Abstract
Activation of the RAS/MAPK pathway is critical in melanoma. Melanoma can be grouped into four molecular subtypes based on their main genetic driver: BRAF-mutant, NRAS-mutant, NF1-mutant, and triple wild-type tumors. The NF1 protein, neurofibromin 1, negatively regulates RAS proteins through GTPase activity. Germline mutations in NF1 cause neurofibromatosis type I, a common genetic tumor syndrome caused by dysregulation of the RAS/MAPK pathway, i.e. RASopathy. Melanomas with NF1 mutations typically occur on chronically sun-exposed skin or in older individuals, show a high mutation burden, and are wild-type for BRAF and NRAS. Additionally, NF1 mutations characterize certain clinicopathologic melanoma subtypes, specifically desmoplastic melanoma. This review discusses the current knowledge of the NF1 gene and neurofibromin 1 in neurofibromatosis type I and in melanoma.
Introduction
In the era of personalized medicine and targeted therapies, molecular subtyping of melanoma is replacing the traditional classification based on clinicopathologic features. Based on exome and genome sequencing studies, cutaneous melanoma can be divided into four distinct molecular subtypes: i) B-Raf proto-oncogene, serine/threonine kinase (BRAF) -mutant, ii) neuroblastoma RAS viral oncogene homolog (NRAS) -mutant, iii) neurofibromin 1 (NF1) -mutant, and iv) BRAF/NRAS/NF1 wild-type (WT) tumors (triple wild-type tumors) 1. This review discusses the current knowledge of the NF1 gene and neurofibromin signaling pathways in neurofibromatosis type I and in melanoma, including prospects for therapeutic targeting.
Neurofibromatosis type I
Neurofibromatosis type I (NF1; Online Mendelian Inheritance in Man (OMIM) database 162200) has been recognized for over a century, with the first report by von Recklinghausen 2 and a thorough characterization by Crowe and colleagues in 1956 3. It is a common genetic syndrome with an incidence of approximately 1 in 3000 live births caused by mutations in the NF1 gene 4–6 predisposing to multiple tumors, most commonly derived from the neural crest. NF1 is an autosomal dominant disorder with a high rate, approximately 50%, of new mutations.
Mutations in the NF1 gene were identified in 1990 by Wallace and coworkers4, who demonstrated translocations and an insertion of this gene in three NF1 patients. Since then, hundreds of mutations have been reported, with over 80% of patients having a nonsense mutation, an insertion, or a deletion predicted to lead to a truncated protein product 7.
As is characteristic for genetic syndrome caused by a mutation in a tumor suppressor gene, tumors in NF1 patients show loss of heterozygosity (LOH), i.e. somatic mutation inactivating the second, remaining allele 8,9.
Pigmentary disorders and neuroectodermal-derived tumors in NF1
The clinical features of NF1 are numerous and affect multiple organ systems. The most common clinical findings involve the skin, as exemplified by the diagnostic criteria of NF1 (Table 1a). These include café-au-lait-macules, axillary and inguinal freckling, neurofibromas, and plexiform neurofibromas, present in 90%, 80%, 60–90%, and 25% of patients, respectively.
Table 1a.
Diagnostic criteria of NF1.
| Clinical diagnosis based on presence of two of the following: |
|---|
| 1. Six or more café-au-lait macules over 5 mm in diameter in prepubertal individuals and over 15 mm in greatest diameter in postpubertal individuals. |
| 2. Two or more neurofibromas of any type or one plexiform neurofibroma. |
| 3. Freckling in the axillary or inguinal regions. |
| 4. Two or more Lisch nodules (iris hamartomas). |
| 5. Optic glioma. |
| 6. A distinctive osseous lesion such as sphenoid dysplasia or thinning of long bone cortex, with or without pseudarthrosis. |
| 7. First-degree relative (parent, sibling, or offspring) with NF-1 by the above criteria. |
Café-au-lait macule (CALM)
Café-au-lait macules (CALMs) are tan to brown sharply demarcated uniformly pigmented macules or patches typically on the trunk and extremities. In NF1, CALMs are the earliest manifestation, typically appearing at 0–3 years of age. One to a couple of CALMs are common in the general population, therefore, six or more CALMs are required to fulfill one diagnostic criteria of NF1 (Table 1a). Histologically, CALMs show hyperpigmentation of the basal layer keratinocytes of the epidermis with normal to slightly increased number of melanocytes. In CALMs of NF1 patients, giant melanin granules (macromelanosomes) are present in melanocytes 10.
Axillary or inguinal freckling
Axillary or inguinal freckling is characterized by multiple tan to brown sharply demarcated uniformly pigmented macules in the axillae or the groin. They typically develop in early childhood. They are similar to CALMs clinically and histologically except smaller in size.
Lisch nodule
Lisch nodules are melanocytic hamartomas of the eye 11. They appear as well-defined, dome-shaped papules on the iris and are clear to yellow or brown. They are the most common manifestation of NF1, present in nearly 100% of patients. Histologically, they show an aggregation of melanocytes, admixed with spindle cells and mast cells. Lisch nodules are not known to result in any ophthalmologic complications.
Neurofibroma
Neurofibromas are benign tumors of nerve sheath origin. Clinically, they present as soft skin colored to pink papules or nodules on the trunk, extremities, or head/neck. Though common in the general population as solitary lesions, their number in NF1 patients can vary from a few to thousands. They typically develop around puberty. Histologically, they are composed of loosely arranged wavy spindled Schwann-like cells, nerve fibers, perineurial cells, and fibroblasts, and infiltrated by mast cells in the dermis or subcutis. Neurofibromas are benign but have a significant impact on quality-of-life mainly due to their appearance 12. A population of stem/progenitor cells that resides in the dermis termed skin-derived precursors are suggested as the cells of origin of neurofibromas 13. In addition, non-neoplastic cells in the tumor microenvironment are proposed to play a role in neurofibroma tumorigenesis 13.
Plexiform neurofibroma
Plexiform neurofibromas are much less frequent than neurofibromas, but are considered pathognomonic of NF1. Additionally, they may serve as precursors for malignant peripheral nerve sheath tumor (MPNST), an aggressive sarcoma typically arising in a pre-existing plexiform neurofibroma 14. Plexiform neurofibromas usually become clinically apparent by 4–5 years of age. Clinically they present as tender, firm subcutaneous nodules often with overlying hyperpigmentation and hypertrichosis. They can infiltrate underlying tissues, cause soft tissue and bony hypertrophy, distortion or compression of adjacent structures, and neurologic deficits. Histologically, they consist of large thick nerves or nerve fibers within a background of neurofibroma.
Optic glioma
Another common tumor in NF1 is optic glioma. Optic gliomas in NF1 are mainly grade I pilocytic astrocytomas, defined as benign tumors that have a favorable prognosis unlike pilocytic astrocytomas in patients without NF1 6.
Malignant peripheral nerve sheath tumor (MPNST)
MPNSTs occur in 3–15% of NF1 patients, with a peak incidence in young adults. Rapid growth, increased pain or a new neurologic deficit may be a sign of malignant transformation of a pre-existing plexiform neurofibroma in NF1 patients. Histologically, the tumor shows tight wavy or interlacing bundles of spindle cells with cellularity and mitoses determining tumor grade. As MPNSTs may resemble desmoplastic melanoma, exclusion of an associated melanocytic precursor lesion (intra-epidermal or intrafollicular melanoma in situ or melanocytic nevus) is required for the diagnosis of MPNST.
Other tumors in NF1
The overall risk of cancer is increased in NF1. Based on epidemiologic studies, cancer incidence in NF1 is approximately 4-fold higher than in general population 15. Other reported neoplasms include juvenile myelomonocytic leukemia, pheochromocytoma, central nervous system tumors other than optic glioma, rhabdomyosarcoma, duodenal carcinoid, somatostatinoma, parathyroid adenoma, and gastrointestinal stromal tumor (Table 1b). Additionally, the risk of breast cancer is 5-fold. Epidemiologic studies have also shown an increased risk of cancer of the esophagus, stomach, colon, liver, lung, bone, thyroid, and ovary, non-Hodgkin’s lymphoma, and chronic myeloid leukemia 16.
Table 1b.
Most common tumor types in NF1.
| Tumor type |
|---|
| Optic glioma (in 10–15%) |
| Malignant peripheral nerve sheath tumor (in 3–15%) |
| Pheochromocytoma |
| Juvenile myelomonocytic leukemia |
| CNS tumors other than optic gliomas |
| Rhabdomyosarcoma (especially of the genitourinary tract) |
| Duodenal carcinoid |
| Somatostatinoma |
| Parathyroid adenoma |
| Gastrointestinal stromal tumors (GIST) |
| Breast cancer (~5-fold increased risk in women <50 years of age) |
Although somatic mutations in NF1 are present in melanoma (Tables 2, 3), the risk of melanoma in individuals with NF1 is only minimally increased, to approximately 3.6-fold 16. Based on a study of 11 NF1 patients with melanoma, a female preponderance, a higher thickness, and a frequent association with a second neoplasia were identified 17. Additionally, a desmoplastic melanoma, a subtype of melanoma sharing some morphologic features with MPNST, has been reported in one individual with NF1 18.
Table 2.
NF1 mutations in cancer.
| Cancer type | Number of tumors with NF1 mutations | % of tumors with NF1 mutations | Reference |
|---|---|---|---|
| Melanoma | 28 of 213 | 13 | 90 |
| 16 of 121 | 13 | 92 | |
| 3 of 25 | 12 | 93 | |
| 9 of 20 | 45 | 97 | |
| 46 of 333 | 14 | 1 | |
| 6 of 34 | 18 | 94 | |
| 14 of 15 | 93 | 96 | |
| 13 of 91 | 14 | 91 | |
| MPNST | 6 of 15 | 40 | 71 |
| Acute lymphoblastic leukemia | 9 of 40 | 23 | 73 |
| Glioblastoma | 31 of 281 | 11 | 75 |
| Non-small cell lung cancer | 137 of 1144 | 12 | 76 |
| Lung squamous cell carcinoma | 21 of 178 | 12 | 77 |
| Lung adenocarcinoma | 29 of 230 | 13 | 78 |
| Bladder urothelial carcinoma | 7 of 50 | 14 | 79 |
| 12 of 109 | 11 | 80 | |
| 13 of 127 | 10 | 81 | |
| Uterine carcinosarcoma | 3 of 22 | 14 | 82 |
| Uterine endometrial carcinoma | 27 of 240 | 11 | 83 |
| Ovarian adenocarcinoma | 37 of 316 | 12 | 84 |
| Pancreatic carcinoma | 12 of 109 | 11 | 85 |
| Metastatic cutaneous squamous cell carcinoma | 3 of 29 | 10 | 86 |
| Gastric adenocarcinoma | 29 of 287 | 10 | 87 |
| 3 of 30 | 10 | 88 |
Table 3.
NF1 mutations in melanoma.
| Reference | Material | Sequencing method | Number of tumors with NF1 mutation | % of tumors with NF1 mutation | Characteristics of NF1-mutant melanomas | Conclusions |
|---|---|---|---|---|---|---|
| 93 | Not stated | WGS1 | 3 of 25 | 12 | Revealed genomic evidence of ultraviolet pathogenesis and discovered a new recurrently mutated gene in melanoma | |
| 92 | Fresh-frozen or short-term cultures (n=121; 15 primary tumors, 30 metastatic tumors, 76 short-term cultures) | WES2 | 16 of 121; 5 of 21 (BRAF/BRAS WT) | 13; 25 (BRAF/NRAS WT) | Development of framework to assess driver and passenger mutations and identification of six novel melanoma genes (PPP6C, RAC1, SNX31, TACC1, STK19, ARID2); provided evidence for direct mutagenic role of UV radiation | |
| 91 | Fresh-frozen or short-term cultures (n=91) | WES | 13 of 91 | 14 | Classification of melanoma into those with high, medium and low mutation count, likely corresponding to chronically exposed, intermittently sun-exposed, and sun-protected lesions, respectively, and identification of new cancer genes such as PPP6C and RAC1 | |
| 94 | Fresh-frozen (n=34) | WES | 6 of 34; 5 of 10 (BRAF/NRAS WT) | 18; 50 (BRAF/NRAS WT) | BRAF/NRAS WT melanomas show extensive UV damage and high mutation load and may require different treatment strategies including combination therapies | |
| 95 | Cell lines | 6 of 61 (BRAF/RAS WT); 1 of 10 (BRAF V600E) | 10 (BRAF/RAS WT); 10 (BRAF V600E) | Loss of NF1 is common in melanoma and associated with RAS activation, MEK-dependence, and resistance to RAF inhibition | ||
| 98 | FFPE3 (desmoplastic melanoma with sarcomatoid differentiation; n=1) | Targeted 230 gene panel | 1 of 1 | 100 | Report of an unusual desmoplastic melanoma with an undifferentiated sarcomatoid nodule, both components sharing a mutational heritage including mutations in NF1 gene | |
| 1 | Fresh-frozen (n=333; 67 primary tumors and 266 metastatic tumors) | WES | 46 of 333 | 14 | Occur in older individuals, show a higher mutation burden | Classification of melanoma into four genomic subtypes: mutant BRAF, mutant NRAS, mutant NF1, and Triple-wild-type tumors; no significant outcome correlation based on genomic classification; however, immune gene expression associated with histologic lymphocytic infiltration associated with improved survival |
| 90 | Fresh-frozen or short-term cultures (n=213) | WES | 28 of 213 | 13 | Significantly more somatic mutations; occurred in older patients; associated with similar overall survival; harbor co-mutations in RASopathy genes RASA2, PTPN11, SOS1, RAF1, SPRED1; 60% of NF1-mutant melanoma cell lines sensitive and 40% resistant to MEK inhibitor selumetinib in vitro | NF1 is a key tumor suppressor lost in melanomas, and that concurrent RASopathy gene mutations may enhance its role in melanomagenesis |
| 97 | Fresh-frozen (desmoplastic melanomas; discovery set; n=20); FFPE (desmoplastic melanomas; validation set; n=42) (total n=62) | WES, WGS (discovery set); targeted 293 gene panel (validation set) | 9 of 20 (discovery set); 34 of 62 (total) | 45 (discovery set); 54 (total) | Desmoplastic melanomas show high mutation burden, UV mutation signature, diverse activation of the MAPK pathway affecting NF1, CBL, ERBB2, MAP2K1, MAP3K1, BRAF, EGFR, PTPN11, MET, RAC1, SOS2, NRAS, PIK3CA | |
| 96 | FFPE (desmoplastic melanomas; n=15); FFPE (non-desmoplastic melanomas; n=20; 18 metastatic tumors, 2 primary tumors) | Targeted 341 gene panel desmoplastic melanomas) | 14 of 15 (desmoplastic melanomas); 4 of 20 (non-desmoplastic melanomas) | 93 (desmoplastic melanomas); 20 (non-associated MIS in 12 of 14; depth 1.5–18 mm; pure (n=6), mixed (n=8) | Median age 71.5 (range 24–82); sites scalp (n=5), face (n=4), arm/shoulder (n=3), neck (n=1), chest (n=1); biology of this melanoma type | High frequency of NF1 mutations in desmoplastic melanomas suggests an important role for NF1 in the |
WGS= whole genome sequencing;
WES= whole exome sequencing;
FFPE= formalin-fixed, paraffin-embedded
The NF1 gene
The NF1 gene was mapped to chromosome 17 through linkage analyses in NF1 kindreds 19–23. Genetic studies were complemented by more refined physical mapping 24. The complete cDNA sequence of the NF1 gene 25 and the structure of the large gene consisting of over 50 exons spanning 300 kb of chromosome 17 26 were later reported.
The NF1 gene at 17q11.2 is now known to consist of 60 exons and to generate several alternatively spliced isoforms. Although identification of mutations in NF1 has been challenging due to the very large size and complexity of the gene, lack of mutational hot spots, and the presence of pseudogenes, hundreds of mutations have been reported 7,27. Through advances in next-generation sequencing technologies, faster and more robust techniques for sequencing of NF1 have been established 28,29.
Most of the mutations in NF1 are loss-of-function mutations 4,5,7,30. These include missense, nonsense, frameshift, and splice site mutations, insertions, deletions, large deletions including NF1 gene and possibly other flanking genes, and translocations 27. Therefore, the gene has been classified as a tumor suppressor. Consistent with a role of a tumor suppressor gene, loss of heterozygosity (LOH) or “second-hit” somatic mutations in the inherited wild-type allele are present in many tumor types in NF1 patients, including neurofibromas, malignant peripheral nerve sheath tumors, pheochromocytomas, and myeloid tumors 31–37.
Wide phenotypic variability exists even among patients within the same family, against a definite genotype-phenotype correlation, although with some exceptions 27,38. A 3-base pair in-frame deletion is associated with the lack of cutaneous neurofibromas, shown in a study of 21 unrelated probands 38. A microdeletion of NF1, the single most common mutation in individuals with NF1, is associated with the development of a large number of neurofibromas at an earlier age and a probable increased risk of MPNSTs 39. Additionally, an association with splice site mutations and the presence of tumors, especially central nervous system gliomas and MPNSTs, has been reported 40.
The phenotypic variability in NF1 could be explained by modifier genes, including protein-coding sequences, microRNA and long non-coding RNA genes, that may affect the NF1 phenotype 41. In support of this, cohort studies have demonstrated that the clinical features tend to be similar in close relatives compared with distant relatives. Proof of modifier genes has been sought from mouse models of NF1, candidate gene studies, or whole genome approaches (reviewed in 41). An array comparative genomic hybridization (aCGH) study of plexiform neurofibromas from NF1 patients showed a recurrent somatic 9p21.3 deletion involving the antisense noncoding RNA in the INK4 locus (ANRIL) and an association between a germline SNP rs2151280 resulting in reduced ANRIL transcript levels and the number of plexiform neurofibromas, suggesting that ANRIL expression mediated plexiform neurofibromas susceptibility 42. Polymorphisms in the adenylate cyclase 8 (ADCY8) gene correlated with glioma risk in NF1 patients in a sex-specific manner, elevating the risk in females and reducing it in males, also strengthening the evidence of a role of cAMP in gliomagenesis in NF1 43. Further studies utilizing genome-wide approaches including genome-wide sequencing appear promising for identification of modifier genes in NF1.
The NF1 protein
NF1 encodes neurofibromin 1, a large protein consisting of over 2800 amino acids 7,44. Neurofibromin 1 contains multiple functional domains. Early studies on the amino acid sequence of neurofibromin 1 demonstrated a 360-residue region with significant similarity to the catalytic domain of GTPase-activating protein (GAP) and a role in the control of cell growth by interaction with RAS was suggested 45.
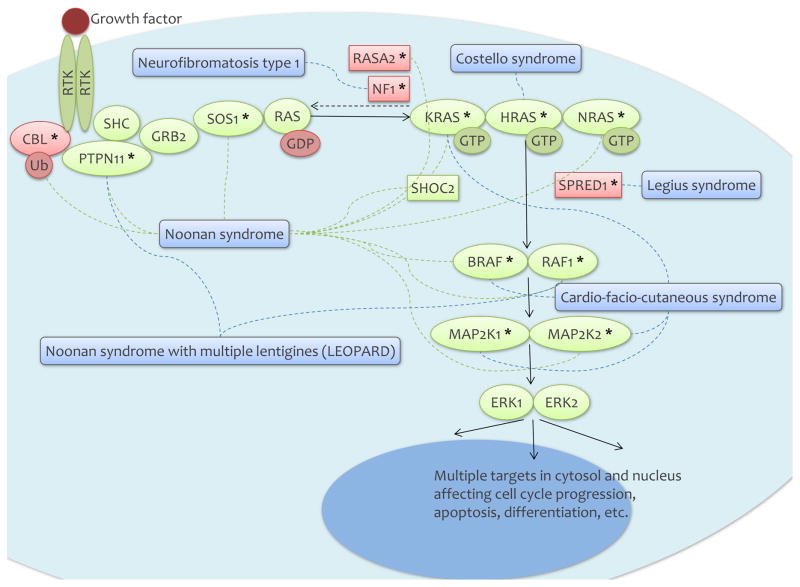
The GTPase-activating protein -related domain is the best studied domain of neurofibromin 1 and is know to negatively regulate RAS by converting the active RAS-guanosine triphosphate (RAS-GTP) to the inactive RAS-guanosine diphosphate (RAS-GDP) thereby inhibiting downstream RAS signaling (Figure 1). In support of the critical role of RAS-GTPase function of neurofibromin 1 in NF1 pathogenesis, some of the missense mutations in NF1 patients selectively abolish RAS-GTPase activity without affecting the protein structure and levels 46. A pleckstrin homology domain interacts with the cytoskeleton and membrane structures. The C-terminal domain can be phosphorylated by protein kinase A (PKA) inhibiting neurofibromin 1 44.
Figure 1.
The RAS/MAPK pathway and RASopathies.
The upstream regulation of neurofibromin 1 includes the granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor, the tyrosine kinase cKIT receptor, the endothelin receptor B (EDNRB), and the tyrosine kinase anaplastic lymphoma kinase (ALK) receptor (6 and references therein). The most studied downstream pathways regulated by neurofibromin 1 are the RAS/mitogen-activated protein kinase (MAPK) (Figure 1) and phosphoinositide 3-kinase (PI3K)/mechanistic target of rapamycin (mTOR) signaling pathways. In addition, neurofibromin 1 is involved in cyclic adenosine monophosphate (cAMP) signaling. Targeting these pathways offer important avenues for therapies (see “Prospects for targeted therapies”).
The RAS/MAPK pathway
The RAS/MAPK pathway plays a critical role in normal development through regulation of cell growth, differentiation, and senescence. RAS genes include NRAS, HRAS, and KRAS, a multigene family encoding guanosine nucleotide bound GTPases. The signaling pathway starts with the interaction between a ligand and a cell surface receptor, either a G-protein coupled receptor or a receptor tyrosine kinase (Figure 1). This leads to activation of RAS by conversion of RAS-GDP into RAS-GTP. Notably, neurofibromin 1 negatively regulates this step through GTPase activating protein activity converting the active RAS-GTP to the inactive RAS-GDP. Active RAS interacts with many downstream mediators, most importantly by binding to the RAS-binding domain of BRAF or Raf-1 proto-oncogene, serine/threonine kinase (RAF1). This results in homo and heterodimerization and activation of RAF that then activates the MAP kinases mitogen-activated protein kinase kinase 1 (MAP2K1) and mitogen-activated protein kinase kinase 2 (MAP2K2) via phosphorylation. These in turn phosphorylate and activate mitogen-activated protein kinase 3 (MAPK3 or ERK1) and/or mitogen-activated protein kinase 1 (MAPK1 or ERK2), the effectors of the pathway that control cell cycle progression, differentiation, and growth.
Dysregulation of the RAS/MAPK pathway is one of the key events in oncogenesis, including melanoma. Most melanomas show activation of the RAS/MAPK pathway and the key genes mutated in melanoma, BRAF, NRAS, NF1, C-KIT, G protein subunit alpha q (GNAQ), and G protein subunit alpha 11 (GNA11), all participate in the RAS/MAPK pathway 47.
RASopathies
RASopathies are genetic syndromes caused by germline mutations in genes encoding components or regulators of the RAS/MAPK pathway 48,49. Given the key role of the RAS/MAPK pathway in normal development, mutations in RAS/MAPK pathway genes in these patients result in developmental abnormalities in multiple organ systems as well as predisposition to cancers. Due to the shared mechanism of RAS/MAPK pathway dysregulation, the syndromes share many clinical features including cutaneous, musculoskeletal, and ocular abnormalities, craniofacial dysmorphology, cardiac malformations, neurocognitive impairment, and increased cancer risk. In addition to NF1, RASopathies include Noonan syndrome caused by activating mutations in protein tyrosine phosphatase, non-receptor type 11 (PTPN11), SOS Ras/Rac guanine nucleotide exchange factor 1 (SOS1), RAF1, KRAS, NRAS, and SHOC2, leucine rich repeat scaffold protein (SHOC2), and inactivating mutations in Cbl proto-oncogene (CBL), Noonan syndrome with multiple lentigines (formerly LEOPARD syndrome) caused by activating mutations in PTPN11, Costello syndrome caused by activating mutations in HRAS, Cardiofacio-cutaneous syndrome caused by activating mutations in BRAF, MAP2K1, and MAP2K2, and Legius syndrome caused by inactivating mutations in sprouty related EVH1 domain containing 1 (SPRED1) (Figure 1). Most of these genes are somatically mutated in melanoma and specifically co-occur with NF1 mutations (see “NF1 mutations in melanoma”).
Cancer risk in these syndromes is increased 50. The most commonly reported cancers include neuroblastoma, acute lymphoblastic leukemia, low grade glioma, and rhabdomyosarcoma in Noonan syndrome and rhabdomyosarcoma, bladder cancer, and neuroblastoma in Costello syndrome. Cancers reported in Cardio-facio-cutaneous syndrome include acute lymphoblastic leukemia, non-Hodgkin lymphoma, hepatoblastoma and rhabdomyosarcoma. In Noonan syndrome with multiple lentigines (formerly LEOPARD syndrome) acute myeloid leukemia, acute lymphoblastic leukemia, neuroblastoma and one melanoma has been reported. In Legius syndrome one occurrence each of non-small cell lung cancer, Wilms tumor, tubular colon adenoma, acute myeloblastic anemia, tenosynovial giant cell tumor, breast cancer and dermoid tumor of the ovary has been reported 51–54.
The PI3K/mTOR, cAMP, and other pathways
The important role of the PI3K/mTOR pathway is supported by the activation of the pathway in NF1-mutant neurofibroma and MPNST cell lines and primary tumors 44,55–58. The cAMP pathway is deregulated in NF1-deficient tumors. In yeast, the NF1 homologues regulate Ras and cAMP signaling. Animal models expressing mutant Nf1, including mouse, Drosophila, and zebrafish, show deregulated cAMP levels in various cell types 59–61. cAMP levels are also altered in human NF1-associated tumors, including gliomas 43. There are many other lesser known neurofibromin 1-regulated effectors downstream of RAS, including afadin, adherens junction formation factor (AFDN), ral guanine nucleotide dissociation stimulator (RALGDS), T-cell lymphoma invasion and metastasis 1 (TIAM1), phospholipase C like 1 (PLCL1), and Ras and Rab interactor 1 (RIN1).
NF1 in the melanocyte lineage
Neurofibromin 1 is proposed to have fundamental functional differences in neural and non-neural tissues, and thus, lead to very different pathologies in different cell lineages 62. In a rat model system, neurofibromin 1 is broadly distributed during embryogenesis including neural crest migration, but postnatal expression is low or absent in non-neural tissues.
Some of the neural crest derivatives include Schwann cells and melanocytes. Schwann cells are glial cells that wrap around axons in the peripheral nervous system and express relatively high amounts of neurofibromin 1. Neurofibromin plays a role in Schwann cell differentiation 63 and Schwann cell-axonal interactions 64. Schwann cells appear vulnerable to defects in NF1, as exemplified by the many Schwann cell –containing tumors in NF1, including neurofibromas, plexiform neurofibromas, and malignant peripheral nerve sheath tumors.
Similar to neurofibromas and other NF1-associated tumors 36, biallelic inactivation of NF1 is observed in melanocytes of CALMs from individuals with NF1 65. The downstream effects of NF1 loss appear cell type dependent. The detailed molecular mechanisms linking loss of NF1 to hyperpigmentation and melanocyte function have been poorly characterized until the recent years (reviewed in 44).
Mice deficient of Nf1 lack pigmentary abnormalities. However, studies using Nf1+/− murine models show that Nf1 deficiency partially reverts a skin pigmentation defect present in Kit and Mitf mutant mice, demonstrating that neurofibromin 1 regulates signaling between Kit and melanogenesis associated transcription factor (Mitf) 66. The Nf1+/− melanocytes also display increased melanogenic gene expression and enhanced Kit-dependent and Kit-independent ERK activity. Additionally, mice with homozygous knock-out of Nf1 in melanocytes show hyperpigmented skin 67.
Primary human melanocytes are derived from the neural crest and grow extremely slowly in vitro. Therefore, recent protocols for human embryonic stem cell (hESC)- and induced pluripotent stem cell (iPSC)-based derivation of melanocytes have been established 68–70. Larribere and co-workers 69 reprogammed patient-derived NF1+/− fibroblasts into NF1+/−iPSCs. The NF1+/− iPSCs showed an active RAS signaling and differential expression of genes associated with the RAS/MAPK signaling pathway compared with WT iPSCs. Furthermore, NF1+/− iPSC-derived melanocytes displayed a senescence phenotype with a lower melanocyte number, altered cell morphology, and reduced proliferative index compared with WT iPSC-derived melanocytes. The authors concluded that NF1 haploinsufficiency through RAS activation promotes a senescence phenotype in the melanocyte lineage.
Allouche and co-workers 70 used NF1+/− hESC-derived melanocytes to study the molecular mechanisms of hyperpigmentation in NF1. NF1+/− hESC-derived melanocytes showed an increase in intracellular melanin content, a greater expression of melanogenic enzymes, and an increase in stage III/IV melanosomes compared with WT hESC-derived melanocytes, recapitulating the in vivo hyperpigmentation phenotype of NF1 in vitro. NF1+/− hESC-derived melanocytes displayed an increase in cAMP activity and ERK signaling, the two downstream pathways dysregulated in NF1. Both cAMP and ERK pathways were blocked successfully in vitro using a mitogen-activated protein kinase kinase (MEK) inhibitor or a PKA/cAMP inhibitor, holding promise for future therapeutic trials.
NF1 mutations in melanoma
NF1 mutations in cancer
Somatic mutations in NF1 are present in sporadic cancers. They are the most frequent in MPNSTs, the most common cancer type in individuals with NF1, being present in 40% of tumors 71. With the expansion of cancer genomics data, online portals offer valuable platforms to explore the available mutation data 72. According to these data, genetic alterations in NF1 were reported in total of 147 studies (http://www.cbioportal.org). Over twenty published studies included more than 15 specimens and showed NF1 mutations in more than 10% of samples (Table 2). Of cancer types other than melanoma, these included 40% of MPNST 71, 23% of acute lymphoblastic leukemia 73, 11–18% of glioblastoma 74,75, 12% of non-small cell lung cancer 76, 12% of lung squamous cell carcinoma 77, 13% of lung adenocarcinoma 78, 10–14% of bladder urothelial carcinoma 79–81, 14% of uterine carcinosarcoma 82, 11–12% of uterine endometrial carcinoma 83, 12% of ovarian serous cystadenocarcinoma 84, 11% of pancreatic carcinoma 85, 10% of metastatic cutaneous squamous cell carcinoma 86, and 10% of gastric adenocarcinoma 87,88.
NF1 mutations in melanoma
Before sequencing data were available, the NF1 locus showed frequent LOH in desmoplastic melanomas, a rare subtype of melanoma 89. Recent large-scale targeted sequencing, whole exome, and whole genome sequencing efforts have established NF1 as one of the key drivers of melanoma (Table 3) 1,90,91. Characteristically for a driver gene, NF1 shows a high frequency of non-silent exonic mutations and a low frequency of synonymous or intronic mutations, as well as being present in a considerable portion of studied melanoma specimens, approximately 12–18% of all melanomas (Table 3) 91–94(67). Mutations in NF1 are more common in melanomas occurring on chronically sun-exposed skin or in older patients 90,94, melanomas with higher mutation burden 90 or wild-type for BRAF and NRAS 92,94,95, and certain clinicopathologic melanoma subtypes, specifically desmoplastic melanoma 96–98.
NF1 mutations in desmoplastic melanoma
NF1 mutations are found in 45–93% of desmoplastic melanomas 96–98. This is not surprising, given the morphologic overlap between desmoplastic melanomas and MPNSTs, which commonly show NF1 mutations 99,100. Desmoplastic melanoma is a rare subtype of melanoma, with distinct clinical and histopathologic features. It typically occurs on chronically sun-exposed skin of elderly individuals. Histologically it shows fibroblast-like spindled melanocytes surrounded by abundant collagen fibers 101. Molecularly, a high mutation burden, UV mutation signature, and a diverse activation of the RAS/MAPK pathway characterize desmoplastic melanoma 96,97.
Interestingly, NF1 loss in melanoma is often associated with concurrent mutations in RASopathy genes 90,102. These include RAS p21 protein activator 2 (RASA2), PTPN11, SOS1, RAF1, and SPRED1, mutated in the germline of individuals with Noonan syndrome (RASA2, PTPN11, SOS1, RAF1), Noonan syndrome with multiple lentigines (PTPN11, RAF1), and Legius syndrome (SPRED1) (Figure 1). It is suggested that the co-occurring mutations may act synergistically in melanomas 102.
Prospects for targeted therapies
As the major consequence of NF1 mutation is the increased RAS/MAPK pathway signaling, therapeutic targeting of this pathway both for melanoma and for syndromic NF1-associated tumors is reasonable and supported by preclinical evidence. Additionally, inhibitors of the PI3K/mTOR and cAMP pathways show promise for targeting NF1-deficient tumors. The downstream impact of NF1 deficiency may be dependent on the cell type, and should be kept in mind in the development of targeted therapies 44.
Preclinical and clinical trials for treatment of NF1-associated tumors, MPNSTs, plexiform neurofibromas, and neurofibromas, include tyrosine kinase inhibitors (imatinib, dasatinib, sunitinib, nilotinib), MEK inhibitors (trametinib, selumetinib), and mTOR inhibitors (rapamycin, sirolimus, everolimus) 103–106. For treatment of melanoma, the U.S. Food and Drug Administration -approved agents affecting the RAS/MAPK pathway include MEK inhibitors trametinib and cobimetinib and the BRAF inhibitors vemurafenib and dabrafenib. Additionally, non-FDA approved inhibitors include RAF-inhibitors sorafenib, encorafenib, XL281, and R05126766, MEK inhibitors selumetinib and bimetinib, and ERK inhibitor ulixertinib, that have been tested mainly in phase I and II studies in melanoma patients 47,107–111. In preclinical models, pan-RAF inhibitors, MEK inhibitors, and ERK inhibitors have shown activity in NRAS-mutant tumors 112–114, but there are no reports to date on these agents specifically in patients with NF1-mutant melanomas.
NF1 may also play a role in driving resistance to RAF/MEK targeted therapies. NF1 loss is thought to mediate resistance to RAF and MEK inhibitors through sustained MAPK pathway activation. Based on murine models, Nf1 mutation suppresses Braf-induced senescence in melanocytes, promoting melanocyte proliferation and enhancing melanoma development. Nf1/Braf-mutant murine tumors are resistant to BRAF inhibitors but sensitive to combined inhibition of MAPK/ERK and mTOR pathways. In human melanoma cell lines, NF1 ablation decreases the sensitivity of melanoma to BRAF inhibitors. Based on data from clinical trials, mutations in NF1 are present in BRAF-mutant tumors intrinsically resistant to BRAF inhibitors as well as from patients showing resistance to BRAF inhibitors 115.
In conclusion, the rapidly increasing understanding of the role of NF1 mutations in neoplasia and neurofibromin 1 in various cellular signaling pathways will likely produce novel treatment approaches relevant for individuals with NF1 and for sporadic tumors with NF1 mutations.
Summary.
The NF1 gene is a tumor suppressor gene mutated in the germline of individuals with neurofibromatosis type 1 (NF1). NF1 is one of the genetic syndromes with mutations in the RAS/MAPK pathway, i.e. RASopathies.
NF1 is characterized by pigmented lesions of the skin and the eye, including café-au-lait macules, axillary freckling, and Lisch nodules. The most common tumors in NF1 include neurofibromas, plexiform neurofibromas, malignant peripheral nerve sheath tumors, and optic gliomas.
NF1 encodes neurofibromin 1 protein, which negatively regulates the RAS/MAPK pathway by converting active RAS-GTP to inactive RAS-GDP. Neurofibromin 1 also functions in the PI3K/mTOR and cAMP signaling.
Somatic mutations in NF1 are common in cancer including melanoma.
Melanomas characterized by NF1 mutations typically occur on chronically sun-exposed skin and in older individuals. They typically show a high mutation rate and UV signature mutations. Desmoplastic melanoma, a rare clinicopathologic subtype of melanoma, displays frequent mutations in NF1.
Targeting neurofibromin 1-regulated pathways offers potential therapeutic options for the treatment of NF1 and melanoma.
Footnotes
Disclosures: Dr. Kiuru’s involvement in this review article was in part supported by the National Cancer Institute, National Institutes of Health, through grant #K12CA138464. No other funding was provided to support this manuscript.
References
- 1.Cancer Genome Atlas Research N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Recklinghausen F. Ueber die multiplen Fibrome der Haut und ihre Beziehung zu den multiplen Neuromen. Hirschwald; 1882. [PubMed] [Google Scholar]
- 3.Crowe FWSW, Neel JV. A Clinical, Pathological and Genetic Study of Multiple Neurofibromatosis. Charles C Thomas; 1956. [Google Scholar]
- 4.Wallace MR, Marchuk DA, Andersen LB, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 5.Viskochil D, Buchberg AM, Xu G, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 6.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen MH, Harper PS, Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1) J Med Genet. 1996;33:2–17. doi: 10.1136/jmg.33.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skuse GR, Kosciolek BA, Rowley PT. Molecular genetic analysis of tumors in von Recklinghausen neurofibromatosis: loss of heterozygosity for chromosome 17. Genes Chromosomes Cancer. 1989;1:36–41. doi: 10.1002/gcc.2870010107. [DOI] [PubMed] [Google Scholar]
- 9.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimbow K, Szabo G, Fitzpatrick TB. Ultrastructure of giant pigment granules (macromelanosomes) in the cutaneous pigmented macules of neurofibromatosis. J Invest Dermatol. 1973;61:300–309. doi: 10.1111/1523-1747.ep12676518. [DOI] [PubMed] [Google Scholar]
- 11.Lubs ML, Bauer MS, Formas ME, Djokic B. Lisch nodules in neurofibromatosis type 1. N Engl J Med. 1991;324:1264–1266. doi: 10.1056/NEJM199105023241807. [DOI] [PubMed] [Google Scholar]
- 12.Wolkenstein P, Zeller J, Revuz J, Ecosse E, Leplege A. Quality-of-life impairment in neurofibromatosis type 1: a cross-sectional study of 128 cases. Arch Dermatol. 2001;137:1421–1425. doi: 10.1001/archderm.137.11.1421. [DOI] [PubMed] [Google Scholar]
- 13.Le LQ, Shipman T, Burns DK, Parada LF. Cell of origin and microenvironment contribution for NF1-associated dermal neurofibromas. Cell Stem Cell. 2009;4:453–463. doi: 10.1016/j.stem.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker T, Wolkenstein P, Revuz J, Zeller J, Friedman JM. Association between benign and malignant peripheral nerve sheath tumors in NF1. Neurology. 2005;65:205–211. doi: 10.1212/01.wnl.0000168830.79997.13. [DOI] [PubMed] [Google Scholar]
- 15.Zoller ME, Rembeck B, Oden A, Samuelsson M, Angervall L. Malignant and benign tumors in patients with neurofibromatosis type 1 in a defined Swedish population. Cancer. 1997;79:2125–2131. [PubMed] [Google Scholar]
- 16.Seminog OO, Goldacre MJ. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. Br J Cancer. 2013;108:193–198. doi: 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillot B, Dalac S, Delaunay M, et al. Cutaneous malignant melanoma and neurofibromatosis type 1. Melanoma Res. 2004;14:159–163. doi: 10.1097/00008390-200404000-00014. [DOI] [PubMed] [Google Scholar]
- 18.Rubinstein TJ, Plesec TP, Singh AD. Desmoplastic melanoma of the eyelid and conjunctival melanoma in neurofibromatosis type 1: a clinical pathological correlation. Surv Ophthalmol. 2015;60:72–77. doi: 10.1016/j.survophthal.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Barker D, Wright E, Nguyen K, et al. Gene for von Recklinghausen neurofibromatosis is in the pericentromeric region of chromosome 17. Science. 1987;236:1100–1102. doi: 10.1126/science.3107130. [DOI] [PubMed] [Google Scholar]
- 20.Barker D, Wright E, Nguyen K, et al. A genomic search for linkage of neurofibromatosis to RFLPs. J Med Genet. 1987;24:536–538. doi: 10.1136/jmg.24.9.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fain PR, Barker DF, Goldgar DE, et al. Genetic analysis of NF1: identification of close flanking markers on chromosome 17. Genomics. 1987;1:340–345. doi: 10.1016/0888-7543(87)90034-6. [DOI] [PubMed] [Google Scholar]
- 22.Goldgar DE, Green P, Parry DM, Mulvihill JJ. Multipoint linkage analysis in neurofibromatosis type I: an international collaboration. Am J Hum Genet. 1989;44:6–12. [PMC free article] [PubMed] [Google Scholar]
- 23.Fain PR, Goldgar DE, Wallace MR, et al. Refined physical and genetic mapping of the NF1 region on chromosome 17. Am J Hum Genet. 1989;45:721–728. [PMC free article] [PubMed] [Google Scholar]
- 24.Fain PR, Wright E, Willard HF, Stephens K, Barker DF. The order of loci in the pericentric region of chromosome 17, based on evidence from physical and genetic breakpoints. Am J Hum Genet. 1989;44:68–72. [PMC free article] [PubMed] [Google Scholar]
- 25.Marchuk DA, Saulino AM, Tavakkol R, et al. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991;11:931–940. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- 26.O’Connell P, Cawthon R, Xu GF, et al. The neurofibromatosis type 1 (NF1) gene: identification and partial characterization of a putative tumor suppressor gene. J Dermatol. 1992;19:881–884. doi: 10.1111/j.1346-8138.1992.tb03799.x. [DOI] [PubMed] [Google Scholar]
- 27.Ko JM, Sohn YB, Jeong SY, Kim HJ, Messiaen LM. Mutation spectrum of NF1 and clinical characteristics in 78 Korean patients with neurofibromatosis type 1. Pediatr Neurol. 2013;48:447–453. doi: 10.1016/j.pediatrneurol.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Maruoka R, Takenouchi T, Torii C, et al. The use of next-generation sequencing in molecular diagnosis of neurofibromatosis type 1: a validation study. Genet Test Mol Biomarkers. 2014;18:722–735. doi: 10.1089/gtmb.2014.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balla B, Arvai K, Horvath P, et al. Fast and robust next-generation sequencing technique using ion torrent personal genome machine for the screening of neurofibromatosis type 1 (NF1) gene. J Mol Neurosci. 2014;53:204–210. doi: 10.1007/s12031-014-0286-7. [DOI] [PubMed] [Google Scholar]
- 30.Cawthon RM, Weiss R, Xu GF, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 31.Glover TW, Stein CK, Legius E, et al. Molecular and cytogenetic analysis of tumors in von Recklinghausen neurofibromatosis. Genes Chromosomes Cancer. 1991;3:62–70. doi: 10.1002/gcc.2870030111. [DOI] [PubMed] [Google Scholar]
- 32.Xu W, Mulligan LM, Ponder MA, et al. Loss of NF1 alleles in phaeochromocytomas from patients with type I neurofibromatosis. Genes Chromosomes Cancer. 1992;4:337–342. doi: 10.1002/gcc.2870040411. [DOI] [PubMed] [Google Scholar]
- 33.Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet. 1993;3:122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- 34.Shannon KM, O’Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 35.Colman SD, Williams CA, Wallace MR. Benign neurofibromas in type 1 neurofibromatosis (NF1) show somatic deletions of the NF1 gene. Nat Genet. 1995;11:90–92. doi: 10.1038/ng0995-90. [DOI] [PubMed] [Google Scholar]
- 36.Sawada S, Florell S, Purandare SM, et al. Identification of NF1 mutations in both alleles of a dermal neurofibroma. Nat Genet. 1996;14:110–112. doi: 10.1038/ng0996-110. [DOI] [PubMed] [Google Scholar]
- 37.Serra E, Rosenbaum T, Winner U, et al. Schwann cells harbor the somatic NF1 mutation in neurofibromas: evidence of two different Schwann cell subpopulations. Hum Mol Genet. 2000;9:3055–3064. doi: 10.1093/hmg/9.20.3055. [DOI] [PubMed] [Google Scholar]
- 38.Upadhyaya M, Huson SM, Davies M, et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c. 2970–2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Raedt T, Brems H, Wolkenstein P, et al. Elevated risk for MPNST in NF1 microdeletion patients. Am J Hum Genet. 2003;72:1288–1292. doi: 10.1086/374821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alkindy A, Chuzhanova N, Kini U, Cooper DN, Upadhyaya M. Genotype-phenotype associations in neurofibromatosis type 1 (NF1): an increased risk of tumor complications in patients with NF1 splice-site mutations? Hum Genomics. 2012;6:12. doi: 10.1186/1479-7364-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasmant E, Vidaud M, Vidaud D, Wolkenstein P. Neurofibromatosis type 1: from genotype to phenotype. J Med Genet. 2012;49:483–489. doi: 10.1136/jmedgenet-2012-100978. [DOI] [PubMed] [Google Scholar]
- 42.Pasmant E, Sabbagh A, Masliah-Planchon J, et al. Role of noncoding RNA ANRIL in genesis of plexiform neurofibromas in neurofibromatosis type 1. J Natl Cancer Inst. 2011;103:1713–1722. doi: 10.1093/jnci/djr416. [DOI] [PubMed] [Google Scholar]
- 43.Warrington NM, Sun T, Luo J, et al. The cyclic AMP pathway is a sex-specific modifier of glioma risk in type I neurofibromatosis patients. Cancer Res. 2015;75:16–21. doi: 10.1158/0008-5472.CAN-14-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larribere L, Utikal J. Multiple roles of NF1 in the melanocyte lineage. Pigment Cell Melanoma Res. 2016;29:417–425. doi: 10.1111/pcmr.12488. [DOI] [PubMed] [Google Scholar]
- 45.Xu GF, O’Connell P, Viskochil D, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 46.Klose A, Ahmadian MR, Schuelke M, et al. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan RJ. The role of mitogen-activated protein targeting in melanoma beyond BRAFV600. Curr Opin Oncol. 2016;28:185–191. doi: 10.1097/CCO.0000000000000271. [DOI] [PubMed] [Google Scholar]
- 48.Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rauen KA, Huson SM, Burkitt-Wright E, et al. Recent developments in neurofibromatoses and RASopathies: management, diagnosis and current and future therapeutic avenues. Am J Med Genet A. 2015;167A:1–10. doi: 10.1002/ajmg.a.36793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kratz CP, Rapisuwon S, Reed H, Hasle H, Rosenberg PS. Cancer in Noonan, Costello, cardiofaciocutaneous and LEOPARD syndromes. Am J Med Genet C Semin Med Genet. 2011;157C:83–89. doi: 10.1002/ajmg.c.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasmant E, Ballerini P, Lapillonne H, et al. SPRED1 disorder and predisposition to leukemia in children. Blood. 2009;114:1131. doi: 10.1182/blood-2009-04-218503. [DOI] [PubMed] [Google Scholar]
- 52.Brems H, Chmara M, Sahbatou M, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 53.Messiaen L, Yao S, Brems H, et al. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA. 2009;302:2111–2118. doi: 10.1001/jama.2009.1663. [DOI] [PubMed] [Google Scholar]
- 54.Denayer E, Chmara M, Brems H, et al. Legius syndrome in fourteen families. Hum Mutat. 2011;32:E1985–1998. doi: 10.1002/humu.21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz D, Lazar A, Lev D. Malignant peripheral nerve sheath tumour (MPNST): the clinical implications of cellular signalling pathways. Expert Rev Mol Med. 2009;11:e30. doi: 10.1017/S1462399409001227. [DOI] [PubMed] [Google Scholar]
- 56.Johannessen CM, Reczek EE, James MF, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- 58.Keng VW, Rahrmann EP, Watson AL, et al. PTEN and NF1 inactivation in Schwann cells produces a severe phenotype in the peripheral nervous system that promotes the development and malignant progression of peripheral nerve sheath tumors. Cancer Res. 2012;72:3405–3413. doi: 10.1158/0008-5472.CAN-11-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hegedus B, Dasgupta B, Shin JE, et al. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1:443–457. doi: 10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 60.Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- 61.Wolman MA, de Groh ED, McBride SM, et al. Modulation of cAMP and ras signaling pathways improves distinct behavioral deficits in a zebrafish model of neurofibromatosis type 1. Cell Rep. 2014;8:1265–1270. doi: 10.1016/j.celrep.2014.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daston MM, Ratner N. Neurofibromin, a predominantly neuronal GTPase activating protein in the adult, is ubiquitously expressed during development. Dev Dyn. 1992;195:216–226. doi: 10.1002/aja.1001950307. [DOI] [PubMed] [Google Scholar]
- 63.Gutmann DH, Tennekoon GI, Cole JL, Collins FS, Rutkowski JL. Modulation of the neurofibromatosis type 1 gene product, neurofibromin, during Schwann cell differentiation. J Neurosci Res. 1993;36:216–223. doi: 10.1002/jnr.490360212. [DOI] [PubMed] [Google Scholar]
- 64.Parrinello S, Noon LA, Harrisingh MC, et al. NF1 loss disrupts Schwann cell-axonal interactions: a novel role for semaphorin 4F. Genes Dev. 2008;22:3335–3348. doi: 10.1101/gad.490608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Schepper S, Maertens O, Callens T, et al. Somatic mutation analysis in NF1 cafe au lait spots reveals two NF1 hits in the melanocytes. J Invest Dermatol. 2008;128:1050–1053. doi: 10.1038/sj.jid.5701095. [DOI] [PubMed] [Google Scholar]
- 66.Diwakar G, Zhang D, Jiang S, Hornyak TJ. Neurofibromin as a regulator of melanocyte development and differentiation. J Cell Sci. 2008;121:167–177. doi: 10.1242/jcs.013912. [DOI] [PubMed] [Google Scholar]
- 67.Deo M, Huang JL, Fuchs H, de Angelis MH, Van Raamsdonk CD. Differential effects of neurofibromin gene dosage on melanocyte development. J Invest Dermatol. 2013;133:49–58. doi: 10.1038/jid.2012.240. [DOI] [PubMed] [Google Scholar]
- 68.Nissan X, Larribere L, Saidani M, et al. Functional melanocytes derived from human pluripotent stem cells engraft into pluristratified epidermis. Proc Natl Acad Sci U S A. 2011;108:14861–14866. doi: 10.1073/pnas.1019070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larribere L, Wu H, Novak D, et al. NF1 loss induces senescence during human melanocyte differentiation in an iPSC-based model. Pigment Cell Melanoma Res. 2015;28:407–416. doi: 10.1111/pcmr.12369. [DOI] [PubMed] [Google Scholar]
- 70.Allouche J, Bellon N, Saidani M, et al. In vitro modeling of hyperpigmentation associated to neurofibromatosis type 1 using melanocytes derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2015;112:9034–9039. doi: 10.1073/pnas.1501032112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones S, Stransky N, McCord CL, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. 2014;5:5006. doi: 10.1038/ncomms6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cancer Genome Atlas Research N. Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li YY, Hanna GJ, Laga AC, et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21:1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 89.Gutzmer R, Herbst RA, Mommert S, et al. Allelic loss at the neurofibromatosis type 1 (NF1) gene locus is frequent in desmoplastic neurotropic melanoma. Hum Genet. 2000;107:357–361. doi: 10.1007/s004390000374. [DOI] [PubMed] [Google Scholar]
- 90.Krauthammer M, Kong Y, Bacchiocchi A, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mar VJ, Wong SQ, Li J, et al. BRAF/NRAS wild-type melanomas have a high mutation load correlating with histologic and molecular signatures of UV damage. Clin Cancer Res. 2013;19:4589–4598. doi: 10.1158/1078-0432.CCR-13-0398. [DOI] [PubMed] [Google Scholar]
- 95.Nissan MH, Pratilas CA, Jones AM, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiesner T, Kiuru M, Scott SN, et al. NF1 Mutations Are Common in Desmoplastic Melanoma. Am J Surg Pathol. 2015;39:1357–1362. doi: 10.1097/PAS.0000000000000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47:1194–1199. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kiuru M, McDermott G, Berger M, Halpern AC, Busam KJ. Desmoplastic melanoma with sarcomatoid dedifferentiation. Am J Surg Pathol. 2014;38:864–870. doi: 10.1097/PAS.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Upadhyaya M, Kluwe L, Spurlock G, et al. Germline and somatic NF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs) Hum Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 100.Bottillo I, Ahlquist T, Brekke H, et al. Germline and somatic NF1 mutations in sporadic and NF1-associated malignant peripheral nerve sheath tumours. J Pathol. 2009;217:693–701. doi: 10.1002/path.2494. [DOI] [PubMed] [Google Scholar]
- 101.Massi G, LeBoit PE. Desmoplastic melanoma. In: Massi G, LeBoit PE, editors. Histological diagnosis of nevi and melanoma. Springer; 2014. pp. 437–444. [Google Scholar]
- 102.Halaban R, Krauthammer M. RASopathy Gene Mutations in Melanoma. J Invest Dermatol. 2016;136:1755–1759. doi: 10.1016/j.jid.2016.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bradford D, Kim A. Current treatment options for malignant peripheral nerve sheath tumors. Curr Treat Options Oncol. 2015;16:328. doi: 10.1007/s11864-015-0328-6. [DOI] [PubMed] [Google Scholar]
- 104.Jousma E, Rizvi TA, Wu J, et al. Preclinical assessments of the MEK inhibitor PD-0325901 in a mouse model of Neurofibromatosis type 1. Pediatr Blood Cancer. 2015;62:1709–1716. doi: 10.1002/pbc.25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karajannis MA, Ferner RE. Neurofibromatosis-related tumors: emerging biology and therapies. Curr Opin Pediatr. 2015;27:26–33. doi: 10.1097/MOP.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karajannis MA, Legault G, Fisher MJ, et al. Phase II study of sorafenib in children with recurrent or progressive low-grade astrocytomas. Neuro Oncol. 2014;16:1408–1416. doi: 10.1093/neuonc/nou059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 109.Banerji U, Camidge DR, Verheul HM, et al. The first-in-human study of the hydrogen sulfate (Hyd-sulfate) capsule of the MEK1/2 inhibitor AZD6244 (ARRY-142886): a phase I open-label multicenter trial in patients with advanced cancer. Clin Cancer Res. 2010;16:1613–1623. doi: 10.1158/1078-0432.CCR-09-2483. [DOI] [PubMed] [Google Scholar]
- 110.Carvajal RD, Sosman JA, Quevedo JF, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014;311:2397–2405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dickson MA, Gordon MS, Edelman G, et al. Phase I study of XL281 (BMS-908662), a potent oral RAF kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs. 2015;33:349–356. doi: 10.1007/s10637-014-0191-5. [DOI] [PubMed] [Google Scholar]
- 112.Nakamura A, Arita T, Tsuchiya S, et al. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer Res. 2013;73:7043–7055. doi: 10.1158/0008-5472.CAN-13-1825. [DOI] [PubMed] [Google Scholar]
- 113.Wong DJ, Robert L, Atefi MS, et al. Antitumor activity of the ERK inhibitor SCH772984 [corrected] against BRAF mutant, NRAS mutant and wild-type melanoma. Mol Cancer. 2014;13:194. doi: 10.1186/1476-4598-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rebecca VW, Alicea GM, Paraiso KH, et al. Vertical inhibition of the MAPK pathway enhances therapeutic responses in NRAS-mutant melanoma. Pigment Cell Melanoma Res. 2014;27:1154–1158. doi: 10.1111/pcmr.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whittaker SR, Theurillat JP, Van Allen E, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]