Lysophosphatidylcholine is associated with neurodegeneration and demyelination. Freeman et al. demonstrate that lysophosphatidylcholine triggers NLRP3- and NLRC4-dependent inflammasome activation, and in a synergistic fashion, NLRP3 and NLRC4 contribute to a cuprizone-induced demyelination model in vivo.
Abstract
Inflammation in the brain accompanies several high-impact neurological diseases including multiple sclerosis (MS), stroke, and Alzheimer’s disease. Neuroinflammation is sterile, as damage-associated molecular patterns rather than microbial pathogens elicit the response. The inflammasome, which leads to caspase-1 activation, is implicated in neuroinflammation. In this study, we reveal that lysophosphatidylcholine (LPC), a molecule associated with neurodegeneration and demyelination, elicits NLRP3 and NLRC4 inflammasome activation in microglia and astrocytes, which are central players in neuroinflammation. LPC-activated inflammasome also requires ASC (apoptotic speck containing protein with a CARD), caspase-1, cathepsin-mediated degradation, calcium mobilization, and potassium efflux but not caspase-11. To study the physiological relevance, Nlrc4−/− and Nlrp3−/− mice are studied in the cuprizone model of neuroinflammation and demyelination. Mice lacking both genes show the most pronounced reduction in astrogliosis and microglial accumulation accompanied by decreased expression of the LPC receptor G2A, whereas MS patient samples show increased G2A. These results reveal that NLRC4 and NLRP3, which normally form distinct inflammasomes, activate an LPC-induced inflammasome and are important in astrogliosis and microgliosis.
Introduction
Neuroinflammation accompanies several diseases with or without an immunological origin, such as multiple sclerosis (MS), stroke, traumatic brain injury, Parkinson’s disease, and Alzheimer’s disease (Wyss-Coray, 2006; Rivest, 2009; Heneka et al., 2010; Kigerl et al., 2014). Such inflammatory response is generally referred to as sterile inflammation because microbial pathogens are not typically involved, but rather, the response is directed by damage-associated inflammatory inducers. Sterile inflammation is linked to a plethora of inflammatory disorders within and outside of the central nervous system (CNS; Rock et al., 2010).
The nucleotide-binding leucine-rich repeat (LRR)–containing (NLR; also known as NOD-like receptors) proteins have emerged as a key family of sensors and regulators responding to pathogen-associated molecular patterns (PAMPs) generated by intracellular pathogen and damage-associated molecular patterns (DAMPs) produced under nonmicrobial inflammatory conditions (Strowig et al., 2012; Broderick et al., 2015; Guo et al., 2015; Broz and Dixit, 2016). There are >20 NLR genes in humans and >30 in mice. NLR genes encode cytoplasmic proteins with a tripartite domain structure consisting of a variable N-terminal effector domain, a central nucleotide-binding domain, and a variable number of C-terminal LRRs. The initial characterization of NLRs showed that many are expressed in cells that contribute to innate immunity such as monocytes, granulocytes, macrophages, and dendritic cells.
A subfamily of NLR proteins mediate the activation of caspase-1, which is referred to as inflammasome activation (Martinon et al., 2002). Multiple NLR proteins have been recognized as mediators of inflammasome activation by the sensing of stimuli (Khare et al., 2012). Some notable examples include NLRP1 (Boyden and Dietrich, 2006), NLRP3 (Kanneganti et al., 2006; Mariathasan et al., 2006; Martinon et al., 2006; Sutterwala et al., 2006), NLRP6 (Anand et al., 2012), NLRP7 (Khare et al., 2012), NLRP12 (Vladimer et al., 2012), NLRC4 (Zhao et al., 2011), NLRC5 (Davis et al., 2011; Triantafilou et al., 2013), and NAIP (NLR family apoptosis inhibitory protein; Kofoed and Vance, 2011; Zhao et al., 2011) or non-NLR proteins (e.g., AIM2; Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Rathinam et al., 2010). Gene mutations in a key family member, NLRP3, lead to several autoinflammatory disorders collectively referred to as the cryopyrin-associated periodic syndromes (Broderick et al., 2015). The association of mutations in inflammasome NLR genes with autoinflammatory diseases underscores an important function of these genes in regulating inflammation in humans.
Among inflammasome NLRs, NLR family CARD (caspase recruitment domain)-containing 4 (NLRC4; initially named Ipaf; Poyet et al., 2001) is well studied and probably better understood in the context of bacterial infection. NLRC4 is a cytosolic sensor of flagellin produced by flagellated pathogens such as Salmonella typhimurium, Legionella pneumophila (Amer et al., 2006), and the type III secretory system (T3SS) from gram-negative pathogens such as S. typhimurium, Burkholderia pseudomallei, Escherichia coli, Shigella flexneri (Suzuki et al., 2007), and Pseudomonas aeruginosa (Sutterwala et al., 2007).
Initial characterization of NLRC4 in human tissues and cell lines demonstrated its direct association with pro–caspase-1 through CARD–CARD interactions (Geddes et al., 2001; Poyet et al., 2001). This interaction causes autocatalytic processing of pro–caspase-1 to caspase-1 (Poyet et al., 2001). Activated caspase-1 can, in turn, cleave >70 substrates, including IL-1β and IL-18 (Shao et al., 2007; Keller et al., 2008). A constitutively active NLRC4 causes autocatalytic processing of pro–caspase-1 leading to caspase-1–dependent apoptosis in transfected cells (Poyet et al., 2001). When an Nlrc4 gene deletion strain was examined, the physiological relevance of this protein showed specificity in caspase-1 activation and IL-1β release caused by S. typhimurium but not by the combination of ATP and LPS (Mariathasan et al., 2004). Because the NLRP3 inflammasome was later found to be activated by ATP and LPS, these results showed that NLRP3 and NLRC4 are activated by different activators. However, NLRC4 was shown to recruit NLRP3 to the inflammasome complex during Salmonella infection (Qu et al., 2016), suggesting that the combined activity is critical to inflammasome activation.
Recent data indicate that the NAIP proteins recognize microbial pathogens and then recruit NLRC4 proteins to the inflammasome. Specifically, NAIP1 and NAIP2 recognize bacterial T3SS rod and needle proteins, whereas NAIP5 and NAIP6 recognize bacterial flagellin. Both sets of NAIP proteins interact with the NLRC4 inflammasome in response to their respective bacterial ligands (Kofoed and Vance, 2011; Zhao et al., 2011; Halff et al., 2012; Rayamajhi et al., 2013; Yang et al., 2013). The use of cryoelectron microscopy has further illuminated how NLRC4 is activated. It was found that one molecule of NAIP2 binding to bacterial PrgJ is sufficient to activate NLRC4, which undergoes a rotation shift, causing polymerization and recruitment of additional NLRC4 molecules into the complex (Hu et al., 2015; Zhang et al., 2015). A similar study of NAIP5/NLRC4 also shows a prominent rotation of the NLRC4 LRR domain upon NAIP5 interaction with flagellin (Diebolder et al., 2015).
Although the role of NLRC4 in bacterial sensing is well studied, the role of NLRC4 in sterile inflammation where the inflammatory source is not microbial in nature remains undefined. In experimental colitis, NLRC4 has been found to provide a protective role, although this finding is not uniformly reported and may be attributed to variable microbiomes because of institutional differences or strain of Nlrc4−/− mice used in the studies (Allen et al., 2010; Hu et al., 2010; Carvalho et al., 2012). Colitis is not considered sterile inflammation because the inflammation associated with colitis is thought to be significantly impacted by the gut microbiome (Elinav et al., 2011; Bauer et al., 2012). More recently, NLRC4 has been found to promote obesity-induced breast cancer, although the DAMPs that activate this pathway were not identified (Kolb et al., 2016).
Inflammation of the CNS is increasingly recognized as a key factor in a plethora of neurological diseases where neuroinflammation is believed to exacerbate demyelinating disease severity. Neuroinflammation may be attributed to lymphocytes and macrophage-myeloid cells associated with the immune system, as well as microglia and astrocytes within the brain parenchyma (Dong and Benveniste, 2001; Carson, 2002). Extensive studies have linked the NLRP3 inflammasome to neurological disorders such as MS and Alzheimer’s disease (Halle et al., 2008; Heneka et al., 2013), whereas AIM2 and NLRP1 have been linked to traumatic brain injury (de Rivero Vaccari et al., 2009; Adamczak et al., 2014). However, NLRC4 has not been extensively studied in these contexts.
In this study, we examine the role of lysophosphatidylcholine (LPC; also known as lysolecithin) as a DAMP relevant to neuroinflammation. LPC, a major component of low-density lipoprotein, is generated by hydrolysis of phosphatidylcholine via phospholipase A2 (PLA2; Schilling et al., 2004) under normal physiological conditions. Normally, LPC is rapidly metabolized or reacetylated; however, under pathological conditions, there is increased activity of PLA2, leading to accumulation of LPC. Such an increase in PLA2 activity is seen in MS, ischemia, epilepsy, Alzheimer’s disease, schizophrenia, and spinal cord injury (Farooqui et al., 2006). LPC is known to cause glial activation resulting in the transcription of proinflammatory cytokines (Sheikh and Nagai, 2011). LPC is also recognized as an inflammatory inducer that activates immune cells, but the underlying mechanism has not been extensively investigated (Kabarowski et al., 2002). With respect to cytokines, LPC is known to activate microglia and lead to IL-1β release in a P2X7R-independent mechanism (Schilling et al., 2004; Stock et al., 2006). Moreover, PLA2 is known to contribute to demyelination and immune cell accumulation in Wallerian degeneration. LPC can enhance demyelination in brain spheroid cultures (Vereyken et al., 2009) and cause demyelination in several animal models (Waxman et al., 1979; Shikishima et al., 1985).
In this study, we find that LPC induces inflammasome activation in a process that is surprisingly dependent on both NLRP3 and NLRC4 through the canonical, but not the noncanonical, inflammasome pathway. We find significant NLRC4 expression during neuroinflammation, especially in astrocytes in mice as well as in human demyelinating disease neurological specimens. The cuprizone mouse model of neuroinflammation and demyelination was used to analyze the roles of these proteins because the neurotoxicant cuprizone leads to robust microglial and astrocyte activation and accumulation in major myelinated nerve tracts, such as the corpus callosum and cerebellar peduncles (Matsushima and Morell, 2001), followed by the death of oligodendrocytes and demyelination (Liu et al., 2010). Disease is easily induced by administering cuprizone through the chow, follows a predictable time course, and generates reproducible pathology. Using this model, we show that NLRC4 and NLRP3 contribute significantly to neuropathology and that deletion of these genes results in reduced neuroinflammation and demyelination.
Results
The inflammasome mediates LPC-induced IL-1β secretion in mouse BMDMs
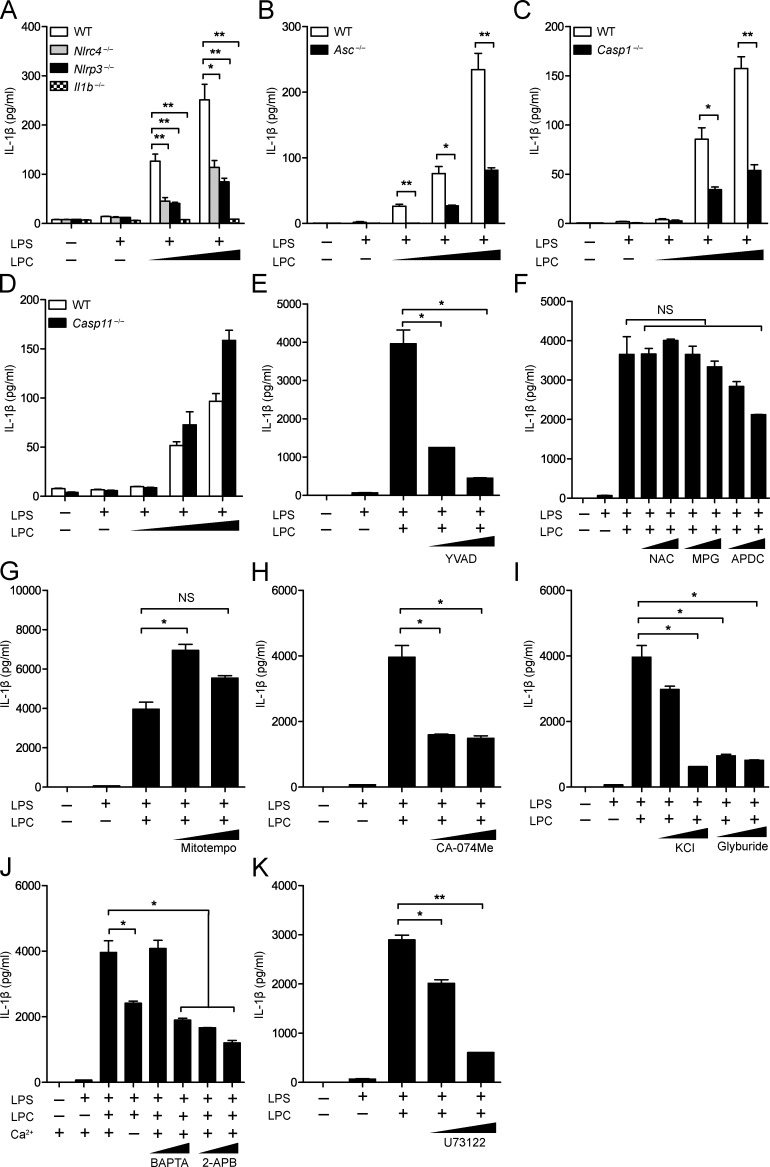
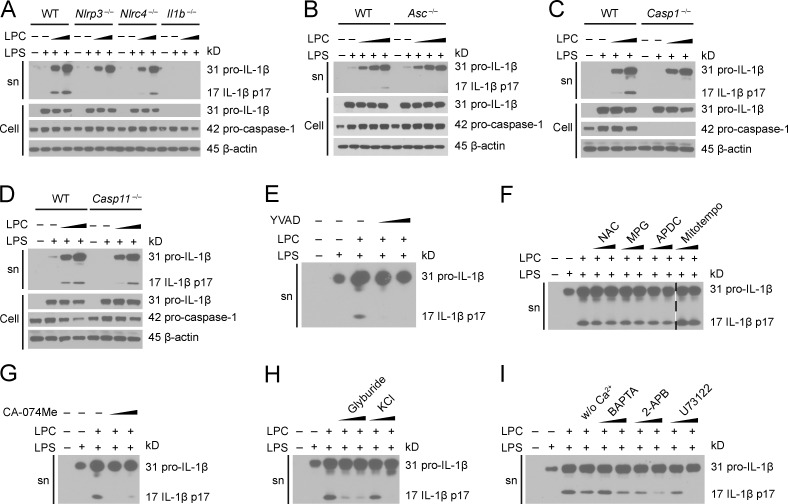
We assessed inflammasome activation by molecules associated with CNS disease by analyzing LPC activity. To assess whether LPC could activate the inflammasome, we generated BMDMs from WT, Nlrp3−/−, Nlrc4−/−, Asc (apoptotic speck containing protein with a CARD)−/−, Casp1−/−, Il1b−/−, and Casp11−/− mice and primed these cells with LPS followed by LPC stimulation. Then, IL-1β release was quantified by ELISA (Fig. 1, A–D) and immunoblotting (Fig. 2, A–D). In these experiments, cells were primed with LPS, which is a well-documented inducer of signal 1 of inflammasome activation, inducing the transcription and translation of inflammasome components, such as pro–IL-1β, pro–caspase-1, and NLRP3. After LPS pretreatment, LPC was tested for signal 2 activity, which normally leads to caspase-1 and IL-1β maturation followed by secretion of IL-1β. A combination of LPS and LPC caused significant IL-1β release in WT BMDMs (Fig. 1, A–D; and Fig. 2, A–D). However, IL-1β secretion in Nlrp3−/−, Nlrc4−/−, Asc−/−, Casp1−/−, and Il1b−/− BMDMs was reduced compared with WT controls but not in Casp11−/− BMDMs (Fig. 1, A–D; and Fig. 2, A–D). These results indicate that inflammasome-associated proteins NLRC4, NLRP3, caspase-1, and ASC mediate LPC-induced IL-1β secretion in BMDMs, whereas the noncanonical inflammasome-associated protein capase-11 is not involved. LPC-induced inflammasome activation was also inhibited by a caspase-1 inhibitor, YVAD (Z-Tyr-Val-Ala-Asp[OMe]-fluoromethylketone; Fig. 1 E and Fig. 2 E).
Figure 1.
The inflammasome mediates LPC-induced IL-1β secretion in mouse BMDMs. (A–D) WT, Nlrc4−/−, Nlrp3−/−, Il1b−/− (A), Asc−/− (B), Casp1−/− (C), and Casp11−/− (D) BMDMs were LPS primed (100 ng/ml) overnight before LPC stimulation at concentrations of 25, 50, and 100 µM for 1 h before IL-1β was collected from supernatants for analysis by ELISA. (E–K) BMDMs were LPS primed (200 ng/ml) for 3 h before stimulation with 50 µM LPC in the presence or absence of the caspase-1 inhibitor YVAD (10 and 50 µM; E); the ROS inhibitors NAC (5 and 25 mM), MPG (1 and 2.5 mM), and APDC (10 and 50 µM; F); the mitochondrial ROS inhibitor Mitotempo (100 and 500 µM; G); the cathepsin B inhibitor (CA-074Me; 10 and 20 µM; H); the potassium efflux inhibitors KCl (50 and 100 mM) and glyburide (100 and 200 µM; I); or the calcium inhibitors BAPTA (10 and 20 µM), 2-APB (20 and 100 µM; J), and U73122 (2 and 10 µM; K). Then, IL-1β was collected from supernatants for analysis by ELISA. Each control and experimental condition was performed in triplicate. All data are representative of at least two independent experiments. Results are displayed as the mean ± SEM. *, P < 0.05; **, P < 0.01; unpaired Student's t test.
Figure 2.
The inflammasome mediates LPC-induced IL-1β maturation in mouse BMDMs. (A–D) WT, Nlrp3−/−, Nlrc4−/−, and Il1b−/− (A), Asc−/− (B), Casp1−/− (C), and Casp11−/− (D) BMDMs were LPS primed (100 ng/ml) overnight before LPC stimulation (25, 50, and 100 µM). Pro–IL-1β and IL-1β in the supernatant (sn) or cell lysate (Cell) and pro–caspase-1 in the cell lysate were assessed by immunoblotting. β-Actin was used as the loading control. (E–I) BMDMs were LPS primed (200 ng/ml) for 3 h and then stimulated with 50 µM LPC in the presence or absence of the caspase-1 inhibitor YVAD (E); the ROS inhibitors NAC, MPG, and APDC and the mitochondrial ROS inhibitor Mitotempo (F); the cathepsin B inhibitor CA-074Me (G); the potassium efflux inhibitors glyburide and KCl (H); and the calcium inhibitors BAPTA, 2-APB, and U73122 (I) as used in Fig. 1. IL-1β in the supernatant was assessed by immunoblotting. The image in F is assembled from two sections of the same blot, and the break between these two sections is indicated by a dash line. The data are representative of two independent experiments. w/o, without.
To further explore how NLRP3 might mediate LPC-induced IL-1β secretion in BMDMs, we pretreated WT BMDMs with inhibitors of intracellular NLRP3 agonists before LPC stimulation and measured IL-1β secretion by ELISA (Fig. 1, F–K) and immunoblotting (Fig. 2, F–I). Inhibitors of reactive oxygen species (ROS; NAC [N-acetyl-l-cysteine], MPG [N-(2-mercaptopropionyl)glycine], and APDC [(2R,4R)-4-aminopyrrolidine-2,4-dicarboxylic acid]) and mitochondrial ROS production (Mitotempo) did not significantly reduce LPC-induced inflammasome activation (Fig. 1, F and G; and Fig. 2 F). Inhibitors of cathepsin B–mediated degradation (CA-074Me), K+ efflux (glyburide), and Ca2+ influx (BAPTA, 2-APB, and U73122) reduced LPC-induced IL-1β secretion in BMDMs (Fig. 1, H–K; and Fig. 2, G–I). This suggests that LPC induction of IL-1β secretion in BMDMs is dependent on cathepsin B, Ca2+ influx, and K+ efflux.
With the observation that LPC induces IL-1β secretion in WT BMDMs, we examined whether NLRP3 and NLRC4 are required for LPC-induced cell death. BMDMs were isolated from WT, inflammasome-associated gene–deficient mice, and noncanonical inflammasome-associated gene–deficient mice, LPS primed, and stimulated with increasing concentrations of LPC. Supernatants were collected, and lactate dehydrogenase (LDH) was measured to assess the level of cell death (Fig. S1). Although there was a decrease in LDH levels in Nlrp3−/−, Nlrc4−/−, Asc−/−, and Casp1−/− BMDMs compared with WT BMDMs, the reduction was modest, suggesting that the inflammasome modestly impacted LPC-induced cell death. There was no difference in LDH levels in Casp11−/− BMDMs compared with WT BMDMs, excluding a role for the noncanonical inflammasome. LPC treatment of Il1b−/− and Il18−/− cells also showed no difference from controls.
Expression of NLRC4 is enhanced in the mouse CNS during neuroinflammation
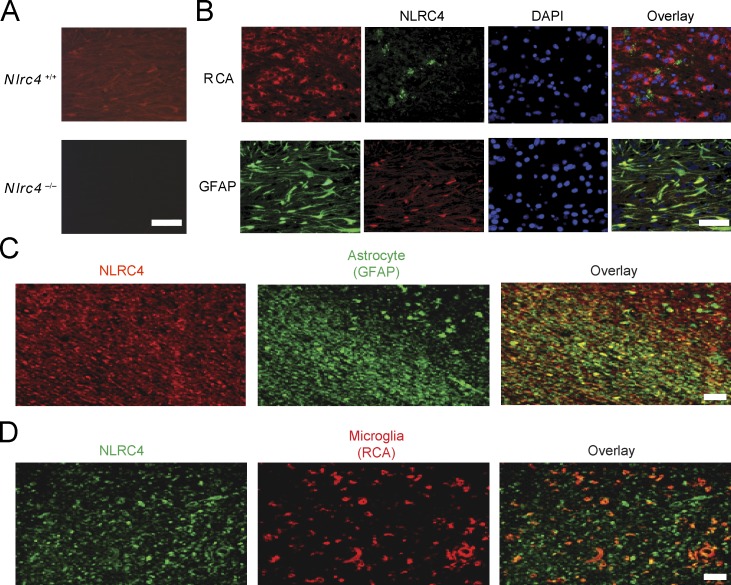
The results in Figs. 1 and 2 revealed that NLRP3 and NLRC4 are important in LPC-mediated inflammasome activation. Although NLRP3 has been studied in several neurodegenerative conditions, the role of NLRC4 has not been extensively explored in the CNS. A previous study has shown that Nlrc4 RNA is expressed in mouse brain as well as in other tissues (Poyet et al., 2001). Because RNA expression does not always correlate with protein abundance, we assessed whether NLRC4 protein is found in the brain in the cuprizone model during peak inflammatory cell accumulation, demyelination, and mature oligodendrocyte death (Hiremath et al., 1998; Arnett et al., 2001). Brains of C57BL/6 mice that had been treated with cuprizone for 4 wk were examined for NLRC4 by immunohistochemistry (IHC) using an anti-NLRC4 antibody. As a specificity control, we showed that NLRC4 expression was detected in the brain of WT mice (Nlrc4+/+; 4-wk cuprizone treated) but not in identically treated Nlrc4−/− mice (Fig. 3 A). The protein was abundant in astrocytes (glial fibrillary acidic protein [GFAP]+) and modestly abundant in microglia expressing the lectin Ricinus communis agglutinin 1 (RCA1+) at the corpus callosum after 4 wk of cuprizone-induced demyelination in WT mice (Fig. 3 B). NLRC4 was not found at detectable levels by NeuN+ neurons and CNPase+ oligodendrocytes (Fig. S2).
Figure 3.
NLRC4 is expressed in mouse CNS during neuroinflammation and in human brain tissue from MS patients. (A) WT mice brains were examined for NLRC4 expression by IHC using a mouse NLRC4 antibody. NLRC4 expression (red) was detected after 4 wk of cuprizone-induced demyelination in WT (Nlrc4+/+) but not in Nlrc4−/− mice. (B) The corpus callosum from 4-wk cuprizone–treated WT mice was stained with anti-NLRC4 (top, green; bottom, red), and RCA (red) was used to detect microglia (top), and GFAP (green) was used to detect astrocytes (bottom) at the corpus callosum. DAPI (blue) was used to label nuclei. Overlay (yellow) indicates colocalization of NLRC4 with the cell-specific biomarkers. (C and D) Chronic MS brain tissues were obtained from the UCLA HBSFRC. The corpus callosum sections were stained for NLRC4 using a human NLRC4 antibody (αNLRC4), as this is the region studied in the cuprizone model. Astrocytes (C) and microglia (D) were detected using GFAP and RCA antibodies. The overlay was performed to detect colocalization of proteins. All data are representative of at least two independent experiments. Bars, 50 µM.
NLRC4 is abundant in lesions and astrocyte-rich regions of human brain tissue from MS patients
To correlate clinical relevance to the cuprizone neuroinflammation model, we examined human tissues with MS lesions and samples without MS plaques, provided by the University of California Los Angeles (UCLA) Human Brain and Spinal Fluid Resource Center (HBSFRC). The clinical synopsis of three controls and three MS samples used for this study is listed in Table 1. Three individuals were diagnosed with chronic active MS and exhibited demyelination, with two having gliosis. Normal control brain tissues were devoid of inflammatory lesions and did not display CNS neuropathology. The plaque location of samples taken from the MS brains is shown in Fig. S3. The three samples shown on the left were diagnosed with MS, whereas the samples on the right (labeled Normal) were not. NLRC4 expression was greatly elevated in the lesion associated with extensive astrogliosis as marked by GFAP as demonstrated in the overlay panel of anti-NLRC4 and anti-GFAP (Fig. 3 C). Microglial accumulation was also evident, although fewer RCA+ microglia cells were detected compared with astrocytes (compare Fig. 3, C and D, middle), and the overlay of anti-NLRC4 and anti-RCA staining showed few NLRC4+RCA+ cells compared with NLRC4+GFAP+ cells (Fig. 3, C and D, right). A summary of Image J (National Institutes of Health) quantification is shown in Tables 2 and 3. The MS samples showed greatly enhanced NLRC4 protein levels in plaque, white matter, and, to a lesser extent, gray matter, whereas normal controls did not express detectable NLRC4 protein. Astrocytes identified by GFAP staining expressed the highest amount of NLRC4, whereas microglial cells identified by RCA staining also expressed the protein but to a lower extent. Neurons and oligodendrocytes did not express detectable levels. These results in human specimens agree with the findings observed in mice in the previous paragraph (Fig. 3, A and B; and Fig. S2).
Table 1. MS patients: clinical synopsis.
| Patient no. | Sample no. | Gender/age | Clinical diagnosis | Lesion type |
|---|---|---|---|---|
| 1 | HSB-2407 | F/58 | Normal | Control |
| 2 | HSB-2803 | M/92 | Dementiaa | Control |
| 3 | HSB-3119 | F/77 | MS diagnosed but not supported by neuropathological diagnosis | Control |
| 4 | HSB-3391 | F/48 | MS; periventricular white matter shows variable demyelination, associated with prominent gliosis and perivascular lymphocytic and macrophage infiltration | Chronic Active MS |
| 5 | HSB-3442 | F/63 | Irregular bilateral periventricular demyelination | Chronic MS |
| 6 | HSB-3704 | F/47 | Extensive white matter demyelination and axonal loss, perivascular lymphocytic cuffing, and extensive gliosis associated with areas of neuronal loss | Chronic Active MS |
The clinical pathology of MS and control brain tissues obtained from the UCLA HBSFRC is described. Each brain was evaluated by neuropathologists at UCLA. Brain tissue from three MS cases was examined along with three control brains from cases without MS. In all cases, the tissue was taken from the region around or at the corpus callosum, as shown in Fig. S2.
Dementia with no distinctive pathology.
Table 2. Distribution of reactivity for NLRC4 in MS lesions.
| Ab | MS brain | Control brain | ||||
|---|---|---|---|---|---|---|
| Plaque | Adjacent white matter | Adjacent gray matter | NAWM | NAGM | ||
| NLRC4 | 39.585 | 28.714 | 1.511 | 0 | 0 | |
Immunofluorescence indicating NLRC4 presence in tissue sections was quantified by National Institutes of Health ImageJ software (Schneider et al., 2012). The images were unstacked. Each image was split into channels, the threshold was set, and then the signal was measured. The numbers represent mean values from the output results. Abbreviations used: Ab, antibody; NAGM, normal-appearing gray matter; NAWM, normal-appearing white matter.
Table 3. Cellular distribution of reactivity for NLRC4 .
| Ab | Cellular distribution | |||
|---|---|---|---|---|
| Astrocytes (GFAP) | Microglia/ Macrophages (RCA) | Oligodendrocytes (O4) | Neurons (NeuN) | |
| NLRC4 | 23.085 | 12.1 | 0 | 0 |
Immunofluorescence indicating NLRC4 presence in various cell types present in the MS lesions was quantified by ImageJ software. The images were unstacked. Each image was split into channels, the threshold was set, and then the signal was measured. The numbers represent mean values from the output results table generated by Image J. Ab, antibody.
Astrocytic NLRC4 and NLRP3 display the same specificity as their macrophage counterparts
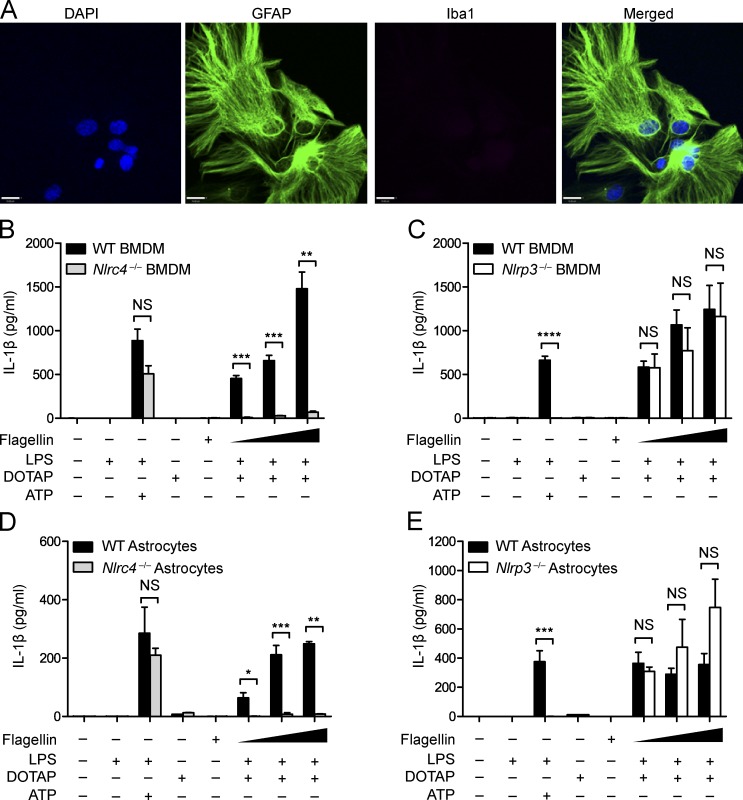
The findings in the previous paragraph led us to compare LPC-induced inflammasome activation in macrophages and astrocytes. Astrocytes were isolated from day-0–2 postnatal pups, and purity was verified by immunocytochemistry (ICC) using the astrocytic marker GFAP for detection of desired astrocytes versus the microglial marker Iba-1 for contaminating microglia (Fig. 4 A). In BMDMs, NLRC4 is known to respond to PAMPs such as bacterial flagellin and T3SS (Ma et al., 2000; Franchi et al., 2006, 2010; Miao et al., 2006, 2010), whereas NLRP3 reacts to a plethora of activators, including ATP (Mariathasan et al., 2006; Duncan et al., 2007). First, we confirmed responses of Nlrc4−/− BMDMs to PAMP stimulation, finding that WT and Nlrc4−/− BMDMs produced similar levels of IL-1β in response to the known NLRP3 stimuli LPS + ATP, which should not be affected by the absence of NLRC4. As expected, IL-1β response to LPS and flagellin was ablated in the Nlrc4−/− BMDMs (Fig. 4 B). Likewise, Nlrp3−/− and WT BMDMs showed no statistical difference in IL-1β production in response to LPS and flagellin, but response to LPS + ATP was ablated in the absence of NLRP3 (Fig. 4 C). This confirmed the specificity of the NLRC4 and NLRP3 inflammasomes in BMDMs. Next, we assessed whether astrocytic NLRC4 displayed the same specificity as BMDM NLRC4 using primary astrocytes isolated from Nlrc4−/− and WT newborn mice (Wu et al., 2010). Once astrocyte purity was confirmed, astrocytes were pretreated with LPS and transfected with S. typhimurium flagellin, and IL-1β release was measured by ELISA. Upon transfection of WT astrocytes with increasing concentrations of flagellin, IL-1β release increased in a dose-dependent fashion. In contrast, Nlrc4−/− astrocytes showed little or no increase in IL-1β release when transfected with increasing concentrations of flagellin (Fig. 4 D). As observed with Nlrp3−/− BMDMs (Fig. 4 C), when Nlrp3−/− astrocytes were transfected with increasing amounts of flagellin, there was no difference in IL-1β release compared with WT astrocytes (Fig. 4 E). Again, response to LPS + ATP was ablated in Nlrp3−/− but not in Nlrc4−/− astrocytes (Fig. 4, D and E). These results show that, similar to BMDMs, NLRC4 in astrocytes is required for inflammasome activation by flagellin, whereas NLRP3 in astrocytes is required for the LPS + ATP response.
Figure 4.
Astrocytic NLRC4 and NLRP3 display the same inflammasome specificity as macrophage NLRs. (A) Immunocytochemical detection of GFAP (green)- but not Iba-1(red)–stained cells from a purified mouse astrocyte culture used in Fig. 4 (D and E). DAPI (blue) was used to label nuclei. Representative images from the experiment performed in triplicate are shown. Bar,16 µM. (B) WT and Nlrc4−/− BMDMs were LPS primed (400 ng/ml) for 4 h and transfected with the transfecting reagent DOTAP and 25, 50, or 100 ng of S. typhimurium flagellin for 1 h before IL-1β was collected from supernatant. WT and Nlrc4−/− LPS-primed BMDMs were also stimulated with 5 mM ATP, as a positive control for inflammasome activation. Cells were transfected with DOTAP alone or stimulated with flagellin alone without LPS as a negative control. Data are representative of two independent experiments. (C) WT and Nlrp3−/− BMDMs were similarly treated as in B. Data are representative of two independent experiments. (D) WT and Nlrc4−/− primary mouse astrocytes were LPS primed (400 ng/ml) for 4 h and transfected with 50, 100, or 200 ng flagellin for 1 h before IL-1β was collected from supernatant. LPS-primed primary mouse WT and Nlrc4−/− astrocytes were also stimulated with 5 mM ATP, as a positive control, and transfected with DOTAP alone or stimulated with flagellin alone as a negative control. Data are representative of three independent experiments. (E) WT and Nlrp3−/− primary mouse astrocytes were similarly treated as described in D. Data are representative of two independent experiments. (D and E) 9–12 mouse pups (0–2 d old) per genotype were used for each experiment. Each control and experimental condition was performed in triplicate. Results are displayed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student’s t test.
Both NLRC4 and NLRP3 are required for LPC-induced IL-1β secretion from primary mouse astrocytes
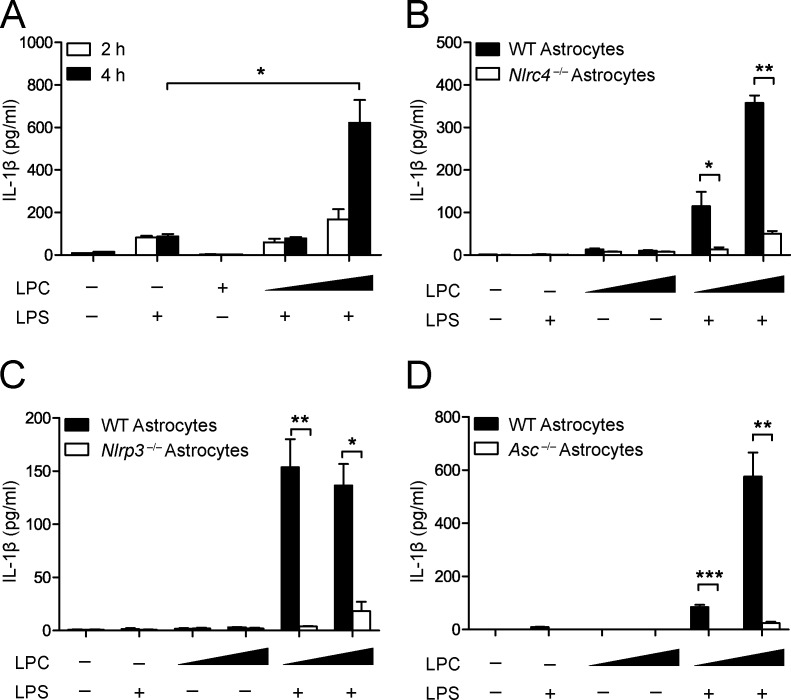
To extend the previous findings of LPC activating the inflammasome in BMDMs, we next examined whether LPC can also activate astrocyte-derived inflammasome activity. To establish inflammasome activation by LPC in astrocytes, we investigated whether LPC stimulation would result in the release of IL-1β in WT mouse astrocytes. A dose-dependent increase of released IL-1β resulted in WT astrocytes treated with increasing concentrations of LPC. Furthermore, when WT astrocytes were stimulated with LPC for 4 h as compared with 2 h, the amount of IL-1β released was enhanced (Fig. 5 A). This suggests that LPC acted to stimulate IL-1β release in astrocytes in a concentration- and time-dependent manner.
Figure 5.
NLRC4 and NLRP3 mediate LPC-induced IL-1β secretion from primary mouse astrocytes. (A) WT astrocytes were LPS primed (1 µg/ml) for 3 h and incubated with 50 and 100 µM LPC for 2 or 4 h, as indicated, before IL-1β was collected from the supernatant for analysis by ELISA. Data are representative of three independent experiments. (B) WT and Nlrc4−/− astrocytes were treated as described in A and assayed for IL-1β secretion at 4 h. Data are representative of three independent experiments. (C and D) WT and Nlrp3−/− astrocytes (C) or WT and Asc−/− astrocytes (D) were treated as described in A and assayed for IL-1β secretion at 4 h. Data are representative of two independent experiments. Each experimental condition was performed in duplicate, and 9–12 mouse pups (0–2 d old) per genotype were used for each experiment. Results are displayed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student's t test.
Next, we investigated whether NLRC4 mediated LPC stimulation of inflammasome activation. Upon 4 h of LPC stimulation, the IL-1β released from Nlrc4−/− astrocytes was attenuated compared with WT astrocytes (Fig. 5 B). Additionally, Nlrp3−/− astrocytes also showed reduced IL-1β release in response to LPC (Fig. 5 C). The adapter protein of NLRP3 and NLRC4 inflammasome ASC was also required for LPC-induced IL-1β production in mouse astrocytes (Fig. 5 D). These data agree with the data generated in BMDMs. This indicates that, in contrast to the flagellin-activated inflammasome, which only requires NLRC4, and the ATP-induced inflammasome, which only requires NLRP3, LPC-induced inflammasome requires both NLRC4 and NLRP3.
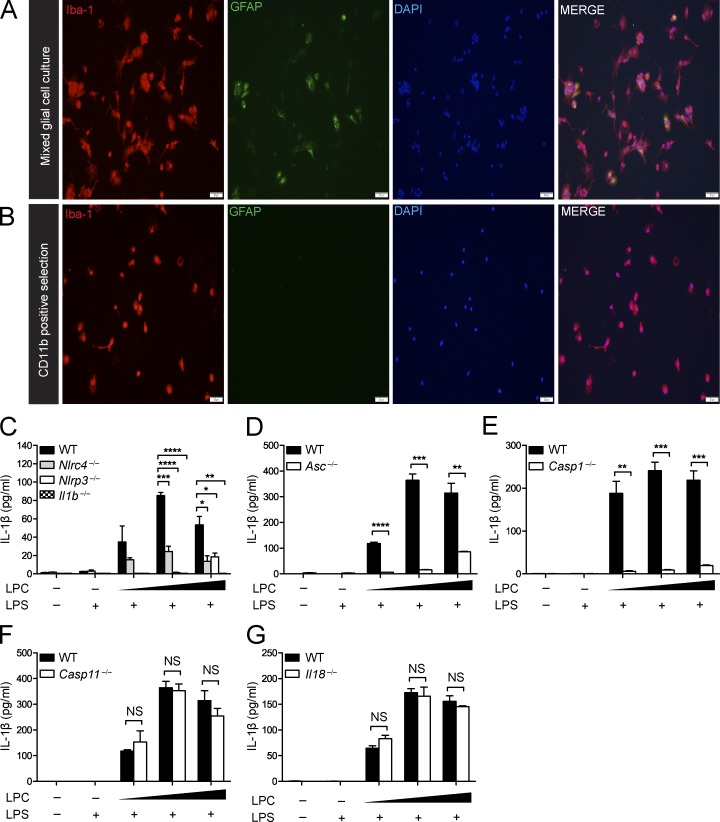
The inflammasome mediates LPC-induced IL-1β secretion in microglia
The NLRP3 inflammasome has been suggested to mediate IL-1β secretion in microglia in neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease and in response to neuropathology-associated markers such as α-synuclein and β-amyloid (Rock et al., 2010; Codolo et al., 2013; Freeman and Ting, 2016). Next, we examined whether LPC induces IL-1β release in microglia and whether this process is dependent on the inflammasome. Microglia were isolated using CD11b-positive selection. ICC (Fig. 6, A and B) was used to assess the separation of microglia (Iba-1+ cells) from GFAP+ astrocytes before and after CD11b selection. Microglia were isolated from brains of WT, Nlrc4−/−, Nlrp3−/−, Il1b−/− (Fig. 6 C), Asc−/− (Fig. 6 D), Casp1−/− (Fig. 6 E), Casp11−/− (Fig. 6 F), and Il18−/− (Fig. 6 G) neonatal mice (postnatal day 3–5). Once microglial purity was confirmed, microglial cells were LPS primed and stimulated with LPC at increasing concentrations (20, 25, and 50 µM). As expected, there was an increase in IL-1β secretion in response to LPC stimulation of WT microglia, but Nlrc4−/−, Nlrp3−/−, Il1b−/−, Asc−/−, and Casp1−/− microglia showed significantly attenuated IL-1β secretion compared with WT microglia. Casp11−/− and Il18−/− samples did not differ from WT controls. These results suggest that NLRC4, NLRP3, caspase-1, and the adapter protein ASC play a critical role in mediating LPC-induced IL-1β secretion in microglia, whereas IL-18 and caspase-11 are not involved.
Figure 6.
The inflammasome mediates LPC-induced IL-1β secretion in microglia. (A and B) ICC of Iba-1 and GFAP staining of mixed glial cell cultures (A) and CD11b-positive selected cells (B) isolated from postnatal day-3–5 WT mouse pups indicates the degree of microglial purity. Bars, 50 µM. (C–G) WT, Nlrc4−/−, Nlrp3−/−, Il1b−/− (C), Asc−/− (D), Casp-1−/− (E), Casp-11−/− (F), and Il18−/− (G) microglia were LPS primed (100 ng/ml) for 3 h and stimulated with LPC (20, 25, and 50 µM) for 1 h. IL-1β in the cell culture supernatant was assessed by ELISA. Data are representative of two independent experiments. Each experimental condition was performed in triplicate. Results are displayed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student's t test.
To determine whether the inflammasome mediates pyroptosis in microglia upon LPC treatment, we isolated microglia from WT and inflammasome-deficient mice. These cells were LPS primed and stimulated with LPC, and supernatants were collected to measure LDH levels (Fig. S4). Although LDH levels were detectable in both WT and inflammasome-deficient microglia, there was no significant difference among any of the canonical or noncanonical inflammasome-deficient microglia (Nlrc4−/−, Nlrp3−/−, Asc−/−, Casp1−/−, Il1b−/−, Il18−/−, Casp1−/−, and Casp11−/−) compared with WT microglia, suggesting that the inflammasome and noncanonical inflammasome do not enhance LPC-induced microglial death.
The inflammasome promotes microglial accumulation and astrogliosis
The in vitro system indicates that NLRC4 and NLRP3 in astrocytes and microglia mediate inflammasome activation by LPC. In parallel with the in vitro studies, we also assessed whether NLRP3 and NLRC4 play a role in the cuprizone model of neuroinflammation. This model consists of feeding mice with chow supplemented with the neurotoxicant cuprizone (0.2%), which results in a CNS disease model comprised of astrogliosis and microglial activation followed by oligodendrocyte cell death and demyelination.
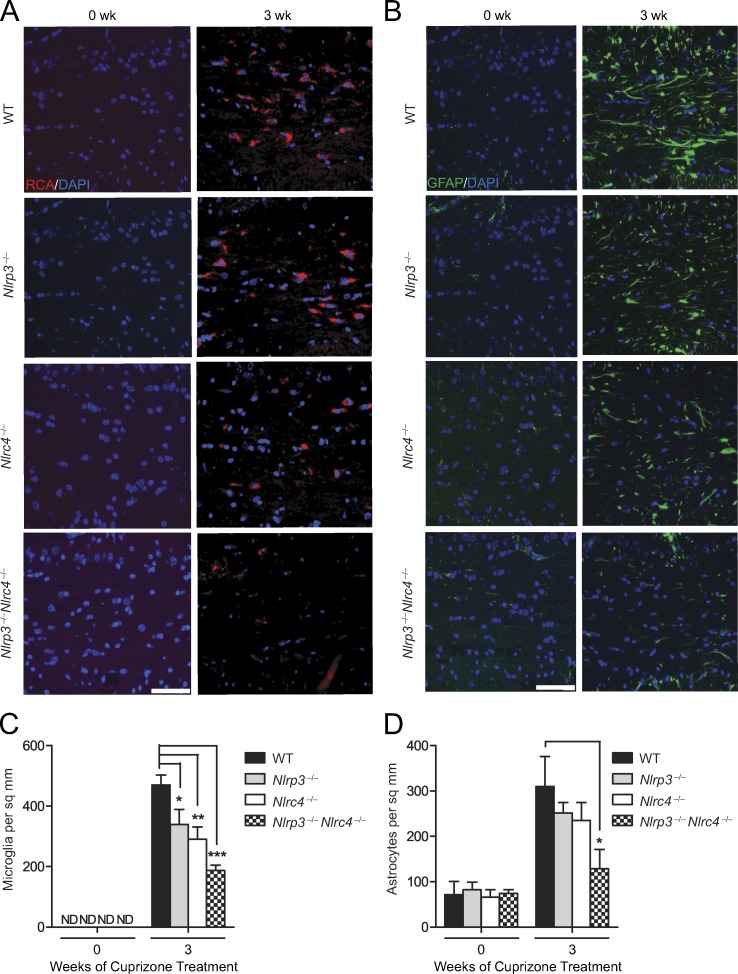
To explore whether the NLRC4 and NLRP3 inflammasomes have a role during cuprizone-induced neuroinflammation, we examined microglial accumulation in WT (control), Nlrc4−/−, Nlrp3−/−, and double KO (DKO) Nlrp3−/−Nlrc4−/− mice (Fig. 7 A). Microglial cells are resident immune cells of the CNS that release cytokines and chemokines and also phagocytose dead cells, cellular debris, and invading pathogens (Hanisch and Kettenmann, 2007; Napoli and Neumann, 2009). The microglia populations at the corpus callosum were quantified by RCA-1 lectin staining (Fig. 7 C). Among the untreated groups (indicated as 0 wk), age-matched WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice showed no difference in numbers of microglia at the corpus callosum, indicating that deletion of Nlrc4 and/or Nlrp3 did not affect microglial accumulation in nondiseased mice at steady state (Fig. 7 C). After 3-wk cuprizone treatment, there was a significant reduction in microglial accumulation in Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice compared with WT controls (Fig. 7 C), with the DKO showing the most significant reduction. These results indicate that NLRC4 and NLRP3 contribute to microglia accumulation at the corpus callosum during demyelination.
Figure 7.
NLRP3 and NLRC4 promote neuroinflammation. (A) Representative images of mouse brain sections stained for the microglial marker, RCA. Samples were obtained from cuprizone-treated WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice. RCA+ (red) microglial cells were measured at the corpus callosum in brains from untreated mice (0 wk) or after 3 wk of cuprizone treatment. DAPI (blue) was used to label nuclei. Bar, 60 µM. (B) Representative images of brain sections stained for the astrocytic marker, GFAP. Samples were obtained as described in A. DAPI (blue) was used to detect nuclei. Bar, 60 µM. (C and D) Quantification of microglial accumulation (C) and astrogliosis (D) at the midline corpus callosum in the indicated strains. Cell counts are averages of six mice per time point. Results are displayed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test. ND, not detectable; sq, square.
Next, we examined astrogliosis in WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice (Fig. 7 B). The astrocyte population at the corpus callosum was studied using GFAP staining as a marker of astrocyte activation. Among the control untreated groups (0 wk), age-matched WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice showed no difference in the number of astrocytes at the corpus callosum (Fig. 7 D), indicating that the basal level of astrocytes is unaffected by deletion of Nlrc4 and/or Nlrp3. After 3 wk of cuprizone treatment, there was a reduction of astrocytes in Nlrc4−/− and Nlrp3−/− compared with WT mice, although the difference did not reach significance. In contrast, there was a significant reduction of astrocytes in Nlrp3−/−Nlrc4−/− DKO mice (Fig. 7 D). These results indicate that NLRC4 and NLRP3 exacerbate astrogliosis in the CNS during demyelination.
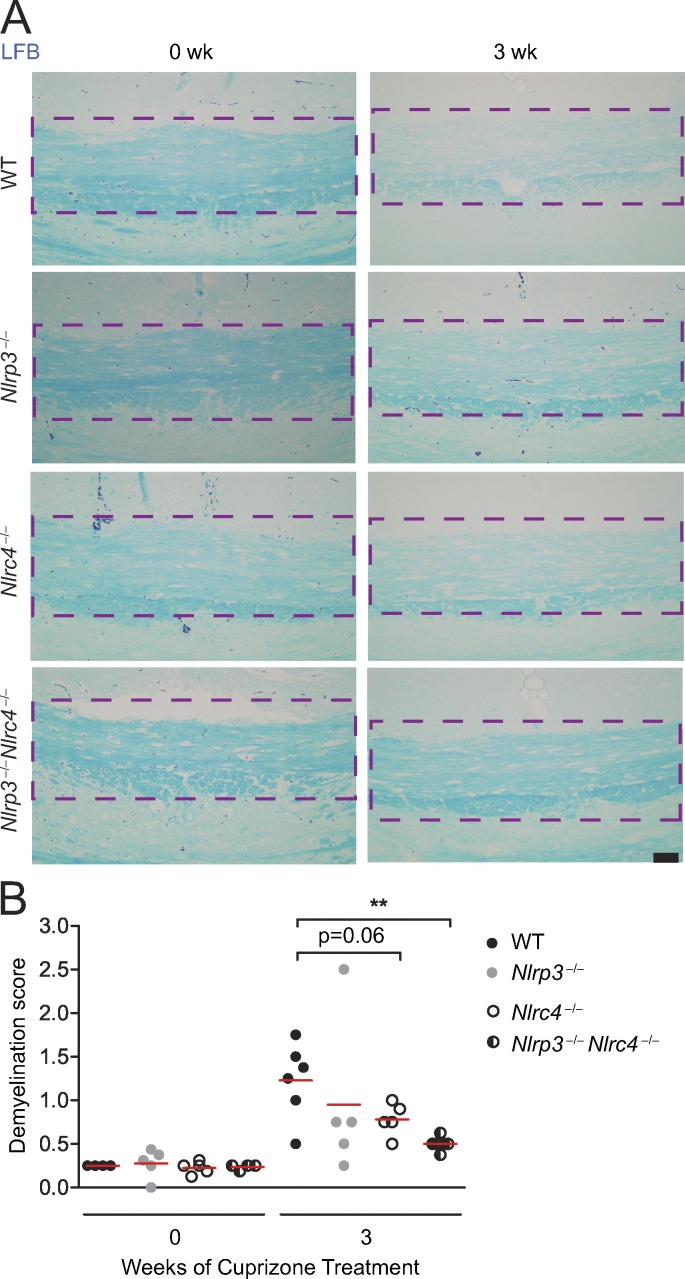
The inflammasome promotes demyelination in mice
In the cuprizone model, overt loss of myelination follows astrogliosis and microglial accumulation (Matsushima and Morell, 2001). To assess whether NLRC4 and NLRP3 play a role in demyelination, WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice were treated with cuprizone. Representative scoring of the extent of demyelination was measured by Luxol Fast blue (LFB) staining for myelin (Fig. 8 A). Slides were read by a reader blinded to the identity of tissues using a scale of 0 (no demyelination) to 3 (complete demyelination), and demyelination was quantified (Fig. 8 B). Cuprizone-treated (3 wk) WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice showed increased demyelination at the corpus callosum (denoted by the purple, bracketed area; Fig. 8 A) compared with untreated (0 wk) WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice. 3-wk cuprizone–treated Nlrc4−/− and Nlrp3−/− mice showed demyelination scores lower than WT mice, although the difference did not reach significance. In contrast, there was a significant reduction of demyelination in Nlrp3−/−Nlrc4−/− mice. This suggests that, in addition to promoting microglial accumulation and astrogliosis, NLRC4 and NLRP3 cooperate to enhance demyelination.
Figure 8.
The inflammasome promotes demyelination. (A) Representative images of brain sections of untreated (0 wk) and 3-wk cuprizone–treated WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice stained for the presence of myelin at the corpus callosum (purple dashed lines) with LFB (blue). Bar, 50 µM. (B) Quantification of LFB-stained WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice brain sections. Each circle represents the mean observed LFB score for one mouse. WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice were scored by a blinded reader on a scale of 0 (no demyelination) to 3 (complete demyelination). The mean value of each dataset is depicted by a red line. All scoring is restricted to the midline corpus callosum (purple dashed lines in A). LFB scores are means of four to six mice per time point. Results are displayed as the mean ± SEM. **, P < 0.01; unpaired Student’s t test.
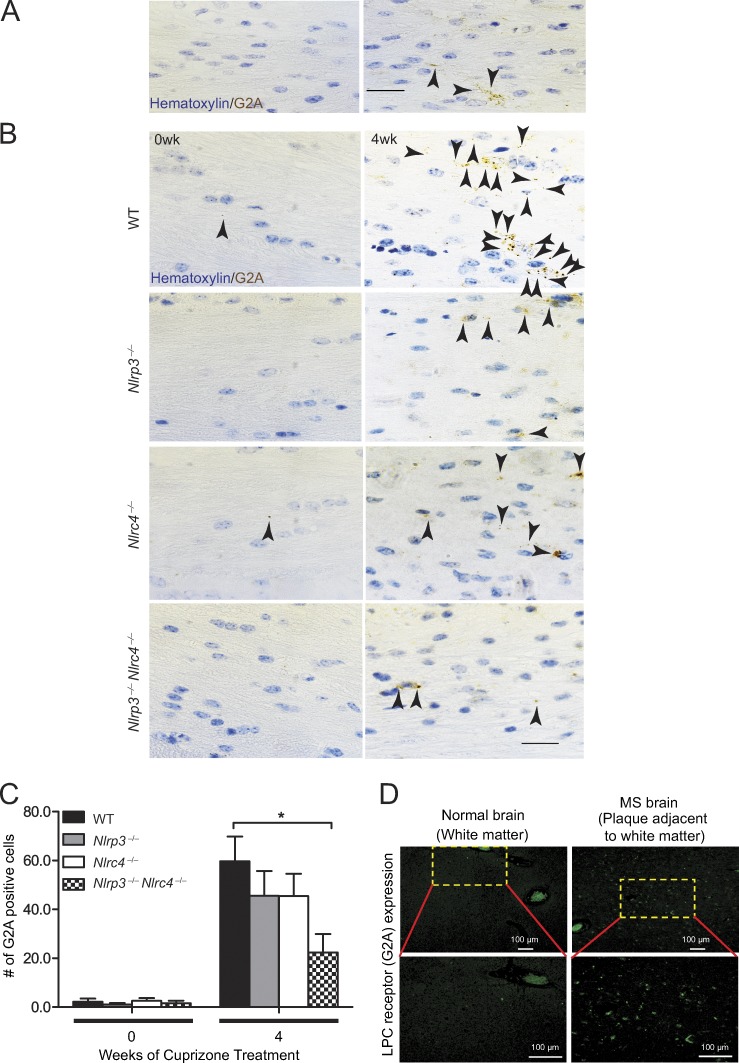
The inflammasome-intact mice have elevated G2A expression
The results thus far indicate that LPC has a proinflammatory effect on microglia, astrocytes, and BMDMs and that the inflammasome mediates LPC-induced IL-1β secretion in all three cell types (Fig. 1, Fig. 5, and Fig. 6, C–G). The results also demonstrate that the NLRP3 and NLRC4 inflammasomes exacerbate microglial accumulation and astrogliosis in vivo. Directly assessing LPC in the brain during cuprizone treatment proved to be technically difficult, with inconsistent results obtained. Alternately, we examined the expression of an LPC receptor, namely G2A, a G-protein–coupled receptor (GPR132) that is known to trigger release of cyclic adenosine monophosphate and intracellular Ca2+ to promote apoptosis and chemotaxis of macrophages, monocytes, and microglia upon binding to LPC (Rikitake et al., 2002; Lin and Ye, 2003). To determine whether G2A expression was detectable during the course of neuroinflammation caused by cuprizone treatment, we examined brains from mice subjected to 4-wk cuprizone treatment. G2A expression was confirmed in cuprizone-treated WT brains (Fig. 9 A). Brain sections from WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− DKO mice were stained for the presence of G2A (denoted by black arrows) with hematoxylin used as a counter stain (Fig. 9 B). Quantification of G2A staining in untreated and cuprizone-treated brains was performed (Fig. 9 C). Untreated (0 wk) brains from WT, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mice showed low levels of G2A expression, with no statistical differences among these genotypes. Brains isolated from cuprizone-treated Nlrp3−/− and Nlrc4−/− mice showed a reduction in G2A+ cells, but the difference did not reach significance when compared with cuprizone-treated WT brains, whereas G2A in the brains of cuprizone-treated Nlrp3−/−Nlrc4−/− DKO mice showed a significant reduction. These data suggest that both NLRP3 and NLRC4 are required for elevated G2A expression under a neuroinflammatory state.
Figure 9.
The inflammasome elevates expression of the LPC receptor G2A. (A) Brain sections from 4-wk cuprizone–treated WT mice were stained for the presence of G2A at the corpus callosum of the brain, as indicated by black arrowheads, with a hematoxylin counter stain. (Left) The negative control (no G2A antibody was used), which shows no brown staining. (Right) G2A-positive cells (brown staining as indicated by black arrowheads). Bar, 20 µm. (B) Representative images of 0-wk and 4-wk cuprizone–treated brains from WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice are shown. G2A-positive cells (as indicated by black arrowheads) are detected within the corpus callosum. Bar, 20 µM. (C) G2A-positive cells within the corpus callosum of cuprizone-treated brains of WT, Nlrc4−/−, Nlrp3−/−, and Nlrc4−/−Nlrp3−/− mice were quantified. Cell counts are means of five to six mice per time point. Results are displayed as the mean ± SEM. *, P < 0.05; unpaired Student’s t test. (D) IHC of G2A-positive cells within white matter brain tissue of normal brain and brain from an MS patient. (Top, left) G2A staining of brain section from control. (Top, right) G2A staining of brain sections from deceased MS patients. The yellow bracketed areas contain G2A-positive cells and are shown magnified to reveal the stained cells in the bottom panels. Bars, 100 µm.
As the cuprizone model represents a mouse model of neuroinflammation and demyelination, we assessed whether G2A expression was elevated in human MS tissue. The presence of G2A in brain tissues from MS patients and patients without MS was analyzed (Fig. 9 D). In normal brain tissue, G2A expression was very low to barely detectable, whereas G2A expression was abundant in brain tissue from MS patients. These data suggest that G2A is elevated in neuroinflammation-associated conditions in both human and mouse tissues.
Discussion
There is increasing evidence that inflammatory processes in the CNS play a large part in neurological disorders. As an understanding of how DAMPs activate neuroinflammation is fundamental to our understanding of CNS disease etiology, this work focuses on LPC, which has been associated with neurodegenerative and demyelination diseases. The results show that LPC serves as a DAMP that induces an atypical pattern of inflammasome activation that is dependent on both NLRP3 and NLRC4, as well as ASC and caspase-1. Conventionally, NLRP3 and NLRC4 mediate inflammasome complexes of distinct and nonoverlapping specificities (Strowig et al., 2012; Broderick et al., 2015; Guo et al., 2015; Broz and Dixit, 2016). Recent studies with Salmonella infection indicate that both NLRP3 and NLRC4 are involved in inflammasome activation by this bacteria, which actually represents a complex pathogen, unlike LPC (Qu et al., 2016). In addition to the in vitro study here, analysis of an animal model of neuroinflammation and demyelination shows that both NLRP3 and NLRC4 exacerbate disease outcome, thus providing an in vivo correlate for in vitro characterization. Finally, analysis of purified astrocyte and microglial preparations show that both NLRP3 and NLRC4 are required for inflammasome activation by LPC in these cells.
Although NLRP3 has been extensively studied, NLRC4 is primarily studied in the context of microbial infection. However, Nlrc4 mRNA is detected in the adult mouse brain (Poyet et al., 2001), is elevated in brain tissues from Alzheimer’s patients, and is shown to mediate palmitate-induced inflammasome activation in astrocytic cultures (Liu and Chan, 2014). Additionally, the absence of NLRC4 reduced brain injury, although this process appears to be independent of IL-1 (Denes et al., 2015). Heightened expression of NLRC4 has been associated with inflammatory diseases such as Kawasaki disease (Ikeda et al., 2010) and atopic dermatitis (Macaluso et al., 2007). Using peripheral blood mononuclear cells from patients, these authors demonstrated increased gene expression of NLRC4 in patients relative to healthy controls, leading the authors to suggest a role for NLRC4 in these inflammatory diseases, although the direct relevance of elevated NLRC4 in these diseases is undefined. A direct link between NLRC4 and inflammation has been determined for recurrent macrophage activation syndrome, where this disorder was associated with an activating NLRC4 mutation (Canna et al., 2014).
Our previous study with Nlrp3−/−, Casp1−/−, Il1β−/−, and Il18−/− mice in the cuprizone model showed a NLRP3-dependent mechanism, leading to demyelination and loss of mature oligodendrocytes (Jha et al., 2010). Moreover, our results in Nlrp3−/− mice showed that NLRP3 deficiency only partially delayed demyelination and neuroinflammation, uncovering the possible involvement of redundant NLR proteins during neuroinflammation and demyelination. This previous work did not analyze the cell types that are involved in inflammasome activation.
The current work provides evidence that NLRC4 is strongly expressed in astrocytes and to a lesser extent in microglia during neuroinflammation. Expression of glial NLRC4 was enhanced in the mouse brain during chronic stages of neuroinflammation in the cuprizone model and in pathological brain sections from MS patients. As the role of the inflammasome in astrocytes is not well understood, the importance of this study lies in the functional delineation of LPC-induced NLRC4 and NLRP3 inflammasome activation in astrocytes. We also show that the canonical inflammasome components NLRP3, NLRC4, ASC, caspase-1, and IL-1β mediate LPC-induced IL-1β secretion in BMDMs and microglia, whereas caspase-11, which represents the noncanonical inflammasome pathway, is not required. These data suggest that inflammasome activation by LPC in macrophages and these two glial populations occurs via indistinguishable pathways. The importance of NLRC4 and NLRP3 in the CNS is further supported by findings in the cuprizone model, where mice lacking individual NLR (Nlrc4−/− or Nlrp3−/−) exhibited modestly reduced astrogliosis, microglial accumulation, and expression of the LPC cognate receptor G2A, whereas Nlrp3−/−Nlrc4−/− DKO mice showed the most significant reduction when compared with WT mice. This suggests that NLRP3 and NLRC4 may function in a synergistic or cooperative fashion during neuroinflammation.
This study also finds that astrocytes express a relatively high level of NLRC4. Astrocytes, the most abundant cell type in the CNS, are increasingly appreciated for their broad regulatory impact on CNS homeostasis and function. Astrocytes perform multiple and diverse roles in the CNS, including the maintenance of extracellular ionic balance, promotion of neuronal survival, formation of synapses, and maintenance of the blood–brain barrier (Molofsky et al., 2012). They also provide nutrients such as glucose to neurons and play a key role in the repair and scarring process in the brain. Activated astrocytes are a major contributor to neuroinflammation and are important for synapse formation. With respect to the inflammasome genes, astrocytes express the MHC class II transactivator (CIITA; Collawn and Benveniste, 1999; Stüve et al., 2002), NOD1 and NOD2, and the inflammasome adapter protein ASC (de Rivero Vaccari et al., 2008). However, inflammasome activity in astrocytes has not been widely studied, so questions of NLRC4 mediating inflammasome functions in astrocytes remain unanswered.
It is commonly understood in the field that different inflammasome NLRs display distinct specificities for agonists/ligands. However, we initially proposed the model wherein NLRs may be able to mix and match, thus expanding the repertoire of PAMP recognition (Ting and Davis, 2005). Indeed, a few pairs of NLRs have been found to display common specificity. For example, the NAIP2–NLRC4 complex physically associates with a T3SS molecule, whereas NAIP5–NLRC4 associates with flagellin (Kofoed and Vance, 2011; Zhao et al., 2011; Halff et al., 2012). Although NLRP3 and NLRC4 display distinct specificities, they are both required for inflammasome activation by Salmonella (Broz et al., 2010), and endogenous NLRP3 and NLRC4 have been colocalized in cells upon Salmonella infection (Man et al., 2014). More recently, analysis of a mutant NLRC4 unexpectedly revealed that this mutant protein can still recruit NLRP3, which then engages ASC to activate the inflammasome upon infection with S. typhimurium (Qu et al., 2016). It will be interesting to examine whether this mutant NLRC4 exhibits intact inflammasome response to LPC.
In summary, this work provides direct evidence for the functional importance of NLRC4 and NLRP3 in astrocytic and microglial inflammasome activation and, furthermore, shows that NLRC4 and NLRP3 mediate inflammasome activity by a CNS-associated DAMP, namely LPC. Considering the expanding importance of astrocytes and microglia in neurological homeostasis and disease states, these findings have broad implications for understanding a plethora of neurological disorders where neuroinflammation is evident.
Materials and methods
Mice
Nlrc4−/− mice on the C57BL/6 background were provided by V. Dixit (Genentech, San Francisco, CA). Nlrp3−/− mice backcrossed on the C57BL/6 background have been previously described (Jha et al., 2010). C57BL/6 mice were purchased from The Jackson Laboratory and were maintained at the University of North Carolina facility. Nlrp3−/− mice were crossed with Nlrc4−/− mice to generate Nlrp3−/−Nlrc4−/− mice. Asc−/−, Casp1−/− (Casp11Tg), and Casp11−/− mice were originally provided by V. Dixit. Il1b−/− mice, originally provided by D. Chaplin (University of Alabama at Birmingham, Birmingham, AL), and Il18−/− mice (The Jackson Laboratory) were also used. All mice were 8–10 wk old before the start of treatment. All mice were allowed to acclimate to the facility for 1–2 wk before cuprizone treatment. C57BL/6, Nlrc4−/−, Nlrp3−/−, and Nlrp3−/−Nlrc4−/− mouse lines were bred and maintained in house. All experiments with animals were conducted under protocols approved by the Institutional Animal Care and Use Committee at University of North Carolina Chapel Hill and the National Institutes of Health Guidelines for Animal Care and Use. All animals were housed under specific pathogen–free conditions in sterile isolated caging in Association for Assessment and Accreditation of Laboratory Animal Care–accredited facilities.
Cuprizone treatment
8–10-wk-old male mice were fed 0.2% (wt/wt) cuprizone (oxalic bis [cyclohexylidenehydrazide]; C9012; Sigma-Aldrich) mixed into ground chow ad libitum for 3–4 wk to induce progressive demyelination (Arnett et al., 2002; Franco-Pons et al., 2007). Untreated control mice were maintained on a diet of normal pellet chow.
Tissue preparation
Mice were anesthetized and perfused with PBS followed by 4% paraformaldehyde (PFA). Brains were removed, postfixed in PFA, and embedded in paraffin. 5-µm coronal sections were cut at the fornix region of the corpus callosum. For frozen sections, mice were perfused and postfixed as described previously (Arnett et al., 2001). Brains were allowed to soak in 30% sucrose in PBS and snap frozen on dry ice in optimal cutting temperature medium. 5- and 20-µm coronal sections were cut at the fornix region of the corpus callosum for analysis by IHC. All analyses were restricted to the midline corpus callosum as described previously (Plant et al., 2007).
Human brain tissue samples
The MS and control tissues were obtained from the UCLA HBSFRC with written informed consent from all donors, and the use of human tissues was reviewed via expedited review procedures (45 CFR 46. 110 b [2]) and approved by the UCLA Institutional Review Board. The clinical pathology of patients is listed in Table 1, with material evaluated by neuropathologists at the UCLA HBSFRC. Brain tissues from three MS cases were examined along with three control brains from cases without MS. Among MS brains, three different samples were obtained from each brain including normal-appearing white matter, normal-appearing gray matter, and plaque. Considering the control brains, two samples were obtained from each brain including normal-appearing white matter and normal-appearing gray matter. In all cases, the tissues were taken from regions around or at the corpus callosum (Fig. S3). The brain samples were obtained in 4% PFA and subsequently embedded in paraffin and sectioned into 5-µm coronal sections.
Histological staining
To examine demyelination, paraffin sections were rehydrated through a graded series of alcohol washes and stained with LFB (S3382; Sigma-Aldrich) as described previously (Arnett et al., 2002). Sections were read by a double-blinded reader and graded on a scale from 0 (no demyelination) to 3 (complete demyelination), with higher scores indicating increased pathology. For detection of microglia/macrophages, sections were rehydrated and permeabilized with 0.1% Triton/PBS for 20 min at room temperature (RT) and then incubated with RCA-1 lectin (1:500; L-1080; Vector Laboratories) at 37°C for 1 h. Only RCA-1+ cells with observable DAPI-stained nuclei were included in the quantification.
IHC
IHC was performed on 5-µm paraffin-embedded sections that were deparaffinized and rehydrated through alcohols as described in the previous paragraph. To detect astrocytes, sections were incubated with 5% normal goat serum in 0.1% Triton/PBS for 20 min at RT. Subsequently, the sections were washed and incubated with GFAP rabbit anti–cow monoclonal antibody (1:100; Z0334; Dako/Agilent Technologies) and goat anti–rabbit FITC-conjugated secondary antibody (1:100; FI-1000; Vector Laboratories). For the detection of NLRC4, the sections were permeabilized with 0.1% Triton/PBS for 10 min and incubated with rabbit anti–mouse NLRC4 antibody (1:100; NBP2-41124; Imgenex/Novus Biologicals) overnight at 4°C. After washing the sections with PBS, the primary antibody was detected by incubation with goat anti–rabbit Alexa Fluor 488–conjugated antibody (A11034; Molecular Probes/Thermo Fisher Scientific) for 1 h at RT. Immunopositive cells with an observable DAPI-stained nucleus were counted twice with sample identities covered from reviewers. Cell counts were averaged for four to seven mice per time point.
G2A staining was performed on paraffin specimens using G2A (M-20) antibody (goat polyclonal; sc-9692; lot 2315; Santa Cruz Biotechnology, Inc.). Antigen retrieval was performed with EDTA-based buffer (950-500; Discovery CC1), pH 8.5, for 64 min at 100◦C and then blocked with Rodent Decloaker (10X; RD913L; Biocare Medical) for 1 h at RT. The samples were given a hydrogen peroxidase block for 8 min at RT and then incubated in the primary antibody (1:50 dilution) for 2 h at RT in a Tris-based diluent (760-212; Discovery PSS Diluent), followed by the secondary antibody (polyclonal rabbit anti–goat immunoglobulin/HRP; P0160; Dako) incubation for 1 h at RT. The samples were treated with diaminobenzidine and Hematoxylin II (790-2208; Ventana) for 12 min, followed by Bluing Reagent (760-2037; Ventana) for 4 min. The slide staining was performed using a Discovery Ultra Automated IHC staining system (Ventana).
ICC
ICC was performed on glial cells plated on polylysine-coated coverslips. To detect the presence of astrocytes, coverslips were permeabilized with Triton X-100 for 20 min and washed with PBS containing 0.05% Tween 20 for 20 min. Coverslips were blocked with 5% normal donkey serum (D9963; Sigma-Aldrich) in Triton X-100 for 30 min. Coverslips were incubated overnight with chicken primary GFAP antibody (ab1334436; Abcam) in 5% normal donkey serum (0.5:1,000) in Triton X-100 overnight. Coverslips were washed four times with PBS containing 0.05% Tween 20 and blocked with 5% normal donkey serum for 30 min. Coverslips were incubated with a donkey anti–chicken Alexa Fluor 488 antibody (703-545-155; Jackson ImmunoResearch Laboratories, Inc.) at 1:500 for 1 h at RT.
To detect the presence of microglia, coverslips were permeabilized with Triton X-100 for 20 min and washed with PBS containing 0.05% Tween 20 for 20 min. Coverslips were blocked with 5% normal goat serum (S-1000; Vector Laboratories) in Triton X-100 for 30 min. Coverslips were incubated overnight with rabbit primary Iba-1 antibody (ab178847; Abcam) in 5% normal goat serum (1:100) in Triton X-100 overnight. Coverslips were washed four times with PBS containing 0.05% Tween 20 and blocked with 5% normal goat serum for 30 min. Coverslips were incubated with a biotinylated goat anti–rabbit antibody (BA-1000; Vector Laboratories) in 5% normal goat serum at 1:100 for 1 h at RT. Then, coverslips were incubated with an avidin-conjugated Alexa Fluor 594 antibody (S32356; Invitrogen) at 1:500 for 1 h at RT. Coverslips were mounted with DAPI media (H-1200; Vector Laboratories) to detect the presence of nuclei.
Imaging
For imaging of IHC, microglial and astrocytic cell counts were taken from the midline of the corpus callosum, confined to images of a 0.033-mm2 area taken with a 50× oil immersion objective, whereas LFB-stained images of the corpus callosum were taken at a magnification of 20. For ICC, images of microglia and astrocytes were taken at 20×. An upright microscope (BX-40; Olympus) with a camera (Optronics) and Scion image acquisition software were used for taking images of the mouse brains at RT. The scale bar was added by Scion image acquisition software. An upright microscope (BX-61; Olympus) with bright-field, dark-field, differential interference contrast, and epifluorescence capability with Velocity software (PerkinElmer), an ORCA RC camera (Hamamatsu Photonics), and a color camera (RETIGA 4000R; QImaging) were used for imaging human brain tissue at RT. Images of a ruler were also taken at 50× by an upright microscope (BX-40) or at 20× by an upright microscope (BX-61), and the scale bars for Fig. 3 and Fig. S2 were generated based on ruler images. Immunofluorescence in tissue sections was quantified by the National Institutes of Health ImageJ software (Schneider et al., 2012).
For imaging of G2A-stained sections, G2A+ cell counts were taken from the midline of the corpus callosum and taken with a 100× oil immersion objective. An inverted microscope (Eclipse Ts2r; Nikon) with a camera (DS-Ri2; Nikon) and NIS-Elements Advanced Research image acquisition software were used for taking images of G2A-stained mouse brains at RT. The scale bar was added by the image acquisition software.
Astroglial isolation and purification
Primary mouse glial cultures were generated from brains as described previously with slight modifications (McCarthy and de Vellis, 1980). Three to four brains from neonatal mice (1–2 d old) were used to seed each T75 flask. In brief, the tissue was triturated and processed with a papain dissociation system according to the manufacturer’s instructions (LK003150; Worthington Biochemical Corporation). Cells obtained as a result of tissue processing were resuspended in DMEM containing 10% FBS (Hyclone), 100 µg/ml streptomycin, and 50 U/ml penicillin and were plated in a T75 flask. These cultures were maintained in a 5%-CO2 humidified incubator at 37°C for 2–3 wk. Media was changed every 2–3 d. Microglial cells growing on top of the confluent astrocyte monolayer were detached by shaking at 260 rpm for 3 h at 37°C. The media was replaced, and the cultures were placed back on shakers at 100 rpm for 24 h at 37°C. A confluent astrocyte layer was detached from the flask using 0.2 g/L EDTA in PBS (Versene; Gibco), and cells were counted and replated as required. Astrocyte purity and potential microglial contamination was assessed by ICC using microglial-specific antibodies (Iba-1; ab107159; Abcam) and astrocyte-specific antibodies (GFAP; 130300; Thermo Fisher Scientific). A microscope (700; ZEISS) with ZEN software was used for taking images of mouse glia at 20×. The scale bar was added using ZEN software.
Microglial isolation and purification
Brains were obtained from postnatal day 3–5 mouse pups and enzymatically treated with a tissue dissociation kit (LK003150). Microglial cells were isolated from mixed glial cell populations through CD11b-positive selection (130-093-634; magnetic-activated cell sorting; Miltenyi Biotec). Microglial purity was confirmed by staining for the presence of a microglial marker (Iba-1) and absence of an astrocyte marker (GFAP) in mixed glial cell cultures and cultures that were subjected to CD11b-positive selection.
Flagellin transfection of astrocytes
Primary glial cultures were harvested from a pool of 0–2-d-old neonatal mouse pup brains as previously described (McCarthy and de Vellis, 1980). Then, astrocytes were replated in a 96-well plate at a density of 50,000 cells per well. Astrocytes were primed with 400 ng/ml LPS (tlrl-3pelps; InvivoGen) in DMEM containing 10% FBS for 4 h. Before transfection, wells were rinsed two to three times with PBS to remove any serum. Then, astrocytes were DOTAP transfected with S. typhimurium flagellin (tlrl-epstfla-5; InvivoGen) in serum-free DMEM media for 1 h at concentrations of 6.5–200 ng per well. Supernatants from each of the wells were collected and then measured for the mouse cytokine IL-1β using an ELISA kit (OptEIA) according to the manufacturer’s instructions (BD).
LPS and LPC stimulation of BMDMs
BMDMs were obtained by flushing femurs of WT, Nlrp3−/−, Nlrc4−/−, Nlrc4−/−Nlrp3−/−, Asc−/−, Casp1−/−, Casp11−/−, Il18−/−, and Il1b−/− mice with DMEM containing 10% FBS and penicillin/streptomycin. Cells were treated with ACK lysis buffer. Cells were cultured with DMEM, supplemented with 30% GM-CSF containing L929 media, 20% FBS, and penicillin/streptomycin, for 5–7 d to allow cells to differentiate into BMDMs. Cells were plated at a density of 100,000 cells per well in a 96-well plate or 1,000,000 cells per well in a 6-well plate. BMDMs were stimulated with DMEM containing 10% FBS and 100 ng/ml LPS overnight. After wells were rinsed twice with PBS, the cells were stimulated with serum free DMEM containing 25, 50, or 100 µM LPC for 1 h. Cells were pelleted by spinning at 400 g for 5 min, and the supernatant was collected and analyzed for IL-1β secretion by ELISA and Western blotting or LDH release for cell toxicity (11644793001; Roche). With regard to inhibitor usage, the inhibitors were added after LPS priming and 30 min before 50-µM LPC stimulation and were maintained in cell culture during LPC stimulation. The caspase-1 inhibitor YVAD (FMK005) was purchased from R&D Systems. NAC (A9165) and MPG (M6635) were purchased from Sigma-Aldrich. APDC (ALX-550-178-M001), Mitotempo (ALX-430-150-M005), Ca-074 Me (BML-PI126-0001), glyburide (BML-KC120-0005), BAPTA-AM (BML-CA411-0025), 2-APB (ALX-400-045-M100), and U73122 (BML-ST391-0005) were purchased from Enzo Life Sciences.
LPS and LPC stimulation of astrocytes
Astrocytes were replated at a density of 50,000 cells per well in a 96-well dish plate. Then, astrocytes were primed with 1 µg/ml LPS in DMEM containing 10% FBS overnight. After rinsing wells with PBS two to three times, then, cells were stimulated with prewarmed serum-free DMEM (37°C) containing LPC concentrations of 50 and 100 µM for 2 and 4 h. Supernatants from each of the wells were collected, and IL-1β levels were measured.
LPS and LPC stimulation of microglia
Microglia were seeded at a density of 50,000 cells per well in a 96-well dish plate. First, microglia were primed with 100 ng/ml LPS in DMEM containing 10% FBS for 3 h before LPC stimulation. After rinsing cells with PBS, cells were then stimulated with prewarmed serum-free DMEM (37°C) containing LPC concentrations of 20, 25, and 50 µM for 1 h. Supernatants from each of the wells were collected and then assessed for mouse cytokine IL-1β or for cytotoxicity by LDH release assay.
Immunoblotting
Cells were resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail, and phosphatase inhibitor cocktail, pH 7.4). Cell lysates were incubated on ice for 30 min, and cell debris was pelleted by centrifugation at 16,000 g for 30 min at 4°C. Clear lysates and cell supernatants were separated by 4–16% Bis-Tris gels (NativePAGE Novex; Thermo Fisher Scientific) and then transferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% milk protein before probing with primary antibodies. Primary antibodies included anti–IL-1β (AF-401-SP; R&D Systems), anti–caspase-1 (AG-20B-0044-C100; Adipogen), and anti–β-actin (sc-1615; HRP conjugate; Santa Cruz Biotechnology, Inc.). Appropriate HRP-conjugated secondary antibodies were used, and proteins were detected using Enhanced Chemiluminescent reagent (Thermo Fisher Scientific).
Statistical analysis
All values are shown as mean ± SEM. Two-tailed, unpaired Student’s t test was used to assess statistical significance in Prism (5.0; GraphPad Software). In all tests, p-values of <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) were considered statistically significant. All figures are representative of at least two independent experiments.
Online supplemental material
Fig. S1 shows LPC-induced toxicity in WT, Nlrc4−/−, Nlrp3−/−, Asc−/−, Casp1−/−, Casp11−/−, Il1b−/−, and Il18−/− BMDMs. Fig. S2 shows negligible expression of NLRC4 in mouse oligodendrocytes or neurons in vivo. Fig. S3 shows sample location and pathology examination of normal and MS patient brain tissues. Fig. S4 shows LPC-induced toxicity in WT, Nlrc4−/−, Nlrp3−/−, Asc−/−, Casp1−/−, Casp11−/−, Il1b−/−, and Il18−/− microglia.
Supplementary Material
Acknowledgments
We thank Janice Weaver and Dawud Hilliard at the Lineberger Animal Histopathology Core for assistance with tissue processing and histology, Victoria Madden and Dr. Robert Bagnell at the Microscopy Services Laboratory for assistance with microscopy, and Drs. Glenn Matsushima, Justin Wilson, and Gregory Robbins for helpful suggestions.
This work was supported by the National Multiple Sclerosis Society (grants CA-1053-A-8 and RG-1785E), National Institutes of Health (grants R37-AI029564 and U19AI109965 to J.P.-Y. Ting, L. Freeman, and H. Guo), and a National Institutes of Health training grant (T32-AIO7273 to C.N. David).
The authors declare no competing financial interests.
Author contributions: L. Freeman, H. Guo, S. Jha, and J.P.-Y. Ting designed the experiments. L. Freeman, H. Guo, S. Jha, and C. David conducted the studies. W.J. Brickey assisted with the experiments and provided intellectual input. J.P.-Y. Ting supervised the study. L. Freeman, H. Guo, S. Jha, W.J. Brickey, and J.P.-Y. Ting interpreted the data and wrote the manuscript.
Footnotes
Abbreviations used:
- CARD
- caspase recruitment domain
- CNS
- central nervous system
- DAMP
- damage-associated molecular pattern
- DKO
- double KO
- GFAP
- glial fibrillary acidic protein
- ICC
- immunocytochemistry
- IHC
- immunohistochemistry
- LDH
- lactate dehydrogenase
- LFB
- Luxol Fast blue
- LPC
- lysophosphatidylcholine
- LRR
- leucine-rich repeat
- MS
- multiple sclerosis
- NAIP
- NLR family apoptosis inhibitory protein
- NLR
- nucleotide-binding LRR-containing
- PAMP
- pathogen-associated molecular pattern
- PFA
- paraformaldehyde
- RCA
- Ricinus communis agglutinin
- ROS
- reactive oxygen species
- RT
- room temperature
References
- Adamczak S.E., de Rivero Vaccari J.P., Dale G., Brand F.J. III, Nonner D., Bullock M.R., Dahl G.P., Dietrich W.D., and Keane R.W.. 2014. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J. Cereb. Blood Flow Metab. 34:621–629. 10.1038/jcbfm.2013.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen I.C., TeKippe E.M., Woodford R.M., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A., Franchi L., Kanneganti T.D., Body-Malapel M., Ozören N., Brady G., Meshinchi S., Jagirdar R., Gewirtz A., Akira S., and Núñez G.. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281:35217–35223. 10.1074/jbc.M604933200 [DOI] [PubMed] [Google Scholar]
- Anand P.K., Malireddi R.K., Lukens J.R., Vogel P., Bertin J., Lamkanfi M., and Kanneganti T.D.. 2012. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 488:389–393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett H.A., Mason J., Marino M., Suzuki K., Matsushima G.K., and Ting J.P.. 2001. TNFα promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 4:1116–1122. 10.1038/nn738 [DOI] [PubMed] [Google Scholar]
- Arnett H.A., Hellendall R.P., Matsushima G.K., Suzuki K., Laubach V.E., Sherman P., and Ting J.P.. 2002. The protective role of nitric oxide in a neurotoxicant-induced demyelinating model. J. Immunol. 168:427–433. 10.4049/jimmunol.168.1.427 [DOI] [PubMed] [Google Scholar]
- Bauer C., Duewell P., Lehr H.A., Endres S., and Schnurr M.. 2012. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig. Dis. 30:82–90. 10.1159/000341681 [DOI] [PubMed] [Google Scholar]
- Boyden E.D., and Dietrich W.F.. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240–244. 10.1038/ng1724 [DOI] [PubMed] [Google Scholar]
- Broderick L., De Nardo D., Franklin B.S., Hoffman H.M., and Latz E.. 2015. The inflammasomes and autoinflammatory syndromes. Annu. Rev. Pathol. 10:395–424. 10.1146/annurev-pathol-012414-040431 [DOI] [PubMed] [Google Scholar]
- Broz P., and Dixit V.M.. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., and Superti-Furga G.. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272. 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Canna S.W., de Jesus A.A., Gouni S., Brooks S.R., Marrero B., Liu Y., DiMattia M.A., Zaal K.J., Sanchez G.A., Kim H., et al. . 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140–1146. 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M.J. 2002. Microglia as liaisons between the immune and central nervous systems: functional implications for multiple sclerosis. Glia. 40:218–231. 10.1002/glia.10145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F.A., Nalbantoglu I., Aitken J.D., Uchiyama R., Su Y., Doho G.H., Vijay-Kumar M., and Gewirtz A.T.. 2012. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol. 5:288–298. 10.1038/mi.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codolo G., Plotegher N., Pozzobon T., Brucale M., Tessari I., Bubacco L., and de Bernard M.. 2013. Triggering of inflammasome by aggregated α-synuclein, an inflammatory response in synucleinopathies. PLoS One. 8:e55375 10.1371/journal.pone.0055375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collawn J.F., and Benveniste E.N.. 1999. Regulation of MHC class II expression in the central nervous system. Microbes Infect. 1:893–902. 10.1016/S1286-4579(99)00228-2 [DOI] [PubMed] [Google Scholar]
- Davis B.K., Roberts R.A., Huang M.T., Willingham S.B., Conti B.J., Brickey W.J., Barker B.R., Kwan M., Taxman D.J., Accavitti-Loper M.A., et al. . 2011. Cutting edge: NLRC5-dependent activation of the inflammasome. J. Immunol. 186:1333–1337. 10.4049/jimmunol.1003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denes A., Coutts G., Lénárt N., Cruickshank S.M., Pelegrin P., Skinner J., Rothwell N., Allan S.M., and Brough D.. 2015. AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3. Proc. Natl. Acad. Sci. USA. 112:4050–4055. 10.1073/pnas.1419090112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rivero Vaccari J.P., Lotocki G., Marcillo A.E., Dietrich W.D., and Keane R.W.. 2008. A molecular platform in neurons regulates inflammation after spinal cord injury. J. Neurosci. 28:3404–3414. 10.1523/JNEUROSCI.0157-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rivero Vaccari J.P., Lotocki G., Alonso O.F., Bramlett H.M., Dietrich W.D., and Keane R.W.. 2009. Therapeutic neutralization of the NLRP1 inflammasome reduces the innate immune response and improves histopathology after traumatic brain injury. J. Cereb. Blood Flow Metab. 29:1251–1261. 10.1038/jcbfm.2009.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebolder C.A., Halff E.F., Koster A.J., Huizinga E.G., and Koning R.I.. 2015. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure. 23:2349–2357. 10.1016/j.str.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Dong Y., and Benveniste E.N.. 2001. Immune function of astrocytes. Glia. 36:180–190. 10.1002/glia.1107 [DOI] [PubMed] [Google Scholar]
- Duncan J.A., Bergstralh D.T., Wang Y., Willingham S.B., Ye Z., Zimmermann A.G., and Ting J.P.. 2007. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. USA. 104:8041–8046. 10.1073/pnas.0611496104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui A.A., Ong W.Y., and Horrocks L.A.. 2006. Inhibitors of brain phospholipase A2 activity: their neuropharmacological effects and therapeutic importance for the treatment of neurologic disorders. Pharmacol. Rev. 58:591–620. 10.1124/pr.58.3.7 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., and Alnemri E.S.. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 458:509–513. 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. . 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Franchi L., Muñoz-Planillo R., Reimer T., Eigenbrod T., and Núñez G.. 2010. Inflammasomes as microbial sensors. Eur. J. Immunol. 40:611–615. 10.1002/eji.200940180 [DOI] [PubMed] [Google Scholar]
- Franco-Pons N., Torrente M., Colomina M.T., and Vilella E.. 2007. Behavioral deficits in the cuprizone-induced murine model of demyelination/remyelination. Toxicol. Lett. 169:205–213. 10.1016/j.toxlet.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Freeman L.C., and Ting J.P.. 2016. The pathogenic role of the inflammasome in neurodegenerative diseases. J. Neurochem. 136:29–38. 10.1111/jnc.13217 [DOI] [PubMed] [Google Scholar]
- Geddes B.J., Wang L., Huang W.J., Lavellee M., Manji G.A., Brown M., Jurman M., Cao J., Morgenstern J., Merriam S., et al. . 2001. Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem. Biophys. Res. Commun. 284:77–82. 10.1006/bbrc.2001.4928 [DOI] [PubMed] [Google Scholar]
- Guo H., Callaway J.B., and Ting J.P.. 2015. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21:677–687. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halff E.F., Diebolder C.A., Versteeg M., Schouten A., Brondijk T.H., and Huizinga E.G.. 2012. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 287:38460–38472. 10.1074/jbc.M112.393512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A., Hornung V., Petzold G.C., Stewart C.R., Monks B.G., Reinheckel T., Fitzgerald K.A., Latz E., Moore K.J., and Golenbock D.T.. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9:857–865. 10.1038/ni.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch U.K., and Kettenmann H.. 2007. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 10:1387–1394. 10.1038/nn1997 [DOI] [PubMed] [Google Scholar]
- Heneka M.T., O’Banion M.K., Terwel D., and Kummer M.P.. 2010. Neuroinflammatory processes in Alzheimer’s disease. J. Neural Transm. (Vienna). 117:919–947. 10.1007/s00702-010-0438-z [DOI] [PubMed] [Google Scholar]
- Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C., et al. . 2013. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 493:674–678. 10.1038/nature11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath M.M., Saito Y., Knapp G.W., Ting J.P., Suzuki K., and Matsushima G.K.. 1998. Microglial/macrophage accumulation during cuprizone-induced demyelination in C57BL/6 mice. J. Neuroimmunol. 92:38–49. 10.1016/S0165-5728(98)00168-4 [DOI] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., and Fitzgerald K.A.. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 458:514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Booth C.J., Strowig T., Jin C., Eisenbarth S.C., and Flavell R.A.. 2010. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc. Natl. Acad. Sci. USA. 107:21635–21640. 10.1073/pnas.1016814108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Yamaguchi K., Tanaka T., Mizuno Y., Hijikata A., Ohara O., Takada H., Kusuhara K., and Hara T.. 2010. Unique activation status of peripheral blood mononuclear cells at acute phase of Kawasaki disease. Clin. Exp. Immunol. 160:246–255. 10.1111/j.1365-2249.2009.04073.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S., Srivastava S.Y., Brickey W.J., Iocca H., Toews A., Morrison J.P., Chen V.S., Gris D., Matsushima G.K., and Ting J.P.. 2010. The inflammasome sensor, NLRP3, regulates CNS inflammation and demyelination via caspase-1 and interleukin-18. J. Neurosci. 30:15811–15820. 10.1523/JNEUROSCI.4088-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabarowski J.H., Xu Y., and Witte O.N.. 2002. Lysophosphatidylcholine as a ligand for immunoregulation. Biochem. Pharmacol. 64:161–167. 10.1016/S0006-2952(02)01179-6 [DOI] [PubMed] [Google Scholar]
- Kanneganti T.D., Ozören N., Body-Malapel M., Amer A., Park J.H., Franchi L., Whitfield J., Barchet W., Colonna M., Vandenabeele P., et al. . 2006. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 440:233–236. 10.1038/nature04517 [DOI] [PubMed] [Google Scholar]
- Keller M., Rüegg A., Werner S., and Beer H.D.. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 132:818–831. 10.1016/j.cell.2007.12.040 [DOI] [PubMed] [Google Scholar]
- Khare S., Dorfleutner A., Bryan N.B., Yun C., Radian A.D., de Almeida L., Rojanasakul Y., and Stehlik C.. 2012. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 36:464–476. 10.1016/j.immuni.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigerl K.A., de Rivero Vaccari J.P., Dietrich W.D., Popovich P.G., and Keane R.W.. 2014. Pattern recognition receptors and central nervous system repair. Exp. Neurol. 258:5–16. 10.1016/j.expneurol.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb R., Phan L., Borcherding N., Liu Y., Yuan F., Janowski A.M., Xie Q., Markan K.R., Li W., Potthoff M.J., et al. . 2016. Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat. Commun. 7:13007 10.1038/ncomms13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., and Ye R.D.. 2003. The lysophospholipid receptor G2A activates a specific combination of G proteins and promotes apoptosis. J. Biol. Chem. 278:14379–14386. 10.1074/jbc.M209101200 [DOI] [PubMed] [Google Scholar]
- Liu L., and Chan C.. 2014. IPAF inflammasome is involved in interleukin-1β production from astrocytes, induced by palmitate; implications for Alzheimer’s Disease. Neurobiol. Aging. 35:309–321. 10.1016/j.neurobiolaging.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Belkadi A., Darnall L., Hu T., Drescher C., Cotleur A.C., Padovani-Claudio D., He T., Choi K., Lane T.E., et al. . 2010. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: relevance to multiple sclerosis. Nat. Neurosci. 13:319–326. 10.1038/nn.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Boone D.L., and Lodolce J.P.. 2000. The pleiotropic functions of interleukin 15. J. Exp. Med. 191:753–756. 10.1084/jem.191.5.753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso F., Nothnagel M., Parwez Q., Petrasch-Parwez E., Bechara F.G., Epplen J.T., and Hoffjan S.. 2007. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp. Dermatol. 16:692–698. 10.1111/j.1600-0625.2007.00589.x [DOI] [PubMed] [Google Scholar]
- Man S.M., Hopkins L.J., Nugent E., Cox S., Glück I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., and Bryant C.E.. 2014. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA. 111:7403–7408. 10.1073/pnas.1402911111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Mariathasan S., Weiss D.S., Newton K., McBride J., O’Rourke K., Roose-Girma M., Lee W.P., Weinrauch Y., Monack D.M., and Dixit V.M.. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232. 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J.. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 440:237–241. 10.1038/nature04516 [DOI] [PubMed] [Google Scholar]
- Matsushima G.K., and Morell P.. 2001. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 11:107–116. 10.1111/j.1750-3639.2001.tb00385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.D., and de Vellis J.. 1980. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85:890–902. 10.1083/jcb.85.3.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., and Aderem A.. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., Krencik R., Ullian E.M., Tsai H.-H., Deneen B., Richardson W.D., Barres B.A., and Rowitch D.H.. 2012. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 26:891–907. 10.1101/gad.188326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I., and Neumann H.. 2009. Microglial clearance function in health and disease. Neuroscience. 158:1030–1038. 10.1016/j.neuroscience.2008.06.046 [DOI] [PubMed] [Google Scholar]
- Plant S.R., Iocca H.A., Wang Y., Thrash J.C., O’Connor B.P., Arnett H.A., Fu Y.X., Carson M.J., and Ting J.P.. 2007. Lymphotoxin beta receptor (LtβR): dual roles in demyelination and remyelination and successful therapeutic intervention using LtβR-Ig protein. J. Neurosci. 27:7429–7437. 10.1523/JNEUROSCI.1307-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet J.L., Srinivasula S.M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E.S.. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309–28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Newton K., Maltzman A., Izrael-Tomasevic A., Arnott D., and Dixit V.M.. 2016. NLRP3 recruitment by NLRC4 during Salmonella infection. J. Exp. Med. 213:877–885. 10.1084/jem.20132234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E., et al. . 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikitake Y., Hirata K., Yamashita T., Iwai K., Kobayashi S., Itoh H., Ozaki M., Ejiri J., Shiomi M., Inoue N., et al. . 2002. Expression of G2A, a receptor for lysophosphatidylcholine, by macrophages in murine, rabbit, and human atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 22:2049–2053. 10.1161/01.ATV.0000040598.18570.54 [DOI] [PubMed] [Google Scholar]
- Rivest S. 2009. Regulation of innate immune responses in the brain. Nat. Rev. Immunol. 9:429–439. 10.1038/nri2565 [DOI] [PubMed] [Google Scholar]
- Rock K.L., Latz E., Ontiveros F., and Kono H.. 2010. The sterile inflammatory response. Annu. Rev. Immunol. 28:321–342. 10.1146/annurev-immunol-030409-101311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T., Lehmann F., Rückert B., and Eder C.. 2004. Physiological mechanisms of lysophosphatidylcholine-induced de-ramification of murine microglia. J. Physiol. 557:105–120. 10.1113/jphysiol.2004.060632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Yeretssian G., Doiron K., Hussain S.N., and Saleh M.. 2007. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 282:36321–36329. 10.1074/jbc.M708182200 [DOI] [PubMed] [Google Scholar]
- Sheikh A.M., and Nagai A.. 2011. Lysophosphatidylcholine modulates fibril formation of amyloid beta peptide. FEBS J. 278:634–642. 10.1111/j.1742-4658.2010.07984.x [DOI] [PubMed] [Google Scholar]
- Shikishima K., Mizuno A., Kawai K., and Matsuzaki H.. 1985. Focal experimental demyelination in monkey optic nerve by lysophosphatidylcholine. Jpn. J. Ophthalmol. 29:429–433. [PubMed] [Google Scholar]
- Stock C., Schilling T., Schwab A., and Eder C.. 2006. Lysophosphatidylcholine stimulates IL-1β release from microglia via a P2X7 receptor-independent mechanism. J. Immunol. 177:8560–8568. 10.4049/jimmunol.177.12.8560 [DOI] [PubMed] [Google Scholar]
- Strowig T., Henao-Mejia J., Elinav E., and Flavell R.. 2012. Inflammasomes in health and disease. Nature. 481:278–286. 10.1038/nature10759 [DOI] [PubMed] [Google Scholar]
- Stüve O., Youssef S., Slavin A.J., King C.L., Patarroyo J.C., Hirschberg D.L., Brickey W.J., Soos J.M., Piskurich J.F., Chapman H.A., and Zamvil S.S.. 2002. The role of the MHC class II transactivator in class II expression and antigen presentation by astrocytes and in susceptibility to central nervous system autoimmune disease. J. Immunol. 169:6720–6732. 10.4049/jimmunol.169.12.6720 [DOI] [PubMed] [Google Scholar]
- Sutterwala F.S., Ogura Y., Szczepanik M., Lara-Tejero M., Lichtenberger G.S., Grant E.P., Bertin J., Coyle A.J., Galán J.E., Askenase P.W., and Flavell R.A.. 2006. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 24:317–327. 10.1016/j.immuni.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., and Flavell R.A.. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245. 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., and Nuñez G.. 2007. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3:e111 10.1371/journal.ppat.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J.P., and Davis B.K.. 2005. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu. Rev. Immunol. 23:387–414. 10.1146/annurev.immunol.23.021704.115616 [DOI] [PubMed] [Google Scholar]
- Triantafilou K., Kar S., van Kuppeveld F.J., and Triantafilou M.. 2013. Rhinovirus-induced calcium flux triggers NLRP3 and NLRC5 activation in bronchial cells. Am. J. Respir. Cell Mol. Biol. 49:923–934. 10.1165/rcmb.2013-0032OC [DOI] [PubMed] [Google Scholar]
- Vereyken E.J., Fluitsma D.M., Bolijn M.J., Dijkstra C.D., and Teunissen C.E.. 2009. An in vitro model for de- and remyelination using lysophosphatidyl choline in rodent whole brain spheroid cultures. Glia. 57:1326–1340. 10.1002/glia.20852 [DOI] [PubMed] [Google Scholar]
- Vladimer G.I., Weng D., Paquette S.W., Vanaja S.K., Rathinam V.A., Aune M.H., Conlon J.E., Burbage J.J., Proulx M.K., Liu Q., et al. . 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 37:96–107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman S.G., Kocsis J.D., and Nitta K.C.. 1979. Lysophosphatidyl choline-induced focal demyelination in the rabbit corpus callosum. Light-microscopic observations. J. Neurol. Sci. 44:45–53. 10.1016/0022-510X(79)90221-1 [DOI] [PubMed] [Google Scholar]
- Wu J., Yoo S., Wilcock D., Lytle J.M., Leung P.Y., Colton C.A., and Wrathall J.R.. 2010. Interaction of NG2+ glial progenitors and microglia/macrophages from the injured spinal cord. Glia. 58:410–422. 10.1002/glia.20932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T. 2006. Inflammation in Alzheimer disease: driving force, bystander or beneficial response?. Nat. Med. 12:1005–1015. 10.1038/nm1484 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. . 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.