ABSTRACT
Among the five serine incorporator (SERINC) family members, SERINC5 (Ser5) was reported to strongly inhibit HIV-1 replication, which is counteracted by Nef. Ser5 produces 5 alternatively spliced isoforms: Ser5-001 has 10 putative transmembrane domains, whereas Ser5-004, -005, -008a, and -008b do not have the last one. Here, we confirmed the strong Ser5 anti-HIV-1 activity and investigated its isoforms' expression and antiviral activities. It was found that Ser5-001 transcripts were detected at least 10-fold more than the other isoforms by real-time quantitative PCR. When Ser5-001 and its two isoforms Ser5-005 and Ser5-008a were expressed from the same mammalian expression vector, only Ser5-001 was stably expressed, whereas the others were poorly expressed due to rapid degradation. In addition, unlike the other isoforms, which are located mainly in the cytoplasm, Ser5-001 is localized primarily to the plasma membrane. To map the critical determinant, Ser5 mutants bearing C-terminal deletions were created. It was found that the 10th transmembrane domain is required for Ser5 stable expression and plasma membrane localization. As expected, only Ser5-001 strongly inhibits HIV-1 infectivity, whereas the other Ser5 isoforms and mutants that do not have the 10th transmembrane domain show very poor activity. It was also observed that the Nef counteractive activity could be easily saturated by Ser5 overexpression. Thus, we conclude that Ser5-001 is the predominant antiviral isoform that restricts HIV-1, and the 10th transmembrane domain plays a critical role in this process by regulating its protein stability and plasma membrane targeting.
IMPORTANCE Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) express a small protein, Nef, to enhance viral pathogenesis in vivo. Nef has an important in vitro function, which is to make virus particles more infectious, but the mechanism has been unclear. Recently, Nef was reported to counteract a novel anti-HIV host protein, SERINC5 (Ser5). Ser5 has five alternatively spliced isoforms, Ser5-001, -004, -005, -008a, and -008b, and only Ser5-001 has an extra C-terminal transmembrane domain. We now show that the Ser5-001 transcripts are produced at least 10-fold more than the others, and only Ser5-001 produces stable proteins that are targeted to the plasma membrane. Importantly, only Ser5-001 shows strong anti-HIV-1 activity. We further demonstrate that the extra transmembrane domain is required for Ser5 stable expression and plasma membrane localization. These results suggest that plasma membrane localization is required for Ser5 antiviral activity, and Ser5-001 is the predominant isoform that contributes to the activity.
KEYWORDS: HIV-1, Nef, SERINC3, SERINC5, infectivity, restriction factor
INTRODUCTION
Nef is a 27- to 32-kDa myristoylated protein produced by all primate lentiviruses (1). It is expressed during the early stage of the viral replication cycle and localizes primarily to the cytoplasm (2). Nef also localizes partially to the plasma membrane via its N-terminal myristoylation site and is enriched in lipid rafts (3, 4). Like the other viral accessory proteins, Nef manipulates the host's cellular environment and promotes infection via three major activities. First, it lowers the T cell activation threshold for viral replication by modulating T cell signaling pathways. Second, it allows the virus to evade the host immune response during infection by downregulation of a number of cell surface molecules. Third, it directly increases the infectivity of virions released from viral producer cells (5).
The mechanism by which Nef increases HIV-1 infectivity has been poorly understood. It was reported ∼20 years ago that nef-deficient (ΔNef) HIV-1 shows lower infectivity in cell culture than its wild-type (WT) counterpart (6–8). Although Nef-mediated CD4 downregulation also promotes HIV-1 replication by preventing CD4/gp120 complex formation (9, 10), the increase of viral infectivity by Nef does not depend on CD4 (11, 12). Levels of infectivity increase by Nef range from 5- to 50-fold depending on the cell types when a single-round viral replication assay is used, and the most significant increase is produced in human Jurkat T cell lines (13). Such a Nef effect is detectable only when Nef is expressed in viral producer cells, but not target cells, and enhances viral replication at an early step of infection (14). In addition, the enhancement is dependent on viral envelope (Env) glycoproteins. When HIV-1 is pseudotyped with the vesicular stomatitis virus envelope glycoproteins (VSV G) (15) or the subgroup A Rouse sarcoma virus (RSV-A) Env glycoproteins (16), but not the amphotropic murine leukemia virus (A-MLV) Env glycoproteins (14) or the RSV-A receptor Tva (16), Nef becomes unable to increase viral infectivity. In addition, Nef becomes much less effective at increasing viral infectivity when viruses are enveloped with HIV-1 Envs from some primary isolates, such as ADA, YU2, and JRFL (17). The Nef activity can be substituted for by a portion of the MLV glycosylated-Gag protein gPr80gag, which retains only the N-terminal cytoplasmic domain, the transmembrane region, and most of the matrix (MA) domain, termed glycoMA (13, 18). Moreover, Nef has been found to interact with the cholesterol biosynthesis pathway to increase HIV-1 infectivity (19, 20). Together, these pioneering works provided valuable insights into how Nef enhances HIV-1 infectivity in viral producer cells, but the precise mechanism was still quite unclear.
Very recently, two of the serine incorporator (SERINC) family members have been reported to inhibit HIV-1 replication in a Nef-sensitive manner. SERINC (Ser) proteins have 5 members, Ser1 to Ser5, with 10 or 11 transmembrane domains, and belong to the type III integral membrane proteins based on the J. S. Singer classification (21). These proteins transport serine molecules into the hydrophobic membrane lipid bilayers and incorporate them into phosphatidylserine and sphingolipids, which are important components of membrane phospholipids essential for cellular functions (21). Although their physiological functions are still unknown, Ser3 and Ser5 were found to reduce HIV-1 infectivity in viral producer cells when Nef was not expressed (22, 23). Ser3 was identified from HIV-1 virions by proteomics, where Ser3 was detectable only in the absence of Nef (23). Ser5 was identified from a panel of human T cell lines by transcriptomic analysis, where Nef-dependent HIV-1 replication positively correlated with Ser5 expression (22). Their antiviral activity and sensitivity to Nef were validated by both ectopic gene expression and gene knockout. They are incorporated into HIV-1 virions and inhibit viral replication at a very early step in target cells, their activity is no longer present if HIV-1 is enveloped with Nef-insensitive HIV-1 Env proteins or pseudotyped with VSV G, their activity is blocked by Nef and glycoMA proteins, and Nef was no longer required for HIV-1 replication after both genes were knocked out from viral producer cells (22, 23). Thus, their antiviral profiles meet expectations for the Nef-targeted host restriction factors. Here, we investigated how Ser5 is expressed in host cells and inhibits HIV-1 replication.
RESULTS
Analysis of Ser family anti-HIV-1 activities.
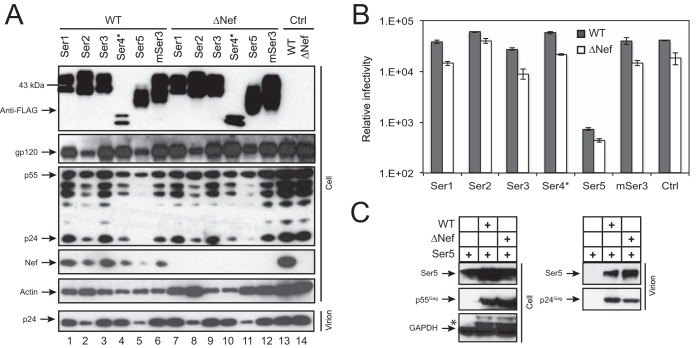
Human Ser1, Ser2, Ser3, Ser4*, and Ser5-001 and murine Ser3 (mSer3) were expressed from the pCMV6-Entry mammalian expression vector. The vector has a human cytomegalovirus (CMV) immediate-early enhancer and promoter to drive target gene expression and adds a Myc tag followed by a FLAG tag to the C termini of expressed proteins. Ser4* is a Ser4 transcriptional variant that bears a large N-terminal deletion and expresses a protein of only 286 amino acids (aa). To compare their anti-HIV activities, 293T cells were transfected with these Ser expression vectors and a WT HIV-1 proviral vector, pNL4-3, or its Nef-deficient mutant, pNL4-3ΔNef. After 48 h, cells or culture supernatants were collected and subjected to further analysis.
First, we determined Ser and viral protein expression in viral producer cells by Western blotting. It was found that all Ser proteins were expressed at similar levels in these 293T cells, except for Ser4* (Fig. 1A, top). In addition, similar levels of the Gag precursor p55 and Env gp120 proteins were detected from cells transfected with HIV-1 proviruses, and Nef was detected only from cells transfected with the WT proviruses. Second, we determined viral infectivity. To verify viral particle release, the collected supernatants were subjected to ultracentrifugation, and the purified particles were analyzed by Western blotting. It was found that the processed p24 Gag proteins were detected from all these samples, confirming that virions were produced from the cells (Fig. 1A, bottom). These virions were then quantified by p24Gag enzyme-linked immunosorbent assay (ELISA). Then, equal numbers of virions were inoculated into the HIV-1 luciferase reporter cell line TZM-b1, and viral infectivity was determined. It was found that Ser5, but not the others, strongly inhibited HIV-1 infectivity, and unexpectedly, Ser5 reduced both WT and ΔNef virus infectivity by ∼50-fold (Fig. 1B).
FIG 1.
Analysis of the anti-HIV-1 activity of Ser family members. (A) 293T cells were transfected with an HIV-1 WT or ΔNef proviral vector in the presence of the indicated pCMV6-Ser expression vectors at a 1:1 ratio. After 48 h, virions were collected from the culture supernatants and purified by ultracentrifugation. The purified virions and cells were lysed and analyzed by Western blotting using the indicted antibodies. (B) After virions produced from 293T cells were quantified by p24Gag ELISA, equal amounts of viruses were used to infect the HIV-1 luciferase reporter cell line TZM-b1 to determine viral infectivity. The error bars represent standard deviations (SD) from three experiments. (C) 293T cells were transfected with pCMV6-Ser5 in the presence or absence of WT or ΔNef HIV-1 vector. Viral particles were purified from culture supernatants by ultracentrifugation. Cell and virion lysates were analyzed by Western blotting. The asterisk indicates remnant protein bands from a previous Western blot. The GenBank accession numbers for the Ser proteins are as follows: NM_020755 (human Ser1), NM_178865 (human Ser2), NM_006811 (human Ser3), NM_001258032 (human Ser4*), NM_001174072 (human Ser5), and NM_012032 (murine Ser3).
To confirm that Ser5 was indeed incorporated into virions, a similar experiment was performed by transfecting 293T cells with Ser5 expression vector in the presence or absence of an HIV-1 proviral vector. After verification of Ser5 and Gag expression in the viral producer cells, it was found that Ser5 was specifically detected in HIV-1 virions, and slightly more Ser5 was found in ΔNef virions than in WT virions (Fig. 1C). Together, we conclude that among the five Ser members, Ser5 strongly inhibits HIV-1 infectivity after being incorporated into virions.
Saturation of HIV-1 Nef activity by Ser5 overexpression.
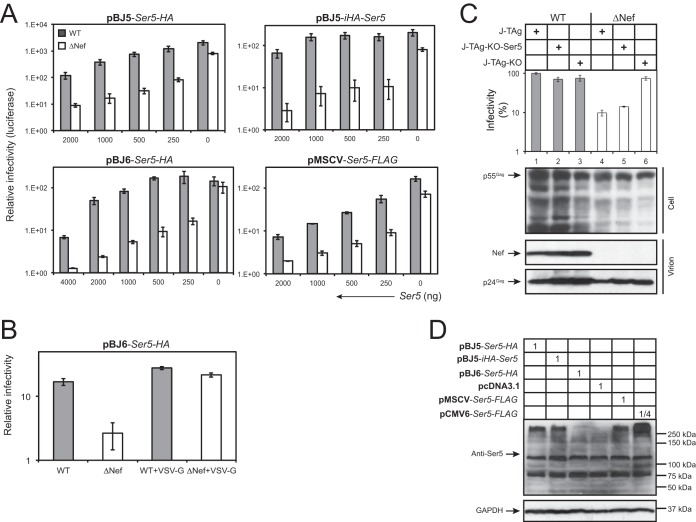
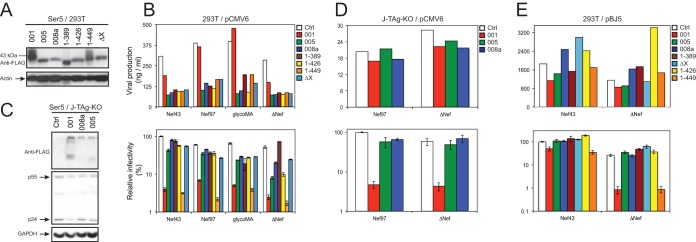
To understand why Nef did not counteract Ser5 in the previous experiment, we expressed Ser5 from other vectors, including pBJ5, pBJ6, and pMSCV. pBJ5 contains a simian virus 40-human T cell leukemia virus (SV40-HTLV) hybrid promoter, pBJ6 contains the same promoter but has the SV40 origin of replication removed, and pMSCV uses the murine stem cell virus long terminal repeat (LTR) as a promoter to express target proteins. Both pBJ5-Ser5-HA and pBJ6-Ser5-HA express Ser5 with a C-terminal hemagglutinin (HA) tag, pBJ5-iHA-Ser5 expresses HA between Ser5 residues 290 and 291, and pMSCV-Ser5-FLAG expresses Ser5 with a C-terminal FLAG tag. When these vectors were used, it was found that, unlike with the pCMV6 vector, Ser5 inhibited ΔNef HIV-1 much more efficiently than WT HIV-1 in a dose-dependent manner; the ratio between WT and ΔNef HIV-1 infectivities in response to Ser5 input reached up to ∼25-fold (Fig. 2A). In addition, the Nef phenotype was strongly suppressed when HIV-1 was pseudotyped with VSV G (Fig. 2B).
FIG 2.
Saturation of HIV-1 Nef counteractive activity by Ser5 overexpression. (A) 293T cells were transfected with a fixed amount of HIV-1 WT or ΔNef proviral vector and the indicated amounts of 4 different Ser5 expression vectors, pBJ6-Ser5-HA, pBJ5-Ser5-HA, pBJ5-iHA-Ser5, and pMSCV-Ser5-FLAG. After 48 h, viruses were collected and normalized by p24Gag ELISA; viral infectivity was measured by infecting TZM-b1 cells. (B) The anti-HIV-1 activity of Ser5 expressed from the pBJ6 vector was determined in the presence or absence of VSV G pseudotyping. (C) J-TAg, J-TAg-KO, and J-TAg-KO-Ser5 cells were infected with WT or ΔNef HIV-1 by spinoculation. The viruses produced were normalized by p24Gag ELISA, and viral infectivity was determined after infection of TZM-b1 cells. Viruses were also purified by ultracentrifugation, and viral protein expression in the Jurkat cells and purified virions was analyzed by Western blotting. (D) The Ser5 expression levels in 293T cells from the different expression vectors were compared by Western blotting using an anti-Ser5 antibody. Numbers in the boxes indicate relative amounts of each vector used for transfection of 293T cells. The error bars represent SD from three experiments.
We then determined the infectivity of viral particles produced from Jurkat cells. Three cell lines were used: WT Jurkat-TAg cells (J-TAg), their Ser3/Ser5 double-knockout (J-TAg-KO), and J-TAg-KO cells transduced with the pMSCV-Ser5-FLAG vector (J-TAg-KO-Ser5). J-TAg cells were reported to express very high levels of Ser3 and Ser5, which selectively inhibit ΔNef HIV-1 replication (22). The cells were infected with WT or ΔNef HIV-1 by spinoculation, and the infectivity of the virions produced was determined after infecting TZM-b1 cells. It was found that viral protein expression was detectable in both viral producer cells and released particles (Fig. 2C, bottom). Importantly, although WT viruses from these cells showed similar levels of infectivity, ΔNef viruses from J-TAg and J-TAg-KO-Ser5 cells showed much lower infectivity than those from J-TAg-KO cells (Fig. 2C, top). Together, these results demonstrate that Nef is able to counteract the endogenous Ser5 and Ser5 expressed from pBJ and pMSCV vectors.
To further understand the differences between pCMV6, pBJ5, pBJ6, and pMSCV, we compared the levels of Ser5 expression from these vectors after transfection of 293T cells by Western blotting using an anti-Ser5 antibody. Even when a much smaller amount of DNA was used, pCVM6 still produced significantly higher levels of Ser5 than pMSCV and pBJ5 (Fig. 2D). In addition, pMSCV and pBJ5 produced similar levels of Ser5, and the Ser5 levels from pBJ6 were lower than from these vectors (Fig. 2D). The Ser5 protein band migrated much more slowly, which was likely due to extensive aggregation. These results demonstrate that pCMV6 expresses much higher levels of Ser5 than the other vectors, which suggests that the Nef counteractive activity can be saturated by Ser5 overexpression.
Analysis of the Ser5 and Ser3 gene expression profile in relevant human cells.
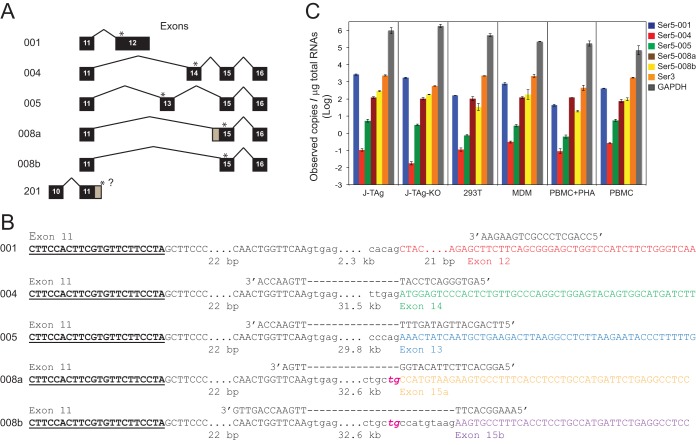
After searching the Ensembl genome browser, we identified six human Ser5 isoforms: Ser5-001, -004, -005, -008a, -008b, and -201 (Fig. 3A). The functions of most alternatively spliced isoforms of any gene are rarely known. Given the importance of Ser5 to the HIV host-pathogen relationship, it is vital to know if these alternatively spliced isoforms are significant for host resistance.
FIG 3.
Detection of human Ser5 gene expression by RT-qPCR. (A) Human Ser5 gene alternatively spliced isoforms. Isoform numbers are shown on the left, and the differentially spliced exons are shown on the right. Exons are shown as black boxes with numerals, but the first 10 exons, which are constitutively spliced, are not shown (except for the putative isoform 201). Exons and introns are not drawn to scale, although exon 12 for isoform 001 is shown as a longer exon. The positions for the stop codons for each isoform are indicated by asterisks. The gray regions for single exons for isoforms 008a and 201 indicate the alternative splice points for the two isoforms. The question mark after exon 11 indicates that the end of the 3′ untranslated region for the isoform has not been identified. (B) Locations of forward and reverse RT-qPCR primers for each Ser5 isoform. The locations of the forward primers (boldface underlined letters) are the same for each primer set. Uppercase letters represent exonic regions, and lowercase letters represent intronic regions. The ellipses indicate sequences that are not shown (the numbers of bases that are not displayed are given below). The dashed lines indicate the intronic regions crossed by the reverse primers. Exons 11, 12, 13, 14, 15a, and 15b are shown in different colors. Instead of the canonical AG normally found for a U2 splice acceptor sequence, 008a and 008b have the rare TG acceptors, which are italicized and shown in magenta. (C) Ser3 and Ser5 copy numbers in J-TAg, J-TAg-KO, and 293T cells; MDMs; PBMCs; and PBMCs activated with PHA. GAPDH was used as a control. The GenBank accession numbers for Ser5-001, -004, -005, -008a, and -008b are NM_001174072.2, BC101280, BC101281, AF498273, and BC101280, respectively. No GenBank accession number was found to be associated with Ser5-201.
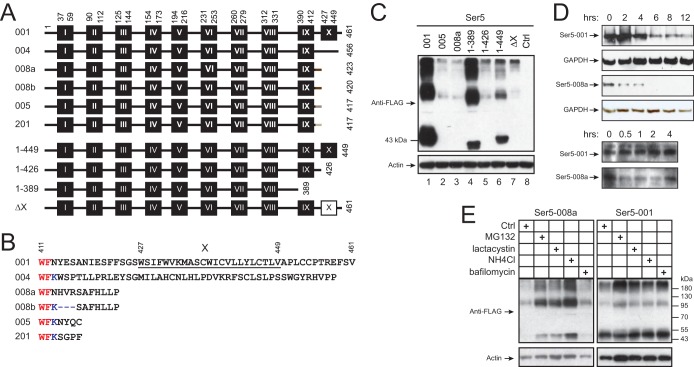
In previous experiments, we used Ser5-001 to study Ser5 anti-HIV-1 activity (Fig. 1 and 2). Ser5-001 has the longest sequence, with 461 aa, and its predicted topology suggests that Ser5-001 should have a 36-aa N-terminal head, 10 transmembrane domains, and a short 12-aa C-terminal tail (Fig. 4A and B). The other five alternative isoforms should have similar topology but differ in their C termini after the 9th transmembrane domain: Ser5-004, -005, -008a, -008b, and -201 do not express the 10th transmembrane domain, and they should have a 47-, 11-, 8-, 5-, or 5-aa C-terminal tail, respectively. No alternative splicing appears to occur in the coding region for Ser3, based upon numerous expressed sequence tags (ESTs) that support a single isoform for the gene, as assessed through the UCSC Genome Browser, and so a similar study was not necessary for the gene.
FIG 4.
Analysis of Ser5 isoform protein expression. (A) Schematic diagram of the putative transmembrane domains of Ser5 isoforms and mutants. The domains were predicted by TransMembrane prediction using hidden Markov models (TMHMM) at the public server (http://www.cbs.dtu.dk/services/TMHMM/) and are shown as black boxes labeled with Roman numerals (not drawn to scale). The Arabic numerals indicate the amino acid positions for the transmembrane domains, as well as the first and the last amino acids of the isoforms. The white box indicates the deleted transmembrane domain. (B) C-terminal amino acid sequences of Ser5 isoforms from residue 411 to the last residue. The 10th transmembrane domain of Ser5-001 is underlined and marked with the Roman numeral X. The red letters indicate the last two residues in the 9th transmembrane domain. (C) The Ser5 proteins were expressed in 293T cells from the pCMV6 vector, and their expression levels were compared by Western blotting. (D) Ser5-001 and Ser5-008a were expressed in 293T cells. The cells were treated with 50 μM cycloheximide, and protein expression was analyzed by Western blotting at the indicated times after treatment. (E) Ser5-001 and Ser5-008a were expressed in 293T cells. The cells were either untreated (control [Ctrl]) or treated with MG132 (2.5 μM), lactacystin (20 μM), NH4Cl (15 mM), or bafilomycin (100 nm). Ser5 expression was analyzed by Western blotting.
Full-length human cDNA clones have been isolated previously for Ser5-001, -004, -005, -008a, and -008b (22–25). One other isoform that could encode a protein is Ser5-201 (Fig. 3A and 4A and B). However, this isoform may be an artifact, because we can find no evidence from full-length cDNAs, ESTs, or transcriptome-sequencing (RNA-seq) data to suggest that it actually exists. Indeed, the listing on the Ensembl website indicates that Ser5-201 has transcript support level 5 (“transcripts that are not supported at all by either an mRNA or an EST”), and at the UCSC Genome Browser (2013 build), the position of the last splice point following exon 11 is listed in the “alternative splicing” track as a “Strangesplice,” the description of which includes the statement “these are usually artifacts of some sort due to sequencing error or polymorphism.” The UCSC Genome Browser also indicates that Ser5-201 “is derived from an Ensembl automatic analysis pipeline.” Although we made several attempts to produce data that would support or refute the existence of the Ser5-201 transcript, the results were inconclusive and so are not reported here.
We then designed specific primers to target Ser5-001, -004, -005, -008a, and -008b and analyzed their expression by real-time quantitative PCR (RT-qPCR) (Fig. 3B; see Fig. S1 in the supplemental material). We also used this strategy to analyze Ser3 expression and used GAPDH (encoding glyceraldehyde-3-phosphate dehydrogenase) as a control. J-TAg, J-TAg-KO, and 293T cells; monocyte-derived macrophages (MDMs); peripheral blood mononuclear cells (PBMCs); and phytohemagglutinin (PHA)-activated PBMCs were used for these analyses.
The quantitative mRNA copy numbers for Ser5 isoforms, Ser3, and GAPDH are shown in Fig. 3C. It was found that Ser5 and Ser3 were expressed at much lower levels than GAPDH. However, when comparisons were made within isoforms, Ser5-001 and Ser3 mRNAs were always present in considerably higher numbers (>10-fold) than Ser5-004, Ser5-005, Ser5-008a, or Ser5-008b; Ser5-008a and Ser5-008b tended to be present at higher copy numbers than either Ser5-004 or Ser5-005. When comparisons were made among different cells, J-TAg cells expressed more Ser5-001 than any of the other cells, which is consistent with the previous report (22). A lower level of expression for Ser5-001 and Ser3 was seen in the J-TAg-KO cell line than in the parental J-TAg cell line. This was expected, because the clustered regularly interspaced short palindromic repeat (CRISPR) knockout mutations cause frameshift mutations and premature stop codons, resulting in decreased, but not absolute, loss of mRNAs caused by nonsense-mediated mRNA decay (26). In addition, MDMs and resting PBMCs expressed much more Ser5-001 and Ser3 than PHA-activated PBMCs, which is consistent with another previous report (23). In contrast, the levels of GAPDH in PBMCs seemed to be increased slightly upon PHA stimulation. Thus, we conclude that Ser5-001 transcripts are expressed at the highest levels among the five Ser5 isoforms, and Ser3 transcripts are also expressed at similarly high levels in these cells.
The Ser5-001 C terminus is required for Ser5 expression.
After understanding how Ser5 transcripts are expressed, we determined how the C terminus could affect Ser5 expression (Fig. 4B). When Ser5-001, -005, and -008a were expressed from the pCMV6 vector, it was found that the steady-state levels of both Ser5-005 and Ser5-008a expression were significantly lower than that of Ser5-001 (Fig. 4C, lanes 1 to 3). To understand whether Ser5-001 is more stable than the others, we compared their protein half-lives. It was found that the Ser5-001 protein half-life was ∼6 h, whereas the Ser5-008a half-life was less than 0.5 h (Fig. 4D). In addition, the Ser5-008a steady-state levels were increased after treatment with MG132, lactacystin, or NH4Cl, and the same treatments did not significantly increase Ser5-001 expression (Fig. 4E). These results strongly suggest that Ser5-001 is much more stable than Ser5-008a and Ser5-005, and its C terminus should play a critical role in Ser5 expression.
To further explore the mechanism, we created 4 Ser5 C-terminal deletion mutants: 1-449 has a deletion of the C-terminal tail, 1-426 has a deletion from the 10th transmembrane domain, 1-389 has a deletion from the 9th transmembrane domain, and ΔX has adeletion of only the 10th transmembrane domain (Fig. 4A). When these mutants were expressed from the pCMV6 vector, it was found that mutants 1-426, 1-449, and ΔX were expressed at low levels, whereas mutant 1-389 was expressed at slightly higher levels (Fig. 4C). These results demonstrate that the l0th transmembrane domain and the C-terminal tail are required for stable Ser5 protein expression.
The 10th transmembrane domain promotes Ser5 localization to the plasma membrane.
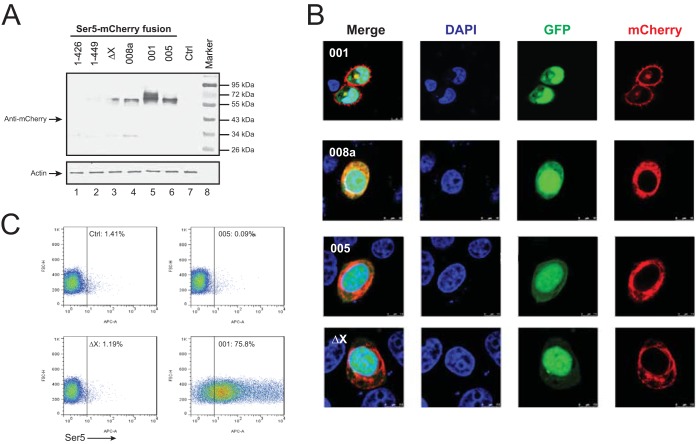
Having understood Ser5 protein expression, we investigated Ser5 subcellular localization using confocal microscopy. Ser5-001, -005, and -008a and mutants 1-426, 1-449, and ΔX were fused to a red fluorescent protein, mCherry, at their C termini and expressed from another CMV-driven pCAGGS vector. When their expression was determined in HeLa cells by Western blotting, it was found that Ser5-001 was expressed at the highest levels; Ser5-005 and -008a and mutant ΔX were also expressed, but at lower levels; the expression of mutants 1-426 and 1-449 was undetectable (Fig. 5A). These results are consistent with those in Fig. 4. We then studied the subcellular distribution of Ser5-001, -005, and -008a and mutant ΔX. Mutants 1-426 and 1-449 were not included because of their low expression. It was found that Ser5-001 exhibited strong plasma membrane distribution, whereas Ser5-005 and -008a and mutant ΔX were largely distributed to the cytoplasm (Fig. 5B).
FIG 5.
Analysis of Ser5 subcellular localization. (A) The indicated Ser5 isoforms and mutants were fused with a C-terminal mCherry tag, and their expression levels were determined by Western blotting. (B) Then, Ser5-001-, 008a-, 005-, and ΔX-mCherry fusion proteins were expressed in HeLa cells in the presence of GFP, and their subcellular localization was determined. (C) Ser5-001, Ser5-005, and Ser5-ΔX with an HA tag inserted between residues 290 and 291 were expressed in 293T cells, and their expression was determined by flow cytometry using an Alexa Fluor 647-labeled anti-HA antibody (BioLegend).
To further confirm these observations, we investigated Ser5 cell surface expression. Ser5-001, Ser5-005, and the ΔX mutant containing an HA tag between residues 290 and 291 were expressed in 293T cells from the pBJ5 vector. Their surface expression was then determined by flow cytometry. It was found that only Ser5-001 was strongly detectable from the cell surface, whereas Ser5-008a and the ΔX mutant were undetectable (Fig. 5C). Together, these results demonstrate that the 10th transmembrane domain is required for Ser5 localization to the plasma membrane.
The 10th transmembrane domain is required for Ser5 anti-HIV-1 activity.
Lastly, we investigated how the C terminus influences Ser5 anti-HIV-1 activity using the above-mentioned isoforms and mutants. Because their protein expression levels varied so much, we first adjusted the amounts of pCMV6 vectors during transfection of 293T cells so that isoforms Ser5-001, -005, and -008a and mutants 1-389, 1-426, 1-449, and ΔX were expressed at similar levels (Fig. 6A). We then used these conditions to express the Ser5 proteins and to produce ΔNef HIV-1 and HIV-1 expressing Nef43, Nef97, or glycoMA in 293T cells. Viral production from these cells was verified by p24Gag ELISA (Fig. 6B, top), and viral infectivity was measured by infecting TZM-b1 cells with equal amounts of viruses (Fig. 6B, bottom). It was found that only Ser5-001 and its mutant 1-449 exhibited strong inhibition of HIV-1 infectivity even in the presence of Nef43, Nef97, and glycoMA, whereas the activity of the other Ser5 proteins was significantly reduced.
FIG 6.
Analysis of Ser5 splicing isoform anti-HIV-1 activities. (A) The expression levels of Ser5 isoforms and mutants from the pCMV6 vector were normalized to similar levels by adjusting the amounts of the expression vectors during transfection of 293T cells and detected by Western blotting. (B) Similar levels of the indicated Ser5 isoforms and mutants from the pCMV6 vector were expressed in 293T cells during production of ΔNef HIV-1 or HIV-1 expressing NL4-3 Nef (Nef43), 97ZA012 Nef (Nef97), or glycoMA. (Top) Virus production was measured by p24Gag ELISA. (Bottom) Then, equal amounts of viruses were used to infect TZM-b1 cells to determine viral infectivity. (C) Similar levels of the indicated Ser5 variants from the pCMV6 vector were expressed in J-TAg-KO cells in the presence of HIV-1, which was verified by Western blotting. (D) After viral production from the Jurkat cells was measured by p24Gag ELISA (top), equal amounts of viruses were used to measure viral infectivity (bottom). (E) The indicated Ser5 isoforms and mutants were cloned into the pBJ5 vector and expressed in 293T cells. Then, viral production and infectivity were determined similarly. The error bars represent SD from three experiments.
To further verify the strong Ser5-001 activity, similar levels of Ser5-001, -005, and -008a were also expressed from the pCMV6 vector in J-TAg-KO cells during production of HIV-1, where Gag and Ser5 expression were verified by Western blotting (Fig. 6C). After quantification of viral production (Fig. 6D, top), viral infectivity was measured as before (Fig. 6D, bottom). Again, it was found that only Ser5-001 potently inhibited both WT and ΔNef HIV-1 infectivity, whereas Ser5-005 and Ser5-008a did not.
To understand whether mutant 1-449 still retains Nef responsiveness, we cloned Ser5-001, -005, and -008a and mutants 1-389, 1-426, 1-449, and ΔX into the pBJ5 vector. WT and ΔNef HIV-1 were produced from 293T cells in the presence of these expression vectors. Viral production was quantified, and viral infectivity was measured as before (Fig. 6E). It was confirmed that Ser-005, Ser-008a, 1-389, 1-426, and ΔX did not inhibit either WT or ΔNef HIV-1 infectivity. However, although WT and ΔNef HIV-1 infectivity was inhibited, ΔNef HIV-1 was much more strongly suppressed than WT HIV-1 by Ser5-001 and the mutant 1-449. Together, these results demonstrate that although both the 10th transmembrane domain and the C-terminal tail are required for Ser5-001 expression, only the 10th transmembrane domain is required for Ser5 anti-HIV-1 activity. In addition, the C-terminal tail does not determine Nef responsiveness.
DISCUSSION
After confirmation of strong Ser5 anti-HIV-1 activity (Fig. 1), we studied the expression of five Ser5 isoforms in relevant human cell lines and primary cells. Alternative splicing occurs in at least half of all human genes (27, 28). In some cases of alternative splicing, the exons are created by the process of exaptation of transposons (29). Although Ser5 exon 13 is derived from nonrepetitive (unique) DNA, alternative exons 14, 15, and 16 are exapted exons and correspond to the recruitment of the following transposable elements: Alu (more specifically, AluSz), an LTR repeat (THE1A), and another LTR (LTR6A), respectively. Preliminary results have suggested that these transposable elements were first inserted near the Ser5 gene following the split of the great apes from other primates. Two other potential isoforms (transcript variant 4 and transcript variant 5) are listed in databases (e.g., Ensembl) and genome browsers (e.g., UCSC Genome Browser). These isoforms are noncoding and are possibly due to sequencing errors causing frameshifts, but even if they are real, they would be very unlikely to be of functional significance, because no protein would be produced.
The data presented here, however, provide evidence that the alternative isoforms are present at less than 1/10 the amount of Ser-001 (Fig. 3C). Ser-004 and Ser-005 may be present at numbers below those of Ser-008a and Ser-008b because they have stop codons that occur before the last intron and are therefore subject to nonsense-mediated mRNA decay (26). Ser-008a and Ser-008b also have stop codons that occur before the last intron, but in these cases, the stop codons are less than 55 bases away from the last exon and are not subject to nonsense-mediated mRNA decay (the “50- to 55-nucleotide rule” [30]). However, we cannot rule out the possibility that the difference in numbers between the various isoforms is partially or predominantly due to differential splicing, in addition to the probable difference in nonsense-mediated mRNA decay. It has been shown that there is a genome-wide correlation between mRNAs and their encoded proteins (31). Although specific antibodies for the different Ser5 isoforms are not available, we believe that it is reasonable to assume that the relative concentrations of the proteins for the various isoforms are reflected to some degree in the relative concentrations of the respective mRNAs. Thus, Ser5-001 should be the most abundantly expressed Ser5 isoform in vivo. Although it might be said, in hindsight, that the report of the predominance of Ser5-001 for anti-HIV activity is inconsequential, without actual experimental determination, a strong influence of the other isoforms would have remained a distinct and unknown possibility. Indeed, exaptation of transposons into mRNA transcripts has been proposed to be an important mechanism of genetic adaptation in evolution (29).
In addition to production of many fewer RNA transcripts, we found that these Ser5 alternative isoforms are poorly translated into functional proteins compared to Ser5-001. When Ser-001, -005, and -008a were expressed from the same pCMV6 vector, Ser5-005 and -008a expressed significantly lower levels of proteins than Ser5-001 (Fig. 4C). When protein stabilities were compared, we found that the protein half-life was reduced from 6 h (Ser5-001) to less than 0.5 h (Ser5-008a) (Fig. 4D). In addition, the steady-state levels of Ser5-008a were increased by treatments with the proteasomal inhibitor MG132 or lactacystin and the lysosomal inhibitor NH4Cl (Fig. 4E). These results demonstrate that Ser5-008a is much less stable than Ser5-001 due to rapid protein degradation. Because Ser5-001 differs from Ser5-008a only in the C-terminal region after residue 412 (Fig. 4B), we created four Ser5 C-terminal deletion mutants, 1-449, 1-426, 1-389, and ΔX, to map the critical determinant for protein stability. We found that, like Ser5-005 and Ser5-008a, all these mutants expressed much less protein than Ser5-001 (Fig. 4C). These results suggest the C-terminal region, including the last transmembrane domain (residues 427 to 449) and the C-terminal tail (residues 450 to 461), stabilizes Ser5 and increases its protein expression. We continued to study Ser5 subcellular expression using confocal microscopy and its cell surface expression using flow cytometry. We found that although Ser5-001 was predominantly localized to the plasma membrane, Ser5-005, Ser5-008a, and the ΔX mutant were largely distributed in the cytoplasm (Fig. 5B). Consistently, only Ser5-001 was detected on the cell surface, whereas Ser5-005 and the ΔX mutant were undetectable (Fig. 5C). Furthermore, we measured their anti-HIV activities after expression from pCMV6 and pBJ5 vectors in 293T and Jurkat cells. We consistently detected strong activity from Ser5-001 and the mutant 1-449, but not from any others (Fig. 6). Collectively, these results demonstrate that plasma membrane localization is required for Ser5 anti-HIV-1 function, and its last transmembrane domain plays an important role in this process. We speculate that in the absence of the 10th transmembrane domain, Ser5 is likely misfolded and subjected to rapid degradation before delivery to the plasma membrane.
We consistently observed that when expressed from the pCMV6 vector, Ser5 inhibits both WT and ΔNef HIV-1 infectivity to similar levels in 293T and Jurkat cells, suggesting that Nef is unable to counteract Ser5 antiviral activity in these experiments (Fig. 1C and 6B and D). In contrast, Nef is able to counteract the endogenous Ser5 in Jurkat cells (Fig. 2C), as well as Ser5 expressed from the pBJ5 and pBJ6 vectors (Fig. 2A and 6E), as reported previously (22, 23). Nef also neutralizes Ser5 expressed from another vector, pMSCV (Fig. 2A and 2C). After comparison of protein expression by Western blotting, we found that pCMV6 expresses much more Ser5 than pBJ5, pBJ6, and pMSCV (Fig. 2D). These results demonstrate that the Nef counteractive activity can be easily saturated by high Ser5 expression. We suggest that the CMV promoter should be avoided for Ser5 expression if experiments are designed to investigate how Nef counteracts Ser5, although it could still be used to study the Ser5 antiviral mechanism.
In summary, we have demonstrated that Ser5-001 expresses at least 10-fold more transcripts than the other isoforms, which are translated into much more stable proteins, resulting in potent inhibition of HIV-1 infectivity. In addition, more Ser5-001 transcripts are expressed in MDMs and resting PBMCs than in activated PBMCs. Thus, we conclude that Ser5-001 is the most important Ser5 isoform and that it should play an important role in defending HIV-1 during primary infection.
MATERIALS AND METHODS
Cells.
The human 293T and HeLa cell lines were obtained from ATCC. TZM-b1 cells were obtained from the NIH AIDS Research and Reference Reagent Program. These cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% bovine calf serum (BCS) (HyClone). The human Jurkat-TAg cell line and its Ser3 Ser5 double-knockout cell line Jurkat-TAg-KO (1) were provided by Heinrich Gottlinger (23). The cells were cultured in RPMI 1640 with 10% fetal bovine serum (FBS) (Sigma).
Human PBMCs were isolated from deidentified source leukocytes (Gulf Coast Regional Blood Center, Houston, TX) by density gradient centrifugation using Ficoll-Paque Plus (GE Healthcare Life Sciences, Pittsburgh, PA). CD14+ monocytes were purified from PBMCs using CD14 antibody-coated magnetic beads (Miltenyi Biotec, Germany). PBMCs and monocytes were cultured in RPMI with 10% FBS. MDMs were generated by stimulating monocytes with 100 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) (R&D Systems) for 7 days. PBMCs were activated by culturing with PHA for 3 days at 3 μg/ml.
Plasmids.
HIV-1 proviral vectors pNL4-3 and pNL4-3ΔNef were described previously (4, 20). The HIV-1 proviral vectors pNL-Nef97ZA012, which has the nef gene precisely replaced by that of the HIV-1 97ZA012.1 strain, and pNL4-3/glycoMA, which has nucleotides 360 to 926 of M-MLV followed by a stop codon inserted between the Nef initiation codon and the unique XhoI site within the nef gene of pNL4-3, were provided by H. Gottlinger (23). pBJ5-Ser5-HA, which has a C-terminal HA, and pBJ5-iHA-Ser5, which has an HA tag inserted between residues 290 and 291 of Ser5, were provided by H. Gottlinger (23). pBJ6-Ser6-HA, which has a C-terminal HA, was provided by M. Pizzato (22).
The pMSCV-Ser5-FLAG vector was constructed by cloning the Ser5 gene with a FLAG tag at the 3′ end into the pMSCVpuro vector (Clontech) via the XhoI/EcoRI sites. The pCMV6-Entry-Ser1, -Ser2, -Ser3, -Ser4*, -Ser5-001, -Ser-008a, and -mSer3 mammalian expression vectors were purchased from Origene. The Ser5 C-terminal deletion mutants were first created by PCR and then cloned back into pCMV6-Entry via BamHI/XhoI digestion. Ser5 and some of its mutants with a C-terminal FLAG tag were also cloned into the pBJ5 vector via XhoI/EcoRI digestion. To express Ser5-mCherry fluorescent fusion proteins, the mCherry gene was first cloned into the CMV-driven pCAGGS mammalian expression vector via EcoRI/XhoI digestion. Then, the vector was linearized by EcoRI digestion, which was used to clone PCR-amplified Ser5 genes containing an 18-bp sequence at the 5′ and 3′ ends homologous to the vector or the 5′ end of mCherry gene sequence, respectively, via homologous recombination using a CloneExpress II one-step cloning kit (Vazyme Biotech, China) following the manufacturer's manual. To express Ser5-005 and ΔX, which have an internal HA tag between residues 290 and 291, the 3′ end or the 10th transmembrane domain of Ser5-001 was deleted accordingly via PCR using the pBJ5-iHA-Ser5 vector as a template and then cloned back into the pBJ5 vector via XbaI/EcoRI digestion. The primers used for these clonings are available upon request.
HIV-1 production and infectivity analysis.
Viruses were produced from 293T cells after transfection as described previously (32). Briefly, 1 μg of HIV-1 proviral vector was transfected with various amounts of Ser expression vectors using polyethylenimine (PEI) as the transfection reagent. Forty-eight h later, viruses were collected from the culture supernatants, and viral production was quantified by p24Gag ELISA. To determine viral infectivity, equal amounts of viruses as normalized to the levels of p24Gag were used to infect the HIV-1 luciferase reporter cell line TZM-bl in a 96-well plate at a density of 1 × 104 per well. After 48 h, each well of cells was lysed with 100 μl 1% NP-40 in phosphate-buffered saline (PBS), and 25 μl was used to determine the luciferase activities using the Bright-Glo luciferase assay system (Promega), which were used to calculate viral infectivity.
Analysis of protein expression by Western blotting.
Western blotting was performed to analyze viral and host protein expression in cell and virion lysates after lysis with 1% NP-40 in PBS. The following HIV-1 antibodies were obtained from the NIH AIDS Research and Reference Reagent Program: catalogue numbers 1513 (Gag), 521 (gp120), 526 (gp41), and 3957 (HIV immunoglobulin). The mouse anti-Nef monoclonal antibody was described previously (20). The mouse anti-FLAG and mouse anti-HA monoclonal antibodies were purchased from Sigma, the rabbit anti-human SERINC5 polyclonal antibody was purchased from Abcam, the mouse anti-GAPDH monoclonal antibody was purchased from Meridian Life Science, and the rabbit anti-human actin polyclonal antibody was purchased from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-conjugated anti-human, -rabbit, and -mouse immunoglobulin G secondary antibodies were purchased from Pierce. The enhanced chemiluminescence detection kit was purchased from Amersham Bioscience. Lactacystin was purchased from Santa Cruz Biotechnology. Bafilomycin, MG132, and NH4Cl were purchased from Sigma.
Confocal microscopy analysis.
HeLa cells were seeded on poly-l-lysine-coated glass slides and transfected with vectors expressing mCherry-tagged Ser5 and green fluorescent protein (GFP). The cells were fixed with 4% paraformaldehyde after 24 h and then incubated with 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining. After washing with PBS, the samples were observed under a confocal microscope (Leica TCS SP5). At least 100 random cells per slide were analyzed, and the most representative image from each slide was selected for presentation.
RT-qPCR analysis.
Total cellular RNAs were extracted from cells using TRIzol reagent (ThermoFisher) following the manufacturer's instructions. One microgram of total RNA was subjected to reverse transcription in a 20-μl volume using a SuperScript III reverse transcriptase kit and oligo(dT)12–18 as a primer (ThermoFisher). Ser3 and Ser5 cDNAs were subjected to RT-qPCR analysis using the SYBR green PCR master mix kit (ThermoFisher) with primers listed in Table 1. Primers were designed in exons 11 and 12 for Ser5-001. For all other Ser5 isoforms, reverse primers were designed to bridge a splice site (Fig. 3B; see Fig. S1 in the supplemental material) to eliminate the chance of any trace genomic-DNA amplification and/or amplification due to repetitive (i.e., Alu or LTR) sequences in other cDNAs (isoforms Ser-004, Ser-005, Ser-008a and Ser-008b are due to exapted SINE or LTR transposon repeat elements; see Discussion for further information). The GAPDH gene was also amplified and analyzed as a control. To prepare standard curves for these analyses, the specific cDNA fragments of Ser5-001, Ser5-004, Ser5-005, Ser5-008a, Ser5-008b, Ser3, and GAPDH were cloned into the pGEM-T Easy vector (Promega) after PCR amplification using the same primers (Table 1) and verified by Sanger sequencing. The clones for each cDNA were quantified by a Quibit dsDNA HS assay kit (ThermoFisher), and their respective copy numbers were calculated based upon the molecular weight of the plasmid for each clone. Copy numbers for each isoform were then calculated based upon threshold cycle (CT) values compared to the RT-qPCR regression line and reported as relative copy numbers within the set of isoforms.
TABLE 1.
PCR primers used to amplify Ser5 isoforms, Ser3, and GAPDH
| Target | Primera |
Tempb (°C) |
Amplicon size (bp) | RT-qPCR |
|||
|---|---|---|---|---|---|---|---|
| Forward | Reverse | F | R | Efficiency (%) | Reg. coef.c | ||
| Ser5-001 | CTTCCACTTCGTGTTCTTCCTA | CCAGCTCCCGCTGAAGAA | 61.1 | 63.1 | 106 | 99.9 | 0.997 |
| Ser5-004 | CTTCCACTTCGTGTTCTTCCTA | AGTGGGACTCCATTTGAACCA | 61.1 | 63.0 | 76 | 102.6 | 0.976 |
| Ser5-005 | CTTCCACTTCGTGTTCTTCCTA | TTCAGCATTGATAGTTTTTGAACCA | 61.1 | 61.5 | 80 | 99.1 | 0.992 |
| Ser5-008a | CTTCCACTTCGTGTTCTTCCTA | AGGCACTTCTTACATGGTTGA | 61.1 | 61.0 | 80 | 94.1 | 0.998 |
| Ser5-008b | CTTCCACTTCGTGTTCTTCCTA | AAAGGCACTTTTGAACCAGTTG | 61.1 | 61.6 | 73 | 103.5 | 0.997 |
| Ser3 | ATTCCACCTCATGCTCTGCT | TTTGCATCAGGGCTGTACCA | 62.9 | 63.4 | 75 | 102.0 | 0.985 |
| GAPDH | ATGACATCAAGAAGGTGGTG | CATACCAGGAAATGAGCTTG | 59.2 | 57.7 | 177 | 94.98 | 0.994 |
Forward primers are identical for all SERINC5 isoforms.
Primer annealing temperatures were determined using IDT OligoAnalyzer with 200 μM deoxynucleoside triphosphates (dNTPs), 50 mM NaCl, and 1.5 mM MgCl2.
reg. coef., regression coefficient.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Gottlinger laboratory and the Pizzato laboratory, as well as the NIH AIDS Research and Reference Reagent Program, for providing various reagents.
X.Z. is supported by a grant (H201602) from the Natural Science Foundation of Heilongjiang, China. Y.H.Z. is supported by grants (AI120189, AI106477, and AI122863) from the National Institutes of Health, USA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00137-17.
REFERENCES
- 1.Basmaciogullari S, Pizzato M. 2014. The activity of Nef on HIV-1 infectivity. Front Microbiol 5:232. doi: 10.3389/fmicb.2014.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geyer M, Fackler OT, Peterlin BM. 2001. Structure–function relationships in HIV-1 Nef. EMBO Rep 2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JK, Kiyokawa E, Verdin E, Trono D. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc Natl Acad Sci U S A 97:394–399. doi: 10.1073/pnas.97.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng YH, Plemenitas A, Linnemann T, Fackler OT, Peterlin BM. 2001. Nef increases infectivity of HIV via lipid rafts. Curr Biol 11:875–879. doi: 10.1016/S0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 5.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol 68:2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med 179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spina CA, Kwoh TJ, Chowers MY, Guatelli JC, Richman DD. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med 179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lama J, Mangasarian A, Trono D. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol 9:622–631. doi: 10.1016/S0960-9822(99)80284-X. [DOI] [PubMed] [Google Scholar]
- 10.Ross TM, Oran AE, Cullen BR. 1999. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol 9:613–621. doi: 10.1016/S0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- 11.Chowers MY, Pandori MW, Spina CA, Richman DD, Guatelli JC. 1995. The growth advantage conferred by HIV-1 nef is determined at the level of viral DNA formation and is independent of CD4 downregulation. Virology 212:451–457. doi: 10.1006/viro.1995.1502. [DOI] [PubMed] [Google Scholar]
- 12.Goldsmith MA, Warmerdam MT, Atchison RE, Miller MD, Greene WC. 1995. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol 69:4112–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizzato M. 2010. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc Natl Acad Sci U S A 107:9364–9369. doi: 10.1073/pnas.1001554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiken C, Trono D. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol 69:5048–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiken C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol 71:5871–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pizzato M, Popova E, Gottlinger HG. 2008. Nef can enhance the infectivity of receptor-pseudotyped human immunodeficiency virus type 1 particles. J Virol 82:10811–10819. doi: 10.1128/JVI.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usami Y, Gottlinger H. 2013. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep 5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Usami Y, Popov S, Gottlinger HG. 2014. The Nef-like effect of murine leukemia virus glycosylated gag on HIV-1 infectivity is mediated by its cytoplasmic domain and depends on the AP-2 adaptor complex. J Virol 88:3443–3454. doi: 10.1128/JVI.01933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van 't Wout AB, Swain JV, Schindler M, Rao U, Pathmajeyan MS, Mullins JI, Kirchhoff F. 2005. Nef induces multiple genes involved in cholesterol synthesis and uptake in human immunodeficiency virus type 1-infected T cells. J Virol 79:10053–10058. doi: 10.1128/JVI.79.15.10053-10058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. 2003. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc Natl Acad Sci U S A 100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inuzuka M, Hayakawa M, Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 22.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA, Mammalian Gene Collection Program Team. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Ji C, Wang L, Cao Y, Dai J, Ye X, Zeng L, Dai J, Wu Q, Xie Y, Mao Y. 2003. Cloning and expression of a novel human C5orf12 gene*, a member of the TMS_TDE family. Mol Biol Rep 30:47–52. doi: 10.1023/A:1022250428015. [DOI] [PubMed] [Google Scholar]
- 26.Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modrek B, Lee C. 2002. A genomic view of alternative splicing. Nat Genet 30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 28.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 29.Nekrutenko A, Li WH. 2001. Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17:619–621. doi: 10.1016/S0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 30.Liu T, Lin K. 2015. The distribution pattern of genetic variation in the transcript isoforms of the alternatively spliced protein-coding genes in the human genome. Mol Biosyst 11:1378–1388. doi: 10.1039/C5MB00132C. [DOI] [PubMed] [Google Scholar]
- 31.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Zhou T, Frabutt DA, Zheng YH. 2016. HIV-1 Vpr increases Env expression by preventing Env from endoplasmic reticulum-associated protein degradation (ERAD). Virology 496:194–202. doi: 10.1016/j.virol.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.