Abstract
Rationale
Direct reprogramming of cardiac fibroblasts to cardiomyocytes has recently emerged as a novel and promising approach to regenerate the injured myocardium. We have previously demonstrated the feasibility of this approach in vitro and in vivo using a combination of four microRNAs (miR-1, miR-133, miR-208 and miR-499) that we named miR combo. However, the mechanism of miR combo mediated direct cardiac reprogramming is currently unknown.
Objective
Here we investigated the possibility that miR combo initiated direct cardiac reprogramming through an epigenetic mechanism.
Methods and Results
Using a qPCR array, we found that histone methyltransferases and demethylases that regulate the tri-methylation of H3K27 (H3K27me3), an epigenetic modification that marks transcriptional repression, were changed in miR combo treated fibroblasts. Accordingly, global H3K27me3 levels were downregulated by miR combo treatment. In particular, the promoter region of cardiac transcription factors showed decreased H3K27me3 as revealed by ChIP-qPCR. Inhibition of H3K27 methyltransferases or of the Polycomb Repressive Complex 2 (PRC2) by pharmaceutical inhibition or siRNA reduced the levels of H3K27me3 and induced cardiogenic markers at the RNA and protein level, similarly to miR combo treatment. In contrast, knockdown of the H3K27 demethylases Kdm6A and Kdm6B restored the levels of H3K27me3 and blocked the induction of cardiac gene expression in miR combo treated fibroblasts.
Conclusions
In summary, we demonstrated that removal of the repressive mark H3K27me3 is essential for the induction of cardiac reprogramming by miR combo. Our data not only highlight the importance of regulating the epigenetic landscape during cell fate conversion but also provide a framework to improve this technique.
Keywords: Chromatin, epigenomics, cellular reprogramming, microRNAs, cardiac myocytes, regeneration
Subject Terms: Cellular Reprogramming, Mechanisms, Epigenetics, Gene Expression and Regulation
INTRODUCTION
Myocardial infarction (MI) is a leading cause of death in humans. In mammals, cardiomyocytes start to exit the cell cycle soon after birth and become terminally differentiated. This dramatic decrease of proliferative potential results in limited regenerative capacity in the adult mammalian heart following cardiac injury.
Over the past decade, various techniques have been employed to regenerate the injured heart with limited success 1, 2. A number of researchers, including ourselves, have employed the direct reprogramming of fibroblasts to cardiomyocytes for efficient cardiac regeneration 3–6. Whilst others have used transcription factors to achieve the direct reprogramming of fibroblasts 7–13 to cardiomyocytes, we have employed microRNAs (miRs). We identified a combination of four miRs (miR-1, miR-133a, miR-208a and miR-499; miR combo) that directly reprogrammed fibroblasts into cardiomyocyte-like cells 14. Importantly, we demonstrated the utility of our strategy in vivo 14, 15. Administration of miR combo into the ischemic myocardium promoted reprogramming; 10 % of the cardiomyocytes in the infarct border zone were from a fibroblast origin two months after MI. This was associated with improved cardiac function post-MI and a dramatic decrease in fibrosis. Recent work from Muraoka et al. showed that miR-133a enhanced cardiac reprogramming mediated by Gata4, Mef2c and Tbx5 (namely GMT) by silencing the fibroblast signature through direct repression of Snai1 16. Despite this advance, the mechanism of direct cardiac reprogramming remains largely unknown.
Direct cardiac reprogramming requires the activation of the cardiogenic transcriptional program in concert with the repression of the fibroblastic transcriptional program. Epigenetic modifications strongly impact cell fate decision during embryonic development and cell differentiation by modulating chromatin accessibility and transcriptional activity. Tri-methylation of the lysine 27 of histone H3 (H3K27me3) is a hallmark of transcriptional repression. It results from the enzymatic activity of the methyltransferases Ezh1 and Ezh2 which associate with Eed and Suz12 to form the Polycomb Repressive Complex 2 (PRC2). Regulation of H3K27 methylation is essential for cardiac development and homeostasis. Conditional knockout of Ezh2 in cardiac progenitors and cardiomyocytes using the Nkx2.5-Cre recombinase leads to lethal congenital heart defects 17, 18. Loss of Ezh2 in cardiomyocytes of the anterior heart field leads to hypertrophy 19. Global gene analysis of conditional Ezh2 mutants demonstrated that Ezh2 promoted the cardiogenic transcriptional program 18, 19. During embryonic stem cell (ESC) differentiation to cardiomyocytes, cardiac gene determinants progressively lose H3K27me3 20, 21. Likewise, GMT-induced cardiomyocytes display low levels of H3K27me3 at the promoter of cardiac markers 8. Removal of H3K27me3 is achieved by the activity of the demethylases Kdm6A (UTX, which is carried on the X chromosome) and Kdm6B (Jmjd3) 22–26. Kdm6A knockout in female mice leads to severe congenital heart defects 27. Similarly, cardiovascular differentiation of ESC is compromised in the absence of Kdm6A and Kdm6B 27, 28 reinforcing the idea that regulation of H3K27 methylation is essential for cardiac fate acquisition. Interestingly, PRC2 and H3K27 methylation also play an important role during somatic reprogramming to pluripotency 29, 30 but their role during direct reprogramming to cardiomyocyte is unknown.
In this study, we examined the possibility that miR combo induces direct reprogramming of fibroblasts into cardiomyocytes by modulating the epigenetic landscape. Using a screening strategy, we identified histone modifiers for H3K27 methylation as potential candidates. MiR combo repressed Ezh2 expression whereas it up-regulated Kdm6A and Kdm6B expressions. Accordingly, H3K27me3 levels were decreased in miR combo transfected fibroblasts, suggesting a global de-repression of transcription in these cells. Using small molecule inhibitor and siRNA mediated knockdown, we demonstrated that demethylation of H3K27 is necessary to initiate cardiac reprogramming by miR combo. Our study has thus identified a mechanism by which miR combo modifies the epigenetic landscape to activate a cardiogenic program in differentiated somatic cells.
METHODS
Cardiac fibroblasts isolation and transfection
Cardiac fibroblasts were isolated from 1-day old C57BL/6 mouse neonates as previously described 31. Briefly, harvested hearts were minced and digested with Collagenase II. To eliminate any cardiomyocytes, the cell mixture was subjected to a Percoll gradient centrifugation. Isolated fibroblasts were plated on 0.2% gelatin-coated flasks and cultured in growth media (DMEM, 15% FBS, Pen/Strep). Cells were passaged twice before being used for transfection.
Transient transfections were performed as previously described 31. Synthetic mimic of pre-miRs (Ambion/Applied Biosystems) and siRNAs (FlexiTube siRNA, Qiagen) were used at a final concentration of 50 nmol/L. A list of the miRs and siRNAs used is provided in Supplementary Material.
DZNep and CHX treatments
For the treatment with 3-Deazaneplanocin A (DZNep), growth media containing 2 μmol/L of DZNep (EMD Millipore) was added 24 hours after transfection, changed daily until day3, then every other day until day14. For the treatment with cycloheximide (CHX), growth media containing 50 mg/L of CHX (Sigma Aldrich, #01810) was added for 12 hours of culture.
RNA isolation and quantitative real-time PCR
Total RNAs were isolated using an RNeasy miniprep kit (Qiagen) according to the manufacturer’s instructions. Reverse Transcription was carried out using a High-Capacity cDNA Synthesis Kit (Thermo-Fisher). Quantitative expression of each gene was assessed using Taqman Gene Expression Assays on a StepOnePlus Real-Time PCR System (Applied Biosystems). A list of the primers used in this study is provided in the Supplementary Material.
Immunoblotting
Histone extracts were isolated from harvested cells using the EpiQuick Total Histone Extraction Kit (Epigentek) according to the manufacturer’s instructions. Nuclear extracts were isolated from harvested cells using standard nuclear fractionation protocol (Abcam). Protein concentrations were determined by the Lowry protein assay and 2 μg of histone or nuclear extract was used per well. Samples were run on 4–12% Bis-Tris gels (NuPAGE, Thermo-Fisher) in MES-SDS running buffer (NuPAGE, Thermo-Fisher). After 90 min of transfer at 30mV, nitrocellulose membranes were stained overnight at 4°C with anti-H3K27me3 (Cell Signaling, #9733; dilution 1/250), anti-H3 (Active Motif, #61475; dilution 1/5000), anti-Ezh2 (Cell Signaling, #5246; dilution 1/500) and anti-TBP (Cell Signaling, #44059; dilution 1/5000) diluted in blocking buffer (TBS, 0.1% Tween, 5% BSA). Protein detection was performed using HRP-coupled antibodies (Cell Signaling) and the ECL Prime detection reagent (GE Healthcare). Membranes were imaged and quantified using the G:BOX Chemi XT4 and associated software (Syngene).
Luciferase reporter assay
Luciferase reporter constructs containing the 3′-UTR of Twf1 or the 3′UTR of Ezh2 were purchased from Genecopoeia (miTarget™ 3′ UTR miRNA Target Clones). The empty vector pEZX-MT05 was used as a control. HEK-293 cells were seeded in duplicate in 24-well plates and co-transfected with miR(s) and luciferase vector using the Lipofectamine LTX and PLUS reagents according to the manufacturer instructions (Thermo-Fisher). Media was collected 48 hours after transfection and the luciferase and alkaline phosphatase activities were measured using the secrete-pair dual luminescence assay kit (Genecopoeia).
ChIP-qPCR
ChIP were performed using the Magnify ChIP system according to the manufacturer’s instructions (Thermo-Fisher). Briefly, 2–3 million cells were cross-linked with 1% paraformaldehyde/PBS for 10 min and sheared using Covaris S220 ultrasonicator (10 cycles of 60 seconds, intensity 2) to obtain an average DNA fragment size of 300bp. The sheared DNA was diluted 10 times and incubated overnight at 4°C with dynabeads protein A/G and 5μg of anti-H3K27me3 (Cell Signaling, #9733) or 1 μg of control IgG (Thermo-Fisher) under constant rotation. The next day, the bound chromatin was washed and eluted by incubating the dynabeads-chromatin complexes with the reverse crosslinking buffer and proteinase K for 1 hour at 55°C and 2 hours at 65°C in a thermocycler. The DNA was purified using magnetic beads and eluted by 1-hour incubation at 55°C with 150 μL of elution buffer. DNA quality/concentration was measured with Qubit dsDNA HS assay kit (Thermo-Fisher).
For qPCR, we used the SYBR Green PCR master mix (Applied Biosystems) and a set of 3 to 5 validated primers for Tbx5, Mef2c and Gata4 32. We also used Gapdh and Pax2 primers as negative and positive controls for H3K27me3 (Active Motif, #71016 and #71020). Results were expressed as a percentage of Input DNA.
Data and statistical analysis
Statistical analysis was performed using Student’s t-test (2-sample equal variance, 2-tailed) unless specified otherwise. P < 0.05 was regarded as significant. Graphs are displayed as mean ± SEM.
RESULTS
MiR combo affects H3K27 methylation and modifying enzymes
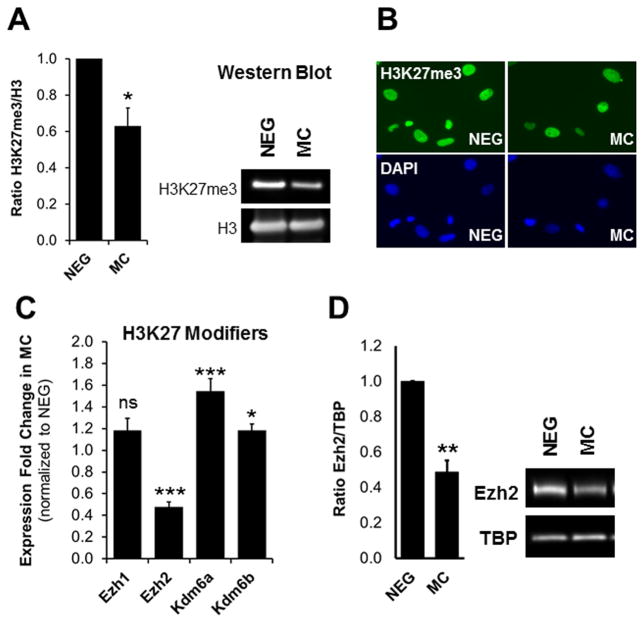
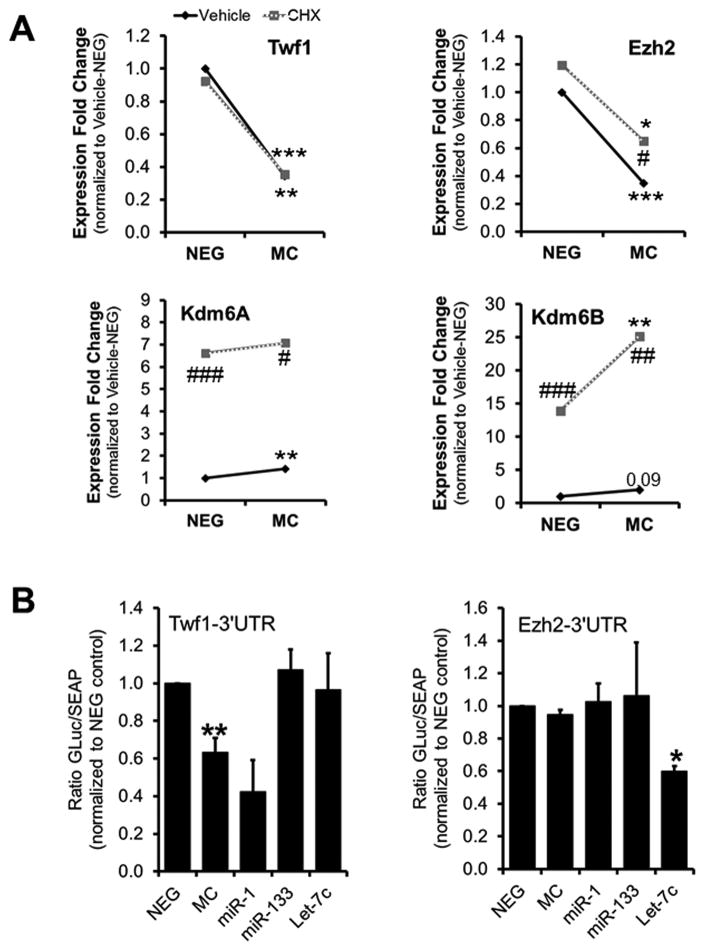
To determine whether miR combo utilizes an epigenetic mechanism to induce the direct reprogramming of fibroblasts into cardiomyocytes we first analyzed the expression of various chromatin modifiers by quantitative PCR array (RT2 Profiler™ PCR Array Mouse Epigenetic Chromatin Modification Enzymes). Since cardiac transcription factor expression is induced within the first 3 days of miR combo treatment, we collected RNA from neonatal cardiac fibroblasts 3 days after transfection. The array profiled the expression of 84 key genes encoding enzymes that are known, or predicted, to regulate DNA methylation as well as enzymes involved in histone acetylation, methylation, phosphorylation, and ubiquitination. Among the different class of histone modifiers, we found that the expression of various histone methyltransferases and demethylases were changed upon miR combo treatment (Online Fig. IA–B). To determine which of these changes were functionally relevant, we analyzed the levels of the histone methylations targeted by these histone modifiers; namely H3K4me1/3, H3K9me3, H4K20me3 and H3K27me3. For this, we used immunoblot and immunocytochemistry analysis of negmiR- and miR combo transfected fibroblasts (Online Fig. IC–D, Fig. 1A–B). Immunocytochemistry was not sensitive enough to detect changes in histone methylations. However, our immunoblot revealed that among the methylated histone marks analyzed only H3K27me3 was significantly altered. Global levels of H3K27me3 were decreased by 40% in miR combo transfected fibroblasts compared to the negmiR controls (Fig. 1A). Interestingly, the qPCR array revealed a 40% increase in Kdm6B expression upon miR combo treatment (Online Fig. IB). Kdm6B (also known as Jmjd3) is one of the two demethylases for H3K27, and its increased expression was consistent with the decrease of H3K27me3 in miR combo treated cells. This observation prompted us to consider the expression levels of all histone modifiers involved in the regulation of H3K27 methylation (Fig. 1C). Using qPCR, we confirmed the increased expression in Kdm6B and found a 50% increase in Kdm6A, the other H3K27 demethylase (Fig. 1C). Notably, this was accompanied by a 50% decrease in Ezh2, the main H3K27 methyltransferase, at the RNA and at the protein level (Fig. 1C–D). To gain more insights on the mechanisms underlying the regulation of the expression of H3K27 modifiers by miR combo, we first assessed which miR of the miR combo was responsible for the downregulation of Ezh2 and the upregulation of Kdm6A and Kdm6B (Online Fig. II). Ezh2 expression decreased in response to miR-1 and miR-133 but not miR-208 or miR-499, suggesting that it may be a direct target of miR-1 and miR-133. We also found that miR-1 and miR-133 were responsible for the upregulation of Kdm6B and Kdm6A expression respectively. To further determine at which moment these changes were first occurring following miR combo transfection we performed a time course analysis (Online Fig. III). Similar to the direct targets of miR combo, Twf1 and Col16a1, the downregulation of Ezh2 expression appeared within the first 48 hours of miR combo treatment. In contrast, the upregulation of Kdm6A occurred 3 to 4 days after transfection with miR combo. To determine whether the effect of miR combo on Ezh2, Kdm6A and Kdm6B expression was direct or required a protein intermediate, we treated negmiR and miR combo transfected fibroblasts with cycloheximide (CHX), an inhibitor of protein synthesis (Fig. 2A). Neonatal cardiac fibroblasts transfected with negmiR or miR combo were treated with 50 mg/L of CHX for 12 hours before being analyzed by qPCR. Similar to the miR-1 direct target Twf1, the downregulation of Ezh2 by miR combo was unaffected by CHX treatment. In contrast, we found that Kdm6A and Kdm6B expression were dramatically increased in the presence of CHX suggesting that Kdm6A and Kdm6B transcription were repressed at baseline. The effect of miR combo treatment on Kdm6A expression was abrogated by CHX (Online Fig. IV). Therefore, miR combo may induce Kdm6A and Kdm6B expressions by targeting a putative transcriptional repressor. So far, our analysis suggested that Ezh2 may be a direct target of miR-1 and miR-133. Despite the absence of putative binding sites for those miRs within Ezh2 3′UTR (Online Fig. V), we performed a luciferase reporter assay to test our hypothesis (Fig. 2B). Luciferase vector containing the Twf1-3′UTR was used as a positive control for the efficiency of miR combo. The microRNA Let-7c which directly targets Ezh2-3′UTR 33 was used as a positive control for Ezh2-3′UTR reporter. As expected, the luciferase activities of the Twf1-3′UTR and of the Ezh2-3′UTR reporters were downregulated by miR combo and by Let-7c respectively. In contrast, neither miR combo, nor miR-1 or miR-133 decreased the luciferase activity of Ezh2-3′UTR reporter, demonstrating that Ezh2 was not a direct target of miR combo.
Figure 1. MiR combo treatment affects H3K27 methylation and the expression of H3K27 modifiers.
(A) Analysis of H3K27me3 levels in negmiR (NEG) and miR combo (MC) treated neonatal cardiac fibroblasts by immunoblot. The graph presents the quantification of H3K27me3 levels normalized to histone H3 levels (N=3). (B) Analysis of H3K27me3 levels in NEG- and MC- treated neonatal cardiac fibroblasts by immunocytochemistry (N=3). (C) Analysis of H3K27 methyltransferase (Ezh1 and Ezh2) and demethylase (Kdm6A and Kdm6B) mRNA levels in MC-treated neonatal fibroblasts by qPCR, 3 days after transfection. Expression data was normalized to NEG-treated cells (N=7). (D) Analysis of Ezh2 protein levels by immunoblot in neonatal cardiac fibroblasts transfected with NEG or MC. TBP was used as loading control and Ezh2/TBP ratios were normalized to NEG controls (N=3).
Asterisks indicate statistical significance between NEG and MC-treated cells determined by standardized T-test (*P ≤0.01, **P ≤0.005 or ***P≤0.0005).
Figure 2. Regulation of the expression of H3K27 modifiers by miR combo.
(A) Analysis of the mRNA levels of Twf1, Ezh2, Kdm6A and Kdm6B by qPCR in NEG or MC transfected cells treated for 12 hours with vehicle (DMSO) or 50 μg/ml of cycloheximide (CHX) to block protein synthesis (N=3–4). (B) Luciferase reporter assays were performed in HEK293 cells by co-transfecting Twf1-3′-UTR vector or Ezh2-3′-UTR vector with negmiR (NEG), miR combo (MC), miR-1, miR-133 or Let7c. The Twf1-3′UTR reporter was used as a positive control for miR combo (Twf1 is a direct target of miR-1). Let7c was used as a positive control for Ezh2-3′UTR (which contains the predicted Let7c binding sequence).
Comparisons were made between NEG and MC (*P ≤0.05, **P ≤0.01 or ***P≤0.005) or between CHX and vehicle-treated cells (# P≤0.05, ## P ≤0.01 or ### P ≤0.005) using paired T-test.
Altogether, our data suggest that removal of H3K27me3 by the coordinated regulation of the demethylases Kdm6A/B and methyltransferase Ezh2 expression may be the mechanism by which miR combo initiated direct cardiac reprogramming.
Inhibition of H3K27 methylation induces fibroblasts to adopt a cardiac fate
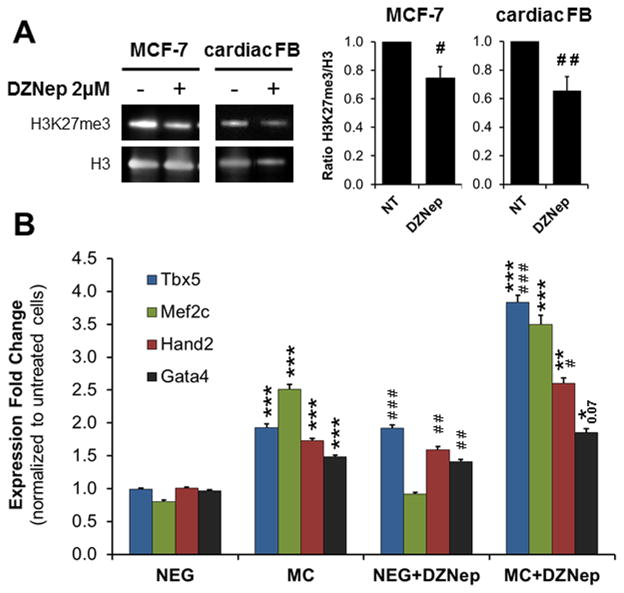
Modulation of H3K27 methylation has been shown to play an important role in cardiac development, cardiac differentiation of ESC and somatic reprogramming to induced pluripotent stem cell (iPSC) 17, 18, 20, 27–30. In order to test the hypothesis that H3K27 demethylation could induce fibroblasts to adopt a cardiac fate, we used the small molecule 3-Deazaneplanocin A (DZNep), an inhibitor of S-adenosylmethionine-dependent methyltransferase. First, we assessed if DZNep treatment was effective in inhibiting H3K27 methyltransferase by analyzing the levels of H3K27me3. We used two cell lines: the cancer cells line MCF-7, for which the role of DZNep has been well documented 34, 35, and primary neonatal cardiac fibroblasts. Treatment with 2 μM of DZNep for three days decreased H3K27me3 levels by 30% in both cell types (Fig. 3A). Following validation of DZNep we then measured cardiac gene expression in fibroblasts treated with the pharmacological inhibitor. Tbx5, Hand2 and Gata4 expression increased significantly in neonatal cardiac fibroblasts treated with DZNep compared to control cells (Fig. 3B). Mef2c expression was unaffected (Fig. 3B). In combination with miR combo, DZNep significantly enhanced the expression of Tbx5 and Hand2 by 1.5- to 2-fold respectively when compared to miR combo alone (Fig. 3B). Using FACS and immunoblot we confirmed that DZNep treatment increased cardiogenic markers at the protein level, similarly to miR combo treatment (Online Fig. VI).
Figure 3. Inhibition of H3K27 methylation induces cardiac transcription factor expression.
(A) Analysis of H3K27me3 levels in MCF7 and neonatal cardiac fibroblasts (FB) cultured for 3 days in growth media containing 2 μmol/L of DZNep or the vehicle (N=3). Immunoblots and quantification are shown as indicated. The H3K27me3/H3 ratio in DZNep treated cells was normalized to the vehicle. (B) Neonatal cardiac fibroblasts were transfected with negmiR (NEG) or miR combo (MC) and cultured in the presence of 2 μmol/L of DZNep or vehicle for 3 days. Tbx5, Mef2c, Hand2 and Gata4 mRNA levels were determined by qPCR (N≥8).
Comparisons were made between NEG and MC (*P ≤0.05, **P ≤0.01 or ***P≤0.0005) or between DZNep and vehicle-treated cells (# P≤0.01, ## P ≤0.005 or ### P ≤0.0005) using a standardized T test
Short treatment with DZNep was sufficient to induce the expression of markers of cardiac commitment in fibroblasts. Therefore, we wondered whether longer treatment could promote the appearance of more mature cardiomyocyte-like cells. Cardiac fibroblasts transfected with negmiR or miR combo were treated for 2 weeks with DZNep and analyzed for their expression of cardiomyocyte markers by qPCR (Online Fig. VIIA). The expression of the late stage cardiac genes Myh6 (α-myosin heavy chain), Tnni3 (cardiac troponin-I), Actn2 (α-sarcomeric actinin) and Scn5a (sodium channel voltage gated type V α subunit) was induced in fibroblasts treated with DZNep alone. Their expression increased 2- to 8-fold compared to untreated cells, a range similar to the one observed with miR combo transfected fibroblasts. Concomitant treatment of fibroblasts with miR combo and DZNep did not significantly increase the level of gene expression when compared to the individual treatments. We next analyzed the effect of DZNep on mature cardiac markers at the protein level by FACS (Online Fig. VIIB). The percentage of Tnnt2 and Myh6 positive cells in the negmiR control group was increased 3-fold by DZNep treatment. However, DZNep did not have significant additive effect when used in combination miR combo. Similarly, Myh6 and Tnni3 positive cells were occasionally observed by immunocytochemistry following 2 weeks of DZNep treatment (Online Fig. VIIC). Overall, the comparison of DZNep effect after 3- and 14 days of treatment suggests that DZNep effect is more robust at the early stage of cardiac conversion than the later stage.
In conclusion, decreasing the level of H3K27me3 by the pharmacological inhibitor DZNep induced cardiac gene expression similarly to miR combo treatment, suggesting that the removal of H3K27me3 is important to initiate direct cardiac reprogramming.
Inhibition of PRC2 function induces fibroblasts to adopt a cardiac fate
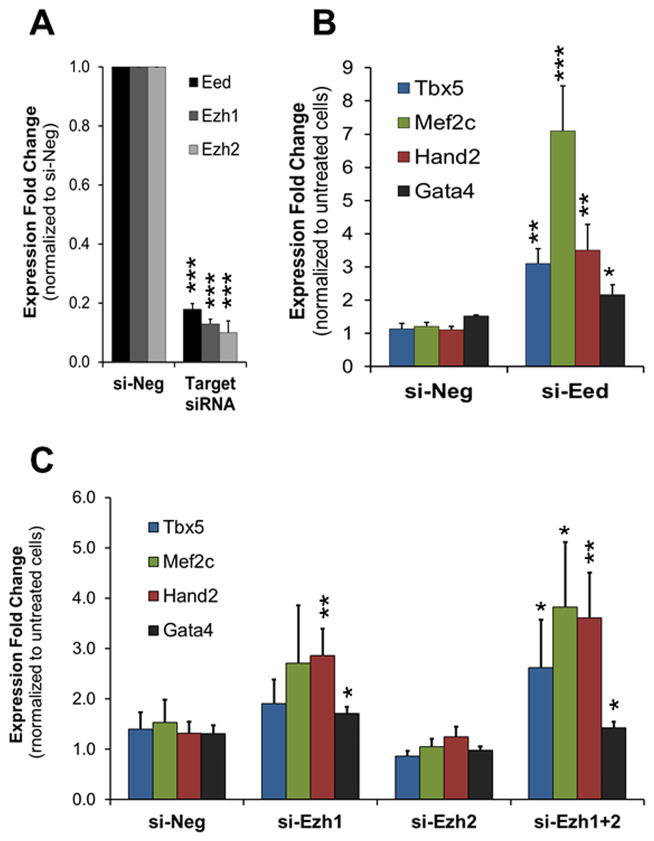
PRC2 promotes H3K27me3 formation which serves as a platform for the repression of target genes. To confirm that reduction of H3K27me3 is important for the induction of cardiac gene expression, we disrupted the formation of PRC2 via the siRNA mediated knockdown of the core component Eed. To achieve high knockdown efficiency, we used a combination of 4 siRNAs targeting Eed (si-Eed). Neonatal cardiac fibroblasts transfected with si-Eed showed an 80% decrease in Eed RNA levels (Fig. 4A). Accordingly, the level of H3K27me3 was reduced by ~60% in these cells (Online Fig. VIIIA). The RNA and protein levels of cardiac transcription factors were strongly increased in cardiac fibroblasts treated with si-Eed (Fig. 4B, Online Fig. VI). Importantly, the increase in RNA levels was higher than the one observed for miR combo treatment, ranging from a 2.5- to 7-fold change when compared to the negative siRNA (si-Neg) control (Fig. 4B). Further experiments combining Eed knockdown with miR combo had no additive effect suggesting that maximal decrease of H3K27me3 was already achieved by si-Eed (Online Fig. IXA). As shown in Figure 1, miR combo dramatically inhibited Ezh2 expression whilst Ezh1 expression was not significantly affected. This suggested that Ezh2 downregulation may be responsible for the decrease in H3K27me3 levels by miR combo. To test this hypothesis and investigate the role of H3K27 methyltransferases in cardiac reprogramming of fibroblasts, we used siRNA-mediated gene knockdown. Three days after transfecting neonatal cardiac fibroblasts with Ezh2 siRNAs (si-Ezh2) RNA levels were decreased by 90% (Fig. 4A) and protein levels were undetectable (Online Fig. VIIIB). Surprisingly, Ezh2 knockdown alone was not sufficient to promote cardiac gene expression (Fig. 4C) and did not enhance the effect of miR combo (Online Fig. IXB). Since Ezh1 could compensate for the loss of Ezh2, we decided to abrogate all H3K27 methyltransferase activity by knocking down both genes using siRNA. We first tested the effect of Ezh1 knockdown alone. The efficiency of Ezh1 knockdown was about 90% in neonatal cardiac fibroblasts (Fig. 4A). Interestingly, Ezh1 knockdown increased the expression of cardiac transcription factors which was significant for Hand2 and Gata4 compared to si-Neg control cells. More importantly, concomitant loss of Ezh1 and Ezh2 (si-Ezh1+2) induced a further increase in Tbx5, Mef2c, Hand2 and Gata4 which reached up to 3- to 4- fold change when compared to si-Neg control cells (Fig. 4C). Increases in Tbx5, Mef2c and Gata4 RNA levels in the si-Ezh1+2 transfected fibroblasts were also observed at the protein level (Online Fig. VI). This was associated to a 50% decrease in H3K27me3 level (Online Fig. VIIIA). Similar to Eed knockdown, combined knockdown of Ezh1 and Ezh2 did not amplify the effect of miR combo (Online Fig. IXB).
Figure 4. Knockdown of PRC2 complex members induces cardiac transcription factor expression.
(A) Expression levels of Eed, Ezh1 and Ezh2 in neonatal cardiac fibroblasts transfected with negative siRNA (si-Neg) or siRNA against Eed (si-Eed), Ezh1 (si-Ezh1) or Ezh2 (si-Ezh2). mRNAs were isolated 3 days after transfection and analyzed by qPCR. Expressions were normalized to si-Neg (N=5). (B and C) Cardiac transcription factor expression was analyzed by qPCR in neonatal cardiac fibroblasts transfected with si-Neg, si-Eed (B), si-Ezh1, si-Ezh2 or si-Ezh1+2 (C), 3 days after transfection. The data was normalized to the si-Neg control (N=5–6).
For all panels, comparisons were made between si-Neg and target siRNA (*P ≤0.05, **P ≤0.01 or ***P ≤0.005).
Altogether, our data demonstrate that disruption of PRC2 function mimics the early effect of miR combo on cardiac gene expression. However, since the single knockdown of Ezh2 did not recapitulate the miR combo effect, it is unlikely that its inhibition by miR combo is sufficient to induce direct reprogramming and suggests a more complex mechanism.
Blockade of H3K27 demethylation inhibits miR combo mediated cardiac reprogramming
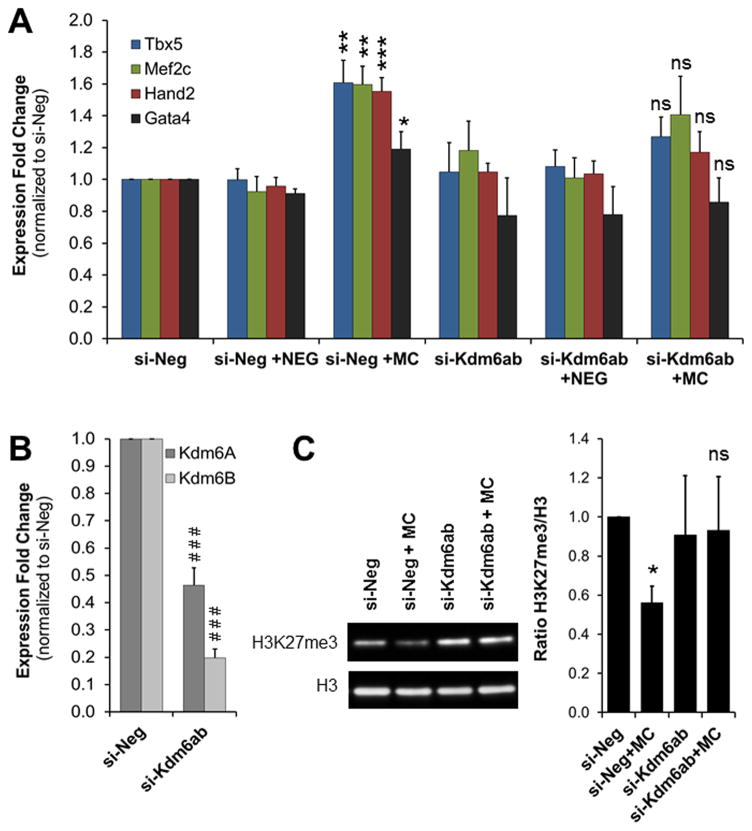
Whilst we have demonstrated an important role for PRC2/H3K27me3 in the induction of the cardiomyocyte fate in fibroblasts, it did not prove that miR combo induced direct cardiac reprogramming by removal of H3K27me3. To test this hypothesis, we decided to knockdown the H3K27 demethylases Kdm6A and Kdm6B which were up-regulated upon miR combo treatment. We reasoned that if removal of H3K27me3 is necessary for miR combo mediated reprogramming then blocking H3K27 demethylation should inhibit the induction of direct cardiac reprogramming by miR combo. To test this hypothesis, we transfected cardiac fibroblasts with negative siRNA (si-Neg) or siRNAs targeting Kdm6A and Kdm6B in the presence of negmiR or miR combo. Tbx5, Mef2c, Hand2 and Gata4 expression were analyzed by qPCR 3 days after transfection. As seen in Figure 4A, in the presence of si-Neg, miR combo-transfected fibroblasts showed an increase in cardiac transcription factor expression, ranging from 1.5- to 2- fold compared to negmiR transfected cells. The efficiencies of Kdm6A- and Kdm6B- knockdown were about 60% and 75% respectively (Fig. 5B). Double knockdown of Kdm6A and Kdm6B did not significantly affect the expression of Tbx5, Mef2c, Hand2 and Gata4. However, in the presence of miR combo, Kdm6A/B knockdown inhibited the induction of cardiac gene expression (Fig. 5A). Importantly, Kdm6A/B knockdown partially blocked the decrease in H3K27me3 induced by miR combo treatment (Fig. 5C).
Figure 5. Inhibition of H3K27me3 demethylases blocks the induction of cardiac reprogramming by miR combo.
(A) Analysis of the expression of cardiac transcription factors in neonatal cardiac fibroblasts transfected with negmiR (NEG) or miR combo (MC), in the presence of negative siRNA (si-Neg) or siRNA targeting Kdm6A and Kdm6B (si-Kdm6A+B). mRNA were isolated 3 days after transfection and analyzed by qPCR. Expression values were normalized to the si-Neg control. (N=5). (B) mRNA levels of Kdm6A and Kdm6B in neonatal cardiac fibroblasts transfected with si-Kdm6A+B compared to control fibroblasts transfected with si-Neg. (N=5). (C) Immunoblot analysis of H3K27me3 levels in neonatal cardiac fibroblasts treated with NEG or MC in combination with si-Neg or si-Kdm6A+B, 3 days after transfection (N=3).
For all panels, comparisons were made between NEG and MC ((*p ≤0.05, **p ≤0.005 or ***p ≤0.0005) or between si-Neg and si-Kdm6A+B (### P≤ 0.0005) respectively.
Therefore, preventing the increase in Kdm6A/B and the decrease in H3K27me3, which are normally induced by miR combo treatment, hindered the reprogramming effect of miR combo.
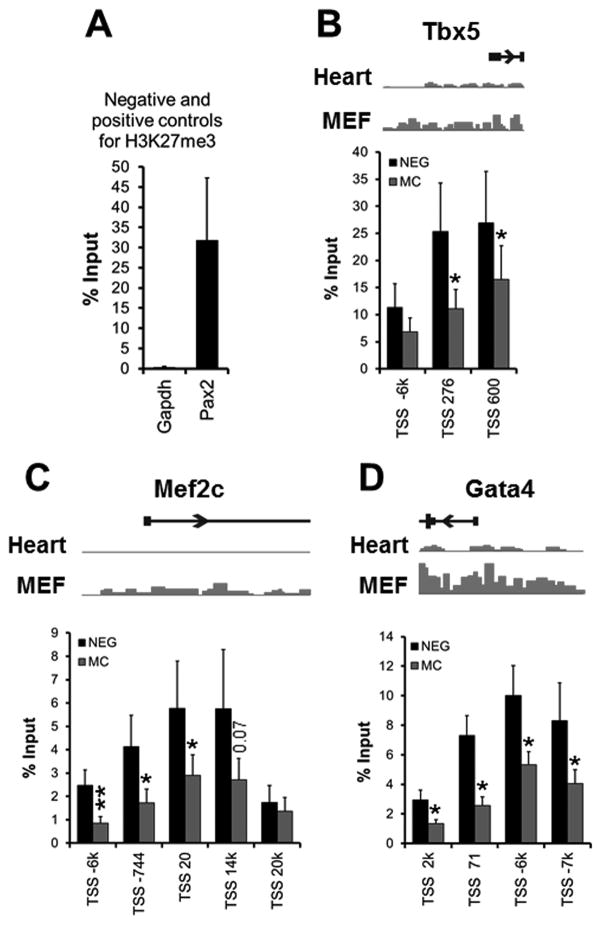
Mir combo treatment reduces H3K27me3 at loci of cardiac transcription factors
Our experiments demonstrated that global demethylation of H3K27me3 plays a critical role in the induction of cardiac reprogramming by miR combo. The first sign of cardiac reprogramming is the induction of cardiac transcription factors which is detectable only 3 days after miR combo treatment. Therefore, we wondered whether the promoter region of these genes was directly affected by the loss of H3K27me3. We performed ChIP-qPCR for Tbx5, Mef2C and Gata4, using 3–5 validated primers for each gene 32. Primers for Gapdh and Pax2 were used as negative and positive controls for H3K27me3 respectively. As expected, no amplification product was observed for Gapdh whilst we saw 35% of input labeled with H3K27me3 for Pax2 (Fig. 6A). In negmiR control, we found that cardiac transcription factors were all marked by H3K27me3 albeit with the lowest levels for Mef2c. Importantly, we found an average 50% decrease in H3K27me3 for Tbx5, Mef2c and Gata4 in the miR combo group compared to negmiR (Fig. 6B). Only a couple of primers for Tbx5 and Mef2c did not show significant decrease in H3K27me3.
Figure 6. Reduction of H3K27me3 at loci of genes encoding cardiac transcription factors.
(A) Analysis of the negative (gapdh) and positive (pax2) controls for H3K27me3, by ChIP-qPCR for H3K27me3 in neonatal cardiac fibroblasts. (B-D) Analysis of the promoter region of Tbx5 (B), Mef2c (C) and Gata4 (D) by ChIP-qPCR for H3K27me3 (N=6). Chromatin samples were isolated from neonatal cardiac fibroblasts transfected with negmiR (NEG) or miR combo (MC) and processed 3 days after transfection. Results were expressed as a percentage of the input control. For each gene, we used a set of 3 to 5 validated primer pairs 32 which were named according to their location relative to the transcription start (TSS). For each gene, a schematic representation of their H3K27me3 profile in heart and in fibroblasts (MEF) is shown.
Comparisons were made between NEG and MC (*p ≤0.05) using 2-tails paired T-test.
This data validates our model and demonstrates that miR combo directly regulates H3K27 methylation at the promoter region of cardiac transcription factors that are essential for cardiomyocyte cell fate.
DISCUSSION
Several reports have looked at epigenetic changes that occur following the addition of reprogramming factors 8, 32. However our study is the first to detect a causal link between removal of H3K27me3 and direct cardiac reprogramming. In this study we demonstrated that miR combo induced direct reprogramming by decreasing the levels of H3K27me3 via the concomitant modulation of H3K27 methyltransferase and demethylase expression. In particular, ChIP analysis showed that cardiogenic loci harbored reduced levels of H3K27me3 in their promoter regions upon miR combo treatment. This data is in agreement with several reports showing the importance of the loss of H3K27me3 for ESC differentiation to cardiomyocytes, somatic reprogramming to iPSCs and more recently GMT-mediated reprogramming to cardiomyocytes 20, 21, 32, 36.
In our study, we found that miR-1 and miR-133 regulate the expression of Ezh2 and Kdm6A/B. We were interested in defining the mechanism for these changes. The downregulation of Ezh2 suggested that it was a direct target of miR-1/133. However, bioinformatic analysis and luciferase assay failed to identify Ezh2 as a direct target of these miRs. Furthermore, the loss of Ezh2 mRNA with miR combo did not require a protein intermediate as demonstrated by the cycloheximide experiment. Since Ezh2 is the direct target of other microRNAs (e.g. let-7 33), it is tempting to speculate that miR combo recruits a microRNA to dowregulate Ezh2 expression. In the case of Kdm6A and Kdm6B, their expression was dramatically increased by cycloheximide indicating that both genes are strongly inhibited at baseline. Importantly, the positive effect of miR combo on Kdm6A expression was lost in the presence of cycloheximide, Altogether, this suggests that miR combo is likely to affect Kdm6A/B expression by targeting a putative transcriptional repressor.
Ezh2 is known to play a critical role in cardiac development and homeostasis 18, 19. Ezh2 expression is directly repressed by Let-7c to promote cardiac-specific gene expression during differentiation of ESC to cardiomyocytes 33. In contrast, reprogramming toward pluripotency is accompanied by an increase in Ezh2 expression 29, 37. This correlates with the essential role of PRC2 in maintaining the pluripotency of ESC through repression of lineage-specific genes 38–40. The role of Ezh2 in somatic reprogramming to iPSC has been controversial. Whilst some groups showed a critical role for Ezh2 in this process 29, 37, the genetic ablation of Ezh2 performed by Fragola and colleagues did not affect iPSC formation 30. The generation of iPSC was only lost when the PRC2 complex was disrupted by genetic knockout of Eed 30. This study may explain in part why we did not observe up-regulation of cardiac genes following Ezh2 knockdown, which we expected to see. Instead, we found that Eed knockdown by siRNA was sufficient to induce cardiac markers at the RNA and protein level in the absence of miR combo, reinforcing the important role of PRC2 in the acquisition of the cardiac cell fate. The contrast observed between the effects of Ezh2 and Eed knockdown on cardiac gene expression suggests that other PRC2 members are likely to be important for reprogramming and/or could compensate for the loss of Ezh2. We provide evidence that Ezh1 may be one of these components since the combined knockdown of Ezh1 and Ezh2 induced the cardiogenic program.
Kdm6A/B knockdown inhibited the ability of miR combo to induce cardiac gene expression. While the demethylation of H3K27 is carried out by Kdm6A/B, these enzymes also have demethylase-independent functions. Uty, the homolog of Kdm6A that is carried by the Y chromosome, has no demethylase activity. Indeed, the presence of Uty in Kdm6A/B mutant male mice rescued the embryonic lethality observed in Kdm6A/B mutant females 41. While the enzymatic activity of Kdm6A was dispensable for embryonic survival, its demethylase function was critical for cardiac differentiation 27.Lee et al. found that Kdm6A bound and activated cardiac gene enhancers by interacting with core cardiac transcription factors and by demethylating H3K27 27. Similarly, cardiovascular differentiation of ESC is compromised in the absence of Kdm6B, a role attributed to the defective demethylation of H3K27 at the Brachyury promoter 28. Our data agrees with these findings in the cardiac field. Loss of Kdm6A/B partially restored H3K27me3 levels in miR combo treated cells to that observed in control fibroblasts, which resulted in the loss of cardiac reprogramming.
Our reprogramming miRs are conserved across species suggesting their applicability for the treatment of myocardial infarction in humans. However, the literature has shown that reprogramming cocktails utilized in mouse fibroblasts have limited effect in human cells and suggest a need for additional factors. Small molecules have recently attracted much interest. They avoid cell-reprogramming techniques that rely on the forced expression of transcription factors that are genetically invasive, not suitable for therapeutic use in patients, and are of low efficiency 42, 43. Various combinations of small molecules, which include modulators of epigenetic modifiers, have been identified for successful reprogramming. Parnate (LSD1 inhibitor), sodium butyrate and valproic acid (HDAC inhibitors), EPZ-004777 (Dot1L inhibitor) and BIX-01294 (G9a inhibitor) have all been used to generate iPSC more efficiently and with fewer exogenous transcription factors 44–47. Direct cardiac reprogramming properties have been reported for some of these modulators 48–50. DZNep is a small molecule that selectively inhibits Ezh2/H3K27me3 35. Here, we showed that treatment of cardiac fibroblasts with DZNep increased the RNA and protein level of cardiac transcription factors and mature cardiomyocyte markers in the absence of miR combo. This suggests that DZNep mimics the effect of miR combo at the genetic level. In support of our findings, the Ezh2 inhibitor GSK126 enhances transcription factor mediated cardiac reprogramming 51. Additionally, Hou and colleagues found that the addition of DZNep to the late phase of reprogramming to iPSC increased the number of Oct4-GFP positive cells by 65 times 52. Interestingly, this role was attributed to the decrease of DNA- and H3K9-methylation at the Oct4 promoter 52. Indeed, seeing that DZNep has been shown to affect other histone methylations such as H4K20me3 and H3K9me3 34, 35, it is possible that the reprogramming effect of DZNep is not solely dependent upon Ezh2/H3K27me3 inhibition. This represents another possibility to explain the difference between siRNA-mediated Ezh2 knockdown and DZNep treatment upon cardiac gene expression. We are currently investigating the role of DZNep regulated marks other than H3K27me3 with respect to their impact on cardiac reprogramming.
In summary, we have shown that the initiation of direct cardiac reprogramming by miR combo requires the demethylation of H3K27me3 at the loci of cardiogenic genes via opposing effects on PRC2 and Kdm6A/B.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
A combination of four microRNAs (miR-1, miR-133, miR-208 and miR-499) named miR combo directly reprograms cardiac fibroblasts into cardiomyocytes in vitro and in vivo.
Modulation of tri-methylation of lysine 27 of histone H3 (H3K27me3), an epigenetic motif for transcriptional repression, is important for cardiac development and homeostasis.
The molecular mechanism of direct cardiac reprogramming is unknown.
What New Information Does This Article Contribute?
Demethylation of H3K27me3 is essential for the induction of direct cardiac reprogramming by miR combo.
MiR combo, which modulates the expression of H3K27me3 modifiers, decreases H3K27me3 levels in the promoter region of cardiac transcription factors.
Remodeling of the epigenetic landscape holds great promise to improve the efficiency of direct cardiac reprogramming.
Direct reprogramming of fibroblasts to cardiomyocytes has emerged as an important strategy to regenerate cardiac muscle that is lost following myocardial infarction. We have achieved direct cardiac reprogramming by using a combination of four microRNAs that we named miR combo. The mechanisms underlying direct reprogramming are unknown. Identifying such mechanisms is necessary to improve the efficiency of this technique. To that end, we have identified an important mechanism by which miR combo modulates the epigenetic landscape to induce direct cardiac reprogramming. In particular, miR combo stimulated the transcription of cardiac transcription factors in fibroblasts by removing the repressive motif H3K27me3 from their promoter regions. Indeed, the forced decrease of H3K27me3 levels induced the expression of cardiogenic genes similarly to miR combo. Moreover, preventing the demethylation of H3K27me3 blocked the induction of direct cardiac reprogramming by miR combo. Our study is the first to highlight a causal link between the induction of direct cardiac reprogramming and the modulation of the epigenetic landscape in response to reprogramming factors. Such mechanistic understanding will help improve the efficiency of direct cardiac reprogramming which is necessary for clinical applications of this technique.
Acknowledgments
We are highly appreciative of the technical input provided by Alan Payne and the assistance of Tilanthi Jayawardena with the qPCR array for epigenetic modifiers.
SOURCES OF FUNDING
Research conducted in these studies was supported by National Heart, Lung, and Blood Institute grants RO1 HL81744, HL72010, and HL73219 (to V.J. Dzau); the Edna and Fred L. Mandel Jr. Foundation (to V.J. Dzau, M. Mirotsou); and the American Heart Association National Scientist Development Award (10SDG4280011, to M. Mirotsou).
Nonstandard Abbreviations and Acronyms
- miR
microRNA
- negmiR
non targeting miRNA
- miR combo
microRNA combination
- GMT
Gata4, Mef2c and Tbx5
- H3K27me3
tri-methylated lysine 27 of histone H3
- H3K4me1/3
mono- and tri-methylated lysine 4 of histone H3
- H3K9me3
tri-methylated lysine 9 of histone H3
- H4K20me3
tri-methylated lysine 20 of histone H3
- PRC2
polycomb repressive complex 2
- qPCR
quantitative polymerase chain reaction
- DZNep
3-Deazaneplanocin A
- si-Neg
negative non-targeting siRNA
- si-Eed
siRNA targeting Eed
- si-Ezh1
siRNA targeting Ezh1
- si-Ezh2
siRNA targeting Ezh2
- si-Kdm6A
siRNA targeting Kdm6A
- si-Kdm6B
siRNA targeting Kdm6B
- ChIP
chromatin immunoprecipitation
Footnotes
In January 2016, the average time from submission to first decision for all original research papers submitted to Circulation Research was 13.77 days.
DISCLOSURES
None.
References
- 1.Lin Z, Pu WT. Strategies for cardiac regeneration and repair. Sci Transl Med. 2014;6:239rv1. doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–41. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dal-Pra S, Mirotsou M. Reprogramming approaches in cardiovascular regeneration. Curr Treat Options Cardiovasc Med. 2014;16:327. doi: 10.1007/s11936-014-0327-0. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkinson CP, Kang MH, Dal-Pra S, Mirotsou M, Dzau VJ. MicroRNAs and Cardiac Regeneration. Circ Res. 2015;116:1700–11. doi: 10.1161/CIRCRESAHA.116.304377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadahiro T, Yamanaka S, Ieda M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ Res. 2015;116:1378–91. doi: 10.1161/CIRCRESAHA.116.305374. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava D, Yu P. Recent advances in direct cardiac reprogramming. Curr Opin Genet Dev. 2015;34:77–81. doi: 10.1016/j.gde.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1:235–47. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, Mercola M, Oshima RG, Willerson JT, Potaman VN, Schwartz RJ. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A. 2012;109:13016–21. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–93. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–32. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, Tohyama S, Yuasa S, Kokaji K, Aeba R, Yozu R, Yamagishi H, Kitamura T, Fukuda K, Ieda M. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A. 2013;110:12667–72. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, Rosenberg PB, Mirotsou M, Dzau VJ. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116:418–24. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, Nishiyama T, Kaneda R, Fukuda T, Takeda S, Tohyama S, Hashimoto H, Kawamura Y, Goshima N, Aeba R, Yamagishi H, Fukuda K, Ieda M. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–81. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Ma Y, Kim EY, Yu W, Schwartz RJ, Qian L, Wang J. Conditional ablation of Ezh2 in murine hearts reveals its essential roles in endocardial cushion formation, cardiomyocyte proliferation and survival. PLoS One. 2012;7:e31005. doi: 10.1371/journal.pone.0031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012;110:406–15. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44:343–7. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–32. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–20. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 25.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 26.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–7. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–62. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 29.Ding X, Wang X, Sontag S, Qin J, Wanek P, Lin Q, Zenke M. The polycomb protein Ezh2 impacts on induced pluripotent stem cell generation. Stem Cells Dev. 2014;23:931–40. doi: 10.1089/scd.2013.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fragola G, Germain PL, Laise P, Cuomo A, Blasimme A, Gross F, Signaroldi E, Bucci G, Sommer C, Pruneri G, Mazzarol G, Bonaldi T, Mostoslavsky G, Casola S, Testa G. Cell reprogramming requires silencing of a core subset of polycomb targets. PLoS Genet. 2013;9:e1003292. doi: 10.1371/journal.pgen.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayawardena T, Mirotsou M, Dzau VJ. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol Biol. 2014;1150:263–72. doi: 10.1007/978-1-4939-0512-6_18. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Chen O, Zheng M, Wang L, Zhou Y, Yin C, Liu J, Qian L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–18. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coppola A, Romito A, Borel C, Gehrig C, Gagnebin M, Falconnet E, Izzo A, Altucci L, Banfi S, Antonarakis SE, Minchiotti G, Cobellis G. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014;12:323–37. doi: 10.1016/j.scr.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–88. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussein SM, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee DS, Cloonan N, Wood DL, Munoz J, Middleton R, Korn O, Patel HR, White CA, Shin JY, Gauthier ME, Le Cao KA, Kim JI, Mar JC, Shakiba N, Ritchie W, Rasko JE, Grimmond SM, Zandstra PW, Wells CA, Preiss T, Seo JS, Heck AJ, Rogers IM, Nagy A. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- 37.Rao RA, Dhele N, Cheemadan S, Ketkar A, Jayandharan GR, Palakodeti D, Rampalli S. Ezh2 mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Sci Rep. 2015;5:8229. doi: 10.1038/srep08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 40.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Federation AJ, Bradner JE, Meissner A. The use of small molecules in somatic-cell reprogramming. Trends Cell Biol. 2014;24:179–87. doi: 10.1016/j.tcb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu C, Liu K, Tang S, Ding S. Chemical approaches to cell reprogramming. Curr Opin Genet Dev. 2014;28:50–6. doi: 10.1016/j.gde.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Scholer HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–20. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–74. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Park G, Yoon BS, Kim YS, Choi SC, Moon JH, Kwon S, Hwang J, Yun W, Kim JH, Park CY, Lim DS, Kim YI, Oh CH, You S. Conversion of mouse fibroblasts into cardiomyocyte-like cells using small molecule treatments. Biomaterials. 2015;54:201–12. doi: 10.1016/j.biomaterials.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 49.Thal MA, Krishnamurthy P, Mackie AR, Hoxha E, Lambers E, Verma S, Ramirez V, Qin G, Losordo DW, Kishore R. Enhanced angiogenic and cardiomyocyte differentiation capacity of epigenetically reprogrammed mouse and human endothelial progenitor cells augments their efficacy for ischemic myocardial repair. Circ Res. 2012;111:180–90. doi: 10.1161/CIRCRESAHA.112.270462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, Ding S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6:951–60. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirai H, Kikyo N. Inhibitors of suppressive histone modification promote direct reprogramming of fibroblasts to cardiomyocyte-like cells. Cardiovasc Res. 2014;102:188–90. doi: 10.1093/cvr/cvu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–4. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.