Significance
RNAi-mediated antiviral defense is well conserved in eukaryotic organisms. Although several RNAi key genes are transcriptionally up-regulated upon virus infection in many hosts, little is known about their induction mechanism. Here, we developed a screening method for identification of the genes mediating robust transcriptional up-regulation upon infection by RNA viruses of the dicer-like 2 (dcl2) gene of an ascomycetous phytopathogen, chestnut blight fungus. We show that two enzymatic activities of histone acetyltransferase and histone deubiquitinase of the universally conserved SAGA (Spt–Ada–Gcn5 acetyltransferase) complex differentially regulate the transcriptional induction of two main antiviral RNA silencing components, dcl2 and argonaute-like 2 (agl2). A virus-specific pattern requirement of AGL2 and DCL2 as positive feedback players for this transcriptional induction is also noted.
Keywords: Cryphonectria parasitica, hypovirus, RNAi, mycovirus, antiviral defense
Abstract
Pathogen recognition and transcriptional activation of defense-related genes are crucial steps in cellular defense responses. RNA silencing (RNAi) functions as an antiviral defense in eukaryotic organisms. Several RNAi-related genes are known to be transcriptionally up-regulated upon virus infection in some host organisms, but little is known about their induction mechanism. A phytopathogenic ascomycete, Cryphonectria parasitica (chestnut blight fungus), provides a particularly advantageous system to study RNAi activation, because its infection by certain RNA viruses induces the transcription of dicer-like 2 (dcl2) and argonaute-like 2 (agl2), two major RNAi players. To identify cellular factors governing activation of antiviral RNAi in C. parasitica, we developed a screening protocol entailing multiple transformations of the fungus with cDNA of a hypovirus mutant lacking the RNAi suppressor (CHV1-Δp69), a reporter construct with a GFP gene driven by the dcl2 promoter, and a random mutagenic construct. Screening for GFP-negative colonies allowed the identification of sgf73, a component of the SAGA (Spt–Ada–Gcn5 acetyltransferase) complex, a well-known transcriptional coactivator. Knockout of other SAGA components showed that the histone acetyltransferase module regulates transcriptional induction of dcl2 and agl2, whereas histone deubiquitinase mediates regulation of agl2 but not dcl2. Interestingly, full-scale induction of agl2 and dcl2 by CHV1-Δp69 required both DCL2 and AGL2, whereas that by another RNA virus, mycoreovirus 1, required only DCL2, uncovering additional roles for DCL2 and AGL2 in viral recognition and/or RNAi activation. Overall, these results provide insight into the mechanism of RNAi activation.
Innate immune responses are the primary defense against pathogens. Many defense-related genes are transcriptionally induced upon pathogen attack (1–4) to minimize the cost of defense, rather than being expressed constitutively. Such genes include those for receptors recognizing molecular patterns of pathogens and downstream components in their signaling pathways in higher eukaryotes. Also included are key genes for RNA silencing, hereafter referred to as RNA interference (RNAi), in many organisms (4–8). RNAi is a small RNA-mediated gene-suppression mechanism primarily working as an innate immune response against viruses and conserved in eukaryotic organisms across kingdoms. Structured or double-stranded (ds) RNA derived from viruses is recognized by Dicer and cleaved into duplex of small-interfering RNAs (siRNAs) of 21–26 nt (9, 10). One of the siRNA strands is incorporated into the Argonaute-involving effector, RNA silencing-induced complex, and guides to its target viral genomic or messenger RNA for degradation.
Fungi provide unique platforms for studying antiviral RNAi. For example, the yeast Saccharomyces cerevisiae, although lacking antiviral RNAi, allows for reconstitution of RNAi against a group of viruses by heterologous transgenic expression of only two components, Dicer-like 1 (DCL1) and Argonaute-like 1 (AGL1), from other yeast species, such as Saccharomyces castellii (11). In filamentous fungi, a simplified antiviral RNAi operates in which much fewer key players are involved than in plants, as exemplified by the presence of 4 DCL and 10 AGO in a model plant, Arabidopsis thaliana (12, 13). The filamentous ascomycete, Cryphonectria parasitica, is not only an important plant pathogen that is destructive to chestnut forests but also a model filamentous fungus that provides opportunities to explore virus/host interactions (13, 14). C. parasitica is known to support replication of diverse viruses, among which Cryphonectria hypovirus 1 [CHV1, with a nonsegmented, positive-sense (+), single-stranded (ss) RNA genome, family Hypoviridae], a member of the expanded picornavirus supergroup (15), and mycoreovirus 1 (MyRV1, with an 11-segmented dsRNA genome, family Reoviridae) can artificially be introduced into the host. Nuss and colleagues (4, 16) identified two major components, dcl2 and agl2, in antiviral RNAi. Of great interest is a high induction of these genes, particularly dcl2, at the transcription level of ∼40-fold upon infections by CHV1 and some dsRNA viruses, such as MyRV1, but not a victorivirus (Rosellinia necatrix victorivirus 1, RnVV1) in C. parasitica (4, 17). However, the mechanism by which dcl2 and agl2 are induced by mycoviruses remains largely unknown.
In this study, we developed a genetic screen protocol to identify host factors involved in the induction of the antiviral RNAi pathway, from the sensing of the virus to transcriptional activation of RNAi-related genes in C. parasitica. This protocol entailed multiple transformations of the fungus by a previously established infectious CHV1 cDNA clone (18), an enhanced green fluorescent protein (eGFP)–based reporter controlled by the dcl2 promoter, and a mutagenic selectable marker cassette (19). Through this, we identified sgf73, a component of the SAGA (Spt–Ada–Gcn5 acetyltransferase) complex, as necessary for virus-induced RNAi activation. SAGA is a universal transcriptional coactivator with a multimodular structure and coactivates transcription of a subset of genes via histone modification (20). A targeted disruption assay shows the histone acetyltransferase (HAT) module of SAGA to be the main regulator of the key RNAi pathway of the filamentous fungus. A virus-specific pattern requirement of AGL2 and DCL2 as positive feedback players for this transcriptional induction is also noted.
Results
Development of a Reporter System to Identify Mutants Defective in the RNAi Activation.
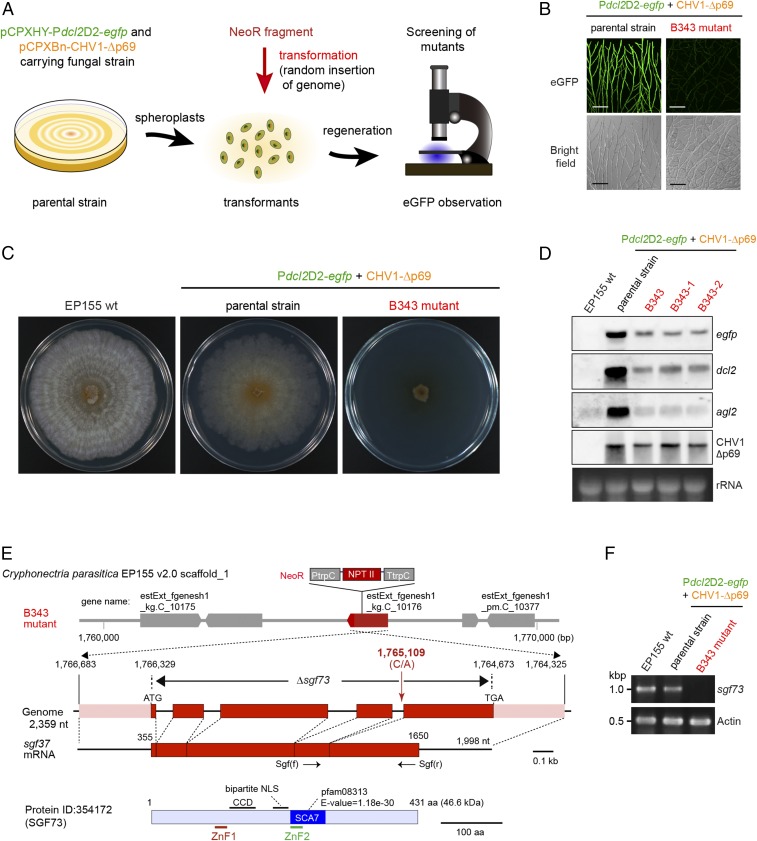
To develop a reporter system to monitor this transcriptional up-regulation, a dcl2 promoter region responsive to virus infection was first identified. The standard C. parasitica strain EP155 was transformed with a series of deletion constructs of the upstream region of dcl2 conjugated with egfp (Fig. 1A, Fig. S1, and Table S1) and subsequently with CHV1-Δp69 cDNA (Fig. 1A) (pCPXBn-CHV1-Δp69). CHV1-Δp69 lacks 88% of the coding domain of p29 that inhibits dcl2 up-regulation and thus enables augmentation of dcl2 transcripts by several 10-fold (4, 17). Full-scale promoter activities were observed with constructs carrying the 3- and 2-kb upstream regions, but not with those carrying the 1-kb upstream region (Fig. S1). In this study, we used the 2-kb construct (pCPXH-Pdcl2D2-egfp) to prepare a reporter fungal strain that theoretically expresses egfp and harbors replicating CHV1-Δp69 in every fungal cell. Note that the reporter transformants green-fluoresced only in the presence of CHV1-Δp69 (Fig. 1B). We also confirmed that the 2-kb dcl2 promoter was responsive to infection by MyRV1 (Fig. 1B). In accordance with the eGFP microscopic assay, egfp and dcl2 were found to be transcriptionally up-regulated only upon infection by CHV1-Δp69 or MyRV1 but not in virus-free transformants (Fig. 1C). Thus, the reporter system with the responsive promoter region was validated.
Fig. 1.
Preparation and validation of the parental fungal strain used in genetic screen. (A) Schematic representation of the reporter construct (pCPXHY- or pCPXNeo-Pdcl2D2-egfp), the dcl2 gene of C. parasitica (16), infectious cDNA of CHV1-Δp69 (pCPXHY- or pCPXBn-CHV1-Δp69) (47), and the cassette for random mutagenesis (NeoR). The dcl2 promoter activities, responsive to CHV1-Δp69, were mapped to an upstream region of ∼2.0 kb using a series of deletion mutants (Fig. S1). (B) Validation of the reporter construct, pCPXHY-Pdcl2D2-egfp. A transformant (Pdcl2D2-egfp) with the reporter construct was retransformed with pCPXBn-CHV1-Δp69 or infected by a reovirus, MyRV1. These fungal strains were tested for green fluorescence together with virus-free Pdcl2D2-egfp and wild-type EP155 strain (Table S1). (Scale bars, 0.1 mm.) (C) Northern analyses of the key fungal strain Pdcl2D2-egfp. Total RNA was isolated from the fungal strains shown in B and analyzed. Probes used in this and subsequent figures specific for the genes shown on the right were DIG-labeled PCR. Ethidium bromide (EtBr)-stained ribosomal RNA (28S rRNA) was used as loading controls in this and all following figures.
Table S1.
Fungal and viral strains used in this study
| Strain | Description | Source |
| Viral | ||
| CHV1-Δp69 | A mutant of CHV1 lacking the entire ORFA encoding a VSR, p29 | (62) |
| MyRV1 | A mycoreovirus with a 11-segmented dsRNA genome | (49) |
| CHV1-EP713 | Prototype of Cryphonectria hypovirus 1 in the family Hypoviridae | (63) |
| Fungal | ||
| EP155 | Standard strain of C. parasitica (virus-free) | ATCC 38755 |
| Pdcl2D2-egfp | EP155 transformant expressing egfp under the control of the dcl2 promoter | Present study |
| Pdcl2D2-egfp+CHV1-Δp69 | Pdcl2D2-egfp cotransformed with an infectious cDNA clone of CHV1-Δp69and serving as the parental strain for random mutagenesis | Present study |
| Δdcl2 | dcl-2 knockout mutant of EP155 (RNA silencing defective, virus-free) | (16) |
| Δagl2 | agl-2 knockout mutant of EP155 (RNA silencing defective, virus-free) | (4) |
| Δdcl2+ Pdcl2D2-egfp | Δdcl2 expressing egfp under the control of the dcl2 promoter | Present study |
| Δagl2+ Pdcl2D2-egfp | Δagl2 expressing egfp under the control of the dcl2 promoter | Present study |
| DK80 | A mutant of EP155 lacking the cpk80 gene involved in nonhomologous recombination | (23) |
| B343 | EP155 carrying a mutagenic cassette with a NeoR gene in the sgf73 locus and an infectious cDNA of CHV1-Δp69 | Present study |
| Δsgf73 | DK80 disrupted for sgf73 encoding a subunit of the DUB module of SAGA | Present study |
| Δsgf73+sgf73 | Δsgf73 complemented with wild-type sgf73 | Present study |
| Δsgf73+sgf73-egfp | Δsgf73 complemented with wild-type sgf73 fused with egfp | Present study |
| Δgcn5 | DK80 disrupted for gcn5 encoding a catalytic subunit of the HAT module of SAGA | Present study |
| Δada2 | DK80 disrupted for ada2 encoding a subunit of the HAT module of SAGA | Present study |
| Δubp8 | DK80 disrupted for ubp8 encoding a subunit of the DUB module of SAGA | Present study |
| Δlzf1 | DK80 disrupted for lzf1 encoding a LYAR-type zinc-finger protein | Present study |
Screening for Mutants Unable to Respond to Virus Infection.
We chose one of the double-transformant strains with pCPXHY-Pdcl2D2-egfp and pCPXBn-CHV1-Δp69 (Pdcl2D2-egfp+CHV1-Δp69) (Table S1) as the parent strain for random mutagenesis with NeoR (Fig. 2A). Approximately 1,000 triple-transformants with the mutagenic construct carrying NeoR were screened under confocal laser scanning microscopy (CLSM). Most triple-transformants tested positive for eGFP fluorescence. One of the transformants, designated as B343 (Table S1), stably manifested an eGFP-negative phenotype (Fig. 2B) and was further analyzed in detail. B343, which was confirmed to be homokaryotic, was reduced severely in growth, compared with the parent or EP155 (Fig. 2C). B343, as well as its single-conidial homokaryotic isolates, B343-1 and B343-2, accumulated less egfp mRNA than the parental strain Pdcl2D2-egfp+CHV1-Δp69 (Fig. 2D). Transcripts of dcl2 in B343 were also much less than in the parental strain, although slightly higher than in the standard EP155 (Fig. 2D), suggesting the disruption of a host factors that is necessary for dcl2 induction. Reduction of agl2 transcripts to the level of EP155 coincided with that of dcl2 transcripts (Fig. 2D). Taken together, the combined results revealed that B343 is able to produce a basal level of transcripts of dcl2 and agl2 but, unlike EP155, unable to respond to virus infection.
Fig. 2.
Genetic screen for host factors involved in the activation of antiviral RNA silencing in C. parasitica. (A) Screening procedure to identify host genes mediating the activation of antiviral RNA silencing. Protoplasts of the parental fungal strain (EP155/Pdcl2D2-egfp+CHV1-Δp69) (Fig. 1A and Table S1) were randomly mutagenized by introducing a cassette (NeoR) carrying a neomycin phosphotransferase II gene (triple transformation) and screened for loss of eGFP fluorescence under confocal laser scanning microscopy. (B) Loss of GFP induction in a selected mutant, B434. The parental fungal strain green-fluoresced, and the mutant B434 carrying mutagenic construct NeoR failed to do so. (Scale bars, 0.1 mm.) (C) One-week-old PDA cultures of the standard EP155 strain, parental strain (Pdcl2D2-egfp+CHV1-Δp69), and B343. (D) Northern analysis of three fungal strains used in A and independent single spore isolates (B343-1 and B343-2) derived from B343. (E) A diagram of the sgf73 gene and NeoR insertion site in B434. The NeoR is shown by fusion primer and nested integrated-PCR (21) to be inserted into one of the four introns of the sgf73 gene (protein ID, 354172; scaffold no. 1), a putative component of the SAGA complex. (See Fig. S5 for the sgf73 sequences.) Map positions in this and subsequent figures are shown according to the genome sequence of C. parasitica strain EP155. The SGF73 is characterized by the presence of domain SCA7 carrying a Zn finger motif (ZnF2), a second Zn finger motif (ZnF2), and a putative bipartite NLS. An sgf73 disruptant (Δsgf73) shown in the following figure lacks the entire coding region. (F) RT-PCR analysis of sgf73 expression in EP155 and B434. ssRNA fractions from virus-free EP155, parental strain (Pdcl2D2-egfp+CHV1-Δp69) and B434 were used in RT-PCR for detection of sgf73 and actin mRNAs (Table S3 for primer sequences) were used.
Identification of the Gene Responsible for the Mutant Phenotype.
Southern blotting revealed multiple integrations, in at least two sites, of the mutagenic construct, NeoR (Fig. 1A and Fig. S2). Taking advantage of a PCR-based method (21), one of the mutated loci was mapped to a gene termed sgf73, homologous to S. cereviciae SGF73 with its product carrying the SCA7 (spinocerebellar ataxia 7) domain (pfam: 08313) (Fig. 2E). Sgf73 is one of the components of SAGA, a transcriptional coactivator (20). The NeoR construct was inserted into the fourth intron of a homolog of sgf73 (Fig. 2E and Table S2). The mutation of sgf73 in B343 was validated by RT-PCR, showing no amplified fragment specific for sgf73 transcripts in B343 that were detected in the standard (EP155) and the parental (Pdcl2D2-egfp+CHV1Δp69) strains (Fig. 2F). sgf73 encodes a protein of 432 amino acids that possesses two zinc-finger motifs (ZnF1 and ZnF2), one of which resides in the SCA7 domain, which is strictly conserved in homologous proteins (22) (Fig. S3A). SGF73 showed closer phylogenetic affinity to counterparts from members of the order Diaporthales, to which C. parasitica belongs (Fig. S3B).
Table S2.
BLASTp analyses for putative SAGA components of C. parasitica
| SAGA module | Query | C. parasitica EP155 v2.0 | ||||
| Yeast* | NCBI ref. sequence no. (JGI gene name) | Protein ID | Location | E-value, identity (%)/coverage (%) | Domain name (accession, E-value) | |
| HAT | yGcn5 | NP_011768 | 346564 | scaffold_5:1504048–1505708 | 2.54e-162, 66.0/68.6 | Bromo_gcn5_like (cd05509, 2.74e-53)Acetyltransf_1 (pfam00583, 4.25e-12)COG5076 (COG5076, 1.94e-68) |
| yAda2 | NP_010736 | 253860 | scaffold_3:2089078–2090760 | 1.84e-92, 49.4/66.5 | ZZ_ADA2 (cd02335, 1.53e-18)SANT (cd00167, 1.45e-04)COG5114 (COG5114, 1.22e-119) | |
| yAda3 | NP_010461 | 216981 | scaffold_5:2405772–2408064 | 2.78e-37, 38/42.7 | Ada3 (pfam10198, 1.38e-40) | |
| DUB | yUbp8 | NP_013950 | 279741 | scaffold_6:2579525–2581768 | 3.57e-41, 42.6/46.9 | zf-UBP (pfam02148, 3.25e-06) UCH (pfam00443, 3.14e-73) |
| ySgf11 | NP_015278 (AN8685)† | no hit (43786) | − (scaffold_5: 3417324–3418640) | − (8.66e-37, 50.9/39.9) | (No hit) | |
| ySgf73 | NP_011449 | 354172 | scaffold_1:1765734–1766624 | 3.08e-18, 65.0/13.9 | SCA7 (pfam08313, 9.13e-31) | |
| ySus1 | NP_878049 (AN7253)† | no hit (no hit) | ||||
| TBP-binding/structure | yAda1 | NP_015069 | 219729 | scaffold_9:343791–345365 | 1.38e-6, 33.7/18.3 | SAGA-Tad1 (pfam12767, 3.59e-38) |
| ySpt3 | NP_010680 | 252945 | scaffold_3:4015565–4016539 | 1.80e-72, 56.7/68.9 | TAF13 (cd07978, 1.80e-31) | |
| ySpt7 | NP_009637 | 332125 | scaffold_6:2446346–2450034 | 5.21e-73, 48.5/26.5 | Bromo_SPT7_like (cd05510, 1.93e-60)TAF9 superfamily (cl22885, 1.03e-06) | |
| ySpt8 | NP_013156 | 270172 | scaffold_11:368325–370370 | 2.29e-85, 45.4/57.8 | WD40 superfamily (cl02567, 2.28e-09) | |
| ySpt20 | NP_014493 (AN0976)† | no hit (346150) | − (scaffold_4:3036013–3040461 (+)) | − (4.82e-40, 46.8/23.0) | (Spt20 (pfam12090, 1.09e-35)) | |
| yTra1 | NP_011967 | 344350 | scaffold_1:5375040–5387168 | 0, 45.3/70.3 | PIKK_TRRAP (cd05163, 4.11e-123) TEL1 (COG5032, 2.77e-158) | |
| TAF | yTaf5 | NP_009757 | 352224 | scaffold_6:613040–617467 | 3.57e-40, 34.4/11.5 | NACHT_N (pfam17100, 1.86e-68) WD40 (cd00200, 2.16e-135) |
| yTaf6 | NP_011403 | 285968 | scaffold_1:6160817 | 3.65e-84, 44.1/59.1 | TAF6 (cd08050,1.06e-141) | |
| yTaf9 | NP_013963 | 221516 | scaffold_5:2434482 | 6.79e-17, 55.2/32.8 | TFIID-31kDa (pfam02291, 7.54e-50) | |
| yTaf10 | NP_010451 | 351499 | scaffold_4:4563748–4564853 | 3.57e-20, 60.4/28.4 | TAF10 superfamily (cl02150, 4.47e-37) | |
| yTaf12 | NP_010429 | 108452 | scaffold_6:142895–145821 | 1.56e-12, 45.7/10.2 | TAF12 (cd07981, 1.21e-18) | |
| Interaction H3K4me | ySgf29 | NP_009917 (AN0668)† | no hit (341987) | − (scaffold_7: 3040365–3041741) | − (4.97e-14, 38.9/21.3) | (DUF1325 (pfam07039, 1.06e-44)) |
| yChd1 | NP_011091 | 260476 | scaffold_5:3305285–3310503 | 0, 54.9/47.3 | HELICc (cd00079, 8.25e-31) DEXDc (cd00046, 3.02e-19) DUF4208 (pfam13907, 7.73e-15) Chromo (pfam00385, 1.62e-10) CHROMO (cd00024, 1.01e-06) | |
S. cerevisiae.
A. nidulans sequence as query.
A disruptant of sgf73 (Δsgf73) was prepared in the background of DK80 that supports efficient homologous recombination-based targeted disruption because DK80 lacks the cpku80 gene necessary for nonhomologous recombination (23) (Table S1). The phenotype of obtained disruptants was indistinguishable from B343, indicating that the sgf73 gene is mainly responsible for the mutant phenotype. Δsgf73 showed a debilitation phenotype, being severely reduced in growth either in the presence or absence of CHV1-Δp69 (Fig. 3B). A closer inspection revealed that mycelia of B343 and Δsgf73 were thicker than those of DK80 (Fig. S4). This observation was slightly surprising to us because the debilitation phenotype for B343 was anticipated to be because of defective RNAi activation and CHV1 Δp69 infection, as previously reported for Δdcl2 infected by CHV1 Δp29 or Δp69 (16, 17).
Fig. 3.
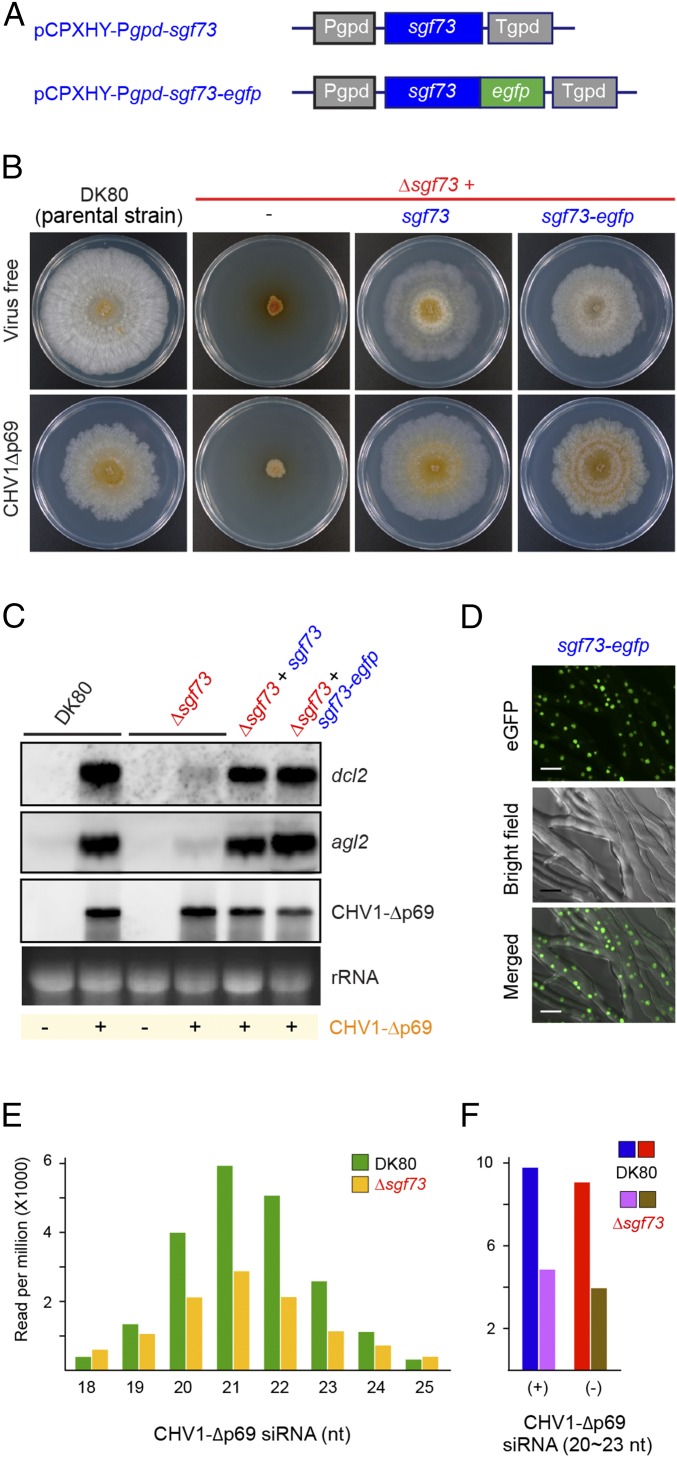
Identification of the gene responsible for loss of induction of antiviral RNA silencing in B343. (A) Schematic representation of constructs used for overexpression of sgf73 or sgf73-egfp. (B) Phenotypic effects (colony morphology) of targeted disruption of sgf73 (Δsgf73) and genetic complementation of Δsgf73 expressing wild-type sgf73 (Δsgf73+sgf73) or egfp-tagged sgf73 (Δsgf73+sgf73-egfp) (Fig. S7 and Table S1). The two complemented strains were fused to EP155 infected CHV1-Δp69 for its horizontal transfer. To obtain Δsgf73 infected with CHV1-Δp69, DK80 was pretransformed with pCPXNeo-CHV1-Δp69 ahead of sgf73 disruption (Materials and Methods). One-week-old PDA cultures of the strains in the absence (virus-free) and presence of CHV1-Δp69 are shown. (C) Northern analyses of the fungal strains DK80, Δsgf73, Δsgf73+sgf73, and Δsgf73+sgf73-egfp. Transcriptional up-regulation of dcl2 observed in DK80 was diminished in Δsgf73. Overexpression of wild-type sgf73 resulted in amendment of the dcl2 induction. (D) Nuclear localization of SGF73. Δsgf73+sgf73-egfp was observed under confocal laser scanning microscopy. (Scale bars, 10 μm.) (E) CHV1-Δp69-derived siRNA size distribution. Viral siRNA reads (20∼30 nt) per million of total small RNAs are shown for the DK80 and Δsgf73 infected with CHV1-Δp69. (F) Comparison of sense and antisense CHV1-Δp69-derived siRNAs from DK80 and Δsgf73.
The disruptant Δsgf73 lost induction of the dcl2 and agl2 transcription upon infection by CHV1-Δp69 (Fig. 3C). Expression of the wild-type sgf73 gene under the control of the C. parasitica glyceraldehyde-3-phosphate dehydrogenase gene promoter (Pgpd) (Fig. 3A) (pCPX-Pgpd-sgf73) restored the growth of colonies to the comparable level of the parental DK80 strain, regardless of presence or absence of CHV1-Δp69 (Fig. 3B and Table S1), and also restored the responsiveness of the RNAi key genes to the virus (Fig. 3C). An eGFP-tagged version of SGF73 appeared to be functional in restoring the fungal growth and RNAi responsiveness to CHV1-Δp69 (Fig. 3 A and C and Table S1) (pCPX-Pgpd-sgf73-egfp). This functional SGF73 was localized in the nucleus as expected from its bipartite nuclear localization signal (NLS) (Figs. 2E and 3D).
A slight phenotypic difference between DK80 and the complemented strain (Δsgf73+sgf73) requires an explanation. Sequence analysis of sgf73 mRNA revealed that at least three variants were produced whose exon/intron maps are shown in Fig. S5. The major variant judged from numbers of cDNA clones obtained is shown in detail (Fig. S5). It is possible that full complementation of disruptants requires the expression of all of the variants at an appropriate ratio. Alternatively, the use of the heterologous promoter (Pgpd) used for complementation may affect the phenotype, as observed occasionally in other organisms.
Next, we tested whether the deletion of sgf73 affected viral siRNA (vsiRNA) profile. To this end, small RNAs from two host strains, DK80 and Δsgf73, were deep-sequenced. Accumulation of CHV1-Δp69–derived siRNA of 20–23 nt in Δsgf73 was decreased to ∼40% of that in DK80 (Fig. 3 E and F), but this rate of reduction was smaller than expected from the relative reduction of dcl2 mRNA (Fig. 3C). Both fungal strains (DK80 and Δsgf73) showed similar patterns for the ratio of sense and antisense vsiRNA (Fig. 3F), vsiRNA distribution along the CHV1-Δp69 genome (Fig. S6A), and 5′-terminal nucleotide preference (Fig. S6B).
Taking these data together, we find the mutation of sgf73 in B343 was shown to be responsible for loss of dcl2 induction leading to reduced vsiRNA production.
SAGA Mediates Virus-Responsive Induction of RNAi Key Genes.
Sgf73 is known as a component of the SAGA complex with at least four modules: HAT, SPT (recruiting the TATA-binding protein TBP), TAF (TBP-associated factor forming a structural core), and DUB (histone H2B deubiquitination) (20) (Table S2). Based on studies of the yeast SAGA complex, Sgf73 is believed to be a component of DUB and link the HAT and TAF modules within the complex (24–27). To determine whether C. parasitica SGF73 works as a component of SAGA to induce dcl2 in response to virus infection, other SAGA component genes representing HAT and DUB were tested. The C. parasitica homologs of yeast GCN5 and ADA2 (for the HAT module) and the homolog of UBP8 (for the DUB module) (Fig. S7 and Table S2), were disrupted in DK80. Although gcn5 encodes the key catalytic enzyme, histone acetyltransferase, ubp8, codes for histone deubiquitinase (20). DK80 infected by CHV-Δp69 was reduced in growth of aerial hyphae while maintaining conidiation and pigmentation compared with virus-free DK80 (Fig. 4A). The phenotype of gcn5 and ada2 disruptants (Δgcn5 and Δada2) was similar to that of Δsgf73, characterized by severely reduced growth that was slightly exacerbated by CHV-Δp69 infection (Fig. 4A). In contrast, the phenotype of disruptants of ubp8 and lzf1 (a SAGA-unrelated gene) (Fig. S7) (Δubp8 and Δlzf1) was not as severely affected as those of Δgcn5 and Δada2 (Fig. 4A). Interestingly, CHV1-Δp69 infection did not cause significant phenotypic change in either Δubp8 or Δlzf1 (Fig. 4A).
Fig. 4.
Mechanism of transcriptional RNAi activation involving SAGA. (A) Colony morphology of disruptants of the four genes, ubp8, gcn5, ada2, and lzf1 (Fig. S7 and Table S1). The first three genes are possible key players in SAGA, and the last one encodes an unrelated zinc-finger protein. The four genes were disrupted in DK80 to obtain Δubp8, Δgcn5, Δada2, and Δlzf1 that were infected or uninfected by CHV1-Δp69. CHV1-Δp69 was introduced into these disruptants by horizontal transfer or pretransformation with viral cDNA (Materials and Methods). (B) Northern analysis of the disruptants. Whereas DK80, Δubp8, and Δlzf1 showed up-regulation of dcl2, no such dcl2 induction was observed in the other two disruptants. (C) Requirement of DCL2 and AGL2 for virus-induced RNAi induction. The dcl2 and agl2 disruptants were retransformed with pCPXNeo-Pdcl2D2-egfp (Table S1). These two strains, together with EP155 transformant (Pdcl2D2-egfp), were fused to EP155 infected CHV1-Δp69 or MyRV1 for its horizontal transfer. Resultant fungal strains were examined for egfp induction by Northern analysis (or green fluorescence by microscopy) (Fig. S9). (D) A model for virus-induced up-regulation of dcl2 and agl2 transcription. The model is an updated version of the model previously proposed by Chiba and Suzuki (17). Virus infection is perceived by an unidentified host sensor (step 1, virus recognition), its signal is transduced and transmitted to the nucleus (step 2, SAGA recruitment) to enhance the transcription of a subset of genes including dcl2 and agl2, in which the histone acetyltransferase activities of SAGA are involved (step 3, transcriptional activation). The DUB module appears to be an independent contributor to the high transcriptional activation of only agl2. The dcl2 is induced by MyRV1, but not by CHV1-Δp69 in the absence of AGL2. This virus-specific requirement of AGL2 for the dcl2 up-regulation adds complexity to the model.
No dcl2 transcriptional induction was observed in Δgcn5 or Δada2 infected with CHV1-Δp69 (Fig. 4B), as in the case for B343 and Δsgf73 (Figs. 2D and 3C). In contrast, dcl2 inducibility was retained in Δubp8 and Δlzf1 (Fig. 4B). Accumulation levels of agl2 in CHV1-Δp69–infected Δgcn5, Δada2, and Δubp8 were between those shown by virus-free DK80 (uninduced state) and CHV1-Δp69–infected DK80 or -Δlzf1 (Fig. 4B), showing partial abrogation of the agl2 up-regulation in these disruptants.
Taken together, these results suggest that the HAT activities in C. parasitica are more influential in the up-regulation of the RNAi genes than DUB activities, and dcl2 and agl2 transcription is differently regulated by SAGA components.
dcl2 Induction by Distinct Viruses Is also Mediated by the SAGA Complex.
Besides CHV1-Δp69, other viruses are known to markedly induce dcl2 induction (17, 28). Thus, it was of interest to investigate whether dcl2 is up-regulated by other viruses in disruptants of SAGA complex components. Two dsRNA viruses, MyRV1 and an alphapartitivirus, Rosellinia necatrix partitivirus 3 (RnPV3), were chosen. As a result, like CHV1-Δp69 [(+) ssRNA virus], the two viruses failed to induce transcription of dcl2 in Δsgf73, indicating the involvement of the SAGA complex in the transcriptional regulation with dsRNA virus infection (Fig. S8).
Virus-Specific and -Nonspecific Requirement of AGL2 and DCL2 for the Full-Scale dcl2 Induction.
It was hypothesized earlier that DCL2 and AGL2 have a positive feedback in their up-regulation (17). First, we tested whether DCL2 plays a role in RNAi activation. Δdcl2 with the background of the standard EP155 fungal strain (16) was transformed with pCPXNeo-Pdcl2D2-egfp (Fig. 1) and then infected with CHV1-Δp69 or MyRV1. Deletion of dcl2 resulted in complete loss of dcl2 transcriptional elevation upon infection by either virus (Fig. 4C). This result was supported by green fluorescence intensity, as assessed by CLSM (Fig. S9).
Similarly, effects of the agl2 deletion on dcl2 transcriptional elevation were examined. Interestingly, different dcl2 induction patterns were observed between Δdcl2 and Δagl2. That is, Δagl2 was able to up-regulate dcl2 upon infection by MyRV1, whereas it was unable to induce dcl2 to the full-scale level upon infection by CHV1-Δp69 (Fig. 4C and Fig. S9). These results indicate that CHV1-Δp69 requires AGL2, and not MyRV1, for dcl2 induction.
Taken together, these results suggest that DCL2 is essential for virus infection-triggered dcl2 transcriptional induction, whereas AGL2 is required for it in a virus-specific manner.
Discussion
Antiviral RNAi is a well-conserved defense mechanism in eukaryotic organisms. Mechanisms of its transcriptional regulation remain largely unknown but appear to be regulated differently in different organisms (5, 6, 29, 30). We have developed a genetic screen protocol using an infectious cDNA clone of a CHV1 mutant, Δp69, which lacks a viral suppressor of RNA silencing (VSR), p29, and induces the key RNAi genes excessively (Fig. 1). This approach will be applicable to the assessment of RNAi activation patterns in different fungi that can support CHV1 replication (31, 32). Given the large number of viruses hosted by C. parasitica (14, 33), the reporter fungal strain (Fig. S8) will be useful for testing RNAi induction by those various viruses.
The genetic screen allowed the identification of sgf73, a homolog of S. cerevisiae SGF73 (Fig. 2). Sgf73 is a component of the highly conserved transcriptional coactivator SAGA that in yeast is involved in the regulation of ∼10% of all genes (34, 35). Extension of the N-terminal polyglutamine tract of its human homolog, ataxin-7, causes a neurodegenerative disorder (36). The SAGA complex is composed of at least four functional modules, HAT, SPT, TAF, and DUB. All homologous components except for Sus1 are searchable in the C. parasitica genome sequence (Table S2). From observations in S. cerevisiae (26), Sgf73 is one of the DUB components that bridges the other two modules (TAF and HAT) via physical interaction with Ada2, Taf12, and possibly other components (25, 37). DUB comprises Sgf11, Sus1, and Sgf73 in addition to Ubp8, whereas HAT consists of Gcn5, Ada2, Ada3, and Sgf29 (Table S2). Our disruption assay of a few components of the two catalytic HAT (gcn5 and ada2) and DUB modules (ubp8 and sgf73) has provided interesting insights. Disruptants of gcn5 and ada2 have been observed to show a similar phenotype (debilitated colony morphology and abolished dcl2 induction) to sgf73 disruptants (Figs. S3 and S4). A discernable difference between them was that agl2 transcription was completely abolished in Δsgf73, whereas it was still enhanced measurably, although not to the level of the parental strain (DK80), in Δgcn5 and Δada2 (Fig. 4B). Disruption of ubp8 had no effects on the dcl2 induction but moderate effects on agl2 transcription as in Δgcn5 and Δada2, whereas that of a SAGA-unrelated gene lzf1 had no effect on dcl2 or agl2 transcriptional induction triggered by CHV1-Δp69 (Fig. 4B). These observations allow us to conclude and suggest a few points as follows.
Our results support the view that high activation of RNAi, via transcription up-regulation of dcl2, is mediated by SAGA as a transcriptional coactivator rather than by SGF73 molecules alone. However, we cannot exclude the possibility that complexes other than SAGA, such as the SAGA-like complex (27, 38), components of which are shared with SAGA, mediate transcriptional regulation of dcl2. Roles of the SAGA complex in abiotic stress responses have been extensively studied in other organisms, particularly in yeast (34, 39, 40), but rarely in biotic stress conditions, including virus infection. This is an example of SAGA’s involvement in an antiviral response. Our results suggest that HAT activities regulate dcl2 induction, whereas agl2 transcriptional augmentation is governed by both HAT and DUB activities in an additive way. Given that Sgf73 bridges the two modules, HAT and DUB, it may not be surprising that disruption of sgf73 leads to inhibition of both enzymatic activities of SAGA, as observed in the deletion mutant of SGF73 in S. cerevisiae (25). In filamentous fungi, two recent reports suggest the SAGA complex of filamentous ascomycetes appears not to include DUB, although Sgf73 is part of the complex (41, 42). Moreover, Sus1, which along with Sgf11 bridges Ubp8 and Sgf73 (22), is not conserved or considerably diverged in filamentous fungi (41, 42). It should also be noted that plants have no SGF73 homolog, suggesting that HAT and DUB are independent structures (40). Therefore, our results with Δsgf73 can be interpreted alternatively. The DUB may act independently of SAGA in filamentous ascomycetous fungi, such as C. parasitica, to up-regulate only agl2 transcription (Fig. 4D). This study provides a framework for understanding possible structural and functional commonalities and differences between SAGAs of filamentous fungi and other eukaryotic organisms.
CHV1-Δp69 stably replicates in RNAi-competent EP155, and its accumulation is greatly enhanced in Δdcl2 (Fig. 4C). Thus, despite lacking the VSR p29, CHV1-Δp69, which is native to C. parasitica, appears to be more tolerant to activated RNAi in C. parasitica than RnVV1, which is a heterologous virus originally isolated from R. necatrix, another phytopathogenic ascomycete (17). Given the marked reduction in transcription of the two key genes of RNAi, antiviral defense was anticipated to be compromised in B343 and Δsgf73. However, CHV1-Δp69 accumulated to the equal or slightly lower level than in the parental strain (Figs. 2D and 3C). These findings suggest that possible mutations simultaneously affect the expression of host factors that support CHV1-Δp69 replication in Δsgf73. The dcl2 transcription was measurably induced upon infection by CHV1-Δp69 in Δsgf73 (Fig. 3C), suggesting that the sgf73-defficient HAT module is still partially functional in the induction. Thus, it is also possible that the slight but measurable dcl2 up-regulation leads to sufficient siRNA production for inhibition, to a certain extent, of CHV1 accumulation.
Debilitated growth in the absence of virus, observed in Δsgf73, Δada2, and Δgcn5 (Figs. 3 and 4), is considered to result from the abolishment of the HAT activity of SAGA. Similar consequences (severe growth defect) after disruption of HAT components have been reported for filamentous fungi and S. cerevisiae (41, 42). Additionally, in Candida albicans, deletion of GCN5 leads to impaired invasive hyphal growth, induction of a thick stubby pseudohyphae-like form, and loss of pathogenesis (43). It is conceivable that a portion of genes regulated by SAGA includes essential genes for normal growth. In contrast, disruption of C. parasitica ubp8 exerts minimal effects on fungal growth (Fig. 4A), as in the case of other fungi (44).
Nuss and colleagues (4, 17) and our group collectively suggested a difference in the requirement of AGL2 for activation of RNAi upon infection by different viruses. This study clearly showed that MyRV1 required DCL2, but not AGL2, for dcl2 full-scale induction (Fig. 4C), whereas AGL2 has an auxiliary positive effect on CHV1-Δp69 infection-mediated RNAi induction (Fig. 4C). Thus, these results revealed additional roles of DCL2 and AGL2, possibly in the steps leading to the recognition of the incoming virus and transcriptional activation (Fig. 4D), which is in line with our earlier model in which DCL2 and AGL2 have a positive feedback in this RNAi activation upon infection by viruses (17). Now added to the model as important players are GCN5, ADA2, SGF73, and UBP8, all of which are expected to work for histone modification to facilitate the entry of transcriptional machinery through relaxing the chromatin structure (Fig. 4D), as supported by the nuclear localization of functional eGFP-tagged SGF73 (Fig. 3D). Moreover, our restriction enzyme-based analysis suggests that no DNA methylation in the promoter and coding region of dcl2 is associated with the alteration of its transcriptional status (Fig. S10), and a recent report suggested DNA methylation to be involved in repression of some transposons and genes in a filamentous fungus (45). However, there are still many missing links between perception of virus infection and transcriptional up-regulation of the RNAi key genes. Their identification using the currently developed screen protocol is underway. In this regard, it is noteworthy that a basic leucine zipper-type and a forkhead box O-type transcription factor have been shown to regulate RNA silencing genes in plant and insect, respectively (29, 46). It will also be of interest to further explore whether SAGA, conserved across eukaryotic organisms from yeast to humans, serves as an RNAi regulator in these organisms and what viral factors determine the virus-specific RNAi activation and requirement of AGL2 for RNAi induction. More studies are needed to address these interesting questions.
Materials and Methods
Viral and Fungal Strains.
All fungal and viral strains used in this study are listed in Table S1. Three viral strains were used: an ORF A mutant of CHV1 with an undivided (+)RNA genome (CHV1-Δp69b, GenBank accession no. M57938 with an internal deletion 496–2,363 nt) lacking the VSR, p29 (47), and a reovirus, MyRV1, with a 11-segmented dsRNA genome (48, 49).
C. parasitica standard strain EP155 and its RNAi-deficient derivatives ∆dcl2 and ∆agl2 (4, 16) were generous gifts from Donald L. Nuss (University of Maryland, College Park, MD). C. parasitica strain DK80, a mutant of EP155 disrupted in the cpk80 gene (23), was a generous gift from Bao-shan Chen, (Guangxi University, Nanning, China). The EP155 strains infected with CHV1-WT, CHV1-∆p69, or MyRV1 (EP155/CHV1-wt, EP155/CHV-∆p69, and EP155/MyRV1) were described earlier (47, 49). Virus horizontal transfer from respective donor strains was successively repeated to avoid heterokaryon formation. Fungal cultures were grown at 22–27 °C on potato dextrose agar (PDA) plates on the bench-top for maintenance and phenotypic observations and in potato dextrose broth liquid media in the dark for RNA preparation.
Plasmid Construction and Transformation of C. parasitica.
Major expression vectors for C. parasitica include pCPXHY2 (50), pCPXBn2 (19), and pCPXNeo (4). A DNA fragment corresponding to CHV-∆p69 cDNA (47) was inserted into the NotI site of pCPXHY3 and pCPXBn2 to produce pCPXH-CHV-∆p69 and pCPXBn-CHV-∆p69, respectively. The dcl2 promoter linked to the egfp coding domain (Pdcl2D2-egfp) fragment was generated by overlapped PCR and cloned between EcoRI and SphI sites of pCPXHY3 and pCPXNeo to produce pCPXHY-Pdcl2D2-egfp and pCPXNeo-Pdcl2D2-egfp, respectively (Fig. 1A). A sgf73 DNA fragment was generated by RT-PCR and inserted between HpaI and SphI sites of pCPXHY3 to produce pCPXHY-sgf73. The sgf73-egfp fragment was generated by overlapped PCR and inserted between HpaI and SphI sites of pCPXHY3 to produce pCPXHY-sgf73-egfp (Fig. 3).
Transformation of C. parasitica protoplasts and subsequent selection with hygromycin B were performed as described previously (18). An approach similar to the one reported by Faruk et al. (19) was taken to screen for genetic factors involved in RNA silencing regulation. First, pCPXBn-CHV1-∆p69 was used to transform the EP155 standard strain of C. parasitica (benomyl selection). Virus autonomous replication is initiated in all transformed cells from CHV1-Δp69 transcripts produced from chromosomally integrated cDNA (18). Homokaryotic transformant strains were retransformed by the pCPXHY-Pdcl2D2-egfp (hygromycin selection). Finally, double transformants (Pdcl2D2-egfp+CHV1-∆p69) were transformed with a neomycin resistance gene cassette (NeoR) amplified by PCR on pCPXNeo for mutagenesis (neomycin selection). Triple transformants grown on neomycin (G418)-containing PDA plates were screened for loss of green fluorescence. The pCPXNeo-Pdcl2D2-egfp plasmid was used to transform ∆dcl2 and ∆agl2 strains (neomycin selection) because these mutant strains already contain a hygromycin resistance gene. For generation of ∆sgf73, ∆gcn5, ∆ada2, ∆ubp8, and ∆lzf1 deletion (knockout) mutants through homologous recombination, DNA fragments, which consist of a NeoR cassette flanked with 700-bp sequences derived from the both upstream and downstream sequences of the coding region of the target gene, were generated by overlapped PCR and were used to transform strain DK80 (neomycin selection). Because ∆sgf73, ∆gcn5, and ∆ada2 mutants were incompetent for viral transmission by hyphal anastomosis, a DK80 strain was first transformed with pCPXHY-CHV1-∆p69 (hygromycin selection) and then used to generate ∆sgf73, ∆gcn5, and ∆ada2 knockout mutants through homologous recombination. The deletion of the target gene was confirmed by genomic PCR. For complementation analysis, pCPXHY-sgf73 and pCPXHY-sgf73-egfp plasmids were used to transform ∆sgf73 mutant strain (hygromycin selection). All knockout mutant strains were subjected to single-conidial spore isolation and representative homokaryon strains were selected for use.
Sequence and Phylogenetic Analyses.
A draft sequence of C. parasitica genome (Cryphonectria parasitica EP155 v2.0, JGI/MycoCosm, genome.jgi.doe.gov/Crypa2/Crypa2.home.html) was screened for yeast SAGA homologs (42). Sequence data were analyzed using GENETYX-MAC (Software Development Co) or Enzyme X v3.3.3 (nucleobytes.com/enzymex/index.html). The conserved protein domains were predicted from the National Center for Biotechnology Information (NCBI) conserved domain database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (51). The putative NLS and coiled-coil domain were determined using cNLS Mapper (nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) (52) and EMBnet COILS (www.ch.embnet.org/software/COILS_form.html) (53), respectively. Multiple sequence alignments and phylogenetic tree construction were performed using MAFFT v7 (54) and PhyML 3.0 (55) with automatic model selection by SMS (Smart Model Selection) (www.atgc-montpellier.fr/phyml-sms/).
eGFP Observation.
eGFP expression in fungal mycelia was observed using an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus) as described previously (14).
RNA and DNA Analyses.
Total RNA and genomic DNA were extracted as described previously (56). Identification of genomic regions flanked a NeoR cassette sequence was performed using fusion primer and nested integrated-PCR method, as described previously (21). DNA blot analysis was performed as described previously (57). sgf73 transcripts were amplified by RT-PCR and sequenced. The 5′ and 3′ ends of sgf73 mRNA were determined using a FirstChoice RLM-RACE Kit (Thermo Fisher Scientific). RNA blotting was performed as described previously (57). Single-stranded RNA fractions obtained from each of the fungal strains were probed by DIG-labeled PCR products specific for genes or virus. All of the primers used in this study are listed in Table S3.
Table S3.
List of the primers used in this study
| Name | Sequence (5′ 3′) | Direction | Used for |
| AJPSF−1 | ATAACTAGTGGCTGGTCGAGAGGGTGGAGGCCAG | Forward | Pdcl2D3-egfp |
| AJPSF−3 | ATAACTAGTTGAGGAGGGTGGGGACAA | Forward | Pdcl2D2-egfp |
| AJPSF−5 | ATAACTAGTGCGATTGAGGGAAGAGAA | Forward | Pdcl2D1-egfp |
| AJPSR | CAGGCCGTTAACGCGGCCGCGCTAGCCTTGCAGCGTCGTACGACAGATCTG | Reverse | Pdcl2D1/2/3-egfp |
| AJPEGFP−F | ATAGCTAGCATGGTGAGCAAGGGCGAG | Forward | Pdcl2D1/2/3-egfp |
| AJPEGFP−R | CCCGCTAGCATTACTTGTACGCTCGTC | Reverse | Pdcl2D1/2/3-egfp |
| NeoR (F) | AGATACCCAGAGTTATCTCACC | Forward | NeoR |
| NeoR (R) | TCATTCCCGGTGTAGGAAGCTG | Reverse | NeoR |
| Sgf (F) | CAACGGGTGTTTGAGGATTAAG | Forward | RT-PCR (sgf73) |
| Sgf (R) | CTAGGCTGGGCCACCCACCGAC | Reverse | RT-PCR (sgf73) |
| CP−Actin (F) | GACCCAGATCGTCTTCGAGACG | Forward | RT-PCR (actin) |
| CP−Actin (R) | CGCACTTCATGATGGAGTTAAA | Reverse | RT-PCR (actin) |
| OL−SGF73−Neo (F) | CGAGGCCTGGATAGCACACCAGATACCCAGAGTTATCTCACCAT | Forward | Δsgf73 |
| OL−SGF73−Neo (R) | CCATCCTGATCCTCCGACTTTCATTCCCGGTGTAGGAAGCTGTT | Reverse | Δsgf73 |
| 5arm−SGF73 (F) | TAGGATTTGTTGCTTCCTAGCC | Forward | Δsgf73 |
| 5arm−SGF73 (R) | TGAGATAACTCTGGGTATCTGGTGTGCTATCCAGGCCTCGTCGC | Reverse | Δsgf73 |
| 3arm−SGF73 (F) | GCTTCCTACACCGGGAATGAAAGTCGGAGGATCAGGATGGCTGA | Forward | Δsgf73 |
| 3arm−SGF73 (R) | TGCCTAGCCGTACAGAATAAAG | Reverse | Δsgf73 |
| OL−UBP8−Neo (F) | CACCGGGCAGGCAATTCAACAGATACCCAGAGTTATCTCACCAT | Forward | Δubp8 |
| OL−UBP8−Neo (R) | GTACTTGCGTCTCGATCTTGTCATTCCCGGTGTAGGAAGCTGTT | Reverse | Δubp8 |
| 5arm−UBP8 (F) | GCCTGCCCGCGGGCTGCTCTGG | Forward | Δubp8 |
| 5arm−UBP8 (R) | TGAGATAACTCTGGGTATCTGTTGAATTGCCTGCCCGGTGATTT | Reverse | Δubp8 |
| 3arm−UBP8 (F) | GCTTCCTACACCGGGAATGACAAGATCGAGACGCAAGTACAATT | Forward | Δubp8 |
| 3arm−UBP8 (R) | TAACGACGCAGTCAAGGCAGTG | Reverse | Δubp8 |
| OL−Ada2−Neo (F) | GCTGCTTGAGCATTGAAACTAGATACCCAGAGTTATCTCACCAT | Forward | Δada2 |
| OL−Ada2−Neo (R) | TCTGTTCATCAGAGCTCTCATCATTCCCGGTGTAGGAAGCTGTT | Reverse | Δada2 |
| 5arm−Ada2 (F) | GGGTTCTTTAGGCTTTGCAGCT | Forward | Δada2 |
| 5arm−Ada2 (R) | TGAGATAACTCTGGGTATCTAGTTTCAATGCTCAAGCAGCAGCC | Reverse | Δada2 |
| 3arm−Ada2 (F) | GCTTCCTACACCGGGAATGATGAGAGCTCTGATGAACAGATCTG | Forward | Δada2 |
| 3arm−Ada2 (R) | GCGCTGTCTTTTCTGCAAAGGC | Reverse | Δada2 |
| OL−GCN5−Neo (F) | AAGTATAGCTGACATGAGCCAGATACCCAGAGTTATCTCACCAT | Forward | Δgcn5 |
| OL−GCN5−Neo (R) | ACAAAGCAGACTTGAGCTTTTCATTCCCGGTGTAGGAAGCTGTT | Reverse | Δgcn5 |
| 5arm−GCN5 (F) | TCGCCTTATTTACCGACACCGC | Forward | Δgcn5 |
| 5arm−GCN5 (R) | TGAGATAACTCTGGGTATCTGGCTCATGTCAGCTATACTTTGTT | Reverse | Δgcn5 |
| 3arm−GCN5 (F) | GCTTCCTACACCGGGAATGAAAAGCTCAAGTCTGCTTTGTAAAG | Forward | Δgcn5 |
| 3arm−GCN5 (R) | CCTTCTGCTTTCTCTTTCCTGA | Reverse | Δgcn5 |
| OL−LZF1−Neo (F) | TCTCTTCTCCCCAAACGCAGAGATACCCAGAGTTATCTCACCAT | Forward | Δ1zf1 |
| OL−LZF1−Neo (R) | CTCTCGTACCAATCGTCAGGTCATTCCCGGTGTAGGAAGCTGTT | Reverse | Δ1zf1 |
| 5arm−LZF1 (F) | TCCAGTCCATCCCAGCCCAACT | Forward | Δ1zf1 |
| 5arm−LZF1 (R) | TGAGATAACTCTGGGTATCTCTGCGTTTGGGGAGAAGAGACGAG | Reverse | Δ1zf1 |
| 3arm−LZF1 (F) | GCTTCCTACACCGGGAATGACCTGACGATTGGTACGAGAGAGTG | Forward | Δ1zf1 |
| 3arm−LZF1 (R) | CACAATACATCATCACATATGT | Reverse | Δ1zf1 |
| HpaIf−Saga1 (F) | GCGGCCAAGCTTGTTAACGCCTCGTCCAGCGCGAAAGCGA | Forward | pCPXHY-sgf73/ pCPXHY-sgf73-egfp |
| SphI−Saga (R) | GGAGATCAGGTCAAGCATGCCTAGGCTGGGCCACCCACCGAC | Reverse | pCPXHY-sgf73-egfp |
| SGF73−Not1 (R) | CCTTGCTCACCATGCGGCCGCCGGCTGGGCCACCCACCGACAGC | Reverse | pCPXHY-sgf73-egfp |
| eGFP−Not1 (F) | GGGTGGCCCAGCCGGCGGCCGCATGGTGAGCAAGGGCGAGGAGC | Forward | pCPXHY-sgf73-egfp |
| eGFP−Sph1 (R) | GGTCAAGCATGCTTACTTGTACAGCTCGTCCATG | Reverse | pCPXHY-sgf7-egfp |
F, forward; R, reverse.
Virus-Derived Small RNA Analysis.
Total RNA was isolated from two fungal strains, DK80 and ∆sgf73, infected with CHV1-∆p69, as described by Eusebio-Cope and Suzuki (56). Small RNA cDNA library construction and sequencing were performed by Macrogen Japan using the Illumina TruSeq Small RNA Library Preparation Kit and the HiSeq 2000 system (50-bp single-end reads). Raw reads were cleaned by trimming off the adaptor sequences and the low-quality reads and <18-nt or >30-nt reads were excluded. The remaining reads were mapped to CHV1-∆p69 by CLC Genomics Workbench (v9.5, CLC Bio). The program MISIS2 (58) was used to view and analyze small RNA maps of viruses.
Supplementary Material
Acknowledgments
We thank Drs. Donald L. Nuss, Bradley I. Hillman, Shin Kasahara, Baoshan Chen, and Satoko Kanematsu for the generous gift of the fungal strains (Δdcl2, Δagl2, Cp9B21, DK80, W1029) and plasmid clones; and Dr. Sotaro Chiba for his helpful comments on the manuscript. This study was supported in part by Yomogi Inc., and Grants-in-Aid for Scientific Research (A) and on Innovative Areas from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI 25252011 and 16H06436) (to N.S. and H.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701196114/-/DCSupplemental.
References
- 1.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Sun Q, Choi GH, Nuss DL. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci USA. 2009;106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Fan B, Chen C, Chen Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc Natl Acad Sci USA. 2001;98:6516–6521. doi: 10.1073/pnas.111440998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosseau C, Moffett P. Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell. 2015;27:1742–1754. doi: 10.1105/tpc.15.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Zhou W, Zhou Y, Wu J, Zhou X. Transcriptome and comparative gene expression analysis of Sogatella furcifera (Horváth) in response to southern rice black-streaked dwarf virus. PLoS One. 2012;7:e36238. doi: 10.1371/journal.pone.0036238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du P, et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7:e1002176. doi: 10.1371/journal.ppat.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 10.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 13.Nuss DL. Mycoviruses, RNA silencing, and viral RNA recombination. Adv Virus Res. 2011;80:25–48. doi: 10.1016/B978-0-12-385987-7.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eusebio-Cope A, et al. The chestnut blight fungus for studies on virus/host and virus/virus interactions: From a natural to a model host. Virology. 2015;477:164–175. doi: 10.1016/j.virol.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV, Choi GH, Nuss DL, Shapira R, Carrington JC. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc Natl Acad Sci USA. 1991;88:10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiba S, Suzuki N. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci USA. 2015;112:E4911–E4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi GH, Nuss DL. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science. 1992;257:800–803. doi: 10.1126/science.1496400. [DOI] [PubMed] [Google Scholar]
- 19.Faruk MI, Eusebio-Cope A, Suzuki N. A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J Virol. 2008;82:740–754. doi: 10.1128/JVI.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, et al. Fusion primer and nested integrated PCR (FPNI-PCR): A new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol. 2011;11:109. doi: 10.1186/1472-6750-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 23.Lan X, et al. Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr Genet. 2008;53:59–66. doi: 10.1007/s00294-007-0162-x. [DOI] [PubMed] [Google Scholar]
- 24.Morgan MT, et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenet Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamata K, et al. C-terminus of the Sgf73 subunit of SAGA and SLIK is important for retention in the larger complex and for heterochromatin boundary function. Genes Cells. 2013;18:823–837. doi: 10.1111/gtc.12075. [DOI] [PubMed] [Google Scholar]
- 28.Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N. A novel betapartitivirus RnPV6 from Rosellinia necatrix tolerates host RNA silencing but is interfered by its defective RNAs. Virus Res. 2016;219:62–72. doi: 10.1016/j.virusres.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Spellberg MJ, Marr MT., 2nd FOXO regulates RNA interference in Drosophila and protects from RNA virus infection. Proc Natl Acad Sci USA. 2015;112:14587–14592. doi: 10.1073/pnas.1517124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D, et al. A petunia ethylene-responsive element binding factor, PhERF2, plays an important role in antiviral RNA silencing. J Exp Bot. 2016;67:3353–3365. doi: 10.1093/jxb/erw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen BS, Chen CH, Bowman BH, Nuss DL. Phenotypic changes associated with wild-type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology. 1996;86:301–310. [Google Scholar]
- 32.Sasaki A, et al. Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol Plant Microbe Interact. 2002;15:780–789. doi: 10.1094/MPMI.2002.15.8.780. [DOI] [PubMed] [Google Scholar]
- 33.Hillman BI, Suzuki N. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv Virus Res. 2004;63:423–472. doi: 10.1016/S0065-3527(04)63007-7. [DOI] [PubMed] [Google Scholar]
- 34.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 35.Lee TI, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 36.Helmlinger D, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 37.Han Y, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker SP, Grant PA. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaupel AC, Begley TJ, Tenniswood M. Gcn5 modulates the cellular response to oxidative stress and histone deacetylase inhibition. J Cell Biochem. 2015;116:1982–1992. doi: 10.1002/jcb.25153. [DOI] [PubMed] [Google Scholar]
- 40.Moraga F, Aquea F. Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front Plant Sci. 2015;6:865. doi: 10.3389/fpls.2015.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgakopoulos P, Lockington RA, Kelly JM. The Spt-Ada-Gcn5 Acetyltransferase (SAGA) complex in Aspergillus nidulans. PLoS One. 2013;8:e65221. doi: 10.1371/journal.pone.0065221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rösler SM, Kramer K, Finkemeier I, Humpf HU, Tudzynski B. The SAGA complex in the rice pathogen Fusarium fujikuroi: Structure and functional characterization. Mol Microbiol. 2016;102:951–974. doi: 10.1111/mmi.13528. [DOI] [PubMed] [Google Scholar]
- 43.Chang P, Fan X, Chen J. Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet Biol. 2015;81:132–141. doi: 10.1016/j.fgb.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Georgakopoulos P, Lockington RA, Kelly JM. SAGA complex components and acetate repression in Aspergillus nidulans. G3 (Bethesda) 2012;2:1357–1367. doi: 10.1534/g3.112.003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon J, et al. Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae. Sci Rep. 2015;5:8567. doi: 10.1038/srep08567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun D, et al. PhOBF1, a petunia ocs element binding factor, plays an important role in antiviral RNA silencing. J Exp Bot. 2017;68:915–930. doi: 10.1093/jxb/erw490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki N, Nuss DL. Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J Virol. 2002;76:7747–7759. doi: 10.1128/JVI.76.15.7747-7759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki N, Supyani S, Maruyama K, Hillman BI. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J Gen Virol. 2004;85:3437–3448. doi: 10.1099/vir.0.80293-0. [DOI] [PubMed] [Google Scholar]
- 49.Hillman BI, Supyani S, Kondo H, Suzuki N. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J Virol. 2004;78:892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craven MG, Pawlyk DM, Choi GH, Nuss DL. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J Virol. 1993;67:6513–6521. doi: 10.1128/jvi.67.11.6513-6521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 55.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 56.Eusebio-Cope A, Suzuki N. Mycoreovirus genome rearrangements associated with RNA silencing deficiency. Nucleic Acids Res. 2015;43:3802–3813. doi: 10.1093/nar/gkv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andika IB, Kondo H, Tamada T. Evidence that RNA silencing-mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol Plant Microbe Interact. 2005;18:194–204. doi: 10.1094/MPMI-18-0194. [DOI] [PubMed] [Google Scholar]
- 58.Seguin J, Otten P, Baerlocher L, Farinelli L, Pooggin MM. MISIS-2: A bioinformatics tool for in-depth analysis of small RNAs and representation of consensus master genome in viral quasispecies. J Virol Methods. 2016;233:37–40. doi: 10.1016/j.jviromet.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 60.Miyazawa N, et al. Human cell growth regulator Ly-1 antibody reactive homologue accelerates processing of preribosomal RNA. Genes Cells. 2014;19:273–286. doi: 10.1111/gtc.12129. [DOI] [PubMed] [Google Scholar]
- 61.Yaegashi H, et al. Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol Ecol. 2013;83:49–62. doi: 10.1111/j.1574-6941.2012.01454.x. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki N, Maruyama K, Moriyama M, Nuss DL. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J Virol. 2003;77:11697–11707. doi: 10.1128/JVI.77.21.11697-11707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shapira R, Choi GH, Nuss DL. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 1991;10:731–739. doi: 10.1002/j.1460-2075.1991.tb08004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.