Abstract
The marine bacterium Vibrio fischeri is the monospecific symbiont of the Hawaiian bobtail squid, Euprymna scolopes, and the establishment of this association involves a number of signaling pathways and transcriptional responses between both partners. We report here the first full RNA-Seq dataset representing host-associated V. fischeri cells from colonized juvenile E. scolopes, as well as comparative transcriptomes under both laboratory and simulated marine planktonic conditions. These data elucidate the broad transcriptional changes that these bacteria undergo during the early stages of symbiotic colonization. We report several previously undescribed and unexpected transcriptional responses within the early stages of this symbiosis, including gene expression patterns consistent with biochemical stresses inside the host, and metabolic patterns distinct from those reported in associations with adult animals. Integration of these transcriptional data with a recently developed metabolic model of V. fischeri provides us with a clearer picture of the metabolic state of symbionts within the juvenile host, including their possible carbon sources. Taken together, these results expand our understanding of the early stages of the squid–vibrio symbiosis, and more generally inform the transcriptional responses underlying the activities of marine microbes during host colonization.
Introduction
Bacterial colonization of host organisms has been studied in many different model systems, and in the context of both pathogenesis and beneficial symbiosis (Bry et al., 1996; Dedeine et al., 2001; Russell and Rychlik, 2001; Hongoh, 2010; Gilbert et al., 2012; Nyholm and Graf, 2012; Bulgarelli et al., 2013; McFall-Ngai et al., 2013; Almagro et al., 2015; Uzal et al., 2015; Kao et al., 2016). Such interactions have strong transcriptional effects upon both host and symbiont, and there is a compelling interest towards better understanding these interactions and, in particular, the chemical dialogue that underlies their initial establishment (Montgomery and McFall-Ngai, 1994; Hughes and Sperandio, 2008; Mullard, 2009; Wier et al., 2010; Shin et al., 2011; Heath-Heckman et al., 2013; Kremer et al., 2013; Kabat et al., 2014; Penterman et al., 2014). The model symbiosis between the marine bioluminescent bacterium Vibrio fischeri and the Hawaiian bobtail squid Euprymna scolopes has served as an insightful window into host–microbe interactions over the past several decades, and provides an excellent system to study both the early and long-term interactions of a mutualistic symbiosis.
V. fischeri is the monospecific symbiont of the E. scolopes light-emitting organ. When first hatched, juvenile E. scolopes are aposymbiotic (i.e., they have no bacterial symbionts), and each generation must obtain their symbiotic bacteria from the surrounding seawater. Colonization of the squid light organ is both rapid and extremely specific, allowing V. fischeri to transition from being a minority member of the bacterioplankton (estimated at 0.01–0.15% of the ambient seawater bacteria) to being the sole resident of the host’s light organ (Ruby and Lee, 1998). This transition from the seawater to the interior of this organ requires passage through several distinct microenvironments within the host’s tissues (Nyholm and McFall-Ngai, 2004). During this process, bacterial cells must contend with chemical challenges (e.g., NO exposure) and recognize chemical cues (e.g., chitobiose, presented as a chemoattractant) used by the host to ensure colonization specificity, and to which V. fischeri is exquisitely adapted (DeLoney et al., 2003; Dunn et al., 2010; Wang et al., 2010; Kremer et al., 2013). Following aggregation on the host’s epithelium, a small number of individual bacteria migrate into the inner tissues of the light organ where they colonize the deep crypts and multiply. This transition into the host leads to the establishment of a mutualistic symbiosis that continues for the duration of the squid’s life, and is directly responsible for a large number of transcriptional and developmental changes within the host (Wier et al., 2010; Heath-Heckman et al., 2013; Kremer et al., 2013).
The functional foundation of this symbiosis is the provision of nutrients and a rich growth environment for the bacterial population, from which the squid host gains a source of light used for anti-predatory behavior at night (Nyholm and McFall-Ngai, 2004). This relationship follows a strong diel rhythm, wherein the host expels up to 95% of the symbiont community each dawn and the population regrows while the animal is buried within the sand during the day. At night the host emerges and the bacterial population has grown sufficiently to luminesce, a process tightly regulated via quorum sensing (Miyashiro and Ruby, 2012). In adult animals, this process is concurrent with a strong transcriptional and metabolic rhythm between the symbionts and host tissues (Weir et al., 2010). During the day the host apparently presents glycerophospholipids to its symbionts, allowing them to undergo anaerobic respiration to maximize growth. However, at night the host withholds these metabolites and, instead, provides chitin, which the bacteria ferment under conditions that promote maximal production of light. This metabolic cycling, however, has been found to be distinct from that occurring during juvenile light-organ development, as host provision of chitin oligosaccharides at night only begins after about four weeks of host development (Schwartzman et al., 2015). These findings are consistent with developmental changes in the host, including within the light organ. Such changes are tightly tied to the initial contact between host and symbiont, with transcriptional responses evident in the light organ’s tissues immediately following initial exposure to bacterial cells (Kremer et al., 2013).
The picture that arises of the establishment of the squid–vibrio symbiosis is one of tightly regulated developmental events within the host, and a well-adapted suite of tools for host colonization on the part of the bacterial symbionts. Prior studies have examined transcriptional patterns within this system in mature host animals, and within the host light organ during initial contact with bacterial symbionts; however, transcriptomics data on the bacteria in the juvenile light organ has thus far been missing because there are <106 symbionts per light organ during this early stage of development, below the input-biomass limit of standard RNA-Seq transcriptomics. In light of the strong and early response by the squid host to the initial presence of the symbiont (Kremer, et al., 2013), we asked what changes occur within the symbionts as they make the transition from the bacterioplankton to the host, a habitat switch similar to that made by most pathogenic Vibrio species. To this end, we performed RNA-Seq experiments on V. fischeri from three environments – (i) cells growing in rich medium, (ii) cells incubated in seawater, and (iii) cells collected immediately after venting from the squid host – to identify transcriptional signals that could be directly associated with the early stages of host colonization (Fig. 1A). In particular, our comparisons of cells in seawater with those that had just exited the host’s light organ revealed a large number of genes that were highly upregulated in response to host contact (Fig. 1B). These data fill an existing gap in our understanding of the early stages of symbiosis between V. fischeri and E. scolopes, addressing questions about both the metabolic state and the environmental responses of these bacteria during host colonization.
Fig. 1.
Schematic cartoons of (A) the colonization of E. scolopes by V. fischeri during the study’s 3-stage transcriptomic comparison, and (B) characteristic metabolic pathways that are differentially expressed in planktonic and squid-associated cells of V. fischeri. TCA cycle intermediates are indicated with one-letter codes: O, oxaloacetate; M, malate; F, fumarate; S, succinate; SCoA, succinyl-CoA; C, citrate; I, isocitrate; K, 2-ketoglutarate; A, aspartate.
Results & Discussion
Robust libraries made from ribo-depleted, low-biomass samples
This study depended upon the reliable measurement of the mRNA levels present in the small numbers of V. fischeri cells expelled from the juvenile squid’s light organ each morning (Lee and Ruby, 1994). The yield of V. fischeri total RNA from 10–100 animals is typically less than 100 ng and, thus, efforts to deplete ribosomal RNA (ribo-depletion) might lead to high collateral loss of mRNA. Tests with the standard Illumina TruSeq library prep have shown reproducible results down to input levels of ~70 ng total RNA but, notably, these protocols do not employ ribo-depletion (Combs and Eisen, 2015). Therefore, we sought to establish the minimal number of juvenile squids necessary to create a reliable symbiont transcriptome. Specifically, we determined the lower limit of ribo-depleted RNA that would produce robust results when following the standard protocol of the TruSeq RNA Sample Preparation Kit (Illumina).
Using a single sample of V. fischeri total RNA from a culture grown in SWT medium, we made TruSeq libraries from three amounts of non-ribo-depleted total RNA (1000, 500, and 100 ng), and nine amounts of ribo-depleted RNA (1000, 500, 100, 50, 25, 10, 5, 2.5, and 1 ng) (Table S1). The 100-ng ribo-depleted sample from this experiment is the same as replicate no. 3 of the cultured-cell treatment in the comparison experiment. Libraries for the 5, 2.5, and 1 ng samples were amplified both with the standard 15 cycles as well as with 17 cycles.
We mapped and counted the reads from each library prep to the V. fischeri ES114 genome as described in the Experimental Procedures. Ribo-depletion using the Ribo-Zero Gold Epidemiology Kit (Epicentre) before library prep with the TruSeq kit reduced the percentage of rRNA in the sample from ~90% to ~1% (Fig. S1). Ribo-depletion did not appreciably affect the relative abundance of individual mRNAs detected, as shown by pairwise scatter plots between ribo-depleted and non-ribo-depleted samples, yielding R-squared values between 0.93 and 0.98 (Fig. S2). Contrasting the conditions represented in these scatter plots using DESeq2 did not identify any genes significantly differentially abundant between ribo-depleted and non-ribo-depleted samples (FDR-adjusted p-value < 0.05).
Having established that ribo-depletion removed ~99% of rRNA from as little as 100 ng total RNA without changing the relative abundance of mRNAs detected in a sample, we then determined how much the input RNA could be lowered, prior to ribo-depletion, without the loss of signal fidelity. Using the 100-ng ribo-depleted sample as a basis of comparison, pairwise scatter plots showed that the relative mRNA composition was maintained down to 10 ng, with R-squared values of 0.98 between the 100-ng and 10-ng samples (Fig. S3). Additional noise was introduced as the level of input RNA was decreased to 1 ng, but the R-squared values remained in the range of 0.91 to 0.95 (Fig. S4). Coverage of the V. fischeri ES114 genome, i.e., the fraction of CDS genes observed one or more times in the sample, was between 98 and 99% with samples as low as 50 ng of input total RNA; below this value, the coverage decreased to ~95% for samples down to 5 ng, and ~85% for input total RNA of 1 ng (Fig. S5A). Increasing the number of library amplification cycles increased the percent coverage somewhat (Fig. S5A); however, it did not necessarily bring the relative mRNA counts closer to those of the 100 ng standard (Fig. S4). As expected, those genes with the lowest relative mRNA abundance (when averaged across all samples from the 15 variations of preparation) were those most likely to be undetected in the low-input RNA samples (Fig. S5B). From these results, we concluded that the relative abundances of mRNA species could be reliably determined from an input level of 100 ng of total RNA (before ribo-depletion). In addition, input total RNA could be (i) reduced to 50 ng without loss of coverage, (ii) reduced further to 10 ng without loss of mRNA relative abundance fidelity, and (iii) reduced further to 2.5 to 5 ng with ~10% reduction in coverage and mRNA relative abundance fidelity (Fig. S5).
Expelled bacteria show transcriptional patterns consistent with previous colonization data
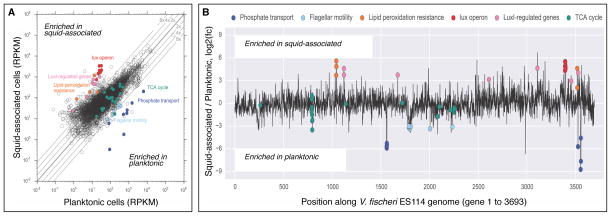
To test how well the mRNA of expelled symbionts represents their transcriptional activity in the light organ, we first asked whether previously identified patterns of specific gene expression were found. Prior studies of the V. fischeri–E. scolopes mutualism have provided evidence of a number of genes that are involved with establishing a successful host colonization. Two such sets of genes include those responsive to quorum signaling, and those encoding flagellar motility (Ruby and Asato, 1993; Visick et al., 2000; Wier et al., 2010; Miyashiro et al., 2011). We found that, when compared to planktonic V. fischeri, the squid-associated symbionts regulate a number of genes consistent with previous physiological and morphological studies of these activities (Table S2). For example, one of the best characterized sets of genes in the squid–vibrio symbiosis is the LuxIR regulon, including both the lux operon, which regulates bacterial light production (Septer and Stabb, 2012), and a set of 21 other quorum signal-induced genes (Antunes et al., 2007). Specifically, relative to either cultured or planktonic cells, we observed at least a 16-fold upregulation of all the genes within the lux operon in the bacteria expelled from the light organ (Fig. 2), consistent with both the high population density within the symbiosis and previous physiological measurements of induced bioluminescence activity (Visick et al., 2000).
Fig. 2.
Scatter plot and genome trace of gene transcript differential abundance between planktonic and squid-associated states, with genes belonging to pathways of interest highlighted. Color coding is from Fig. 1B. (A) Scatter plot showing log-scale differences in relative (RPKM) transcript abundances between planktonic (pre-squid colonization) and vented (post-squid colonization) states. Parallel lines show unity (log2 of fold change, log2(fc) = 0), 2x fold change (log2(fc) = ±1), 4x fold change (log2(fc) = ±2), and 8x fold change (log2(fc) = ±3). (B) Genome trace of log2(fc) between squid-associated and planktonic states. Genes are plotted in their order on the V. fischeri ES114 genome, which consists of Chromosome 1 (genes 1–2527), Chromosome 2 (genes 2528–3641), and a conjugative plasmid (genes 3642–3693).
In contrast, while flagellar elaboration and motility are both critical during the initial step of host colonization, flagellation disappears once colonization has been established (Ruby and Asato, 1993; Graf et al. 1994), although no molecular evidence has indicated its control at the transcriptional level. Thus, we looked at genes involved in the assembly of both the flagellum and flagellar motor of V. fischeri (Brennan et al., 2013), and observed significantly lower expression (|log2(fc)| > 1|, padj < .01) of 30 of 42 of these genes among the squid-associated symbionts (Table S2; Fig. 2). The focus here is on the habitat transition between the transcriptomes of planktonic and squid-associated bacteria, as these are the conditions most relevant to the life history of the symbiosis. However, the comparison of the SWT-cultured and squid-associated samples corroborates many of these results, and serves as a control for the rich growth conditions within the light organ (Graf and Ruby, 1998). Taken together, the expression of genes involved in these two symbiosis-driven physiological activities (Table S2) is both consistent with previous studies, and provides evidence that our samples of expelled symbionts are likely to faithfully report transcriptional activity within the host light organ.
Insight into symbiont metabolism within the juvenile host
Prior work has demonstrated that the symbiosis between V. fischeri and E. scolopes follows a well-regulated diel metabolic cycle (Wier et al., 2010). These studies also demonstrated that at least some of the metabolic pathways characteristic of symbionts in adult light organs are not established until the host has matured to approximately four weeks of age (Schwartzman et al., 2015). Most notably, a major nocturnal activity of symbionts in adult animals is the fermentation of chitin and its monomer N-acetylglucosamine (GlcNAc), while during the day they appear to anaerobically respire glycerol and glycerol-3-phosphate (G3P) using nitrate as the terminal electron acceptor (Wier et al., 2010). However, regardless of the time of day, GlcNAc is not a significant nutrient source for symbionts of juvenile squid; i.e., even when defective in GlcNAc catabolism, V. fischeri cells maintain a robust population until the symbiosis has matured, around 28 days (Miyashiro et al., 2011; Schwartzman et al., 2015). Thus, while the primary catabolic activity(s) of bacteria in the juvenile crypts has not been established, the transcriptomic signature of the released symbionts may provide some clues.
One such clue comes from an analysis of tricarboxylic acid (TCA) cycle gene expression within the squid-associated bacteria, which revealed distinct regulatory patterns consistent with fermentation rather than respiration. Specifically, we observed a significant down-regulation confined to the cycle’s sucABCD and acnB genes, encoding the enzymes responsible for converting citrate to succinate (Table 1). This weakening of the oxidative side of the TCA cycle (Fig. 1B) leaves the reductive side in a position to support fermentation (Neidhardt, 1996); in addition, the resulting buildup in citrate is expected to enhance bioluminescence (Septer et al., 2015). Consistent with, and perhaps responsible for, this signature of fermentative activity is an upregulation of VF_1068, a homolog of the regulatory protein pirin (Table 1), whose induction switches the flow of glycolysis-derived pyruvate from respiration to fermentation (Soo et al., 2007; Hansen et al., 2012).
Table 1.
Differentially expressed V. fischeri genes indicating symbiont metabolism in the juvenile host
| Gene* | Vnt/Plk log2(fc)† | FDR-adjusted p-value† | Description |
|---|---|---|---|
| Tricarboxylic Acid (TCA) cycle interruption | |||
| sucA (VF_0823) | −1.5 | 2.2 E-03 | α-ketoglutarate dehydrogenase E1 |
| sucB (VF_0824) | −2.3 | 1.1 E-06 | dihydrolipoamide succinyltransferase |
| sucC (VF_0825) | −3.5 | 1.1 E-05 | succinyl-CoA synthetase subunit beta |
| sucD (VF_0826) | −2.2 | 1.9 E-03 | succinyl-CoA synthetase subunit alpha |
| acnB (VF_2158) | −1.7 | 1.3 E-02 | bifunctional aconitate hydratase |
| Respiration/fermentation switch regulator | |||
| VF_1068 | 3.5 | 9.5 E-09 | pirin |
| Phospholipid/G3P catabolism | |||
| pldA (VF_1556) | 1.8 | 7.7 E-03 | outer membrane phospholipase A |
| glpD (VF_A0239) | 3.1 | 1.3 E-04 | glycerol-3-phosphate (G3P) dehydrogenase |
| glpC (VF_A0248) | 0.5 | 0.69 | anaerobic G3P dehydrogenase subunit C |
| glpB (VF_A0249) | 2.3 | 8.4 E-03 | anaerobic G3P dehydrogenase subunit B |
| glpA (VF_A0250) | 1.7 | 1.8 E-02 | anaerobic G3P dehydrogenase subunit A |
| glpQ (VF_A0958) | 3.2 | 2.8 E-21 | glycerophosphoryl diester phosphodiesterase GlpQ |
| Formate dehydrogenase subunits | |||
| fdnI (VF_1358) | −2.1 | 3.2 E-05 | formate dehydrogenase N subunit gamma |
| fdnH (VF_1359) | −2.7 | 1.3 E-03 | formate dehydrogenase N subunit beta |
| fdnG (VF_1360) | −2.9 | 5.9 E-08 | formate dehydrogenase N subunit alpha |
| fdhD (VF_1366) | −0.6 | 0.34 | formate dehydrogenase subunit delta |
| fdhF (VF_A0251) | −1.7 | 2.1 E-02 | formate dehydrogenase-H |
| Phosphate transport | |||
| pstB (VF_1983) | −0.3 | 0.52 | phosphate ABC transporter ATP-binding protein |
| pstA (VF_1984) | −1.2 | 4.8 E-02 | phosphate ABC transporter permease |
| pstC (VF_1985) | −1.2 | 1.6 E-02 | phosphate ABC transporter permease |
| pstS (VF_1986) | 1.8 | 1.3 E-05 | phosphate-binding protein |
| phoR (VF_1987) | 2.1 | 1.7 E-07 | phosphate regulon sensor |
| phoB (VF_1988) | 2.5 | 1.8 E-02 | DNA-binding response regulator with PhoR |
| pstS2 (VF_1610) | −5.7 | 1.8 E-24 | phosphate-binding protein |
| pstC2 (VF_1611) | −5.9 | 2.3 E-24 | phosphate ABC transporter permease |
| pstA2 (VF_1612) | −5.3 | 2.5 E-16 | phosphate ABC transporter permease |
| pstB2 (VF_1613) | −5.5 | 1.3 E-41 | phosphate ABC transporter ATP-binding protein |
| VF_A0555 | −1.7 | 1.8 E-04 | phosphate-binding protein |
 : upregulated;
: upregulated;
 : down-regulated in squid relative to seawater
: down-regulated in squid relative to seawater
Genes listed showed differential expression between squid-associated (Vnt) and planktonic (Plk) V. fischeri of at least abs(log2(fc) > 1) and FDR-adjusted p-value < 0.01.
In addition, genes associated with the extracellular breakdown of glycerophosphodiester to G3P and an alcohol (i.e., glpQ), and the subsequent catabolism of G3P (i.e., glpAB/glpD) were upregulated in the squid-associated samples (Table 1). This transcriptional pattern is reminiscent of the morning upregulation of genes associated with the anaerobic respiration of glycerol/G3P observed in the symbionts of adult squid (Wier et al., 2010); however, other, anaerobic respiration-linked genes upregulated by these symbionts (e.g., those encoding formate dehydrogenase; Table 1) were instead down-regulated in the symbionts of juveniles. These results indicate that, within the pre-dawn juvenile host, V. fischeri is likely to be fermenting G3P, distinct from its daytime respiration and, the nighttime fermentation of GlcNAc, in the mature symbiosis (Schwartzman et al., 2015).
To further investigate which nutrients might be used by the symbionts of juvenile animals, we applied flux coupling analysis (Burgard et al., 2004) using a new genome-scale metabolic model for V. fischeri. This analysis can identify metabolic genes that are linked to the utilization of specific carbohydrates, such that flux through the reactions catalyzed by these genes will indicate that a particular substrate is being used by the bacteria. This flux coupling analysis identified N-acetylneuraminic acid (NANA) as a possible carbon source for V. fischeri in juvenile animals based on an apparent upregulation of several genes involved in NANA uptake and utilization (Table S3). Independent support for NANA as a symbiont substrate comes from a high-throughput insertion sequencing study (Brooks et al., 2014) predicting genes encoding the putative transporter and lyase of NANA (nanT and nanA) to be colonization factors. In addition, NANA is a component of squid mucus, and a chemoattractant of V. fischeri (DeLoney-Marino et al., 2003; Schwartzman et al., 2015). The analysis also suggested glycerophosphodiester as a potential carbon source for V. fischeri in juvenile animals based on the upregulation in the symbionts of glpQ, as mentioned above. In the analysis, glpQ is the only gene specifically connected to the utilization of glycerophosphodiesters, because other genes involved in glycerol and G3P catabolism also play a role in the use of other nutrients. Overall, these results support the conclusion that nutrients such as NANA and glycerophosphodiester may be used as carbon sources by symbionts in the juvenile light organ (Pan et al., 2015). Future metabolomics studies aimed at detecting these and other potential carbon sources in the host will help refine comparisons between our flux coupling analysis and transcriptomic results.
Finally, while no specific mechanism has yet been established to explain what limits the growth of bacterial symbionts as they repopulate the light organ each day (Ruby, 1996), one hypothesis has been that the host controls symbiont growth by limiting access to a specific nutrient, such as phosphate. However, we observed that genes encoding for phosphate transporters were consistently more highly transcribed in both fast-growing cultured and slow-growing planktonic cells than in the squid-associated cells (Table 1), inconsistent with the notion that the symbiont population is phosphate-limited.
Clues to other conditions within the host
We observed several examples of genes that were relatively upregulated in the squid-associated bacteria, and that could represent responses to two specific classes of host-derived biochemical stress: reactive oxygen species (ROS) and antimicrobial peptides. The symbiont transcriptome indicated a relatively high expression of several genes predicted to be involved in the detoxification of peroxidized membrane lipids (Table 2; Fig. 2), a condition arising from exposure to ROS. Two of these genes are predicted to be glutathione S-transferases (GST). While the enzymatic activity of these putative GSTs has not been characterized, one (yfcG, VF_1082) shows strong sequence and domain homology to yfcG from Escherichia coli (blastp E-value < 10−54, amino acid identity = 46%), a GST with an important role in ROS resistance (Kanai et al., 2006; Stourman et al., 2011). Furthermore, we also observed significant upregulation of the N-ethylmaleimide reductase gene, nemA. This gene has similarly been linked to lipid-peroxidation stress in E. coli (Miura et al., 1997; Williams and Bruce, 2002). The NemA homolog of V. fischeri ES114 shows significant identity to YqjM from Bacillus subtilis (blastp E-value < 10−43, amino acid identity = 32%), which is involved in the oxidative stress response induced by hydrogen peroxide (Fitzpatrick et al., 2003). These results lead to the conclusion that V. fischeri is exposed to ROS stress within its eukaryotic host, a notion that is supported by previous analyses of the host tissue and symbiont genetics (Visick and Ruby, 1998; Ruby and McFall-Ngai, 1999; Mone et al., 2014; Schwartzman and Ruby, 2016).
Table 2.
Differentially expressed V. fischeri genes indicating potential stress in the juvenile symbiosis
| Gene* | Vnt/Plk log2(fc)† | FDR-adjusted p-value† | Description |
|---|---|---|---|
| Detoxification of peroxidized lipids | |||
| acrR (VF_1081) | 3.7 | 3.5 E-13 | TetR family transcriptional regulator |
| yfcG (VF_1082) | 5.6 | 2.9 E-28 | glutathione S-transferase |
| yghU (VF_1083) | 4.9 | 1.8 E-38 | glutathione S-transferase YghU |
| nemA (VF_A1049) | 4.6 | 2.5 E-11 | N-ethylmaleimide reductase FMN-linked |
| VF_A1050 | 2.1 | 1.8 E-02 | ABC transporter domain protein |
| RND family exporters | |||
| VF_1161 | 4.6 | 5.9 E-19 | periplasmic protein of efflux system |
| tolC (VF_1162) | 3.7 | 1.1 E-10 | outer membrane protein TolC |
| VF_1163 | 2.0 | 5.9 E-06 | export ABC transporter permease |
| VF_1164 | 1.5 | 1.9 E-04 | export ABC transporter permease |
| macB (VF_1165) | 2.8 | 3.1 E-06 | macrolide ABC transporter ATP-binding membrane protein |
| VF_A0742 | 2.7 | 7.6 E-03 | hypothetical protein |
| VF_A0743 | 3.0 | 1.7 E-03 | multidrug resistance protein A |
| VF_A0904 | 2.0 | 0.08 | transporter |
| VF_A0905 | 3.8 | 1.9 E-03 | hypothetical protein |
| VF_A0906 | 3.0 | 7.7 E-03 | acriflavin resistance periplasmic protein |
| VF_A0907 | 2.2 | 1.2 E-02 | acriflavin resistance plasma membrane |
| VF_A0044 | 3.8 | 3.8 E-04 | acriflavin resistance periplasmic protein |
| Colonization regulators | |||
| tcpP (VF_A0473) | 3.3 | 1.8 E-05 | transcriptional regulatory protein TcpP |
| tcpH (VF_A0474) | 4.9 | 3.5 E-05 | transcriptional regulatory protein TcpH |
| Outer membrane proteins | |||
| ompU (VF_0475) | 1.6 | 6.5 E-03 | outer membrane protein U porin |
| ompU2 (VF_A0487) | −6.7 | 2.9 E-33 | outer membrane protein U paralog |
 : upregulated;
: upregulated;
 : down-regulated in squid relative to seawater
: down-regulated in squid relative to seawater
Genes listed showed differential expression between squid-associated (Vnt) and planktonic (Plk) V. fischeri of at least abs(log2(fc) > 1) and FDR-adjusted p-value < 0.01; p-value >0.05 in black italics.
It was of particular interest to find three sets of genes that were annotated as encoding RND-family multidrug efflux pumps (Table 2). Such proteins share a domain structure predicted to be involved in the extrusion of antimicrobial small molecules and peptides (Koronakis et al., 2000), and are required by Vibrio cholerae for effective colonization (Bina et al., 2008). The relatively high expression of these genes suggests that V. fischeri cells are exposed to antimicrobial compounds that the symbionts are actively pumping out while they are within the light-organ crypts (Heath-Heckman et al., 2014). One locus (VF_1161–1165) has homology to a polyketide efflux system, and is regulated via quorum signaling (Antunes et al., 2007); thus, its induction is likely driven by the high density the symbiont population achieves within the crypts (McFall-Ngai, 2014). The basis for the 4- to 14-fold increase in expression of the two other putative RND-family gene sets (VF_A0742–0743) and (VF_ A0044; A0904–0907) is not clear; however, in V. cholerae there is a linkage between antimicrobial resistance and the increased expression of the membrane-bound regulatory proteins TcpP/H (Bina et al., 2008); these regulators are 8- and 30-fold induced within the light organ relative to seawater (Table 2). This high level of expression led us to ask whether tcpP/H plays a role in early colonization, perhaps by regulating important symbiosis determinants. Interestingly, while a tcpP/H deletion mutant colonized about as well as wild-type parent over the first 48 h, it increasingly lost the ability to compete over time (Fig. 3). Although V. fischeri doesn’t encode ToxT, the best-known target of TcpP/H in V. cholerae, we predict that the regulon of TcpP/H in V. fischeri includes important colonization genes, whose identities are under investigation.
Fig. 3.
Colonization defect of a tcpPH deletion mutant of V. fischeri. Newly hatched juvenile squid were placed into seawater inoculated with an equal number of wild-type and mutant cells. After 2, 3 and 4 days, the composition of the symbiont population in the light organ was determined and calculated as a relative colonization index (RCI). The tcpPH mutant was increasingly out-competed over the course of the colonization. Data are from two independent trials.
Finally, the major outer membrane porin OmpU senses and protects V. cholerae from bactericidal/permeability-increasing peptides (Mathur and Waldor, 2004; Mathur et al., 2007), like those induced by V. fischeri during colonization of its squid host (Krasity et al., 2011). Much as in V. cholerae, the 3- to 4-fold increase in ompU expression by symbiotic V. fischeri (Table 2) may be triggered by the surfeit of amino acids in the crypt matrix (Graf and Ruby, 1998; Mey et al., 2012). Taken together, these patterns of symbiont gene expression indicate that the environment of the light-organ crypt presents V. fischeri with the ongoing challenge of host-produced antimicrobial compounds. The recognition that OmpU is a protecting colonization factor for both pathogenic (Duperthuy et al., 2010) and beneficial (Aeckersberg et al., 2001) species of Vibrio emphasizes the context-dependency of host-microbe interaction (Hentschel et al., 2000)
Recent work with other symbioses has revealed that host-derived stresses are a common theme, not only between V. fischeri and its pathogenic congeners, but also among the microbiota of several vertebrate and invertebrate mutualisms (Schwartzman and Ruby, 2016). In each of these other associations, by presenting a particular mixture of antimicrobial peptides on its epithelial surfaces, the host apparently manages the composition of its epithelium-associated microbiota, selecting for particular communities of beneficial microbes while excluding pathogens (Duerkop, 2009; Vaishnava, 2011; Franzenburg, 2013). Perhaps the synergistic activity of these kinds of RND multidrug effluxers (Bohnert et al., 2011) induced by the symbionts (Table 2) allow them to target the squid’s specific antimicrobial peptide, galaxin (Heath-Heckman et al., 2014), and/or other compounds, contributing to the strict specificity of the symbiosis (McFall-Ngai, 2014).
Conclusions
We have reported here the first full RNA-Seq datasets of Vibrio fischeri, including one that is representative of the bacteria during colonization of the juvenile host. The gene expression patterns of bacteria making the habitat transition from seawater to host tissue have provided previously unavailable information suggesting that, once within the juvenile host, V. fischeri switches from a respiratory metabolism to one characteristic of G3P fermentation. Further, a flux coupled analysis using these transcriptional datasets and a newly developed genome-scale metabolic model for V. fischeri (Pan et al., in review) have also provided us with predictive details about symbiont metabolism that would be inaccessible through conventional gene-expression analysis alone. Finally, this study identified the V. cholerae virulence determinants tcpPH as candidate regulators during the habitat transition of V. fischeri, and went on to experimentally demonstrate their importance to the success of a beneficial microbial symbioses.
Experimental procedures
Bacterial strains and growth conditions
The wild-type bacterium used in this study was V. fischeri strain ES114, isolated from an E. scolopes light organ (Boettcher and Ruby, 1990). Mutant V. fischeri cells were derived from strain ES114. The Escherichia coli cells used for cloning were strain DH5α λpir (Hanahan, 1983). Deletion mutants and fluorescently labeled strains were constructed as described previously (Stabb and Ruby, 2002; Dunn et al., 2006; Le Roux et al., 2007; Shibata and Visick, 2012). Primers were designed to generate homology regions upstream and downstream of the tcpPH locus (tcpPH_H1_Fwd: 5′-TCA GCC GTC GAC TGT TTA CTT CAA ATT AAT GAA GAG; tcpPH_H1_Rev: 5′-TCA GCC GCA TGC AAC CAT ATC ATC CTG ACT TAA; tcpPH_H2_Fwd: 5′-TCA GCC GCA TGC AAA TAC AAT AAT TAA TTT ATT TTC AAT AAT TTG; tcpPH_H2_Rev: 5′-TCA GCC ACT AGT ATT AAG ATA TGA TTT GAA AAA CAA ATT TTC), and the genotype of the mutant was confirmed via PCR (tcpPH_Conf_Fwd: 5′-ATT GCC AAA AAT CGG TTT; tcpPH_Conf_Rev: 5′-AAC CAT CCA CTT TGA TTT CAT). V. fischeri cells were grown at 28 °C with shaking in either Luria-Bertani salt (LBS) broth (per liter, 10 g tryptone, 5 g yeast extract, and 20 g NaCl) or a dilute artificial seawater-based tryptone (SWT) broth (5 g tryptone, 3 g yeast extract, 3 mL glycerol, 700 mL Instant Ocean (IO; Aquarium Systems, Mentor, OH, USA) at 33 ppt, and 300 mL water) as indicated. E. coli cells were grown in Luria-Bertani (LB) broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 37 °C with shaking. Planktonic conditions were simulated by culturing V. fischeri cells to mid-log phase in SWT medium, then inoculating filter-sterilized IO (FSIO) with approximately 2×108 CFU, and allowing the suspension to incubate at room temperature (~23 °C) for 18 h. Antibiotics were used at the following concentrations when appropriate: erythromycin at 5 mg/mL for V. fischeri and 150 mg/mL for E. coli, and chloramphenicol at 2.5 mg/mL for V. fischeri and 25 mg/mL for E. coli.
Collection of bacterial RNA
Bacterial RNA was obtained from V. fischeri cells either grown in SWT to an OD600 of ~0.5 (‘cultured’), or cultured in SWT to an OD600 of ~0.5, then incubated in artificial seawater for 18 h (‘planktonic’), as described above (Fig. 1A). In both cases, the bacteria were collected by centrifugation at 12,000 rpm for 2 min (Model 5254R, Eppendorf) immediately prior to RNA extraction and purification using a ZR RNA MicroPrep Kit (Zymo Research).
To obtain squid-associated bacteria, freshly hatched juvenile squid from the University of Wisconsin-Madison aquarium facility were collected into V. fischeri-free IO. Animals were subsequently exposed for 3 h to approximately 5000 CFU of V. fischeri per mL, and then transferred to fresh V. fischeri-free IO. Animals were maintained on a 12:12 hour day:night cycle in batches of approximately 30 to 35 animals to optimize uniformity of colonization timing between individual animals. After ~36 h, colonization of the animals was verified by determining their production of bioluminescence using a luminometer (Turner TD 20/20). At approximately 48 h post-colonization, equivalent to the end of night time, animals were collected in batches of 100 into 80 mL of filter-sterilized IO, and exposed to light for 15 min to induce the normal dawn-cued expulsion of their bacterial population (Boettcher and Ruby, 1990). Squid were subsequently removed, and the expelled bacteria were collected by centrifugation at 12,000 rpm for 10 min at room temperature in an SS-34 rotor (Thermo Fisher Scientific). The supernatant was discarded, and total bacterial RNA was immediately extracted and purified using a ZR RNA MicroPrep Kit. In all cases, the time from initial light exposure to RNA extraction was < 30 min.
RNA-Seq library preparation and sequencing
Three (3) replicate RNA samples for each condition (squid-associated, planktonic, or cultured) were processed for RNA-Seq transcriptome analysis. Starting with total RNA, ribosomal RNA was removed using the Ribo-Zero Gold Epidemiology Kit (Epicentre), which removed prokaryotic (V. fischeri) rRNA as well as any contaminating eukaryotic (E. scolopes) rRNA. The resulting ribo-depleted RNA was then used for library preparation using the TruSeq RNA sample preparation kit (Illumina). Libraries were sequenced using Illumina HiSeq 2500 with high output, V4 chemistry, and 100-bp single-end reads. Fastq files were de-multiplexed, yielding on average 18M reads per sample (range: 5 to 26M reads per sample). Per-sample raw fastq files and processed CDS and rRNA count tables have been submitted to NCBI Gene Expression Omnibus (GEO) with accession number GSE80607.
Sequence read processing and mapping
Single-end fastq files were trimmed with Trimmomatic v.0.33, using the parameters ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 (the file TruSeq3-SE.fa contained TruSeq3 indexed and universal adapters as well as poly-A and poly-T sequences) (Bolger et al., 2014). Reads from each sample were mapped to the V. fischeri ES114 genome (NC_006840.2) using bwa v.0.6.2 (Burrows–Wheeler Aligner), applying commands index, aln, and samse. The resulting SAM files were processed with SAMtools v.0.1.19 to generate BAM files (Li et al., 2009). The numbers of reads mapping to protein-coding (CDS) or rRNA genes were calculated using the htseq-count command of HTSeq v.0.6.1p1 (Anders et al., 2015). Ribo-depleted samples for the primary three-treatment comparison yielded an average of 10.0M CDS reads mapped per sample; ribo-depleted samples for the low-biomass comparison yielded an average of 5.3M CDS reads mapped per sample; in addition, ~34,000 rRNA reads were mapped per sample across both sets of ribo-depleted samples.
Differential expression analysis
Detection of genes differentially expressed (i.e., relative transcript abundance) between conditions was performed using the R packages DESeq2 and NOISeq from the Bioconductor program (Anders and Huber, 2010; Tarazona et al., 2011). Low counts were removed using the NOISeq command filtered.data, with norm = FALSE, depth = NULL, method = 1, cv.cutoff = 10, and cpm = 5. After loading data into a data frame, DESeq2 was run using the commands DESeqDataSetFromMatrix, estimateSizeFactors, estimateDispersions, and nbinomWaldTest. The three conditions (squid-associated, planktonic, and cultured) were contrasted pairwise for all genes, and results exported as Benjamini–Hochberg adjusted p-values and log2(fold change). Differentially expressed genes were identified using tiered cutoffs of these values, with the most stringent cutoff being an adjusted p-value < 0.001 and abs(log2(fold change)) > 3.0 (three replicates per condition). All code used in data analysis and manuscript preparation is available at https://github.com/cuttlefishh/papers/tree/master/vibrio-fischeri-transcriptomics.
Light-organ colonization assay
To determine the relative colonization effectiveness of the tcpP/H deletion mutant, inocula were prepared that contained a 1:1 mixture of wild-type and mutant cells, differentially labeled with either green- (GFP) or red-fluorescent protein (RFP) (Dunn et al., 2006). The inocula were prepared by growing each strain in SWT medium to mid-exponential phase (OD600 ~0.3) before diluting them in FSIO. Newly hatched squid were then transferred into the inoculated FSIO containing a pair of strains, with a total inoculum of around 5,000 colony-forming unit (CFU) per mL. After 3 h of exposure to the inoculum, each squid was removed to a vial containing 4 mL of bacteria-free FSIO; at 24 h the FSIO was changed, and at 48, 72 and 96 h, 20 squids were homogenized, dilutions spread on LBS agar, and the number of GFP and RFP CFUs were counted using a fluorescence dissecting scope. The extent of a colonization defect in the tcpP/H mutant was determined by a relative competitive index (RCI), calculated as the ratio of the mutant to wild type in the light organ divided by the ratio in the inoculum (RCI = log[(CFU mutant)/(CFU wild type)]/[(inoculum CFU mutant)/(inoculum CFU wild type)]).
Flux-coupling analysis of symbiont sugar uptake
We analyzed the transcriptome data using a constraint-based genome-scale metabolic model developed for V. fischeri ES114 (Pan et al., in review). Flux-coupling analysis was performed on the model to identify which reactions (and their associated genes) are fully, partially, or directionally coupled to nutrient uptake (Burgard et al., 2004). These types of reaction–nutrient coupling indicate that if the coupled reaction is active (i.e., carries a non-zero flux) then the nutrient must be consumed. As a result, the genes associated with these coupled reactions might serve as biomarkers for nutrient uptake. Flux-coupling analysis was performed assuming all sugars, amino acids, nucleotides and inorganic nutrients were available (by making the lower limits for all the exchange reactions in the model negative). The genes associated with reactions coupled to only one nutrient were compared to the list of differentially regulated genes to allow us to determine which nutrients could be used by V. fischeri. This metabolic model is included within the supplemental materials (Table S4).
Supplementary Material
Effectiveness of ribo-depletion for enrichment of CDS transcripts over rRNA transcripts. (A) Proportions of reads assigned to CDS and rRNA in ribo-depleted and non-ribo-depleted samples. (B) Distribution of read assignments to CDS and rRNA among “sum good alignments”, “not aligned”, “ambiguous”, and “no feature” groupings.
Pairwise scatter plots of CDS transcript counts between ribo-depleted and non-ribo-depleted samples, in relative abundance units (counts per million total CDS counts).
Pairwise scatter plots of low-biomass (between 10 and 1000 ng) total-RNA library-prep counts versus counts from the 100-ng total-RNA sample. All samples shown were ribo-depleted. Transcript counts are in relative abundance units (counts per million total CDS counts).
Pairwise scatter plots of low-biomass (between 1 and 5 ng) total-RNA library-prep counts versus counts from the 100-ng total RNA sample. All samples shown were ribo-depleted. Transcript counts are in relative abundance units (counts per million total CDS counts).
Effectiveness of low-biomass analyses to detect rare transcripts. (A) Percent coverage of the V. fischeri genome as function of total RNA input and the number of library amplification cycles. (B) Fraction of library preps a transcript is detected in (out of 15 total) as a function of total RNA input amount.
Summary of V. fischeri samples analyzed by RNA-Seq. For each sample name the following information is provided: accession number (NCBI GEO SRA), description and source name (low-biomass test or replicate #1–3 of the planktonic, SWT or vented treatments), organism, growth medium, total RNA input, ribo-depletion status, library PCR amplification cycles, replicate number, and molecule type. Full study data including CDS and rRNA count tables are available using the main GEO accession number GSE80607 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80607). Note: the sample for the 100-ng ribo-depleted test (i.e., 100ng_Ribodpl) is identical to SWT replicate 3 (Swt3_Ribodpl).
Relative expression levels of bioluminescence and motility genes in V. fischeri. Genes listed showed differential expression between squid-associated (Vnt) and planktonic (Plk) V. fischeri of at least abs(log2(fc) > 1) and FDR-adjusted p-value < 0.01
Differentially expressed genes appearing during both the comparisons of squid-associated V. fischeri (Vnt), seawater-suspended V. fischeri (Plk) and SWT medium-grown V. fischeri(SWT). Differential expression cutoffs used |log2(fc)| > 3 and FDR-adjusted p-value < .001. Those genes whose relative level of expression is in the same direction for Vnt/Plk and Vnt/SWT comparisons are indicated by a check mark.
Originality-Significance Statement.
This is the first whole-genome transcriptional investigation of symbionts that have made the habitat transition from seawater into their juvenile host. Performance of this study required development of a transcriptional analysis procedure that was sensitive enough to measure and validate the mRNA signature of the few hundred thousand bacteria present in the juvenile symbiosis, amidst a large background signal chiefly composed of host and symbiont ribosomal RNA. The significance and novelty of this work is that it identifies which genes are likely candidates for driving specific bacterial activities early in the Vibrio fischeri–squid symbiosis. Previously undescribed transcriptional patterns that were detected include responses to biochemical stresses inside the host, and metabolic patterns distinct from those previously described in the symbionts of adult animals. The sole similarities to the only previous transcriptional study performed on symbionts in the (adult) host were the presence of high luciferase and low flagellin gene expression. One unexpected discovery, the induction of the regulatory locus tcpPH in juvenile symbionts, led to experiments that showed these genes, which are virulence determinants in Vibrio cholerae, are required for beneficial colonization of the juvenile host.
Acknowledgments
We thank members of the Ruby, McFall-Ngai, and Reed labs for their input and critical review of this work and manuscript, K Jepsen, M Khosroheidari, H Matsui, and J Huntley for technical assistance in RNA-Seq, P Richmond for assistance in read mapping and counting, and J Morton for advice on statistical analyses. This work was funded by Gordon and Betty Moore Foundation grants #3396 to EGR and JLR, and MSN156910 to RK. Animal experiments were supported by NIH grant OD011024 to EGR. KN was supported by a Ruth L. Kirschstein National Research Service Award from the NIGMS (F32GM112214).
References
- Aeckersberg F, Lupp C, Feliciano B, Ruby EG. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J Bacteriol. 2001;183:6590–6597. doi: 10.1128/JB.183.22.6590-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S, Pruss K, Taylor RK. Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog. 2015;11:e1004787. doi: 10.1371/journal.ppat.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG, Greenberg EP. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol. 2007;189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Provenzano D, Nguyen N, Bina JE. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun. 2008;76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert JA, Szymaniak-Vits M, Schuster S, Kern WV. Efflux inhibition by selective serotonin reuptake inhibitors in Escherichia coli. J Antimicrob Chemother. 2011;66:2057–2060. doi: 10.1093/jac/dkr258. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. MicrobiologyOpen. 2013;2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A. 2014;111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14:301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs PA, Eisen MB. Low-cost, low-input RNA-seq protocols perform nearly as well as high-input protocols. PeerJ. 2015;3:e869–e869. doi: 10.7717/peerj.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Bouletreau M. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci USA. 2001;98:6247–6252. doi: 10.1073/pnas.101304298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloney-Marino CR, Wolfe AJ, Visick KL. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol. 2003;69:7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Karr EA, Wang Y, Batton AR, Ruby EG, Stabb EV. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol Microbiol. 2010;77:44–55. doi: 10.1111/j.1365-2958.2010.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperthuy M, Binesse J, LeRoux F, Romestand B, Caro A, Got P, Givaudan A, Mazel D, Bachere E, Destoumieux-Garzon D. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol. 2010;12:951–963. doi: 10.1111/j.1462-2920.2009.02138.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Macheroux P. Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J Biol Chem. 2003;278:19891–19897. doi: 10.1074/jbc.M211778200. [DOI] [PubMed] [Google Scholar]
- Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci USA. 2013;110:E3730–8. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Q Rev Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Ruby EG. Characterization of the nutritional environment of a symbiotic light organ using bacterial mutants and biochemical analyses. Proc Natl Acad Sci USA. 1998;95:1818–1822. [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hansen GA, Ahmad R, Hjerde E, Fenton CG, Willassen NP, Haugen P. Expression profiling reveals Spot 42 small RNA as a key regulator in the central metabolism of Aliivibrio salmonicida. BMC Genomics. 2012;13:37. doi: 10.1186/1471-2164-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EA, Gillette AA, Augustin R, Gillette MX, Goldman WE, McFall-Ngai MJ. Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ Microbiol. 2014;16:3669–3682. doi: 10.1111/1462-2920.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio. 2013;4(2):e00167–13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Steinert M, Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000;8:226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- Hongoh Y. Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci Biotechnol Biochem. 2010;74(6):1145–1151. doi: 10.1271/bbb.100094. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35:507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T, Takahashi K, Inoue H. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J Biochem. 2006;140:703–711. doi: 10.1093/jb/mvj199. [DOI] [PubMed] [Google Scholar]
- Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39:14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans. 2011;39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol. 2007;73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Davis BM, Waldor MK. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol. 2007;63:848–858. doi: 10.1111/j.1365-2958.2006.05544.x. [DOI] [PubMed] [Google Scholar]
- Mathur J, Waldor MK. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect Immun. 2004;72:3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun. 2012;80:518–528. doi: 10.1128/IAI.05851-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Tomioka Y, Hoshi Y, Suzuki H, Yonezawa M, Hishinuma T, Mizugaki M. The effects of unsaturated fatty acids, oxidizing agents and Michael reaction acceptors on the induction of N-ethylmaleimide reductase in Escherichia coli: possible application for drug design of chemoprotectors. Methods Find Exp Clin Pharmacol. 1997;19:147– 151. [PubMed] [Google Scholar]
- Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. The N-acetyl-D-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol. 2011;82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Ruby EGE. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone Y, Monnin D, Kremer N. The oxidative environment: a mediator of interspecies communication that drives symbiosis evolution. Proc R Soc B. 2014;281:20133112. doi: 10.1098/rspb.2013.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai M. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- Mullard A. Microbiology: Tinker, bacteria, eukaryote, spy. Nature. 2009;459:159–161. doi: 10.1038/459159a. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. [Google Scholar]
- Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai M. The winnowing: establishing the squid–vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Pan M, Schwartzman JA, Dunn AK, Lu Z, Ruby EG. A Single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. mBio. 2015;6:e00811–15. doi: 10.1128/mBio.00811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Abo RP, De Nisco NJ, Arnold MF, Longhi R, Zanda M, Walker GC. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci USA. 2014;111:3561–3566. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Lee KH. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- Russell JB, Rychlik JL. Factors that alter rumen microbial ecology. Science. 2001;292:1119–1122. doi: 10.1126/science.1058830. [DOI] [PubMed] [Google Scholar]
- Schwartzman JA, Koch E, Heath-Heckman EA, Zhou L, Kremer N, McFall-Ngai MJ, Ruby EG. The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci USA. 2015;112:566–571. doi: 10.1073/pnas.1418580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman JA, Ruby EG. Stress as a normal cue in the symbiotic environment. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septer AN, Stabb EV. Coordination of the arc regulatory system and pheromone-mediated positive feedback in controlling the Vibrio fischeri lux operon. PLoS ONE. 2012;7:e49590. doi: 10.1371/journal.pone.0049590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septer AN, Bose JL, Lipzen A, Martin J, Whistler C, Stabb EV. Bright luminescence of Vibrio fischeri aconitase mutants reveals a connection between citrate and the Gac/Csr regulatory system. Mol Microbiol. 2015;95:283–296. doi: 10.1111/mmi.12864. [DOI] [PubMed] [Google Scholar]
- Shibata S, Visick KL. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol. 2012;194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Soo PC, Horng YT, Lai MJ, Wei JR, Hsieh SC, Chang YL, et al. Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J Bacteriol. 2007;189:109–118. doi: 10.1128/JB.00710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stourman NV, Branch MC, Schaab MR, Harp JM, Ladner JE, Armstrong RN. Structure and function of YghU, a nu-class glutathione transferase related to YfcG from Escherichia coli. Biochemistry. 2011;50:1274–1281. doi: 10.1021/bi101861a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzal FA, McClane BA, Cheung JK, Theoret J, Garcia JP, Moore RJ, Rood JI. Animal models to study the pathogenesis of human and animal Clostridium perfringens infections. Vet Microbiol. 2015;179:23–33. doi: 10.1016/j.vetmic.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIII-gamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci USA. 2010;107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RE, Bruce NC. ‘New uses for an Old Enzyme’–the Old Yellow Enzyme family of flavoenzymes. Microbiology. 2002;148:1607–1614. doi: 10.1099/00221287-148-6-1607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effectiveness of ribo-depletion for enrichment of CDS transcripts over rRNA transcripts. (A) Proportions of reads assigned to CDS and rRNA in ribo-depleted and non-ribo-depleted samples. (B) Distribution of read assignments to CDS and rRNA among “sum good alignments”, “not aligned”, “ambiguous”, and “no feature” groupings.
Pairwise scatter plots of CDS transcript counts between ribo-depleted and non-ribo-depleted samples, in relative abundance units (counts per million total CDS counts).
Pairwise scatter plots of low-biomass (between 10 and 1000 ng) total-RNA library-prep counts versus counts from the 100-ng total-RNA sample. All samples shown were ribo-depleted. Transcript counts are in relative abundance units (counts per million total CDS counts).
Pairwise scatter plots of low-biomass (between 1 and 5 ng) total-RNA library-prep counts versus counts from the 100-ng total RNA sample. All samples shown were ribo-depleted. Transcript counts are in relative abundance units (counts per million total CDS counts).
Effectiveness of low-biomass analyses to detect rare transcripts. (A) Percent coverage of the V. fischeri genome as function of total RNA input and the number of library amplification cycles. (B) Fraction of library preps a transcript is detected in (out of 15 total) as a function of total RNA input amount.
Summary of V. fischeri samples analyzed by RNA-Seq. For each sample name the following information is provided: accession number (NCBI GEO SRA), description and source name (low-biomass test or replicate #1–3 of the planktonic, SWT or vented treatments), organism, growth medium, total RNA input, ribo-depletion status, library PCR amplification cycles, replicate number, and molecule type. Full study data including CDS and rRNA count tables are available using the main GEO accession number GSE80607 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80607). Note: the sample for the 100-ng ribo-depleted test (i.e., 100ng_Ribodpl) is identical to SWT replicate 3 (Swt3_Ribodpl).
Relative expression levels of bioluminescence and motility genes in V. fischeri. Genes listed showed differential expression between squid-associated (Vnt) and planktonic (Plk) V. fischeri of at least abs(log2(fc) > 1) and FDR-adjusted p-value < 0.01
Differentially expressed genes appearing during both the comparisons of squid-associated V. fischeri (Vnt), seawater-suspended V. fischeri (Plk) and SWT medium-grown V. fischeri(SWT). Differential expression cutoffs used |log2(fc)| > 3 and FDR-adjusted p-value < .001. Those genes whose relative level of expression is in the same direction for Vnt/Plk and Vnt/SWT comparisons are indicated by a check mark.