ABSTRACT
Amoebae are unicellular eukaryotes that consume microbial prey through phagocytosis, playing a role in shaping microbial food webs. Many amoebal species can be cultivated axenically in rich media or monoxenically with a single bacterial prey species. Here, we characterize heterolobosean amoeba LPG3, a recent natural isolate, which is unable to grow on unicellular cyanobacteria, its primary food source, in the absence of a heterotrophic bacterium, a Pseudomonas species coisolate. To investigate the molecular basis of this requirement for heterotrophic bacteria, we performed a screen using the defined nonredundant transposon library of Vibrio cholerae, which implicated genes in corrinoid uptake and biosynthesis. Furthermore, cobalamin synthase deletion mutations in V. cholerae and the Pseudomonas species coisolate do not support the growth of amoeba LPG3 on cyanobacteria. While cyanobacteria are robust producers of a corrinoid variant called pseudocobalamin, this variant does not support the growth of amoeba LPG3. Instead, we show that it requires cobalamin that is produced by the Pseudomonas species coisolate. The diversity of eukaryotes utilizing corrinoids is poorly understood, and this amoebal corrinoid auxotroph serves as a model for examining predator-prey interactions and micronutrient transfer in bacterivores underpinning microbial food webs.
IMPORTANCE Cyanobacteria are important primary producers in aquatic environments, where they are grazed upon by a variety of phagotrophic protists and, hence, have an impact on nutrient flux at the base of microbial food webs. Here, we characterize amoebal isolate LPG3, which consumes cyanobacteria as its primary food source but also requires heterotrophic bacteria as a source of corrinoid vitamins. Amoeba LPG3 specifically requires the corrinoid variant produced by heterotrophic bacteria and cannot grow on cyanobacteria alone, as they produce a different corrinoid variant. This same corrinoid specificity is also exhibited by other eukaryotes, including humans and algae. This amoebal model system allows us to dissect predator-prey interactions to uncover factors that may shape microbial food webs while also providing insight into corrinoid specificity in eukaryotes.
KEYWORDS: amoeba, corrinoids, microbial interactions, vitamin B12
INTRODUCTION
Unicellular organisms dominate the eukaryotic phylogenetic tree of life. Although protists make up a significant part of natural microbial communities, they have been largely overlooked and are understudied as simple model eukaryotes (1, 2). While culture-independent metagenomic studies provide insight into the abundance and natural diversity of protists in environmental settings (3), in-depth characterization requires the isolation and cultivation of individual species. A large range of eukaryotic microalgal species have been cultivated and studied, particularly as a source of renewable biofuel compounds in recent years (4, 5); however, besides microalgae, there are only a few protist microbes developed as model systems. Amoebae are a morphologically defined group of phylogenetically diverse single-celled eukaryotes that generally feed by phagocytosis of other microbes. They make up the Amoebozoa supergroup, the Heterolobosea class of the Excavata supergroup, and the Rhizaria of the SAR (stramenopiles, alveolates, and Rhizaria) supergroup (6). Model amoebal species, including Dictyostelium discoideum and various Acanthamoeba species, are contained within the Amoebozoa supergroup. These model amoebae can be grown axenically in rich undefined media but can also be cultivated monoxenically with a range of different bacterial prey species as a food source. Phagotrophic protists play an important role in shaping microbial communities (7, 8), and insight into their role in nutrient turnover will facilitate our understanding of microbial food webs.
Prime examples of micronutrient concentration up food webs are B vitamins (9), which include corrinoids that are only synthesized de novo by certain prokaryotes, including some archaeal species (10). Corrinoids are a family of related molecules that include vitamin B12 and are obligatorily or facultatively utilized by many organisms, including animals, algae, and bacteria (10, 11). Corrinoids comprise a porphyrin ring that coordinates a central cobalt ion (12) with variable upper and lower axial ligands (13, 14). The most well-studied corrinoid variants are cobalamins, with dimethylbenzimidazole (DMB) as the lower axial ligand, and which include vitamin B12 (or cyanocobalamin, Cbl), which has a CN group as the upper axial ligand. Major sites of corrinoid production are soil and aquatic habitats and within animal gut environments (15). Heterotrophic bacteria, cyanobacteria, and archaea are major sources of corrinoids in marine environments (9, 16–18). Certain cyanobacterial strains were recently demonstrated to produce the corrinoid variant pseudocobalamin de novo (19), with adenine as the lower axial ligand, while other cyanobacterial strains require corrinoid supplementation for growth (20, 21).
Cyanobacteria are significant primary producers in marine and freshwater ecosystems. As such, grazing on cyanobacteria is a major process at the base of food webs in these environments, and amoebae are among the natural grazers of cyanobacteria (22–24). As part of our efforts to understand the grazing process, we recently isolated a diverse set of amoebae that can be propagated using cyanobacterial species as prey (25). Characterization of one of these isolates showed that cyanobacterial surface properties affected amoebal grazing (26). While most of these amoebal isolates can be grown monoxenically on cyanobacteria (Synechococcus elongatus or Leptolyngbya sp. strain BL0902), an exception was the heterolobosean amoeba LPG3. It is phylogenetically distant from model amoebal species and is placed within the Heterolobosea class of the Excavata supergroup, which also includes Naegleria pathogens. Despite attempts to establish monoxenic growth with only cyanobacteria, heterotrophic bacteria were always present on stock plates of amoeba LPG3. Here, we show that amoeba LPG3 requires both the heterotrophic bacterial coisolate and cyanobacteria for growth. Using bacterial genetics and supplementation studies, we demonstrate the requirement of amoeba LPG3 for the corrinoid variant cobalamin produced by heterotrophic bacterial prey and, furthermore, that the pseudocobalamin variant produced by the cyanobacterial prey cannot support growth of amoeba LPG3. This description of a corrinoid requirement by an amoeba reflects the intricate nature of protist grazing in complex multispecies environments and contributes to our understanding of micronutrient transfer in food webs.
RESULTS
Amoeba LPG3 requires cyanobacteria and heterotrophic bacteria for growth.
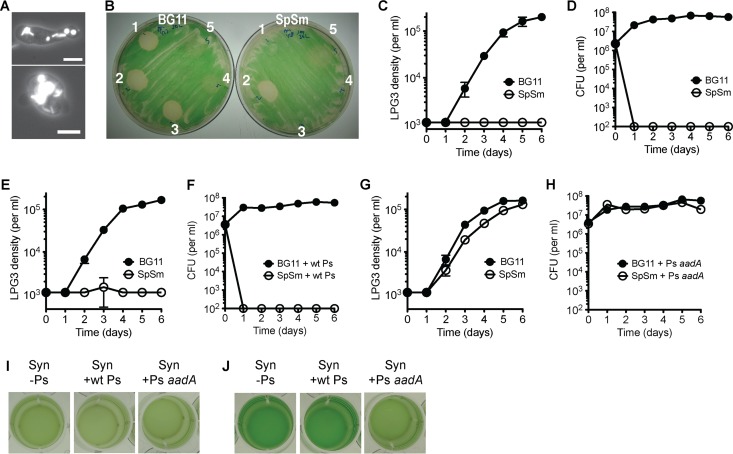
Amoeba LPG3 was isolated using unicellular Synechococcus elongatus PCC 7942 as a food source (Fig. 1A), and cultures harbored a persistent heterotrophic bacterial coisolate that was required for the propagation of LPG3 on S. elongatus lawns. This bacterium was isolated by streaking to obtain a single colony and was grown in pure culture. To further characterize the bacterial coisolate, the genome was sequenced. The bacterial coisolate was determined to be a Pseudomonas species, closely related to Pseudomonas mendocina and Pseudomonas pseudoalcaligenes (see Fig. S1 in the supplemental material). This Pseudomonas isolate was named LPH1 for lily pond heterotroph 1. To determine whether Pseudomonas isolate LPH1 was sufficient to support the growth of amoeba LPG3 on S. elongatus lawns, it was transformed with a plasmid conferring resistance to the antibiotics streptomycin and spectinomycin (aadA). Upon spotting amoeba LPG3 with Pseudomonas isolate LPH1 expressing aadA onto S. elongatus lawns (Fig. 1B, spot 2), amoebal plaque formation occurred on solid media with and without supplemented antibiotics, and the presence of amoebal cells was confirmed by microscopy. However, when amoeba LPG3 alone was spotted on cyanobacterial lawns or with wild-type (wt) Pseudomonas isolate LPH1 (Fig. 1B, spots 1 and 3), plaque formation occurred only on the medium without antibiotics. Wild-type Pseudomonas isolate LPH1 and Pseudomonas isolate LPH1 expressing aadA spotted alone did not cause plaque formation (Fig. 1B, spots 4 and 5), although there was initially a slight growth inhibition of S. elongatus. This demonstrates that the bacterial coisolate carried over with LPG3 cells is required for amoebal replication on cyanobacterial lawns and that, in the presence of antibiotics, the addition of the Pseudomonas coisolate expressing aadA was sufficient for supporting LPG3 growth.
FIG 1.
Natural amoebal isolate LPG3 requires both cyanobacterial and heterotrophic bacterial prey for optimal growth. (A) Fluorescence images of LPG3 cells showing ingested S. elongatus (Syn) cells, visible by chlorophyll autofluorescence. Scale bar, 10 μm. (B) Amoebal plaque formation on Syn lawns without antibiotics (left, BG11) and with antibiotics streptomycin and spectinomycin (right, SpSm) spotted with (1) LPG3 only, (2) LPG3 and Pseudomonas isolate LPH1 expressing aadA (Ps aadA), (3) LPG3 and wild-type Pseudomonas isolate LPH1 (wt Ps), (4) wt Ps only, and (5) Ps aadA only. (C to H) Amoebal growth curves and Pseudomonas CFU with Syn in BG11 medium with and without supplemented antibiotics (SpSm), with no added Pseudomonas (C, D), the addition of wild-type Pseudomonas (E, F), or Ps aadA (G, H). Values are averages of triplicate wells plus or minus standard errors of the mean (SEM). (I, J) Images showing Syn density at day 7 in BG11 (I) or BG11 with SpSm (J).
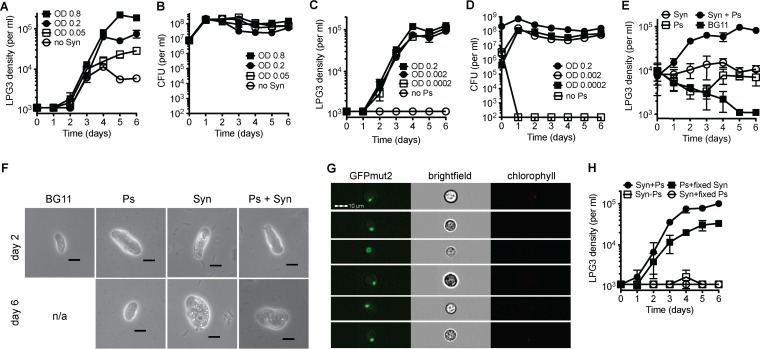
This requirement of amoeba LPG3 for Pseudomonas isolate LPH1 was also examined in liquid culture growth experiments, where the Pseudomonas cells carried over with the LPG3 cells can survive and support amoebal growth only in the absence of antibiotics (Fig. 1C and D). Wild-type LPH1 that was added to the wells could not survive antibiotic treatment and failed to support amoebal growth in the presence of antibiotics (Fig. 1E and F), while LPH1 expressing aadA survived antibiotic treatment and supported amoebal growth in the presence and absence of antibiotics (Fig. 1G and H). While the cyanobacterial culture had a noticeable decrease in density in the wells with growing LPG3 cells compared to wells without amoebal growth (Fig. 1I and J), the concentration of surviving Pseudomonas isolate LPH1 was steady over the course of the experiment. This suggests that the major food source of amoeba LPG3 is the S. elongatus cyanobacterium and not the Pseudomonas heterotroph. Indeed, by altering cyanobacterial concentrations with fixed Pseudomonas isolate LPH1 concentrations, amoebal growth varied (Fig. 2A and B). Various Pseudomonas isolate LPH1 concentrations with fixed S. elongatus concentrations did not affect amoebal growth rates (Fig. 2C); however, this result is confounded by the rapid growth of Pseudomonas isolate LPH1 to steady-state concentrations (Fig. 2D).
FIG 2.
S. elongatus is the primary food source for amoeba LPG3. Amoebal growth curves and Pseudomonas isolate LPH1 aadA (Ps) cell density in CFU in BG11 containing SpSm, with various S. elongatus (Syn) concentrations and a fixed Ps concentration (A, B) and various Ps concentrations with a fixed Syn concentration (C, D). (E) LPG3 growth with the addition of Syn only, Ps aadA only, both, or BG11 control. (F) LPG3 cells cultured on Syn only, Ps aadA only, both (Syn + Ps), or none (BG11) and imaged on day 2 and 6. In BG11 samples, no LPG3 cells were recovered on day 6. Scale bar, 10 μm. (G) Amoeba LPG3 was coincubated with GFPmut2-expressing Pseudomonas in the absence of Syn, and images were acquired using an imaging flow cytometer. Images in the chlorophyll channel are also shown to ensure that the GFPmut2 signal is not due to cyanobacterial autofluorescence. Scale bar, 10 μm. (H) Amoebal growth with the addition of formaldehyde-fixed Syn or formaldehyde-fixed Ps aadA. Values are averages of triplicate wells plus or minus SEM.
Amoeba LPG3 was evaluated for survival on single bacterial species. When provided with S. elongatus or Pseudomonas isolate LPH1 alone, it could only grow up to or survive at densities of up to 104 per milliliter (Fig. 2E), and attempts at long-term culturing on either strain alone failed. LPG3 cells maintained their size when grown on S. elongatus alone, which is comparable to cells grown with both S. elongatus and Pseudomonas isolate LPH1, and there was an increase in the number of small bright vacuoles (Fig. 2F). Despite being able to ingest Pseudomonas isolate LPH1 (Fig. 2G), LPG3 cells became smaller when grown on Pseudomonas isolate LPH1 alone (Fig. 2F), which is comparable to cells that were cultured in only BG11 medium. Additionally, formaldehyde-fixed Pseudomonas isolate LPH1 could not support amoebal growth, while formaldehyde-fixed S. elongatus cells could (Fig. 2H), suggesting that Pseudomonas isolate LPH1 provides amoeba LPG3 with a critical factor that is sensitive to fixation or that must be synthesized continuously.
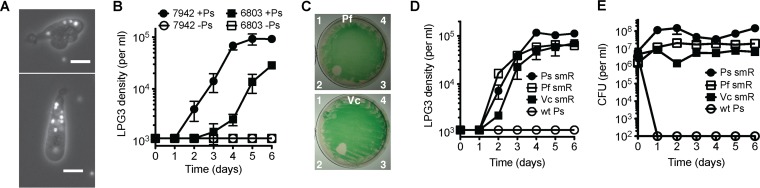
To determine whether the requirement of amoeba LPG3 for both cyanobacteria and heterotrophic bacteria was due to a particular deficiency in S. elongatus or a unique trait in Pseudomonas isolate LPH1, additional cyanobacterial and heterotrophic bacterial species were tested. Synechocystis sp. PCC 6803 is another unicellular freshwater cyanobacterial species that is a widely used model organism. Amoeba LPG3 can ingest PCC 6803 cells (Fig. 3A) and grow on PCC 6803 as a food source, and this also required the presence of Pseudomonas isolate LPH1 (Fig. 3B). Examination of other heterotrophic bacterial species showed that Pseudomonas fluorescens and Vibrio cholerae can also support LPG3 growth on S. elongatus lawns and in liquid culture (Fig. 3C and D). Similar to Pseudomonas isolate LPH1, concentrations of P. fluorescens and V. cholerae remained relatively steady over the course of the experiment (Fig. 3E).
FIG 3.
Amoeba LPG3 growth supported by other cyanobacterial and heterotrophic bacterial species. (A, B) Synechocystis sp. PCC 6803 supported the growth of amoeba LPG3. (A) Amoeba LPG3 fluorescence images showing chlorophyll autofluorescence from ingested PCC 6803 cells. Scale bar, 10 μm. (B) LPG3 growth curves with Synechocystis PCC 6803 and S. elongatus PCC 7942 controls with and without the addition of Pseudomonas isolate LPH1 aadA (Ps). (C to E) P. fluorescens and V. cholerae also support LPG3 growth. (C) S. elongatus lawns on BG11 streptomycin plates were spotted with (1) LPG3 only, (2) LPG3 with streptomycin-resistant P. fluorescens (Pf smR, top) or streptomycin-resistant V. cholerae (Vc smR, bottom), (3) BG11 medium only, and (4) Pf smR only (top) or Vc smR only (bottom). (D) Amoebal growth with S. elongatus in liquid culture with the addition of Pf smR, Vc smR, or Ps smR controls with (E) corresponding CFU of heterotrophic bacteria. Values are averages of triplicate wells plus or minus SEM.
Genetic screen identifies requirement of corrinoid biosynthesis in heterotrophic bacteria to support amoebal growth.
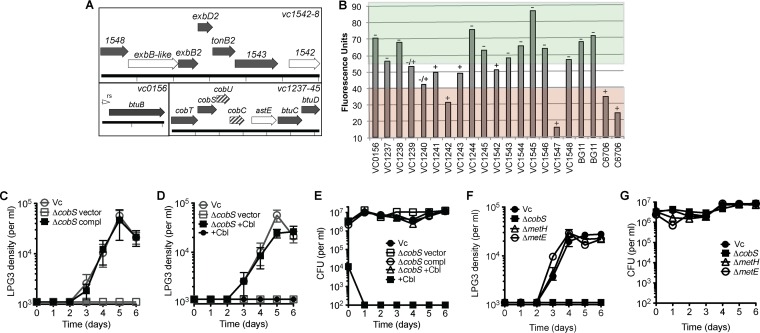
To investigate the molecular basis for the requirement of amoeba LPG3 for heterotrophic bacteria, a screen was performed using a defined nonredundant transposon library of V. cholerae (27). Mutants that could not support amoebal growth on solid medium and in liquid culture were identified (Fig. 4A and B). Transposon insertions clustered to corrinoid uptake and biosynthesis genes. A transposon mutant with a disrupted corrinoid receptor gene btuB was unable to support amoebal growth as were transposon insertion mutants in associated inner membrane importer genes btuCD. BtuBCD requires TonB to provide energy to import molecules, and transposon insertion mutants in tonB2 and associated genes exbBD were also impaired. Also identified in the screen were corrinoid biosynthesis genes cobCUST, with partial phenotypes for cobC and cobU. Cobalamin synthase catalyzes the last step of corrinoid biosynthesis through addition of the lower ligand to the aminopropanol arm of the corrin ring. We constructed a V. cholerae deletion mutant of cobalamin synthase (cobS in Salmonella nomenclature), and this mutant was deficient for supporting amoebal growth. The mutation could be complemented genetically, and examination of bacterial CFU showed concentrations comparable to the wild type (Fig. 4C and E). Amoebal growth with the ΔcobS strain could also be rescued through supplementation of the medium with the corrinoid compound cyanocobalamin (Cbl, vitamin B12) with no effects on the growth of the ΔcobS mutant, but supplementation with cyanocobalamin alone did not rescue amoebal growth (Fig. 4D and E). V. cholerae carries metH and metE, corrinoid-dependent and corrinoid-independent methionine synthases, respectively. To determine whether MetH activity is required to support amoebal growth, ΔmetH and ΔmetE strains of V. cholerae were constructed. Analysis of these mutants showed that they both fully support amoebal growth in the presence of S. elongatus (Fig. 4F and G).
FIG 4.
Corrinoid uptake and remodeling genes are required for V. cholerae support of amoebal growth. (A) V. cholerae genes required (filled arrows), partially required (striped arrows), and not required (open arrows) for supporting amoebal growth on S. elongatus. Cobalamin biosynthesis gene names are assigned using Salmonella nomenclature. vc1241 and vc1243 (flanking astE, not depicted) are not required, and the predicted riboswitch is indicated (rs). (B) Amoeba LPG3 liquid culture screen with V. cholerae transposon mutant library. Cyanobacterial autofluorescence was measured, and LPG3 cell density was scored as follows: −, few or no LPG3 cells; −/+, intermediate number of LPG3 cells; and +, many LPG3 cells. Wells with fluorescence values above 55 did not support amoebal growth (green), wells with fluorescence values below 40 fully supported amoebal growth (red), and wells with intermediate values were scored based on the density of LPG3 cells. Data for two replicates (each) are shown for BG11 and C6706. (C to E) Rescue of V. cholerae cobalamin synthase deletion mutant (vc1238, ΔcobS) for supporting amoebal growth on S. elongatus by genetic complementation (C) and supplementation with cyanocobalamin (Cbl) (D), with corresponding CFU of V. cholerae strains (E). Gray indicates the same growth curve controls. (F) Amoebal growth curves with S. elongatus and V. cholerae mutants of B12-dependent methionine synthase (ΔmetH) and B12-independent methionine synthase (ΔmetE) with corresponding CFU from assay wells (G). Values are averages of triplicate wells plus or minus SEM.
Amoebal growth is supported by cobalamin but not pseudocobalamin.
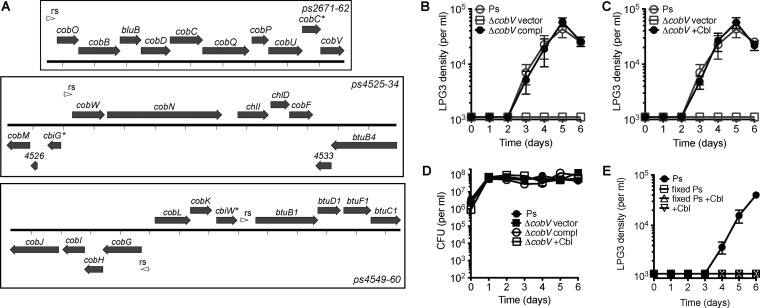
In contrast to V. cholerae, the genome of Pseudomonas isolate LPH1 contains all of the genes necessary for de novo corrinoid biosynthesis (Fig. 5A). We constructed a deletion mutant of the cobalamin synthase gene (cobV in the Pseudomonas denitrificans nomenclature) in Pseudomonas isolate LPH1, and this strain was deficient in supporting amoebal growth. This mutant can be complemented genetically (Fig. 5B and D), and amoebal growth was rescued through supplementation with cyanocobalamin (Fig. 5C). As was the case for the V. cholerae ΔcobS strain, supplementation with cyanocobalamin alone was not sufficient to support LPG3 growth on S. elongatus. To determine whether additional factors from Pseudomonas isolate LPH1 were required, amoebal growth curves were performed in the presence of S. elongatus, supplemented cyanocobalamin, and fixed Pseudomonas isolate LPH1. However, the addition of fixed Pseudomonas isolate LPH1 was not sufficient to rescue amoebal growth on S. elongatus with supplemented cyanocobalamin (Fig. 5E).
FIG 5.
Pseudomonas isolate LPH1 corrinoid biosynthesis gene is required for support of amoebal growth. (A) Pseudomonas isolate LPH1 (Ps) cobalamin biosynthesis genes using Pseudomonas denitrificans nomenclature with predicted riboswitches indicated. cobC*, cbiG*, and cbiW* refer to homologs of Salmonella enterica. For clarity, ps4553 (flanked by a predicted riboswitch and cobL) is not depicted. (B to D) Rescue of Pseudomonas isolate LPH1 cobalamin synthase deletion mutant (ps2662::aacC1, ΔcobV) for supporting amoebal growth on S. elongatus by genetic complementation (B) and supplementation with cyanocobalamin (Cbl) (C), with corresponding CFU from assay wells (D). Gray indicates the same growth curve controls. (E) Amoebal growth curves with S. elongatus, with the addition of supplemented cyanocobalamin (+Cbl) and fixed Pseudomonas isolate LPH1. Values are averages of triplicate wells plus or minus SEM.
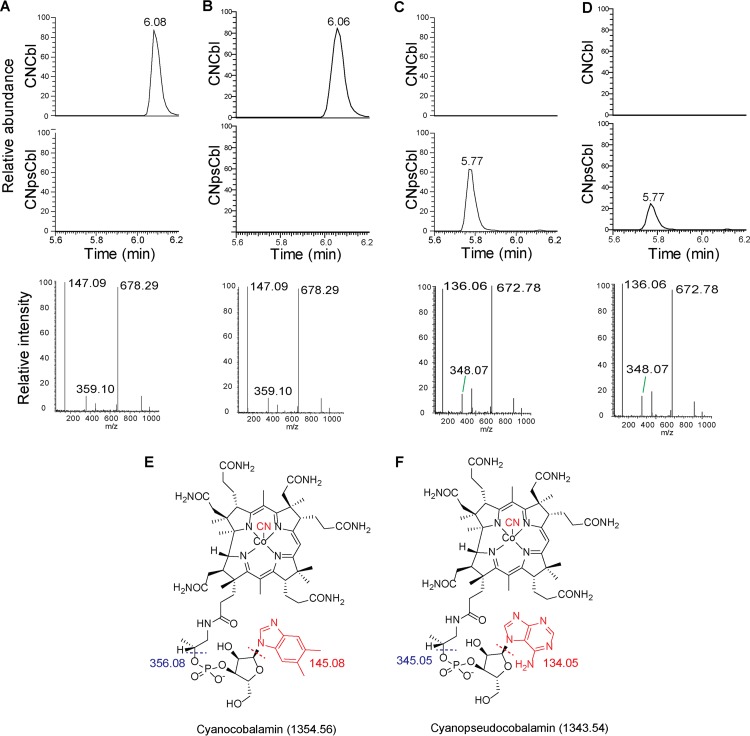
Analysis of lysates of Pseudomonas isolate LPH1 by ultra-high-performance liquid chromatography mass spectrometry (UPLC-MS) showed that it produces cobalamin, with DMB as the lower axial ligand (Fig. 6A and E), which is consistent with the purchased standard (Fig. 6B). In contrast to Pseudomonas isolate LPH1, analysis of lysates by UPLC-MS showed that S. elongatus PCC 7942 (Fig. 6C and F) and Synechocystis sp. PCC 6803 (Fig. 6D) produce pseudocobalamin, with adenine as the lower axial ligand as has been previously observed (19).
FIG 6.
Pseudomonas isolate LPH1 and cyanobacteria produce different corrinoid variants. (A to D) UPLC-MS analysis of Pseudomonas isolate LPH1 (A), cyanocobalamin standard (B), S. elongatus (C), and Synechocystis sp. PCC 6803 (D). UPLC traces corresponding to cyanocobalamin (CNCbl, top) or cyanopseudocobalamin (CNpsCbl, middle), with respective mass spectra (bottom). Structures of cyanocobalamin (E) and cyanopseudocobalamin (F) with predicted molecular weights of associated fragments. For cyanocobalamin mass spectra, the 678.29 peak corresponds to doubly charged intact cyanocobalamin (678.29 × 2 − 2 = 1,354.58), the 147.09 peak corresponds to a protonated DMB base fragment (147.09 − 2 = 145.09), and the 359.10 peak corresponds to a protonated DMB base and linker (359.10 − 3 = 356.10). For cyanopseudocobalamin, the 672.78 peak corresponds to doubly charged intact cyanopseudocobalamin (672.78 × 2 − 2 = 1,343.56), the 136.06 peak corresponds to a protonated adenine base fragment (136.06 − 2 = 134.06), and the 348.07 peak corresponds to a protonated adenine base and linker (348.07 − 3 = 345.07). These observed m/z values have been reported (19, 43).
DISCUSSION
This work describes an amoebal model system that requires cyanobacterial and heterotrophic bacterial prey species to sustain the growth of the amoebal grazer, with corrinoids playing a major role. Corrinoids act as cofactors in specific enzymatic reactions (28) and regulate gene expression through a variety of mechanisms, including modulating translation in human cells (29), transcriptional repression via a photoreceptor (30), and riboswitches in bacteria (31). Approximately half of all cultured eukaryotic algal species from marine and freshwater environments are corrinoid auxotrophs (11). Many algal species from diverse lineages, including green algae, diatoms, and alveolates, are associated with bacteria that supply corrinoids to them (32, 33). Analysis of sequenced algal genomes showed a strong correlation between corrinoid auxotrophy and loss of corrinoid-independent methionine synthase METE, such that these algae require corrinoids to support the essential activity of corrinoid-dependent methionine synthase METH (34). While it is unknown if the coisolation of Pseudomonas isolate LPH1 with amoeba LPG3 is simply incidental, it has been proposed in the “foraging-to-farming” hypothesis that grazers may evolve mutualistic relationships with their prey, resulting in an obligate association (33). Indeed, farming behavior of some natural D. discoideum isolates has been described (35).
Critical to our work was the identification of corrinoid biosynthesis genes through the screening of a V. cholerae transposon library. Although V. cholerae does not contain a complete de novo corrinoid biosynthesis pathway, the cob genes that it does carry are associated with corrinoid salvaging and remodeling, in which incomplete or complete corrinoids are imported and modified through the addition of the lower axial ligand. It appears that V. cholerae can import mature corrinoids or their precursors produced by cyanobacteria and remodel them, thereby supporting amoebal growth. Methionine synthesis via B12-dependent MetH is a widely conserved corrinoid-dependent process. As with many organisms, the only corrinoid-dependent enzyme identified in V. cholerae and Pseudomonas isolate LPH1 is MetH. Inactivation of metH in a V. cholerae deletion mutant still supported amoebal growth, suggesting that amoeba LPG3 requires cobalamin itself and not products of corrinoid-dependent processes within the heterotrophic bacteria. While supplementation with cyanocobalamin in the absence of the heterotrophic bacteria was not sufficient to relieve the corrinoid auxotrophy of amoeba LPG3, additional factors produced by the heterotrophic bacteria may be required by amoeba LPG3. The addition of fixed Pseudomonas isolate LPH1 to cobalamin supplementation studies did not rescue amoebal growth, although the putative additional factors may be sensitive to fixation or may require continuous synthesis. Alternatively, the failure of cobalamin supplementation in rescuing the growth of amoeba LPG3 may be due to the bacterivorous strategy of nutrient acquisition in amoebae.
After ingestion of bacterial prey by amoebal cells, the phagocytic vacuoles mature into digestive food vacuoles, and nutrients must be extracted through the vacuolar membrane to the amoebal cell cytosol. This is reminiscent of cobalamin trafficking within humans, in which circulating cobalamin bound to transcobalamin II is recognized by cellular receptors and gets endocytosed and released within the lysosome, followed by transport across the lyso-endosomal membrane (36, 37). In amoebae, however, nutrients like cobalamin may not be transported directly from the external medium due to the lack of specific cellular receptors. Indeed, the axenic growth of the well-studied amoebal model D. discoideum is facilitated by mutations that promote macropinocytosis, while the parental strain was unable to grow in the absence of bacterial prey (38). Because the recently isolated LPG3 requires heterotrophic bacterial prey for growth in addition to cyanobacterial prey, it may require other factors produced by Pseudomonas isolate LPH1 and V. cholerae in addition to cobalamin. However, the requirement for heterotrophic bacteria in the cobalamin supplementation assays is likely due to the amoebal nutrient trafficking pathway in which bacteria that can produce or import cobalamin must be ingested through phagocytosis in order for amoeba LPG3 to gain access to the required cobalamin.
Our analysis of corrinoid variant production in Pseudomonas isolate LPH1 and cyanobacteria reveals molecular specificity of amoeba LPG3 for cobalamin over pseudocobalamin. Differences in corrinoid biosynthesis in Pseudomonas isolate LPH1 and freshwater cyanobacteria provide insight into the amoebal growth phenotypes observed. The canonical pathway for aerobic production of cobalamins, with DMB as the lower axial ligand, is based on the Pseudomonas denitrificans pathway (39) that includes the bluB (40) and cobU genes (homologous to Salmonella cobT) (41), which produce and activate DMB substrates, respectively, for addition as the lower axial ligand in cobalamin production. While S. elongatus contains most of the same corrinoid biosynthesis genes as Pseudomonas isolate LPH1, many of which are essential to the cyanobacterium (42), it critically lacks bluB and cobU, suggesting that it is does not produce cobalamins. Indeed, it has recently been shown that marine and freshwater cyanobacteria produce a corrinoid variant called pseudocobalamin, with adenine as the lower axial ligand (19). Although LPG3 grazes on these cyanobacterial species that robustly produce pseudocobalamin, its requirement for heterotrophic bacteria that produce cobalamin suggests that it has specificity for cobalamin and cannot utilize pseudocobalamin. While pseudocobalamin is produced by many bacterial species, including cyanobacteria (19), Lactobacillus reuteri (43), and Clostridium cochlearium (44), other eukaryotes that utilize corrinoids also have specificity for cobalamin over pseudocobalamin. The human intrinsic factor protein binds cobalamin and facilitates absorption across the small intestine and has specificity for cobalamin over pseudocobalamin (45). While most supplementation studies are performed with cobalamin, recent analyses were performed with pseudocobalamin supplementation to eukaryotic algae (18, 19). Pseudocobalamin generally supported growth poorly compared to cobalamin, although the difference was slight for some organisms. Bacterial specificity for different corrinoid variants is also likely to occur and may be satisfied through corrinoid remodeling pathways for lower ligand exchange (46). Remarkably, corrinoid remodeling of pseudocobalamin to cobalamin was also observed in certain eukaryotic algal species when supplied with DMB (19). There are likely many more examples of corrinoid specificity in a variety of organisms in a range of biological settings, and these molecular details can be elucidated through the high-precision analysis of corrinoid variants of organisms grown in well-defined culturing conditions.
This work contributes to our understanding of the breadth of eukaryotic organisms that require corrinoids and further indicates that the isolation and cultivation of amoebal species using single bacterial prey species may obscure complex multispecies interactions, preventing cultivation of amoebal strains with certain auxotrophies. Amoeba LPG3 consumes primary producing cyanobacteria as its main food source and heterotrophic bacteria to provide corrinoid micronutrients. While vitamin B12 (cyanocobalamin) is typically included in vitamin supplements for axenic cultivation of amoebae in defined medium preparations (47), it is often not required by the amoebae (48, 49). Additionally, model amoebal species D. discoideum and Acanthamoeba castellanii can be cultivated monoxenically with bacterial strains that do not produce cobalamins de novo (50, 51), although trace amounts of cobalamins may be present. While there are likely many uncultured organisms with corrinoid auxotrophies, the facultative utilization of corrinoids is beginning to be appreciated in model eukaryotic organisms. Facultative corrinoid utilization may occur in D. discoideum because it carries all three known eukaryotic vitamin B12-dependent enzymes: methionine synthase METH, methylmalonyl coenzyme A (CoA) mutase, and a class II ribonucleotide reductase (52). Corrinoid utilization was only recently discovered in the well-studied model organism Caenorhabditis elegans, in which the vitamin accelerates development although at the cost of decreased fertility (53). Because corrinoids are known to be synthesized de novo by only some prokaryotic species, they are excellent compounds for examining the cross-feeding of mature and incomplete vitamin cofactors in bacterial interspecies (54) and microbial interactions. The identification of an amoeba that is auxotrophic for corrinoids adds to our understanding of the breadth of corrinoid usage and specificity in eukaryotes, and future work identifying corrinoid-dependent enzymes in LPG3 will provide insight into its usage of cobalamin.
MATERIALS AND METHODS
Strain cultivation.
Pseudomonas isolate LPH1, Vibrio cholerae C6706, Pseudomonas fluorescens NCIMB 10586, and Escherichia coli strains DH5α λpir and SM10 λpir were grown in LB medium at 37°C. Antibiotics were supplemented to LB as necessary at the following concentrations: ampicillin at 100 μg/ml, streptomycin at 100 μg/ml, streptomycin and spectinomycin at 20 μg of each per ml, and gentamicin at 25 μg/ml. Cyanobacterial strains Synechococcus elongatus PCC 7942 and Synechocystis sp. PCC 6803 were grown in BG11 medium (55) at 30°C under continuous illumination at 50 to 100 μmol photons m−2 s−1. Cyanobacterial stocks were routinely plated to BG11 Omni plates (BG11 with 0.04% glucose and 5% LB) and tested negative for contamination. Amoeba LPG3 was isolated and maintained on S. elongatus PCC 7942 lawns in the absence of antibiotics as described previously (25).
Strain construction.
The V. cholerae in-frame deletion mutants of cobalamin synthase (cobS, vc1238), corrinoid-dependent methionine synthase (metH, vc0390), and corrinoid-independent methionine synthase (metE, vc1704) were constructed through allelic exchange (56) using suicide plasmid pWM91. A complementation plasmid was constructed in the pBAD24 vector. The Pseudomonas isolate LPH1 cobalamin synthase (cobV) deletion mutant was constructed through gene replacement with an aacC1 cassette (57). A complementation plasmid was constructed in an RSF1010 vector bearing an aadA cassette (conferring resistance to streptomycin and spectinomycin) and a PconII promoter with translational regulation via theophylline riboswitch variant E (58). Both complementing plasmids have baseline expression under noninducing conditions, and this expression was sufficient for complementation of cobalamin synthase mutations. All plasmids were constructed using Seamless (Thermo Fisher) or HiFi (New England BioLabs) DNA assembly kits.
Amoebal growth assays.
For amoebal growth assays, LPG3 cells were harvested directly from stocks maintained on S. elongatus lawns by scraping cells at the plaque interface, or LPG3 cells were first pregrown in liquid culture in 6-well microtiter dishes containing BG11 and wild-type S. elongatus. BG11 medium was supplemented with 5 μg/ml streptomycin and spectinomycin or 5 μg/ml streptomycin alone. Agar plates or microtiter dishes were incubated at 30°C under continuous illumination at 2 to 3 μmol photons m−2 s−1. For assays on solid media, cyanobacterial lawns were plated on 10-cm BG11 agar plates supplemented with antibiotics or control plates lacking antibiotics. One hundred microliters of S. elongatus expressing aadA concentrated to an optical density at 750 nm (OD750) of 5 was spread per plate. LPG3 cells were added at 900 to 2,000 per spot, and 5 μl of heterotrophic bacteria (Pseudomonas isolate LPH1 with and without aadA expression, streptomycin-resistant V. cholerae C6706, or streptomycin-resistant P. fluorescens NCIMB 10586) normalized to an OD600 of 0.01 was spotted. After 10 to 14 days, plates were imaged over a white light box using a Sony Cyber-shot camera. LPG3 growth experiments were performed in liquid culture in 24-well flat-bottom plates with a 2-ml volume per well of BG11 supplemented with antibiotics or control experiments lacking antibiotics. S. elongatus PCC 7942 expressing aadA or Synechocystis sp. PCC 6803 expressing aadA were normalized to an OD750 of 0.2 in BG11 medium. LPG3 cells were inoculated at 1 × 103 to 2 × 103 cells per ml for growth curves or 1 × 104 to 2 × 104 cells per ml for survival curves. Heterotrophic bacteria were added to wells to a final OD600 of 0.002 unless otherwise noted. Positive control wells with Pseudomonas strain LPH1 expressing aadA and negative-control wells without added heterotrophic bacteria were included for all experiments. For fixation studies, S. elongatus and Pseudomonas isolate LPH1 were fixed with 10% neutral buffered formalin for 30 min at room temperature and washed with BG11 three times. Fixed cells were plated to BG11 or LB agar plates to confirm fixation. Supplementation with cyanocobalamin (Sigma-Aldrich) was performed at a 50 μM final concentration. Wells were resuspended and sampled daily to enumerate the CFU of heterotrophic bacteria on LB plates and to assay LPG3 growth. CFU plots are on log scale and all zero counts were set at the limit of detection. Amoebal cells were counted using a Hausser 3200 counting chamber (Hausser Scientific). A total volume of 3 × 3 × 0.1 mm was counted per sample to increase the limit of detection. Growth curves were plotted on log scale and all zero counts were set at the limit of detection. All experimental groups were assayed in triplicate, and all experiments were repeated at least twice. All experiments were subject to statistical analysis by one-way analysis of variance (ANOVA) tests with Tukey posttesting (GraphPad Prism version 5.0b) using day 6 values. All groups that did not support amoebal growth were significantly different than groups that did support growth, with a P value of <0.05 for Fig. 2H, a P value of <0.01 for Fig. 4C and D, and a P value of <0.001 for all remaining growth curves.
V. cholerae screen.
The library was first screened using spotting assays on 15-cm BG11 agar plates supplemented with 5 μg/ml streptomycin. Five microliters of master mix containing amoebal cells and cyanobacteria was first spotted to plates at densities of 4 × 105 LPG3 cells per ml and S. elongatus expressing aadA at an OD750 of 0.5. The V. cholerae transposon mutant library was grown in 96-well plates containing LB supplemented with 25 μg/ml streptomycin, and 3 μl of overnight culture was spotted on top of amoebal and cyanobacterial spots. All plates included C6706-positive controls and BG11 medium-negative controls. The library was also spotted in the absence of LPG3 and cyanobacteria to verify the growth of the library on BG11 agar. Plates were incubated at 30°C under continuous illumination at 2 to 3 μmol photons m−2 s−1 for 10 to 14 days. Spots with cyanobacterial growth corresponded to V. cholerae transposon mutant insertion strains that could not support amoebal growth. Hits identified in the primary screen and transposon insertion strains that were part of the same operons were streaked for a single colony and retested by spotting assay on BG11 agar plates. These strains were additionally tested for supporting amoebal growth in liquid culture as described above, and 100 μl was sampled on day 6 for assays. LPG3 cell density was qualitatively determined by examination under an inverted microscope, and chlorophyll fluorescence was measured in a black-walled 96-well plate using a SpectraMax fluorescence plate reader (excitation, 425 nm; emission, 680 nm).
Pseudomonas isolate LPH1 genome sequencing and analysis.
To characterize the heterotrophic bacterial coisolate and to facilitate strain construction, the genome was sequenced. DNA was extracted from Pseudomonas isolate LPH1 through phenol-chloroform extraction and purified using the DNeasy blood and tissue kit (Qiagen). Genome sequencing was conducted at the Institute for Genomic Medicine (IGM) Genomics Center at the University of California, San Diego (UCSD). Eight micrograms of DNA was sheared to an average size of 15 kb using a Covaris g-Tube. Sequencing libraries were generated using SMRTbell template preparation reagent kits (Pacific Biosciences). Size selection with a 10-kb cutoff was performed using a BluePippin (Sage Science). Libraries were sequenced via 240-min movies on two SMRT cells using P6/C5 sequencing chemistry on a PacBio RS II sequencer. Following removal of adaptor sequences, de novo genome assembly was performed using Celera Assembler 8.3rc1. The genome was annotated using rapid annotation using the Subsystem Technology server version 2.0 (59), and riboswitch motifs were identified (60).
Corrinoid analysis.
Four-hundred-milliliter cultures of cyanobacteria grown in BG11 and 200-ml cultures of Pseudomonas isolate LPH1 grown in LB were harvested for corrinoid analysis by UPLC-MS as described previously (46, 61, 62). For exchange of the corrinoid upper ligand, cell pellets were resuspended in 1 mg/ml KCN in methanol and incubated at 60°C for 1.5 h. Dried samples were resuspended in water, purified using C18 Sep-Pak columns (Waters), and eluted with methanol. Cyanocobalamin (Sigma-Aldrich) was used as a standard. Corrinoids were analyzed on a Thermo Scientific Dionex UltiMate 3000 quaternary UHPLC coupled to a Thermo Scientific Q Exactive Plus Quadrupole-Orbitrap mass spectrometer in positive ion mode with a heated electrospray ionization source in an Ion Max source housing. Liquid chromatography separation was performed on a Waters XBridge C18 2.1- by 100-mm, 3.5-μm column maintained at 60°C. Solvent A was aqueous 0.1% acetic acid, and solvent B was 0.1% acetic acid in acetonitrile. Corrinoids were separated using a 0.4 ml/min solvent gradient (A/B) as follows: 0 to 2 min, 95/5; 2 to 3 min, 80/20; 3 to 5 min, linear ramp to 0/100; 5 to 7 min, 0/100; and subsequent reconditioning of the column. On the Q Exactive Plus, a full scan was performed from 500 to 2,000 m/z at 70,000 resolution and tandem mass spectrometry (MS-MS) at 50 to 2,000 m/z at 17,500 resolution. Data were processed in Xcalibur (Thermo). The UPLC plots were normalized to 4 × 106 for experimental samples and 1 × 107 for the standard.
Image acquisition.
Transmitted light and fluorescence microscopy images of LPG3 cells were acquired using an Axioplan microscope (Zeiss) through a 100× oil immersion objective and a Spot Pursuit camera and Spot Advanced software version 5.1 (Spot Imaging Solutions) as described previously (25). For imaging ingestion of Pseudomonas isolate LPH1, washed LPG3 cells were seeded to a 24-well plate and incubated for 1 day with Pseudomonas at an OD600 of 0.2 harboring pCV0001 (57), an RSF1010 plasmid for constitutive expression of GFPmut2. LPG3 cells were washed and harvested for acquisition of bright-field and fluorescence images using an ImageStream imaging flow cytometer (Amnis). For fluorescence images, a 488-nm laser was used with imaging in channel 2 for GFPmut2 (505- to 560-nm emission) and in channel 5 for chlorophyll (660- to 745-nm emission). Images were analyzed using IDEAS Software version 6.1 (Amnis).
Accession number(s).
The genome of Pseudomonas isolate LPH1 was deposited into GenBank under accession number CP017290. ps2671 to ps2662 correspond to locus tags BHQ29_13485 to BHQ29_13440, ps4525 to ps4534 correspond to locus tags BHQ29_22625 to BHQ29_22670, and ps4549 to ps4560 correspond to locus tags BHQ29_22745 to BHQ29_22795.
Supplementary Material
ACKNOWLEDGMENTS
We thank Douglas Bartlett (Scripps Institution of Oceanography, UCSD) for use of his biosafety level 2 lab space, John J. Mekalanos (Harvard Medical School) for the V. cholerae transposon library, Nathaniel Snyder (Drexel University) for assistance with and use of the mass spectrometer, Orna Cook and Mark Hildebrand (Scripps Institution of Oceanography, UCSD) for assistance with and use of the ImageStream flow cytometer, and Joshua Earl (Drexel University College of Medicine) for advice regarding bacterial phylogeny.
This work was funded by the Department of Energy Consortium for Algal Biofuels Commercialization grant DE-EE0003373. A.T.M. was supported by the Life Sciences Research Foundation, sponsored by the Howard Hughes Medical Institute.
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00035-17.
REFERENCES
- 1.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. 2009. Protists are microbes too: a perspective. ISME J 3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 2.Montagnes D, Roberts E, Lukes J, Lowe C. 2012. The rise of model protozoa. Trends Microbiol 20:184–191. doi: 10.1016/j.tim.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R, Fierer N. 2013. Global biogeography of highly diverse protistan communities in soil. ISME J 7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgianna DR, Mayfield SP. 2012. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- 5.Benemann J. 2013. Microalgae for biofuels and animal feeds. Energies 6:5869–5886. doi: 10.3390/en6115869. [DOI] [Google Scholar]
- 6.Burki F. 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adl MS, Gupta VS. 2006. Protists in soil ecology and forest nutrient cycling. Can J For Res 36:1805–1817. doi: 10.1139/x06-056. [DOI] [Google Scholar]
- 8.Edgcomb VP. 2016. Marine protist associations and environmental impacts across trophic levels in the twilight zone and below. Curr Opin Microbiol 31:169–175. doi: 10.1016/j.mib.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Sanudo-Wilhelmy SA, Gomez-Consarnau L, Suffridge C, Webb EA. 2014. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci 6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 10.Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 11.Croft MT, Warren MJ, Smith AG. 2006. Algae need their vitamins. Eukaryot Cell 5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep 19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 13.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. 2014. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe 15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Men Y, Seth EC, Yi S, Crofts TS, Allen RH, Taga ME, Alvarez-Cohen L. 2015. Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ Microbiol 17:4873–4884. doi: 10.1111/1462-2920.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degnan PH, Taga ME, Goodman AL. 2014. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab 20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO, Post AF, Sipler RE, Spackeen JL, Xu K, Bronk DA, Hutchins DA, Allen AE. 2015. Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci U S A 112:9938–9943. doi: 10.1073/pnas.1501615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. 2015. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9:461–471. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote-Maestas W, Hmelo LR, Moffett JW, Devol AH, Armbrust EV, Stahl DA, Ingalls AE. 2017. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc Natl Acad Sci U S A 114:364–369. doi: 10.1073/pnas.1608462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Krautler B, Scanlan DJ, Warren MJ, Smith AG. 2016. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol 26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez AA, Liu Z, Rodionov DA, Li Z, Bryant DA. 2016. Complementation of cobalamin auxotrophy in Synechococcus sp. strain PCC 7002 and validation of a putative cobalamin riboswitch in vivo. J Bacteriol 198:2743–2752. doi: 10.1128/JB.00475-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez AA, Rodionov DA, Bryant DA. 2016. Identification and regulation of genes for cobalamin transport in the cyanobacterium Synechococcus sp. strain PCC. 7002. J Bacteriol 198:2753–2761. doi: 10.1128/JB.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dryden RC, Wright SJL. 1987. Predation of cyanobacteria by protozoa. Can J Microbiol 33:471–482. doi: 10.1139/m87-080. [DOI] [Google Scholar]
- 23.Van Wichelen J, Van Gremberghe I, Vanormelingen P, Debeer AE, Leporcq B, Menzel D, Codd GA, Descy JP, Vyverman W. 2010. Strong effects of amoebae grazing on the biomass and genetic structure of a Microcystis bloom (cyanobacteria). Environ Microbiol 12:2797–2813. [DOI] [PubMed] [Google Scholar]
- 24.Wright SJL, Redhead K, Maudsley H. 1981. Acanthamoeba castellanii, a predator of cyanobacteria. J Gen Microbiol 125:293–300. [Google Scholar]
- 25.Ma AT, Daniels EF, Gulizia N, Brahamsha B. 2016. Isolation of diverse amoebal grazers of freshwater cyanobacteria for the development of model systems to study predator-prey interactions. Algal Res 13:85–93. doi: 10.1016/j.algal.2015.11.010. [DOI] [Google Scholar]
- 26.Simkovsky R, Daniels EF, Tang K, Huynh SC, Golden SS, Brahamsha B. 2012. Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc Natl Acad Sci U S A 109:16678–16683. doi: 10.1073/pnas.1214904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giedyk M, Goliszewska K, Gryko D. 2015. Vitamin B12 catalysed reactions. Chem Soc Rev 44:3391–3404. doi: 10.1039/C5CS00165J. [DOI] [PubMed] [Google Scholar]
- 29.Oltean S, Banerjee R. 2005. A B12-responsive internal ribosome entry site (IRES) element in human methionine synthase. J Biol Chem 280:32662–32668. doi: 10.1074/jbc.M501964200. [DOI] [PubMed] [Google Scholar]
- 30.Jost M, Fernandez-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY, Kang G, Padmanabhan S, Elias-Arnanz M, Drennan CL. 2015. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526:536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nahvi A, Barrick JE, Breaker RR. 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 33.Kazamia E, Helliwell KE, Purton S, Smith AG. 2016. How mutualisms arise in phytoplankton communities: building eco-evolutionary principles for aquatic microbes. Ecol Lett 19:810–822. doi: 10.1111/ele.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. 2011. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 35.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee R. 2006. B12 trafficking in mammals: a case for coenzyme escort service. ACS Chem Biol 1:149–159. doi: 10.1021/cb6001174. [DOI] [PubMed] [Google Scholar]
- 37.Seetharam B, Bose S, Li N. 1999. Cellular import of cobalamin (vitamin B-12). J Nutr 129:1761–1764. [DOI] [PubMed] [Google Scholar]
- 38.Bloomfield G, Traynor D, Sander SP, Veltman DM, Pachebat JA, Kay RR. 2015. Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium. eLife 4:e04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 40.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazra AB, Tran JL, Crofts TS, Taga ME. 2013. Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin B(12). Chem Biol 20:1275–1285. doi: 10.1016/j.chembiol.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A, Golden SS. 2015. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci U S A 112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos F, Vera JL, Lamosa P, de Valdez GF, de Vos WM, Santos H, Sesma F, Hugenholtz J. 2007. Pseudovitamin B(12) is the corrinoid produced by Lactobacillus reuteri CRL1098 under anaerobic conditions. FEBS Lett 581:4865–4870. doi: 10.1016/j.febslet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann B, Oberhuber M, Stupperich E, Bothe H, Buckel W, Konrat R, Kräutler B. 2000. Native corrinoids from Clostridium cochlearium are adeninylcobamides: spectroscopic analysis and identification of pseudovitamin B12 and factor A. J Bacteriol 182:4773–4782. doi: 10.1128/JB.182.17.4773-4782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stupperich E, Nexo E. 1991. Effect of the cobalt-N coordination on the cobamide recognition by the human vitamin B12 binding proteins intrinsic factor, transcobalamin and haptocorrin. Eur J Biochem 199:299–303. doi: 10.1111/j.1432-1033.1991.tb16124.x. [DOI] [PubMed] [Google Scholar]
- 46.Yi S, Seth EC, Men Y-J, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl Environ Microbiol 78:7745–7752. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuster FL. 2002. Cultivation of pathogenic and opportunistic free-living amebas. Clin Microbiol Rev 15:342–354. doi: 10.1128/CMR.15.3.342-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke J, Kessin R. 1977. A defined minimal medium for axenic strains of Dictyostelium discoideum. Proc Natl Acad Sci U S A 74:2157–2161. doi: 10.1073/pnas.74.5.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nerad TA, Visvesvara GS, Daggett P-M. 1983. Chemically defined media for the cultivation of Naegleria: pathogenic and high temperature tolerant species. J Protozool 30:383–387. doi: 10.1111/j.1550-7408.1983.tb02935.x. [DOI] [PubMed] [Google Scholar]
- 50.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watts DJ, Guest JR. 1975. Studies on the vitamin nutrition of the cellular slime mould Dictyostelium discoideum. J Gen Microbiol 86:333–342. doi: 10.1099/00221287-86-2-333. [DOI] [PubMed] [Google Scholar]
- 52.Crona M, Avesson L, Sahlin M, Lundin D, Hinas A, Klose R, Soderbom F, Sjoberg BM. 2013. A rare combination of ribonucleotide reductases in the social amoeba Dictyostelium discoideum. J Biol Chem 288:8198–8208. doi: 10.1074/jbc.M112.442434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJ. 2014. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156:759–770. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seth EC, Taga ME. 2014. Nutrient cross-feeding in the microbial world. Front Microbiol 5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. [Google Scholar]
- 56.Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 57.Taton A, Unglaub F, Wright NE, Zeng WY, Paz-Yepes J, Brahamsha B, Palenik B, Peterson TC, Haerizadeh F, Golden SS, Golden JW. 2014. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res 42:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma AT, Schmidt CM, Golden JW. 2014. Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Appl Environ Microbiol 80:6704–6713. doi: 10.1128/AEM.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrick JE. 2009. Predicting riboswitch regulation on a genomic scale. Methods Mol Biol 540:1–13. doi: 10.1007/978-1-59745-558-9_1. [DOI] [PubMed] [Google Scholar]
- 61.Crofts TS, Seth EC, Hazra AB, Taga ME. 2013. Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem Biol 20:1265–1274. doi: 10.1016/j.chembiol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 62.Hazra AB, Han AW, Mehta AP, Mok KC, Osadchiy V, Begley TP, Taga ME. 2015. Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc Natl Acad Sci U S A 112:10792–10797. doi: 10.1073/pnas.1509132112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.