Significance
Our study addresses two fundamental questions regarding the effect of random mutations on fitness: First, do fitness effects differ between species when controlling for demographic effects? Second, what are the responsible biological factors? We show that amino acid-changing mutations in humans are, on average, more deleterious than mutations in Drosophila. We demonstrate that the only theoretical model that is fully consistent with our results is Fisher’s geometrical model. This result indicates that species complexity, as well as distance of the population to the fitness optimum, modulated by long-term population size, are the key drivers of the fitness effects of new amino acid mutations. Other factors, like protein stability and mutational robustness, do not play a dominant role.
Keywords: distribution of fitness effects, mutational robustness, protein stability, Fisher’s geometrical model, Poisson random field
Abstract
The distribution of fitness effects (DFE) of new mutations plays a fundamental role in evolutionary genetics. However, the extent to which the DFE differs across species has yet to be systematically investigated. Furthermore, the biological mechanisms determining the DFE in natural populations remain unclear. Here, we show that theoretical models emphasizing different biological factors at determining the DFE, such as protein stability, back-mutations, species complexity, and mutational robustness make distinct predictions about how the DFE will differ between species. Analyzing amino acid-changing variants from natural populations in a comparative population genomic framework, we find that humans have a higher proportion of strongly deleterious mutations than Drosophila melanogaster. Furthermore, when comparing the DFE across yeast, Drosophila, mice, and humans, the average selection coefficient becomes more deleterious with increasing species complexity. Last, pleiotropic genes have a DFE that is less variable than that of nonpleiotropic genes. Comparing four categories of theoretical models, only Fisher’s geometrical model (FGM) is consistent with our findings. FGM assumes that multiple phenotypes are under stabilizing selection, with the number of phenotypes defining the complexity of the organism. Our results suggest that long-term population size and cost of complexity drive the evolution of the DFE, with many implications for evolutionary and medical genomics.
The distribution of fitness effects (DFE) represents the distribution of selection coefficients, s, of random mutations in the genome. Here, s quantifies the relative change in fitness due to the mutation. The DFE plays a fundamental role in evolutionary genetics because it quantifies the amount of deleterious, neutral, and adaptive mutations entering a population (1). Despite the importance and considerable study of the DFE (1–3), the extent to which the DFE in terms of s differs across species has yet to be systematically quantified. Furthermore, the biological factors determining the DFE in different species remain elusive. Several theoretical models propose different mechanisms for the evolution of the DFE (Fig. 1) (4–8). Although each of these models has a reasonable theoretical basis as well as some support from experimental evolution studies or microbial studies, which model best explains differences in the DFE between species has not yet been determined. Nor have these models been tested with genetic variation data from natural populations in higher organisms. Because experimental evolution studies in laboratory organisms often use a homogeneous environment and genetically homogeneous organisms, they may better satisfy some of the assumptions of these theoretical models. However, natural populations may provide different qualitative results due to increased resolution to measure weakly deleterious mutations and the unnatural selection pressure in the laboratory (2, 9).
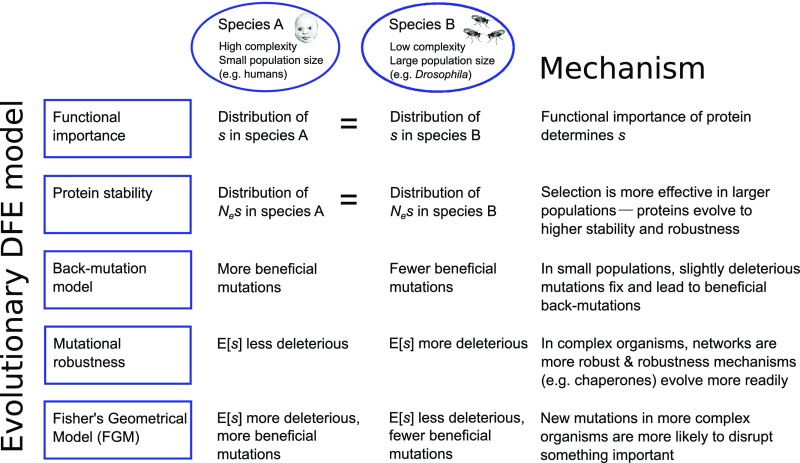
Fig. 1.
Overview of the main predictions of five theoretical models regarding DFE differences between two species. Here, E[s] is the average selection coefficient of a new mutation, and Ne is the effective population size. See SI Appendix, Text S5, for more details.
Importantly, five theoretical models for the evolution of the DFE predict that the DFE will differ between species with different levels of organismal complexity and long-term population size (Fig. 1). These models are the functional importance model, the protein stability model, the back-mutation model, the mutational robustness model, and Fisher’s geometrical model (FGM). For example, the mutational robustness model predicts that more complex species will have more robust regulatory networks that will better buffer the effects of deleterious mutations, leading to less deleterious selection coefficients (10). Here, we leverage these predictions of how the DFE is expected to differ across species to test which theoretical model best explains the evolution of the DFE by comparing the DFE in natural populations of humans, Drosophila, yeast, and mice. We find that humans have more strongly deleterious mutations than Drosophila and that the average selection coefficient becomes more deleterious with increasing species complexity. Furthermore, genes showing greater pleiotropy, as inferred from breadth of gene expression, tend to have a DFE that is less variable than that of less pleiotropic genes. Of the theoretical models outlined in Fig. 1, only FGM can explain these patterns.
Results and Discussion
Mutations Are More Deleterious in Humans than in Drosophila.
We used polymorphism data of a sample of 112 individuals from Yoruba in Ibadan, Nigeria (YRI) from the 1000 Genomes Project (11) and 197 African Drosophila melanogaster lines from the Drosophila Population Genomics Project (12). We summarize the polymorphism data by the folded site frequency spectrum (SFS), which represents the number of variants at different minor allele frequencies in the sample (SI Appendix, Fig. S1A). Because population history can also affect patterns of polymorphism, we first use the synonymous SFS to estimate demographic models separately in each species (SI Appendix, Text S1 and S2). We infer that the population size of YRI and Drosophila expanded 2.3-fold 6,000 generations ago and 2.7-fold 500,000 generations ago, respectively (SI Appendix, Table S1). Note that demographic estimates from synonymous sites are biased by selection affecting linked neutral sites (13, 14), but that this bias does not affect our ability to infer the DFE (14, 15) (SI Appendix, Text S3).
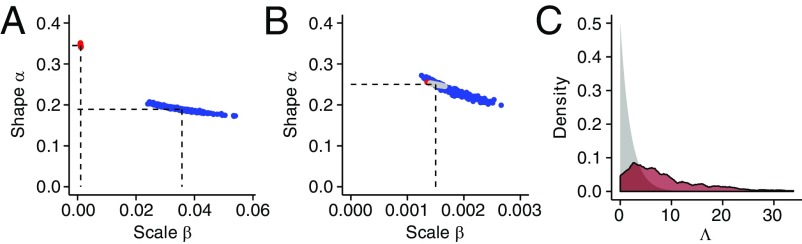
Conditional on the estimated demographic parameters, we estimate the DFE for new nonsynonymous mutations in both species using the nonsynonymous SFS (SI Appendix, Table S2). In short, our approach uses the fact that more deleterious mutations segregate in lower numbers and at lower frequencies than less deleterious or neutral mutations (2, 3, 16, 17). We next compare the estimates of the DFE from the two species in a likelihood ratio test (LRT) framework that accounts for differences in recent demographic history between the two species (Materials and Methods; SI Appendix, Text S2). Briefly, we assume that the DFE follows a gamma distribution and find that a model where each species has its own shape and scale parameters fits the SFSs for the two species significantly better than a model where the parameters are constrained to be the same in both species (LRT statistic Λ = 12,012; df = 2, P < 10−16; Fig. 2 A–C). This result holds even when making different assumptions about the mutation rate, as well as when omitting singleton variants (SI Appendix, Text S3 and Table S2).
Fig. 2.
Testing the null hypothesis of the same distribution of s in both species. The log-likelihood surface for the shape and scale parameters of a gamma-distributed DFE(s) for (A) humans, (B) Drosophila, and (C) both datasets combined (constrained model). Colors from yellow to red indicate the difference in log-likelihood of that set of parameter values compared with the MLE (see color scale). For example, orange indicates parameters ∼100 log-likelihood units below the MLE. Proportions of mutations for various ranges of |s| are computed from the estimated (D) gamma distribution, (E) mixture of gamma distribution with neutral point mass, and (F) log-normal distribution. The gray bars indicate the proportions under the null hypothesis of the same distribution of s in both species (constrained model). Darker colors in E reflect the estimated proportions of neutral mutations.
Examination of the maximum-likelihood gamma distribution shows that Drosophila have a much higher proportion of weakly deleterious mutations with selection coefficient |s| < 10−4 than do humans (Fig. 2D). The proportion of strongly deleterious mutations with |s| > 10−3 is significantly larger in humans (55%) than in Drosophila (5%). Thus, our results provide statistical support for humans and Drosophila having different DFEs (of s) that cannot be explained by differences in population size or demography between the species.
Robustness of Our Results.
To evaluate the robustness of our finding to the assumed functional form of the DFE, we tested a range of different distributions such as a log-normal, shifted gamma, mixture of gamma with point mass at neutrality, as well as a nonparametric discretized distribution. We also tested whether differences in the DFE between species are caused by analyzing different sets of genes in the two species. We filtered for genes that are strictly orthologous between humans and Drosophila, and also required gene sets to have similar expression profiles. In all cases, we consistently find that mutations are on average more deleterious in humans than in Drosophila (SI Appendix, Text S3, Figs. S1B, S2, S5, S8A, S12A, and S14, and Tables S2, S3, and S4).
In Drosophila, as much as 22% of synonymous sites could be under strong purifying selection (18). We next tested whether this type of selection could confound our inferences of the DFE. To do this, we computed a synonymous SFS that would be expected under neutrality given this extreme estimate of selection on synonymous sites (see SI Appendix, Text S3, for details). We further assumed that the SFS from short introns has a neutral shape (19, 20). We only see a negligible effect on the DFE inference using this predicted neutral SFS compared with estimates using the observed synonymous SFS (SI Appendix, Fig. S7). Thus, our inferences are robust to extreme selection occurring on synonymous sites.
Because a variety of demographic, statistical, and numerical biases can confound LRTs using the SFS, we evaluated the performance of our statistical approach by analyzing simulated datasets. Specifically, we performed forward-in-time simulations that include realistic levels of linkage disequilibrium and background selection (SI Appendix, Text S4). When we estimated the DFE from the simulations of the full and constrained models, the estimates were unbiased (Fig. 3 A and B). This suggests that the size change model fit to synonymous polymorphisms successfully controls for the effects of background selection (SI Appendix, Fig. S3; see also refs. 14 and 15). As expected, the null distribution of Λ derived from simulations under the constrained model is broader than the χ2 distribution with 2 df (Fig. 3C). However, all of the 300 Λ values that we simulated were smaller than 34, suggesting the probability of seeing a Λ value bigger than 12,012 is substantially less than 0.33% under the null. Because selective sweeps were suggested to be a major determinant of genetic diversity in Drosophila (21), we also examined the effect of recurrent selective sweeps on our inference. We found that selective sweeps do not significantly bias our DFE estimates when correcting for the effect of demography using the observed SFS at neutral sites (SI Appendix, Fig. S9 and Text S3). In summary, a combination of confounding factors cannot account for our finding of different DFEs between humans and Drosophila (SI Appendix, Text S3).
Fig. 3.
Estimates of the shape and scale parameters of a gamma DFE from 300 simulations of human (blue) and Drosophila (red) data. (A) Estimates from simulations under the alternative hypothesis (H1), that is, assuming maximum likelihood parameters in both species (dashed lines). Results show that we can retrieve the true parameters. (B) Estimates from simulations under the null hypothesis (H0), that is, assuming a single set of parameters in both species (dashed lines). In gray are the estimation results using data from both species simultaneously, assuming H0 is correct. Results show that, under H0, we correctly retrieve the same set of parameters for both species. (C) The expected (gray) and simulated (dark red) null distribution of the test statistic Λ = –2*log(LConstrained,max/LFull,max) for testing the null hypothesis of no difference in shape and scale parameters between humans and Drosophila.
Testing Models of the Evolution of the DFE.
The most basic null model for the evolution of the DFE is that it remained constant over evolutionary time. This may be expected under a model where protein function is the main driver of fitness effects of mutations, and where protein function does not systematically change between species (SI Appendix, Text S5). Having established that humans and Drosophila have significantly different DFEs, we can reject this functional importance model. We next examined which of the four remaining theoretical models (Fig. 1) can explain the differences in the DFE across species. The second model, the protein stability model, predicts that much of the selection pressure involves maintaining the thermodynamic stability of proteins. This model predicts that the distribution of Nes is gamma distributed (22) and independent of the effective population size (Ne) when at equilibrium (7) (see SI Appendix, Text S5, for specific assumptions). Thus, this model predicts that Nes is the same across taxa. However, in contrast to this prediction, we found that a model with different Nes distributions in each species fits the data significantly better than a model where Nes is constrained to be the same in both species (Λ = 21,734, P < 10−16; SI Appendix, Figs. S4 and S6), consistent with previous results (23). Comparing this LRT statistic to the null distribution obtained from forward simulations similar to those discussed above suggests that such a large LRT statistic is highly incompatible with a model that assumes the same gamma (or lognormal) Nes distribution in both species (P < 0.0033). Thus, our data do not support protein stability models as the driving force in the evolution of the DFE between species.
The third model, the back-mutation model, predicts that there is a category of weakly advantageous mutations that restore fitness after deleterious mutations become fixed (24). The back-mutation model predicts that, in small populations, the proportion of slightly beneficial mutations is greater than in large populations, because more slightly deleterious mutations can become fixed in small populations, leading to more opportunities for new beneficial back-mutations (SI Appendix, Text S5). Using this logic, Piganeau and Eyre-Walker (25) derived a formula for the equilibrium DFE as a function of population size [see also Rice et al. (6)]. When we estimate the parameters in this model on our data using our inference framework, we found an unrealistically large effective population size in Drosophila (5.2 × 1019). Importantly, the back-mutation model predicts that the average effect size of mutations (i.e., the absolute value of the selection coefficient) will be the same in both species, and that differences in the DFE are solely attributable to long-term differences in population size. To test this prediction, we inferred distinct parameters of the effect size distribution (the distribution of |s|) in the two species (SI Appendix, Table S4). In contrast to the predictions of the back-mutation model, we found that the average effect size E[|s|] of a mutation in humans is about 55-fold larger in humans than in Drosophila (SI Appendix, Fig. S8B). Although the Piganeau and Eyre-Walker model fits well within both species, it falls short in providing an evolutionary or mechanistic explanation for a large difference in E[|s|] between species.
The fourth model, the mutational robustness model, postulates that more robust, or complex, organisms have, on average, less deleterious mutations (4, 5, 10). Here, more complex organisms have a greater ability to compensate and buffer the effects of deleterious mutations (SI Appendix, Text S5). Note that complexity can be hard to define and quantify in a biologically and evolutionarily meaningful way. However, a number of biological factors suggest that humans are more complex than Drosophila. Such factors include a larger genome, a larger number of genes, a larger number of proteins and protein–protein interactions (26), and likely also a larger number of cell types (27) in humans than in Drosophila. Mutational robustness models predict greater mutational robustness in humans than in Drosophila because of the higher complexity and the smaller effective population size of humans compared with Drosophila. However, inconsistent with this prediction, we have shown that E[s] is 70- to 110-fold more deleterious in humans than in Drosophila, and humans have a larger proportion of strongly deleterious mutations with |s| > 0.001 (Fig. 2 D–F). Furthermore, robustness models predict that less pleiotropic mutations are more deleterious, because the smaller effective complexity of such mutations impedes the evolution of robustness (28). Assuming that broadly expressed genes are more pleotropic than tissue-specific genes, we observe that tissue-specific genes have less negative estimates of E[s] than broadly expressed genes (SI Appendix, Fig. S12A). In other words, more pleiotropic mutations tend to be more deleterious. This finding is inconsistent with predictions from the robustness model. However, although our results suggest that mutational robustness mechanisms are not the main driver of differences in the DFE across species, this finding is not necessarily at odds with previous work on these models. The clearest empirical evidence for an increase of mutational robustness by selection comes from experimental evolution studies of viruses and bacteria (29, 30). Viruses and bacteria have large mutation rates and population sizes. The specific mechanism that promotes robustness in such organisms may not be applicable to higher organisms with smaller population mutation rates (31). Our results suggest that if mutational robustness mechanisms play a role in shaping the DFE of higher organisms, they do not compensate for other factors that increase the deleteriousness of mutations in humans compared with Drosophila.
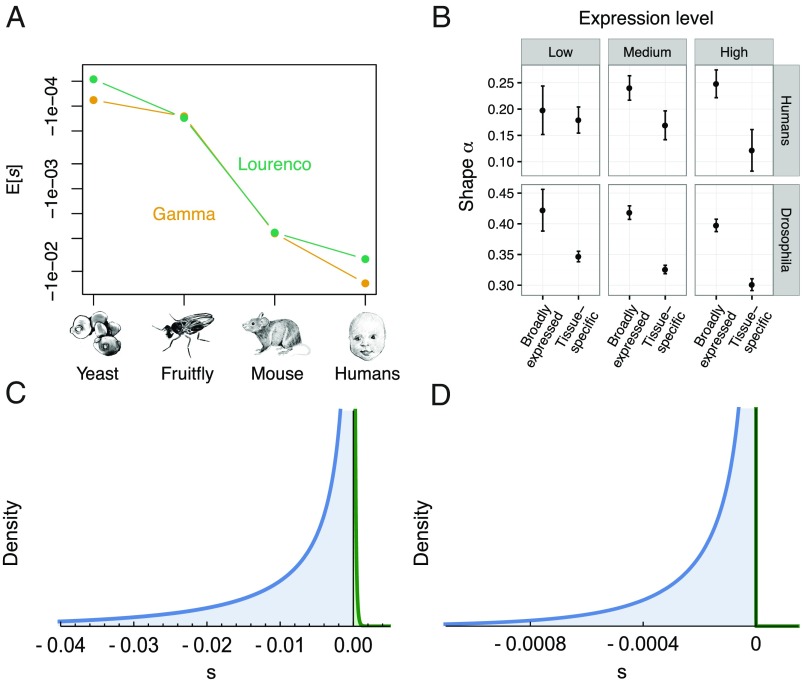
The fifth model, FGM, represents phenotypes as points in a multidimensional phenotype space, and fitness is a decreasing function of the distance from the optimal phenotype (4). The dimensionality of the phenotype space is termed “complexity.” FGM makes three predictions that we test with our data (SI Appendix, Text S5). The first prediction of FGM is that mutations in more complex organisms, like humans, are on average more deleterious than in Drosophila, because mutations are more likely to disrupt something important in a complex organism than in a simple one (32) (see SI Appendix, Text S7, for assumptions that go into this prediction). Indeed, this prediction is well supported by our data because the average selection coefficient E[s] is estimated to be 70- to 110-fold more deleterious in humans than in Drosophila (Fig. 2). To further validate this finding in a larger phylogenetic context, we analyzed polymorphism data from mouse (Mus musculus castaneus) and yeast (Saccharomyces paradoxus). Although sample size is one order of magnitude smaller, we replicate the pattern of increasing deleteriousness of mutations with increasing complexity (Fig. 4A; SI Appendix, Table S5). In principle, differences in population size across species could explain some of this pattern, because more complex organisms also tend to have smaller population sizes. However, our results suggest that variation in population size by itself cannot account for the pattern shown in Fig. 4A because we have shown that models only including population size effects for determining the DFE (e.g., the back-mutation model and the protein stability model) do not fit the data (see above and SI Appendix, Figs. S8 and S10 and Table S4). Furthermore, our analyses of the DFEs in yeast and mice are not consistent with the predictions of the functional importance, mutational robustness, or protein stability models (SI Appendix, Table S5), providing further evidence that these models cannot explain the patterns in the data.
Fig. 4.
Empirical support for FGM. (A) Both under the gamma DFE and the Lourenço et al. DFE, estimated average deleteriousness of mutations increases as a function of organismal complexity. (B) The shape parameter of the gamma DFE depends on the breadth of gene expression. Tissue-specific genes have a smaller shape parameter (α) than broadly expressed genes, supporting FGM. This pattern is consistent across overall expression levels. (C and D) By fitting the DFE of Lourenço et al., we can model slightly beneficial mutations in the DFE (green) that are thought to compensate for fixed deleterious mutations in species with small population size. We find support for a larger proportion of slightly beneficial mutations in the DFE of (C) humans than in (D) Drosophila.
The second prediction of FGM is that smaller populations are predicted to have a larger proportion of beneficial mutations due to increased fixation of deleterious mutations in smaller populations when populations are in equilibrium [drift load (33)]. Note that population size here refers to long-term effective population size; thus, it could be affected by background selection and selective sweeps as well as demographic processes. To test this prediction, we estimated the parameters for the DFE based on FGM. Formulas have been derived for the DFE assuming the population is at an arbitrary distance from the optimal phenotype [equation 8 in Lourenço et al. (33) and equation 5 in Martin and Lenormand (34)], or assuming mutation–selection–drift equilibrium [equation 15 in Lourenço et al. (33)]. We found that the equilibrium DFE fits just as well or better than the nonequilibrium versions (SI Appendix, Table S4). This result suggests that, in both populations, most genes are close to equilibrium and are not affected by environmental perturbations of the phenotypic optimum. Furthermore, in humans, the equilibrium Lourenço DFE shows a significantly better fit over the plain gamma DFE (SI Appendix, Table S4), with a Ne,long-term of 2,476 (95% CI: 1,805–3,146). Note that this value of Ne,long-term is of the same order of magnitude as the ancestral population size estimated from synonymous sites (7,070). This is surprising because the estimate of Ne,long-term is not based on neutral genetic diversity, but on the degree of maladaptation due to drift load that results in some proportion of beneficial compensatory mutations in the DFE. Thus, it is estimated from the predicted effect of drift load on the nonsynonymous SFS and likely reflects a much larger time span than the estimate from the synonymous SFS. In Drosophila, fitting the equilibrium Lourenço model led to a similar fit as the plain gamma DFE (SI Appendix, Table S4). Furthermore, the large Ne,long-term (8.4 × 107) estimated here is also similar to that estimated from the neutral synonymous sites (2.8 × 106). The fact that long-term population sizes inferred under FGM are consistent with previous estimates from genetic variation data suggests that this prediction of FGM is satisfied by our data.
The third prediction of FGM is that more pleotropic mutations will show smaller variation in s. As before, we use gene expression breadth as a proxy for pleiotropy. We found that the shape parameter (α) of the gamma distribution is smaller for tissue-specific genes than for broadly expressed genes (Fig. 4B). The shape parameter is inversely related to the coefficient of variation (CV) of the selection coefficient: CV(s) = 1/sqrt(α). Thus, the smaller shape parameter indicates a larger CV(s) and is consistent with the idea that mutations in tissue-specific genes are less pleiotropic than in broadly expressed genes. Similar conclusions were derived by explicitly estimating pleiotropy from fitting the Lourenço DFE to the data (SI Appendix, Fig. S13). Note that genes within a species evolve under the same population size. Thus, this pattern supports an effect of complexity on the DFE that is not confounded by population size differences, providing further evidence that the pattern shown in Fig. 4A is not solely driven by differences in population size. In sum, all three predictions made by FGM are supported by our data.
Conclusions
We conclude that FGM is a viable model to explain differences in the DFE between species and genes. Under this model, complexity as well as distance of the population to the fitness optimum, modulated by long-term population size, are the key drivers of the DFE of new amino acid mutations. Note that many essential elements of protein evolution are captured by FGM (35), where many molecular phenotypes (not just protein stability) are under stabilizing selection (36). Thus, although we reject a simple protein stability model determining the DFE, this should not be taken to mean that general principles of protein evolution do not play a role in determining the DFE. Further note that for testing the back-mutation and protein stability models, we assume that populations are in mutation–selection–drift equilibrium. Fluctuating population size or environments can move a population out of equilibrium and thereby change the expected DFE (7, 34). Fitting a nonequilibrium FGM model did not support a deviation from mutation–selection–drift equilibrium (SI Appendix, Table S4); however, a more thorough theoretical and empirical examination of nonequilibrium models is warranted.
Our findings have implications for important aspects of evolutionary genetics. First, FGM allows us to estimate the proportion of new mutations that are adaptive. When assuming FGM, we estimate that 14% of new nonsynonymous mutations in humans are beneficial. The majority (98%) of these beneficial mutations have small selection coefficients, with s < 0.0005 (Fig. 4C). In Drosophila, however, the model including positive selection had a similar fit to the data as the plain gamma DFE (SI Appendix, Table S4), and only 1.5% of new mutations are beneficial (Fig. 4D). This finding is qualitatively in the opposite direction compared with what has been seen in previous studies of adaptive evolution in these two species. The proportion of amino acid substitutions that are beneficial was estimated to be larger in Drosophila (50%) than in humans (10–20%), using a McDonald–Kreitman (MK) approach (3, 37–40). More generally, our results suggest that inferences of the amount of adaptive evolution considering fixed substitutions may be qualitatively different from those considering new mutations. One explanation for this apparent difference might be that a small proportion of strongly beneficial mutations contribute substantially to substitutions in Drosophila but only rarely show up as polymorphisms. Our approach provides limited information about the class of strongly beneficial mutations because it only uses polymorphism data. Second, the larger proportion of amino acid substitutions fixed by positive selection in large populations like Drosophila could also be driven by an overall decrease in the total amount of nonsynonymous divergence in this species due to a decreased rate of fixation of nearly neutral and mildly deleterious mutations due to more efficient purifying selection (41, 42). Alternative measures of adaptive evolution that are not affected by varying efficiency in purifying selection were found to be only weakly (42) or not at all (41) correlated with population size. Additionally, the amount of positive selection in the human genome has been recently debated (43, 44). After controlling for background selection, Enard et al. (43) found that, in humans, estimates of the amount of adaptive evolution from MK approaches may be severe underestimates. Their results instead argue that there may be many small-scale adaptive steps in humans, that is, many weak selective sweeps that are only detectable when averaging across many instances. Such a mode of adaptation is in fact predicted by FGM for organisms with high complexity (45).
Second, a varying DFE over phylogenetic timescales has implications for understanding the overdispersed molecular clock (46). The substitution rate of deleterious mutations is a function of the compound parameter Nes (47). Thus, not only phylogenetic changes in Ne but also changes in s may contribute to overdispersion. Our results suggest that changes in the distribution of s are coupled with changes in population size and complexity. For example, the larger complexity of humans is supposed to reduce the nonsynonymous divergence along the human lineage to lower values than what would be expected from the two orders-of-magnitude population size difference to Drosophila. Accurate characterization of the DFE from many species across the tree of life will enable a direct test of the contribution of changing DFEs to the dispersion of the molecular clock.
Last, our results have implications for assessing the biological function of sequences using evolutionary information. The comparative genomics paradigm postulates that biologically important regions of the genome are constrained across long evolutionary times (48). This implies that s for a particular sequence is determined by the biological importance of the sequence and that s remains constant over time. If, as our work suggests, selection coefficients change over time as a consequence of species complexity and long-term population size, this could result in important sequences not showing the prototypical signatures of conservation, leading to such sequences being missed by comparative approaches. Furthermore, it suggests that complexity and population size are important factors to consider when deciding which species to use in future comparative genomic studies.
Materials and Methods
Data.
We used published next-generation sequencing datasets to extract the synonymous and nonsynonymous SFS (SI Appendix, Text S1). For humans, we used the sample of 112 individuals from YRI from the 1000 Genomes Project (11). For Drosophila melanogaster, we used the Drosophila Population Genomics Project phase 3 data of a sample of 197 lines originating from Zambia, Africa (12). To infer the DFE in Mus musculus castaneus (mouse) and Saccharomyces paradoxus (yeast), we used data from Gossmann et al. (42). To study the effect of gene expression, we used two recent tissue-specific gene expression datasets from humans (49) and Drosophila (50) (SI Appendix, Text S1).
Statistical Test for Different DFEs Between Two Species.
We used the SFS from polymorphism data from two species, A and B, to test whether the DFE differs between these two species. We used the software ∂a∂i (51) to infer the parameters of a single size change model from the synonymous SFS of each species independently (denoted and ), and conditional on the estimated size change model, we infer the DFE from the nonsynonymous SFS (SI Appendix, Text S2). Specifically, we initially assumed that the DFE in both species follows a gamma distribution with the shape parameter and scale parameter . We used a Poisson composite likelihood function (3), where the SFS at nonsynonymous SNPs in species A is treated as being independent of that from species B, which is reasonable for distantly related species (52). Then, the likelihood function for the parameters is as follows:
Here, n and m are the sample size of species A and species B, respectively, and refers to the number of SNPs at frequency i in species A. We test whether the shape () and scale () parameters in species A differ from those in species B. To do this, we propose the following LRT:
The null hypothesis (constrained model) is that and . The full model allows for and . We optimized the likelihood function under both the null and full models (SI Appendix, Text S2). In all cases, we conditioned on the demographic parameters in each population (, thus accounting for differences in population history. Asymptotically, follows a χ2 distribution with 2 df, due to the two additional free parameters in the full model compared with the constrained model. Simulations were used to test how well the usual asymptotic theory applies in this situation (SI Appendix, Text S4). This test can be extended to any DFE distribution, and any number of species. Here, we also tested the parameters of a gamma+neutral and a log-normal distribution. The degree of freedom of the χ2 null distribution is p*k − p, where p is the number of parameters of the distribution, and k is the number of species. We test the robustness of the inferred DFE parameters to a number of potential confounding factors (SI Appendix, Text S3).
Supplementary Material
Acknowledgments
We thank Bridgett vonHoldt, Tanya Phung, and Sebastian Matuszewski for comments that greatly improved the manuscript; Toni I. Gossmann for providing access to site frequency spectrum data from Mus musculus castaneus and Saccharomyces paradoxus; and Maria T. Huber for the drawings in Figs. 1 and 4. C.D.H., C.D.M., and K.E.L. were supported by a Searle Scholars Fellowship, an Alfred P. Sloan Research Fellowship in Computational and Molecular Biology, and NIH Grant R35GM119856 (to K.E.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619508114/-/DCSupplemental.
References
- 1.Loewe L, Hill WG. The population genetics of mutations: Good, bad and indifferent. Philos Trans R Soc Lond B Biol Sci. 2010;365:1153–1167. doi: 10.1098/rstb.2009.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 3.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenaillon O. The utility of Fisher’s geometric model in evolutionary genetics. Annu Rev Ecol Evol Syst. 2014;45:179–201. doi: 10.1146/annurev-ecolsys-120213-091846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegal ML, Leu J-Y. On the nature and evolutionary impact of phenotypic robustness mechanisms. Annu Rev Ecol Evol Syst. 2014;45:496–517. doi: 10.1146/annurev-ecolsys-120213-091705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice DP, Good BH, Desai MM. The evolutionarily stable distribution of fitness effects. Genetics. 2015;200:321–329. doi: 10.1534/genetics.114.173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein RA. Population size dependence of fitness effect distribution and substitution rate probed by biophysical model of protein thermostability. Genome Biol Evol. 2013;5:1584–1593. doi: 10.1093/gbe/evt110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M, Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewe L. A framework for evolutionary systems biology. BMC Syst Biol. 2009;3:27. doi: 10.1186/1752-0509-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M. Model of effectively neutral mutations in which selective constraint is incorporated. Proc Natl Acad Sci USA. 1979;76:3440–3444. doi: 10.1073/pnas.76.7.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lack JB, et al. The Drosophila genome nexus: A population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics. 2015;199:1229–1241. doi: 10.1534/genetics.115.174664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messer PW, Petrov DA. Frequent adaptation and the McDonald-Kreitman test. Proc Natl Acad Sci USA. 2013;110:8615–8620. doi: 10.1073/pnas.1220835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim BY, Huber CD, Lohmueller KE. 2016. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. bioRxiv:071431.
- 15.Tataru P, Mollion M, Glemin S, Bataillon T. 2016. Inference of distribution of fitness effects and proportion of adaptive substitutions from polymorphism data. bioRxiv:062216. [DOI] [PMC free article] [PubMed]
- 16.Sawyer SA, Hartl DL. Population genetics of polymorphism and divergence. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson SH, et al. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci USA. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrie DS, Messer PW, Hershberg R, Petrov DA. Strong purifying selection at synonymous sites in D. melanogaster. PLoS Genet. 2013;9:e1003527. doi: 10.1371/journal.pgen.1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemente F, Vogl C. Unconstrained evolution in short introns?—an analysis of genome-wide polymorphism and divergence data from Drosophila. J Evol Biol. 2012;25:1975–1990. doi: 10.1111/j.1420-9101.2012.02580.x. [DOI] [PubMed] [Google Scholar]
- 20.Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, Andolfatto P. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27:1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serohijos AWR, Shakhnovich EI. Contribution of selection for protein folding stability in shaping the patterns of polymorphisms in coding regions. Mol Biol Evol. 2014;31:165–176. doi: 10.1093/molbev/mst189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlesworth J, Eyre-Walker A. The other side of the nearly neutral theory, evidence of slightly advantageous back-mutations. Proc Natl Acad Sci USA. 2007;104:16992–16997. doi: 10.1073/pnas.0705456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piganeau G, Eyre-Walker A. Estimating the distribution of fitness effects from DNA sequence data: Implications for the molecular clock. Proc Natl Acad Sci USA. 2003;100:10335–10340. doi: 10.1073/pnas.1833064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stumpf MPH, et al. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valentine JW, Collins AG, Meyer CP. Morphological complexity increase in metazoans. Paleobiology. 1994;20:131–142. [Google Scholar]
- 28.Gros P-A, Tenaillon O. Selection for chaperone-like mediated genetic robustness at low mutation rate: Impact of drift, epistasis and complexity. Genetics. 2009;182:555–564. doi: 10.1534/genetics.108.099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masel J, Maughan H. Mutations leading to loss of sporulation ability in Bacillus subtilis are sufficiently frequent to favor genetic canalization. Genetics. 2007;175:453–457. doi: 10.1534/genetics.106.065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloom JD, et al. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 2007;5:29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orr HA. The population genetics of adaptation: The distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- 33.Lourenço J, Galtier N, Glémin S. Complexity, pleiotropy, and the fitness effect of mutations. Evolution. 2011;65:1559–1571. doi: 10.1111/j.1558-5646.2011.01237.x. [DOI] [PubMed] [Google Scholar]
- 34.Martin G, Lenormand T. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution. 2006;60:893–907. [PubMed] [Google Scholar]
- 35.Weinreich DM, Knies JL. Fisher’s geometric model of adaptation meets the functional synthesis: Data on pairwise epistasis for fitness yields insights into the shape and size of phenotype space. Evolution. 2013;67:2957–2972. doi: 10.1111/evo.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 37.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 38.Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 39.Fay JC, Wyckoff GJ, Wu C-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 40.Sawyer SA, Kulathinal RJ, Bustamante CD, Hartl DL. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J Mol Evol. 2003;57:S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- 41.Galtier N. Adaptive protein evolution in animals and the effective population size hypothesis. PLoS Genet. 2016;12:e1005774. doi: 10.1371/journal.pgen.1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gossmann TI, Keightley PD, Eyre-Walker A. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol Evol. 2012;4:658–667. doi: 10.1093/gbe/evs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enard D, Messer PW, Petrov DA. Genome-wide signals of positive selection in human evolution. Genome Res. 2014;24:885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez RD, et al. 1000 Genomes Project Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lourenço JM, Glémin S, Galtier N. The rate of molecular adaptation in a changing environment. Mol Biol Evol. 2013;30:1292–1301. doi: 10.1093/molbev/mst026. [DOI] [PubMed] [Google Scholar]
- 46.Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- 47.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29:33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Alföldi J, Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23:1063–1068. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li JJ, Huang H, Bickel PJ, Brenner SE. Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data. Genome Res. 2014;24:1086–1101. doi: 10.1101/gr.170100.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrie DS, Petrov DA. Comparative population genomics: Power and principles for the inference of functionality. Trends Genet. 2014;30:133–139. doi: 10.1016/j.tig.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.