previously reported methods were followed (13, 15). Briefly, CK1-phosphorylated HMR-1(cyto80) (pHMR-1(cyto80)) was separated from unphosphorylated samples using a MonoQ ion exchange column, and mixed with purified HMP-2(13–678). The complex of HMP-2(13–678) and pHMR-1(cyto80) was purified by Superdex 200 gel filtration column, mixed with purified HMP-1(FL) and the ternary complex purified by Superdex 200 gel filtration chromatography using GF buffer.
GST pulldown assay
Pulldown assays were performed by incubating purified 10 μm GST-tagged protein on G-agarose beads (GST-vinculin D1 and GST-DEB-1 C206S D1) with 10 μm HMP-1 M domain (HMP-1(270–386) and HMP-1(270–646)) in GF buffer. As a negative control, each HMP-1 M domain was incubated with G-agarose beads in the absence of GST-tagged protein. After a 1-h incubation at room temperature, beads were washed with GF buffer three times. Bound HMP-1 M domain was visualized by Coomassie-stained SDS-PAGE.
Actin cosedimentation assay
Rabbit G-actin (Cytoskeleton) was incubated in actin polymerization buffer (20 mm Tris-Cl, pH 7.5, 50 mm KCl, 2 mm MgCl2, 1 mm ATP) for 1 h at room temperature to prepare F-actin. Four different constructs of HMP-1 (HMP-1(FL), HMP-1(12–646), HMP-1(672–863), and HMP-1(672–927)) and HMP-1·HMP-2 binary complex were tested for their ability to cosediment with F-actin. 4 μm each HMP-1 protein was mixed with 4 μm F-actin in F-actin buffer (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 2 mm MgCl2, 1 mm ATP, 1 mm DTT) and incubated for 30 min at room temperature. To control for background sedimentation, HMP-1 proteins were incubated in the absence of F-actin and centrifuged. After centrifugation of each reaction mixture at 135,700 × g for 20 min in a TLA-110 rotor (Beckman), supernatant and pellet were separated and diluted into equal volumes of SDS sample buffers. These were loaded on a SDS-PAGE gel and visualized by Coomassie Blue staining. To calculate the binding affinity of HMP-1 to F-actin, cosedimentation assays were performed at different HMP-1 concentrations, from 0.1 to 15 μm and 2 μm F-actin. The pelleted HMP-1 band intensity of each SDS-PAGE gel lane was measured and quantified with ImageLab (Bio-Rad). To determine the fraction of HMP-1 bound to F-actin, background sedimentation of HMP-1 (no F-actin control) was subtracted first and each band intensity was then normalized to the pelleted F-actin. The amount of bound protein was calculated using the standard curve plotted by running known amounts of starting materials on a separate SDS-PAGE gel and measuring the band intensity. The same procedure was performed using HMP-1·HMP-2 binary and HMP-1·HMP-2·pHMR-1 ternary complexes. All binding data were processed with GraphPad Prism 7.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were performed at 25 °C using a Nano ITC (TA Instruments, Inc.). For measurement of binding affinity of HMP-1(FL) with HMP-2(13–678) in the absence and presence of pHMR-1(cyto80), 5–8 μm HMP-1(FL) was loaded in the cell and 80–150 μm HMP-2(13–678) or HMP-2(13–678)·pHMR-1(cyto80) complex was loaded into the injector. Each titration experiment was carried out in GF buffer by injecting 6∼7-μl samples 30–40 times with 200-s intervals. To determine the affinity between HMP-1 M domain and vinculin D1, 160 μm vinculin D1 was loaded onto the injector and 10 μm HMP-1(270–386), HMP-1(270–504), or HMP-1(270–646) was loaded onto the cell. Titration experiments were performed as described above. Thermodynamic parameters for each reaction were calculated by NanoAnalyze software.
Small angle X-ray scattering
SAXS data from the HMP-1 head domain were collected at SSRL beamline 4-2 using HMP-1(12–646) at 4 different concentrations (1, 2.5, 5, and 9 mg ml−1). SAXS data from HMP-1(FL) at concentrations of 1.5, 3.5, and 7.5 mg ml−1 were measured at Pohang Accelerator Laboratory (PAL) beamline 4C. Data collection statistics are summarized in Table 3. Each data set was analyzed after subtraction of background scattering using the ATSAS software package (27). The radius of gyration (Rg) and the maximum particle size (Dmax) for each protein were calculated using PRIMUS and GNOM, respectively (28, 29). Molecular envelopes of HMP-1 head and HMP-1(FL) were generated with the program DAMAVER by averaging the output of 10 GASBOR runs (30, 31). A full-length HMP-1 model was built with crystal structures of the HMP-1 M domain and the N-terminal and the C-terminal domains of αN-catenin (PDB codes 4P9T and 4K1O), using the crystal structure of dimeric αE-catenin as a guide.
Crystal structure determination and refinement of HMP-1 M domain
Crystallization of the HMP-1 M domain and X-ray diffraction data collection statistics were reported previously (18). The structure was solved by molecular replacement with the program PHASER, using the M domain structure of αE-catenin (276–631) (PDB code 4IGG) as a search model. Two copies were found in the asymmetric unit of the P212121 crystal. Several cycles of refinement with PHENIX (32) and manual rebuilding with COOT (33) were run to generate the final model, which consists of one copy with residues 270–352, 356–526, and 538–641 and the other with residues 270–347, 355–526, 540–606, and 609–640. The final model was validated by MolProbity (Table 1) (34). The figures were generated with PyMOL (17).
Author contributions
H. K. and I. B. conducted most of the experiments, including protein purification, X-ray crystallographic and SAXS experiments, ITC experiments, GST pulldown, and actin pelleting assays. K. S. J. assisted SAXS data collection and analysis. B. L. and J. L. conducted cloning and mutagenesis of DEB-1. X. S., J. A. H., and A. V. K. conducted actin pelleting assays and analyzed the data with H. K. and I. B. A. V. K., W. J. N., J. H., W. I. W., and H. J. C. outlined the manuscript and W. I. W. and H. J. C. wrote the paper. H. J. C. conceived and directed the study. All authors discussed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Jae Sung Woo for access to a nanoITC machine (IBS, Seoul National University) and beamline 4C and 7A staffs at Pohang Accelerator Laboratory (PAL) and Thomas Weiss at beamline 4-2 of SSRL for help on SAXS data collection. Experiments at PLS-II were supported in part by MSIP and POSTECH. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by DOE Office of Biological and Environmental Research, and by National Institutes of Health, NIGMS Grant P41GM103393.
This work was supported by a TJ Park Science Fellowship (to H. J. C.), Basic Research Laboratory Grant NRF-2014R1A4A10052590 from the National Research Foundation of Korea (to H. J. C. and J. L.), and National Institutes of Health Grants GM094663 (to W. J. N. and W. I. W.), GM114462 (to W. I. W.), GM058038 (to J. H.), and HL127711 (to A. V. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S6 and Table S1.
The atomic coordinates and structure factors (code 5H5M) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ABD
- actin-binding domain
- r.m.s. deviation
- root mean square deviation
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- SAXS
- small angle X-ray scattering
- TEV
- tobacco etch virus.
References
- 1. Ladoux B., Nelson W. J., Yan J., and Mege R. M. (2015) The mechanotransduction machinery at work at adherens junctions. Integr. Biol. (Camb.) 7, 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rangarajan E. S., and Izard T. (2013) Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller P. W., Pokutta S., Ghosh A., Almo S. C., Weis W. I., Nelson W. J., and Kwiatkowski A. V. (2013) Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J. Biol. Chem. 288, 22324–22332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drees F., Pokutta S., Yamada S., Nelson W. J., and Weis W. I. (2005) α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 123, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamada S., Pokutta S., Drees F., Weis W. I., and Nelson W. J. (2005) Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wickline E. D., Dale I. W., Merkel C. D., Heier J. A., Stolz D. B., and Kwiatkowski A. V. (2016) αT-Catenin is a constitutive actin-binding α-catenin that directly couples the cadherin·catenin complex to actin filaments. J. Biol. Chem. 291, 15687–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckley C. D., Tan J., Anderson K. L., Hanein D., Volkmann N., Weis W. I., Nelson W. J., and Dunn A. R. (2014) The minimal cadherin-catenin complex binds to actin filaments under force. Science 346, 1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M. (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 [DOI] [PubMed] [Google Scholar]
- 9. Twiss F., Le Duc Q., Van Der Horst S., Tabdili H., Van Der Krogt G., Wang N., Rehmann H., Huveneers S., Leckband D. E., and De Rooij J. (2012) Vinculin-dependent cadherin mechanosensing regulates efficient epithelial barrier formation. Biol. Open 1, 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maki K., Han S. W., Hirano Y., Yonemura S., Hakoshima T., and Adachi T. (2016) Mechano-adaptive sensory mechanism of α-catenin under tension. Sci. Rep. 6, 24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao M., Qiu W., Liu R., Efremov A. K., Cong P., Seddiki R., Payre M., Lim C. T., Ladoux B., Mège R. M., and Yan J. (2014) Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5, 4525. [DOI] [PubMed] [Google Scholar]
- 12. Costa M., Raich W., Agbunag C., Leung B., Hardin J., and Priess J. R. (1998) A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi H. J., Loveless T., Lynch A. M., Bang I., Hardin J., and Weis W. I. (2015) A conserved phosphorylation switch controls the interaction between cadherin and β-catenin in vitro and in vivo. Dev. Cell 33, 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi H. J., Huber A. H., and Weis W. I. (2006) Thermodynamics of β-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 281, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 15. Kwiatkowski A. V., Maiden S. L., Pokutta S., Choi H. J., Benjamin J. M., Lynch A. M., Nelson W. J., Weis W. I., and Hardin J. (2010) In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 14591–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller P. W., Clarke D. N., Weis W. I., Lowe C. J., and Nelson W. J. (2013) The evolutionary origin of epithelial cell-cell adhesion mechanisms. Curr. Top. Membr. 72, 267–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeLano W. L. (2002) The PyMOL Molecular Graphics System, version 1.8 Schrodinger, LLC, New York [Google Scholar]
- 18. Kang H., Bang I., Weis W. I., and Choi H. J. (2016) Purification, crystallization and initial crystallographic analysis of the α-catenin homologue HMP-1 from Caenorhabditis elegans. Acta Crystallogr. F Struct. Biol. Commun. 72, 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J., Newhall J., Ishiyama N., Gottardi C., Ikura M., Leckband D. E., and Tajkhorshid E. (2015) Structural determinants of the mechanical stability of α-catenin. J. Biol. Chem. 290, 18890–18903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi H. J., Pokutta S., Cadwell G. W., Bobkov A. A., Bankston L. A., Liddington R. C., and Weis W. I. (2012) αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U.S.A. 109, 8576–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishiyama N., Tanaka N., Abe K., Yang Y. J., Abbas Y. M., Umitsu M., Nagar B., Bueler S. A., Rubinstein J. L., Takeichi M., and Ikura M. (2013) An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barstead R. J., and Waterston R. H. (1989) The basal component of the nematode dense-body is vinculin. J. Biol. Chem. 264, 10177–10185 [PubMed] [Google Scholar]
- 23. Callaci S., Morrison K., Shao X., Schuh A. L., Wang Y., Yates J. R. 3rd, Hardin J., and Audhya A. (2015) Phosphoregulation of the C. elegans cadherin-catenin complex. Biochem. J. 472, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shibahara T., Hirano Y., and Hakoshima T. (2015) Structure of the free form of the N-terminal VH1 domain of monomeric α-catenin. FEBS Lett. 589, 1754–1760 [DOI] [PubMed] [Google Scholar]
- 25. Pokutta S., Choi H. J., Ahlsen G., Hansen S. D., and Weis W. I. (2014) Structural and thermodynamic characterization of cadherin·β-catenin·α-catenin complex formation. J. Biol. Chem. 289, 13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huber A. H., and Weis W. I. (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 27. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 [Google Scholar]
- 29. Semenyuk A. V., and Svergun D. I. (1991) Gnom: a program package for small-angle scattering data-processing. J. Appl. Cryst. 24, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.