Summary
Melanoma patients with BRAFV600E-mutant tumors display striking responses to BRAF inhibitors (BRAFi); however, almost all invariably relapse with drug-resistant disease. Here we report that microRNA-125a (miR-125a) expression is upregulated in human melanoma cells and patient tissues upon acquisition of BRAFi resistance. We show that miR-125a induction confers resistance to BRAFV600E melanoma cells to BRAFi by directly suppressing pro-apoptotic components of the intrinsic apoptosis pathway, including BAK1 and MLK3. Apoptotic suppression and prolonged survival favor reactivation of the MAPK and AKT pathways by drug-resistant melanoma cells. We demonstrate that miR-125a inhibition suppresses the emergence of resistance to BRAFi and, in a subset of resistant melanoma cell lines, leads to partial drug re-sensitization. Finally, we show that miR-125a upregulation is mediated by TGFβ signaling. In conclusion, the identification of this novel role for miR-125a in BRAFi resistance exposes clinically relevant mechanisms of melanoma cell survival that can be exploited therapeutically.
Keywords: melanoma, BRAF inhibitor, resistance, microRNA, apoptosis
Introduction
Melanoma is the leading cause of death from skin cancer, with incidence that continues to rise (Ferlay et al., 2015). Somatic, activating mutations in BRAF, predominantly V600E, occur in ~40-50% of cutaneous malignant melanomas (Akbani et al., 2015, Krauthammer et al., 2015); and BRAF inhibitors (BRAFi) have become important therapeutic agents in the treatment of metastatic BRAFV600E melanoma (Bollag et al., 2010, Chapman et al., 2011, Flaherty et al., 2010, Sosman et al., 2012). Initial responses to BRAFi are exceptional with metastatic tumors that routinely vanish in clinical imaging of treated patients; however, tumor cells are not completely eradicated and resistance of melanoma cells to these inhibitors occurs in nearly all patients, resulting in progression with treatment refractory disease. Numerous molecular mechanisms involved in the acquisition of BRAFi resistance have been reported. Most resistance mechanisms involve reactivation of the MAPK pathway, commonly through NRAS mutation (Nazarian et al., 2010), BRAF splicing changes (Poulikakos et al., 2011) or amplification (Shi et al., 2012), but also through less frequent alterations, such as MEK1/2 mutation (Emery et al., 2009), COT hyperactivation (Johannessen et al., 2010), or RTK/EGF receptor upregulation (Girotti et al., 2013). Alternatively, the PI3K/AKT pathway (i.e. PTEN loss (Paraiso et al., 2011), AKT hyperactivation (Shao and Aplin, 2010), NF1 loss (Whittaker et al., 2013), PIP3 loss (Ye et al., 2013), IGF1R upregulation (Villanueva et al., 2010)) or additional mechanisms (Haq et al., 2013b, Hilmi et al., 2008, Smith et al., 2014, Straussman et al., 2012, Shen et al., 2016) become hyper-activated in BRAFi-resistant melanoma. In addition to these described mechanisms, up to 40% of BRAFi-resistant tumors harbor unknown mechanisms of resistance (Rizos et al., 2014, Johnson et al., 2015), and not all can be explained by genetic/genomic changes (Hugo et al., 2015).
Common BRAFi resistance mechanisms, which reactivate MAPK or activate PI3K signaling, are typically thought to be acquired molecular alterations as opposed to selection of pre-existing tumor clones (Lackner et al., 2012). Development of such mechanisms likely requires activation of cellular survival pathways to evade BRAFi-induced cell death until permanent resistance mechanisms are acquired.
The involvement of non-genomic alterations in the acquisition of BRAFi resistance has not been fully explored. MicroRNA (miRNA), which are modulators of gene expression and molecular pathways, play central roles in a variety of normal and pathological cellular processes (Lujambio and Lowe, 2012). A few recent studies show involvement of miRNA in BRAFi resistance of melanoma. Vergani et al. demonstrate that a set of three miRNA (miR-34a, miR-100 and miR-125b) confers BRAFi resistance downstream of the chemokine CCL2 (Vergani et al., 2016), whereas Liu et al. identify miR-200c as a sensitizer of melanoma cells to BRAFi treatment (Liu et al., 2015). miR-514a may contribute to BRAFi resistance, as its expression promotes survival of melanoma cells treated with BRAFi (Stark et al., 2015); however, evidence of miR-514a modulation in clinical samples or models of BRAFi resistance has not been reported. Finally, Sun et al. found miR-7 downregulation in a model of BRAFi resistance and reported that its re-expression sensitized resistant cells to BRAFi treatment (Sun et al., 2016).
We hypothesized that specific miRNA can directly confer BRAFi resistance or contribute to the establishment of other resistance mechanism(s) that lead to MAPK and/or PI3K/Akt activation. To identify miRNA candidates that may contribute to BRAFi resistance in melanoma, we performed miRNA expression profiling of BRAFi resistant cell clones and their respective parental cells. miR-125a was consistently overexpressed upon acquisition of resistance to BRAF inhibition. Upregulation of miR-125a was also observed in clinical BRAFi-treated tumors relative to paired, pre-treatment tumor samples, further supporting its potential contribution to BRAFi therapeutic resistance. Mechanistically, we show that miR-125a facilitates BRAFi resistance by suppressing the intrinsic apoptotic pathway. Our findings support the possibility to use anti-miRNA based approaches to prevent or overcome BRAFi resistance.
Results
MiR-125a is overexpressed in BRAFi resistant melanoma
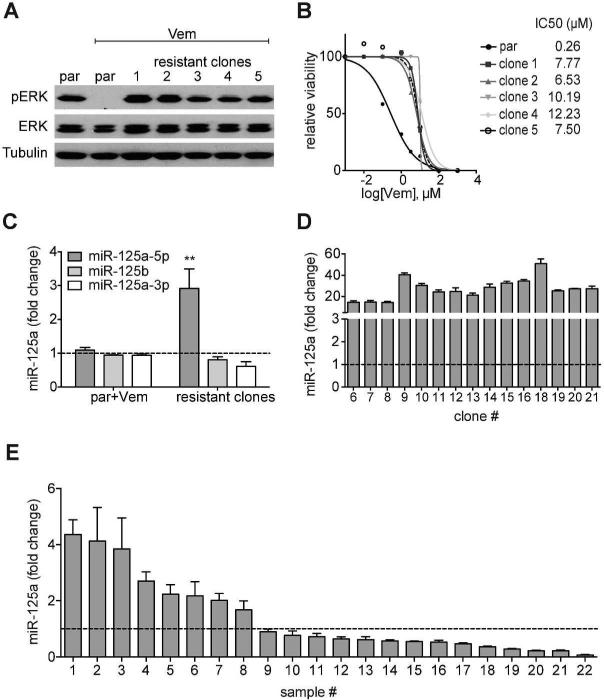
To identify miRNA that may contribute to BRAFi resistance, we conducted miRNA expression profiling of BRAFV600E mutant SK-MEL-239 cells (BRAFi sensitive cells) treated with DMSO or Vemurafenib (Vem) for 24h, and a panel of BRAFi-resistant cell clones generated through prolonged exposure to 2μM Vemurafenib (Poulikakos et al., 2011). As previously reported, resistant clones universally reactivated the MAPK pathway (Fig. 1A) and exhibited higher IC50 values compared to their parental counterpart (Fig. 1B). We conducted expression profiling of 800 miRNA by Nanostring of 5 BRAFi-resistant clones, DMSO-treated SK-MEL-239 cells, and SK-MEL-239 cells treated with Vemurafenib for 24 hours (Table S1) We observed miR-125a-5p consistently upregulated in all BRAFi-resistant clones analyzed (n=5, Fig. 1C), a finding confirmed by RT-qPCR (Fig. S1A). MiR-125a-5p induction was observed in multiple additional resistant SK-MEL-239 clones (Fig. 1D) and other resistant cell lines (Fig. S1B), as well as in BRAFi plus MEK inhibitor (MEKi) double resistant cell clones (Fig. S1C). Importantly, we evaluated miR-125a expression in 22 paired human melanoma clinical samples acquired pre-treatment and after BRAFi treatment. We observed miR125a-5p levels to be upregulated in 8 of 22 (36%) tumor samples while on therapy with BRAFi (Fig. 1E), supporting the potential relevance of miR-125a induction to development of therapeutic resistance to BRAFi in melanoma patients.
Fig. 1. Increased miR-125a levels in BRAFi resistant melanoma cells and human tissues.
A. Western Blot of pERK (Thr202/Tyr204) and ERK in parental SK-MEL-239 cells (par), parental cells treated with 2μM Vemurafenib for 24h and five resistant clones kept on 1μM Vemurafenib (representative of n=3). B. IC50 curves of SKMEL-239 cells and five resistant clones using increasing amounts of Vemurafenib (0.001μM-100μM) (representative of n=3) C. Average miR-125a-5p, miR-125b and miR125a-3p measurements (normalized to untreated SK-MEL-239 cells (par)), as detected by the nCounter NanoString platform, of parental SK-MEL-239 cells treated with Vemurafenib (n = 2) or resistant clones (n = 5 independent clones). Error bars represent standard deviation of biological replicates. Statistical analysis performed by Student's T test. D. Fold change of miR-125a-5p expression (by RT-qPCR) in a panel of additional SK-MEL-239-derived resistant clones (n = 16) compared to untreated parental SK-MEL-239 cells. Error bars represent standard deviation of technical replicates. E. Fold change of miR-125a-5p (by RT-qPCR) in 22 human melanoma tissue samples post-Vemurafenib treatment compared to their respective paired pre-treated sample (dotted line).
MiR-125a suppresses apoptosis in the presence of BRAFi and promotes BRAFi resistance
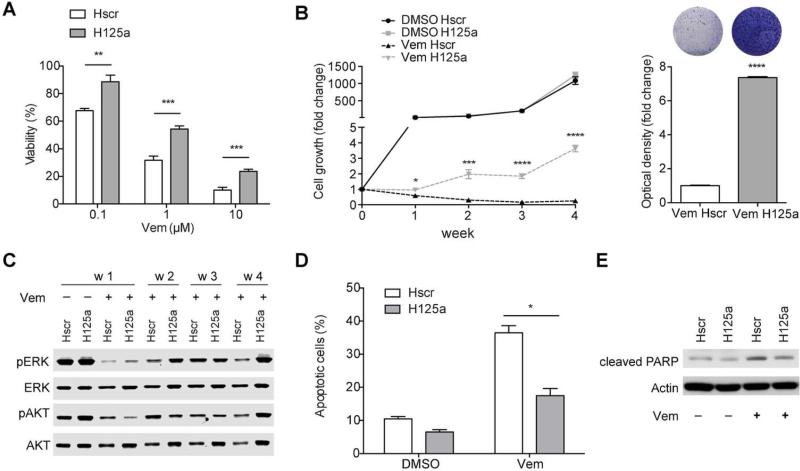
We sought to determine if miR-125a upregulation functionally contributes to the acquisition of resistance to BRAFi by melanoma cells. We stably overexpressed miR-125a in SK-MEL-239 cells by lentiviral transduction (Fig. S2). MiR-125a overexpressing cells displayed increased viability in the presence of Vemurafenib at all tested doses (Fig. 2A). Moreover, miR-125a overexpression allowed melanoma cells to grow in the presence of BRAFi (Fig. 2B). Interestingly, we observed that this resistance phenotype was not immediately accompanied by reactivation of the MAPK or AKT pathways (week1, Fig. 2C), but resulted in enhanced reactivation of those pathways at later time points (weeks 2-4, Fig. 2C). Since Vemurafenib exerts its anti-tumor effect predominantly by triggering apoptosis (Lee et al., 2010), we examined the role of miR-125a on apoptosis induction of Vemurafenib-treated melanoma cells. Ectopic expression of miR-125a in Vemurafenib-treated cells significantly decreased the number of Annexin+ apoptotic cells (Fig. 2D) and suppressed PARP cleavage (Fig. 2E). Moreover, miR-125a overexpression also conferred increased resistance to the combined treatment of BRAFi plus MEKi (Fig. S3A, B). Collectively, these data indicate that miR-125a aids melanoma cell survival in the presence of BRAFi and promotes BRAFi resistance, likely through suppression of apoptosis.
Fig. 2. Ectopic expression of miR-125a confers resistance to SK-MEL-239 cells through evasion of apoptosis.
A. Viability of scrambled- (Hscr) or miR-125- (H125a) transduced SK-MEL-239 cells using increasing Vemurafenib concentrations measured after 4 days of drug treatment. B. Long term growth of Hscr or H125a SK-MEL-239 cells treated with DMSO or 2μM Vemurafenib for 4 weeks (left panel). Optical density of crystal violet staining after 4 weeks of Vemurafenib treatment (right panel). C. Western Blots for pERK (Thr202/Tyr204), ERK, pAKT (Ser473) and AKT of protein extracts from Hscr and H125a SK-MEL-239 cells treated with DMSO or 2μM Vemurafenib (week1-4). D. Apoptotic cells detected through FACS analysis of AnnexinV/PI staining after one week of DMSO or 2μM Vemurafenib treatment of Hscr and H125a SK-MEL-239 cells. E. Western Blot of cleaved PARP in Hscr and H125a SK-MEL-239 cells after one week of DMSO or 2μM Vemurafenib treatment. The results in A are an average of three independent experiments, BE are representative of three independent experiments ± standard deviation (SD). *P<0.05, **P<0.001, ***P<0.0001, ****P<0.00001.
MiR-125a inhibition suppresses the emergence of resistance to BRAFi and partially restores sensitivity to BRAFi in a subset of resistant cell lines
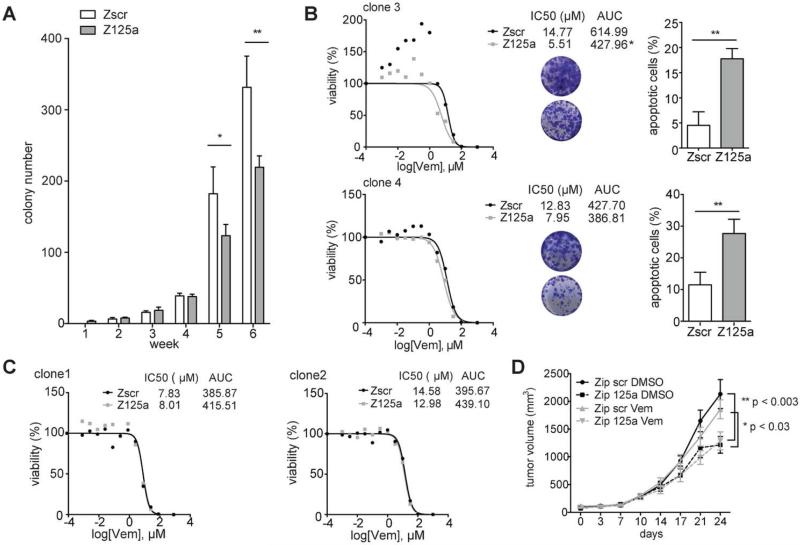
Because miR-125a promotes melanoma cell survival during BRAFi treatment and its up-regulation is maintained in BRAFi-resistant cells, we hypothesized that its inhibition may delay or prevent the emergence of resistance to BRAFi, suppress growth and survival of resistant cells, and/or re-sensitize resistant cells to BRAF inhibition. We stably downregulated miR-125a with a lentiviral vector expressing an antisense sequence (Fig. S4A). MiR-125a suppression in BRAFi-sensitive parental SK-MEL-239 cells did not have an effect on their viability in vitro (Fig. S4B, S4C), but yielded a significant reduction in the emergence of BRAFi resistant colonies after prolonged exposure to Vemurafenib (Fig. 3A). Next, we examined if the miR-125a upregulation in established BRAFi resistant cell clones is required to maintain resistance to BRAFi. Interestingly, in vitro, some resistant cell clones were partially re-sensitized to BRAF inhibition by miR-125a depletion (Fig. 3B, left panels), by increasing Vemurafenib induced apoptosis (Fig. 3B, right panels); however, other resistant clones, which also overexpress miR-125a, were not re-sensitized to BRAF inhibition by miR-125a suppression (Fig. 3C). In vivo, we observed that miR-125a depletion suppressed tumor growth, irrespective of Vemurafenib treatment, of a BRAFi resistant clone (Fig. 3D) that, in vitro, was ‘re-sensitized’ to BRAF inhibition by miR-125a depletion (see Fig. 3B). In addition, miR-125a inhibition partially restored BRAFi sensitivity to non-clonal resistant A375 cells in vitro, further supporting the role of miR-125a in maintenance of BRAFi resistance (Fig. S4D). Collectively, these findings highlight that miR-125a is important for establishment and, in some cases, for maintenance of BRAFi resistance in melanoma, suggesting these cells have become partially addicted to miR-125a for their proliferation.
Fig. 3. Inhibition of miR-125a suppresses emergence of resistance and partially restores BRAFi sensitivity of some resistant clones.
A. Number of colonies formed by SK-MEL-239 cells transduced with Zip scr and Zip 125a constructs and treated with 2μM Vemurafenib for 6 weeks (average of quintuplicates). B. Viability curves of resistant clones transduced with Zip scr and Zip 125a and treated with increasing Vemurafenib concentrations (0.001μM-100μM) for 4 days (left panel). Tables indicate IC50 value and area under the curve (AUC). Photo inserts of crystal violet staining of Zip scr (upper circle) or Zip 125a (lower circle) transduced resistant clones after 8 days (clone 4) or 10 days (clone 3) of treatment with 2μM Vemurafenib. Apoptosis detected through FACS analysis of AnnexinV/PI staining after 8 days (clone 4) or 10 days (clone 3) in Zip scr and Zip 125a resistant clones treated with DMSO or 2μM Vemurafenib (right panel). C. Viability curves of resistant clones transduced with Zip scr and Zip 125a and treated with increasing Vemurafenib concentrations for 4 days. D. Tumor growth curves mock- (DMSO) or vemurafenib-treated (vem) BRAFi-resistant SK-MEL-239 clone transduced with Zip scr or Zip 125a. For every assay in A-C, a representative experiment of a triplicate is shown along with SD. AUC: area under the curve, clone3: p≤0.05, clone 4, 1, 2: n.s. *P≤0.05, **P<0.001.
MiR-125a suppresses the apoptotic program in BRAFi treated melanoma cells. BAK1 and MLK3 are direct miR-125a targets
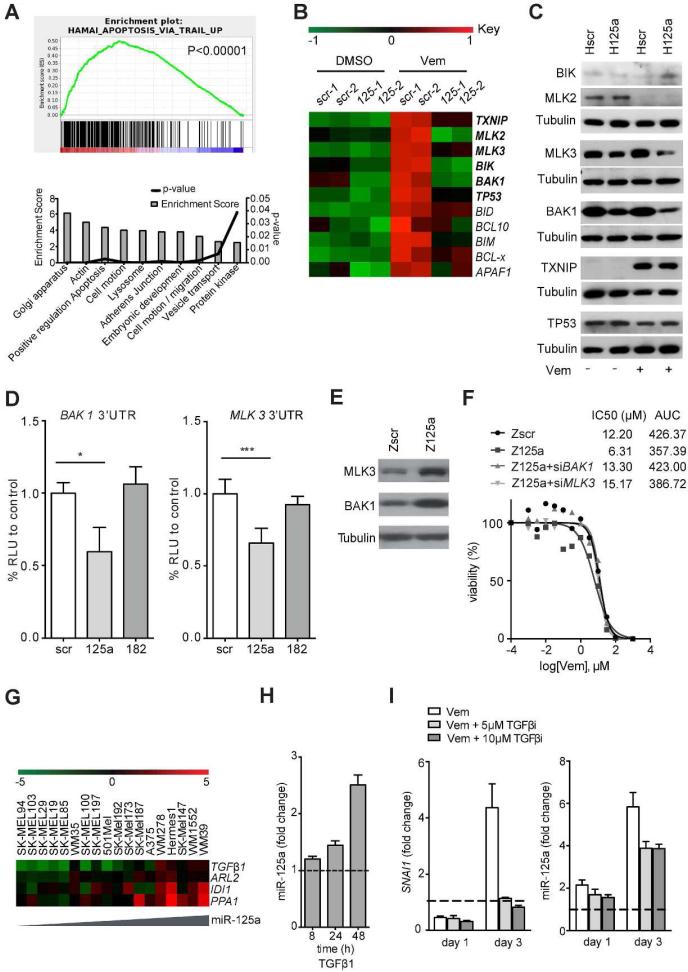
To elucidate miR-125a effectors involved in BRAFi resistance, we performed RNA sequencing of SK-MEL-239 cells overexpressing miR-125a or a scrambled control, in the presence of Vemurafenib or vehicle. Gene Set Enrichment Analysis (GSEA) of deregulated genes (p<0.05) in Vemurafenib-treated miR-125a overexpressing cells versus Vemurafenib treated scramble control cells revealed apoptosis as the main differentially regulated gene category (p<0.00001) (Fig 4A, upper graph). Since miRNA are negative regulators of gene expression we conducted GO analysis (http://david.abcc.ncifcrf.gov/) specifically for genes downregulated by miR-125a overexpression, which again identified ‘apoptotic regulation’ among the most significantly affected programs (p<0.003) (Fig. 4A, lower graph). In order to identify direct targets, which might mediate miR-125a pro-survival effects in the presence of BRAFi, we overlapped the list of 1999 downregulated genes (p<0.05, FC<0.8) with a combined list of 2018 putative targets from three publicly available databases (TargetScan (Friedman et al., 2009), Starbase (Li et al., 2014), and miRWalk (Dweep et al., 2011)). Within the overlapping list of 503 putative targets downregulated by miR-125a over-expression in the presence of Vemurafenib were several components of the intrinsic pro-apoptotic pathway (TXNIP, MLK2, MLK3, BIK1, BAK1, TP53; Fig. 4B). Validation through Western Blot analysis revealed regulation of MLK3 and BAK1 by miR-125a (Fig.4C), which was particularly evident upon BRAFi treatment. 3'UTR luciferase reporter assays confirmed that miR-125a directly targets both MLK3 and BAK1 (Fig. 4D). Accordingly, resistant clones showed increased MLK3 and BAK1 protein levels after miR-125a inhibition (Fig. 4E). Interestingly, miR-125a-induced repression of MLK3 did not affect the apoptotic JNK/p38 pathway, which MLKs are known to regulate (Fig. S5A).
Fig. 4. Downstream targets and upstream regulators of miR-125a.
A. Gene Set Enrichment Analysis (GSEA) of deregulated genes (upper graph) and DAVID pathway analysis of downregulated genes (lower graph) based on RNA sequencing data of Hscr or H125a SK-MEL-239 cells treated with 2μM Vemurafenib for 1 week. B. Heatmap of apoptosis-related genes in Hscr and H125a cells treated with DMSO or 2μM Vemurafenib. In bold are predicted targets of miR-125a. C. Western Blot of predicted miR-125a apoptotic targets in Hscr and H125a cells treated with DMSO or 2μM Vemurafenib for one week. Results are representative of three independent experiments. Loading control (Tubulin) is shown for each blot performed (below experimental protein(s)) D. Luciferase activity of 293T cells transfected with 3'UTR luciferase construct of BAK1 and MLK3 and miR-scr, miR-125a or miR-182 mimic. Results show the average of three independent experiments ± SD. *P<0.05, ***P<0.0001. E. Western Blot of BAK1 and MLK3 in a resistant clone transduced with Zip scr or Zip 125a and treated with Vemurafenib for one week. Results are representative of three independent experiments. F. Viability curves of a resistant clone transduced with Zip scr or Zip 125a and transfected with 100nM siBAK1 or siMLK3 for 4 days and treated with increasing Vemurafenib concentrations. IC50 and AUC values are indicated for each condition (0.001μM-100μM). G. Heatmap of transcription factors that show positive correlation with miR-125a expression in melanoma cell lines, based on gene expression profiling (GSE22306). H. MiR-125a levels in SK-MEL-239 cells treated with 5ng/ml recombinant TGFβ1. I. SNAI1 mRNA (left panel) and miR-125a levels (right panel) in SK-MEL-239 cells treated with 2μM Vemurafenib with or without indicated concentrations of Galunisertib, a TGFβ I receptor inhibitor, for indicated times compared to untreated parental cells (dashed line). The results in D are an average of three independent experiments, F, H-I are representative of three independent experiments ± SD. *P<0.05, ***P<0.0001.
Individual and combined silencing of BAK1 and MLK3 in SK-MEL-239 cells was unable to mimic the ability of miR-125a to promote BRAFi resistance (Fig. S5B-C), suggesting that miR-125a effects require the combined suppression of multiple apoptosis suppressors. However, in resistant cells sensitized to BRAFi through miR-125a inhibition, depletion of either BAK1 or MLK3 restored BRAFi resistance (Fig. 4F, Fig S5D). To examine whether downregulation of BAK1 and/or MLK3 correlated with resistance to BRAFi therapy in clinical samples, we analyzed their expression in a previously published profile (Rizos et al., 2014) of paired biopsies isolated pre-treatment and at disease progression (BRAFi-resistance) from 21 metastatic melanoma patients. There was evidence of downregulation (relative to the patient-matched pre-treatment samples) of BAK1 or MLK3 in 6 (~29%) and 10 (~43%) of 21 patients, respectively (Fig. S5E). These findings support that BAK1 and MLK3 downstream of miR-125a might be involved in clinical resistance to BRAFi therapy. Overall, these results suggest that regulation of pro-apoptotic targets might critically mediate miR-125a effects on melanoma cell resistance to BRAFi.
MiR-125a expression is regulated by TGFβ signaling
MiR-125a expression was induced after short-term Vemurafenib exposure of SK-MEL-239 cells (Fig. S6A) and rapidly returned to basal levels after drug removal (Fig. S6B), suggesting that it is likely regulated by transient, reversible mechanisms. To examine if this regulation occurs transcriptionally, we tested the effect of Vemurafenib on the expression of the miR-125a primary transcript, which includes miR-125a, miR-99 and let-7b, and found it upregulated in resistant clones (Fig. S6C). Surprisingly, of the miRNA expressed in this cluster, only mature miR-125a was found significantly upregulated (Fig. S6D), suggesting that differential processing may add further specificity to the induction of miR-125a expression by BRAF inhibition.
To identify potential upstream regulators of miR-125a expression, we correlated miR-125a with coding mRNA abundance from a previously published gene expression profiling of a panel of melanoma cell lines (Rose et al., 2011). Strikingly, the most highly correlated mRNA was TGFβ1 (r2 = 0.8, p<0.00001) (Fig 4G), which has previously been implicated in acquisition of BRAFi resistance (Sun et al., 2014). Moreover, in silico analysis of the miR-125a promoter region revealed three putative binding sites of the TGFβ induced gene EGR-1 (Chen et al., 2006) and one putative Smad binding site. Treatment of BRAFi sensitive melanoma cells with recombinant TGFβ1 protein induced miR-125a expression (Fig 4H). Importantly, TGFβ signaling is induced upon Vemurafenib treatment (Fig. 4I, left panel, Fig S7), and treatment with the TGFβ receptor I inhibitor Galunisertib suppressed miR-125a induced by BRAF inhibition (Fig. 4I, right panel). Overall, our data indicate that TGFβ signaling contributes to miR-125a induction in response to BRAF inhibition.
Discussion
A major hurdle for targeted therapy in melanoma remains the widespread development of BRAFi resistance. Whereas most of the described resistance mechanisms are based on genomic alterations, the contributions of non-genomic alterations to resistance are only beginning to be explored (Hugo et al., 2015). Delineating the molecular mechanisms involved in the acquisition and maintenance of BRAFi resistance may provide valuable insights into novel therapeutic strategies to more effectively treat patients with BRAFV600E-mutant melanoma. Here, in a model that recapitulates clinical BRAFi resistance (Poulikakos et al., 2011) and additional cell line models, we consistently observed upregulation of miR-125a in BRAFi-resistant clones. Moreover, miR-125a was upregulated in human clinical melanoma samples from patients treated with BRAFi compared to their pre-treatment biopsies. In addition to these observations, we experimentally demonstrated that miR-125a expression favors acquisition of BRAFi resistance by suppressing the induction of apoptosis via direct targeting of the intrinsic apoptosic pathway. Finally, we demonstrate that TGFβ signaling is required for miR-125a induction during BRAFi treatment. Collectively, our data document a critical role of miR-125a supporting the acquisition of BRAFi resistance in melanoma.
MiR-125a is a member of the miR-10 family, which also includes two homologs, hsa-miR-125b-1, hsa-miR-125b-2, with which miR-125a shares an identical seed region. MiR-125 homologs play crucial roles in many different cellular and molecular processes, such as differentiation, proliferation, apoptosis, and regulation of matrix-metalloproteases (Bi et al., 2012, Xu et al., 2012, Balakrishnan et al., 2012, Bousquet et al., 2008, Sun et al., 2013, Ge et al., 2011).
For the different miR-125 homologs, contrasting properties have been reported in different cancer types; they may contribute to the initiation and progression of cancers by acting as either tumor suppressors or oncogenes (Cowden Dahl et al., 2009, Jiang et al., 2011, Jiang et al., 2010). The opposing properties of the miR-125 homologs in different solid tumors demonstrate that they can have context-dependent functions in cancer pathogenesis, progression and treatment resistance. The underlying direct target mediators and molecular mechanisms in different cell contexts require further investigation.
Although miR-125a has not yet been described as a mediator of acquired BRAFi resistance of melanoma cells, previous studies have documented the ability of miR-125b to increase resistance of cancer cell models to anticancer agents (Sun et al., 2013, Zhou et al., 2010). For instance, Sun et al. demonstrated a role for miR-125b in Taxol resistance in breast cancer (Sun et al., 2013). Moreover, recently, Vergani et al. reported that miR-125b (and other miRNAs) was upregulated in response to CCL2 induction during development of BRAFi resistance in melanoma (Vergani et al., 2016). Similar to our findings for miR-125a, miR-125b blocked apoptotic induction, though the molecular mechanisms downstream of this phenotype are not yet elucidated. They did not observe miR-125a upregulation in their model of BRAFi resistance used for miRNA profiling, nor did we observe miR-125b induction in our models. It is tempting to speculate that context-dependent increase of one of the miR-125 homologs is required in the early stages of BRAFi treatment.
To our knowledge, our study provides the first evidence of miR-125a promoting therapeutic resistance in melanoma. Importantly, our findings identify a targetable step that may improve the efficacy of BRAF inhibition and/or BRAFi/MEKi combination therapy by blocking a cell survival pathway activated early in the treatment of metastatic BRAF mutant melanoma patients.
The prevalent BRAFV600E mutation provides melanoma cells with enhanced proliferation and survival signals. The therapeutic effects of BRAF inhibition stem from the activation of intrinsic apoptosis mechanisms. Our data suggest a mechanism of BRAFi mediated cell death evasion, carried out by miR-125a and its direct targeting of the pro-apoptotic factors MLK3 and BAK1. MLKs commonly modulate apoptosis through the JNK/p38 pathway. In this study, we demonstrate that downregulation of the specific member MLK3 by miR-125a plays a role in resistance through increasing cell viability in a JNK/p38-independent manner. A possibility is that the low expression of MLK3 provides a survival advantage through the inactivation of the intrinsic apoptotic pathway via AMPK regulation (Luo et al., 2015) or other related factors. BAK1 functions as a pro-apoptotic regulator by localizing to mitochondria and accelerating the release of cytochrome c (Westphal et al., 2014). Downregulation of BAK1 by miR-125a therefore leads to a decrease in mitochondrial apoptotic signaling, effectively resulting in increased cell survival. Thus, the direct suppression of apoptosis by miR-125a provides a mechanism by which melanoma cells may evade BRAF inhibition. This general survival benefit enacted by miR-125a may permit tumor cells to acquire secondary, ‘permanent’ resistance mechanisms resulting in eventual reactivation of the MAPK and AKT pathways. Therefore, miR-125a upregulation likely coexists with other mechanisms of resistance, playing a supporting role for cell survival. This is supported by our observation that miR-125a inhibition suppressed tumor growth of BRAFi resistant cells independent of BRAFi treatment. Whether miR-125a upregulation favors a particular subsequent resistance mechanism remains an area of active investigation.
Since miRNA can have pleiotropic effects, suppression of apoptosis might not be the only mechanism by which miR-125a contributes to resistance to BRAFi. Cells overexpressing the miRNA and treated with BRAFi exhibited phenotypic changes, such as lipid-like accumulations (data not shown), suggesting that alterations in cell metabolism might also be involved. In support of this notion, IPA analysis of genes modulated by miR-125a overexpression in the presence of BRAFi revealed ‘Cholesterol metabolism’ among the top dysregulated categories. As cholesterol availability is altered in cancer cells and influences oncogenic networks (Ura et al., 1994), investigating metabolic reprogramming of melanoma cells in relation to BRAFi resistance may provide further valuable insight. Accordingly, recent work has identified a metabolic shift towards oxidative phosphorylation as another response of melanoma cells to BRAFi (Haq et al., 2013a).
Due to miR-125a targeting of pro-apoptotic factors, uncovering upstream miR-125a regulators in human tissues will likely provide relevant therapeutic avenues and is an area that warrants further investigation. In our system, the quick and reversible change in miR-125a expression levels upon BRAFi modulation suggests that it results from transcriptional and/or post-transcriptional mechanisms (rather than genetic amplification or mutation). We observed that TGFβ signaling, induced by Vemurafenib treatment, contributes to the regulation of miR-125a expression and therefore to resistance. Previous reports have linked activated TGFβ signaling to the acquisition of BRAFi resistance (Sun et al., 2014) and miR-125a induction may critically contribute to those effects.
While the upregulation of miR-125a provides a mechanism for drug resistance, the ability to target this miRNA opens the possibility to interfere with development of resistance. Indeed, depletion of this miRNA during BRAFi treatment in vitro led to a decreased emergence of resistant clones and partially re-sensitized a subset of BRAFi resistant cells. These findings suggest potential benefit of establishing clinical approaches for miRNA suppression in combination with BRAFi or BRAFi/MEKi therapy in the treatment of BRAFV600E melanoma. Thus, the identification of miR-125a's involvement in BRAFi resistance elucidates clinically relevant mechanisms of melanoma cell survival and has important therapeutic implications.
Methods
Cells and cell culture
SK-MEL-239 parental and some Vemurafenib-resistant cell lines (single clones) were a kind gift of Dr. Poulikos Poulikakos and Dr. Emily Bernstein. Vemurafenib-resistant A375R and 451LuR cells (pooled clones) cells were derived from their respective parental cell lines by treatment with 1μM Vemurafenib for several weeks. A375 and 451Lu cells were cultured in Dulbecco's modified Eagle's medium (DMEM), resistant single clones were derived from the SK-MEL-239 cell line through prolonged treatment with 2μM Vemurafenib (single resistance) or 1μM Vemurafenib + 2μM PD0325901 (double resistance) in RPMI-1640 medium, supplemented with 10% fetal bovine serum and penicillin/streptomycin. The resistant cell lines were cultured in the presence of 1μM Vemurafenib. For TGFβ experiments cells were treated with human TGFβ1 protein (R&D Systems) or TGFβ receptor I inhibitor (Galunisertib, Selleckchem).
Cell line authentication by short tandem repeat (STR) analysis was performed for SK-MEL-239 at ATCC and confirmed that these cells didn't match with any of the cell lines deposited in the cell bank (this cell line isn't present in any of the main cell line repositories and thus there is no ‘baseline’ STR profile from which to compare). STR profiling is ongoing for A375 and 451Lu. Cell lines were previously analyzed by morphologic assessment and gene expression profiling for lineage-specific gene expression (TYR, TRYP1, DCT, TRYP1B, MITF, EDNRB, KIT) by microarray.
MiRNA expression profiling
Total RNA was extracted from SK-MEL-239 cells and derived resistant clones using QIAzol reagent (Qiagen). 100ng RNA per sample was used for miRNA profiling using the NanoString nCounter expression assay of 800 human miRNAs according to manufacturer's protocol (Nanostring nCounter® miRNA Expression Assay User Manual). Raw expression values were normalized to those of the 100 highest expressed miRNAs and expression profiles were compared to the sensitive parental cells applying a differential expression cut-off of 2-fold. Student's t-test analysis was conducted and miRNAs with p-values <0.05 were selected for further analyses.
Pre-miRNA or anti-miRNA stable transduction
Lentiviral vectors carrying miRNA precursors (H miRNA) and antisense miRNA sequences (Zip miRNA) were purchased from Dharmacon. Pre-miR- and anti-miR-scramble sequences were used as negative controls. Lipofectamine 2000 (Invitrogen) was used for transfection of 293T cells with pre-miRNAs or Zip miRNAs for virus production. 48h after transfection virus was collected and titrated. Infection of melanoma cells was performed for 6h using 4μg/ml polybrene.
Oligonucleotide transient transfection
miRIDIAN oligonucleotides mimics, anti-miRNAs or the SMARTpool siRNAs (mixture of 4 siRNA provided as a single reagent) (all Dharmacon) were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. Transfection efficiency was monitored using Block-iT Fluorescent Oligo (Invitrogen).
Real-time quantitative PCR
Total RNA was isolated from cultured cells using the miRNeasy Mini Kit (Qiagen). MiRNA and pri-miRNA expression analysis was performed using miRNA-specific TaqMan MicroRNA Assay Kits (Applied Biosystems). 12.5ng of total RNA were reverse-transcribed using the corresponding RT primer and the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems). PCR was performed on 1.33μl of RT products by adding Taqman PCR primers and Taqman Universal Master Mix (Applied Biosystems). RNU44 small RNA was used for normalization of input RNA/cDNA levels.
MLK3 and BAK1 mRNA expression analysis was performed by synthesizing cDNA from 1μg of total RNA using Reverse Transcriptase cDNA Synthesis kit (Applied Biosystems). For quantitative PCR, cDNA was mixed with SYBR Green PCR Master Mix (Applied Biosystems) and various sets of gene-specific primers and then subjected to RT-PCR quantification by using the iQ5 real time PCR system (Bio-Rad). The sequences of the primers used were as follows: MLK3 5’ primer (5’-ATGCCACTCGACTTCAAGCA-3’) and 3’ primer (5’-GACGTTTCTCCTCCGGTCAA-3’), BAK1 5’ primer (5’-CGGCAGAGAATGCCTATGAGT -3’) and 3’ primer (5’-AAACAGGCTGGTGGCAATCTT-3’). GAPDH was used for normalization of input RNA/cDNA.
FACS analysis
Cells were pretreated with Annexin V Pacific Blue (BioLegend) and Propidium Iodide (PromoKine) and analyzed with a BD LSR II (BD Biosciences) and FACS Express Software (De Novo Software). After an initial gating on forward-versus-side scatter plots, apoptotic cells were identified through gating for the AnnexinV+/PI+ and AnnexinV+ cells.
Western blotting
Cell lysates were harvested using a commercial cell lysis buffer (Cell Signaling) supplemented with protease and phosphatase inhibitors (Roche). Cell lysates (25-30μg) were resolved in Tris-/glycine or Bis-Tris SDS/PAGE gels (Invitrogen) and transferred to nitrocellulose or PVDF membranes (Invitrogen). Membranes were blocked with 5% non-fat milk or 5% BSA in TBS-T for 60min and probed with primary antibodies overnight at 4°C for BIK (Abcam), MLK2 (Abcam), MLK3 (Abcam), Tubulin (Sigma), TXNIP (VDUP-1) (Invitrogen), BAK1, TP53, JNK, pJNK (Thr183/Tyr185), p38, p-p38 (Thr180/Tyr182), ERK, pERK (Thr202/Tyr204), AKT, pAKT (Ser473) (all Cell Signaling). Membranes were exposed to HRP- or fluorescence-conjugated secondary antibodies (1:10,000) in TBS-T with 1% blocking agent for 60min. Following extensive washing with TBS-T, western membranes were digitally imaged (fluorescent, Licor) or developed with the ECL Western blotting detection kit (HRP, Merck Millipore) and exposed on film (Denville Scientific).
Luciferase assay
A plasmid containing the luciferase cDNA conjugated to the 3'UTR of MLK3 was purchased from SwitchGear. A plasmid containing the luciferase cDNA conjugated to the 3'UTR of BAK1 was purchased from GeneCopeia. HEK293T cells were seeded into a 96-well plate and co-transfected with 3'UTR reporter vectors and indicated amounts of miRIDIAN miR-125a or miR-182 mimics or mimic negative control (Dharmacon). Luciferase activity was measured using the Dual Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to corresponding firefly luciferase activity and plotted as a percentage of the control.
Cell viability assay
Cells were seeded into a 96-well plate and treated with indicated concentrations of Vemurafenib (Plexxikon) (0.001μM-100μM). 96h after treatment cell viability was measured using the CellTiter Glo Luminescent Cell Viability Assay (Promega) according to manufacturer's instructions.
Short-term proliferation assay
Cells were seeded into a 96-well plate and treated with 2μM Vemurafenib (Plexxikon) for indicated times. Cells were then fixed using 1% glutaraldehyde and stained with crystal violet. Dye was then solubilized using 15% acetic acid and read at OD590.
Long-term proliferation assay
Cells were seeded in triplicate into 6 well plates and treated with DMSO or 2μM Vemurafenib (Plexxikon) for the indicated time points. At each time point cells were counted and cell numbers were referred to initial seeding at day 0.
In vivo xenograft experiment with resistant cells
1×106 cells (resistant clone 4 transduced with Zscr or Z125a) were injected in 1 flank/mouse of 20 mice per group. Tumors began to form at 1 week after injection. Drug treatment was initiated at 100mm3 tumor volume. 10 mice per group were injected with 25mg/kg Vemurafenib or DMSO through intraperitoneal injection twice per day for 24 days.
Clinical specimens
Patients with BRAFV600-mutant metastatic melanoma were treated with a BRAFi. After obtaining appropriate Institutional Review Board approved protocol, 7μm formalin-fixed and paraffin-embedded (FFPE) sections were received from tumors biopsied before treatment or at the time of progression.
FFPE RNA Isolation
Human metastatic melanoma areas were identified through H&E staining by an experienced pathologist (Dr. Farbod Darvishian). Tumor areas were micro-dissected using a precision scalpel. Depending on tumor size, 3 to 5 sections per sample, each 7μM thick were used per RNA extraction. RNA was extracted using the QIAGEN miRNeasy FFPE Tissue Kit (Qiagen) according to manufacturers protocol.
RNA sequencing
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen). Total RNA quality and quantity were determined using Agilent 2100 Bioanalyzer and Nanodrop ND-1000. Expression profiling of the duplicate experimental sample groups (SK-MEL-239 cells with lentiviral scramble or miR-125a constructs after 7 days of DMSO or 2μM Vemurafenib treatment) was performed using Illumina HiSeq 2500 System. Samples were subject to PolyA selection using oligo-dT beads and the resulting RNA samples were then used as input for library construction according to the manufacturer's instructions (Illumina TruSeq RNA Sample Prep Kit v2). RNA libraries were then sequenced on the Illumina HiSeq2500 using paired end sequencing with v4 technology.
RNA sequencing data analysis
Sequencing results were demultiplexed and converted to FASTQ format using Illumina Bcl2FastQ software. Paired-end reads were aligned to the human genome (build hg19/GRCh37) using the splice-aware STAR aligner (Dobin et al., 2013). PCR duplicates were removed using the Picard toolkit (http://broadinstitute.github.io/picard/). HTSeq package (Anders et al., 2015) was utilized to generate counts for each gene based on how many aligned reads overlap its exons. These counts were then used to test for differential expression using negative binomial generalized linear models implemented by the DESeq2 R package (Love et al., 2014).
The functional annotations of the gene lists were performed using the NIH web based tool DAVID (Database for Annotation, Visualization and Integrated Discovery) (Huang da et al., 2009) and/or Gene Set Enrichment Analysis (GSEA, http://www.broadinstitute.org/gsea/) (Subramanian et al., 2008).
Statistical methodologies
Unless otherwise indicated, mean values ± SD are representative of one of three independent experiments. Statistical significance was determined by Student's t test (GraphPad Prism Software). Of note, *P < 0.05; **P < 0.001; and ***P < 0.0001, ****P < 0.00001.
Supplementary Material
Significance.
Emergence of resistance to BRAFi therapy is a prevalent problem in the treatment of patients with metastatic BRAFV600E mutant melanoma. We document TGFβ-mediated upregulation of miR-125a as a pro-survival mechanism that favors the acquisition of BRAFi resistance. Our data support the possibility to target miR-125a or its upstream regulators to limit the emergence of BRAFi resistance in melanoma. Moreover, as a pro-survival mechanism, miR-125a upregulation could contribute to development of resistance to other therapeutic agents, thus our findings might be applicable to the broader cancer field.
Acknowledgements
We thank Adriana Heguy and members of the New York University Cancer Institute Genome Technology Center (GTC) for microRNA profiling, RNA sequencing and analysis, as well as the services of the NYU Experimental Pathology Core Facilities. We are grateful to Dr. Poulikos Poulikakos and to Taniya Panda (Icahn School of Medicine at Mount Sinai) for generating and providing us with some of the resistant SK-MEL-239 clones, and to members of the Hernando lab for discussions and technical assistance. This work was funded by NIH-NCI R01CA155234 grant.
Footnotes
Conflict of Interest
The authors disclose no potential conflicts of interest.
References
- AKBANI R, NG PK, WERNER HM, SHAHMORADGOLI M, ZHANG F, JU Z, LIU W, YANG JY, YOSHIHARA K, LI J, LING S, SEVIOUR EG, RAM PT, MINNA JD, DIAO L, TONG P, HEYMACH JV, HILL SM, DONDELINGER F, STADLER N, BYERS LA, MERIC-BERNSTAM F, WEINSTEIN JN, BROOM BM, VERHAAK RG, LIANG H, MUKHERJEE S, LU Y, MILLS GB. Corrigendum: A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2015;6:4852. doi: 10.1038/ncomms5852. [DOI] [PubMed] [Google Scholar]
- ANDERS S, PYL PT, HUBER W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALAKRISHNAN A, STEARNS AT, PARK PJ, DREYFUSS JM, ASHLEY SW, RHOADS DB, TAVAKKOLIZADEH A. Upregulation of proapoptotic microRNA mir-125a after massive small bowel resection in rats. Ann Surg. 2012;255:747–53. doi: 10.1097/SLA.0b013e31824b485a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BI Q, TANG S, XIA L, DU R, FAN R, GAO L, JIN J, LIANG S, CHEN Z, XU G, NIE Y, WU K, LIU J, SHI Y, DING J, FAN D. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7:e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLLAG G, HIRTH P, TSAI J, ZHANG J, IBRAHIM PN, CHO H, SPEVAK W, ZHANG C, ZHANG Y, HABETS G, BURTON EA, WONG B, TSANG G, WEST BL, POWELL B, SHELLOOE R, MARIMUTHU A, NGUYEN H, ZHANG KY, ARTIS DR, SCHLESSINGER J, SU F, HIGGINS B, IYER R, D'ANDREA K, KOEHLER A, STUMM M, LIN PS, LEE RJ, GRIPPO J, PUZANOV I, KIM KB, RIBAS A, MCARTHUR GA, SOSMAN JA, CHAPMAN PB, FLAHERTY KT, XU X, NATHANSON KL, NOLOP K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUSQUET M, QUELEN C, ROSATI R, MANSAT-DE MAS V, LA STARZA R, BASTARD C, LIPPERT E, TALMANT P, LAFAGE-POCHITALOFF M, LEROUX D, GERVAIS C, VIGUIE F, LAI JL, TERRE C, BEVERLO B, SAMBANI C, HAGEMEIJER A, MARYNEN P, DELSOL G, DASTUGUE N, MECUCCI C, BROUSSET P. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAPMAN PB, HAUSCHILD A, ROBERT C, HAANEN JB, ASCIERTO P, LARKIN J, DUMMER R, GARBE C, TESTORI A, MAIO M, HOGG D, LORIGAN P, LEBBE C, JOUARY T, SCHADENDORF D, RIBAS A, O'DAY SJ, SOSMAN JA, KIRKWOOD JM, EGGERMONT AM, DRENO B, NOLOP K, LI J, NELSON B, HOU J, LEE RJ, FLAHERTY KT, MCARTHUR GA, GROUP B-S. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN SJ, NING H, ISHIDA W, SODIN-SEMRL S, TAKAGAWA S, MORI Y, VARGA J. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J Biol Chem. 2006;281:21183–97. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- COWDEN DAHL KD, DAHL R, KRUICHAK JN, HUDSON LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–15. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, JHA S, BATUT P, CHAISSON M, GINGERAS TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWEEP H, STICHT C, PANDEY P, GRETZ N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- EMERY CM, VIJAYENDRAN KG, ZIPSER MC, SAWYER AM, NIU L, KIM JJ, HATTON C, CHOPRA R, OBERHOLZER PA, KARPOVA MB, MACCONAILL LE, ZHANG J, GRAY NS, SELLERS WR, DUMMER R, GARRAWAY LA. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERLAY J, SOERJOMATARAM I, DIKSHIT R, ESER S, MATHERS C, REBELO M, PARKIN DM, FORMAN D, BRAY F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- FLAHERTY KT, PUZANOV I, KIM KB, RIBAS A, MCARTHUR GA, SOSMAN JA, O'DWYER PJ, LEE RJ, GRIPPO JF, NOLOP K, CHAPMAN PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDMAN RC, FARH KK, BURGE CB, BARTEL DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GE Y, SUN Y, CHEN J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIROTTI MR, PEDERSEN M, SANCHEZ-LAORDEN B, VIROS A, TURAJLIC S, NICULESCU-DUVAZ D, ZAMBON A, SINCLAIR J, HAYES A, GORE M, LORIGAN P, SPRINGER C, LARKIN J, JORGENSEN C, MARAIS R. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–67. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAQ R, SHOAG J, ANDREU-PEREZ P, YOKOYAMA S, EDELMAN H, ROWE GC, FREDERICK DT, HURLEY AD, NELLORE A, KUNG AL, WARGO JA, SONG JS, FISHER DE, ARANY Z, WIDLUND HR. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013a;23:302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAQ R, YOKOYAMA S, HAWRYLUK EB, JONSSON GB, FREDERICK DT, MCHENRY K, PORTER D, TRAN TN, LOVE KT, LANGER R, ANDERSON DG, GARRAWAY LA, DUNCAN LM, MORTON DL, HOON DS, WARGO JA, SONG JS, FISHER DE. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci U S A. 2013b;110:4321–6. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILMI C, LARRIBERE L, GIULIANO S, BILLE K, ORTONNE JP, BALLOTTI R, BERTOLOTTO C. IGF1 promotes resistance to apoptosis in melanoma cells through an increased expression of BCL2, BCL-X(L), and survivin. J Invest Dermatol. 2008;128:1499–505. doi: 10.1038/sj.jid.5701185. [DOI] [PubMed] [Google Scholar]
- HUANG DA W, SHERMAN BT, LEMPICKI RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- HUGO W, SHI H, SUN L, PIVA M, SONG C, KONG X, MORICEAU G, HONG A, DAHLMAN KB, JOHNSON DB, SOSMAN JA, RIBAS A, LO RS. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG F, LIU T, HE Y, YAN Q, CHEN X, WANG H, WAN X. MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer. 2011;11:425. doi: 10.1186/1471-2407-11-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG L, HUANG Q, ZHANG S, ZHANG Q, CHANG J, QIU X, WANG E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHANNESSEN CM, BOEHM JS, KIM SY, THOMAS SR, WARDWELL L, JOHNSON LA, EMERY CM, STRANSKY N, COGDILL AP, BARRETINA J, CAPONIGRO G, HIERONYMUS H, MURRAY RR, SALEHI-ASHTIANI K, HILL DE, VIDAL M, ZHAO JJ, YANG X, ALKAN O, KIM S, HARRIS JL, WILSON CJ, MYER VE, FINAN PM, ROOT DE, ROBERTS TM, GOLUB T, FLAHERTY KT, DUMMER R, WEBER BL, SELLERS WR, SCHLEGEL R, WARGO JA, HAHN WC, GARRAWAY LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON DB, MENZIES AM, ZIMMER L, EROGLU Z, YE F, ZHAO S, RIZOS H, SUCKER A, SCOLYER RA, GUTZMER R, GOGAS H, KEFFORD RF, THOMPSON JF, BECKER JC, BERKING C, EGBERTS F, LOQUAI C, GOLDINGER SM, PUPO GM, HUGO W, KONG X, GARRAWAY LA, SOSMAN JA, RIBAS A, LO RS, LONG GV, SCHADENDORF D. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51:2792–9. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRAUTHAMMER M, KONG Y, BACCHIOCCHI A, EVANS P, PORNPUTTAPONG N, WU C, MCCUSKER JP, MA S, CHENG E, STRAUB R, SERIN M, BOSENBERG M, ARIYAN S, NARAYAN D, SZNOL M, KLUGER HM, MANE S, SCHLESSINGER J, LIFTON RP, HALABAN R. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LACKNER MR, WILSON TR, SETTLEMAN J. Mechanisms of acquired resistance to targeted cancer therapies. Future Oncol. 2012;8:999–1014. doi: 10.2217/fon.12.86. [DOI] [PubMed] [Google Scholar]
- LEE JT, LI L, BRAFFORD PA, VAN DEN EIJNDEN M, HALLORAN MB, SPROESSER K, HAASS NK, SMALLEY KS, TSAI J, BOLLAG G, HERLYN M. PLX4032, a potent inhibitor of the B-Raf V600E oncogene, selectively inhibits V600E-positive melanomas. Pigment Cell Melanoma Res. 2010;23:820–7. doi: 10.1111/j.1755-148X.2010.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI JH, LIU S, ZHOU H, QU LH, YANG JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU S, TETZLAFF MT, WANG T, YANG R, XIE L, ZHANG G, KREPLER C, XIAO M, BEQIRI M, XU W, KARAKOUSIS G, SCHUCHTER L, AMARAVADI RK, XU W, WEI Z, HERLYN M, YAO Y, ZHANG L, WANG Y, ZHANG L, XU X. miR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015;28:431–41. doi: 10.1111/pcmr.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOVE MI, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUJAMBIO A, LOWE SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO L, JIANG S, HUANG D, LU N, LUO Z. MLK3 phophorylates AMPK independently of LKB1. PLoS One. 2015;10:e0123927. doi: 10.1371/journal.pone.0123927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAZARIAN R, SHI H, WANG Q, KONG X, KOYA RC, LEE H, CHEN Z, LEE MK, ATTAR N, SAZEGAR H, CHODON T, NELSON SF, MCARTHUR G, SOSMAN JA, RIBAS A, LO RS. Melanomas acquire resistance to BRAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARAISO KH, XIANG Y, REBECCA VW, ABEL EV, CHEN YA, MUNKO AC, WOOD E, FEDORENKO IV, SONDAK VK, ANDERSON AR, RIBAS A, PALMA MD, NATHANSON KL, KOOMEN JM, MESSINA JL, SMALLEY KS. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POULIKAKOS PI, PERSAUD Y, JANAKIRAMAN M, KONG X, NG C, MORICEAU G, SHI H, ATEFI M, TITZ B, GABAY MT, SALTON M, DAHLMAN KB, TADI M, WARGO JA, FLAHERTY KT, KELLEY MC, MISTELI T, CHAPMAN PB, SOSMAN JA, GRAEBER TG, RIBAS A, LO RS, ROSEN N, SOLIT DB. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIZOS H, MENZIES AM, PUPO GM, CARLINO MS, FUNG C, HYMAN J, HAYDU LE, MIJATOV B, BECKER TM, BOYD SC, HOWLE J, SAW R, THOMPSON JF, KEFFORD RF, SCOLYER RA, LONG GV. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–77. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- ROSE AE, POLISENO L, WANG J, CLARK M, PEARLMAN A, WANG G, VEGA YSDMEC, MEDICHERLA R, CHRISTOS PJ, SHAPIRO R, PAVLICK A, DARVISHIAN F, ZAVADIL J, POLSKY D, HERNANDO E, OSTRER H, OSMAN I. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;71:2561–71. doi: 10.1158/0008-5472.CAN-10-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAO Y, APLIN AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–81. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN CH, KIM SH, TROUSIL S, FREDERICK DT, PIRIS A, YUAN P, CAI L, GU L, LI M, LEE JH, MITRA D, FISHER DE, SULLIVAN RJ, FLAHERTY KT, ZHENG B. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med. 2016;22:1056–61. doi: 10.1038/nm.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHI H, MORICEAU G, KONG X, LEE MK, LEE H, KOYA RC, NG C, CHODON T, SCOLYER RA, DAHLMAN KB, SOSMAN JA, KEFFORD RF, LONG GV, NELSON SF, RIBAS A, LO RS. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH MP, SANCHEZ-LAORDEN B, O'BRIEN K, BRUNTON H, FERGUSON J, YOUNG H, DHOMEN N, FLAHERTY KT, FREDERICK DT, COOPER ZA, WARGO JA, MARAIS R, WELLBROCK C. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 2014;4:1214–29. doi: 10.1158/2159-8290.CD-13-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOSMAN JA, KIM KB, SCHUCHTER L, GONZALEZ R, PAVLICK AC, WEBER JS, MCARTHUR GA, HUTSON TE, MOSCHOS SJ, FLAHERTY KT, HERSEY P, KEFFORD R, LAWRENCE D, PUZANOV I, LEWIS KD, AMARAVADI RK, CHMIELOWSKI B, LAWRENCE HJ, SHYR Y, YE F, LI J, NOLOP KB, LEE RJ, JOE AK, RIBAS A. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STARK MS, BONAZZI VF, BOYLE GM, PALMER JM, SYMMONS J, LANAGAN CM, SCHMIDT CW, HERINGTON AC, BALLOTTI R, POLLOCK PM, HAYWARD NK. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015;6:17753–63. doi: 10.18632/oncotarget.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRAUSSMAN R, MORIKAWA T, SHEE K, BARZILY-ROKNI M, QIAN ZR, DU J, DAVIS A, MONGARE MM, GOULD J, FREDERICK DT, COOPER ZA, CHAPMAN PB, SOLIT DB, RIBAS A, LO RS, FLAHERTY KT, OGINO S, WARGO JA, GOLUB TR. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUBRAMANIAN S, LUI WO, LEE CH, ESPINOSA I, NIELSEN TO, HEINRICH MC, CORLESS CL, FIRE AZ, VAN DE RIJN M. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- SUN C, WANG L, HUANG S, HEYNEN GJ, PRAHALLAD A, ROBERT C, HAANEN J, BLANK C, WESSELING J, WILLEMS SM, ZECCHIN D, HOBOR S, BAJPE PK, LIEFTINK C, MATEUS C, VAGNER S, GRERNRUM W, HOFLAND I, SCHLICKER A, WESSELS LF, BEIJERSBERGEN RL, BARDELLI A, DI NICOLANTONIO F, EGGERMONT AM, BERNARDS R. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- SUN X, LI J, SUN Y, ZHANG Y, DONG L, SHEN C, YANG L, YANG M, LI Y, SHEN G, TU Y, TAO J. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016 doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN YM, LIN KY, CHEN YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- URA H, OBARA T, NISHINO N, TANNO S, OKAMURA K, NAMIKI M. Cytotoxicity of simvastatin to pancreatic adenocarcinoma cells containing mutant ras gene. Jpn J Cancer Res. 1994;85:633–8. doi: 10.1111/j.1349-7006.1994.tb02406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERGANI E, DI GUARDO L, DUGO M, RIGOLETTO S, TRAGNI G, RUGGERI R, PERRONE F, TAMBORINI E, GLOGHINI A, ARIENTI F, VERGANI B, DEHO P, DE CECCO L, VALLACCHI V, FRATI P, SHAHAJ E, VILLA A, SANTINAMI M, DE BRAUD F, RIVOLTINI L, RODOLFO M. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. 2016;7:4428–41. doi: 10.18632/oncotarget.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VILLANUEVA J, VULTUR A, LEE JT, SOMASUNDARAM R, FUKUNAGAKALABIS M, CIPOLLA AK, WUBBENHORST B, XU X, GIMOTTY PA, KEE D, SANTIAGO-WALKER AE, LETRERO R, D'ANDREA K, PUSHPARAJAN A, HAYDEN JE, BROWN KD, LAQUERRE S, MCARTHUR GA, SOSMAN JA, NATHANSON KL, HERLYN M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WESTPHAL D, KLUCK RM, DEWSON G. Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 2014;21:196–205. doi: 10.1038/cdd.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITTAKER SR, THEURILLAT JP, VAN ALLEN E, WAGLE N, HSIAO J, COWLEY GS, SCHADENDORF D, ROOT DE, GARRAWAY LA. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU N, ZHANG L, MEISGEN F, HARADA M, HEILBORN J, HOMEY B, GRANDER D, STAHLE M, SONKOLY E, PIVARCSI A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287:29899–908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YE Y, LI Q, HU WL, TSENG HY, JIN L, ZHANG XD, ZHANG LJ, YANG S. Loss of PI(4,5)P2 5-Phosphatase A Contributes to Resistance of Human Melanoma Cells to RAF/MEK Inhibitors. Transl Oncol. 2013;6:470–81. doi: 10.1593/tlo.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU M, LIU Z, ZHAO Y, DING Y, LIU H, XI Y, XIONG W, LI G, LU J, FODSTAD O, RIKER AI, TAN M. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.