Abstract
Bafilomycins are an important subgroup of polyketides with diverse biological activities and possible applications as specific inhibitors of vacuolar H+-ATPase. However, the general toxicity and structural complexity of bafilomycins present formidable challenges to drug design via chemical modification, prompting interests in improving bafilomycin activities via biosynthetic approaches. Two bafilomycin biosynthetic gene clusters have been identified, but their post-polyketide synthase (PKS) tailoring steps for structural diversification and bioactivity improvement remain largely unknown. In this study, the post-PKS tailoring pathway from bafilomycin A1 (1)→C1 (2)→B1 (3) in the marine microorganism Streptomyces lohii was elucidated for the first time by in vivo gene inactivation and in vitro biochemical characterization. We found that fumarate is first adenylated by a novel fumarate adenylyltransferase Orf3. Then, the fumaryl transferase Orf2 is responsible for transferring the fumarate moiety from fumaryl-AMP to the 21-hydroxyl group of 1 to generate 2. Last, the ATP-dependent amide synthetase BafY catalyzes the condensation of 2 and 2-amino-3-hydroxycyclopent-2-enone (C5N) produced by the 5-aminolevulinic acid synthase BafZ and the acyl-CoA ligase BafX, giving rise to the final product 3. The elucidation of fumarate incorporation mechanism represents the first paradigm for biosynthesis of natural products containing the fumarate moiety. Moreover, the bafilomycin post-PKS tailoring pathway features an interesting cross-talk between primary and secondary metabolisms for natural product biosynthesis. Taken together, this work provides significant insights into bafilomycin biosynthesis to inform future pharmacological development of these compounds.
Keywords: antibiotics, microbiology, natural product biosynthesis, polyketide, secondary metabolism
Introduction
Plecomacrolides are a subgroup of polyketides that contain a 16- or 18-membered macrolactone ring connected to a 6-membered hemiacetal ring via a three-carbon linker. Among the growing number of plecomacrolides such as hygrolidin, concanamycins, bafilomycins, leucanicidin, and formamicin (1), bafilomycins with a 16-membered ring have attracted significant attention due to their diverse bioactivities such as antifungal (2), antibacterial (3), antitumor (4), immunosuppressive (5), antiparasitic (5), and anti-osteoporotic (6) potency.
Bafilomycins are mainly produced by Streptomyces. For example, bafilomycin A1 (1), C1 (2), and B1 (3) were first isolated from Streptomyces griseus sp. sulfurus (TÜ 1922) in 1983 (7). Bafilomycin D and E were isolated from S. griseus TÜ 2599 (i.e. DSM 2610) in 1985 (8). Later, an endophytic microorganism Streptomyces sp. YIM56209 was reported to produce 9-hydroxy-bafilomycin D, 29-hydroxy-bafilomycin D, and a number of known bafilomycin compounds (9). From 2004 to 2011, bafilomycin F-K and other novel bafilomycin derivatives were discovered from different Streptomyces spp. (10–13).
Among these natural bafilomycin derivatives, 1 is of particular importance because it is the structural core of all bafilomycins. This compound has been characterized to be a specific inhibitor of vacuolar H+-ATPase (V-ATPase), which has been known as an important drug target for both osteoporosis and tumor metastasis (6, 14, 15). However, the high toxicity of 1 prevents it from clinical application (14). Despite continuous efforts on total synthesis and semi-synthesis of bafilomycin derivatives (16–18), their general toxicity has not been substantially reduced so far. One important reason is that the structural complexity (multiple chiral centers and many functional groups with similar reactivity) of bafilomycins largely limits the extensive chemical modification of their structures to explore more comprehensive structure-activity relationship. The existence of dozens of natural bafilomycin derivatives, together with the recent success of using a sugar-O-methyltransferase (LeuA) and a glycosyltransferase (LeuB) to decorate the structure of 1 (19), demonstrates the effectiveness of biosynthetic systems in structural diversification of these polyketides (6). To take advantage of the highly selective and eco-friendly biocatalysts to generate novel bafilomycin analogues for drug development, it is demanded to elucidate the bafilomycin biosynthetic pathway and understand the characteristics of all tailoring enzymes.
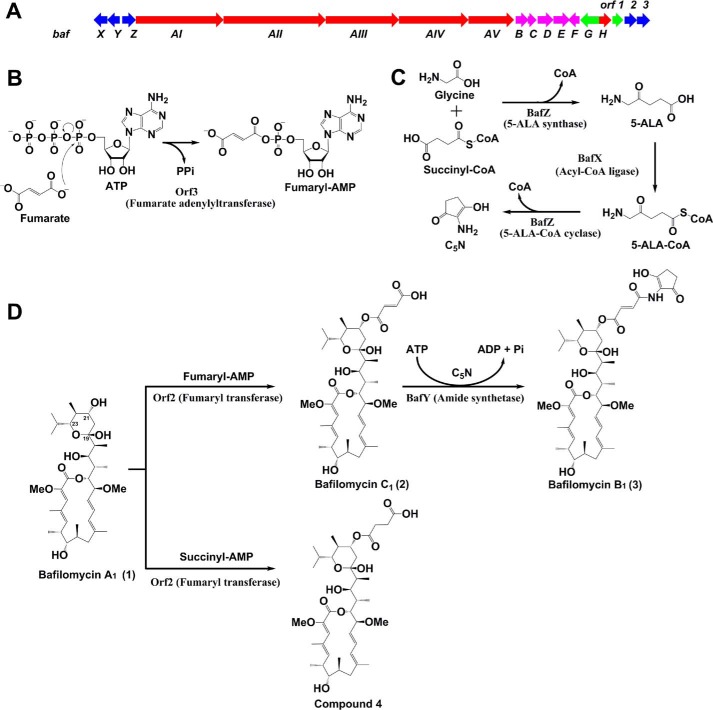
In previous studies (20, 21), our laboratory and others have identified two bafilomycin biosynthetic gene clusters from Streptomyces lohii ATCC BAA-1276 and S. griseus DSM 2608, respectively. As proposed, 1 is assembled by five type I polyketide synthases (PKSs)2 (BafAI-AV) sequentially. Subsequently, a fumarate moiety is transferred to the C21-hydoxyl group of 1 to afford 2. Finally, the 2-amino-3-hydroxycyclopent-2-enone (C5N) unit derived from glycine and succinyl-CoA is linked to the fumarate moiety via an amide bond giving rise to 3 (Fig. 1). However, all these putative biosynthetic steps lack of experimental evidence and the responsible enzymes remain largely unknown. The only exception is that we identified BafZ to be a 5-aminolevulinate (5-ALA) synthase in a previous study (20).
Figure 1.
A, the bafilomycin biosynthetic gene cluster (baf) of S. lohii (red, PKS and TEII genes responsible for 1 formation; blue, tailoring enzyme genes; magenta, the genes for synthesis of the methoxymalonyl-CoA extender unit; green, regulatory genes). B, the adenylation of fumarate by Orf3. C, the biosynthesis of C5N by BafX and BafZ. D, the post-PKS tailoring steps in bafilomycin biosynthetic pathway.
Herein, we elucidate the complete post-PKS tailoring steps of bafilomycin biosynthetic pathway for the first time through in vivo gene inactivation and in vitro reconstitution of enzyme activities. Biochemically, Orf3 is characterized as a novel fumarate adenylyltransferase showing a substrate preference for fumarate over a variety of other short chain organic acids; Orf2 with high substrate specificity is responsible for transferring the fumarate moiety from fumaryl-AMP to 1, giving rise to 2; BafX and BafZ cooperatively synthesize the C5N unit, and BafY catalyzes the amide bond formation between 2 and C5N, finishing the bafilomycin biosynthetic pathway of S. lohii.
Results
Re-interrogation of the baf gene cluster for post-PKS tailoring enzymes
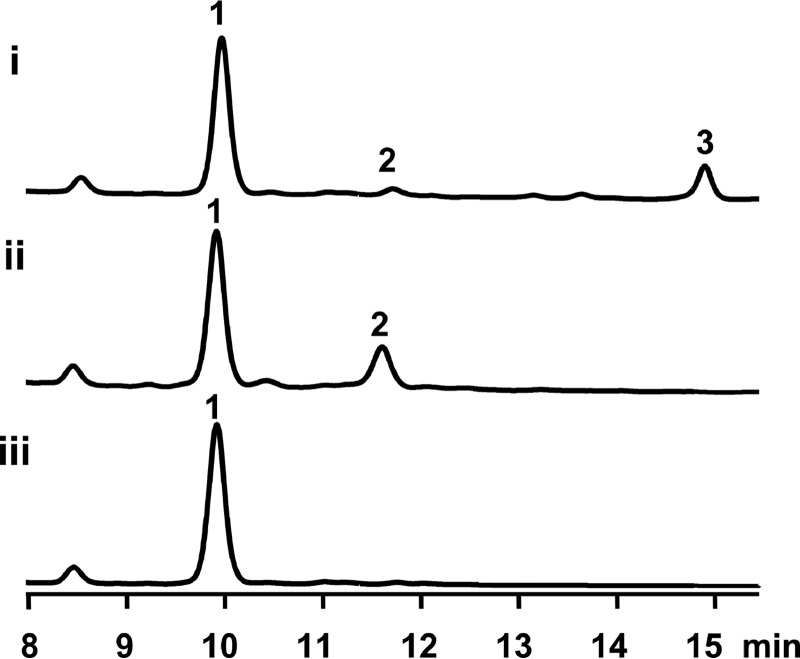
In our previous study (20), because we did not identify any apparent candidate ORFs that could encode an enzyme mediating the attachment of fumarate moiety, it was surmised that BafY might be a bifunctional enzyme, which not only mediates the fumarate attachment via an ester bond formation (1→2), but also catalyzes the amide bond formation between 2 and C5N (2→3). To test this hypothesis, the suicide knock-out vector pCIMt002-ΔbafY was constructed and used to transform S. lohii by interspecies conjugation to replace bafY in-frame with the apramycin resistance gene aac(IV) by following the protocol of Chen et al. (22) (supplemental Fig. S1A), the double-crossover mutants with bafY removed were readily screened by picking up white colonies from blue colonies indicative of undesired single-crossover mutants. After PCR confirmation of the genotype (supplemental Fig. S1B), the bafY inactivation mutant (ΔbafY) was cultured in parallel with the wild-type strain. Unexpectedly, the mutant strain ΔbafY only lost the ability to produce 3 (Fig. 2, trace i and ii), strongly suggesting that BafY should only be involved in the transformation from 2 to 3, but unrelated to the fumarate transferring step.
Figure 2.
HPLC analysis (254 nm) of the fermentation culture of wild-type and mutant S. lohii strains. Trace i, wild-type; trace ii, ▵bafY; trace iii, ▵orf2&orf3.
Generally, the genes involved in the biosynthesis of a given Streptomyces secondary metabolite are organized in a gene cluster. To find out the enzyme responsible for the initial tailoring step, we reanalyzed the baf gene cluster (Fig. 1A) with the GenBank accession number of GU390405.1, and located two uncharacterized genes including orf2 and orf3 at the 3′ end of this cluster, which were previously proposed to encode a malonyl transferase and a CoA ligase, respectively, simply based on bioinformatics analysis (20, 21). Because the transformation from 1 to 2 likely requires a transferase activity, we hypothesize that Orf3 could activate fumarate by ligating a CoA moiety, and Orf2 might be responsible for transferring the fumaryl group to the C21 hydroxyl group of 1. Orf1 is homologous to the LuxR-family transcription regulator, therefore, it is unlikely to be an enzyme. Thus, we in-frame replaced the orf2 plus orf3 region with aac(IV) (supplemental Fig. S1C). The mutant stain Δorf2&3 only produced 1 (Fig. 2, trace iii) without any accumulation of 2 and 3. This result indicates that Orf2 and/or Orf3 are required for the initial tailoring of 1. However, their exact functionality requires further biochemical investigation (see below).
Protein expression and purification of Orf2, Orf3, BafX, BafY, and BafZ
To characterize the in vitro activity of Orf2, Orf3, BafX, BafY, and BafZ, and reconstitute the entire bafilomycin post-PKS tailoring pathway, the coding sequences of the four genes (except bafZ that was cloned previously (20)) were PCR amplified and inserted into the pET28b vector, respectively. The recombinant proteins of Orf2, BafX, BafY, and BafZ with the N-terminal His6 tag were successfully overexpressed in Escherichia coli BL21 (DE3), and purified by nickel-nitrilotriacetic acid chromatography (Fig. 3A). However, the recombinant Orf3 turned out to form inclusion bodies. Significant efforts, including optimization of expression conditions, making different expression constructs, use of molecular chaperones, and different E. coli host cells carrying extra rare codon tRNA genes, failed to solve the problem of insolubility (data not shown). This led to a reconsideration of the orf3 annotation. Careful BLAST analysis of Orf3 and its homologous proteins revealed that several putative proteins, such as BafCI from S. griseus DSM 2608 (21) with >90% protein sequence identity to Orf3, have approximately 20 more amino acids at the N terminus compared with the original Orf3 (supplemental Fig. S2). This finding suggests that orf3 is likely to be mis-annotated previously, and the N-terminal truncation is probably the reason for the insolubility of Orf3. Re-inspection of the 5′ upstream sequence of orf3 resulted in the discovery of another possible start codon, which is 81 bp upstream of the previously assigned start codon. Interestingly, this GTG start codon overlaps with the TGA stop codon of Orf2 (Fig. 3B), which suggests Orf2 and Orf3 are likely to be co-transcribed, co-translated, and functionally related. When the re-annotated orf3 was cloned into pET28b and expressed, Orf3 became soluble and was successfully purified by His tag affinity chromatography (Fig. 3A). Gel filtration results indicated that Orf3, BafX, and BafZ predominantly dimerize in solution, whereas Orf2 and BafY form monomers (supplemental Fig. S3).
Figure 3.
A, SDS-PAGE analysis of recombinant BafX, BafY, BafZ, Orf2, and Orf3 purified from E. coli BL21 (DE3). B, the re-annotation of the coding sequence of Orf3. Note: the black and green letters denote partial C-terminal coding sequence of Orf2 and partial N-terminal coding sequence of Orf3, respectively. The stop codon of Orf2 is marked in red. The underlined GTG codons in green and blue represent the corrected and the previously mis-annotated start codons of Orf3, respectively.
In vitro characterization of Orf2 and Orf3
According to the in vivo orf2&3 knock-out result (Fig. 2) and the fact that these two genes overlap each other, Orf2 and Orf3 are likely co-responsible for the transformation from 1 to 2. To dissect their catalytic activities, we decided to investigate these two enzymes separately. We and others previously assigned Orf3 to be a putative CoA-ligase based on BLAST searches (20, 21). In this study, more detailed bioinformatics analysis found that the protein sequence of Orf3 shows >30% identity with a number of identified arylcarboxylate adenylating enzymes including GriC (23), EsmB1 (24), and EphF (25). Protein sequence alignment of these enzymes (supplemental Fig. S4) revealed that Orf3 exhibits many hallmarks as a typical adenylating enzyme of the ANL family, which includes CoA ligases, adenylation domains of nonribosomal peptide synthetases, and firefly luciferase (26). Specifically, the crucial nucleotide recognition sites of adenylating enzymes (27, 28) are well conserved in Orf3, including Asp327, Ile345, Tyr259, and the motif 234SGT236. The carboxyl group of Asp327 may coordinate with the 2′- and 3′-hydroxyl groups of the nucleotide sugar. Residues 97SGGTTGTP104 of Orf3 likely forms the p-loop (phosphate-binding loop) that is a conserved motif in many ATP-binding enzymes (26). The substrate-binding pocket analogous to EphF is also present in Orf3: the conserved stretches of 259YGNT262 and 145HLIG148 are expected to be involved in substrate binding, whereas several arginine residues including Arg121, Arg122, Arg127, and Arg152 might make polar contacts with the carboxylate group of fumarate (24). Importantly, there does not exist any CoA-binding sites in the protein sequence of Orf3, which suggests this enzyme is more likely to be a fumarate adenylyltransferase rather than a fumaryl-CoA ligase.
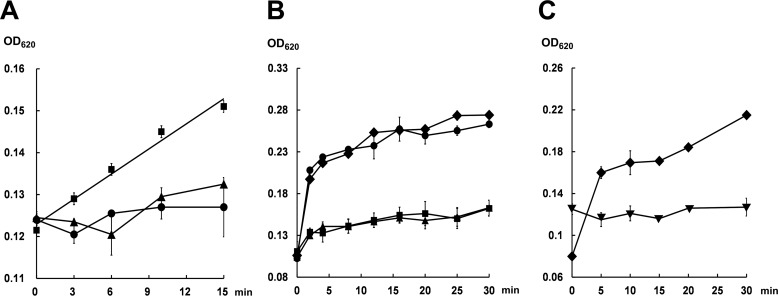
To test the in vitro activity of Orf3, the reaction mixture containing 10 μm Orf3, 5 mm disodium fumarate, 10 mm MgCl2, 0.4 units/ml of inorganic pyrophosphatase in Tris-HCl buffer, pH 7.4, was initiated by adding 5 mm ATP. Upon a 15-min incubation at 28 °C, 32.0 ± 0.5 μm phosphate (Pi) released from pyrophosphate (PPi) by pyrophosphatase was detected using the malachite green phosphate assay (Fig. 4A), corresponding to presumable formation of 16.0 μm fumaryl-AMP (29), which is impractical to be purified because of the high instability. Indeed, AMP (presumably derived from spontaneous hydrolysis of fumaryl-AMP) instead of fumaryl-AMP was detected in an Orf3 reaction mixture (supplemental Fig. S5). According to previous study (30), the low turnover number is likely due to the slow release of fumaryl-AMP from Orf3 when the downstream acceptor enzyme is absent. Notably, no significant monophosphate release was observed when pyrophosphatase or disodium fumarate was omitted in the assay (Fig. 4A), suggesting that Pi was solely derived from PPi as the co-product of fumaryl-AMP.
Figure 4.
Activity measurements (620 nm) of Orf3 and BafY using the malachite green assay. A, the reaction containing 10 μm Orf3, 5 mm disodium fumarate, 5 mm ATP, 10 mm MgCl2, and 0.4 units/ml of inorganic pyrophosphatase in Tris-HCl buffer, pH 7.4 (28 °C) (■); the control reaction with no addition of disodium fumarate (●); and the control reaction without pyrophosphatase (▴). B, the reaction containing 5 μm BafY, 200 μm 2, 5 mm ATP, 10 mm MgCl2, and 0.4 units/ml of inorganic pyrophosphatase in Tris-HCl buffer, pH 7.4 (28 °C) (■); the reaction with no addition of 2 (●); the reaction in the absence of pyrophosphatase (▴); and the reaction without both 2 and pyrophosphatase (♦). C, the same BafY reactions in the presence of 5 mm ATP (♦) or 5 mm ADP (▾) and 10 mm MgCl2.
To determine the substrate specificity of Orf3, a select group of organic acids including malonate, succinate, butyrate, maleate, and glutarate were kinetically evaluated using fumarate as control (Table 1, supplemental Fig. S6-S10). As expected, Orf3 showed a strong substrate preference toward fumarate with the kcat/Km value of 59.4 × 10−3 mm−1 min−1. Succinate was determined to be the suboptimal substrate (kcat/Km = 3.9 × 10−3 mm−1 min−1) perhaps due to its high structural similarity to fumarate. Malonate, glutarate, maleate, and butyrate were all poor substrates, suggesting that the carbon chain length, the configuration of the C=C double bond, and the dicarboxylic acid feature are crucial for substrate recognition of Orf3. Collectively, Orf3 is a novel fumarate adenylyltransferase.
Table 1.
Steady-state kinetic parameters of Orf3 toward different substrates
| Substrate | Km | kcat × 10−2 | kcat/Km× 10−3 |
|---|---|---|---|
| mm | min−1 | mm−1 min−1 | |
| Fumarate | 3.2 ± 0.6 | 19.0 ± 1.0 | 59.4 |
| Succinate | 19.2 ± 2.2 | 7.5 ± 0.3 | 3.9 |
| Malonate | 24.1 ± 3.8 | 4.4 ± 0.3 | 1.8 |
| Glutarate | 32.5 ± 8.7 | 3.4 ± 0.4 | 1.0 |
| Maleate | 29.7 ± 4.9 | 3.5 ± 0.2 | 1.2 |
| Butyrate | NDa | ND | ND |
a ND, not determined due to extremely low activity.
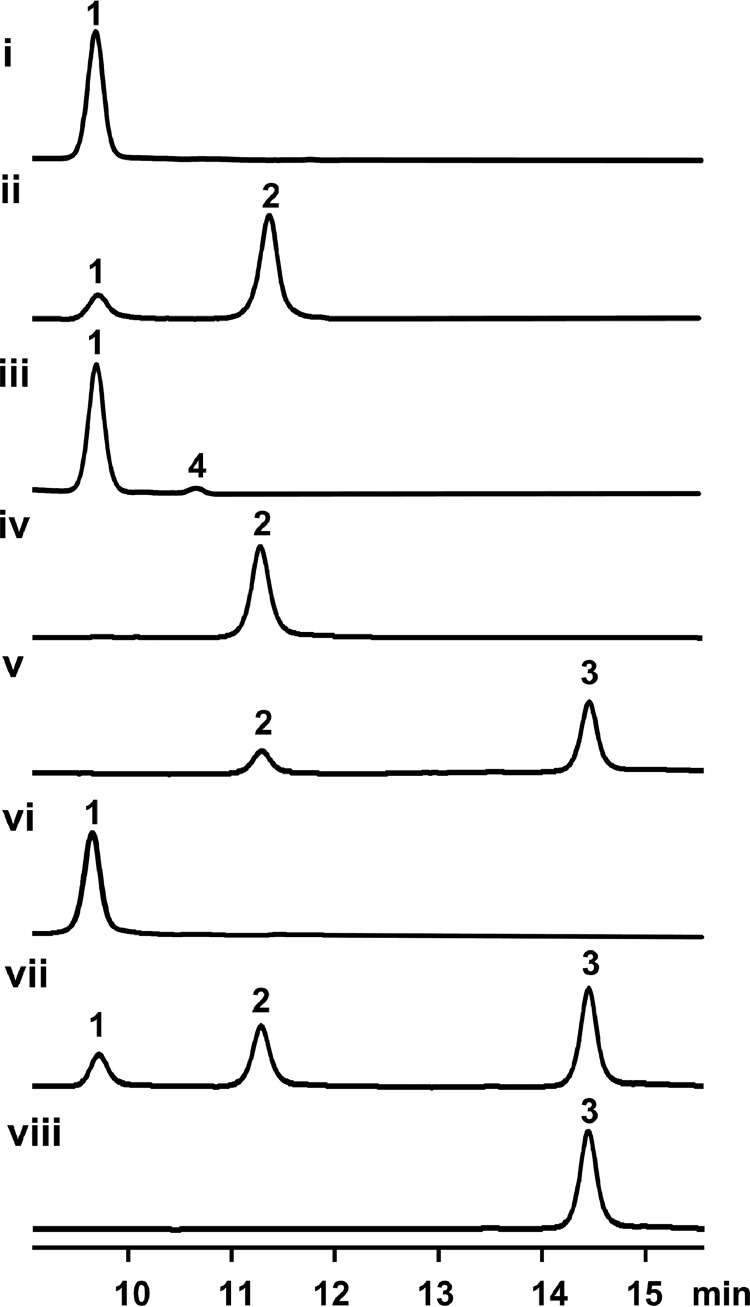
Because fumaryl-AMP is the product of Orf3, we hypothesized that 1 and fumaryl-AMP could be co-substrates for the transferase Orf2. Thus, the in vitro transformation from 1 to 2 was set up to contain 10 μm Orf2 and Orf3, 80 μm 1, 5 mm disodium fumarate, 5 mm ATP, and 10 mm MgCl2 in Tris-HCl buffer, pH 7.4. Upon an incubation at 28 °C for 4 h, 83.1 ± 3.2% (calculated from substrate consumption) of 1 was converted into 2 (Fig. 5, trace ii). This result clearly indicates that Orf2 and Orf3 cooperatively synthesize 2. Moreover, it was proven that Orf3 is not a fumaryl-CoA ligase as previously proposed (20, 21) because the conversion remained unaffected with or without CoA, and fumaryl-CoA was not observed by LC-high resolution mass spectra (HRMS) (data not shown).
Figure 5.
HPLC analysis (254 nm) of in vitro biochemical conversions of bafilomycins. Trace i, the negative control for trace ii, in which Orf2 and Orf3 were boiling inactivated; trace ii, in vitro transformation from 1 to 2 by Orf2 and Orf3; trace iii, in vitro conversion from 1 to 4 by Orf2 and Orf3; trace iv, the negative control for trace v, in which BafX, BafY, and BafZ were boiling inactivated; trace v, in vitro transformation from 2 to 3 by BafX, BafY, and BafZ; trace vi, the negative control for trace vii, in which all five tailoring enzymes were boiling inactivated; trace vii, one-pot transformation from 1→2→3 by Orf2, Orf3, BafX, BafY, and BafZ. Trace viii, 3 authentic standard. Note: all reactions were carried out at 28 °C for 4 h. The concentration of all used enzymes was 10 μm.
Next, we assessed the substrate specificity of Orf2 by determining if this enzyme is capable of incorporating alternative organic acids into the bafilomycin macrolactone scaffold. In the same assay (see above) using Orf2 and Orf3 as biocatalysts, fumarate was substituted by malonate, succinate, butyrate, maleate, or glutarate. As results, except for a small amount (2.2 ± 0.1% conversion rate) of succinyl derivative (4) (Figs. 1D and 5, trace iii, supplemental Fig. S11 and Table S1), no other derivatives were detected by LC-HRMS. Enhanced amounts of Orf2/Orf3, increased concentrations of alternative organic acids, and prolonged reaction time did not generate any detectable novel bafilomycin derivatives (data not shown). Because Orf3 is able to convert all these acids into corresponding adenylated intermediates in varying efficiency (Table 1), we deduce that Orf2 should be a strict fumaryl transferase with marginal succinyl transferase activity.
In vitro characterization of the ATP dependence of BafY
The in vivo inactivation of bafY led to accumulation of 2 (Fig. 2, trace ii), which strongly suggests that BafY is involved in the final step of bafilomycin biosynthesis (Fig. 1D). According to bioinformatics analysis (supplemental Fig. S12), BafY is proposed as an ATP-dependent amide synthetase because it shows >41/56% protein sequence identity/similarity to several identified ATP-dependent amide synthetases including MoeB4 (31), SimL (32), AusD1 (33), and Orf33 (34). It was reported that these enzymes activate their substrates via AMPylation (i.e. adenylation). Because direct LC-MS detection of the activated form of 2 was unsuccessful likely due to the high instability, the in vitro activity of BafY was indirectly measured using the malachite green phosphate assay. Specifically, 5 μm BafY was incubated with 200 μm 2, 5 mm ATP, 10 mm MgCl2, and 0.4 units/ml of inorganic pyrophosphatase at 28 °C for 30 min. As expected, Pi was continuously released as reflected by the increasing A620 values. Strikingly, the amounts of released Pi at different time points seemed to be unaffected by the addition of pyrophosphatase (Fig. 4B), strongly suggesting there was no PPi released. This is further supported by no detection of AMP during the BafY reaction (supplemental Fig. S5). Instead, ADP accumulation was observed in the reaction mixture (supplemental Fig. S5), and ADP cannot be further hydrolyzed by BafY (Fig. 4C). These results clearly indicate that BafY should activate 2 by forming an acyl phosphate intermediate as does a glutamine synthetase (35, 36). Interestingly, BafY indeed shows reasonable protein sequence identity/similarity (32/47%) to the glutamine synthetase of Streptomyces coelicolor (37, 38). In the absence of 2, again, the Pi releasing rates in the presence and absence of pyrophosphatase are quite similar to each other (Fig. 4B). Remarkably, these rates were significantly higher than those in the presence of 2, demonstrating a better ATPase activity of BafY than its phosphoryl transferase activity. This could result from the slow dissociation of the phosphorylated intermediate from BafY when C5N (see below) is absent. Taken together, BafY is confirmed to be an ATP-dependent amide synthetase with both phosphoryl transferase and ATPase activity (supplemental Fig. S13).
In vitro conversion of bafilomycin C1 (2) to bafilomycin B1 (3)
The transformation from 2 to 3 is analogous to the post-PKS tailoring of ECO-02301, which has been elucidated to be co-mediated by Orf33, Orf34, and Orf35 (34). Because BafX (acyl-CoA ligase), BafY (amide synthetase), and BafZ (dually function as 5-ALA synthase and 5-ALA-CoA cyclase) are 62/72%, 40/55%, 68/80% identical/similar to Orf35, Orf33, and Orf34, respectively, we propose a biosynthetic route leading to 3 (Fig. 1, C and D), the end product of bafilomycin pathway. First, the pyridoxal 5-phosphate-dependent 5-ALA synthase BafZ catalyzes the condensation of succinyl-CoA and glycine to yield 5-ALA, which has been confirmed in our previous work (20). Second, 5-ALA is activated by the acyl-CoA ligase BafX to form 5-ALA-CoA. Third, BafZ acts again to cyclize 5-ALA-CoA to afford C5N, avoiding the non-enzymatic cyclization to the 6-membered ring 2,5-piperidinedione (34). Finally, the ATP-dependent amide synthetase BafY is responsible for mediating the phosphorylation of the carboxylic group of 2 and the following condensation with the amino group of C5N to afford 3 (20).
To accomplish the above proposed biotransformation, the in vitro assay containing 10 μm BafX, BafY, and BafZ, 80 μm 2, 2.5 mm CoA, 2.5 mm 5-ALA, 150 μm pyridoxal 5-phosphate, and 5 mm ATP was carried out at 28 °C for 4 h. As expected, 56.1 ± 4.3% (based on substrate consumption) of 2 was converted to 3 (Fig. 5, trace v). Finally, we attempted to in vitro reconstitute the whole pathway from 1 to 3 by setting up a one-pot reaction, which contains 10 μm of all involved enzymes including Orf2, Orf3, BafX, BafY, and BafZ, 80 μm 1, and all required co-substrates and cofactors. As expected, 2 and 3 were successfully detected by HPLC analysis (Fig. 5, trace vii).
Discussion
Based on the structures of 1, 2, and 3, the bafilomycin post-PKS tailoring pathway of 1→2→3 can be readily proposed (Fig. 1). However, it has not been confirmed. More importantly, the involved biosynthetic enzymes remain unclear. In this work, these problems are solved with no ambiguity.
The post-PKS tailoring pathway is physiologically important because 2 and 3 demonstrate enhanced antifungal and antibacterial activities compared with 1 (3). According to the structure-activity relationship, the fumarate moiety leads to the improvement of bioactivities, and 3 acquires further potency from the C5N appendix. These facts may provide a reasonable explanation for the fumarate preference of Orf3 and the high substrate specificity of the fumaryl transferase Orf2, without which a variety of other organic acids could be installed onto 1. This would divert the metabolic flux to multiple bafilomycin derivatives, thereby lowering the yield of 3 that holds the highest biological activity. To derivatize bafilomycin structures for drug development, the substrate spectrum of Orf3 and Orf2 needs to be expanded. This requires understanding of the structural basis for substrate selectivity of the fumarate adenylyltransferase Orf3 and the fumaryl transferase Orf2, which is currently ongoing in our laboratories.
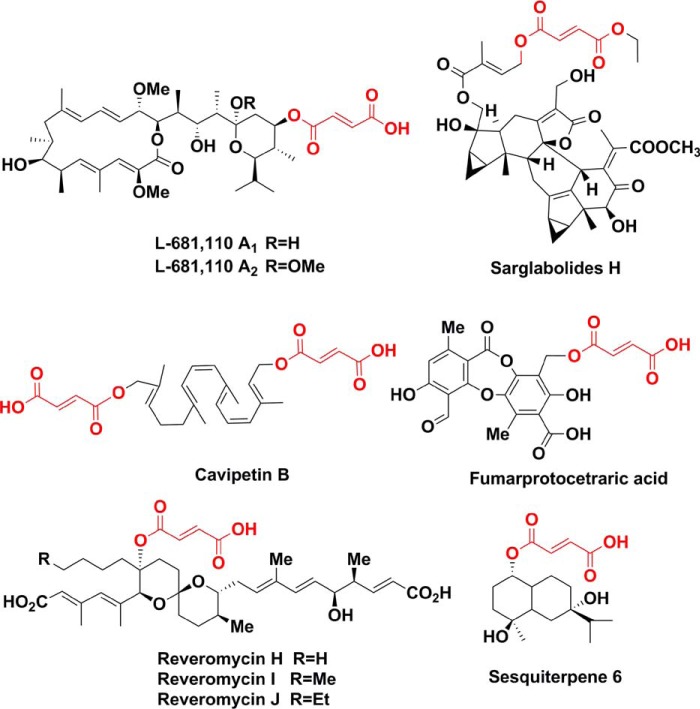
Interestingly, the bafilomycin post-PKS tailoring pathway directly recruits multiple primary metabolites including glycine, succinyl-CoA, and fumarate to decorate the secondary scaffold, which features an interesting cross-talk between the primary and secondary metabolisms. Glycine plays a vital role in supporting the growth and reproduction of microorganisms. It is also often seen in the scaffolds of secondary metabolites, such as alkaloids, ribosomal and nonribosomal peptides (39, 40). Succinyl-CoA, as an important intermediate of the tricarboxylic acid (TCA) cycle, normally takes part in biosynthesis of polyketides indirectly through generating several PKS extender units (41). Fumarate, another important intermediate of the TCA cycle, is present in dozens of natural products, such as sarglabolides H (42), sesquiterpene 6 (43), cavipetin B (44), fumarprotocetraric acid (45), reveromycins (46), and other bafilomycin derivates (8, 47, 48) (Fig. 6). However, the biosynthetic mechanisms for fumarate incorporation are largely unknown. Recently, the fumarate linking process of sesquiterpene 6 was proposed as follows: malate might be transferred to the C1 hydroxyl group of sesquiterpene 8 and followed by a dehydration step to generate the fumaryl side chain (43). The results of this study apparently suggest an alternative pathway for sesquiterpene 6 production.
Figure 6.
Selected natural products with the fumarate element highlighted in red.
Nonetheless, it requires more experimental evidence to examine if the Orf2/Orf3 co-mediated transformation is a general strategy for fumarate attachment in natural products. Interestingly, a carboxylic acid AMP ligase often closely cooperates with another enzyme to accomplish the substrate transformation in natural product biosynthesis because the carboxyl-AMP intermediate is highly unstable. Thus, direct uptake of carboxyl-AMP from the ligase by a downstream enzyme becomes essential. In addition to Orf2/Orf3, GriC/GriD (23) and EphF/EphG (25) are paired to serve the biosynthesis of grixazone and phenazine, respectively. This feature may suggest a useful strategy for discovery of natural products containing the fumarate moiety. Briefly, the coding sequences of orf2 and orf3, especially the latter gene encoding the fumaryl transferase with strict substrate specificity, could be used as probes to mine the big data of genomes. Next, novel fumaryl derivatives and their association with corresponding orf2/orf3-like genes could be discovered.
In summary, through in vivo gene inactivation and in vitro enzymatic activity reconstitution, we have completely elucidated the post-PKS tailoring pathway of bafilomycin. Five tailoring enzymes including Orf2 (fumaryl transferase), Orf3 (fumarate adenylyltransferase), BafX (acyl-CoA ligase), BafY (amide synthetase), and BafZ (5-ALA synthase as well as 5-ALA-CoA cyclase) have been biochemically characterized. This pathway incorporates multiple primary metabolites including glycine, succinyl-CoA, and fumarate into the structural core of secondary metabolites. These new findings highlight the role of the cross-talk between primary and secondary metabolisms in structural diversification and bioactivity improvement of natural products.
Experimental procedures
General materials
All chemicals and antibiotics used in this study were purchased from Sigma, Solarbio, or Sinopharm Chemical Reagent, unless otherwise specified. Fast-digest restriction endonucleases were bought from Thermo Fisher Scientific. T4 DNA ligase, PrimeSTAR GXL high-fidelity DNA polymerase were obtained from Takara. Isolation of plasmid DNA from E. coli was performed using the E.Z.N.A. Plasmid Mini Kit I from Omega. Purification of DNA fragments from PCR or agarose gels was performed using a Wizard SV Gel and PCR Clean-up System (Promega). ClonExpress II One-Step Cloning Kit was purchased from Vazyme. Malachite Green Phosphate Assay Kit is product of Bioassay System, and the inorganic pyrophosphatase was bought from Sigma. Bradford Protein Assay Kit was bought from Beyotime Biotechnology. The FlexiRunTM pre-mixed gel solution for SDS-PAGE was obtained from MDBio. Primer synthesis and DNA sequencing were performed by Tsingke.
Construction of knock-out vectors
The homologous arms (2–3 kb) of suicide vectors pCIMt002-Δorf2&3 and pCIMt002-ΔbafY were amplified from the genome of S. lohii using the primers as follows (supplemental Table S2): the left homologous arm of pCIMt002-Δorf2&3 (forward primer Δorf2&3-left arm-FP and reverse primer Δorf2&3-right arm-RP); the right homologous arm of pCIMt002-Δorf2&3 (forward primer Δorf2&3-right arm-FP and reverse primer Δorf2&3-right arm-RP); the left homologous arm of pCIMt002-ΔbafY (forward primer ΔbafY-left arm-FP and reverse primer ΔbafY-left arm-RP); the right homologous arm of pCIMt002-ΔbafY (forward primer ΔbafY-right arm-FP and reverse primer ΔbafY-right arm-RP). The left homologous arms were inserted into the NcoI restriction site and the right homologous arms were inserted into the NheI restriction site of pCIMt002 using the One-Step Cloning Kit to generate pCIMt002-ΔbafY and pCIMt002-Δorf2&3.
Construction of expression vectors for BafX, BafY, Orf2, and Orf3
The bafX, bafY, orf2, and orf3 genes were amplified from gDNA of S. lohii using the primers as shown in supplemental Table S3. For bafX, the forward primer was BafX-EcoRI-FP and the reverse primer was BafX-HindIII-RP; for bafY, the forward primer was BafY-EcoRI-FP and the reverse primer was BafY-HindIII-RP; for orf2, the forward primer was Orf2-NdeI-FP and the reverse primer was Orf2-EcoRI-RP; for orf3, the forward primer was Orf3-NdeI-FP and the reverse primer was Orf3-NdeI-RP. The PCR fragments of bafX, bafY, and orf2 were double digested and ligated into the expression vector pET28b. The PCR fragment of orf3 was cloned into pET28b using a One-Step Cloning Kit. The recombinant expression vectors were confirmed by restriction enzyme digestions and DNA sequencing. The positive plasmids were used to transform E. coli BL21 (DE3).
Gene disruption
Gene disruption in S. lohii was performed by following the protocol of Chen et al. (22). The suicide vectors pCIMt002-Δorf2&3 and pCIMt002-ΔbafY were transferred by interspecies conjugation from E. coli ET12567 to S. lohii (49). After treatment of 1.25 mg of apramycin and 0.5 mg of nalidixic acid for each plate for about 3–5 days at 28 °C, the blue colonies on plates were undesired single-crossover mutants due to production of indigoidine, whereas the white colonies indicative of double-crossover mutants were picked up for genomic DNA preparation according to standard Streptomyces protocols (49) and subsequent PCR confirmation (supplemental Fig. S1, B and D).
PCR confirmation of S. lohii mutants
The primers for PCR confirmation are shown in supplemental Table S2. The forward primer ΔbafY-KO-FP and the reverse primer ΔbafY-KO-RP were used for screening the ΔbafY mutants. The expected length of the PCR fragment from wild-type and the ΔbafY mutant is 1804 and 2337 bp, respectively (supplemental Fig. S1B). The forward primer Δorf2&3-KO-FP and the reverse primer Δorf2&3-KO-RP were used for screening the Δorf2&3 mutants. The expected length of the PCR fragment from wild-type and the Δorf2&3 mutant is 1957 and 1097 bp, respectively (supplemental Fig. S1D).
Fermentation and HPLC analysis of S. lohii and its mutants
The MS medium agar (mannitol, 20 g; soybean flour, 20 g; and agar, 20 g, per liter) was used for sporulation of wild-type S. lohii (ATCC BAA-1276) and its mutant strains. S. lohii strains were grown in 30 ml of 2× YT medium (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter) at 28 °C, 220 rpm. After 2 days, 3 ml of seed culture was inoculated into 30 ml of fermentation medium (soybean oil, 60 g; glucose, 20 g; soybean flour, 20 g; corn syrup, 1.5 g; NZ-amine, 2 g; yeast extract, 1 g; NaNO3, 8 g; NaCl, 5 g; (NH4)2SO4, 6 g; K2HPO4, 0.3 g; and CaCO3, 8 g, per liter, pH 7.1), and cultured at 28 °C, 250 rpm for another 7 days. Next, the fermentation broth was extracted by adding 3-fold volume of methanol, vortexed, and centrifuged. The organic extracts were directly used for HPLC analysis. HPLC analysis was performed on a Thermo C-18 column (4.6 × 150 mm) with a linear gradient of 60–100% CH3CN over 15 min, 100% CH3CN for 8 min, and 100–60% CH3CN over 2 min in H2O (+ 0.1% trifluoroacetic acid) at a flow rate of 1 ml/min.
Protein expression and purification
The single colony of E. coli BL21 (DE3) carrying pET28b-bafX, pET28b-bafY, pET28b-orf2, or pET28b-orf3 was grown at 37 °C in 10 ml of LB medium with 50 μg/ml of kanamycin at 37 °C overnight. Then, the seed culture was used to inoculate 1 liter of LB medium containing 50 μg/ml of kanamycin with the ratio of 1:100 at 37 °C. Isopropyl β-d-thiogalactopyranoside was added to the final concentration of 0.15 mm to initiate protein overexpression when A600 values reached 0.6–1.0. The cells were cultured at 16–18 °C for an additional 24 h, collected by centrifugation (6,000 rpm, 10 min, 4 °C), resuspended in 50 ml of lysis buffer (50 mm Tris-HCl, 0.5 m NaCl, and 5 mm imidazole, pH 8.0), and lysed by sonication on ice. After high-speed centrifugation (12,000 rpm, 30 min, 4 °C), 1 ml of nickel-nitrilotriacetic acid-agarose resin (Qiagen) was added to the supernatant, and mixed at 4 °C for 1 h. The resin with bound His6-tagged proteins was loaded onto a gravity flow column, washed with 500 ml of wash buffer (50 mm Tris-HCl, 0.5 m NaCl, and 20 mm imidazole, 10% glycerol, pH 8.0), and eluted by elution buffer (50 mm Tris-HCl, 0.5 m NaCl, and 250 mm imidazole, 10% glycerol, pH 8.0). The purified proteins (BafX, BafY, BafZ, Orf2, and Orf3) were concentrated using Amicon Ultra filters (30 K) and exchanged into desalting buffer (50 mm Tris-HCl, 10% glycerol, pH 7.4) using PD-10 columns (GE Healthcare). The purified proteins were flash frozen in liquid nitrogen and stored at −80 °C for later use. The protein concentrations were measured using a Bradford Assay Kit with bovine serum albumin (BSA) as standard. The expected molecular masses of BafX, BafY, BafZ, Orf2, and Orf3 are 60.4, 58.6, 45.7, 36.0, and 42.4 kDa (Fig. 3A), respectively.
The ATP-PPi/ATP-Pi release assay
The PPi or Pi released from ATP by Orf3 or BafY was measured using the Malachite Green Phosphate Assay Kit (29). For qualitative analysis of Orf3, the reaction contained 10 μm Orf3, 5 mm disodium fumarate, 5 mm ATP, 10 mm MgCl2, and 0.4 units/ml of inorganic pyrophosphatase in Tris-HCl buffer, pH 7.4, at 28 °C (29, 34). For BafY, 5 μm BafY was incubated with 200 μm 2, 5 mm ATP, 10 mm MgCl2, and 0.4 units/ml of inorganic pyrophosphatase in Tris-HCl buffer, pH 7.4, at 28 °C. All PPi/Pi releasing reactions were carried out in triplicate and the negative controls were set up using boiling inactivated Orf3 or BafY. For kinetic analysis of Orf3, a standard assay contained 10 mm MgCl2, 5 mm ATP, varying concentrations of selected organic acids (0.5–100 mm), an appropriate amount of Orf3, and 0.4 units/ml of inorganic pyrophosphatase in 100 μl 50 mm Tris-HCl buffer, pH 7.4. Each reaction was initiated by adding 5 mm ATP and carried out at 28 °C for 0, 3, 6, 10, 15, 20, and 30 min on a 96-well plate. The reaction mixtures were 20-fold diluted, and 80-μl diluents were added into wells pre-containing 20 μl of malachite green. Upon an incubation at room temperature for 15 min, the absorbance was monitored at 620 nm on an Infinite M200 PRO plate reader (TECAN). At a certain substrate concentration, the velocity of Pi release (μm/min, derived from ΔA620/min using the standard curve of 0–50 μm Na3PO4), which is twice the rate of PPi formation, was calculated from the slope of a linear fitting curve (seven data points) with the control slope subtracted. The triplicated data were fitted to the Michaelis-Menten equation for calculating the kcat and Km values of each substrate using Origin 8.5.
Isolation and purification of bafilomycin A1 (1), bafilomycin C1 (2), and bafilomycin B1 (3)
5 Liters of fermentation broth of S. lohii was centrifuged at 6,000 rpm for 10 min. The aqueous phase containing very few bafilomycins was discarded. The mycelia and oil phase was extracted by a 2-fold volume of methanol twice and concentrated by vacuum rotary evaporation. The crude extract was dissolved in methanol/dichloromethane (v/v = 1:1), and the soluble constituents were separated with petroleum ether/ethyl acetate from the 10:1 (v/v) to 3:1 gradient on a silica gel column. The eluents from 4:1 to 2:1 (petroleum ether/ethyl acetate) were collected and combined for semi-preparative HPLC on a Waters XBridgeTM C-18 column (10 × 250 mm) with a linear gradient of 65–100% acetonitrile over 30 min, 100% acetonitrile for 5 min in H2O at a flow rate of 2.5 ml/min. The structures of 1, 2, and 3 were confirmed by HRMS analysis (supplemental Fig. S14–S16) and comparison of their NMR spectra (supplemental Fig. S17–S22) with previous reports (7, 50, 51).
LC-HRMS analysis
The LC analysis was performed on a Waters symmetry column (4.6 × 150 mm, RP18) with a linear gradient of 60–100% acetonitrile over 40 min, and 100% acetonitrile for 10 min in H2O (+0.1% formic acid) at a flow rate of 0.5 ml/min. The HRMS were recorded on a Dionex Ultimate 3000 coupled to a Bruker Maxis Q-TOF.
Author contributions
Z. Li, D. H. S., S. G., H. X., and S. Li conceived this study, analyzed the results, and wrote the manuscript. Z. Li, L. D., W. Z., X. Z., Y. J., K. L., P. M., J.L. F., and B. Y. conducted experiments. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Prof. Yihua Chen and Dr. Pengwei Li at Institute of Microbiology, Chinese Academy of Sciences, for providing the plasmids and protocols for the efficient blue-white screening-based gene inactivation system for Streptomyces. We thank two anonymous reviewers for detailed and constructive suggestions. We also thank Fali Bai and Dr. Shaohua Huang at Qingdao Institute of Bioenergy and Biotechnology, Chinese Academy of Sciences, and Prof. Dehai Li at Ocean University of China for assistance in LC-MS and NMR data collection and analysis.
Note added in proof
During the review process, another paper was published relating to the topics in this article (52).
This work was supported by National Natural Science Foundation of China Grant NSFC 21472204, Shandong Provincial Natural Science Foundation Grant JQ201407, Primary Research & Development Plan of Shandong Province Grant 2016GSF121001, Applied Basic Research Program of Qingdao Grant 15-9-1-93-jch, China Postdoctoral Science Foundation Grant 2016M590669, and the Youth Innovation Promotion Association of CAS Grant 2015166. This work was also supported by National Institutes of Health Grant R35GM118101 (to D. H. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S22 and Tables S1–S3.
- PKS
- polyketide synthase
- C5N
- 2-amino-3-hydroxycyclopent-2-enone
- 5-ALA
- 5-aminolevulinic acid
- PPi
- pyrophosphate
- Pi
- monophosphate
- TCA
- tricarboxylic acid
- LC-HRMS
- liquid chromatography-high resolution mass spectrometry.
References
- 1. Dai W. M., Guan Y., and Jin J. (2005) Structures and total syntheses of the plecomacrolides. Curr. Med. Chem. 12, 1947–1993 [DOI] [PubMed] [Google Scholar]
- 2. Kinashi H., Someno K., and Sakaguchi K. (1984) Isolation and characterization of concanamycins A, B and C. J. Antibiot. 37, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 3. Werner G., Hagenmaier H., Drautz H., Baumgartner A., and Zähner H. (1984) Metabolic products of microorganisms 224: bafilomycins, a new group of macrolide antibiotics: production, isolation, chemical structure and biological activity. J. Antibiot. 37, 110–117 [DOI] [PubMed] [Google Scholar]
- 4. Wilton J. H., Hokanson G. C., and French J. C. (1985) PD 118,576: a new antitumor macrolide antibiotic. J. Antibiot. 38, 1449–1452 [DOI] [PubMed] [Google Scholar]
- 5. Goetz M. A., McCormick P. A., Monaghan R. L., Ostlind D. A., Hensens O. D., Liesch J. M., and Albers-Schonberg G. (1985) L-155,175: a new antiparasitic macrolide: fermentation, isolation and structure. J. Antibiot. 38, 161–168 [DOI] [PubMed] [Google Scholar]
- 6. Bowman E. J., Siebers A., and Altendorf K. (1988) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. 85, 7972–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Werner G., Hagenmaier H., Albert K., and Kohlshorn H. (1983) The structure of the bafilomycins, a new group of macrolide antibiotics. Tetrahedron Lett. 24, 5193–5196 [Google Scholar]
- 8. Kretschmer A., Dorgerloh M., Deeg M., and Hagenmaier H. (1985) The structures of novel insecticidal macrolides: bafilomycins D and E, and oxohygrolidin. Agric. Biol. Chem. 49, 2509–2511 [Google Scholar]
- 9. Yu Z., Zhao L. X., Jiang C. L., Duan Y., Wong L., Carver K. C., Schuler L. A., and Shen B. (2011) Bafilomycins produced by an endophytic actinomycete Streptomyces sp. YIM56209. J. Antibiot. 64, 159–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J., Lu C., and Shen Y. (2010) Macrolides of the bafilomycin family produced by Streptomyces sp. CS. J. Antibiot. 63, 595–599 [DOI] [PubMed] [Google Scholar]
- 11. Zhang D. J., Wei G., Wang Y., Si C. C., Tian L., Tao L. M., and Li Y. G. (2011) Bafilomycin K, a new antifungal macrolide from Streptomyces flavotricini Y12–26. J. Antibiot. 64, 391–393 [DOI] [PubMed] [Google Scholar]
- 12. Carr G., Williams D. E., Díaz-Marrero A. R., Patrick B. O., Bottriell H., Balgi A. D., Donohue E., Roberge M., and Andersen R. J. (2010) Bafilomycins produced in culture by Streptomyces spp. isolated from marine habitats are potent inhibitors of autophagy. J. Nat. Prod. 73, 422–427 [DOI] [PubMed] [Google Scholar]
- 13. Lu C., and Shen Y. (2004) Two new macrolides produced by Streptomyces sp. CS. J. Antibiot. 57, 597–600 [DOI] [PubMed] [Google Scholar]
- 14. Dröse S., and Altendorf K. (1997) Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200, 1–8 [DOI] [PubMed] [Google Scholar]
- 15. Pérez-Sayáns M., Somoza-Martín J. M., Barros-Angueira F., Rey J. M., and García-García A. (2009) V-ATPase inhibitors and implication in cancer treatment. Cancer Treat. Rev. 35, 707–713 [DOI] [PubMed] [Google Scholar]
- 16. Scheidt K. A., Bannister T. D., Tasaka A., Wendt M. D., Savall B. M., Fegley G. J., and Roush W. R. (2002) Total synthesis of (−)-bafilomycin A1. J. Am. Chem. Soc. 124, 6981–6990 [DOI] [PubMed] [Google Scholar]
- 17. Toshima K., Jyojima T., Yamaguchi H., Noguchi Y., Yoshida T., Murase H., Nakata M., and Matsumura S. (1997) Total synthesis of bafilomycin A1. J. Org. Chem. 62, 3271–3284 [DOI] [PubMed] [Google Scholar]
- 18. Gagliardi S., Gatti P. A., Belfiore P., Zocchetti A., Clarke G. D., and Farina C. (1998) Synthesis and structure-activity relationships of bafilomycin A1 derivatives as inhibitors of vacuolar H+-ATPase. J. Med. Chem. 41, 1883–1893 [DOI] [PubMed] [Google Scholar]
- 19. Wu C., Medema M. H., Läkamp R. M., Zhang L., Dorrestein P. C., Choi Y. H., and van Wezel G. P. (2016) Leucanicidin and endophenasides result from methyl-rhamnosylation by the same tailoring enzymes in Kitasatospora sp. MBT66. ACS Chem. Biol. 11, 478–490 [DOI] [PubMed] [Google Scholar]
- 20. Zhang W., Fortman J. L., Carlson J. C., Yan J., Liu Y., Bai F., Guan W., Jia J., Matainaho T., Sherman D. H., and Li S. (2013) Characterization of the bafilomycin biosynthetic gene cluster from Streptomyces lohii. ChemBioChem 14, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hwang J. Y., Kim H. S., Kim S. H., Oh H. R., and Nam D. H. (2013) Organization and characterization of a biosynthetic gene cluster for bafilomycin from Streptomyces griseus DSM 2608. AMB Express 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li P., Li J., Guo Z., Tang W., Han J., Meng X., Hao T., Zhu Y., Zhang L., and Chen Y. (2015) An efficient blue-white screening based gene inactivation system for Streptomyces. Appl. Microbiol. Biotechnol. 99, 1923–1933 [DOI] [PubMed] [Google Scholar]
- 23. Suzuki H., Ohnishi Y., and Horinouchi S. (2007) GriC and GriD constitute a carboxylic acid reductase involved in grixazone biosynthesis in Streptomyces griseus. J. Antibiot. 60, 380–387 [DOI] [PubMed] [Google Scholar]
- 24. Rui Z., Ye M., Wang S., Fujikawa K., Akerele B., Aung M., Floss H. G., Zhang W., and Yu T.-W. (2012) Insights into a divergent phenazine biosynthetic pathway governed by a plasmid-born esmeraldin gene cluster. Chem. Biol. 19, 1116–1125 [DOI] [PubMed] [Google Scholar]
- 25. Bera A. K., Atanasova V., Gamage S., Robinson H., and Parsons J. F. (2010) Structure of the d-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes. Acta Crystallogr. D Biol. Crystallogr. 66, 664–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gulick A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reger A. S., Wu R., Dunaway-Mariano D., and Gulick A. M. (2008) Structural characterization of a 140° domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry 47, 8016–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. May J. J., Kessler N., Marahiel M. A., and Stubbs M. T. (2002) Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. 99, 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McQuade T. J., Shallop A. D., Sheoran A., Delproposto J. E., Tsodikov O. V., and Garneau-Tsodikova S. (2009) A nonradioactive high-throughput assay for screening and characterization of adenylation domains for nonribosomal peptide combinatorial biosynthesis. Anal. Biochem. 386, 244–250 [DOI] [PubMed] [Google Scholar]
- 30. Rusnak F., Faraci W. S., and Walsh C. T. (1989) Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry 28, 6827–6835 [DOI] [PubMed] [Google Scholar]
- 31. Ostash B., Campbell J., Luzhetskyy A., and Walker S. (2013) MoeH5: a natural glycorandomizer from the moenomycin biosynthetic pathway. Mol. Microbiol. 90, 1324–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galm U., Schimana J., Fiedler H.-P., Schmidt J., Li S.-M., and Heide L. (2002) Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tü 6040. Arch. Microbiol. 178, 102–114 [DOI] [PubMed] [Google Scholar]
- 33. Rui Z., Petrícková K., Skanta f., Pospísil S., Yang Y., Chen C.-Y., Tsai S.-F., Floss H. G., Petrícek M., and Yu T.-W. (2010) Biochemical and genetic insights into asukamycin biosynthesis. J. Biol. Chem. 285, 24915–24924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang W., Bolla M. L., Kahne D., and Walsh C. T. (2010) A three enzyme pathway for 2-amino-3-hydroxycyclopent-2-enone formation and incorporation in natural product biosynthesis. J. Am. Chem. Soc. 132, 6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kowalsky A., Wyttenbach C., Langer L., and Koshland D. (1956) Transfer of oxygen in the glutamine synthetase reaction. J. Biol. Chem. 219, 719–725 [PubMed] [Google Scholar]
- 36. Krishnaswamy P., Pamiljans V., and Meister A. (1962) Studies on the mechanism of glutamine synthesis: evidence for the formation of enzyme-bound activated glutamic acid. J. Biol. Chem. 237, 2932–2940 [Google Scholar]
- 37. Wray L. V. Jr., and Fisher S. H. (1988) Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene 71, 247–256 [DOI] [PubMed] [Google Scholar]
- 38. Fisher S. H., and Wray L. V. Jr. (1989) Regulation of glutamine synthetase in Streptomyces coelicolor. J. Bacteriol. 171, 2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bewley C. A., Debitus C., and Faulkner D. J. (1994) Microsclerodermins A and B: antifungal cyclic peptides from the lithistid sponge Microscleroderma sp. J. Am. Chem. Soc. 116, 7631–7636 [Google Scholar]
- 40. Zhuang Y., Teng X., Wang Y., Liu P., Li G., and Zhu W. (2011) New quinazolinone alkaloids within rare amino acid residue from coral-associated fungus, Aspergillus versicolor LCJ-5–4. Org. Lett. 13, 1130–1133 [DOI] [PubMed] [Google Scholar]
- 41. Chan Y. A., Podevels A. M., Kevany B. M., and Thomas M. G. (2009) Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26, 90–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang P., Luo J., Zhang Y. M., and Kong L. Y. (2015) Sesquiterpene dimers esterified with diverse small organic acids from the seeds of Sarcandra glabra. Tetrahedron 71, 5362–5370 [Google Scholar]
- 43. Zhao F., Sun C., Ma L., Wang Y. N., Wang Y. F., Sun J. F., Hou G. G., Cong W., Li H. J., Zhang X. H., Ren Y., and Wang C. H. (2016) New sesquiterpenes from the rhizomes of Homalomena occulta. Fitoterapia 109, 113–118 [DOI] [PubMed] [Google Scholar]
- 44. Toyota M., and Hostettmann K. (1990) Antifungal diterpenic esters from the mushroom Boletinus cavipes. Phytochemistry 29, 1485–1489 [Google Scholar]
- 45. Elix J. A., and Wardlaw J. H. (1999) The structure of chalybaeizanic acid and quaesitic acid, two new lichen depsidones related to salazinic acid. Aust. J. Chem. 52, 713–716 [Google Scholar]
- 46. Fremlin L., Farrugia M., Piggott A. M., Khalil Z., Lacey E., and Capon R. J. (2011) Reveromycins revealed: new polyketide spiroketals from Australian marine-derived and terrestrial Streptomyces spp. a case of natural products vs. artifacts. Org. Biomol. Chem. 9, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 47. Uyeda M., Kondo K. I., Ito A., Yokomizo K., and Kido Y. (1995) A new antiherpetic agent produced by Streptomyces sp. strain no. 758. J. Antibiot. 48, 1234–1239 [DOI] [PubMed] [Google Scholar]
- 48. Huang L., Albers-Schonberg G., Monaghan R. L., Jakubas K., Pong S. S., Hensens O. D., Burg R. W., Ostlind D. A., Conroy J., and Stapley E. O. (1984) Discovery, production and purification of the Na+, K+ activated ATPase inhibitor, L-681,110 from the fermentation broth of Streptomyces sp. MA-5038. J. Antibiot. 37, 970–975 [DOI] [PubMed] [Google Scholar]
- 49. Kieser T. (2000) Practical streptomyces genetics, John Innes Foundation, Norwick, United Kingdom [Google Scholar]
- 50. Kleinbeck F., and Carreira E. M. (2009) Total synthesis of bafilomycin A1. Angew. Chem. Int. Ed. Engl. 48, 578–581 [DOI] [PubMed] [Google Scholar]
- 51. Moon S.-S., Hwang W.-H., Chung Y. R., and Shin J. (2003) New cytotoxic bafilomycin C1-amide produced by Kitasatospora cheerisanensis. J. Antibiot. 56, 856–861 [DOI] [PubMed] [Google Scholar]
- 52. Nara A., Hashimoto T., Komatsu M., Nishiyama M., Kuzuyama T., and Ikeda H. (2017) Characterization of bafilomycin biosynthesis in Kitasatospora setae KM-6054 and comparative analysis of gene clusters in Actinomycetales microorganisms. J. Antibiot. 10.1038/ja.2017.33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.