The E3 ubiquitin ligase Neuralized is shown to interact with a subset of the Stardust isoforms to regulate the endocytosis of the apical protein Crumbs and thereby promote epithelial remodeling during Drosophila development.
Abstract
Crumbs (Crb) is a conserved determinant of apical membrane identity that regulates epithelial morphogenesis in many developmental contexts. In this study, we identify the Crb complex protein Stardust (Sdt) as a target of the E3 ubiquitin ligase Neuralized (Neur) in Drosophila melanogaster. Neur interacts with and down-regulates specific Sdt isoforms containing a Neur binding motif (NBM). Using a CRISPR (clustered regularly interspaced short palindromic repeats)-induced deletion of the NBM-encoding exon, we found that Sdt is a key Neur target and that Neur acts via Sdt to down-regulate Crb. We further show that Neur promotes the endocytosis of Crb via the NBM-containing isoforms of Sdt. Although the regulation of Crb by Neur is not strictly essential, it contributes to epithelium remodeling in the posterior midgut and thereby facilitates the trans-epithelial migration of the primordial germ cells in early embryos. Thus, our study uncovers a novel regulatory mechanism for the developmental control of Crb-mediated morphogenesis.
Introduction
Epithelia form continuous sheets of interconnected and polarized cells that act as protective boundaries to separate inner from outer milieus and promote selective exchanges. While keeping these boundary functions over time, epithelia often undergo dynamic remodeling through changes in the spatial arrangement of cell–cell contacts. The maintenance of epithelial polarity and the stabilization of cell–cell contacts during epithelial remodeling require the apical polarity protein Crumbs (Crb; Tepass et al., 1990; Grawe et al., 1996; Tepass, 1996; Bulgakova and Knust, 2009; Campbell et al., 2009; St Johnston and Ahringer, 2010; Thompson et al., 2013).
The protein Crb has a large extracellular region and a short intracellular tail with a PDZ95/Dlg/ZO-1 (PDZ) binding motif that recruits the scaffolding protein Stardust (Sdt in flies, Pals1 in mammals; Li et al., 2014). Sdt/Pals1 is a membrane-associated guanylate kinase family protein (Bachmann et al., 2001; Hong et al., 2001; Li et al., 2014) that also interacts with Patj (protein associated with tight junctions) to form a Crb–Sdt/Pals1–Patj complex (Bulgakova and Knust, 2009; Thompson et al., 2013). One function of Sdt is to stabilize Crb at the plasma membrane, in part by inhibiting the endocytosis of Crb (Lin et al., 2015). Crb and Sdt are required to stabilize epithelia (Grawe et al., 1996; Tepass, 1996; Bachmann et al., 2001; Hong et al., 2001; Campbell et al., 2009) and to negatively regulate actomyosin during tissue remodeling (Röper, 2012; Flores-Benitez and Knust, 2015). Consistent with this, changes in Crb protein levels, subcellular localization, and/or gene expression correlated with various epithelial remodeling events (Campbell et al., 2011; Röper, 2012; Sherrard and Fehon, 2015). Additionally, the crb and sdt genes encode multiple isoforms in Drosophila melanogaster. This complexity could contribute to the tissue-specific regulation of Crb and may also confer functional diversity (Hong et al., 2001; Berger et al., 2007; Campbell et al., 2009; Bulgakova et al., 2010; Vichas et al., 2015).
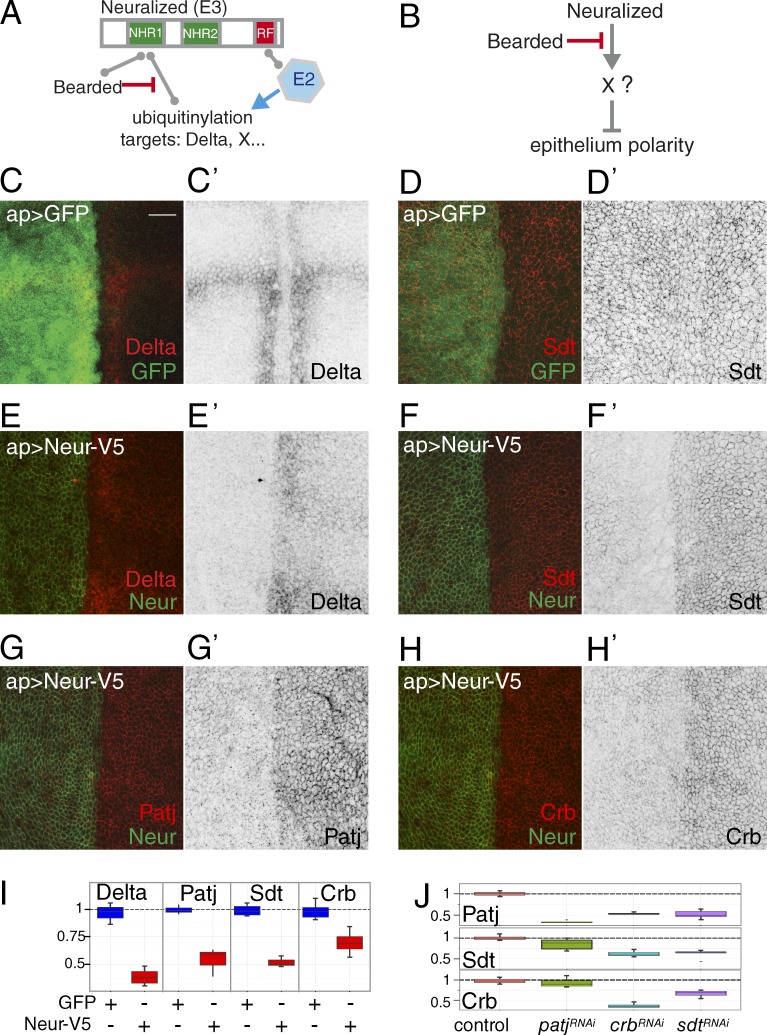
Neuralized (Neur) is a conserved membrane-associated E3 ubiquitin ligase (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001; Yeh et al., 2001). It contains a C-terminal RING domain that is predicted to bind an E2 enzyme that transfers ubiquitin to protein substrates bound to Neur. The best known target of Neur is the Notch ligand Delta (Dl; Fig. 1 A; Lai et al., 2001; Pavlopoulos et al., 2001; Le Borgne and Schweisguth, 2003; Bardin and Schweisguth, 2006; Commisso and Boulianne, 2007; Fontana and Posakony, 2009; Daskalaki et al., 2011). The ubiquitination of Dl by Neur regulates its endocytosis, and Neur-mediated endocytosis of Dl promotes the trans-activation of Notch (Weinmaster and Fischer, 2011). In Drosophila, this activity of Neur proteins is antagonized by proteins of the Bearded (Brd) family that compete with Dl for interaction with Neur (Fig. 1 A; Lai et al., 2000; Bardin and Schweisguth, 2006; De Renzis et al., 2006; Chanet et al., 2009; Fontana and Posakony, 2009). Interestingly, the synthetic deletion of all Brd family genes was found to result in embryonic lethality associated with a loss of epithelium integrity similar to the one seen in crb mutant embryos. This Brd mutant phenotype was suppressed by the loss of neur activity, hence revealing that Neur negatively regulates epithelial polarity (Chanet and Schweisguth, 2012). This activity of Neur was independent of Dl-Notch signaling. Thus, Neur must regulate epithelial morphogenesis via other molecular targets (Fig. 1 B). The molecular identity of these targets and the mechanism whereby Neur antagonizes epithelial polarity are unknown.
Figure 1.
Neur targets core Crb complex components. (A) Molecular activity and regulation of Neur. The E3 ubiquitin ligase Neur interacts with an E2 enzyme via its RING Finger (RF) and recruits its molecular targets, e.g., Dl, via its NHRs. Brd family proteins also bind the NHR and inhibit Neur–target interactions. (B) Neur negatively regulates epithelium polarity via an unknown target or targets. Brd antagonizes this activity of Neur. (C–H’) Ectopic Neur triggered the down-regulation of Dl (C and E), Sdt (D and F), Patj (G), and Crb (H). GFP (C and D) and Neur (V5 tag; green in E–H) were expressed in the dorsal cells of third instar wing disks using apterous (ap)-Gal4 (see Fig. S1 for negative GFP controls for G and H). Dorsal is left. Bar, 10 µm. (I) Box plots showing the dorsal/ventral (d/v) intensity ratios measured in controls (ap>GFP, blue; n = 5 disks) and upon ectopic Neur (ap>Neur, red; n = 5). For all proteins, similar levels were measured in dorsal and ventral cells, i.e., a ratio of ∼1, in control disks. Upon Neur expression, lower levels were seen in dorsal cells, resulting in a ratio of <1. In these and all other Tukey box plots, whiskers extend to the highest and lowest values that are within a 1.5 interquartile range. (J) Box plots as in I showing the d/v intensity ratios for Patj, Sdt, and Crb upon silencing of patj, crb, and sdt (in all cases, n = 5; also see Fig. S1).
In this study, we identify Sdt as a molecular target of Neur. We find that a specific subset of the Sdt isoforms physically interacts with Neur via a Neur binding motif (NBM). We provide evidence that the NBM-containing isoforms of Sdt are key in vivo targets of Neur and that Neur acts via these isoforms to promote the endocytosis of Crb. We further show that this regulation of Sdt and Crb by Neur contributes to the rapid remodeling of the posterior midgut (PMG) epithelium in the early embryo. Thus, Neur destabilizes epithelia by down-regulating Crb through its direct interaction with specific Sdt isoforms.
Results
Neur down-regulates Crb complex proteins
We previously showed that the inhibition of Neur by the Brd proteins is required to stabilize the primary epithelium during germband extension. Indeed, high Neur activity in Brd mutant embryos resulted in the rapid loss of the apical Crb, Sdt, Patj, Par6, and atypical PKC (aPKC) proteins at stages 6–8 (Chanet and Schweisguth, 2012). This suggested that the apical Crb complex is a target, direct or indirect, of Neur. To further investigate the regulation of Crb by Neur, we used an overexpression assay in third instar wing imaginal disks. Neur was ectopically expressed in the dorsal wing pouch cells that do not express Neur. As expected, ectopic Neur led to a down-regulation of Dl. Interestingly, it also led to a strong down-regulation of Crb, Sdt, and Patj (Fig. 1, C–I). Neur also had a weak effect on Par6 and no significant effect on aPKC, epithelial cadherin (E-Cad), and Bazooka (Baz; Fig. S1). Although these data confirmed that Crb, Sdt, and Patj are candidate targets of Neur, it remained unclear which of these proteins, if any, is a direct target. This is because Crb, Sdt, and Patj are mutually dependent for their stable accumulation at the cortex, at least in some epithelia (Bachmann et al., 2001; Hong et al., 2001; Bulgakova et al., 2008; Pénalva and Mirouse, 2012; Sen et al., 2012; Zhou and Hong, 2012; Lin et al., 2015). Indeed, lowering the levels of one was sufficient to lower the levels of the two others (Figs. 1 J and S1). Nevertheless, because the silencing of Patj had a weaker effect on the levels of Sdt and Crb than ectopic Neur (Fig. 1 J), Patj may only be an indirect target of Neur. This would imply that Neur may act on Crb via Sdt or vice versa. Because ectopic Neur had a stronger effect on Sdt than on Crb (Fig. 1 I), we decided to further investigate whether Sdt could be a direct target of Neur.
Neur interacts with the NBM-containing isoforms of Sdt
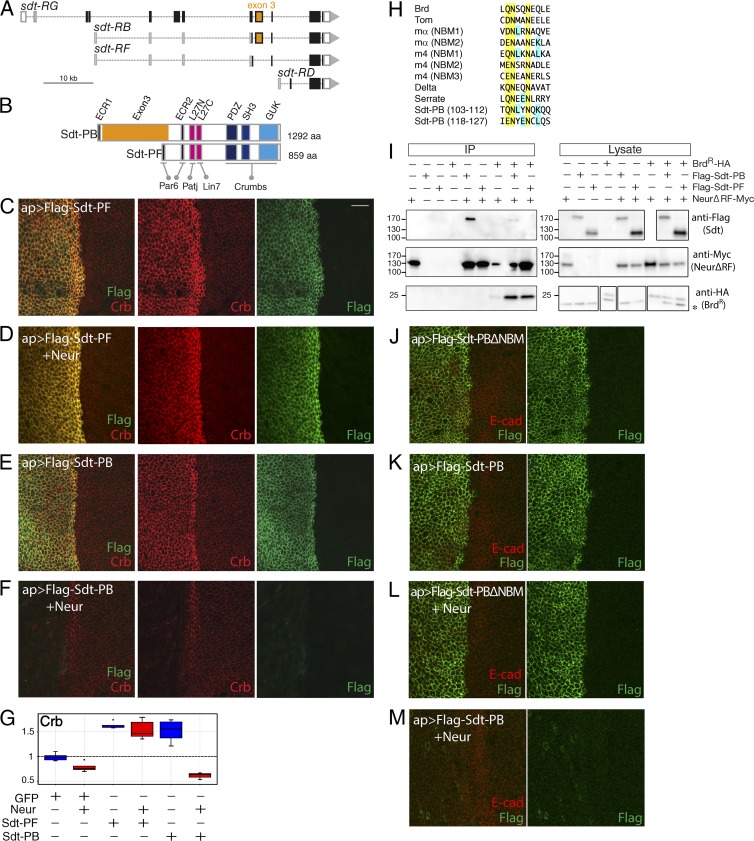
The sdt gene encodes 12 predicted protein isoforms produced by alternative splicing (Fig. 2 A; Berger et al., 2007; Bulgakova et al., 2010). Two classes of Sdt isoforms can be defined by the alternative exon 3 (numbering according to Hong et al. [2001] and Berger et al. [2007]) that encodes 433 amino acids with no clearly identifiable motifs or domains (region noted in this study, Exon3). Isoforms containing exon 3 are represented in this study by Sdt-PB (Bulgakova et al., 2010; noted as Sdt-A in Berger et al., 2007), whereas the other class is represented by Sdt-PF (noted as Sdt-B1 in Fig. 2 B; Berger et al., 2007). When overexpressed in wing imaginal disk epithelia, both Sdt-PB and Sdt-PF stabilized Crb (Fig. 2, C, E, and G). This effect is consistent with both isoforms interacting via their SH3-GUK-PDZ domains with the PDZ binding motif of Crb (Li et al., 2014). In contrast, we found that Neur down-regulated Sdt-PB but not Sdt-PF (Fig. 2, D, F, and G). This indicated that the down-regulation of Sdt by Neur required the presence of exon 3.
Figure 2.
Neur interacts with a subset of the Sdt isoforms. (A) Genomic organization of the sdt locus. Of the 12 isoforms, only four transcripts (RG, RB, RF, and RD) are shown here. Exons are shown as boxes (ORF, black; exon 3, orange). (B) Domain structure of Sdt-PB and Sdt-PF encoded by the RB and RF transcripts, respectively. Interaction domains and motifs with Par6, Patj, Lin7, and Crb are shown. The Exon3 region (orange) separates the motifs ECR1 and ECR2, which are predicted to mediate the interaction with Par6. (C–F) Neur induced the down-regulation of Sdt-PB, but not of Sdt-PF (Flag, green), in dorsal cells that expressed Sdt-PB, Sdt-PF, and/or Neur under the control of ap-Gal4. Crb (red) was stabilized upon expression of Sdt-PB and Sdt-PF (C and E). Coexpression of Sdt-PB and Neur led to a very strong loss of Crb (F). In contrast, Sdt-PF stabilized Crb even in the presence of Neur (D). (G) Box plots showing the d/v intensity ratios for Crb upon expression of GFP, Neur, Sdt-PF, and/or Sdt-PB, as indicated below (n = 5 for each genotype). (H) Sequence alignment with known NBMs suggesting that Exon3 contains two putative NBMs (yellow, conserved amino acids; blue, amino acids of Sdt-PB found in other NBMs). (I) Coimmunoprecipitation with NeurΔRF (anti-Myc) indicated that Sdt-PB (anti-Flag,) but not Sdt-PF, physically interacts with NeurΔRF (anti-Myc) and that this interaction is completed by BrdR (anti-HA). Total lysates and anti-Myc immunoprecipitates were analyzed by Western blots using anti-Flag, anti-Myc, and anti-HA. A background anti-HA band (asterisk) was detected in the lysates. The low Neur signal in the immunoprecipitates (IP) from cells coexpressing Brd was not reproducibly observed, i.e., Brd does not antagonize Neur immunoprecipitation. The positions of the molecular mass (kD) markers are shown on the left. (J–M) NBM-dependent down-regulation of Sdt. Neur induced the down-regulation of Sdt-PB but not of Sdt-PBΔNBM (Flag is in green, and E-Cad is in red) in cells expressing Neur, Sdt-PB, and/or Sdt-PBΔNBM. Bar, 10 µm. ap, apterous.
In these experiments, the down-regulation of Crb by Neur was maximal when Neur was coexpressed with Sdt-PB as compared with when Neur was expressed alone or together with Sdt-PF (Fig. 2 G). Thus, Sdt-PB appeared to synergize with Neur to down-regulate Crb, suggesting that Neur acts together with Sdt-PB to down-regulate Crb. We also observed that Sdt-PF stabilized Crb despite ectopic Neur (Fig. 2 D). This indicated that Sdt-PF can protect Crb from down-regulation by Neur. We suggest that Sdt-PF may compete for the binding of Crb and the Sdt isoforms that mediate the effect of Neur (e.g., Sdt-PB), thereby blocking the down-regulation of Crb.
We next investigated how Neur discriminated between Sdt-PB and Sdt-PF. One hypothesis is that Neur specifically interacts with Sdt-PB via the Exon3. To test this possibility, we used a coimmunoprecipitation assay in Drosophila S2R+ cells that were cotransfected to express Flag-tagged Sdt-PB (or Sdt-PF) and a Myc-tagged version of Neur deleted of its RING Finger domain (NeurΔRF; deletion of the RING Finger domain resulted in increased levels of Neur; Lai et al., 2001). In this assay, Neur physically interacted with Sdt-PB but not Sdt-PF (Fig. 2 I). Moreover, this interaction was reduced when a stabilized version of the Brd protein, BrdR, was coexpressed (Fig. 2 I). Thus, Brd appeared to compete with Sdt-PB for interaction with Neur. Both Brd proteins and Dl interact with the Neur homology repeat (NHR) domain of Neur via related NBMs, thus providing a simple molecular explanation for the competitive binding of Brd and Dl to Neur (Bardin and Schweisguth, 2006; Fontana and Posakony, 2009; He et al., 2009; Daskalaki et al., 2011; Gupta et al., 2013).
Because Brd appeared to compete with Sdt-PB for Neur binding, we wondered whether Exon3 contained an NBM. Sequence analysis of Exon3 suggested the existence of two putative NBMs close to each other (Figs. 2 H and S2 A). Consistent with a role of these NBMs in Neur binding, deletion analysis of Exon3 indicated that the region encompassing these two NBMs is important for the Neur–Sdt interaction (Fig. S2, B and C). Additionally, deletion of these NBMs (in Sdt-PBΔNBM) abolished the effect of Neur on Sdt accumulation both in vivo (Fig. 2, J–M) and in transfected S2 cells (Fig. S2, D and E). Collectively, these data strongly indicated that Neur directly interacts with the NBM-containing isoforms of Sdt and that this interaction is required for the down-regulation of Sdt by Neur.
Regulation of endogenous Sdt by Neur
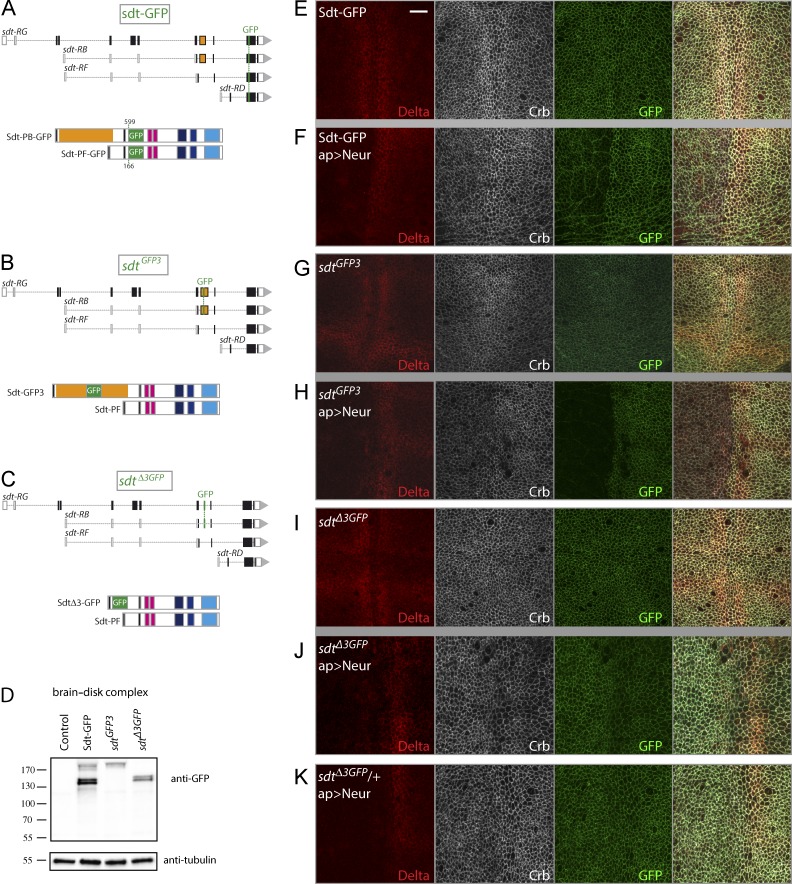
We next addressed whether Neur targets specific isoforms of endogenous Sdt. However, to specifically detect the NBM-containing isoforms of Sdt, new genetic tools were needed. In this study, three GFP-tagged versions of Sdt were produced as bacterial artificial chromosome (BAC) transgenes (Sdt-GFP) and knock-in alleles (Sdt-GFP3 and SdtΔ3-GFP). First, to detect all Sdt isoforms, GFP was inserted within an exon shared between all isoforms in the context of a 69-kb BAC transgene to produce Sdt-GFP (Fig. 3 A). Genomic rescue showed that this BAC encodes a fully functional version of the sdt gene (see the Transgenes section of Materials and methods). Second, to specifically detect all NBM-containing isoforms, we used CRISPR (clustered regularly interspaced short palindromic repeats)-mediated homologous recombination (HR) to insert GFP within exon 3 at the locus. This produced sdtGFP3 flies that were viable and fertile (Fig. 3 B). We collectively refer to the GFP-tagged isoforms of Sdt produced by these flies as Sdt-GFP3. Of note, Flp recombination target (FRT) sites were also inserted in the introns flanking exon 3, allowing for conditional deletion of exon 3 (Fig. S3). Third, exon 3 was replaced at the locus by an exon encoding GFP using CRISPR-mediated HR to produce the sdtΔ3GFP allele (Fig. 3 C). These sdtΔ3GFP mutant flies were viable and fertile. In these flies, the splicing events incorporating exon 3 into mature mRNAs generate GFP-tagged proteins lacking the sequence of exon 3, hence lacking the NBM. These are collectively referred to as SdtΔ3-GFP. Western blot analysis indicated that the predicted GFP-tagged isoforms were produced from these modified loci (Fig. 3 D). Sdt-GFP, Sdt-GFP3, and SdtΔ3-GFP were detected at the apical cortex in wing imaginal disks (Fig. 3, E, G, and I), where they colocalized with aPKC, apical to E-Cad (Fig. S3, C–F). These results indicate that NBM-containing isoforms of Sdt are endogenously present in wing imaginal cells and that the apical localization of Sdt does not depend on exon 3.
Figure 3.
Isoform-specific down-regulation of Sdt by Neur. (A) Structure of Sdt-GFP isoforms. GFP was inserted into a common exon (top) at amino acid position 599 in Sdt-PB and 166 in Sdt-PF (bottom). (B and C) Structure of the sdt alleles produced by CRIPSR-mediated HR. In sdtGFP3, GFP was inserted at amino acid 188 of Sdt-PB within Exon3 to produce Sdt-GFP3 (B). In sdtΔ3GFP, GFP replaced the sequence of exon 3, leading to the synthesis of SdtΔ3-GFP (C). These two alleles should not modify the synthesis of Sdt-PF. (D) Western blot analysis of brain–disk complexes from Sdt-GFP, Sdt-GFP3, and SdtΔ3-GFP larvae (control: wild-type larvae). As expected, Sdt-GFP3 and SdtΔ3-GFP comprise a subset of the Sdt-GFP isoforms. Molecular mass (kD) markers are shown on the left. (E–K) Neur induced the down-regulation of Sdt-GFP (GFP, green; E and F) and Sdt-GFP3 (G and H) but not SdtΔ3-GFP (I and J). Although Neur targeted Dl (red) for degradation independently of the Sdt isoforms, low Crb levels were detected in sdt-GFP and sdtGFP3 larvae but not in sdtΔ3GFP larvae. Thus, Neur regulates Crb indirectly via Sdt. Ectopic Neur down-regulated Dl, whereas Crb levels remained unchanged in heterozygous sdtΔ3GFP/+ larvae (K) as in sdtΔ3GFP larvae (J), despite the presence of endogenous Neur-sensitive Sdt from the wild-type locus. Thus, Neur-resistant SdtΔ3-GFP appeared to be sufficient to stabilize Crb. Bar, 10 µm. ap, apterous.
We then tested whether Neur specifically targets the endogenous NBM-containing isoforms of Sdt. Ectopic Neur strongly down-regulated Sdt-GFP and Sdt-GFP3 but had no significant effect on SdtΔ3-GFP (Fig. 3, E–J; and Fig. S3 A). Of note, a weak Sdt-GFP signal remained detectable at the cortex, whereas Sdt-GFP3 was not detectable. We interpret this weak Sdt-GFP signal as reflecting the persistence of Sdt isoforms lacking an NBM, which therefore seemed to be expressed at low levels in this tissue. Conversely, the strong loss of Sdt-GFP signal (marking all isoforms) upon Neur expression was interpreted to suggest that NBM-containing isoforms are the predominant isoforms in this tissue. Collectively, these data indicated that Neur specifically targets the NBM-containing isoforms of Sdt in vivo.
We next examined the levels of Crb. First, we observed that ectopic Neur down-regulated Crb in the presence of the NBM-containing isoforms but not in their absence (Fig. 3, H and J; and Fig. S3 B). This showed that the effect of Neur on Crb is indirect and is mediated by the NBM-containing isoforms of Sdt. We therefore conclude that the NBM-containing isoforms of Sdt are direct in vivo targets of Neur and that Neur acts via these Sdt isoforms to down-regulate Crb. Second, in the absence of Neur, we observed similar levels of Crb in sdtGFP3 and sdtΔ3GFP wing imaginal cells (Fig. 3, G and I). Similarly, no change in Crb, Sdt, and Par6 levels were seen upon Flp-out recombination of the exon 3 at the sdtGFP3 locus, i.e., upon converting exon 3–containing isoforms into exon 3–lacking isoforms (Fig. S3, G–J). Thus, the presence of exon 3 appeared to have no significant effect on the ability of Sdt to stabilize Crb in the absence of Neur.
Because Neur specifically targets the NBM-containing isoforms, and because all Sdt isoforms can stabilize Crb, Neur might have limited effects on Crb levels in cells expressing both types of Sdt isoforms because Neur-resistant Sdt would stabilize Crb. To test this idea, we examined the effect of Neur overexpression in wing disks from heterozygous sdtΔ3GFP larvae. In this context, NBM-containing isoforms are produced from the wild-type allele, whereas isoforms lacking exon 3 are generated from the sdtΔ3GFP knock-in allele. Interestingly, we found that ectopic Neur down-regulated Dl but not Crb (Figs. 3 K and S3 B). This indicated that the presence of Neur-resistant Sdt isoforms was sufficient to stabilize Crb. Sdt-PB can, however, override the stabilization activity of the Neur-resistant isoforms when overexpressed, likely by competing with endogenous Sdt isoforms for Crb binding, thereby leading to a nearly complete loss of Crb in the presence of Neur (Fig. 2, F and G). Collectively, these data strongly suggest that Neur can down-regulate Crb in epithelial cells only when the NBM-containing isoforms of Sdt are the predominant molecular species.
Neur regulates the endocytosis of Crb via the NBM-containing isoforms of Sdt
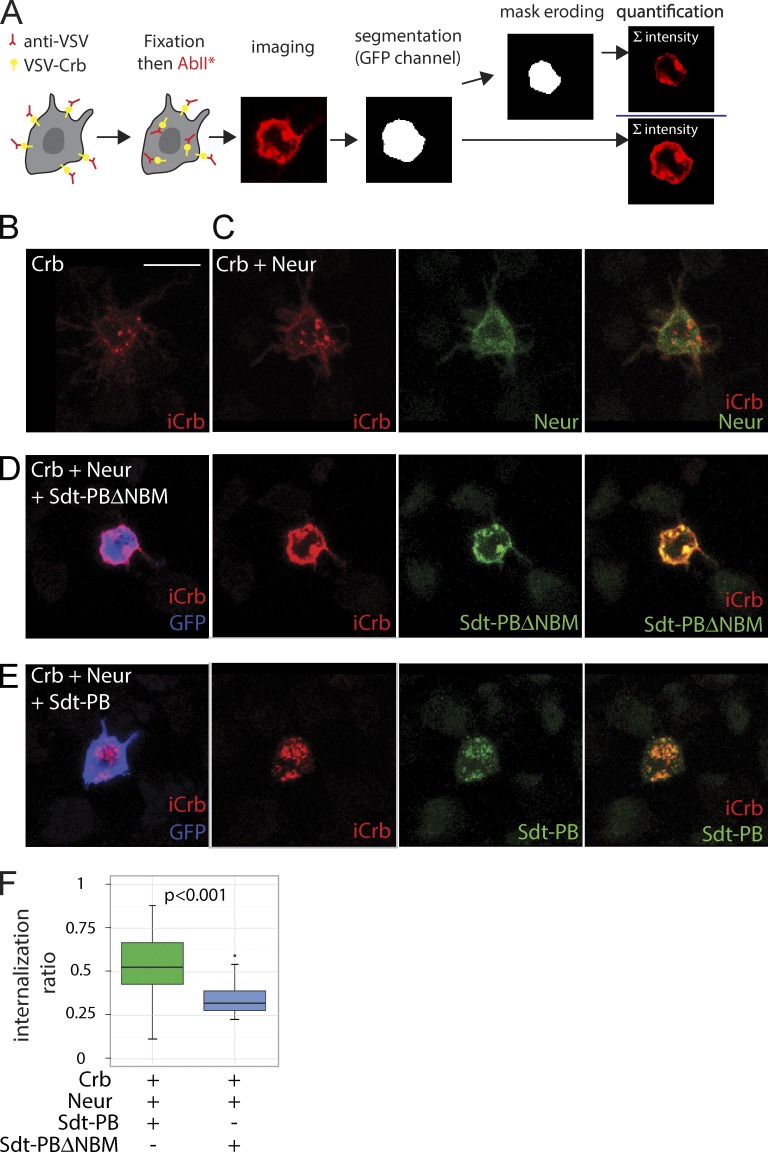
How does Neur regulate negatively apical Crb levels? Because Neur is known to regulate the endocytosis of Dl (Lai et al., 2001; Pavlopoulos et al., 2001; Le Borgne and Schweisguth, 2003), one possibility is that Neur acts via Sdt to promote the endocytosis of Crb. To test this, we used an antibody uptake assay to monitor the endocytosis of a tagged version of Crb in S2R+ cells. In this assay, the extracellular domain of Crb was deleted and replaced by a vesicular stomatitis virus (VSV) G tag (Médina et al., 2002), and the endocytosis of Crb was monitored by the internalization of anti-VSV antibodies. The latter was measured as the signal intensity ratio between internalized and total anti-VSV signals in individual cells. The potential role of Neur on the endocytosis of Crb was examined in cells expressing full-length Sdt-PB or Sdt-PBΔNBM. To ensure coexpression of Neur and Sdt at the single-cell level, we used a multicistronic expression system based on the viral-derived 2A-like peptides system (González et al., 2011). Using this assay, we found that Crb was efficiently endocytosed in cells coexpressing Sdt-PB and Neur. In contrast, significantly lower levels of endocytosis were seen in cells expressing Neur only, Sdt-PB only, or coexpressing Neur and Sdt-PBΔNBM (Fig. 4). These results therefore indicate that Neur can act via NBM-containing isoforms to up-regulate the endocytosis of Crb.
Figure 4.
Neur promotes the internalization of Crb via Sdt. (A) Crb internalization assay. S2R+ cells transfected with VSV-Crb were pulsed with anti-VSV antibodies, briefly chased, and then fixed and stained to detect both surface-bound and internalized anti-VSV antibodies (iCrb signal). After image segmentation to detect cell contours (using the GFP signal), an internalization ratio was calculated for each transfected GFP+ cell as the ratio between intracellular and total anti-VSV signals. (B–E) Representative S2R+ cells showing iCrb (red), Flag-tagged Neur (green in C), Flag-tagged Sdt (green in D and E), and GFP (blue). In the absence of Sdt (B and C), iCrb localized mostly into intracellular dots independently of Neur. While iCrb colocalized with Sdt-PB into intracellular dots (E), it colocalized with Sdt-PBΔNBM at the cell cortex. Bar, 10 µm. (F) Box plots showing the internalization ratios measured as described in A. Sdt-PBΔNBM (n = 22 cells), but not Sdt-PB (n = 45), was able to stabilize VSV-Crb at the cell surface in the presence of Neur. A Shapiro test (for normality) and a Wilcoxon rank sum test (for statistical significance) were performed.
Neur acts via Sdt to disrupt epithelial integrity in Brd mutants
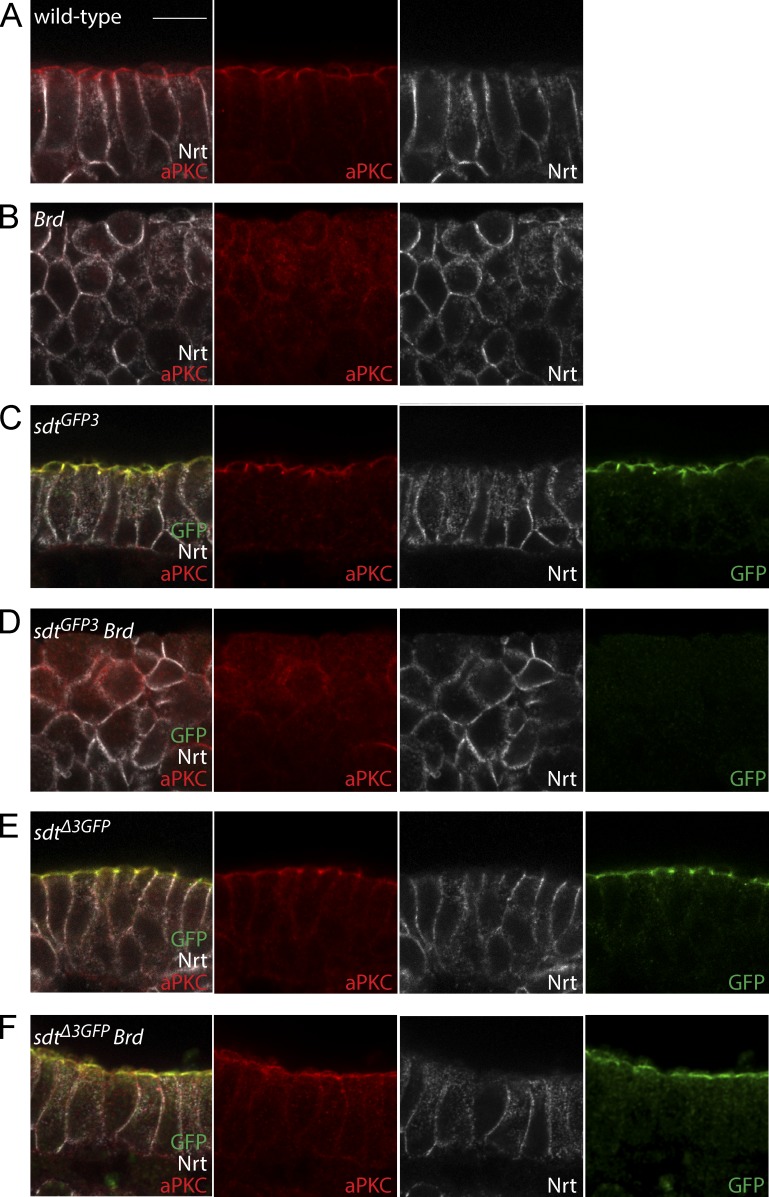
We have previously shown that the increased activity of Neur in Brd mutant embryos led to a rapid loss of epithelial integrity associated with a strong reduction in the level of Crb, Sdt, and Patj (Fig. 5, A and B; Chanet and Schweisguth, 2012). Our aforementioned data suggest that these effects of Neur on epithelial polarity could be mediated by the NBM-containing isoforms of Sdt. Consistent with this possibility, we found that Sdt-GFP3 was expressed during germband extension (Fig. 5 C), indicating that NBM-containing isoforms are endogenously expressed in the early embryo. Moreover, the accumulation of Sdt-GFP3 was severely reduced in Brd mutant embryos (Fig. 5 D), indicating that increased Neur activity led to the down-regulation of these NBM-containing isoforms. In contrast, SdtΔ3-GFP localized apically in wild-type embryos and was not detectably down-regulated in Brd mutant embryos (Fig. 5, E and F). This confirmed that the Sdt isoforms that lack an NBM are insensitive to increased Neur activity. Finally, if the down-regulation of Sdt by Neur caused the Brd mutant phenotype, deletion of the exon 3 should suppress this phenotype in sdtΔ3GFP Brd double mutant embryos. We therefore compared Brd mutant embryos with sdtΔ3GFP Brd double mutant embryos and found that epithelial integrity and apical localization of aPKC were remarkably restored upon deletion of the exon 3 (Fig. 5, B and F). We therefore conclude that the down-regulation of the NBM-containing isoforms of Sdt by Neur caused the loss of epithelial polarity in Brd mutant embryos. This demonstrated that the NBM-containing isoforms of Sdt are key in vivo targets of Neur.
Figure 5.
Loss of epithelial integrity in Brd mutants is caused by Neur-mediated down-regulation of Sdt. (A and B) Apical–basal polarity (aPKC, red; Nrt, white) and epithelial integrity were lost in Brd mutant embryos at stages 9–10 (B; wild-type control shown in A). (C–F) The loss of epithelial polarity (aPKC, red; Nrt, white) seen in Brd mutants was suppressed by the deletion of exon 3 in Brd sdtΔ3GFP double mutant embryos (F; control sdtΔ3GFP embryo shown in E). In contrast, a strong loss of epithelial integrity was observed in Brd sdtGFP3 double mutant embryos (D; control sdtGFP3 embryo shown in C). Although both Sdt-GFP3 and SdtΔ3-GFP (green) localized at the apical cortex in control embryos (C and E), only SdtΔ3-GFP remained apical in Brd mutants (F), whereas Sdt-GFP3 was not detected (D). Bar, 10 µm.
Sdt regulation during eye morphogenesis
We next studied the regulation of Sdt by Neur in Brd+ flies. As mentioned in the Regulation of endogenous Sdt by Neur section, sdtΔ3GFP mutant flies were viable and fertile. Similarly, deletion of the exon 3 at the sdtGFP3 locus using FLP-mediated excision of the engineered FRT–exon 3–FRT cassette produced viable and fertile sdtΔ3 flies. This showed that the NBM-containing isoforms of Sdt are not essential and can be replaced by isoforms lacking exon 3. Also, the deletion of exon 3 had no effect on the localization of Neur at the apical cortex, indicating that the interaction of Neur with Sdt is not key for its apical localization (Fig. S3, K and L). Thus, the Sdt–Neur interaction and the potential regulation of Sdt by Neur appeared to be largely dispensable during development.
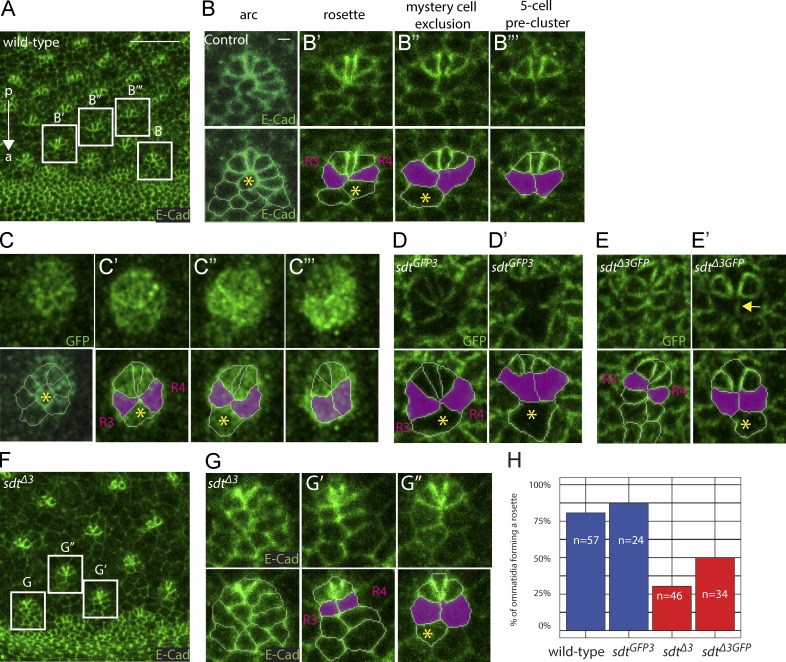
Nevertheless, we tested whether endogenous Neur targets Sdt during eye development. In eye imaginal disks, regularly spaced clusters of photoreceptors called ommatidia form posterior to the morphogenetic furrow that travels from posterior to anterior across the eye neuroepithelium. Cell–cell rearrangements underlie a stereotyped multicellular patterning process resulting in ommatidia morphogenesis posterior to the furrow (Fig. 6 A; Wolff and Ready, 1991; Pichaud, 2014). During this process, cells first organize into arcs with a central R8 photoreceptor cell flanked by six to seven cells, which then progressively resolve into five-cell preclusters via stereotyped cell–cell rearrangements whereby the photoreceptor cells R3 and R4 come into direct contact and thereby exclude the mystery cell (Fig. 6, B–B′′′; Wolff and Ready, 1991; Pichaud, 2014). Using a BAC-encoded GFP-tagged Neur (GFP-Neur; unpublished data), we found that Neur is dynamically expressed throughout this process (Fig. 6, C–C′′′; and Fig. S4; del Alamo and Mlodzik, 2006). Interestingly, the R3/R4 cells that express high Neur levels (Fig. S4 B) exhibited lower levels of Sdt-GFP3 relative to SdtΔ3-GFP (Fig. 6, D–E′), indicating that Sdt is down-regulated in an exon 3–dependent manner. Finally, multicellular patterning defects were observed in sdtΔ3 mutant discs (Fig. 6, F–H), suggesting that this cell-specific down-regulation of Sdt contributes to multicellular patterning in the developing retina. These defects were, however, transient, and a regular pattern of ommatidia eventually developed. Collectively, our data suggest that Neur targets Sdt in the developing eye.
Figure 6.
Neur targets Sdt in the developing eye. (A–B’’’) Multicellular patterning in the developing eye. E-Cad (green) staining of an eye imaginal disk showing the emergence of a stereotyped pattern of ommatidia posterior to the morphogenetic furrow (anterior [a]–posterior [p] axis indicated with an arrow). Cells first organize into arcs with a central R8 cell (B) that resolve into a six-cell rosette (B’) and a five-cell precluster (B’’’) via the exclusion of the central mystery cell (B’’; yellow asterisk) so that the R3 and R4 cells (magenta) come into contact. (C–E’) Dynamics of Neur (C–C’’’) and Sdt (C–E’) expression. GFP-Neur (green in C–C’’’) was detected in the posterior-most cells of the arc (C) and in all rosette cells but at a lower level in the mystery cell (C’ and C’’). High levels of GFP-Neur were eventually detected in R3/R4 of the five-cell preclusters (C’’’). Sdt-GFP3 (green in D and D’) accumulated at lower levels in the future ommatidial cells (D) and in the R3/R4 cells (D’) that express high levels of Neur (Fig. S4 B). In contrast, SdtΔ3-GFP (green in E and E’) was detected in all ommatidial cells, albeit at lower levels in R3/R4 (E and E’; the arrow points to the R3/R4 interface exhibiting low SdtΔ3-GFP levels; mystery cells, yellow asterisks). (F–H) Deletion of the exon 3 in sdtΔ3 flies led to cell–cell rearrangement defects as arcs resolved into elongated structures favoring early R3/R4 contacts at the expense of six-cell rosettes (F–G’’). This phenotype was quantified as the percentage of ommatidia (n = 24–57, as indicated in the panel) located in row 2 posterior to the morphogenetic furrow, which formed a rosette. (H) Loss of NBM-containing Sdt led to defective rosette formation. Bars: (A) 10 µm; (B) 1 µm.
Neur-dependent down-regulation of Sdt in the PMG
We next investigated the role of Neur in the remodeling of the PMG epithelium in early embryos, as this process was shown to depend on zygotic neur activity (Chanet and Schweisguth, 2012). During gastrulation, cells of the posterior endoderm invaginate to form the PMG (Fig. 7 A). As endoderm cells invaginate, they carry along the primordial germ cells (PGCs) that adhere to their apical surface. Once in the lumen of the PMG, PGCs migrate across this epithelium at stage 10 to eventually colonize the gonads. The timely migration of PGCs across the endoderm requires both the activation of a G protein–coupled receptor in PGCs (Kunwar et al., 2008) and the remodeling of the endodermal epithelium (Seifert and Lehmann, 2012) associated with a neur-dependent loss of several apical markers, including Crb, Par6, and aPKC (Chanet and Schweisguth, 2012). In neur mutant embryos, both loss of apical Crb and migration of the PGCs appeared to be delayed (Chanet and Schweisguth, 2012). Moreover, a loss of apical Crb appeared to be both necessary and sufficient to promote epithelial remodeling and PGC migration (Campbell et al., 2011; Seifert and Lehmann, 2012). These data led to a model whereby Neur destabilizes the PMG epithelium to facilitate the trans-epithelium migration of the PGCs (Chanet and Schweisguth, 2012). Concomitant to this epithelium remodeling, a subset of PMG cells called interstitial precursor cells (ICPs) that are marked by Scute and Asense are specified through Notch signaling and are apically extruded (Tepass and Hartenstein, 1995; Seifert and Lehmann, 2012). In this study, we found that PGC migration in early stage 10 embryos was detected before the onset of Scute and Asense expression in the PMG (Fig. S5, A–F). Additionally, ICPs are specified in excess numbers in neur mutant embryos but remained epithelial at stage 10 (Fig. S5, G and H). Thus, delayed epithelium remodeling was seen in neur mutant embryos showing an excess number of ICPs caused by the loss of Notch signaling. This further supports the notion that Neur has a Notch-independent activity in the regulation of epithelium morphogenesis (Chanet and Schweisguth, 2012).
Figure 7.
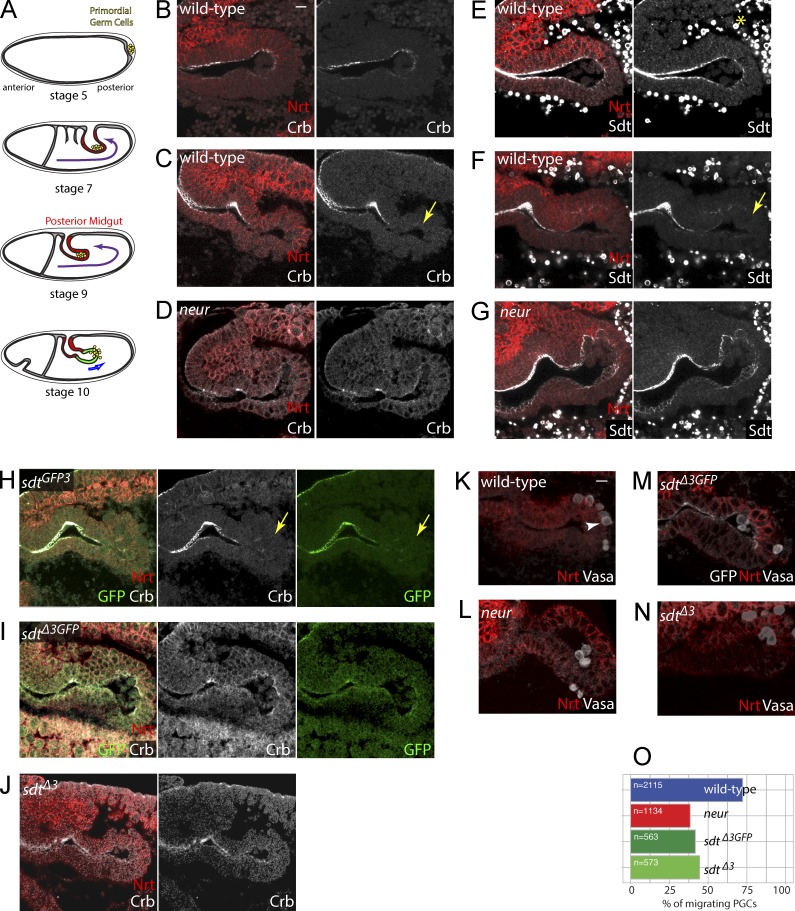
Epithelial remodeling by the Neur-mediated regulation of Crb. (A) PGCs (yellow) form at the posterior pole of the embryo (stage 5) and adhere to the surface of the midgut primordium as it invaginates at stages 7–9. PGCs located in the lumen of the PMG migrate across the epithelium at stage 10. (B–G) Crb (white, B–D; Nrt, red) and Sdt (white, E–G; note that anti-Sdt cross-reacted with yolk granules; yellow asterisk) were detected apically all along the PMG at stage 9 (B and E). At stage 10, Crb (C) and Sdt (F) were no longer detected in distal PMG cells (yellow arrows). In neur mutants, both Crb and Sdt remained detectable at the apical cortex of distal PMG cells (D and G). (H–J) Crb (white) and Sdt-GFP3 (green) were not detected at the apical cortex of distal PMG cells (yellow arrows) in stage 10 sdtGFP3 embryos (H), whereas both Crb and SdtΔ3-GFP (green) remained apical in sdtΔ3GFP embryos (I), indicating that the neur-dependent down-regulation of Crb required the NBM-containing isoforms of Sdt. Similarly, Crb (white) remained apical in distal PMG cells in sdtΔ3 mutant embryos (J). (K–O) PGC migration was quantified at stage 10 by counting the number of PGCs (arrowhead; Vasa, white in K–N) in the lumen, within the epithelium, or inside the embryo (Nrt, red; SdtΔ3-GFP, white; also shown in M). Migrating PGCs correspond to the last two categories (n = 563–2,115 PGCs; for each genotype, n is the total number of PGCs scored from >30 embryos). PGC migration across the PMG epithelium appeared to be similarly delayed in neur, sdtGFP3, and sdtΔ3 mutant embryos (O). Bars, 10 µm.
We next tested whether Neur down-regulates Crb via Sdt in the PMG. First, the levels of apical Crb and Sdt were reduced in distal PMG cells at stage 10, and this down-regulation was neur dependent (Fig. 7, B–G). Similarly, the NBM-containing isoforms of Sdt showed reduced levels at stage 10 (Fig. 7 H), and this down-regulation was isoform specific (Fig. 7 I). In contrast, Crb remained detectable at the apical cortex of distal PMG cells in sdtΔ3GFP and sdtΔ3 embryos (Fig. 7, I and J). We therefore conclude that Neur prevents the apical accumulation of Sdt in an NBM-dependent manner and that the NBM-containing isoforms of Sdt are required for the neur-dependent down-regulation of Crb.
We next studied the trans-epithelial migration of PGCs across the endoderm epithelium. In both sdtΔ3GFP and sdtΔ3 mutant embryos, fewer PGCs had migrated through the PMG epithelium at stage 10 (Fig. 7, K–O). This migration delay appeared to be similar to the one seen in neur mutant embryos (Fig. 7 O). We therefore conclude that the NBM-containing isoforms of Sdt are key targets of Neur and that the Sdt-mediated regulation of Crb by Neur contributes to epithelial remodeling in the PMG.
Discussion
Crb is a key determinant of apical membrane identity that controls epithelial morphogenesis in a tissue-specific manner (Tepass et al., 1990; Grawe et al., 1996; Tepass, 1996; Bulgakova and Knust, 2009; Campbell et al., 2009; St Johnston and Ahringer, 2010; Thompson et al., 2013). In this study, we have identified a molecular pathway that regulates its activity during Drosophila development. Indeed, we have shown that Neur directly interacts with a subset of the Sdt isoforms and thereby targets apical Crb for endocytosis in cells expressing these isoforms. Although not strictly essential, this regulation of Crb by Neur contributes to epithelial remodeling during development.
Several lines of evidence indicate that specific Sdt isoforms are key molecular targets of Neur. First, Neur physically interacts with a subset of the Sdt isoforms via an NBM encoded by an alternatively spliced exon. Second, using an endocytosis assay in S2R+ cells, we found that Neur regulates the endocytosis of Crb via Sdt in an NBM-dependent manner. Third, overexpression of Neur led to reduced levels of Sdt and Crb in vivo, and deletion at the sdt locus of the NBM-coding exon showed that this effect of Neur on Crb required the presence of NBM-containing Sdt isoforms. Thus, Neur regulates Crb via Sdt. This regulation may account for the secondary down-regulation of other apical proteins, implying that Sdt is the key target of Neur. Fourth, the loss of epithelial integrity resulting from high Neur activity in Brd mutant embryos required the presence of NBM-containing isoforms of Sdt. This demonstrates that Sdt is the only key target of Neur in Brd mutant embryos. Consistent with this view, Brd appeared to inhibit the Neur–Sdt interaction. We therefore propose that Neur physically interacts with a subset of the Crb complexes, i.e., those comprising the NBM-containing Sdt isoforms, to target Crb toward endolysosomal degradation, a major regulatory mechanism for Crb in both flies and mammals (Pocha et al., 2011; Schottenfeld-Roames et al., 2014; Rodríguez-Fraticelli et al., 2015; Sherrard and Fehon, 2015). Whether Neur ubiquitinates Sdt, Crb, and/or another Sdt-associated protein remains to be determined.
Although Sdt and, indirectly, Crb are key targets of Neur, this regulation is not essential, as flies lacking the NBM-containing isoforms of Sdt showed no major developmental defects. In this regard, we interpret the lethality associated with nonsense mutations located within exon 3 as resulting from a loss of Sdt activity caused by nonsense-mediated mRNA decay, a surveillance pathway that eliminates mRNA containing premature translation–termination codons, rather than from a loss of an essential isoform-specific function. We speculate that the regulation of Sdt and Crb by Neur may be redundant with other regulatory processes during morphogenesis. Consistent with this, key morphogenetic processes often involve redundant regulatory inputs to achieve efficient and timely output (Kölsch et al., 2007). Thus, other regulatory mechanisms likely act in parallel with the regulation of Crb by Neur to promote the remodeling of the PMG epithelium, as exemplified by the transcriptional regulation of the crb gene by Srp (Campbell et al., 2011).
Previous studies have highlighted functional diversity at the protein and RNA levels among the various Sdt isoforms (Berger et al., 2007; Horne-Badovinac and Bilder, 2008; Bulgakova et al., 2010). In this study, using CRISPR-mediated deletion of exon 3, we showed that the exon 3–containing isoforms could be substituted by isoforms lacking it. This isoform switch had no significant effect on apical Crb and Par6 levels in wing epithelial cells that do not express Neur. In contrast, changes in Crb levels were detected in cells with active Neur in the embryonic PMG and Brd mutant embryos. Thus, isoform diversity at the sdt locus provides a basis for a Neur-mediated and cell-specific regulation of Crb.
In mammals and flies, Neur regulates synaptic plasticity and memory (Pavlopoulos et al., 2008, 2011). Neur is expressed at high levels in the brain, where it regulates dendritic spine morphogenesis. In the mouse, this function of Neur is in part mediated by the ubiquitination of CPEB3, which stimulates the local synthesis of AMPA receptors at the synapse (Pavlopoulos et al., 2011). Because the mouse homologue of Sdt, Pals1, regulates dendritic spine morphogenesis (Zhang and Macara, 2008), our study raises the possibility that Neur may have a conserved function in regulating the activity of Sdt/Pals1 in the brain.
Finally, our analysis suggests an intriguing connection between Notch signaling and apical–basal polarity. Indeed, in analogy with the inhibition of Neur by Brd, one may wonder whether the Sdt–Neur interaction may similarly compete with the Dl–Neur interaction. Consistent with this possibility, increased Dl endocytosis, and hence Notch activity, was observed in crb mutant eyes (Richardson and Pichaud, 2010), a phenotype that might result from increased Neur activity caused by the loss of Sdt in crb mutant cells. It is also conceivable that Sdt might positively contribute to Neur-mediated Dl signaling by promoting the apical localization of Neur. Reciprocally, high Neur in signal-sending cells might be associated with decreased epithelial stability and partial or complete delamination of neural cells as seen in neuroblasts. Conversely, high Notch, and hence high Brd, might contribute to stabilize epithelial polarity through Neur inhibition. Interestingly, a related regulatory mechanism was recently proposed to exist in zebrafish. In this context, the E3 ubiquitin ligase Mindbomb 1 (Mib1) targets both DeltaD, a Notch ligand, and Ebp41l5, a FERM (Band 4.1, Ezrin, Radixin, and Moesin) domain protein that regulates junction disassembly to coordinate Notch-mediated fate specification with delamination in a neuroepithelium (Matsuda et al., 2016). Future studies will address the interplay between Notch signaling and Neur-dependent regulation of morphogenesis in epithelia.
Materials and methods
Transgenes
The BAC CH321-22E06 (Venken et al., 2009) was used to generate the Sdt-GFP BAC transgene using recombineering-mediated gap repair (Venken et al., 2006). First, a 34-kb fragment located 3′ to the sdt gene was deleted using the following primers: Sdt3′partrpsl forward, 5′-AAATACAATCAGAAATGCGAATGATGGGGGGACGGGACCTGGCCTGGTGATGATGGCGGG-3′; Sdt3′partneo reverse, 5′-ACCGAAATCAGCCTGAGTGAGAGAGAGAGAGAGAGCGCTATCAGAAGAACTCGTCAAGAA-3′; and Sdt3′part forward, 5′-AGAAATGCGAATGATGGGGGGACGGGACCTTAGCGCTCTCTCTCTCTCTCTCACTCAGGC-3′.
The sequence of the superfolder GFP was then inserted within the sdt ORF at amino acid 599. This site was chosen because it displayed low sequence conservation among various fly species. This strategy should help maintain function in the fusion protein (Couturier et al., 2012, 2013). The following primers were used: Sdtrpsl forward, 5′-GGTAAAGCGGTTGCCCAGCAGCAGCAGCAGGGCCTGGTGATGATGGCGGG-3′; Sdtneo reverse, 5′-CACGCCAACCACCTGGGTGGCCAGCGTTGCTCAGAAGAACTCGTCAAGAA-3′; Sdt-GFP forward, 5′-GGTAAAGCGGTTGCCCAGCAGCAGCAGCAGGGGGTTGGTATGGTGTCCAAGGGCGAGGAGC-3′; and Sdt-GFP reverse, 5′-CACGCCAACCACCTGGGTGGCCAGCGTTGCACCAACACCGGATCCCTTGTACAGCTCATCC-3′.
The resulting BAC was verified by sequencing of the recombined regions before phiC31-mediated integration at the M[3×P3-RFP attP]ZH (51C) site. Injection was performed by BestGene Inc. The sdtK85 mutant flies carrying one copy of the corresponding BACs were viable and fertile.
The DNA fragments encoding Sdt-PB/PF (blunted NotI–XhoI fragments from pUASt-Sdt-PB/-PF; gifts from E. Knust, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) were subcloned into pPac5.1 (opened at the blunted EcoRI and XhoI sites) to generate pPac-3×FLAG-Std-PB/-PF. These plasmids were used to express FLAG-tagged isoforms under the control of the actin5C promoter in S2R+ cells. Starting from pPac-3×FLAG-Std-PB, a deletion analysis of the sequence encoded by the exon 3 was performed by the sequence and ligation independent cloning method (Li and Elledge, 2012) using PCR fragments generated with the common primers 5′-CCCCAAATTGAATCACATGCCGCAACTGATAG-3′ and 5′-GCTTCCTGGAGCCACGAAGACTGTTCC-3′ in combination with the deletion-specific primers pPac-3×FLAG-Std-PBΔa, 5′-GTTGTTCATTTCGAAGCTTTGTGGTGGTGGTGGTGGTGTTG-3′ and 5′-CACCACCACCACAAAGCTTCGAAATGAACAACACGAACAAC-3′; pPac-3×FLAG-Std-PBΔb, 5′-GTTGGTGTTGCTAAGCTTTGTTGCTGCCATTGCGGTTG-3′ and 5′-CAATGGCAGCAACAAAGCTTAGCAACACCAACAGCAAC-3′; pPac-3×FLAG-Std-PBΔc, 5′-CAGCTGTTTCAACCAAGCTTCATTTGGTTGCAACGCCCGTG-3′; and pPac-3×FLAG-Std-PBΔd, 5′-GGCGAAGGCGCAAGCTTCGTTGCAACCAAATGTTGTTGC-3′ and 5′-TTGGTTGCAACGAAGCTTGCGCCTTCGCCTGTCACTGC-3′.
pPac-3×FLAG-Std-PBΔNBM was obtained from pPac-3×FLAG-Std-PBΔa by replacing a HindIII fragment with a PCR fragment generated with the primers 5′-CCCAAGCTTATCACAGCTTTGACTAACAATGATC-3′ and 5′-GGGAAGCTTTTCCGATTGCAGGCATTGTGTGGCCACATCG-3′.
The EcoRI–NotI DNA fragments encoding 3×FLAG-Std-PB (from pPac-3×FLAG-Std-PB) and 3×FLAG-Sdt-PBΔNBM (from pPac-3×FLAG-Std-PBΔNBM) were subcloned into a pUASp-attB vector. These two transgenes were integrated at the PB[attP-3B y+]VK0033 (65B2) site. The pPac-BrdR-3×HA plasmid was produced by gene synthesis (to replace all 10 lysines, K, of the Brd ORF by arginine, R, hence producing the BrdR mutant version of Brd) and subcloning as an EcoRI–XhoI fragment into the pPac5.1 vector. The BrdR mutant protein was found to be more active than wild-type Brd.
CRISPR-mediated HR
CRSIPR-mediated HR was achieved by injecting three plasmids, one corresponding to the donor template and two encoding the guide RNAs (gRNAs), into Cas9-expressing embryos. The following gRNAs were selected using the Optimal Target Finder tool (Gratz et al., 2014): gRNA1, 5′-TTCCATTGGAGTCCACTAAG-3′, and gRNA2, 5′-TCCGGCGGTGCAGGTCCGAC-3′.
Corresponding oligonucleotides were cloned into pU6-BbsI-chiRNA (45946; Addgene) as described by Gratz et al. (2014). The same gRNAs were used for both sdtΔ3GFP and sdtGFP3. Donor templates for HR were produced by BAC recombineering in Escherichia coli using the sdt BAC CH321-22E06 as described by Venken et al. (2006). In the first steps, the protospacer-adjacent motif sequences targeted by the gRNAs were mutated in the donor templates to avoid their Cas9-mediated cleavage, and the 3×P3-RFP marker flanked by loxP sites, which was produced by gene synthesis (Bivik et al., 2016), was inserted in the intron downstream of exon 3 using the following primers: sdtgRNA3′rpsl forward, 5′-TTAACCCCGCATCATACGCATTGATTCCCTTTCTTGTATTCGGCGGTGCAGGCCTGGTGATGATGGCGGG-3′; sdtgRNA3′neo reverse, 5′-GGTTGGTTGGATACCGGGGTCTATGGGTCAACTGGAGGCTGAAACGGCTCTCAGAAGAACTCGTCAAGAAGG-3′; sdtmutgRNA3′ forward, 5′-ACGCATTGATTCCCTTTCTTGTATTCGGCGGTGCATTGAATCAATTGAGCCGTTTCAGCCTCCAGTTGACCCATAGACCCC-3′; sdtloxPK7rpsl forward, 5′-GAAATATTCCCAAAATTGTAATGCCAATGCTCAGTTCATTTACATAACAAGGCCTGGTGATGATGGCGGG-3′; sdtloxPK7neo reverse, 5′-TTCAACTTAATGTGGTTATTGTTTTTAATTTAAGGGGTTTCCATTGGAGTTCAGAAGAACTCGTCAAGAAGG-3′; sdtloxPK7 forward, 5′-AAAATTGTAATGCCAATGCTCAGTTCATTTACATAACAATTATCCGACAAATAACTTCGTATAATGTATGCTATACGAAGTTATGTCGACGAATTCGCGG-3′; and sdtloxPK7 reverse, 5′-TTCAACTTAATGTGGTTATTGTTTTTAATTTAAGGGGTTTCCATTGGAGTATAACTTCGTATAGCATACATTATACGAAGTTATACTAGAGAGCTTCGCA-3′. The sequence of the superfolder GFP was inserted in frame to precisely replace the sequence of Exon3 in sdtΔ3GFP using the following primers: sdtDelex19rpsl forward, 5′-CCACCACCAACACCACCACCACATGCACCACTTAACCCACACGCACACAGGGCCTGGTGATGATGGCGGG-3′; sdtDelex19neo reverse, 5′-CCAGTGAAAGGCTGAGACAGATAGCGAGTACAAAATGCAATGTGGTATACTCAGAAGAACTCGTCAAGAAGG-3′; sdtDelex19GFP forward, 5′-CCACCACCAACACCACCACCACATGCACCACTTAACCCACACGCACACAGGGGGTTGGTATGGTGTCCAAGGGCGAGG-3′; and sdtDelex19GFP reverse, 5′-CCAGTGAAAGGCTGAGACAGATAGCGAGTACAAAATGCAATGTGGTATACACCAACACCGGATCCCTTGTACAGCTC-3′.
In sdtGFP3, GFP was inserted at amino acid 188 of Sdt-PB, and the sequence of exon 3 was flanked with two FRT sites. Thus, GFP tags all six NBM-containing isoforms (Sdt-PB, -PD, -PE, -PG, -PM, and -PN) as well as three isoforms that contain only part of Exon3 excluding the NBM (Sdt-PI, -PJ, and -PL; see http://www.flybase.org for a detailed description of the 12 predicted isoforms of Sdt). To do so, we used the following primers: sdtex19FRT1rpsl forward, 5′-CCGCATTTGTCCTTAACTAACAATGACTACAACCACCAACCAAAAGACCCGGCCTGGTGATGATGGCGGG-3′; sdtex19FRT1neo reverse, 5′-ACGCACAGGAAATCATCGGTGATGCTGCTGTTGTTGCTGCTGCTGCTGCTTCAGAAGAACTCGTCAAGAAGG-3′; FRT1sdtex19Gfp forward, 5′-CCGCATTTGTCCTTAACTAACAATGACTACAACCACCAACCAAAAGACCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAAAAAAAAAAACGTATTTTTTAATC-3′; FRT1sdtex19Gfp reverse, 5′-ACGCACAGGAAATCATCGGTGATGCTGCTGTTGTTGCTGCTGCTGCTGCTACCAACACCGGATCCCTTGTACAGCTCATCC-3′; sdtFRT1Gfp forward, 5′-GCAACAACAATAGCCACATAGTTGGCATCAGCGGGGTTGGTATGGTGTCCAAGGGCGAGGAGC-3′; and sdtFRT1Gfp reverse, 5′-GCTCCTCGCCCTTGGACACCATACCAACCCCGCTGATGCCAACTATGTGGCTATTGTTGTTGC-3′.
The DNA fragments of interest were then fetched from these recombineered BACs into multicopy vectors using the following primers: pCRIIsdtloxP forward, 5′-TTGGTAAGCCAATGGCCAATGGCCATTGACCGTCTCCAGTTAGCTCCAAACTGTAATCATCACAAGCGGCTTGGTTAAAAAATGAGCTGA-3′; and pCRIIsdtloxP reverse, 5′-ATCACCAGCTCCAGCTCCATCTTCAGTTTTCCCTTCTGGAATCCCTTTGGGTAATGATGAACGTCAGTGGCTTCCGCTTCCTCGCTCACT-3′.
Left and right homology arms flanking the target sites were 1.5 kb long. All recombineered fragments were verified by sequencing.
A mix of donor template (300 ng/µl) and gRNA plasmids (150 ng/µl) was injected into 900–1,200 embryos from the PBac{vas-Cas9}VK00027 stock (BL-51324). One to three independent RFP+ founder flies were selected. In both cases, proper recombination was checked by PCR.
Flies
Wild-type controls were w1118 flies. The Brd mutant genotype corresponds to the combination of two deficiencies, Df(3)Brd-C1 and Df(3)E(spl)δ-6, as described by Chanet et al. (2009) and Chanet and Schweisguth (2012). These were maintained over a TM3 hb-lacZ balancer for embryo genotyping. A GFP knock-in allele of the scute gene (scuteGFP) was generated using CRISPR-mediated HR and used here to detect the expression of Scute in the PMG (Corson et al., 2017). A BAC-encoded version of GFP-tagged Neur (GFP-Neur) was used here to examine the distribution of Neur. Otherwise, the following stocks were used: sdtK85 (a gift from E. Knust), neurIF65 maintained over a TM3 hb-lacZ balancer for embryo genotyping, UASt-FLAG-Sdt-PB and UASt-FLAG-Sdt-PF (described as UAS-Sdt-A and UAS-Sdt-B1 by Berger et al. [2007]; gifts from E. Knust), UASt-Neur13.4 (Neur-PA isoform; a gift from C. Delidakis, IMBB, Heraklion, Greece), UAS–double-stranded RNA (dsRNA) sdt (BL-33909), UAS-dsRNA crb (BL-40969), and UAS-dsRNA patj (BL-35547).
Immunostaining
Dissection of third instar larva, fixation, and antibody staining of imaginal disks were performed using standard procedures. Staged embryos (3–6 h after egg laying) were fixed in 4% formaldehyde and processed using standard procedures. The following antibodies were used: rabbit aPKC (1:500; Santa Cruz Biotechnology, Inc.), rabbit Patj (1:100; a gift from K. Choi, Korea Advanced Institute of Science and Technology, Daejon, South Korea), rat Crb (1:1,000; a gift from U. Tepass, University of Toronto, Toronto, Canada), rabbit Baz (1:500; a gift from A. Wodarz, University of Cologne, Cologne, Germany), anti-DlICD mAb 10D5 intracellular Dl (1:1,000; a gift from M. Rand, University of Rochester Medical Center, Rochester, NY), anti-DlECD mAb extracellular Dl (1:1,000; Developmental Studies Hybridoma Bank), N2 7A1 mAb Arm (1:500; Developmental Studies Hybridoma Bank), goat β-gal (1:500; Biogenesis), rabbit Vasa (1:1,000; a gift from P. Lasko, McGill University, Montreal, Canada), guinea pig Par6 (1:1,000; a gift from A. Wodarz), rat DCAD2 E-Cad (1:100; Developmental Studies Hybridoma Bank), and mouse BP106 neurotactin (Nrt; 1:500; Developmental Studies Hybridoma Bank). All secondary antibodies were Alexa Fluor 488–, Cy3-, and Cy5-coupled antibodies from Jackson ImmunoResearch Laboratories, Inc. Eye disks were stained for actin using atto647N-phalloidin (1:1,000; Sigma-Aldrich). After secondary antibody incubation and washes, disks were mounted in 4% N-propylgalate and 80% glycerol. Immunostained embryos were genotyped using the hb-lacZ marker of the TM3 balancer and anti–β-gal detection, precisely staged under a binocular scope, sectioned using a sharp scalpel blade, and mounted flat on a slide in 4% N-propylgalate in 80% glycerol. Images were acquired using a confocal microscope (LSM780; ZEISS) with a 63× Plan Apochromat 1.4 NA differential interference contrast M27 objective. Selected three to five planes from confocal z stacks (Δz = 0.6 µm) were used for maximal projections using Fiji (ImageJ; National Institutes of Health; Schindelin et al., 2012). The GFP-Neur signal was quantified using Fiji (Schindelin et al., 2012), and mean intensity values per cell were measured in eye disks after image segmentation using the actin signal (Aigouy et al., 2010).
Coimmunoprecipitation and Western blot analysis
S2R+ cells (a gift from C. Saleh, Pasteur Institute, Paris, France) were grown in Schneider S2 medium (Gibco). Plasmids directing the expression of FLAG-tagged Sdt, Myc-tagged NeurΔRF (a gift from E. Lai; Lai et al., 2001), and HA-tagged BrdR were cotransfected in S2R+ cells using FuGENE (Promega). Total DNA concentration was kept constant using an empty vector. 48 h after transfection, cells were lysed in 330 µl of 0.5% Triton X-100 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, 0.5 mM DTT, and 1× EDTA-free protease inhibitor cocktail [Roche]) for 15 min on ice. Extracts were cleared by centrifugation at 13,000 rpm for 20 min. 5 µl of polyclonal rabbit anti-Myc (EMD Millipore) and mouse anti-Myc 9E10 (Roche) was added to the extract and incubated for 1 h. This was followed by the addition of 25 µl of washed protein A beads (Roche) for 2 h at 4°C. Beads were washed seven times with 0.5% Triton X-100 buffer (the last two washes were 500-mM NaCl). Western blots were performed using mouse anti-Flag (1:1,000; Flag M2; Sigma-Aldrich), mouse anti-Myc 9E10 (1:1,000), and mouse anti-HA (1:500; 12CA5; Roche) and then were revealed using ECL+ substrate (Thermo Fisher Scientific).
GFP-tagged Sdt was studied by Western blot analysis using brain–disk complexes dissected from third instar larvae (five larvae per well). Protein extracts were prepared in the 0.5% Triton X-100 buffer (see previous paragraph) and loaded on 4–20% precast Miniprotean TGX gels (Bio-Rad Laboratories) for SDS-PAGE. Proteins were transferred onto 0.2-µm nitrocellulose membranes (Bio-Rad Laboratories). HRP-coupled anti-GFP antibodies (1:5,000; Abcam) were used for detection with SuperSignal WestFemto (Thermo Fisher Scientific).
To achieve coexpression of Sdt and full-length Neur in S2R+ cells, we used the 2A system (González et al., 2011). We used sequence and ligation independent cloning (Li and Elledge, 2012) to assemble DNA fragments obtained by restriction enzyme digestion with XhoI, XbaI, and SmaI from pAc5-STABLE2-blast (32426; Addgene), Flag-Sdt-PB/Flag-Sdt-PBΔNBM, and Neur cDNA constructs to produce intermediate plasmids. These were further modified to restore the ORF and/or remove Neur, hence producing pAc5-FlagSdtB-2A-GFP-2A-Blast, pAc5-FlagSdtB-2A-Neur-2A-GFP-2A-Blast, pAc5-FlagSdtBΔNBM-2A-GFP-2A-Blast, and pAc5-FlagSdtBΔNBM-2A-Neur-2A-GFP-2A-Blast using the primers 5′-GGCGCGATCCCGCGGTAAGGACGG-3′, 5′-CCCCTCGCGGCCGCCTGACTCGTCGCGATTGTTGTG-3′, 5′-GGCGCGATCCCGCGGTAAGGACGG-3′, 5′-TTCACCGGTTGACTCGTCGCGATTGTTGTGC-3′, 5′-CGAGTCAACCGGTGAAGGACGCGGCAGCCTACTGAC-3′, and 5′-CTCCCCTCGCGGCCGCCCGTGGTGTAG-3′.
Cell lysates of cells transfected with these plasmids were analyzed by Western blots using anti-FLAG and anti-GFP antibodies (as described above) using rabbit anti-FLAG (1:1,000; Sigma-Aldrich) and HRP-coupled anti-GFP antibodies (1:5,000; Abcam). Relative protein accumulation levels were studied in a ratiometric manner using GFP as an internal control. Signals were quantified using a My ECL Imager (Thermo Fisher Scientific).
Endocytosis assay
These plasmids as well as the control pAc-Myc-Neur plasmid (a gift from E. Lai; Lai et al., 2001) were cotransfected with a plasmid directing the expression of VSV-Crb (pMT-VSV-Crb; a gift from A. Le Bivic; described by Médina et al. [2002]) in S2R+ cells using FuGENE. Expression of VSV-Crb was induced 16 h after transfection using 0.5 mM Cu2+. Crb endocytosis was assayed using anti-VSV antibody uptake 5 h after induction. Cells deposited on slides coated with poly-l-lysine were incubated for 5 min at 25°C in S2 medium containing mouse anti-VSV (1:1,000; P5D4 mAb; pulse period; Sigma-Aldrich) and then washed and further incubated without antibodies for 15 min (chase period) at 25°C. Cells were then fixed in 4% paraformaldehyde (20 min at room temperature), stained using rabbit anti-Flag (1:1,000; Sigma-Aldrich) and goat anti-GFP (1:2,000; Abcam), and mounted in 4% N-propylgalate and 80% glycerol. Images were acquired using an LSM780 confocal microscope with a 40× Plan Apochromat 1.4 NA differential interference contrast M27 objective. Segmentation and measure of signal intensity were done using Fiji (Schindelin et al., 2012). Data analysis was performed under the R environment.
Statistical analysis
Shapiro tests (for normality) and Wilcoxon rank sum tests (for statistical significance) were performed.
Online supplemental material
Results showing the effect of ectopic Neur on the steady-state levels of apical polarity proteins are presented in Fig. S1. The identification of the NBM of Sdt and the analysis of its functional significance for Sdt localization and Neur-dependent regulation are shown in Figs. S2 and S3. Evidence for the Sdt-independent localization of Neur is shown in Fig. S4. Results showing that the trans-epithelial migration of PGCs precedes the specification of IPCs are shown in Fig. S5.
Supplementary Material
Acknowledgments
We thank K. Choi, C. Delidakis, J. Knoblich, E. Knust, E. Lai, A. Le Bivic, P. Lasko, M. Rand, C. Saleh, U. Tepass, A. Wodarz, the Bloomington Drosophila Stock Center, the Developmental Studies Hybridoma Bank, and Flybase for flies, antibodies, and other resources. We thank S. Chanet for her initial input. We thank L. Couturier for technical help. We thank A. Bardin and S. Chanet for critical reading and laboratory members for discussion.
This work was funded by an Agence Nationale de la Recherché grant (ANR12-BSV2-0010-01). G. Perez-Mockus received fellowships from the Ministére de l’Education Nationale de la Recherche et de la Technologie and the Fondation pour la Recherché Médicale.
The authors declare no competing financial interests.
Author contributions: G. Perez-Mockus performed all in vivo and microscopy experiments. V. Roca performed all S2 cell experiments and produced several DNA constructs. K. Mazouni generated BAC transgenes and all molecular reagents for CRISPR-HR. G. Perez-Mockus and F. Schweisguth analyzed the data and wrote the paper.
Footnotes
Abbreviations used:
- aPKC
- atypical PKC
- BAC
- bacterial artificial chromosome
- CRISPR
- clustered regularly interspaced short palindromic repeats
- d/v
- dorsal/ventral
- E-Cad
- epithelial cadherin
- FRT
- Flp recombination target
- gRNA
- guide RNA
- HR
- homologous recombination
- ICP
- interstitial precursor cell
- NBM
- Neur binding motif
- NHR
- Neur homology repeat
- PGC
- primordial germ cell
- PMG
- posterior midgut
- VSV
- vesicular stomatitis virus
References
- Aigouy B., Farhadifar R., Staple D.B., Sagner A., Röper J.-C., Jülicher F., and Eaton S.. 2010. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 142:773–786. 10.1016/j.cell.2010.07.042 [DOI] [PubMed] [Google Scholar]
- Bachmann A., Schneider M., Theilenberg E., Grawe F., and Knust E.. 2001. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature. 414:638–643. 10.1038/414638a [DOI] [PubMed] [Google Scholar]
- Bardin A.J., and Schweisguth F.. 2006. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 10:245–255. 10.1016/j.devcel.2005.12.017 [DOI] [PubMed] [Google Scholar]
- Berger S., Bulgakova N.A., Grawe F., Johnson K., and Knust E.. 2007. Unraveling the genetic complexity of Drosophila stardust during photoreceptor morphogenesis and prevention of light-induced degeneration. Genetics. 176:2189–2200. 10.1534/genetics.107.071449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivik C., MacDonald R.B., Gunnar E., Mazouni K., Schweisguth F., and Thor S.. 2016. Control of neural daughter cell proliferation by multi-level Notch/Su(H)/E(spl)-HLH signaling. PLoS Genet. 12:e1005984 10.1371/journal.pgen.1005984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova N.A., and Knust E.. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122:2587–2596. 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- Bulgakova N.A., Kempkens O., and Knust E.. 2008. Multiple domains of Stardust differentially mediate localisation of the Crumbs-Stardust complex during photoreceptor development in Drosophila. J. Cell Sci. 121:2018–2026. 10.1242/jcs.031088 [DOI] [PubMed] [Google Scholar]
- Bulgakova N.A., Rentsch M., and Knust E.. 2010. Antagonistic functions of two stardust isoforms in Drosophila photoreceptor cells. Mol. Biol. Cell. 21:3915–3925. 10.1091/mbc.E09-10-0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K., Knust E., and Skaer H.. 2009. Crumbs stabilises epithelial polarity during tissue remodelling. J. Cell Sci. 122:2604–2612. 10.1242/jcs.047183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K., Whissell G., Franch-Marro X., Batlle E., and Casanova J.. 2011. Specific GATA factors act as conserved inducers of an endodermal-EMT. Dev. Cell. 21:1051–1061. 10.1016/j.devcel.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Chanet S., and Schweisguth F.. 2012. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nat. Cell Biol. 14:467–476. 10.1038/ncb2481 [DOI] [PubMed] [Google Scholar]
- Chanet S., Vodovar N., Mayau V., and Schweisguth F.. 2009. Genome engineering-based analysis of Bearded family genes reveals both functional redundancy and a nonessential function in lateral inhibition in Drosophila. Genetics. 182:1101–1108. 10.1534/genetics.109.105023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C., and Boulianne G.L.. 2007. The NHR1 domain of Neuralized binds Delta and mediates Delta trafficking and Notch signaling. Mol. Biol. Cell. 18:1–13. 10.1091/mbc.E06-08-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson F., Couturier L., Rouault H., Mazouni K., and Schweisguth F.. 2017. Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila. Science. 10.1126/science.aai7407 [DOI] [PubMed] [Google Scholar]
- Couturier L., Vodovar N., and Schweisguth F.. 2012. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14:131–139. 10.1038/ncb2419 [DOI] [PubMed] [Google Scholar]
- Couturier L., Mazouni K., and Schweisguth F.. 2013. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr. Biol. 23:588–593. 10.1016/j.cub.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Daskalaki A., Shalaby N.A., Kux K., Tsoumpekos G., Tsibidis G.D., Muskavitch M.A., and Delidakis C.. 2011. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J. Cell Biol. 195:1017–1031. 10.1083/jcb.201105166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre G.A., Lai E.C., and Kintner C.. 2001. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell. 1:795–806. 10.1016/S1534-5807(01)00091-0 [DOI] [PubMed] [Google Scholar]
- del Alamo D., and Mlodzik M.. 2006. Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 fates in the Drosophila eye. Dev. Cell. 11:887–894. 10.1016/j.devcel.2006.09.016 [DOI] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., and Wieschaus E.. 2006. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell. 10:257–264. 10.1016/j.devcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Flores-Benitez D., and Knust E.. 2015. Crumbs is an essential regulator of cytoskeletal dynamics and cell-cell adhesion during dorsal closure in Drosophila. eLife. 4:e07938 10.7554/eLife.07398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J.R., and Posakony J.W.. 2009. Both inhibition and activation of Notch signaling rely on a conserved Neuralized-binding motif in Bearded proteins and the Notch ligand Delta. Dev. Biol. 333:373–385. 10.1016/j.ydbio.2009.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M., Martín-Ruíz I., Jiménez S., Pirone L., Barrio R., and Sutherland J.D.. 2011. Generation of stable Drosophila cell lines using multicistronic vectors. Sci. Rep. 1:75 10.1038/srep00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M., and O’Connor-Giles K.M.. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 196:961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawe F., Wodarz A., Lee B., Knust E., and Skaer H.. 1996. The Drosophila genes crumbs and stardust are involved in the biogenesis of adherens junctions. Development. 122:951–959. [DOI] [PubMed] [Google Scholar]
- Gupta D., Beaufils S., Vie V., Paboeuf G., Broadhurst B., Schweisguth F., Blundell T.L., and Bolanos-Garcia V.M.. 2013. Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized. Advances in Enzyme Research. 1:61–75. 10.4236/aer.2013.13007 [DOI] [Google Scholar]
- He F., Saito K., Kobayashi N., Harada T., Watanabe S., Kigawa T., Güntert P., Ohara O., Tanaka A., Unzai S., et al. 2009. Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway. J. Mol. Biol. 393:478–495. 10.1016/j.jmb.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Hong Y., Stronach B., Perrimon N., Jan L.Y., and Jan Y.N.. 2001. Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature. 414:634–638. 10.1038/414634a [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S., and Bilder D.. 2008. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet. 4:e8 10.1371/journal.pgen.0040008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch V., Seher T., Fernandez-Ballester G.J., Serrano L., and Leptin M.. 2007. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 315:384–386. 10.1126/science.1134833 [DOI] [PubMed] [Google Scholar]
- Kunwar P.S., Sano H., Renault A.D., Barbosa V., Fuse N., and Lehmann R.. 2008. Tre1 GPCR initiates germ cell transepithelial migration by regulating Drosophila melanogaster E-cadherin. J. Cell Biol. 183:157–168. 10.1083/jcb.200807049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C., Bodner R., Kavaler J., Freschi G., and Posakony J.W.. 2000. Antagonism of notch signaling activity by members of a novel protein family encoded by the bearded and enhancer of split gene complexes. Development. 127:291–306. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Deblandre G.A., Kintner C., and Rubin G.M.. 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 1:783–794. 10.1016/S1534-5807(01)00092-2 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., and Schweisguth F.. 2003. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell. 5:139–148. 10.1016/S1534-5807(03)00187-4 [DOI] [PubMed] [Google Scholar]
- Li M.Z., and Elledge S.J.. 2012. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852:51–59. 10.1007/978-1-61779-564-0_5 [DOI] [PubMed] [Google Scholar]
- Li Y., Wei Z., Yan Y., Wan Q., Du Q., and Zhang M.. 2014. Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc. Natl. Acad. Sci. USA. 111:17444–17449. 10.1073/pnas.1416515111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Currinn H., Pocha S.M., Rothnie A., Wassmer T., and Knust E.. 2015. AP-2-complex-mediated endocytosis of Drosophila Crumbs regulates polarity by antagonizing Stardust. J. Cell Sci. 128:4538–4549. 10.1242/jcs.174573 [DOI] [PubMed] [Google Scholar]
- Matsuda M., Rand K., Palardy G., Shimizu N., Ikeda H., Dalle Nogare D., Itoh M., and Chitnis A.B.. 2016. Epb41l5 competes with Delta as a substrate for Mib1 to coordinate specification and differentiation of neurons. Development. 143:3085–3096. 10.1242/dev.138743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médina E., Williams J., Klipfell E., Zarnescu D., Thomas G., and Le Bivic A.. 2002. Crumbs interacts with moesin and βHeavy-spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158:941–951. 10.1083/jcb.200203080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E., Pitsouli C., Klueg K.M., Muskavitch M.A., Moschonas N.K., and Delidakis C.. 2001. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell. 1:807–816. 10.1016/S1534-5807(01)00093-4 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E., Anezaki M., and Skoulakis E.M.. 2008. Neuralized is expressed in the α/β lobes of adult Drosophila mushroom bodies and facilitates olfactory long-term memory formation. Proc. Natl. Acad. Sci. USA. 105:14674–14679. 10.1073/pnas.0801605105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E., Trifilieff P., Chevaleyre V., Fioriti L., Zairis S., Pagano A., Malleret G., and Kandel E.R.. 2011. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 147:1369–1383. 10.1016/j.cell.2011.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénalva C., and Mirouse V.. 2012. Tissue-specific function of Patj in regulating the Crumbs complex and epithelial polarity. Development. 139:4549–4554. 10.1242/dev.085449 [DOI] [PubMed] [Google Scholar]
- Pichaud F. 2014. Transcriptional regulation of tissue organization and cell morphogenesis: the fly retina as a case study. Dev. Biol. 385:168–178. 10.1016/j.ydbio.2013.09.031 [DOI] [PubMed] [Google Scholar]
- Pocha S.M., Wassmer T., Niehage C., Hoflack B., and Knust E.. 2011. Retromer controls epithelial cell polarity by trafficking the apical determinant Crumbs. Curr. Biol. 21:1111–1117. 10.1016/j.cub.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Richardson E.C., and Pichaud F.. 2010. Crumbs is required to achieve proper organ size control during Drosophila head development. Development. 137:641–650. 10.1242/dev.041913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Fraticelli A.E., Bagwell J., Bosch-Fortea M., Boncompain G., Reglero-Real N., García-León M.J., Andrés G., Toribio M.L., Alonso M.A., Millán J., et al. 2015. Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat. Cell Biol. 17:241–250. 10.1038/ncb3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. 2012. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Dev. Cell. 23:939–953. 10.1016/j.devcel.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottenfeld-Roames J., Rosa J.B., and Ghabrial A.S.. 2014. Seamless tube shape is constrained by endocytosis-dependent regulation of active Moesin. Curr. Biol. 24:1756–1764. 10.1016/j.cub.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J.R., and Lehmann R.. 2012. Drosophila primordial germ cell migration requires epithelial remodeling of the endoderm. Development. 139:2101–2106. 10.1242/dev.078949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Nagy-Zsvér-Vadas Z., and Krahn M.P.. 2012. Drosophila PATJ supports adherens junction stability by modulating Myosin light chain activity. J. Cell Biol. 199:685–698 (published correction appears in J. Cell Biol. 2013. 200:853). 10.1083/jcb.201206064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard K.M., and Fehon R.G.. 2015. The transmembrane protein Crumbs displays complex dynamics during follicular morphogenesis and is regulated competitively by Moesin and aPKC. Development. 142:1869–1878. 10.1242/dev.115329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., and Ahringer J.. 2010. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 141:757–774. 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Tepass U. 1996. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev. Biol. 177:217–225. 10.1006/dbio.1996.0157 [DOI] [PubMed] [Google Scholar]
- Tepass U., and Hartenstein V.. 1995. Neurogenic and proneural genes control cell fate specification in the Drosophila endoderm. Development. 121:393–405. [DOI] [PubMed] [Google Scholar]
- Tepass U., Theres C., and Knust E.. 1990. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 61:787–799. 10.1016/0092-8674(90)90189-L [DOI] [PubMed] [Google Scholar]
- Thompson B.J., Pichaud F., and Röper K.. 2013. Sticking together the Crumbs - an unexpected function for an old friend. Nat. Rev. Mol. Cell Biol. 14:307–314. 10.1038/nrm3568 [DOI] [PubMed] [Google Scholar]
- Venken K.J., He Y., Hoskins R.A., and Bellen H.J.. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 314:1747–1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Venken K.J., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P., et al. 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 6:431–434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichas A., Laurie M.T., and Zallen J.A.. 2015. The Ski2-family helicase Obelus regulates Crumbs alternative splicing and cell polarity. J. Cell Biol. 211:1011–1024. 10.1083/jcb.201504083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G., and Fischer J.A.. 2011. Notch ligand ubiquitylation: what is it good for? Dev. Cell. 21:134–144. 10.1016/j.devcel.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T., and Ready D.F.. 1991. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 113:841–850. [DOI] [PubMed] [Google Scholar]
- Yeh E., Dermer M., Commisso C., Zhou L., McGlade C.J., and Boulianne G.L.. 2001. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr. Biol. 11:1675–1679. 10.1016/S0960-9822(01)00527-9 [DOI] [PubMed] [Google Scholar]
- Zhang H., and Macara I.G.. 2008. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev. Cell. 14:216–226. 10.1016/j.devcel.2007.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., and Hong Y.. 2012. Drosophila Patj plays a supporting role in apical-basal polarity but is essential for viability. Development. 139:2891–2896. 10.1242/dev.083162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.