ABSTRACT
Clostridium saccharoperbutylacetonicum N1-4 is well known as a hyper-butanol-producing strain. However, the lack of genetic engineering tools hinders further elucidation of its solvent production mechanism and development of more robust strains. In this study, we set out to develop an efficient genome engineering system for this microorganism based on the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated 9 (CRISPR-Cas9) system. First, the functionality of the CRISPR-Cas9 system previously customized for Clostridium beijerinckii was evaluated in C. saccharoperbutylacetonicum by targeting pta and buk, two essential genes for acetate and butyrate production, respectively. pta and buk single and double deletion mutants were successfully obtained based on this system. However, the genome engineering efficiency was rather low (the mutation rate is <20%). Therefore, the efficiency was further optimized by evaluating various promoters for guide RNA (gRNA) expression. With promoter PJ23119, we achieved a mutation rate of 75% for pta deletion without serial subculturing as suggested previously for C. beijerinckii. Thus, this developed CRISPR-Cas9 system is highly desirable for efficient genome editing in C. saccharoperbutylacetonicum. Batch fermentation results revealed that both the acid and solvent production profiles were altered due to the disruption of acid production pathways; however, neither acetate nor butyrate production was eliminated with the deletion of the corresponding gene. The butanol production, yield, and selectivity were improved in mutants, depending on the fermentation medium. In the pta buk double deletion mutant, the butanol production in P2 medium reached 19.0 g/liter, which is one of the highest levels ever reported from batch fermentations.
IMPORTANCE An efficient CRISPR-Cas9 genome engineering system was developed for C. saccharoperbutylacetonicum N1-4. This paves the way for elucidating the solvent production mechanism in this hyper-butanol-producing microorganism and developing strains with desirable butanol-producing features. This tool can be easily adapted for use in closely related microorganisms. As also reported by others, here we demonstrated with solid data that the highly efficient expression of gRNA is the key factor determining the efficiency of CRISPR-Cas9 for genome editing. The protocol developed in this study can provide essential references for other researchers who work in the areas of metabolic engineering and synthetic biology. The developed mutants can be used as excellent starting strains for development of more robust ones for desirable solvent production.
KEYWORDS: CRISPR-Cas9, Clostridium saccharoperbutylacetonicum, buk, butanol, gRNA, genome engineering, pta
INTRODUCTION
Biofuels produced from renewable biomass provide a feasible solution for addressing problems such as environmental pollution and climate change resulting from fossil fuel consumption. Thus, biofuels are considered appropriate substitutes for fossil fuels at their depletion (1). Biobutanol produced through acetone-butanol-ethanol (ABE) fermentation possesses various advantages over ethanol as a fuel source and is considered one of the most promising biofuel candidates for the future (2). Additionally, butanol manifests an even higher commodity value when it is considered as the feedstock for numerous industries (3). There are generally four primary solventogenic clostridial species for ABE production: Clostridium acetobutylicum, Clostridium beijerinckii, Clostridium saccharobutylicum, and Clostridium saccharoperbutylacetonicum (2). Among them, undoubtedly, C. acetobutylicum and C. beijerinckii have been the most extensively investigated for their ABE fermentation physiology, genomics, genetics, and mutant development (especially in recent years) (4–9). C. saccharoperbutylacetonicum N1-4 (ATCC 13564) was isolated in 1959 in Japan and was used for industrial butanol production from then until the late 1980s (10, 11). As a well-known hyper-butanol-producing strain (12), C. saccharoperbutylacetonicum N1-4 has been broadly studied for its fermentation characteristics, including desirable fermentation media and fermentation conditions (13–16). Recently, the genome sequence of C. saccharoperbutylacetonicum N1-4 ATCC 27021 (a lysogenic derivative strain of ATCC 13564) has been published (17, 18). However, compared to C. acetobutylicum and C. beijerinckii, the study of the genomics and genetics of C. saccharoperbutylacetonicum has lagged. In addition, there were rare reports concerning attempts at metabolic engineering of this species. So far, to our best knowledge, there are only two reports related to genetic engineering in this strain; one is about the development of a host-vector system and the expression of an amylase gene in C. saccharoperbutylacetonicum N1-4 (19), and the other is about the downregulation of the hydrogenase gene cluster using the antisense RNA strategy (20). Both reports were published about 10 years ago from the same research group. During the preparation of thi article, Herman et al. reported the development of a transformation protocol and the metabolic engineering of C. saccharoperbutylacetonicum using plasmid-based overexpression and nonreplicative vector-based allelic exchange (21). However, clearly, more efficient and versatile genetic engineering tools for C. saccharoperbutylacetonicum are highly desirable in order to elucidate its complex fermentation phenotype and regulation mechanism and develop better strains for butanol production.
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) system is an RNA-mediated immune system in bacteria and archaea that can efficiently protect the host from invading foreign DNAs, such as phages or plasmids (22). Recently, the CRISPR system has been engineered as a cutting-edge genome engineering tool for both eukaryotic and prokaryotic cells, for which the type II CRISPR-Cas9 system from Streptococcus pyogenes has been mostly employed (23–28). In the CRISPR-Cas9 system, the CRISPR RNA (crRNA) and the trans-activating CRISPR RNA (tracrRNA) form a complex to make a dual tracrRNA-crRNA, directing the Cas9 to the site-specific DNA sequence with the protospacer-adjacent motif (PAM). The endonuclease activity of Cas9 can then lead to double-strand breakage (DSB) (29). Genetic mutations can be introduced through nonhomologous end joining (NHEJ) or homology-directed repair (HDR) by providing a DNA editing template (30). Jinek et al. engineered the dual tracrRNA-crRNA into a single chimeric guide RNA (gRNA) which demonstrated comparable efficiency for the DNA targeting purpose, making the CRISPR-Cas9 system much easier to implement (29). In the past few years, numerous successes in genome editing based on the CRISPR-Cas9 system in various bacteria have been reported, including Escherichia coli (31, 32), S. pneumoniae (23), Streptomyces species (33–35), Tatumella citrea (32), C. pasteurianum (36), C. beijerinckii (8, 26, 34, 37), and C. acetobutylicum (38, 39). It was demonstrated that although the CRISPR-Cas9-based genome editing also relies on the targeted DSB from Cas9, the DSB more likely serves as a selection power for the edited mutant cells against the unedited background cells (23, 26).
During the biphasic ABE fermentation, the production of acids is indispensable and plays important roles in the metabolic switch for solvent production (40). In the acidogenic phase, acetate is produced from acetyl coenzyme A (acetyl-CoA) through the action of phosphotransacetylase (PTA) and acetate kinase (ACK), while butyrate is generated through catalysis by phosphotransbutyrylase (PTB) and butyrate kinase (BUK). Subsequently, during the solventogenic phase, both acids are reassimilated for the production of acetone, butanol, and ethanol (41). Disruption of acid formation pathways has been achieved previously in C. acetobutylicum and C. beijerinckii; the results demonstrated that the production patterns of both acids and solvents were altered by such manipulation (42–44).
Therefore, in this study, we developed an efficient CRISPR-Cas9-based genome editing system for C. saccharoperbutylacetonicum. Based on this system, the primary pta gene, buk gene, or both were deleted, and the fermentation features of the mutants compared with those of the wild-type strain were characterized. The developed CRISPR-Cas9 system provided a versatile genome engineering tool for C. saccharoperbutylacetonicum, allowing easy elucidation of the metabolism of this microorganism and development of robust strains for biobutanol production. Additionally, this study provides essential references for researchers developing customized CRISPR-Cas9 genome engineering tools for other microorganisms.
RESULTS
pta deletion in C. saccharoperbutylacetonicum N1-4.
In previous work, Wang et al. developed a customized CRISPR-Cas9 system for efficient genome editing in C. beijerinckii (26). Here, we first evaluated whether we could apply this tool directly for genome engineering in C. saccharoperbutylacetonicum. We selected pta as our first target gene because it was demonstrated that the deletion of this gene is achievable in other solventogenic clostridial strains and also because the deletion of this gene can potentially lead to enhanced butanol production (40, 41). The plasmid pYW34-deltpta was constructed using the “general” CRISPR-Cas9 vector pYW34 for C. beijerinckii as the mother vector (26). It was transformed into C. saccharoperbutylacetonicum through electroporation, with a transformation efficiency of 1.6 × 104 CFU/μg DNA. After 24 h of cultivation, single colonies were picked and subcultured in tryptone-glucose-yeast extract (TGY) liquid medium plus erythromycin (TGYE). In order to evaluate the genome engineering efficiency in C. saccharoperbutylacetonicum of the CRISPR-Cas9 that was customized for C. beijerinckii, we started to induce the expression of the Cas9 from the second generation of the transferring culture. That is, starting from the second generation, while the culture continued to be transferred in TGYE medium, it was also transferred into TGYE liquid medium supplemented with lactose (TGYLE). Colony PCR (cPCR) was performed with primers YW1044 and YW1045 using the liquid culture in TGYLE as the template to test whether the PCR band from the pta deletion mutant could be detected. In this case, the pta deletion mutant would generate a band of 2,278 bp, compared to a band of 3,280 bp that would be generated from the wild type. If the mutant band was visible, then the culture from TYGLE liquid medium was spread onto TGYLE agar plates and the same cPCR was performed to identify the mutant from the colonies. Interestingly, from the second generation of the subculture, we started to detect the mutant band (beside the wild-type band) from the liquid culture (see Fig. S3 in the supplemental material). However, no mutant was able to be identified from the colony grown on the plates from either the second or the third generation of culture (for each generation, at least 48 colonies were randomly screened), while for the fourth generation, three colonies out of 16 were detected with the mutant band (see Fig. S4 in the supplemental material).
As described by Wang et al., vector integration events (VIE) could occur during the genome editing of the bacterial chromosome using the CRISPR-Cas9 system (26). Here, we further detected VIE using primer pairs YW1044/YW200 and YW847/YW1045, and the expected PCR bands for VIE were detected for both ends in all the three mutant colonies (see Fig. S5 in the supplemental material). One of the three colonies was picked and respread onto new TGYLE agar plates. Further cPCR for detecting VIE was performed, and two out of 36 screened colonies showed no PCR bands for VIE. These two mutant colonies were then inoculated into TGY liquid medium and subcultured in the same medium for 7 generations for plasmid curing. The culture was then spread onto TGY plates, and cPCRs with YW880 and YW881 were conducted to detect the existence of the plasmid. It was shown that none of the tested colonies showed a visible PCR band, thus confirming the curing of the plasmid. Selected colonies (tested as mutants without a plasmid) were spread onto the TGYE plates, and no colonies were grown after 48 h of cultivation, further verifying the loss of the plasmid. Finally, the mutation in the strain was further verified through cPCR using primers YW1044 and YW1045 (Fig. 1A). The clean mutant strain with the whole pta open reading frame (ORF) deleted was named C. saccharoperbutylacetonicum deltpta.
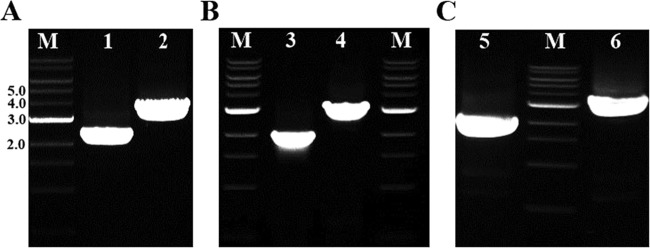
FIG 1.
Colony PCR (cPCR) results confirm the deletion of the pta and buk genes. (A) cPCR results using primers YW1044 and YW1045 flanking the upstream and downstream sequences of the homologous recombination region of the pta gene in C. saccharoperbutylacetonicum deltpta (lane 1, 2,278 bp) and the wild type (lane 2, 3,280 bp). (B) cPCR results using primers YW953 and YW954 flanking the upstream and downstream sequences of the homologous recombination region of the buk gene in C. saccharoperbutylacetonicum deltbuk (lane 3, 2,061 bp) and the wild type (lane 4, 3,119 bp). (C) cPCR results using primers YW953 and YW954 flanking the upstream and downstream sequences of the homologous recombination region of the buk gene in C. saccharoperbutylacetonicum deltptabuk (lane 5, 2,061 bp) and the C. saccharoperbutylacetonicum deltpta strain (lane 6, 3,119 bp). The NEB 1-kb DNA ladder was used as the marker (lane M), with numbers on the left representing the band length in kb.
buk deletion in C. saccharoperbutylacetonicum N1-4.
In a similar manner, we attempted to delete buk in C. saccharoperbutylacetonicum N1-4. The plasmid pYW34-deltbuk was transformed into wild-type C. saccharoperbutylacetonicum N1-4, and the transformed cells were plated onto TGYE plates. A transformation efficiency of 1.5 × 104 CFU/μg DNA was obtained. Single colonies were picked and cultivated successively in TGYE liquid medium. From the second generation, cultures were also transferred into TGYLE for induction of Cas9 expression, followed by plating on TGYLE plates. cPCR was performed with primers YW953 and YW954 to detect the mutation in the TGYLE liquid culture or on the plates. No mutant band was observed from the PCR using the liquid culture as the template until the seventh generation. For the cPCR with the colonies from the seventh generation, two colonies out of 32 tested demonstrated double bands (2,061 bp for the mutant and 3,119 bp for the wild type) (data not shown). The colony with the brighter mutant band was picked and replated onto a new TGYLE plate. Further screening of colonies from the replating indicated that 11 out of 16 colonies demonstrated double bands and 1 out of 16 colonies showed the pure mutant band (see Fig. S6 in the supplemental material). VIE were also detected in this mutant using primers YW953/YW200 and YW847/YW954 (data not shown). Through further replating and screening, the colonies without VIE were identified. Plasmid curing was then carried out, and finally a clean buk deletion mutant was obtained (Fig. 1B, lane 3). The mutant was designated C. saccharoperbutylacetonicum deltbuk.
buk deletion in C. saccharoperbutylacetonicum deltpta.
In a further step, we tried to achieve a double knockout, that is, knocking out buk using C. saccharoperbutylacetonicum deltpta as the starting strain. The plasmid pYW34-deltbuk was transformed into C. saccharoperbutylacetonicum deltpta. Through the same procedure as discussed above, the cPCR did not show visible mutant bands using the second to sixth generations of the TGYLE liquid culture as the template. In the liquid culture of the seventh generation, both the mutant and the wild-type bands were observed. For the cPCR with the colonies plated from the seventh-generation culture, four out of 32 tested colonies exhibited double bands (2,061 bp for the mutant and 3,119 bp for the wild type) (data not shown). Further screening of colonies from the replating indicated that two out of 16 colonies showed the pure mutant band (see Fig. S7 in the supplemental material). VIE were also detected in both colonies. Through further replating, screening, and subculturing (for plasmid curing), the clean pta buk double knockout mutant was obtained (Fig. 1C, lane 5). The mutant was designated C. saccharoperbutylacetonicum deltptabuk.
Optimization of CRISPR-Cas9 efficiency by evaluating various promoters for gRNA expression.
With the customized CRISPR-Cas9 system, nearly 100% genome engineering efficiency could be achieved in C. beijerinckii (26). Here, when we applied this system directly to C. saccharoperbutylacetonicum, desirable results for genome engineering purposes were achieved, but the efficiency was rather low (the mutation rate was less than 20% following the induction of Cas9 expression). We hypothesized that this might be because the selection power (against the unedited background cells) from the CRISPR-Cas9 was not very effective in C. saccharoperbutylacetonicum. For the functionality of the CRISPR-Cas9 system, a high level of gRNA expression is essential and thus this could be the limiting factor in many cases, as indicated by many previous studies (45, 46). Therefore, we decided to enhance the CRISPR-Cas9 efficiency in C. saccharoperbutylacetonicum by identifying a promoter for more efficient gRNA expression. For this purpose, the veg promoter from Bacillus subtilis (Pvegb) (47), the hypothetical veg promoter from C. saccharoperbutylacetonicum (Pvegc), and the E. coli promoter J23119 (48) were selected and evaluated. In addition, for easy comparison, we still targeted pta deletion and used exactly the same 20-nucleotide (nt) guiding sequence fused to the small RNA (sRNA) promoter (PsRNA) and the same homology arms. The corresponding vectors pSHW1-pta, pSHW2-pta, and pSHW3-pta were constructed and transformed into C. saccharoperbutylacetonicum N1-4. Following the same procedure, the pta deletion mutant was generated and the genome editing efficiency was evaluated.
As illustrated in Table 1, for the genome engineering with CRISPR-Cas9 using four different promoters for gRNA expression, mutant bands all started to be detected from the second generation of the TGYLE liquid culture. However, for the colonies grown from the replating of the second generation of liquid culture, mutants were identified (with cPCR screening for at least 48 colonies for each, unless mutants were detected with screening on fewer colonies) only from the transformations with pSHW1-pta and pSHW3-pta, with mutation rates of 12.5% and 75%, respectively, while for the transformation with pSHW2-pta, no mutant was identified until the seventh generation, where one mutant was obtained out of 16 screened colonies (representing a mutation rate of 6.25%). These results demonstrated that the various promoters for gRNA expression in CRISPR-Cas9 led to genome engineering efficiency in C. saccharoperbutylacetonicum in the order PJ23119 > Pvegb > PsRNA > Pvegc. Therefore, the CRISPR-Cas9 system with the J23119 promoter for gRNA expression (and the lactose-inducible promoter for Cas9 expression) constructed here could be used as an efficient genome editing tool in C. saccharoperbutylacetonicum. Mutants can be easily obtained after one generation of cultivation in TGYE and one additional generation of incubation in TGYLE for the induction of Cas9 expression. Of course, the efficiency could well be dependent on the chromosomal target and the exact genome engineering purpose, which warrants further investigation in the future.
TABLE 1.
Comparison of genome editing efficiencies using CRISPR-Cas9 with different promoters for gRNA expression
| Plasmid used for transformation | Promotera | Generation for: |
No. of mutants/no. of colonies screened (mutation rate, %) | |
|---|---|---|---|---|
| Mutant band starting to be detected from TGYLE liquid culture using cPCR | Obtainment of mutant strain | |||
| pYW34-deltpta | PsRNA | Second | Fourth | 3/16 (18.75) |
| pSHW1-deltpta | Pvegb | Second | Second | 2/16 (12.5) |
| pSHW2-deltpta | Pvegc | Second | Seventh | 1/16 (6.25) |
| pSHW3-deltpta | PJ23119 | Second | Second | 12/16 (75) |
PsRNA, small RNA promoter from C. beijerinckii; Pvegb, veg promoter from B. subtilis; Pvegc, hypothetical veg gene promoter from C. saccharoperbutylacetonicum; PJ23119, J23119 promoter from E. coli.
As a summary, the general strategy for gene deletion and mutant screening is shown in Fig. S1 in the supplemental material. After transformation of the plasmid DNA containing the desirable construct for deleting the targeted gene (pta or buk in this case), recombinant strains were subcultured successively to permit efficient homologous recombination. During this process, both double crossovers (Fig. S1, A1) and single crossovers (Fig. S1, B1 and C1) could possibly occur, generating a chromosome with a clean gene deletion (Fig. S1, A2) and a chromosome with vector integrated (Fig. S1. B2 and C2). Moreover, some recombinant strains might still be the wild type containing the transformed plasmid (Fig. S1, D1). Lactose-containing medium was then used to induce the expression of Cas9, which would lead to the DSB on the chromosome wherever the target site (the 20-nt guiding sequence in gRNA) still existed (Fig. S1, B2, C2, and D2). Finally, only the mutant with the clean deletion of pta or buk survived from the selection and could be screened through cPCR.
Fermentation of the mutants in P2 medium.
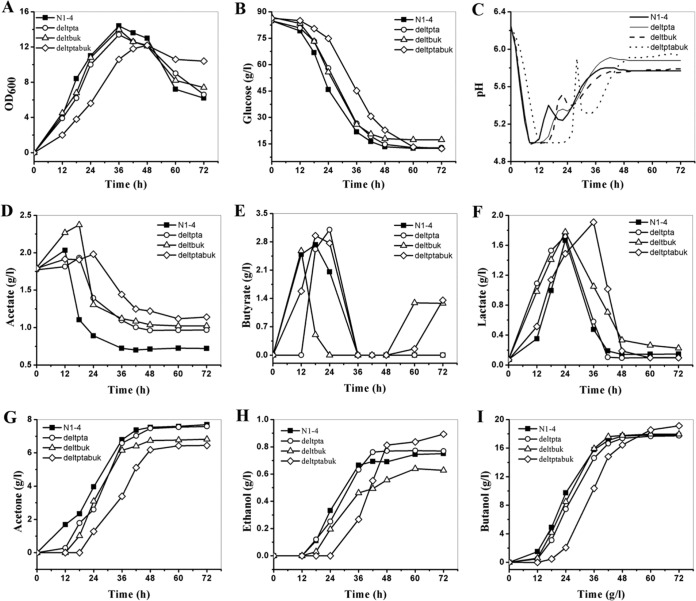
The kinetics of batch fermentation in P2 medium with mutants as well as the wild type are demonstrated in Fig. 2. Though a lower growth rate was observed when the pta or buk gene was deleted (Table 2), both C. saccharoperbutylacetonicum deltpta and C. saccharoperbutylacetonicum deltbuk displayed growth kinetics similar to those of the wild-type strain (Fig. 2A). The slowest growth was observed in the mutant with the deletion of both pta and buk. It showed a much longer lag phase and reached a maximum optical density at 600 nm (OD600) of about 5.5% to 14.4% lower than those of the other three strains. As indicated in Fig. 2B, C. saccharoperbutylacetonicum deltptabuk also showed the slowest glucose consumption, with 73.1 g/liter consumed in 60 h, while 70.2, 69.5, and 71.5 g/liter glucose were consumed within 48 h in C. saccharoperbutylacetonicum deltpta and deltbuk and the wild-type strain, respectively. The pH profiles are illustrated in Fig. 2C. In all strains, with the production of acids from the beginning of the fermentation, the pH dropped quickly to 5.0 (and was then controlled at ≥5.0); corresponding to the initiation of solventogenesis, the pH increased to a peak, which was followed by another slight decrease, and finally rose back to a high plateau between 5.7 and 6.0. In comparison, mutants with single gene deletion (pta or buk) dropped to pH 5.0 at about 8 h and 9 h, respectively, similar to the wild type (8 h), but stayed at 5.0 for a longer time (5 h for deltpta and 6 h for deltbuk) than the wild type (3 h) before increasing to the higher level. For the double knockout mutant, deltptabuk, corresponding to the slowest growth, it took longer (about 12 h) for the pH to decrease to pH 5.0, and it stayed at this point for more than 10 h before increasing to pH 5.9, generating the highest peak (Fig. 2C).
FIG 2.
Batch fermentation profiles of the C. saccharoperbutylacetonicum mutants compared to the wild type in P2 medium. (A) Cell growth; (B) glucose consumption; (C) pH profiles; (D) acetate production; (E) butyrate production; (F) lactate production; (G) acetone production; (H) ethanol production; (I) butanol production. N1-4, C. saccharoperbutylacetonicum N1-4; deltpta, C. saccharoperbutylacetonicum deltpta; deltbuk, C. saccharoperbutylacetonicum deltbuk; deltptabuk, C. saccharoperbutylacetonicum deltptabuk. Fermentation was carried out in replicates, and results from one batch are reported here as representative.
TABLE 2.
Summary of fermentation results for the C. saccharoperbutylacetonicum mutants compared to the wild-type strain N1-4
| Characteristic | Result (mean ± SD) in medium: |
|||||||
|---|---|---|---|---|---|---|---|---|
| P2 |
MP2 |
|||||||
| N1-4 | deltpta | deltbuk | deltptabuk | N1-4 | deltpta | deltbuk | deltptabuk | |
| Growth rate (/h) | 0.42 ± 0.01 | 0.38 ± 0.02 | 0.36 ± 0.03 | 0.29 ± 0.01 | 0.66 ± 0.01 | 0.41 ± 0.01 | 0.56 ± 0.03 | 0.35 ± 0.02 |
| Maximum OD600 | 14.6 ± 0.2 | 13.2 ± 0.2 | 13.8 ± 0.5 | 12.5 ± 0.3 | 16.0 ± 0.2 | 14.7 ± 0.15 | 13.8 ± 0.4 | 11.8 ± 0.2 |
| Peak acetate (g/liter)a | 2.1 ± 0.03 | 1.9 ± 0.04 | 2.4 ± 0.01 | 2.0 ± 0.01 | 0.69 ± 0.01 | 0.59 ± 0.02 | 1.9 ± 0.01 | 0.67 ± 0.02 |
| Peak butyrate (g/liter) | 2.7 ± 0.02 | 3.1 ± 0.04 | 2.5 ± 0.02 | 2.9 ± 0.01 | 2.5 ± 0.05 | 3.8 ± 0.05 | 2.0 ± 0.03 | 3.6 ± 0.05 |
| Peak lactate (g/liter) | 1.6 ± 0.05 | 1.7 ± 0.03 | 1.7 ± 0.01 | 1.9 ± 0.01 | 1.2 ± 0.05 | 2.2 ± 0.15 | 1.5 ± 0.05 | 1.3 ± 0.1 |
| Butanol (g/liter)b | 17.7 ± 0.1 | 17.7 ± 0.2 | 17.9 ± 0.2 | 19 ± 0.1 | 18.4 ± 0.02 | 18.7 ± 0.05 | 17.7 ± 0.1 | 18.8 ± 0.15 |
| Butanol yield (g/g) | 0.24 ± 0.01 | 0.24 ± 0.01 | 0.25 ± 0.01 | 0.25 ± 0.01 | 0.23 ± 0.01 | 0.26 ± 0.02 | 0.24 ± 0.02 | 0.26 ± 0.01 |
| Acetone (g/liter) | 7.7 ± 0.05 | 7.5 ± 0.1 | 6.8 ± 0.05 | 6.5 ± 0.05 | 7.7 ± 0.02 | 4.4 ± 0.05 | 6.8 ± 0.05 | 4.7 ± 0.1 |
| Ethanol (g/liter) | 0.72 ± 0.02 | 0.78 ± 0.01 | 0.6 ± 0.04 | 0.87 ± 0.02 | 0.72 ± 0.01 | 0.61 ± 0.01 | 0.73 ± 0.02 | 0.44 ± 0.01 |
| Total ABE (g/liter) | 25.9 ± 0.16 | 25.8 ± 0.06 | 25.2 ± 0.08 | 26.3 ± 0.03 | 26.8 ± 0.02 | 23.6 ± 0.11 | 25.2 ± 0.17 | 23.8 ± 0.22 |
| Butanol selectivityc (g/g, %) | 68.2 ± 0.05 | 68.3 ± 0.05 | 71.3 ± 0.01 | 72.4 ± 0.25 | 68.5 ± 0.1 | 79 ± 0.2 | 70.3 ± 0.1 | 78.7 ± 0.1 |
There was approximately 1.7 g/liter of acetate preadded in the P2 medium.
The reported titers were the maximum values after the solvent production reached a plateau.
Ratio of butanol to total ABE.
As illustrated in Fig. 2D and E, deletion of the whole ORF of the pta or buk gene did not completely eliminate the formation of acetate or butyrate, indicating that additional acetate- and butyrate-forming pathways were probably present in C. saccharoperbutylacetonicum. Compared to that in the wild type, pta deletion decreased the acetate production slightly, by 0.2 g/liter and 0.1 g/liter in C. saccharoperbutylacetonicum deltpta and C. saccharoperbutylacetonicum deltptabuk, respectively (Fig. 2D; Table 2). To make up energy (ATP) loss caused by pta deletion, C. saccharoperbutylacetonicum deltpta and C. saccharoperbutylacetonicum deltptabuk increased the production of butyrate by 0.4 g/liter (14.8%) and 0.2 g/liter (7.4%), respectively, compared to that in the wild type (Fig. 2E; Table 2). Similarly, in the mutant deltbuk with a buk deletion, butyrate production was decreased by 7.4%, while acetate production was increased significantly by 0.3 g/liter (a 14.3% increase compared to the wild type). In addition to acetate and butyrate, in a similar manner, lactate was also produced (the maximum was ≥1.6 g/liter) and reassimilated in all the strains (Fig. 2F). Deletion of pta or buk led to the increase of lactate in the mutants, especially in C. saccharoperbutylacetonicum deltptabuk, in which lactate reached a peak level of 1.9 g/liter, representing an 18.8% improvement compared to the wild type.
ABE started to be produced in the early exponential phase in all mutants and the wild type (Fig. 2G, H, and I). ABE production from the mutants and the wild type is compared in Table 2. Compared to the wild type, C. saccharoperbutylacetonicum deltpta, except for a later initiation for solvent production, demonstrated very similar acetone, butanol and ethanol, production profiles and also similar butanol and ABE yields as well as butanol selectivity (butanol/ABE ratio). C. saccharoperbutylacetonicum deltbuk generated an amount of butanol comparable to that for the wild type but decreased production of acetone (by 11.7%) and ethanol (16.7%), resulting in a slight increase in the selectivity (71.3% versus 68.2%). Compared to the wild type, C. saccharoperbutylacetonicum deltptabuk produced 15.6% less acetone but 20.8% more ethanol. This strain also produced much more butanol (1.3 g/liter more than the wild type, reaching 19.0 g/liter) with a slightly higher butanol yield. The butanol selectivity was also higher (72.4% versus 68.2%).
Fermentation of the mutants in MP2 medium.
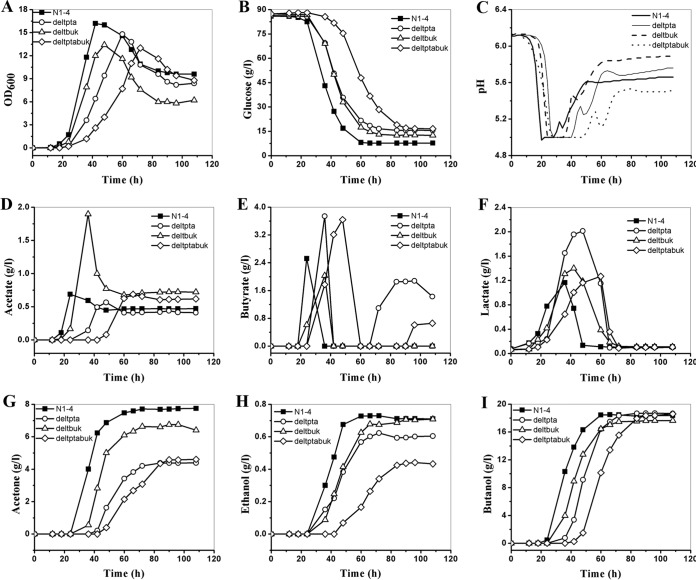
To exclude the interference of acetate preadded in P2 medium for the fermentation kinetics, MP2 medium was used to carry out further fermentation with the mutants as well as the wild type (Fig. 3). Compared to the fermentation with P2 medium, a longer lag phase of around 12 h in MP2 was observed for the wild type and C. saccharoperbutylacetonicum deltbuk, and a prolonged lag phage of around 24 h was observed for both C. saccharoperbutylacetonicum deltpta and deltptabuk (Fig. 3A). In the exponential phase, all the mutants grew more slowly and reached much lower maximal ODs than the wild type (Table 2). Corresponding to the slow growth, all mutants consumed glucose more slowly, especially when both the pta and buk genes were deleted (Fig. 3B). Within 60 h, 77.8 g/liter, 64.6 g/liter, and 69.9 g/liter glucose were consumed by the wild type, C. saccharoperbutylacetonicum deltpta, and C. saccharoperbutylacetonicum deltbuk, respectively, while in C. saccharoperbutylacetonicum deltptabuk, it took 84 h to consume 68.3 g/liter glucose.
FIG 3.
Batch fermentation profiles of the C. saccharoperbutylacetonicum mutants compared to the wild type in MP2 medium. (A) Cell growth; (B) glucose consumption; (C) pH profiles; (D) acetate production; (E) butyrate production; (F) lactate production; (G) acetone production; (H) ethanol production; (I) butanol production. N1-4, C. saccharoperbutylacetonicum N1-4; deltpta, C. saccharoperbutylacetonicum deltpta; deltbuk, C. saccharoperbutylacetonicum deltbuk; deltptabuk, C. saccharoperbutylacetonicum deltptabuk. Fermentation was carried out in replicates, and results from one batch are reported here as representative.
As illustrated in Fig. 3C, corresponding well with the long lag phase in cell growth, it took a long time for the pH to drop to pH 5.0 (about 20 h, 28 h, 24 h, and 28 h for the wild type, deltpta, deltbuk, and deltptabuk, respectively). Similarly, the pH was held at 5.0 for a much longer time (10 to 20 h) than in P2 medium before it increased to high levels. Production profiles for acetate and butyrate are demonstrated in Fig. 3D and E. Without presupplemented acetate in MP2, the deletion of the major acetate-producing gene pta led to an obvious decrease of acetate production by 14.5% and a significant increase of butyrate production by 52.0% in C. saccharoperbutylacetonicum deltpta compared to the wild type, while when the buk gene, responsible for butyrate production, was knocked out, a 20.0% decrease of butyrate production and a 175.4% increase of acetate production were observed in C. saccharoperbutylacetonicum deltbuk compared to the wild type. In C. saccharoperbutylacetonicum deltptabuk, the peak acetate production was comparable to that in the wild type, but the peak butyrate production was 44.0% more than that in the wild type (Table 2). Besides lactate and butyrate, high-level lactate was also detected in all the strains with MP2 (Fig. 3F). Compared with the lactate production in the wild type, improvements of 25.0% and 8.3% were observed in C. saccharoperbutylacetonicum deltbuk and deltptabuk, respectively, while C. saccharoperbutylacetonicum deltpta demonstrated the highest lactate production, with an 83.3% increase compared to the wild type (Table 2).
The ABE production profiles are shown in Fig. 3G to I and also summarized in Table 2. Compared to the wild type (18.4 g/liter), butanol production was slightly improved to 18.7 g/liter and 18.8 g/liter, respectively, in C. saccharoperbutylacetonicum deltpta and deltptabuk. However, the butanol yield was improved remarkably by 13.0% in these two mutants, due to their decreased consumption of sugars. The butanol production in C. saccharoperbutylacetonicum deltbuk decreased by 0.7 g/liter compared with that in the wild type; however, a butanol yield similar to that in the wild type was still obtained in C. saccharoperbutylacetonicum deltbuk, also due to the decreased consumption of sugar. Compared to the wild type, lower levels of acetone production were observed in all three mutants, with 42.9%, 11.7%, and 39.0% decreases in C. saccharoperbutylacetonicum deltpta, deltbuk, and deltptabuk, respectively. C. saccharoperbutylacetonicum deltbuk generated the same ethanol production as the wild type, but the ethanol production was reduced by 15.2% and 38.9% in C. saccharoperbutylacetonicum deltpta and deltptabuk, respectively. The butanol selectivity was improved by ∼10% in both C. saccharoperbutylacetonicum deltpta and deltptabuk, while that in deltbuk increased slightly.
DISCUSSION
C. saccharoperbutylacetonicum N1-4 is well known as a hyper-butanol-producing microorganism and was used for commercial butanol production in Japan from the 1960s until the shutdown of the butanol production facilities (11, 12, 49). As the type strain of the species C. saccharoperbutylacetonicum, strain N1-4 is very different from other extensively studied solventogenic clostridial strains, such as C. acetobutylicum (ATCC 824) and C. beijerinckii (NCIMB 8052), in terms of both genotype and phenotype (50, 51). C. saccharoperbutylacetonicum (N1-4) has the largest genome (6.7 Mb) among all the solventogenic clostridia characterized so far (49). It also has a 136-kb megaplasmid which contains no genes apparently related to solvent formation. This is very different from the case for C. beijerinckii, which does not have a plasmid, and C. acetobutylicum, which contains a 192-kb megaplasmid whose loss leads to strain degeneration (52). DNA transformation in C. saccharoperbutylacetonicum is difficult, possibly demonstrating a methylome very different from those previously reported (53–55). An amenable transformation protocol was reported very recently (21). On the other hand, C. saccharoperbutylacetonicum demonstrates very robust cell growth (reaching an OD600 of up to 16.0 as observe in this study, which has never been seen in batch fermentation with either C. beijerinckii or C. acetobutylicum), a very high butanol production capability, and high butanol tolerance and selectivity. It also has a comparatively low sporulation frequency and can perform ABE fermentation using a wide range of carbohydrates (12, 51, 56). However, compared with other ABE-producing species such as C. acetobutylicum and C. beijerinckii, the study of the genomics and genetics of C. saccharoperbutylacetonicum is lagging. To elucidate the complex fermentation metabolism in C. saccharoperbutylacetonicum and further develop improved strains for solvent production, the development of a versatile genetic engineering system is highly desirable. In this study, an efficient genome editing toolkit was developed based on the CRISPR-Cas9 system, paving the way for efficient genomic engineering in C. saccharoperbutylacetonicum. Based on this system, single and double knockout mutants with mutations related to the acid production pathways were generated.
First, we evaluated the CRISPR-Cas9 system that was customized for C. beijerinckii for genome engineering in C. saccharoperbutylacetonicum (37). The pta deletion mutant was obtained in the fourth generation of subculture with a mutation rate of 18.75%, and the buk deletion mutant was obtained in the seventh generation of subculture with a mutation rate of 6.25% (12.5% for buk deletion in generating the pta buk double deletion mutant). The varied efficiency in deleting different genes indicated that the CRISPR-Cas9 genome engineering efficiency could be affected by different target sites and homology arm sequences, as previously reported (46). Notwithstanding, the efficiency was much lower than that observed when the same system was used for genome engineering in C. beijerinckii (in that case, generally nearly a 100% mutation rate was observed for various genome engineering purposes) (26). This result was not surprising considering the differences between these two strains. Obviously, this customized high-efficiency genome engineering system for C. beijerinckii does not function well in C. saccharoperbutylacetonicum.
As indicated by previous reports (26, 57), CRISPR-Cas9-based genome engineering in bacterial strains very likely relies on the selection power of the Cas9 endonuclease against the background cells induced by DSB. gRNA expression is especially essential for the functionality of the whole system (45, 46). Therefore, we further set out to identify a more powerful promoter for gRNA expression. The veg promoter of the Gram-positive B. subtilis (Pvegb) has been employed for efficiency of gRNA expression in B. subtilis (47). Therefore, we decided to evaluate its efficiency for gRNA expression in C. saccharoperbutylacetonicum; meanwhile, the endogenous hypothetical veg promoter in C. saccharoperbutylacetonicum (Pvegc) was also selected. Additionally, the E. coli promoter PJ23119 has proven to be effective for gRNA expression in E. coli and recently in solventogenic C. acetobutylicum and C. beijerinckii (33, 38, 48, 58). Therefore, this promoter was also selected to drive the gRNA expression in this study. As shown in Table 1, by following the same procedure to obtain the pta deletion mutant, it was determined that the order of the promoters for efficient gRNA expression (and thus genome editing efficiency) is PJ23119 > Pvegb > PsRNA > Pvegc. With PJ23119, a high mutation rate of 75% was obtained through screening the colonies from the second generation of subculture. This represents a very desirable genome engineering efficiency. Though the mutation rate was not as high as that obtained in C. beijerinckii (100%), mutants can be easily obtained through screening after one generation of subculturing in TGYE followed by one generation of incubation in TGYLE without serial transfers as done in C. beijerinckii (26). Comparatively, lower efficiency was obtained with Pvegc for gRNA expression. It is worthwhile to point out that the total 325 bp including the whole Pvegc region was used in this test because the exact transcription start site of the veg gene was unknown. It is possible that an improved efficiency could be achieved if we did a careful selection of the Pvegc sequence to be used for gRNA expression (especially, the ribosome binding site [RBS] was likely redundant and might have a negative impact since gRNA is a noncoding RNA). It has been reported that the strong expression of gRNA is essential for the functionality of the CRISPR-Cas9 system (45, 46). Along this line, in this study, we provided solid data to verify that the achievement of potent gRNA expression is the critical step to develop a high-efficiency CRISPR-Cas9 system for genome engineering.
As reported for other Clostridium strains (44, 59), the inactivation of the pta gene did not completely eliminate the acetate formation, indicating that additional acetate-forming pathways might exist in C. saccharoperbutylacetonicum. It needs to be pointed out that the acid production pathways in many of the previously characterized mutants were disrupted with TargeTron-based intron insertion approaches (43, 44). It is suspected that residual activities of the enzymes encoded by disrupted genes can still exist in the mutant. In this study, clean and complete deletion of the primary acid production genes was obtained using the CRISPR-Cas9 system; the corresponding acid production in the mutant was not eliminated either. In the fermentation with P2 medium, the deletion of pta resulted in a slight decrease of acetate production in C. saccharoperbutylacetonicum deltpta and deltptabuk (Fig. 2D and Table 2). Since ATP is the fundamental demand for cell growth and other metabolisms (60) and more ATP per glucose can be generated through the acetate formation pathway than through the butyrate formation pathway, the deletion of pta imposed a severe metabolic burden on the cells (61). To compensate for the energy loss and avoid a significant decline in overall cell growth, more carbon flow was directed to butyrate formation, leading to a >7% increase of butyrate production in both C. saccharoperbutylacetonicum deltpta and deltptabuk. Similarly, the deletion of the buk gene led to a 14.3% increase of acetate production in C. saccharoperbutylacetonicum deltbuk and a slight decrease of butyrate production. Production of butyrate was still observed in C. saccharoperbutylacetonicum deltbuk and deltptabuk, suggesting the existence of other active butyrate formation pathway genes. Besides buk (GenBank accession no. AGF54071.1), which was deleted in this study, there are two other butyrate kinase-encoding genes in the C. saccharoperbutylacetonicum N1-4 genome, which are denoted butyrate kinase 1 (WP_015392233.1) and butyrate kinase 2 (WP_015395384.1). Further study is in progress in our lab to evaluate the contributions of these three butyrate kinases to butyrate production in this microorganism.
In MP2 medium without preadded acetate, long lag phases were observed in all the mutants as well as in the wild type (Fig. 3A), indicating a positive effect of acetate on cell growth, as also reported previously (62, 63). The mutants showed lower growth rates than the wild type (Table 2); the disruption of acid production pathways might lead to the decline in energy generation and thus the metabolic burden on cell growth (59). In MP2 medium with the elimination of the interference from the preadded acetate, C. saccharoperbutylacetonicum deltpta clearly demonstrated reduced acetate production and enhanced butyrate production to compensate for the ATP generation. Similarly, the buk deletion in C. saccharoperbutylacetonicum deltbuk led to an apparent decrease of butyrate production and a significant increase of acetate production to balance the ATP generation. These results confirmed that the deleted pta (GenBank accession no. AGF55073.1) and buk (GenBank accession no. AGF54071.1) are the primary acid formation pathway genes for acetate and butyrate production, respectively, although other annotated homologous genes exist. A comprehensive transcriptomic analysis is desirable to further enhance the genome annotation and assess the function of each specific gene (64, 65).
As reported previously, lactate dehydrogenase in solventogenic clostridia shows remarkable activities only under certain growth conditions, such as iron limitation (66). However, we observed that the wild-type C. saccharoperbutylacetonicum N1-4 produced 1.6 g/liter lactate (at the peak level) in P2 medium and 1.2 g/liter in MP2 medium, which was even more than the production of acetate, while under similar conditions, less than 0.2 g/liter lactate was produced by C. beijerinckii 8052 (44) and less than 0.1 g/liter lactate was observed in C. acetobutylicum ATCC 824 (66). Therefore, the lactate formation pathway may be more active in C. saccharoperbutylacetonicum N1-4. Moreover, there are six genes annotated as lactate dehydrogenase genes in C. saccharoperbutylacetonicum; the action of these genes and the mechanism for lactate production and reutilization in C. saccharoperbutylacetonicum need to be further elucidated. Similar to the results in C. beijerinckii (44), deletion of pta or buk led to the improvement of lactate production; especially in the pta deletion mutant, the lactate production was increased by 83.3% compared to that of the wild type in MP2 medium. It was reported that the lactate formation pathway can serve as a less efficient method for energy generation and NADH oxidation when the pathway for the disposal of protons and electrons by the generation of molecular hydrogen is inhibited (44). The increase of lactate production may help mitigate the decreased ATP production in the mutants.
In P2 medium, the deletion of the buk gene in C. saccharoperbutylacetonicum deltbuk slightly increased the butanol production and butanol yield while significantly decreasing the production of acetone (by 11.7%) and ethanol (by 16.7%), resulting in a higher butanol selection. In C. saccharoperbutylacetonicum deltptabuk, the acetone production was decreased by 15.6%, and the butanol production was improved by 1.3 g/liter, reaching 19.0 g/liter. Therefore, a much higher butanol selectivity (72.4%, compared to 68.2% in the wild type) was observed. The butanol production reported here represents one of the highest ever reported from batch fermentations for biobutanol production (43). However, a significant improvement in solvent production in the deltptabuk strain compared to the wild type was not observed, as previously reported in the mutant of C. acetobutylicum with the same homologous genes disrupted (43). This from another angle indicated that C. saccharoperbutylacetonicum is different from the extensively studied canonical C. acetobutylicum in terms of the regulation of solvent production metabolism. Butanol-producing strains can rarely tolerate >2% butanol, and thus the butanol production titer in the regular batch fermentation can hardly exceed 20.0 g/liter (67). Butanol tolerance is considered one of the major bottlenecks in improving butanol production in the host. Therefore, the further enhancement of butanol tolerance through systematic metabolic engineering might be a rational strategy in order to further improve the final butanol production in C. saccharoperbutylacetonicum. With MP2 medium, the butanol yield was improved by 13.0% in both C. saccharoperbutylacetonicum deltpta and deltptabuk compared to that from the wild type. Furthermore, the butanol selectivity was also improved by ∼10% in C. saccharoperbutylacetonicum deltpta and deltptabuk compared to the wild type in the fermentation with MP2 medium. With these desirable butanol-producing features, the mutants constructed in this study can be used as excellent starting strains to construct more robust ones for butanol production through metabolic engineering. Considering that the ABE fermentation is medium dependent (as shown with P2 and MP2 media) (44), fermentation conditions can be further optimized for the mutants to achieve more desirable solvent production.
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and oligonucleotides.
All strains and plasmids used in this study are listed in Table 3, and DNA primers are listed in Table S1 in the supplemental material. E. coli strain DH5α was used for DNA cloning. It was grown aerobically at 37°C in Luria-Bertani (LB) medium supplemented with 100 μg/ml of ampicillin (Amp) as needed. C. saccharoperbutylacetonicum N1-4 was grown in an anaerobic chamber (N2-CO2-H2 with a volume ratio of 85:10:5) at 35°C in tryptone-glucose-yeast extract (TGY) medium containing 30 g/liter of tryptone, 20 g/liter of glucose, 10 g/liter of yeast extract, and 1 g/liter of l-cysteine (44). TGY supplemented with 40 mM lactose (TGYL) was used to induce the Cas9 expression. When appropriate, 25 μg/ml of erythromycin (Erm) was added to either TGY or TGYL to make the TGYE or TGYLE medium, respectively, for C. saccharoperbutylacetonicum cultivation.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | NEB |
| C. beijerinckii NCIMB 8052 | ATCC 51743 | ATCC |
| C. saccharoperbutylacetonicum | ||
| N1-4 | ATCC 13564, wild-type strain | ATCC |
| deltpta | N1-4 Δpta | This work |
| deltbuk | N1-4 Δbuk | This work |
| deltptabuk | N1-4 Δpta Δbuk | This work |
| Plasmids | ||
| pYW34 | CAK1 ori Ampr Ermr Plac::Cas9 gRNA | 26 |
| pYW34-pta | pYW34 PsRNA::20-nt gRNA targeting pta | This work |
| pYW34-deltpta | pYW34-pta::homology arm sequences | This work |
| pYW34-buk | pYW34 PsRNA::20-nt gRNA targeting buk | This work |
| pYW34-deltbuk | pYW34-buk::homology arm sequences | This work |
| pSHW1-pta | pYW34 Pvegb::20-nt gRNA targeting pta | This work |
| pSHW2-pta | pYW34 Pvegc::20-nt gRNA targeting pta | This work |
| pSHW3-pta | pYW34 PJ23119::20-nt gRNA targeting pta | This work |
| pSHW1-deltpta | pSHW1-pta::homologous arm sequences | This work |
| pSHW2-deltpta | pSHW2-pta::homologous arm sequences | This work |
| pSHW3-deltpta | pSHW3-pta::homologous arm sequences | This work |
PsRNA, small RNA promoter from C. beijerinckii; Pvegb, veg promoter from B. subtilis; Pvegc, hypothetical veg gene promoter from C. saccharoperbutylacetonicum; PJ23119, J23119 promoter from E. coli.
Plasmid construction.
Plasmid pYW34 (26) carrying the Cas9 open reading frame (ORF) from S. pyogenes under the control of a lactose-inducible promoter along with the chimeric gRNA sequence was used as the mother vector to construct plasmids for disrupting the pta (GenBank accession no. AGF55073.1) and buk (GenBank accession no. AGF54071.1) genes. To delete pta, the small RNA (sRNA) (sCbei_5830) promoter fused with the 20-nt guiding sequence (5′-TCTGGAGCAGTTCATACAAC-3′) was amplified from C. beijerinckii 8052 genomic DNA using primers YW484 and YW1041 and inserted into the BtgZI site of pYW34 with Gibson Assembly, generating pYW34-pta. To achieve the desirable homologous recombination around the pta ORF, two homology arms (1,076 bp and 1,078 bp) flanking at both sides of the pta ORF were amplified from C. saccharoperbutylacetonicum genomic DNA with primers YW1035 and YW1036 and primers YW1037 and YW1038, respectively, and then these two fragments were fused together with primers YW1039 and YW1040 through splicing by overlapping extension PCR (SOE-PCR), and inserted into the NotI site of pYW34-pta, generating pYW34-deltpta.
Similarly, for targeting on buk, the fragment containing the sRNA promoter and 20-nt guiding sequence (5′-GGAATGCTTAAGCCAGTAGA-3′) was amplified with primers YW484 and YW1053 and then inserted into the BtgZI site of pYW34, generating pYW34-buk. One 1-kb homology arm sequence upstream of the start codon (ATG) of buk was amplified using primers YW949 and YW950, and another 1-kb homology arm sequence downstream of the stop codon (TAA) of buk was amplified using primers YW951 and YW952. The whole 2-kb homology arm was then obtained through SOE-PCR with primers YW949 and YW952 and inserted into the NotI site of pYW34-buk, generating the plasmid pYW34-deltbuk.
To improve the genomic editing efficiency in C. saccharoperbutylacetonicum using the CRISPR-Cas9 system, other strong constitutive promoters were evaluated for gRNA expression (targeting the same 20-nt sequence in pta as described above), including the veg promoter from B. subtilis (Pvegb) (47), the hypothetical veg (CSPA_RS02900) gene promoter (Pvegc) from C. saccharoperbutylacetonicum, and the E. coli promoter PJ23119 (48). The Pvegb promoter (65 bp) from B. subtilis with the 20-nt guiding sequence was amplified using primer pairs YW1340 -YW1342 and YW1341-YW1342 in two steps and inserted into the BtgZI and NotI sites of pYW34, generating pSHW1-pta. The Pvegc promoter fused with the 20-nt guiding sequence was amplified from C. saccharoperbutylacetonicum N1-4 with primers YW1042 and YW1043 and inserted into the BtgZI site of pYW34, generating pSHW2-pta, while the PJ23119 promoter fused with the 20-nt guiding sequence was amplified using primer pairs YW1338-YW1342 and YW1339-YW1342 in two steps and inserted into the BtgZI and NotI sites of pYW34, generating pSHW3-pta. Subsequently, the fragment containing the two homology arm sequences for pta deletion was inserted into the NotI site of pSHW1-pta, pSHW2-pta, and pSHW3-pta, generating pSHW1-deltpta, pSHW2-deltpta, and pSHW3-deltpta, respectively.
All cloning PCR was carried out using high-fidelity DNA polymerases, either Phusion (New England BioLabs Inc., Ipswich, MA) or PrimeSTAR (TaKaRa Bio USA, Inc., Mountain View, CA). All constructs were verified through Sanger sequencing.
DNA transformation and mutant screening.
Transformation of C. saccharoperbutylacetonicum N1-4 was carried out with electroporation following the protocol for the transformation of C. beijerinckii as described by Wang et al. with minor modifications (44). C. saccharoperbutylacetonicum N1-4 was cultivated anaerobically at 35°C until the optical density of the culture at 600 nm (OD600) reached 0.8 to 1.0. The cells were harvested through centrifugation at 4,200 × g at 4°C for 10 min. The cell pellets were washed once with the same volume (as the original cell culture) of ice-cold 15% glycerol and then resuspended in 1/20 volume of ice-cold 15% glycerol. Immediately, 1.0 μg of plasmid DNA was added to 400 μl of competent cells, and then the mixture was transferred into a precooled 0.2-cm electroporation cuvette. Electroporation was carried out at a voltage of 1,500 V, a capacitance of 25 μF, and a resistance of 300 Ω using a Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA) which was connected to the anaerobic chamber. Subsequently, the cells were transferred into 1.6 ml of TGY medium and recovered anaerobically at 35°C (generally for 10 to 12 h until signs of cell growth were observed). The recovered cells were spread onto TGYE agar plates and incubated at 35°C for about 24 h under anaerobic conditions until colonies appeared.
Following the outgrowth on selective plates, single colonies were picked randomly for colony PCR (cPCR) using primers YW880 and YW881 to confirm the presence of plasmid. The transformants were then incubated in TGYE liquid medium and subcultured successively to permit efficient homologous recombination (37). Starting from the second generation, while the culture continued to be transferred in TGYE medium, it was also transferred into TGYLE liquid medium for the induction of Cas9 expression and further spread onto TGYLE plates when necessary (that is, when double bands from both the mutant and the wild type were detected through cPCR with TGYLE liquid culture as the template). Mutants were screened through cPCR using primers (YW1044 and YW1045 for detecting pta deletion and YW953 and YW954 for detecting buk deletion) annealing to the chromosomal loci beyond the homologous recombination regions (and thus the primers can only anneal to the chromosome and not to the homology arms on the plasmids).
Due to the existence of plasmids harboring homologous sequences within the mutant strain, vector integration events (VIE) could occur during the bacterial genome engineering (26). To detect VIE, pairs of primers (one annealing to the plasmid and the other annealing to the chromosome) were designed. Specifically, primer pairs YW1044-YW200 and YW847-YW1045 were used for detecting VIE of pYW34-deltpta both upstream and downstream of the potential VIE, while YW953-YW200 and YW847-YW 954 were used for detecting VIE of pYW34-delbuk in a similar manner (see Fig. S2 in the supplemental material).
Plasmid curing.
To cure the donor plasmid pYW34-deltpta or pYW34-deltbuk, mutants that were identified without vector integration were subcultured in TGY medium without antibiotics as described previously (44, 68). After around five cycles of subculturing, the culture was spread onto TGY plates to form colonies. cPCR was performed to confirm the curing of plasmids using primers YW880 and YW881. Colonies lacking PCR products were spread onto TGYE plates to further confirm the loss of plasmids. Finally, cPCR was conducted to further verify the existence of the desirable mutation.
Fermentation.
Bach fermentation was performed in BioFlo 115 benchtop bioreactors (New Brunswick Scientific Co., Enfield, CT) with a working volume of 1.5 liters. P2 medium or modified P2 (MP2) medium (44) was used as the fermentation medium, and 80 g/liter glucose was used as the sole carbon source. To generate an anaerobic condition, oxygen-free nitrogen was sparged through the fermentation broth starting several hours before the inoculation until the cell culture initiated its own gas production.
Mutant strains as well as the wild type (stored as glycerol stocks) were propagated in TGY medium in anaerobic chambers until the OD600 reached 0.8. The culture was then inoculated into the reactor at a 5% (vol/vol) inoculum ratio. Fermentation was performed at 30°C with agitation at 50 rpm. The pH was controlled above 5.0 throughout the fermentation by adding 1 M NaOH. All fermentations were performed in duplicate.
Analytical methods.
Cell density (OD600) was measured with an Ultrospec 10 cell density meter (Amersham Biosciences Corp., Piscataway, NJ). The NBS BioCommand software (New Brunswick Scientific Co, Inc., Edison, NJ) was used to record pH profiles in real time throughout the fermentation. Concentrations of acetone, butanol, and ethanol (ABE), acids (acetate, butyrate, and lactate), and glucose were determined using high-performance liquid chromatography (Agilent Technologies 1260 Infinity series) with a refractive index detector (RID) and a diode array UV detector (DAD) equipped with an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA). The column was eluted with 0.005 N H2SO4 with a flow rate of 0.6 ml/min at 25°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hans Blaschek (from the University of Illinois at Urbana-Champaign) for providing the plasmids. We also thank Jason Peters (from Carol Gross' group at the University of California—San Francisco) for sharing the Pvegb sequence information with us.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00233-17.
REFERENCES
- 1.Jang YS, Lee J, Malaviya A, Seung DY, Cho JH, Lee SY. 2012. Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. Biotechnol J 7:186–198. doi: 10.1002/biot.201100059. [DOI] [PubMed] [Google Scholar]
- 2.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. 2008. Fermentative butanol production by Clostridia. Biotechnol Bioeng 101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 3.Buehler EA, Mesbah A. 2016. Kinetic study of acetone-butanol-ethanol fermentation in continuous culture. PLoS One 11:e0158243. doi: 10.1371/journal.pone.0158243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croux C, Lee J, Raynaud C, Saint-Prix F, Gonzalez-Pajuelo M, Meynial-Salles I, Soucaille P. 2016. Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum. Biotechnol Biofuels 9:23. doi: 10.1186/s13068-015-0423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo M, Bestel-Corre G, Croux C, Riviere A, Meynial-Salles I, Soucaille P. 2015. A quantitative system-scale characterization of the metabolism of Clostridium acetobutylicum. mBio 6:e01808-15. doi: 10.1128/mBio.01808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Xu S, Chai C, Yang S, Jiang W, Minton NP, Gu Y. 2016. Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum. FEMS Microbiol Lett 363:fnw065. doi: 10.1093/femsle/fnw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellido C, Infante C, Coca M, González-Benito G, Lucas S, García-Cubero MT. 2015. Efficient acetone-butanol-ethanol production by Clostridium beijerinckii from sugar beet pulp. Bioresour Technol 190:332–338. doi: 10.1016/j.biortech.2015.04.082. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang ZT, Seo SO, Lynn P, Lu T, Jin YS, Blaschek HP. 2016. Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol Bioeng 113:2739–2743. doi: 10.1002/bit.26020. [DOI] [PubMed] [Google Scholar]
- 9.Dash S, Ng CY, Maranas CD. 2016. Metabolic modeling of clostridia: current developments and applications. FEMS Microbiol Lett 363:fnw004. doi: 10.1093/femsle/fnw004. [DOI] [PubMed] [Google Scholar]
- 10.Green EM. 2011. Fermentative production of butanol—the industrial perspective. Curr Opin Biotechnol 22:337–343. doi: 10.1016/j.copbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Hongo M, Murata A. 1965. Bacteriophages of Clostridium saccharoperbutylacetonicun. I. Some characteristics of the twelve phages obtained from the abnormally fermented broths. II. Enumeration of phages by the application of the plaque-count technique and some factors influencing the plaque formation. Agric Biol Chem 29:1135–1145. [Google Scholar]
- 12.Motoyoshi H. July 1960. Process for producing butanol by fermentation. US patent 2945786 A.
- 13.Al-Shorgani NKN, Kalil MS, Yusoff WMW. 2012. Fermentation of sago starch to biobutanol in a batch culture using Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564). Ann Microbiol 62:1059–1070. doi: 10.1007/s13213-011-0347-x. [DOI] [Google Scholar]
- 14.Tashiro Y, Shinto H, Hayashi M, Baba Si Kobayashi G, Sonomoto K. 2007. Novel high-efficient butanol production from butyrate by non-growing Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564) with methyl viologen. J Biosci Bioeng 104:238–240. doi: 10.1263/jbb.104.238. [DOI] [PubMed] [Google Scholar]
- 15.Tashiro Y, Takeda K, Kobayashi G, Sonomoto K, Ishizaki A, Yoshino S. 2004. High butanol production by Clostridium saccharoperbutylacetonicum N1-4 in fed-batch culture with pH-stat continuous butyric acid and glucose feeding method. J Biosci Bioeng 98:263–268. doi: 10.1016/S1389-1723(04)00279-8. [DOI] [PubMed] [Google Scholar]
- 16.Thang VH, Kanda K, Kobayashi G. 2010. Production of acetone-butanol-ethanol (ABE) in direct fermentation of cassava by Clostridium saccharoperbutylacetonicum N1-4. Appl Biochem Biotechnol 161:157–170. doi: 10.1007/s12010-009-8770-1. [DOI] [PubMed] [Google Scholar]
- 17.del Cerro C, Felpeto-Santero C, Rojas A, Tortajada M, Ramón D, García JL. 2013. Genome sequence of the butanol hyperproducer Clostridium saccharoperbutylacetonicum N1-4. Genome Announc 1:e00070-13. doi: 10.1128/genomeA.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keis S, Bennett CF, Ward VK, Jones DT. 1995. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int J Syst Evol Microbiol 45:693–705. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama S, Irie R, Kosaka T, Matsuura K, Yoshino S, Furukawa K. 2007. New host-vector system in solvent-producing Clostridium saccharoperbutylacetonicum strain N1-4. J Gen Appl Microbiol 53:53–56. doi: 10.2323/jgam.53.53. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama S-I, Kosaka T, Hirakawa H, Matsuura K, Yoshino S, Furukawa K. 2008. Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1-4. Appl Microbiol Biotechnol 78:483–493. doi: 10.1007/s00253-007-1323-z. [DOI] [PubMed] [Google Scholar]
- 21.Herman NA, Li J, Bedi R, Turchi B, Liu X, Miller MJ, Zhang W. 11 November 2016. Development of a high-efficiency transformation method and implementation of rational metabolic engineering for the industrial butanol hyper-producer Clostridium saccharoperbutylacetonicum strain N1-4. Appl Environ Microbiol doi: 10.1128/AEM.02942-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang ZT, Seo SO, Lynn P, Lu T, Jin YS, Blaschek HP. 2016. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth Biol 5:721–732. doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- 27.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doudna JA, Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 29.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutschner T, Hämmerle M, Genovese G, Draetta GF, Chin L. 2016. Cas9 protein engineering for cell cycle-specific genome editing to enhance homology directed repair. Cancer Res 76:71–71. doi: 10.1158/1538-7445.AM2016-71. [DOI] [Google Scholar]
- 31.Chung ME, Yeh I, Sung LY, Wu MY, Chao YP, Ng IS, Hu YC. 2017. Enhanced integration of large DNA into E. coli chromosome by CRISPR/Cas9. Biotechnol Bioeng 114:172–183. doi: 10.1002/bit.26056. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Zheng G, Jiang W, Hu H, Lu Y. 2015. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin 47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Cobb R, Zhao H. 2016. High-efficiency genome editing of Streptomyces species by an engineered CRISPR/Cas system. Methods Enzymol 575:271–284. doi: 10.1016/bs.mie.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Cobb RE, Wang Y, Zhao H. 2015. High-efficiency multiplex genome editing of streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. 2016. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep 6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Zhang ZT, Seo SO, Choi K, Lu T, Jin YS, Blaschek HP. 2015. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol 200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Chen J, Minton NP, Zhang Y, Wen Z, Liu J, Yang H, Zeng Z, Ren X, Yang J. 2016. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol J 11:961–72. doi: 10.1002/biot.201600053. [DOI] [PubMed] [Google Scholar]
- 39.Bruder MR, Pyne ME, Moo-Young M, Chung DA, Chou CP. 2016. Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in the genus Clostridium. Appl Environ Microbiol 82:6109–6119. doi: 10.1128/AEM.02128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hüsemann MH, Papoutsakis ET. 1988. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic acid and proton concentrations. Biotechnol Bioeng 32:843–852. doi: 10.1002/bit.260320702. [DOI] [PubMed] [Google Scholar]
- 41.Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol Rev 50:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann D, Hönicke D, Ehrenreich A, Schmidt M, Weuster-Botz D, Bahl H, Lütke-Eversloh T. 2012. Modifying the product pattern of Clostridium acetobutylicum. Appl Microbiol Biotechnol 94:743–754. doi: 10.1007/s00253-011-3852-8. [DOI] [PubMed] [Google Scholar]
- 43.Jang YS, Lee JY, Lee J, Park JH, Im JA, Eom MH, Lee J, Lee SH, Song H, Cho JH. 2012. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio 3:e00314-12. doi: 10.1128/mBio.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Li X, Milne CB, Janssen H, Lin W, Phan G, Hu H, Jin YS, Price ND, Blaschek HP. 2013. Development of a gene knockout system using mobile group II introns (Targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii. Appl Environ Microbiol 79:5853–5863. doi: 10.1128/AEM.00971-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu T, Li Y, Van Nostrand JD, He Z, Zhou J. 2014. Cas9-based tools for targeted genome editing and transcriptional control. Appl Environ Microbiol 80:1544–1552. doi: 10.1128/AEM.03786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. 2013. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CH, Koo BM, Marta E. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poehlein A, Krabben P, Dürre P, Daniel R. 2014. Complete genome sequence of the solvent producer Clostridium saccharoperbutylacetonicum strain DSM 14923. Genome Announc 2:e01056-14. doi: 10.1128/genomeA.01056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson J, Toth J, Santiwatanakul S, Chen JS. 1997. Cultures of “Clostridium acetobutylicum” from various collections comprise Clostridium acetobutylicum, Clostridium beijerinckii, and two other distinct types based on DNA-DNA reassociation. Int J Syst Evol Microbiol 47:420–424. [DOI] [PubMed] [Google Scholar]
- 51.Keis S, Shaheen R, Jones DT. 2001. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int J Syst Evol Microbiol 51:2095–2103. doi: 10.1099/00207713-51-6-2095. [DOI] [PubMed] [Google Scholar]
- 52.Cornillot E, Nair RV, Papoutsakis ET, Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J Bacteriol 179:5442–5447. doi: 10.1128/jb.179.17.5442-5447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mermelstein L, Papoutsakis E. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 59:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lesiak JM, Liebl W, Ehrenreich A. 2014. Development of an in vivo methylation system for the solventogen Clostridium saccharobutylicum NCP 262 and analysis of two endonuclease mutants. J Biotechnol 188:97–99. doi: 10.1016/j.jbiotec.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2014. Expansion of the genetic toolkit for metabolic engineering of Clostridium pasteurianum: chromosomal gene disruption of the endogenous CpaAI restriction enzyme. Biotechnol Biofuels 7:163. doi: 10.1186/s13068-014-0163-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaheen R, Shirley M, Jones DT. 2000. Comparative fermentation studies of industrial strains belonging to four species of solvent-producing clostridia. J Mol Microbiol Biotechnol 2:115–124. [PubMed] [Google Scholar]
- 57.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiktor J, Lesterlin C, Sherratt DJ, Dekker C. 2016. CRISPR-mediated control of the bacterial initiation of replication. Nucleic Acids Res doi: 10.1093/nar/gkw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Zhu Y, Yang S-T. 2006. Butyric acid and hydrogen production by Clostridium tyrobutyricum ATCC 25755 and mutants. Enzyme Microb Technol 38:521–528. doi: 10.1016/j.enzmictec.2005.07.008. [DOI] [Google Scholar]
- 60.Liu J, Guo T, Wang D, Shen X, Liu D, Niu H, Liang L, Ying H. 2016. Enhanced butanol production by increasing NADH and ATP levels in Clostridium beijerinckii NCIMB 8052 by insertional inactivation of Cbei_4110. Appl Microbiol Biotechnol 100:4985–4996. doi: 10.1007/s00253-016-7299-9. [DOI] [PubMed] [Google Scholar]
- 61.Zhu Y, Liu X, Yang ST. 2005. Construction and characterization of pta gene-deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid fermentation. Biotechnol Bioeng 90:154–166. doi: 10.1002/bit.20354. [DOI] [PubMed] [Google Scholar]
- 62.Chen C-K, Blaschek HP. 1999. Effect of acetate on molecular and physiological aspects of Clostridium beijerinckii NCIMB 8052 solvent production and strain degeneration. Appl Environ Microbiol 65:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C-K, Blaschek H. 1999. Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl Microbiol Biotechnol 52:170–173. doi: 10.1007/s002530051504. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Li X, Mao Y, Blaschek HP. 2011. Single-nucleotide resolution analysis of the transcriptome structure of Clostridium beijerinckii NCIMB 8052 using RNA-Seq. BMC Genomics 12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y. 2012. Transcriptomic analyses of Clostridium beijerinckii NCIMB 8052 during transition from acidogenesis to solventogenesis and under butyrate supplemented conditions. Ph.D. thesis. University of Illinois, Urbana-Champaign, IL. [Google Scholar]
- 66.Bahl H, Gottwald M, Kuhn A, Rale V, Andersch W, Gottschalk G. 1986. Nutritional factors affecting the ratio of solvents produced by Clostridium acetobutylicum. Appl Environ Microbiol 52:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu S, Qureshi N. 2009. How microbes tolerate ethanol and butanol. N Biotechnol 26:117–121. doi: 10.1016/j.nbt.2009.06.984. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M, Liao JC, Schadt CW, Guss AM, Yang Y, Graham DE. 2012. Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol Biofuels 5:2. doi: 10.1186/1754-6834-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.