Supplemental Digital Content is available in the text
Keywords: EPIYA, gastric diseases, Helicobacter pylori
Abstract
Background:
Increasingly, studies have focused on the relationship between Helicobacter pylori (H pylori) cytotoxin associated gene A protein (CagA) Glu-Pro-Ile-Tyr-Ala (EPIYA)-D motifs or multiple EPIYA-C phosphorylation sites and peptic ulcer disease (PUD) or gastric cancer (GC) risk. However, the conclusions have been inconsistent. The aim of this meta-analysis was to evaluate whether 1 CagA EPIYA-D motif or multiple EPIYA-C phosphorylation sites were associated with PUD or GC risk.
Materials and methods:
A literature search was performed in PubMed, Web of Science, Wanfang Data, Excerpt Medica Database, and the Chinese National Knowledge Infrastructure database to identify eligible research. We analyzed the odds ratios (OR) and 95% confidence intervals (CI) to assess the strength of association.
Results:
Compared with 1 EPIYA-C motif in Asian populations, 1 EPIYA-D site was associated with an increased GC risk (OR=1.91, 95% CI=1.19–3.07, P = .008). However, 1 EPIYA-D motif was not significantly associated with PUD (OR = 0.90, 95% CI = 0.46–1.76, P = .764), gastric ulcer (GU) (OR = 0.85, 95% CI = 0.27–2.63, P = .771), or duodenal ulcer (DU) (OR = 0.89, 95% CI = 0.25–3.16, P = .859) risk. Compared with no more than 1 EPIYA-C motif, multiple motifs were associated with increased PUD (OR = 2.33, 95% CI = 1.29–4.20, P = .005) and DU (OR = 2.32, 95% CI = 1.08–5.00, P = .031) risk in Asia and GC risk in the United States and Europe (OR = 3.28, 95% CI = 2.32–4.64, P < .001). Multiple EPIYA-C sites were not associated with GU risk (OR = 4.54, 95% CI = 0.95–21.83, P = .059). There was no publication bias identified in these comparisons.
Conclusions:
In Asia, 1 EPIYA-D motif was significantly associated with increased GC risk. Multiple EPIYA-C motifs were associated with increased PUD and DU risk, particularly in Asia. In the United States and Europe, multiple EPIYA-C motifs were associated with increased GC risk. Therefore, detection of polymorphic CagA EPIYA motifs may improve clinical prediction of disease risk.
1. Introduction
Helicobacter pylori (H pylori) is a Gram-negative bacteria that colonizes the human gastric mucosa of different races and from different regions. More than half of the world's population is infected.[1] Persistent H pylori infection in the stomach may cause chronic gastritis, peptic ulcer disease (PUD) and gastric cancer (GC).[2,3] However, the clinical outcomes following infection vary in severity in different populations. This is likely to be a combined result of host genetic susceptibility, environmental factors and differing H pylori pathogenicity.[4–6]
Many H pylori virulence factors, including CagA, lipopolysaccharide, peptidoglycan, vacuolating cytotoxin, and gamma glutamyl transpeptidase, have been associated with gastric disease. Among them, CagA is a virulence factor encoded by the terminal cagA gene of the type IV secretion system.[7,8] CagA can be translocated into the host cells by this secretion system, where the C-terminal repeated EPIYA tyrosine can be rapidly phosphorylated by Src Family Kinases (SFKs). This may eventually lead to abnormal gastric epithelial cell proliferation, cytoskeletal abnormalities[9–15] or even activation of cellular oncogenes.[16–21] However, not all studies have indicated that the CagA protein performs the aforementioned functions. Polymorphisms in the EPIYA sequence determine differences in CagA protein function. Based on the EPIYA motifs, H pylori was subcategorized as Western or East Asian strains. Western H pylori strains contain EPIYA-A, EPIYA-B, and EPIYA-C segments, and the EPIYA-C site is often repeated. The common EPIYA polymorphic types are primarily ABC, ABCC, and ABCCC. In contrast, East Asian H pylori strains contain EPIYA-A, EPIYA-B, and EPIYA-D motifs. The most common EPIYA polymorphic type is ABD.[22,23] It has been previously demonstrated that phosphorylation of the CagA protein EPIYA motif is closely associated with the existence and number of C or D, but not A or B, sites.
EPIYA-C or -D site phosphorylation plays an important role in activating downstream molecular events. For example, Vianna et al[24] analyzed the association between H pylori strains with multiple EPIYA-C sites and PUD and compared that to H pylori strains with no more than 1 EPIYA-C motif. The authors observed that multiple C sites were associated with increased PUD risk. In contrast, Torres et al[25] determined that multiple EPIYA-C sites did not increase PUD risk. Ferreira et al[26] analyzed the relationship between H pylori strains carrying multiple EPIYA-C sites and GC and concluded that multiple C sites were associated with increased GC risk. However, Chomvarin[27] determined that multiple EPIYA-C sites were not associated with GC risk. Similarly, several studies have contradictory conclusions regarding the relationship between EPIYA-D and GC. For example, Li et al.[28] found that H pylori strains carrying 1 EPIYA-D site are associated with increased GC risk. However, Xia et al.[29] determined that 1 D site did not increase the risk of GC. Taken together, the conclusions regarding the association of EPIYA-C or -D phosphorylation sites with gastric diseases are inconsistent. Therefore, to further clarify the association between CagA EPIYA polymorphisms and gastric disease, we proposed a systematic review to explore the association of the existence and number of EPIYA-C or -D phosphorylation sites with gastric disease. Our meta-analysis will help clarify the diverse clinical outcomes in patients with H pylori and detect H pylori infection based on EPIYA classification. Furthermore, this study will allow for earlier eradication of H pylori carrying risky EPIYA types and ultimately reduce the development of H pylori-associated gastric diseases.
2. Methods
2.1. Ethics statement: N/A
2.1.1. Identification and eligibility of relevant studies
Literature in electronic databases, including PubMed, Embase, Web of Knowledge, Wanfang Data, and CNKI, were systematically searched using the terms, “cytotoxin associated gene A/CagA,” “EPIYA,” and “H pylori.” The corresponding Chinese terms were used when searching Chinese databases. Furthermore, references that were cited in each included study were also searched manually to identify potential additional relevant studies. When the information provided in the article was unclear, we contacted the author for detailed raw data. If data were overlapping, we adopted the most recent and comprehensive research for this meta-analysis. The last search date was March 28, 2016.
2.1.2. Inclusion and exclusion criteria
The inclusion criteria were as follows: studies investigating the association of CagA EPIYA-D or EPIYA-C phosphorylation sites with PUD and GC risk; studies with sufficient raw data to estimate ORs and 95% CIs; and studies with gastritis or functional dyspepsia (FD) as a control group. Exclusion criteria included: reviews or meta-analyses; animal or cytology experiments; duplicate publications; studies not involving PUD or GC; and studies published neither in English nor Chinese.
2.1.3. Data extraction
From the included studies, data, including first author, year of publication, population ethnicity, population age, number of cases and controls, detection methods of CagA EPIYA-D, and EPIYA-C phosphorylation sites and H pylori isolate source, were carefully extracted by 2 authors (Qiuping Li and Yuehua Gong) independently. Inconsistencies were resolved following discussion, and a consensus was reached for all extracted data.
2.1.4. Evaluation of the validity of the included studies
The Newcastle–Ottawa scale (NOS) with 8 items was used to estimate the validity of the included studies.[30] We evaluated the studies on a 9 star scale based on selection (4 stars maximum), comparability (2 stars maximum) and exposure (3 stars maximum). NOS scores of 1–3, 4–6, and 7–9 were considered low, medium, and high quality, respectively (Table S1).
2.2. Statistical analysis
We analyzed the association of 1 CagA EPIYA-D or multiple EPIYA-C phosphorylation sites with PUD and GC risk using Stata software (version 11.0; StataCorp, College Station, TX). Cumulative ORs and the corresponding 95% CIs were used to measure the strength of associations. All P values were 2 sided, and P < .05 was considered statistically significant. Heterogeneity across the studies was assessed using a Q statistic (considered significant heterogeneity if P < .10) and an I-squared (I2) value.[31] When significant heterogeneity was detected, a random-effects model based on the DerSimonian and Laird method[32] was used to perform the meta-analysis. Otherwise, a fixed-effects model using the Mantel–Haenszel method was performed.[33] A sensitivity analysis was performed to explore heterogeneity when significant heterogeneity was indicated. Subgroup analysis was used to explore the effect of geographical region and GU or DU. Moreover, publication bias was evaluated quantitatively using Begg's[34] and Egger's tests.[35] Significant publication bias was indicated if P value < .05.
3. Results
3.1. Characteristics of the included studies
The flow chart of included studies is summarized in Fig. 1. A systematic search through electronic databases yielded 593 citations after duplicate removal. After reviewing the titles, abstracts and full texts, articles that were not relevant to this analysis, animal experiments and reviews or cytology experiments were removed, resulting in the exclusion of 553 records. The remaining 40 full-text articles were further assessed for eligibility. Finally, 23 full-text articles that met the inclusion criteria were included in this meta-analysis.[24–29,36–53] Among these articles, 7 (8 studies) compared 1 EPIYA-C with 1 EPIYA-D site (6 studies: PUD vs gastritis/FD; 3 studies: GU vs gastritis/FD; 3 studies: DU vs gastritis/FD; 7 studies: GC vs gastritis/FD). Additionally, 19 articles (19 studies) compared no more than 1 EPIYA-C with multiple EPIYA-C sites (15 studies: PUD vs gastritis/FD; 5 studies: GU vs gastritis/FD; 9 studies: DU vs gastritis/FD; 14 studies: GC vs gastritis/FD).
Figure 1.
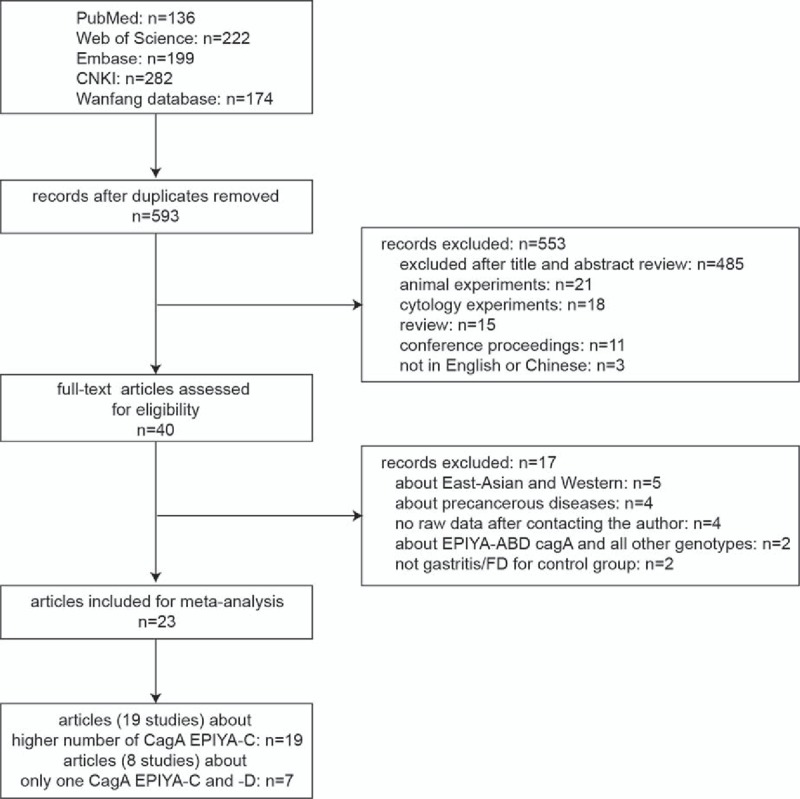
Flow chart of literature search and study selection. DU = duodenal ulcer, FD = functional dyspepsia, GC = gastric cancer, GU = gastric ulcer, PUD = peptic ulcer disease.
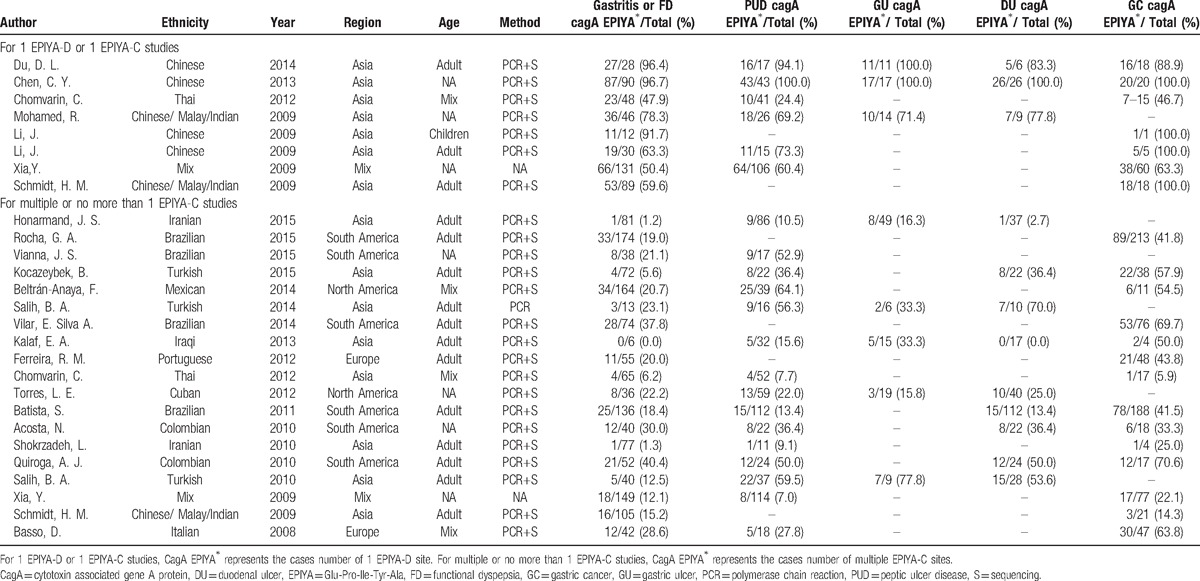
The primary characteristics of all studies included in this meta-analysis are summarized in Table 1. For the meta-analysis comparing 1 EPIYA-D with 1 EPIYA-C site, all the populations were from Asia. EPIYA motif types were all detected by PCR-based sequencing. Only 1 article[28] divided the population into adults and children. The detailed analysis of 1 EPIYA-D and 1 EPIYA-C motif and the total number of subjects is displayed in Table 1. For the meta-analysis comparing multiple EPIYA-C sites with no more than 1 site, data were compiled from 4 geographical regions, including Asia, South America, North America, and Europe. The number of EPIYA-C motifs was detected by PCR-based sequencing, except for the study by Salih et al[36] that used PCR. The details of this analysis and the total number of subjects are also displayed in Table 1. Analysis of 1 EPIYA-D and 1 EPIYA-C motif revealed that the EPIYA polymorphisms included D, ABD, and ABABD types when 1 D site was present and AC, BC, ABC, AABC, and ABBC types when 1 C site was present. Comparison of multiple EPIYA-C sites with no more than 1 EPIYA-C site revealed that the EPIYA polymorphisms included ABCC, ABCCC, ABBCCC, and ABCCCCC types when multiple C sites were present and AB, AC, BC, ABC, AABC, and ABBC types when no more than 1 C site was present.
Table 1.
Characteristics of selected studies for cagA EPIYA-D or EPIYA-C phosphorylation sites analysis.
3.2. Association between 1 EPIYA-D or EPIYA-C site and PUD and GC
First, we analyzed the relationship between 1 EPIYA-D or EPIYA-C phosphorylation site and PUD. With gastritis and FD as the control, compared with 1 EPIYA-C site, 1 EPIYA-D site was not associated with increased PUD risk (OR = 0.90, 95% CI: 0.46–1.76, P = .764; Table 2 and Fig. 2). However, there was heterogeneity in these studies (I2 = 46.80%, P = .094). To further investigate the sources of heterogeneity, we conducted a sensitivity analysis (Fig. S1). After removing the most obvious outlying study by Chomvarin[27] (OR = 0.35), no significant heterogeneity remained (I2 = 0.00%, P = .586). In the remaining studies, 1 EPIYA-D site was still not associated with increased PUD risk (OR = 1.32, 95% CI: 0.86–2.03, P = .201). Similarly, with gastritis and FD as controls, 1 EPIYA-D motif was not associated with increased risk of GU (OR = 0.85, 95% CI: 0.27–2.63, P = .771; Table 2) or DU (OR = 0.89, 95% CI: 0.25–3.16, P = .859; Table 2).
Table 2.
Meta-analysis results for association between cagA EPIYA-D or EPIYA-C phosphorylation sites and PUD and GC.
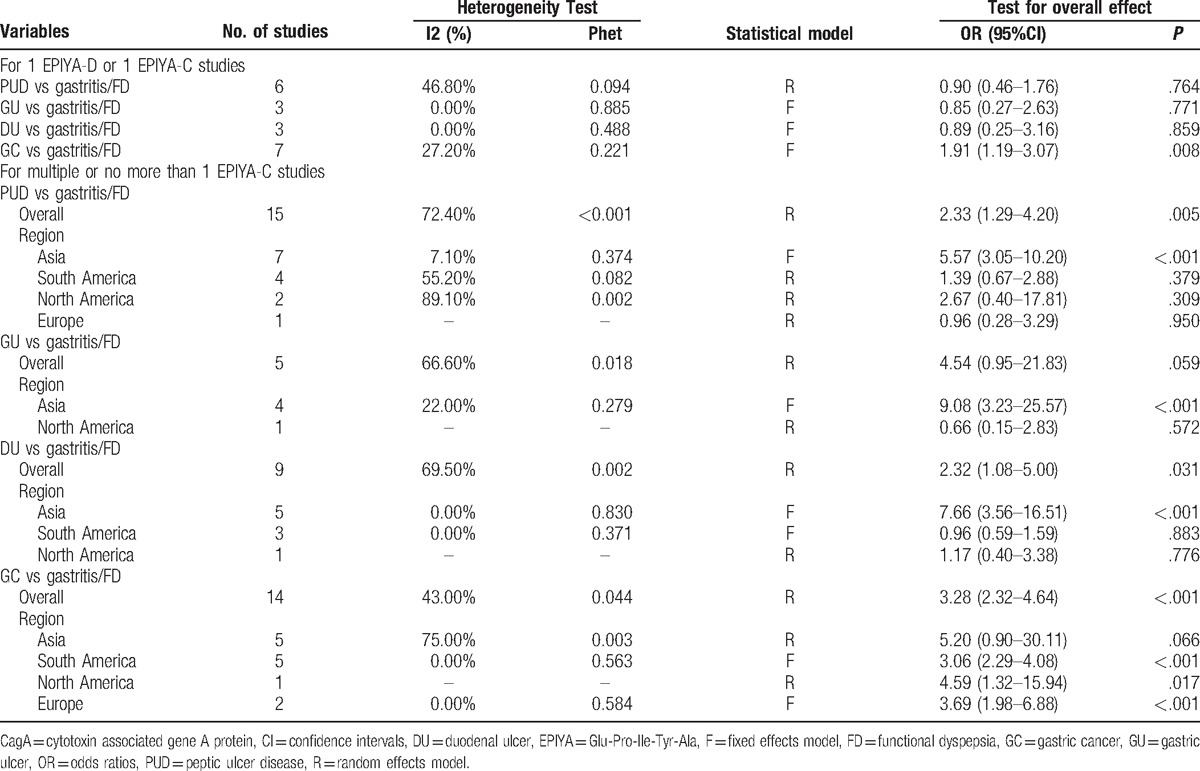
Figure 2.
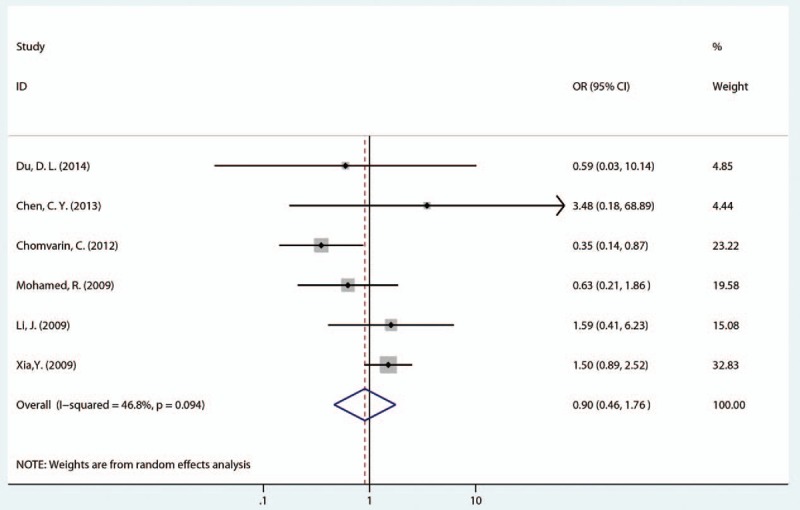
Forest plot of the association between 1 EPIYA-D phosphorylation site and PUD risk. EPIYA = Glu-Pro-Ile-Tyr-Ala, PUD = peptic ulcer disease.
Next, we evaluated the relationship between 1 EPIYA-D or EPIYA-C phosphorylation site and GC. Compared with 1 EPIYA-C site, 1 EPIYA-D site was significantly associated with increased GC risk (controls: gastritis and FD; OR = 1.91, 95% CI: 1.19–3.07, P = .008; Table 2 and Fig. 3). There was no significant heterogeneity in these studies (I2 = 27.20%, P = .221).
Figure 3.
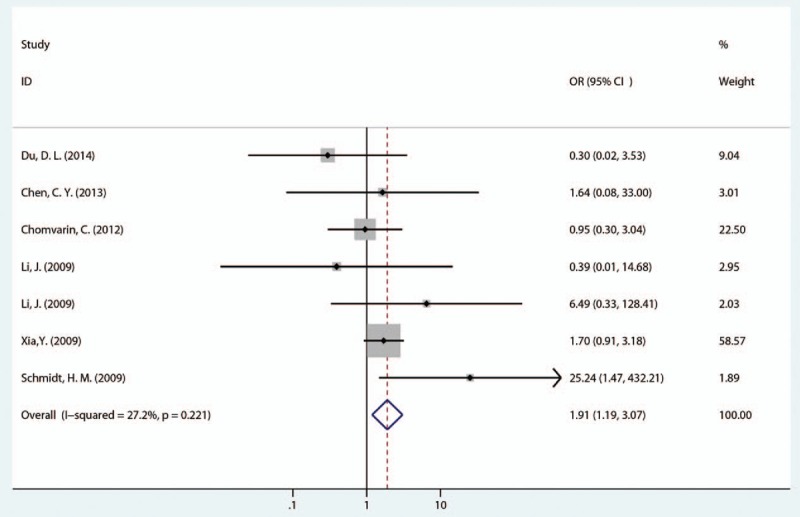
Forest plot of the association between 1 EPIYA-D phosphorylation site and GC risk. EPIYA = Glu-Pro-Ile-Tyr-Ala, GC = gastric cancer.
3.3. Association between multiple EPIYA-C sites or no more than 1 EPIYA-C site and PUD and GC
First, we evaluated the association between multiple EPIYA-C phosphorylation sites or no more than 1 EPIYA-C phosphorylation site and PUD. Multiple EPIYA-C motifs were significantly associated with increased PUD risk (control: gastritis and FD; OR = 2.33, 95% CI: 1.29–4.20, P = .005; Table 2 and Fig. 4). However, significant heterogeneity existed between these studies (I2 = 72.40%, P < .001). Thus, we conducted a sensitivity analysis to further identify the sources of heterogeneity (Fig. S2). After omitting the most obvious outlying study by Xia et al[29] (OR = 0.55), heterogeneity still remained (I2 = 68.80%, P < .001). In the remaining studies, the conclusion was still significant (OR = 2.64, 95% CI: 1.46–4.76, P = .001). Subgroup differences and study design or quality could also not explain the heterogeneity source. In the subgroup analysis according to geographical region, multiple EPIYA-C phosphorylation sites were significantly associated with increased PUD risk in Asia (OR = 5.57, 95% CI: 3.05–10.20, P < .001; Table 2) but not in South America, North America, or Europe (South America: OR = 1.39, 95% CI: 0.67–2.88, P = .379; North America: OR = 2.67, 95% CI: 0.40–17.81, P = .309; Europe: OR = 0.96, 95% CI: 0.28–3.29, P = .95; Table 2). Compared with no more than 1 EPIYA-C site, multiple EPIYA-C phosphorylation sites were not associated with GU risk (OR = 4.54, 95% CI: 0.95–21.83, P = .059; Table 2). Furthermore, the subgroup analysis revealed that multiple EPIYA-C phosphorylation sites were significantly associated with increased GU risk in Asian (OR = 9.08, 95% CI: 3.23–25.57, P < .001; Table 2) but not North American populations (OR = 0.66, 95% CI: 0.15–2.83, P = .572; Table 2). Multiple EPIYA-C phosphorylation sites were significantly associated with DU risk (OR = 2.32, 95% CI: 1.08–5.00, P = .031; Table 2). Subgroup analysis revealed that multiple EPIYA-C phosphorylation sites were associated with increased DU risk in Asia (OR = 7.66, 95% CI: 3.56–16.51, P < .001; Table 2) but not in South America or North America (South America: OR = 0.96, 95% CI: 0.59–1.59, P = .883; North America: OR = 1.17, 95% CI: 0.40–3.38, P = .776; Table 2).
Figure 4.
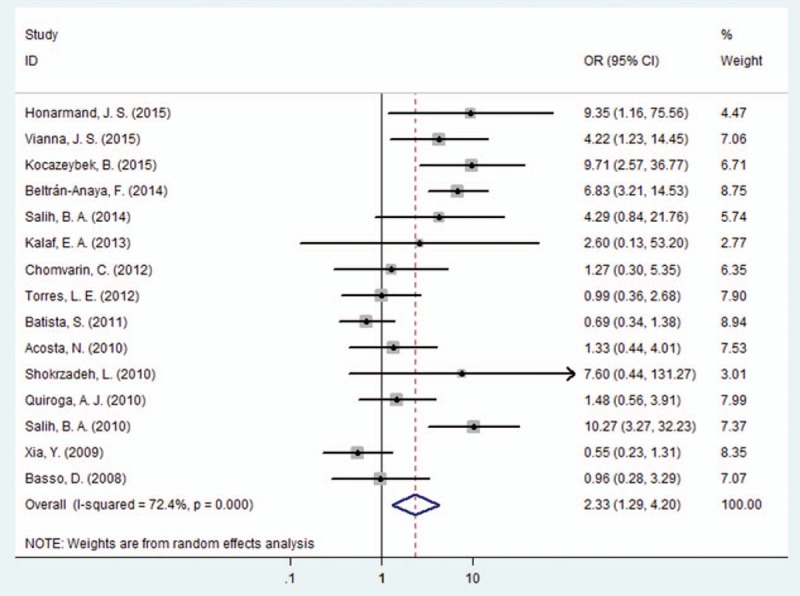
Forest plot of the association between multiple EPIYA-C phosphorylation sites and PUD risk. EPIYA = Glu-Pro-Ile-Tyr-Ala, PUD = peptic ulcer disease.
Next, we evaluated the correlation between EPIYA-C phosphorylation sites and GC. Compared with no more than 1 EPIYA-C site, multiple C phosphorylation sites were significantly associated with increased GC risk (controls: gastritis and FD; OR = 3.28, 95% CI: 2.32–4.64, P < .001; Table 2 and Fig. 5). Because there was significant heterogeneity among these studies (I2 = 43.00%, P = .044), a sensitivity analysis was performed (Fig. S3). After omission of the most obvious outlying study by Kocazeybe et al[37] (OR = 23.38), the heterogeneity was no longer significant (I2 = 0.00%, P = .462). However, multiple EPIYA-C phosphorylation sites were still associated with increased GC risk in the remaining studies (OR = 2.95, 95% CI: 2.34–3.72, P < .001). A subgroup analysis of different regions was performed. In the Asian subgroup, we observed no significant association of multiple EPIYA-C phosphorylation sites with GC (controls: gastritis and FD; OR = 5.20, 95% CI: 0.90–30.11, P = .066; Table 2). However, there was a statistically significant increased GC risk identified in South American, North American and European subgroups (South America: OR = 3.06, 95% CI: 2.29–4.08, P < .001; North America: OR = 4.59, 95% CI: 1.32–15.94, P = .017; Europe: OR = 3.69, 95% CI: 1.98–6.88, P < .001, Table 2).
Figure 5.
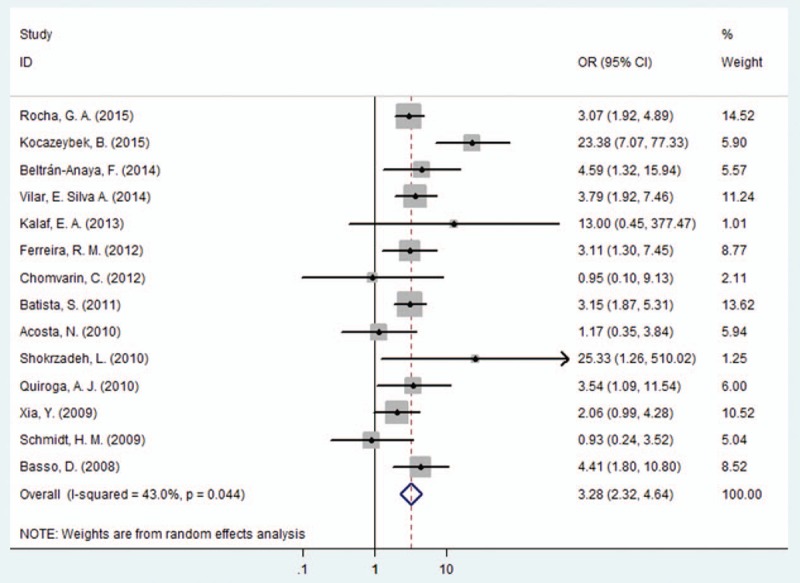
Forest plot of the association between multiple EPIYA-C phosphorylation sites and GC risk. EPIYA = Glu-Pro-Ile-Tyr-Ala, GC = gastric cancer.
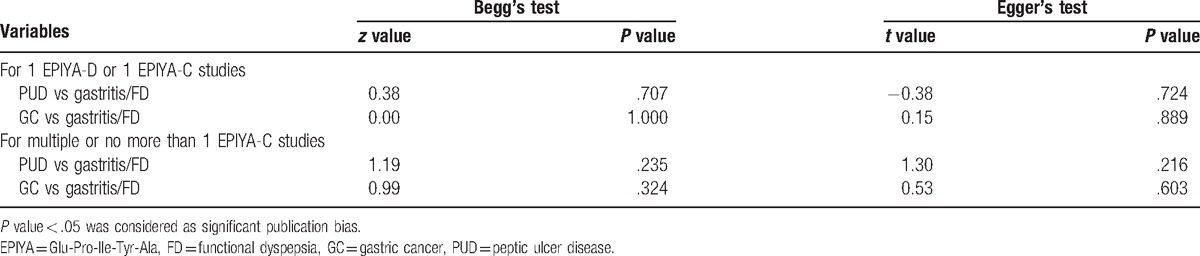
3.4. Publication bias
We performed Begg's and Egger's tests to quantitatively evaluate the publication bias of the association of 1 CagA EPIYA-D or multiple CagA EPIYA-C phosphorylation sites with PUD and GC risk. Publication bias observed in this meta-analysis was not significant (Table 3, Figs. S4 and S5).
Table 3.
Publication bias.
4. Discussion
The type and number of CagA C-terminal EPIYA motifs impact protein size, function and polymorphisms, resulting in bacterial virulence (pathogenicity) differences.[21,54] Therefore, the correlation of CagA EPIYA motifs with clinical results has become of great interest in H pylori research. However, research investigating whether H pylori CagA EPIYA-D or -C phosphorylation sites are related to increased PUD and GC risk has been inconsistent. In this meta-analysis, we determined that 1 EPIYA-D site, compared with 1 EPIYA-C segment, was significantly associated with an increased GC risk in Asia. Moreover, we observed that, compared with no more than 1EPIYA-C site, multiple EPIYA-C sites were associated with increased PUD risk (controls: gastritis and FD). Furthermore, in Asia, multiple EPIYA-C sites were not only associated with increased PUD risk but also with increased risk of GU and DU. Additionally, multiple EPIYA-C segments were also associated with increased GC risk, particularly in South America, North America, and Europe. To the best of our knowledge, this is the first meta-analysis investigating the relationship between EPIYA-D or -C phosphorylation sites and PUD and GC.
For the comparison between 1 EPIYA-D and 1 EPIYA-C site, the entire study population was from Asia. For the pooled analysis, 1 EPIYA-D segment was not significantly associated with PUD risk (controls: gastritis and FD). Similarly, 1 EPIYA-D segment was not related to an increased risk of GU or DU. However, we did identify a significant association between 1 EPIYA-D segment and increased GC risk. These results suggest that the presence of only 1 EPIYA-D segment in Asian populations was closely associated with increased risk of GC but not PUD. Phosphorylated CagA binds the Src homology 2 (SH2) domain of Src homology 2 phosphatase (SHP-2), altering SHP-2 conformation and interfering with host cell signaling pathways, ultimately resulting in epithelial structure disorder.[55,56] In addition, the phosphorylated CagA and SHP-2 complex can active the ERK signaling pathway in a Ras-dependent or -independent manner. De Souza et al.[57] demonstrated that SHP-2 domains recognize phosphopeptide motifs composed of phosphotyrosine (pY) followed by several CagA C-terminal residues. Amino acid differences at the C-terminal residues within the CagA EPIYA-D and -C segments can result in different SHP-2 affinities. As a result, SHP-2 binds EPIYA-D more tightly than EPIYA-C. Furthermore, Higashi et al[23] determined in vitro that the East Asian EPIYA-D strain bound SHP-2 more tightly, compared with the Western EPIYA-C strain, and induced epithelial cell morphology transformation. This may explain why the EPIYA-D site has a greater pathogenicity than the EPIYA-C site.
The studies comparing multiple EPIYA-C sites with no more than 1 EPIYA-C site included populations from Asia, North and South America, and Europe. For the pooled analysis of the EPIYA-C phosphorylation site, we determined that multiple EPIYA-C motifs were associated with increased PUD risk. However, significant heterogeneity among studies was identified, and we, therefore, conducted further sensitivity analyses. Though the most obvious outliers were eliminated, heterogeneity was still significant, and subgroup analysis could not explain the heterogeneity source. The geographical stratification analysis demonstrated that, in Asia, multiple EPIYA-C phosphorylation sites were associated with increased PUD risk. However, this association was not identified in North America, South America, or Europe. Additional disease and regional subgroup analyses revealed that multiple EPIYA-C phosphorylation sites were significantly associated with increased GU and DU risk in Asia. The pooled estimate also demonstrated that multiple EPIYA-C phosphorylation sites were associated with increased GC risk in Europe and America. Some mechanistic studies have partially explained the association between multiple EPIYA-C phosphorylation sites and PUD and GC. Higashi et al[23] found that when strains carrying multiple EPIYA-C sites were co-cultured with human gastric adenocarcinoma cell (AGS), each EPIYA-C site could be equally phosphorylated. Thus, phosphorylation was associated with the number of EPIYA-C sites. Naito et al[58] also confirmed that CagA with multiple EPIYA-C or EPIYA-D motifs bound SHP-2 more robustly than CagA with single EPIYA-C or EPIYA-D motif, and the former had a stronger ability to activate SHP-2. Higashi et al[23] suggested that the ability of CagA to bind SHP-2 was dependent on the number and sequence of tyrosine phosphorylation sites. Additionally, EPIYA-C sites increased CagA protein phosphorylation, which significantly increased CagA-SHP-2 complex formation and enhanced the ability of CagA to induce cellular phenotypic changes. Therefore, the number of CagA EPIYA-C sites is the key factor affecting the ability of CagA to interfere with intracellular signal transduction. In addition, CagA with EPIYA-ABCCC and CagA with EPIYA-D have the same carcinogenic potential.[18] Hence, strains with multiple EPIYA-C sites are associated with PUD and GC risk. It is possible that regional differences in the association between EPIYA-C and PUD and GC may be attributed to fewer studies investigating multiple EPIYA-C sites and PUD in Europe and America. Thus, it is necessary to further expand the sample size to confirm the above conclusions. Alternatively, these findings may be due to a different distribution of Eastern and Western strains. Most Eastern strains carry EPIYA-D, while Western strains tend to harbor the EPIYA-C site. Therefore, multiple EPIYA-C motifs were more relevant to GC risk in Europe and America but not in Asia.
This meta-analysis had some limitations. First, we only included studies written in English or Chinese. Thus, selection bias might exist. Second, the sample size of the studies included was relatively small, resulting in an even smaller sample size for the stratified analyses. This may be related to the stringent requirements for H pylori culture technology or the high cost of EPIYA sequencing; both limit the acquisition of large sample sizes. Third, in Asian populations, because there were fewer articles investigating the relationship between multiple EPIYA-D or single EPIYA-C motifs (or multiple EPIYA-C motifs) and PUD and GC, we only compared the association of a single EPIYA-D or C motif with PUD and GC. We concluded that a single D motif was related to increased GC risk, which still needs further verification. Fourth, there was significant heterogeneity in the analysis between multiple EPIYA-C motifs and PUD. Moreover, subgroup and sensitivity analyses could not explain the source of heterogeneity. Furthermore, the limited number of relevant studies also prevented us from performing meta-regression analysis to further explore the sources of heterogeneity.
5. Conclusions
This meta-analysis demonstrated that a single EPIYA-D phosphorylation site, compared with a single EPIYA-C phosphorylation site, was associated with increased GC risk in Asia. Multiple EPIYA-C phosphorylation sites, compared with no more than 1 EPIYA-C site, are associated with increased PUD, GU and DU risk in Asia and are related to increased GC risk in Europe and America. H pylori carrying a single EPIYA-D motif or multiple EPIYA-C motifs may be a potential marker for predicting PUD or GC risk. In addition, it is necessary to expand the sample size of studies investigating the association between CagA EPIYA polymorphisms and PUD and GC to confirm our meta-analysis conclusions.
Supplementary Material
Footnotes
Abbreviations: CagA = cytotoxin associated gene A protein, CI = confidence intervals, DU = duodenal ulcer, EPIYA = Glu-Pro-Ile-Tyr-Ala, F = fixed effects model, FD = functional dyspepsia, GC = gastric cancer, GU = gastric ulcer, H pylori = Helicobacter pylori, NOS = Newcastle–Ottawa scale, OR = odds ratios, PCR = Polymerase Chain Reaction, PUD = peptic ulcer disease, R = random effects model, S = sequencing.
Funding: This study is supported by grants from the Natural Science Foundation of Liaoning Province (Ref No. 201602822).
Competing Interests: The authors have declared that no competing interests exist.
Author contributions Conceived and designed the experiments: YHG. Performed the experiments: QPL and JWL. Analyzed the data: QP, YHG. Wrote the manuscript: QPL, YHG, YY.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest 2004;113:321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2002;2:28–37. [DOI] [PubMed] [Google Scholar]
- [3].Palli D, Masala G, Del Giudice G, et al. CagA+ Helicobacter pylori infection and gastric cancer risk in the EPIC-EURGAST study. Int J Cancer 2007;120:859–67. [DOI] [PubMed] [Google Scholar]
- [4].El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 2000;404:398–402. [DOI] [PubMed] [Google Scholar]
- [5].Machado JC, Figueiredo C, Canedo P, et al. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 2003;125:364–71. [DOI] [PubMed] [Google Scholar]
- [6].Figueiredo C, Machado JC, Pharoah P, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Nat Cancer Inst 2002;94:1680–7. [DOI] [PubMed] [Google Scholar]
- [7].Censini S, Lange C, Xiang Z, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA 1996;93:14648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Odenbreit S, Puls J, Sedlmaier B, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science (New York, NY) 2000;287:1497–500. [DOI] [PubMed] [Google Scholar]
- [9].Segal ED, Cha J, Lo J, et al. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA 1999;96:14559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA 2000;97:1263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mimuro H, Suzuki T, Tanaka J, et al. Grb2 is a key mediator of helicobacter pylori CagA protein activities. Mol Cell 2002;10:745–55. [DOI] [PubMed] [Google Scholar]
- [12].Hirata Y, Maeda S, Mitsuno Y, et al. Helicobacter pylori CagA protein activates serum response element-driven transcription independently of tyrosine phosphorylation. Gastroenterology 2002;123:1962–71. [DOI] [PubMed] [Google Scholar]
- [13].Hatakeyama M. Deregulation of SHP-2 tyrosine phosphatase by the Helicobacter pylori virulence factor CagA. Keio J Med 2002;51suppl 2:26–32. [DOI] [PubMed] [Google Scholar]
- [14].Bagnoli F, Buti L, Tompkins L, et al. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci USA 2005;102:16339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kurashima Y, Murata-Kamiya N, Kikuchi K, et al. Deregulation of beta-catenin signal by Helicobacter pylori CagA requires the CagA-multimerization sequence. Int J Cancer 2008;122:823–31. [DOI] [PubMed] [Google Scholar]
- [16].Murata-Kamiya N. Pathophysiological functions of the CagA oncoprotein during infection by Helicobacter pylori. Microbes Infect 2011;13:799–807. [DOI] [PubMed] [Google Scholar]
- [17].Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: the master key hypothesis. Helicobacter 2010;15:163–76. [DOI] [PubMed] [Google Scholar]
- [18].Higashi H, Tsutsumi R, Muto S, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science (New York, NY) 2002;295:683–6. [DOI] [PubMed] [Google Scholar]
- [19].Suzuki M, Kiga K, Kersulyte D, et al. Attenuated CagA oncoprotein in Helicobacter pylori from Amerindians in Peruvian Amazon. J Biol Chem 2011;286:29964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yamazaki S, Yamakawa A, Ito Y, et al. The CagA protein of Helicobacter pylori is translocated into epithelial cells and binds to SHP-2 in human gastric mucosa. J Infect Dis 2003;187:334–7. [DOI] [PubMed] [Google Scholar]
- [21].Suzuki M, Mimuro H, Kiga K, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 2009;5:23–34. [DOI] [PubMed] [Google Scholar]
- [22].Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci 2011;102:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Higashi H, Tsutsumi R, Fujita A, et al. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA 2002;99:14428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vianna JS, Ramis IB, Halicki PC, et al. Detection of Helicobacter pylori CagA EPIYA in gastric biopsy specimens and its relation to gastric diseases. Diagn Microbiol Infect Dis 2015;83:89–92. [DOI] [PubMed] [Google Scholar]
- [25].Torres LE, Gonzalez L, Melian K, et al. EPIYA motif patterns among Cuban Helicobacter pylori CagA positive strains. Biomedica 2012;32:23–31. [DOI] [PubMed] [Google Scholar]
- [26].Ferreira RM, Machado JC, Leite M, et al. The number of Helicobacter pylori CagA EPIYA C tyrosine phosphorylation motifs influences the pattern of gastritis and the development of gastric carcinoma. Histopathology 2012;60:992–8. [DOI] [PubMed] [Google Scholar]
- [27].Chomvarin C, Phusri K, Sawadpanich K, et al. Prevalence of cagA EPIYA motifs in Helicobacter pylori among dyspeptic patients in northeast Thailand. Southeast Asian J Trop Med Public Health 2012;43:105–15. [PubMed] [Google Scholar]
- [28].Li J, Ou Z, Wang F, et al. Distinctiveness of the cagA genotype in children and adults with peptic symptoms in South China. Helicobacter 2009;14:248–55. [DOI] [PubMed] [Google Scholar]
- [29].Xia Y, Yamaoka Y, Zhu Q, et al. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS One 2009;4:e7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [31].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [32].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [33].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Nat Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [34].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [35].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical Research ed) 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Salih BA, Guner A, Karademir A, et al. Evaluation of the effect of cagPAI genes of Helicobacter pylori on AGS epithelial cell morphology and IL-8 secretion. Antonie Van Leeuwenhoek 2014;105:179–89. [DOI] [PubMed] [Google Scholar]
- [37].Kocazeybek BS, Caliskan R, Erdamar Cetin S, et al. Patterns of EPIYA motifs among cagA-positive Helicobacter pylori strains: a case-control study in a Turkish population with Eurasian geographical features. J Med Microbiol 2015;64:1117–23. [DOI] [PubMed] [Google Scholar]
- [38].Vilar ESA, Junior MR, Vinagre RM, et al. Evaluation of the pattern of EPIYA motifs in the Helicobacter pylori cagA gene of patients with gastritis and gastric adenocarcinoma from the Brazilian Amazon region. Int J Bacteriol 2014;2014:418063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shokrzadeh L, Baghaei K, Yamaoka Y, et al. Analysis of 3′-end variable region of the cagA gene in Helicobacter pylori isolated from Iranian population. J Gastroenterol Hepatol 2010;25:172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schmidt HM, Goh KL, Fock KM, et al. Distinct cagA EPIYA motifs are associated with ethnic diversity in Malaysia and Singapore. Helicobacter 2009;14:256–63. [DOI] [PubMed] [Google Scholar]
- [41].Salih BA, Bolek BK, Arikan S. DNA sequence analysis of cagA 3′ motifs of Helicobacter pylori strains from patients with peptic ulcer diseases. J Med Microbiol 2010;59(pt 2):144–8. [DOI] [PubMed] [Google Scholar]
- [42].Rocha GA, Rocha AM, Gomes AD, et al. STAT3 polymorphism and Helicobacter pylori CagA strains with higher number of EPIYA-C segments independently increase the risk of gastric cancer. BMC Cancer 2015;15:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Quiroga AJ, Huertas A, Combita AL, et al. Variation in the number of EPIYA-C repeats in CagA protein from Colombian Helicobacter pylori strains and its ability middle to induce hummingbird phenotype in gastric epithelial cells. Biomedica 2010;30:251–8. [PubMed] [Google Scholar]
- [44].Kalaf EA, Al-Khafaji ZM, Yassen NY, et al. Study of the cytoxin-associated gene a (CagA gene) in Helicobacter pylori using gastric biopsies of Iraqi patients. Saudi J Gastroenterol 2013;19:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Honarmand-Jahromy S, Siavoshi F, Malekzadeh R, et al. Multiple repeats of Helicobacter pylori CagA EPIYA-C phosphorylation sites predict risk of gastric ulcer in Iran. Microb Pathog 2015;89:87–92. [DOI] [PubMed] [Google Scholar]
- [46].Beltran-Anaya FO, Poblete TM, Roman-Roman A, et al. The EPIYA-ABCC motif pattern in CagA of Helicobacter pylori is associated with peptic ulcer and gastric cancer in Mexican population. BMC Gastroenterol 2014;14:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Batista SA, Rocha GA, Rocha AM, et al. Higher number of Helicobacter pylori CagA EPIYA C phosphorylation sites increases the risk of gastric cancer, but not duodenal ulcer. BMC Microbiol 2011;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Basso D, Zambon CF, Letley DP, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 2008;135:91–9. [DOI] [PubMed] [Google Scholar]
- [49].Acosta N, Quiroga A, Delgado P, et al. Helicobacter pylori CagA protein polymorphisms and their lack of association with pathogenesis. World J Gastroenterol 2010;16:3936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schmidt HM, Goh KL, Fock KM, et al. Distinct cagA EPIYA motifs are associated with ethnic diversity in Malaysia and Singapore. Helicobacter 2009;14:256–63. [DOI] [PubMed] [Google Scholar]
- [51].Mohamed R, Hanafiah A, Rose IM, et al. Helicobacter pylori cagA gene variants in Malaysians of different ethnicity. Eur J Clin Microbiol Infect Dis 2009;28:865–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen CY, Wang FY, Wan HJ, et al. Amino acid polymorphisms flanking the EPIYA-A motif of Helicobacter pylori CagA C-terminal region is associated with gastric cancer in east China: experience from a single center. J Dig Dis 2013;14:358–65. [DOI] [PubMed] [Google Scholar]
- [53].Du DLS, Sun WJ, Liu H, et al. Analysis of the 3′ region of cytotoxin-associated gene A (cagA) in Helicobacter pylori isolates from Weihai. J Pathogen Biol 2014;9:783–7. [Google Scholar]
- [54].Argent RH, Zhang Y, Atherton JC. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J Clin Microbiol 2005;43:791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Higashi H, Nakaya A, Tsutsumi R, et al. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J Biol Chem 2004;279:17205–16. [DOI] [PubMed] [Google Scholar]
- [56].Schneider N, Krishna U, Romero-Gallo J, et al. Role of Helicobacter pylori CagA molecular variations in induction of host phenotypes with carcinogenic potential. J Infect Dis 2009;199:1218–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].De Souza D, Fabri LJ, Nash A, et al. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry 2002;41:9229–36. [DOI] [PubMed] [Google Scholar]
- [58].Naito M, Yamazaki T, Tsutsumi R, et al. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 2006;130:1181–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


