Abstract
To improve the precision of molecular diagnosis and to develop and guide targeted therapies of breast cancer, it is essential to determine mechanisms that underlie the specific tumor phenotypes. To this end, the application of a snapshot of gene expression profile for breast cancer diagnosis and prognosis is fundamentally challenged since the tissue-based data are derived from heterogonous cell types and are not likely to reflect the dynamics of context-dependent tumor progression and drug sensitivity. The intricate network of epithelial differentiation program can be concertedly controlled by tumor suppressor maspin, a homologue of clade B serine protease inhibitors (serpin), through its multifaceted molecular interactions in multiple subcellular localizations. Unlike most other serpins that are expressed in multiple and cell types, maspin is epithelial specific and has distinct roles in luminal and myoepithelial cells. Endogenously expressed maspin has been found in the nucleus and cytoplasm, and detected on the surface of cell membrane. It is also secreted free and as an exosomal cargo protein. Research in the field has led to the identification of the maspin targets and maspin-associated molecules, as well as the structural determinants of its suppressive functions. The current review discusses the possibility for maspin to serve as a cell type-specific and context-sensitive marker to improve the precision of breast cancer diagnosis and prognosis. These advancements further suggest a new window of opportunity for designing novel maspin-based chemotherapeutic agents with improved anti-cancer potency.
Introduction
Breast cancer is the most common cancer in women worldwide. In the United States, according to American Cancer Society statistics, about 250,000 women were newly diagnosed with breast cancer, and more than 40,000 women succumbed to the disease in 2016. Currently based on the tumor grade and the expression profile of Her2/neu oncogene, estrogen receptor (ER) and progesterone receptor (PR), breast cancer can be classified into at least four subtypes: triple negative (Her2−/ER−/PR−), luminal A (Her2−/ER+, and tumor grade 1 or 2), luminal B (ER+ with Her2+ or Her2−, and higher tumor grade), and Her2 type (Her2+/ER−/PR−, and higher tumor grade) [Perou et al., 2000; Sorlie et al., 2003]. To improve the precision of molecular diagnosis of breast cancer to develop and guide targeted therapies, researchers identified additional multiple subtypes of breast cancer based on genome-wide expression profiling [Curtis et al., 2012]. However, to date, the hierarchical order of control of gene expression profiles that underlie specific tumor phenotypes remain elusive. The specific epigenetic expression profiles that confer tumor cell propensity to invade and metastasize are also unclear. It is worth noting, the application of a snapshot of gene expression profile for breast cancer diagnosis and prognosis is fundamentally challenged since the tissue-based expression profiles are derived from heterogonous cell types and are not likely to reflect the dynamics of context-dependent tumor progression and drug sensitivity.
Maspin, also known as Serpin B5, is a homologue of clade B serine protease inhibitors (serpin) [Zou et al., 1994]. Two independent studies showed that Maspin global knockout is embryonically lethal [Dzinic et al., 2016; Gao et al., 2004]. Unlike most other serpins that are expressed in multiple and cell types, maspin is epithelial specific. Endogenously expressed maspin has been found in the nucleus and cytoplasm, and detected on the surface of cell membrane. It is also secreted free [Bodenstine et al., 2014; Dean et al., 2017; Pemberton et al., 1997; Sternlicht et al., 1997] and as an exosomal cargo protein [Dean et al., 2017]. Accumulated evidence demonstrates that maspin is a multifaceted tumor suppressor. Importantly, maspin is a potential sensitive marker for tracking breast tumor cell plasticity and may be used as a surrogate marker for tumor response to changes of tumor microenvironments and drug treatment. The current review summarizes the existing literature and discusses the potential utility of maspin as a sensitive marker for more precise stratification of different subtypes of breast cancer, and the potential of maspin-associated molecular mechanisms as novel therapeutic targets.
Distinct Patterns of Maspin Expression in Different Subtypes of Breast Epithelial Cells in Development and Carcinogenesis
In mammalian mammary glands (human, dog and mouse), maspin is expressed by nearly 100% of the myoepithelial cells which are involved in mammary gland morphogenesis and mammary duct contractile function during lactation. The myoepithelial layer also comprises bipotent progenitors that give rise to luminal epithelial cells. According to histone and DNA modification profiles with cells freshly isolated from human tissue specimens, there are luminal progenitor-specific enhancers that are intermediate between the myoepithelial and luminal cells [Pellacani et al., 2016]. The signature maspin expression in myoepithelial cells may be directly activated by p63 [Popnikolov et al., 2003]. Accumulated evidence indicates that maspin has distinct roles in the outer myoepithelial layer and in the luminal epithelial cells of the mammary gland branching ductal network [Dzinic et al., 2016; Espinosa de los Monteros et al., 2005; Jones et al., 2004; Sternlicht et al., 1997].
In myoepithelial cell lesions, maspin expression can be used to ascertain their myoepithelial lineage. For example, strong maspin immunoreactivity was observed in the myoepithelial components in sclerosing papilloma (SP), tubular adenomyoepithelioma (TA), adenoid cystic carcinoma (ACC), epithelial-myoepithelial carcinoma of the breast (EMC), and malignant adenomyoepithelioma (MA). In all these cases, maspin immunoreactivity was confined to the nucleus and cytoplasm of the myoepithelial-like cells. No stromal, neural or vascular components were immunostained for maspin. Interestingly, metaplastic carcinomas of the breast (MCBs), an unusual neoplasm characterized by an admixture of glandular epithelial components, were shown to express myoepithelial markers as well as maspin [Reis-Filho et al., 2003; Reis-Filho et al., 2002; Reis-Filho et al., 2001].
In the progression of breast adenocarcinoma, a hallmark of tumor invasion is the loss of the myoepithelial cells. The loss of myoepithelial cells in invasive duct carcinoma (IDC) may directly contribute to epithelial tumor induction and/or progression [Runswick et al., 2001]. Based on a comprehensive gene expression profiling study, it is postulated that perturbation of normal myoepithelial cell differentiation may actually underlie the malignant progression of breast cancer [Polyak, 2010]. Consistently, in a preclinical study, enriched normal mouse myoepithelial cells suppressed the growth of breast tumor cells in a mouse syngeneic model of mammary adenocarcinoma [Farhanji et al., 2015]. Cancer-derived myoepithelial cells are still capable of polarizing luminal epithelial cells [Gudjonsson et al., 2002]. A recent study by Rohilla et al. [Rohilla et al., 2015] characterized the expression of maspin in the myoepithelial cells of pure ductal carcinoma in situ (DCIS), the DCIS component of IDC, and the adjacent normal breast tissue of both groups of human specimens. It was noted that the level of maspin expression was decreased in 20% of pure DCIS and 26.6% of the DCIS component of IDC. These data suggest that the tumor-associated myoepithelial cells undergo progressive reduction in maspin-mediated tumor suppressive activity before they are eliminated altogether.
Benign luminal epithelial cells are predominantly maspin-negative, except during pregnancy and lactation [Zhang et al., 1997]. A multiphasic differential expression pattern of maspin has been observed in mammary hyperplasia and adenocarcinoma. Luminal epithelial cells of atypical ductal hyperplasia, which is thought to have a high risk of progression to breast cancer, are maspin-negative [Umekita and Yoshida, 2003]. Maspin is upregulated in DCIS in the nuclei and the cytoplasm [Maass et al., 2001; Umekita et al., 2002; Umekita and Yoshida, 2003]. It was shown that luminal epithelial cells in low grade breast tumor that express maspin in the nucleus specifically correlated with a better overall survival [Jones et al., 2004; Machowska et al., 2014]. Tumor cells in IDC tumors appear to be more heterogeneous with regard to maspin expression. Compared to tumor cells in DCIS, IDC with predominant intraductal components features further increased level of maspin expression. The subset of infiltrating carcinomas that show maspin expression is significantly associated with a lower rate of lymph node metastasis at the time of diagnosis. In addition, in a study of 1068 invasive breast tumor specimens, nuclear staining of maspin correlated with ER and PR positivity. Conversely, cytoplasmic staining of maspin was associated with ER and PR negativity, high cell cycle S-phase fraction and aneuploidy [Mohsin et al., 2003]. A significant stepwise decrease in maspin expression was observed in the sequence DCIS, to IDC, and to lymph node metastasis. Maspin expression in luminal cells of both luminal type A and type B breast cancer correlated with less lymph node metastasis [Strien et al., 2017]. Reduction of maspin expression correlated with breast tumor brain metastasis [Stark et al., 2010]. Conversely, maspin expression detected in bone marrow micrometastases correlated with a statistically significant improved regression free survival of breast cancer patients [Ferrucci et al., 2007].
It is not clear whether the subset of maspin-positive breast tumor detected at metastatic sites have never lost maspin expression in the process of disease progression, or, as suggested by Barsky et al, reverted to a maspin-expressing state as a result of establishment of metastasis. Approximately 20% of breast cancer lymph node metastases revert to a DCIS growth pattern [Barsky et al., 1997]. The revertant growths have intact and circumferential basement membrane, can be distinguished by the re-expression of maspin, S-100 and smooth muscle actin, and exhibited complete concordance with the primary DCIS pattern. These data highlight the value of maspin for tracking the dynamics of breast tumor cell phenotypic plasticity.
In addition to the intrinsic propensity of breast carcinoma cells for phenotypic plasticity, accumulated evidence reveals complex lineage-specific subtypes of breast cancer based on the gene expression signature and histopathological characteristics [Curtis et al., 2012; Perou et al., 2000; Sorlie et al., 2003]. Clinical studies suggest that the differential expression patterns of maspin may be distinct in different subtypes of breast cancer. In the case of triple negative breast cancer, maspin upregulation seems to be specifically associated with basal cell markers, shorter relapse-free survival and overall survival [Umekita et al., 2011]. This finding is not entirely surprising as a significant fraction of triple negative breast cancer bears significant resemblance to that of stem-like basal cells, which may reside in the maspin-positive myoepithelial layer [Foulkes et al., 2010]. Importantly, however, this finding raises a concern over the data interpretation with the attempt to correlate the total level of, rather than cell type-specific, maspin expression with the histological or molecular subyptes of breast cancer.
It is clear that future studies need to address the value of maspin as a breast cancer marker in a cell-type and tumor stage- specific manner. For example, the association of maspin with ER (α- or β-) seems inconclusive. While the detection of maspin in IDC or triple negative breast cancer may suggest an inverse correlation between maspin and ER or PR [Lee et al., 2006; Umekita et al., 2002], experimental evidence seems to suggest that maspin expression may be either activated or repressed by ER [Khalkhali-Ellis et al., 2004; Liu et al., 2004; Stark et al., 2010; Wang et al., 2010]. To this end, as the so called normal breast epithelial cells used in laboratory research do not represent any freshly isolated normal human mammary cells [Pellacani et al., 2016], the current cellular models used may not recapitulate the specific biological and molecular characteristics of various subtypes of breast cancer in vivo. Despite the biphasic maspin differential expression in breast tumor progression, the 60 human breast cancer cell lines included in the National Cancer Institute (NCI) for anticancer drug screening (NCI60/NCBI profile: GDS4296/204855_at/) lost maspin expression.
Multifaceted Biological Functions of Maspin in Mammary Gland Development and Carcinogenesis
According to a phylogenetic analysis, maspin is one of the most evolutionarily conserved serpins [Benarafa and Remold-O’Donnell, 2005]. In development, maspin plays an essential role in embryogenesis, and a critical role in epithelial differentiation [Dzinic et al., 2016]. Although the biological functions of maspin may differ in distinct types of normal epithelial cells, accumulated clinical and preclinical evidence demonstrate several common tumor suppressive properties of maspin as it undergoes differential expression in multiple types of carcinomas including breast cancer.
Normal mammary myoepithelial cells that express maspin at a high level and predominantly in the nucleus are widely known to resist malignant transformation [Pandey et al., 2010]. When the Maspin gene is conditionally knocked out in oogenesis by Zp3-Cre mediated recombination, the virgin mammary glands of the resulting animals display myoepithelial hyperplasia and increased mammary duct branching while the luminal epithelial cell morphology remains largely unchanged [Dzinic et al., 2016]. In early stages of breast carcinogenesis, the myoepithelial layer is believed to play a critical tumor suppressive role, which is attributed, at least in part, to the high level of maspin secretion. For example, myoepithelial cells and derived cell lines (HMS 1–6) secrete maspin that can exert anti-invasive [Sternlicht et al., 1997] and anti-proliferative [Shao et al., 1998] paracrine activity. Interestingly, treatment of myoepithelial cells with tamoxifen, but not 17beta-estradiol, increases both maspin secretion and invasion-blocking ability, suggesting an additional benefit of tamoxifen in treating early stage breast cancer or in breast cancer chemoprevention. It is noted that myoepithelial cells lack ER-α but express ER-β. Tamoxifen may increase activator protein 1 (AP-1) mediated gene transcription of maspin [Shao et al., 2000].
Although the expression of maspin is very low or negligible in virgin mammary luminal epithelial cells, it can be upregulated in pregnancy, lactation or cancer. Targeted overexpression of maspin under the whey acidic protein (WAP) promoter, which is activated in lactating mammary epithelial cells, reduced the mammary duct branching and increased epithelial apoptosis [Zhang et al., 1999]. Consistent with the notion that down-regulation of maspin in invasive breast cancer is a gain of function, maspin overexpression in the mammary glands of the WAP-Tag/WAP-maspin bitransgenic mice blocked tumorigenesis and metastasis [Zhang et al., 2000a]. Systemic delivery of mouse Maspin gene by nonviral liposome blocked the proliferation and metastasis of orthotopically implanted breast tumor in a syngeneic metastatic model [Shi et al., 2002]. Conversely, maspin is down-regulated when the spontaneous breast adenocarcinoma of MMTV-TGF-α transgenic mice becomes invasive [Reddy et al., 2001].
Since the first report that ectopically expressed maspin inhibits breast tumor cell invasion in vitro and blocks tumor metastasis in vivo [Zou et al., 1994], preclinical evidence with established mammary epithelial cell lines in vivo and in vitro demonstrates that the collective function of maspin is to support the differentiated phenotype of epithelial cells, which includes polarized and stable organization through cell-cell and cell-matrix interaction, the sensitivity to contact-inhibition of proliferation, strong defense against stress and lack of invasiveness. It is well documented that normal breast epithelial cells MCF-10A can form acini in three dimensional extracellular matrix (ECM) such as Matrigel. In an earlier study, we have reported the generation of MCF-10A-derived maspin knock- down stable clones [Dean et al., 2017]. In contrast to the acini formation by the parental MCF-10A and scrambled siRNA-transfected control (NC1), maspin knock-down clones (siMas8 and siMas11) were more proliferative, formed disorganized cellular aggregates with increased infiltration into the Matrigel (Figure 1,). Conversely, under suspension conditions designed to evaluate cell stemness, the high maspin expressing MCF10-A and NC1 cells yielded irregularly shaped cell aggregates whereas the maspin knock down clones grew into compact circumferential tumorspheres indicative of a stem-like phenotype. These data are consistent with the observation that maspin re-expression in prostate tumor cells DU145 rendered epithelial acini formation in an in vivo xenograft model for tumor bone metastasis [Cher et al., 2003] and in an organotypic 3D collagen I model system [Bernardo et al., 2015; Bernardo et al., 2011]. Moreover, these results are in agreement with a previous report [Bernardo et al., 2015] that maspin re-expression in prostate tumor cells averts stemness. Maspin, therefore, appears to play a critical role in the maintenance of normal breast epithelium terminal differentiation.
Figure 1. Maspin down regulation in breast epithelial cells is transformative.
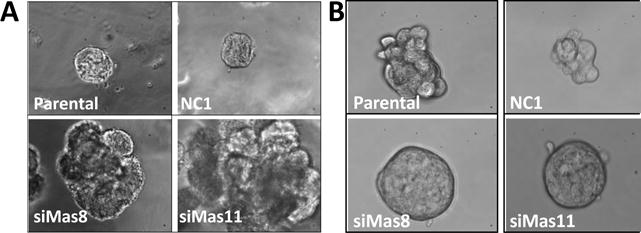
Representative phase contrast images of MCF-10A cells parental and maspin knock down clones obtained by transfection with a shRNA against maspin (siMas8 and siMas11) or with a scrambled sequence (NC1) cultured embedded in 3D Matrigel (×200, day 12) (A) and in suspension (×400, day 3) (B).
Laboratory investigations further demonstrate that the multifaceted tumor suppressive activities of maspin may derive from its multiple sub-cellular compartmentalization. Maspin can be secreted via dual non-classical secretory pathways as both a soluble protein and as an exosomal cargo protein [Dean et al., 2017]. Soluble maspin has been shown to inhibit tumor cell invasion and motility [Sheng et al., 1996; Sheng et al., 1994; Zhang et al., 1997], increase cell adhesion to ECM [Bernardo et al., 2011], block tumor-associated ECM degradation [Cher et al., 2003], and retard the detachment of cancer cells from established focal adhesion sites [Yin et al., 2006]. Extracellular maspin may further regulate the tumor microenvironment. For example, recombinant maspin directly blocks endothelial cell migration toward basic fibroblast growth factor and vascular endothelial growth factor in vitro, and blocks angiogenesis in vivo [Zhang et al., 2000b]. Exosomal maspin produced by normal immortalized breast epithelial cells suppresses the motility of fibroblasts [Dean et al., 2017].
As indicated by the clinical evidence, in breast tumor progression, maspin is dysregulated not only at the level of gene expression, but also at the level of subcellular partitioning between the nucleus and the cytoplasm. Three studies thus far have aimed to address the relative functional significance of nuclear vs cytoplasmic maspin. In the study by Goulet and colleagues [Goulet et al., 2011], breast cancer and epidermoid cancer cell lines ectopically expressing maspin that is fused to a nuclear export signal lost the tumor suppressive effects both in vitro and in vivo. Machowska et al. [Machowska et al., 2014] transiently transfected normal breast epithelial cells and multiple breast cancer cell lines with green-fluorescent protein-fused maspin with a nuclear localization sequence. The resulting cancer cells exhibited a significant reduction of proliferation, whereas the transfected normal cells were not affected. It is likely that normal cells that express a high background level of maspin both in the nucleus and the cytoplasm may not be significantly affected by the additional ectopically expressed recombinant maspin. In the third study, we showed that amino acid D346 in maspin is critical for its translocation from the nucleus to the cytoplasm in prostate cancer cells [Dzinic et al., 2013]. Conservative site-directed substation of Asp346 (D346) by Glu (E) (MaspinD346E) rendered maspin exclusively nuclear. In 3D Matrigel and Collagen I matrices, infection of SUM159 triple negative breast anaplastic tumor cells by adenovirus encoding for wild-type maspin (MaspinWT) or for the D346E mutation (MaspinD346E) reverted those cells to a better differentiated state with a highly organized structures, relative to control cells infected with empty vector adenovirus (MaspinEV), (Figure 2). Further, nuclear maspin sub-cellular localization correlated with the least invasive phenotype as evidenced by the lack of infiltration into ECM. Conversely, in suspension, cells devoid of maspin yielded compact tumorspheres whereas cells infected to express maspin, particularly in the nucleus, assembled in loosely attached aggregates.
Figure 2. Maspin re-expression and nuclear localization induces re-differentiation of breast tumor cells.
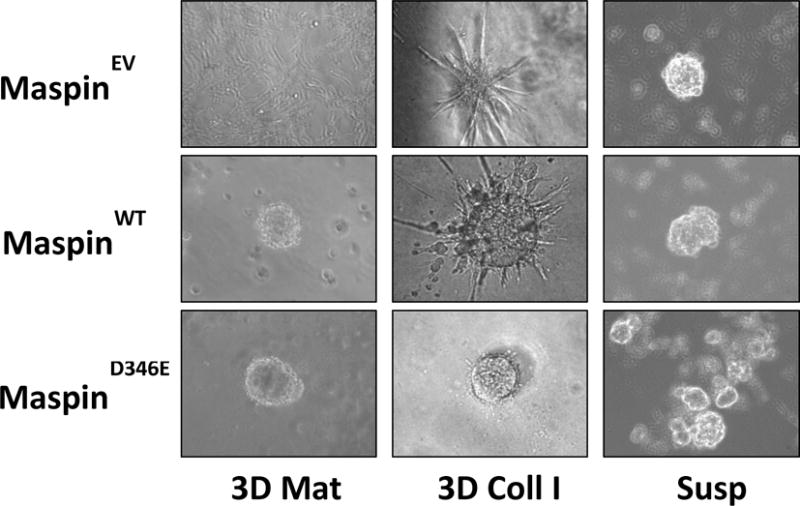
Representative phase contrast images of SUM159 cells seeded in 3D Matrigel, 3D Collagen I and in suspension at day 12. (×200)
The Molecular Mechanisms Underlying the Context-dependent Maspin Functions
Through the studies with recombinant maspin and endogenous maspin in various established cell lines, several serine-protease- like molecules or serine protease zymogens in different subcellular compartments have been identified as the enzymatic targets of maspin, including single-chain tissue type plasminogen activator (sc-tPA) [Sheng et al., 1998] pro-urokinase plasminogen activator (pro-uPA) [Biliran and Sheng, 2001; McGowen et al., 2000; Yin et al., 2006], cathepsin D [Khalkhali-Ellis and Hendrix, 2007], and HDAC1 [Li et al., 2006; Yin et al., 2005]. While pro-uPA is proteolytically activated upon binding to its cell surface receptor uPAR [Dano et al., 2005], tPA is secreted as sc-tPA with very low enzymatic activity in plasminogen activation in the absence of co-factors such as fibrin and heparin [Blasi and Sidenius, 2010; Castellino and Ploplis, 2005]. The substrate binding surface of HDACs has a conformation similar to that of a serine protease [Finnin et al., 1999; Min et al., 2001; Somoza et al., 2004]. Although cathepsin D is classified as an aspartic protease, it can be cross-inhibited by serine protease inhibitors [Guo et al., 2015]. In addition to serine protease-like targets, maspin was also shown to interact with heat shock protein 90 (HSP90), glutathione S-transferase (GST), heat shock protein 70 (HSP70) [Yin et al., 2005], interferon regulatory factor 6 (IRF6) [Bailey and Hendrix, 2008; Bailey et al., 2005], integrin β1 that is complexed with uPA/uPAR system [Cella et al., 2006; Endsley et al., 2011], and collagen type I and III [Blacque and Worrall, 2002].
A serpin molecule typically presents its reactive center loop (RCL) P1P1′ peptide bond as a pseudo-substrate to the catalytic pocket of the cognate target enzyme [Gettins and Olson, 2016; Huntington, 2011; Silverman et al., 2001]. The X-ray crystallographic analysis of maspin demonstrates that maspin cannot undergo the conformational changes required for the inhibitory activity against a typical serine protease. Consistent with accumulated evidence, the structure of maspin may support its interaction with serine protease-like targets [Al-Ayyoubi et al., 2004; Law et al., 2005]. Not surprisingly, maspin depends on the RCL for its inhibitory effects on breast tumor cell invasion and motility ([Sheng et al., 1996; Sheng et al., 1994]. Substitution of maspin RCL P1 position Arg340 (R340) by Gln (Q) abolished the ability of maspin to induce ECM adhesion and to inhibit invasion of breast cancer cells [Ngamkitidechakul et al., 2003]. We have shown that substitution of R340 by Ala (A) abolished the maspin interaction with pro-uPA [Yin et al., 2006]. Further, maspin depends on its R340 to inhibit HDAC1-mediated deacetylation of histones 3 and 4 [Li et al., 2006]. It is worth noting that the catalytic pocket of HDAC has a unique narrow “tunnel” extending into the center of the molecule. At the bottom of the “tunnel” is a Zn2+ ion chelated by two Asp (N)-His (H) charge-relay systems. In HDAC1-mediated protein deacetylation, a H2O molecule and a ε-acetyl-lysine side chain bind to Zn2+, leading to a series of proton transfers and lysine deacetylation. The maspin RCL P1 residue R340 has a similar structure as ε-acetyl-lysine [Finnin et al., 1999; Min et al., 2001; Somoza et al., 2004]. The exposed maspin RCL with R340 at the center appears to act as a natural cap to block the entry of HDAC1 substrates.
Maspin does not have a leader sequence for classical secretion. However, it is secreted independently free and as an exosome cargo protein via endosome-dependent mechanisms. Interestingly, maspin secretion seems to be disproportionally decreased, relative to the level of maspin transcription, when it undergoes nuclear to cytoplasmic translocation [Dean et al., 2017]. An earlier study showed that the first 50 N-terminal amino acids of maspin were important for its association with secretory vesicles [Pemberton et al., 1997]. In addition, mutation of 86 amino acids located between amino acids 139 and 225 of maspin diminished the adhesion of MCF- 10A cells to their own ECM, and may encompass the binding site to integrin β1 [Cella et al., 2006; Endsley et al., 2011]. Efforts to elucidate what intrinsic signals in maspin are involved in its partition to different subcellular compartments identified D346 to be critical in its nuclear retention [Dzinic et al., 2013]. Conservative substitution of D346 by E rendered maspin exclusively nuclear and increased the maspin interaction with HDAC1 in tumor cells that otherwise expressed maspin predominantly in the cytoplasm. The current experimental evidence supports a working model that D346 may be involved in the interaction with a nuclear protein that may be down regulated in tumor progression. Interestingly, maspin amino acids 240–250 in the G-helix support a novel conformational switch [Law et al., 2005] which may serve as an additional point of contact for other protein-protein interaction and has been shown to play a role in the maspin effects on cell migration and adhesion [Ravenhill et al., 2010].
Maspin protein may be subject to regulation by post-translational modifications. Although the cysteine sulfhydryl groups of purified recombinant maspin are all in the reduced state [Fitzpatrick et al., 1996], it is reported that maspin binds to GST [Yin et al., 2005], which is important to keep maspin in the reduced state when breast epithelial cells are under oxidative stress [Nawata et al., 2011]. Maspin has been shown to undergo phosphorylation. Specifically, maspin is found to be phosphorylated on tyrosine moiety(ies) in normal mammary epithelial cells [Odero-Marah et al., 2002]. While inhibition of tyrosine phosphatases increased maspin protein levels and leads to its cytoplasmic accumulation [Tamazato Longhi and Cella, 2012], epidermal growth factor receptor (EGFR) signaling is shown to lead to maspin tyrosine phosphorylation and increase maspin nuclear localization in MCF-10A cells [Tamazato Longhi et al., 2016]. In addition, proteins secreted by mammary epithelial cells may be phosphorylated at multiple cysteine sites. Currently, the functional implication of the specific maspin phosphorylation modifications remains unclear [Narayan et al., 2011].
The identification of the aforementioned maspin targets and associated molecules is in line with the biological functions of maspin in epithelial differentiation and tumor suppression, and supports the notion that the net biological function of maspin derives from its coordinated activities in different subcellular compartments. The effects of maspin on the maintenance of epithelial differentiation are consistent with its nuclear inhibitory activity of HDAC1. Consistently, binding of nuclear maspin to chromatin is essential for suppressing invasion and metastasis of breast cancer cell lines [Goulet et al., 2012]. It is important to note that maspin is the only endogenous polypeptide HDAC1 inhibitor identified thus far. It has been shown that maspin commonly regulates only a small set of HDAC1 target genes that are functionality clustered to regulate epithelial cell stemness and differentiation [Bernardo et al., 2015; Bernardo et al., 2011]. Bioinformatics analysis suggests that these genes may be specifically responsive to the regulation by TGFβ signaling. Indeed, proteomic analysis of MCF-10A cell lysates confirmed a specific connection of maspin with TGFβ signaling [Friedman et al., 2007]. Interestingly, another nuclear maspin-associated protein, IRF6, is thought to regulate epithelial differentiation by restricting cell cycle entry into the G(0) phase [Bailey and Hendrix, 2008; Bailey et al., 2009]. Regarding the underlying mechanisms of IRF6 regulation by maspin, it has been shown that IRF5, a close homologue of IRF6 [Taniguchi et al., 2001] directly binds to both histone acetyl-transferases (HAT) and HDACs to tightly control gene expression [Feng et al., 2010]. Based on the evidence of Dean et al. [Dean et al., 2017], it is intriguing to further speculate that maspin delivered by exosomal particles into stromal cells, e.g. fibroblasts, may also function as de novo synthesized maspin to regulate HDAC1- and/or IRF6-dependent gene expression.
The effects of maspin on pericellular ECM degradation involved in tumor invasion and angiogenesis may occur through inhibition of the plasminogen activation-dependent proteolytic cascade and/or the enzymatic activity of cathepsin D [Biliran and Sheng, 2001; Khalkhali-Ellis and Hendrix, 2007; McGowen et al., 2000]. The localized pericellular anti-proteolytic activity of maspin may be further modulated directly by its interaction of with integrin β1 [Endsley et al., 2011] and collagen [Blacque and Worrall, 2002], or indirectly by integrin β1 through the integrin β1/uPAR complex. The dynamics of maspin-mediated ECM degradation may be subsequently propelled by the robust internalization of the maspin/pro-uPA/uPAR complex [Yin et al., 2006]. Prior to its lysosome-mediated turnover [Yin et al., 2006], internalized maspin by carcinoma cells has been shown to transiently block the activation of the small GTPase RAC1 to block tumor cell motility [Odero-Marah et al., 2002; Sheng, 2003].
Mspin is upregulated by multiple cellular stress signals [Bailey et al., 2006; Lockett et al., 2006]. The current evidence suggests that the biological functions of cytoplasmic maspin may be directly involved in epithelial stress response. For example, maspin enhances GST-mediated cellular resistance to the generation of reactive oxygen species [Yin et al., 2005]. While the biological effects and underlying mechanisms of the maspin interaction with HSP90 and HSP70 need to be further interrogated, it is worth noting that both HSP90 and HSP70 are chaperone proteins designed to facilitate cellular stress response. Cytoplasmic maspin in MCF- 10A cells was also associated with the endoplasmic reticulum (ER), and displayed a mutual exclusive relationship with ER chaperone protein glucose-regulated protein 78 (GRP78, [Dzinic et al., 2013]). It is possible that maspin blocks the deacetylation of GRP78. The acetylated GRP78, in turn, is dissociated from ER. Interestingly, although cathepsin D can be secreted, it also exists in three forms in the cytoplasm. Pro-cathepsin D is in the Golgi complex, intermediate and mature cathepsin Ds are enzymatically active and localized in the endosomes and lysosomes, respectively. Mature cathepsin D has been shown to play a crucial role in autophagy and apoptosis. Future studies are needed to address whether maspin specifically binds to the pro-intermediate and mature forms of cathepsin D [Nicotra et al., 2010].
Concluding Remarks
Studies of maspin in breast cancer demonstrate an intricate network of cellular functions that can be concertedly controlled by maspin through distinct molecular interactions in multiple subcellular localizations. These critical insights suggest an exciting possibility for maspin to serve as a cell type-specific and context-sensitive marker to improve the precision of breast cancer diagnosis and prognosis. The identification of the maspin targets and maspin-associated molecules, as well as the structural determinants of the tumor suppressive functions of maspin opens a new window of opportunity for designing novel chemotherapeutic agents with improved anti-cancer potency.
Acknowledgments
This work was supported by NIH grants (R01CA127735 and R01CA084176 to Sheng, S and R21CA154319 to Sheng S in a Dual PI plan), the Ruth Sager Memorial Fund (to Sheng S), WSU UROP grant to Matta MJ, and NIH Cancer Center Support Grant CA022453 to Gerold Bepler of the Karmanos Cancer Institute at Wayne State University (with Sheng S as a Program Leader).
Abbreviations
- ACC
adenoid cystic carcinoma
- AP-1
activator protein 1
- DCIS
ductal carcinoma in situ
- ECM
extracellular matrix
- EGFR
epidermal growth factor
- EMC
epithelial-myoepithelial carcinoma
- ER
endoplasmic reticulum
- ER
estrogen receptor
- GRP78
glucose regulated protein 78
- GST
glutathione S-transferase
- HAT
histone acetyltransferase
- HDAC1
histone deacetylase 1
- HSP70
heat shock protein 70
- HSP90
heat shock protein 90
- IDC
invasive duct carcinoma
- MA
malignant adenomyoepithelioma
- MCB
metaplastic carcinomas of the breast
- PR
progesterone receptor
- RCL
reactive center loop
- sct-PA
single chain tissue plasminogen activator
- SP
sclerosing papilloma
- TA
tubular adenomyoepithelioma
- u-PA
urokinase plasminogen activator
- u-PAR
urokinase plasminogen activator receptor
- WAP
whey acidic protein
References
- Al-Ayyoubi M, Gettins PG, Volz K. Crystal structure of human maspin, a serpin with antitumor properties: reactive center loop of maspin is exposed but constrained. J Biol Chem. 2004;279:55540–4. doi: 10.1074/jbc.M409957200. [DOI] [PubMed] [Google Scholar]
- Bailey CM, Hendrix MJ. IRF6 in development and disease: a mediator of quiescence and differentiation. Cell Cycle. 2008;7:1925–30. doi: 10.4161/cc.7.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Khalkhali-Ellis Z, Kondo S, Margaryan NV, Seftor RE, Wheaton WW, Amir S, Pins MR, Schutte BC, Hendrix MJ. Mammary serine protease inhibitor (Maspin) binds directly to interferon regulatory factor 6: identification of a novel serpin partnership. J Biol Chem. 2005;280:34210–7. doi: 10.1074/jbc.M503523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJ. Biological functions of maspin. J Cell Physiol. 2006;209:617–24. doi: 10.1002/jcp.20782. [DOI] [PubMed] [Google Scholar]
- Bailey CM, Margaryan NV, Abbott DE, Schutte BC, Yang B, Khalkhali-Ellis Z, Hendrix MJ. Temporal and spatial expression patterns for the tumor suppressor Maspin and its binding partner interferon regulatory factor 6 during breast development. Dev Growth Differ. 2009;51:473–81. doi: 10.1111/j.1440-169X.2009.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsky SH, Doberneck SA, Sternlicht MD, Grossman DA, Love SM. ‘Revertant’ DCIS in human axillary breast carcinoma metastases. J Pathol. 1997;183:188–94. doi: 10.1002/(SICI)1096-9896(199710)183:2<188::AID-PATH898>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Benarafa C, Remold-O’Donnell E. The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc Natl Acad Sci U S A. 2005;102:11367–72. doi: 10.1073/pnas.0502934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo MM, Kaplun A, Dzinic SH, Li X, Irish J, Mujagic A, Jakupovic B, Back JB, Van Buren E, Han X, Dean I, Chen YQ, Heath E, Sakr W, Sheng S. Maspin Expression in Prostate Tumor Cells Averts Stemness and Stratifies Drug Sensitivity. Cancer Res. 2015;75:3970–9. doi: 10.1158/0008-5472.CAN-15-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo MM, Meng Y, Lockett J, Dyson G, Dombkowski A, Kaplun A, Li X, Yin S, Dzinic S, Olive M, Dean I, Krass D, Moin K, Bonfil RD, Cher M, Sakr W, Sheng S. Maspin reprograms the gene expression profile of prostate carcinoma cells for differentiation. Genes Cancer. 2011;2:1009–22. doi: 10.1177/1947601912440170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biliran H, Jr, Sheng S. Pleiotrophic inhibition of pericellular urokinase-type plasminogen activator system by endogenous tumor suppressive maspin. Cancer Res. 2001;61:8676–82. [PubMed] [Google Scholar]
- Blacque OE, Worrall DM. Evidence for a direct interaction between the tumor suppressor serpin, maspin, and types I and III collagen. J Biol Chem. 2002;277:10783–8. doi: 10.1074/jbc.M110992200. [DOI] [PubMed] [Google Scholar]
- Blasi F, Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923–30. doi: 10.1016/j.febslet.2009.12.039. [DOI] [PubMed] [Google Scholar]
- Bodenstine TM, Seftor RE, Seftor EA, Khalkhali-Ellis Z, Samii NA, Monarrez JC, Chandler GS, Pemberton PA, Hendrix MJ. Internalization by multiple endocytic pathways and lysosomal processing impact maspin-based therapeutics. Mol Cancer Res. 2014;12:1480–91. doi: 10.1158/1541-7786.MCR-14-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thromb Haemost. 2005;93:647–54. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- Cella N, Contreras A, Latha K, Rosen JM, Zhang M. Maspin is physically associated with [beta]1 integrin regulating cell adhesion in mammary epithelial cells. FASEB J. 2006;20:1510–2. doi: 10.1096/fj.05-5500fje. [DOI] [PubMed] [Google Scholar]
- Cher ML, Biliran HR, Jr, Bhagat S, Meng Y, Che M, Lockett J, Abrams J, Fridman R, Zachareas M, Sheng S. Maspin expression inhibits osteolysis, tumor growth, and angiogenesis in a model of prostate cancer bone metastasis. Proc Natl Acad Sci U S A. 2003;100:7847–52. doi: 10.1073/pnas.1331360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M. Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dano K, Behrendt N, Hoyer-Hansen G, Johnsen M, Lund LR, Ploug M, Romer J. Plasminogen activation and cancer. Thromb Haemost. 2005;93:676–81. doi: 10.1160/TH05-01-0054. [DOI] [PubMed] [Google Scholar]
- Dean I, Dzinic SH, Bernardo MM, Zou Y, Kimler V, Li X, Kaplun A, Granneman J, Mao G, Sheng S. The secretion and biological function of tumor suppressor maspin as an exosome cargo protein. Oncotarget. 2017;8:8043–8056. doi: 10.18632/oncotarget.13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzinic SH, Bernardo MM, Li X, Fernandez-Valdivia R, Ho YS, Mi QS, Bandyopadhyay S, Lonardo F, Vranic S, Oliveira DS, Bonfil RD, Dyson G, Chen K, Omerovic A, Sheng X, Han X, Wu D, Bi X, Cabaravdic D, Jakupovic U, Wahba M, Pang A, Harajli D, Sakr WA, Sheng S. An Essential Role of Maspin in Embryogenesis and Tumor Suppression. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzinic SH, Kaplun A, Li X, Bernardo M, Meng Y, Dean I, Krass D, Stemmer P, Shin N, Lonardo F, Sheng S. Identification of an intrinsic determinant critical for maspin subcellular localization and function. PLoS One. 2013;8:e74502. doi: 10.1371/journal.pone.0074502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endsley MP, Hu Y, Deng Y, He X, Warejcka DJ, Twining SS, Gonias SL, Zhang M. Maspin, the molecular bridge between the plasminogen activator system and beta1 integrin that facilitates cell adhesion. J Biol Chem. 2011;286:24599–607. doi: 10.1074/jbc.M111.235788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa de los Monteros A, Millan MY, Ramirez GA, Ordas J, Reymundo C, Martin de las Mulas J. Expression of maspin in mammary gland tumors of the dog. Vet Pathol. 2005;42:250–7. doi: 10.1354/vp.42-3-250. [DOI] [PubMed] [Google Scholar]
- Farhanji B, Latifpour M, Alizadeh AM, Khodayari H, Khodayari S, Khaniki M, Ghasempour S. Tumor suppression effects of myoepithelial cells on mice breast cancer. Eur J Pharmacol. 2015;765:171–8. doi: 10.1016/j.ejphar.2015.08.023. [DOI] [PubMed] [Google Scholar]
- Feng D, Sangster-Guity N, Stone R, Korczeniewska J, Mancl ME, Fitzgerald-Bocarsly P, Barnes BJ. Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression. J Immunol. 2010;185:6003–12. doi: 10.4049/jimmunol.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci PF, Rabascio C, Gigli F, Corsini C, Giordano G, Bertolini F, Martinelli G. A new comprehensive gene expression panel to study tumor micrometastasis in patients with high-risk breast cancer. Int J Oncol. 2007;30:955–62. doi: 10.3892/ijo.30.4.955. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–93. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick PA, Wong DT, Barr PJ, Pemberton PA. Functional implications of the modeled structure of maspin. Protein Eng. 1996;9:585–9. doi: 10.1093/protein/9.7.585. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multivariable difference gel electrophoresis and mass spectrometry: a case study on transforming growth factor-beta and ERBB2 signaling. Mol Cell Proteomics. 2007;6:150–69. doi: 10.1074/mcp.D600001-MCP200. [DOI] [PubMed] [Google Scholar]
- Gao F, Shi HY, Daughty C, Cella N, Zhang M. Maspin plays an essential role in early embryonic development. Development. 2004;131:1479–89. doi: 10.1242/dev.01048. [DOI] [PubMed] [Google Scholar]
- Gettins PG, Olson ST. Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance. Biochem J. 2016;473:2273–93. doi: 10.1042/BCJ20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet B, Chan G, Chambers AF, Lewis JD. An emerging role for the nuclear localization of maspin in the suppression of tumor progression and metastasis. Biochem Cell Biol. 2012;90:22–38. doi: 10.1139/o11-053. [DOI] [PubMed] [Google Scholar]
- Goulet B, Kennette W, Ablack A, Postenka CO, Hague MN, Mymryk JS, Tuck AB, Giguere V, Chambers AF, Lewis JD. Nuclear localization of maspin is essential for its inhibition of tumor growth and metastasis. Lab Invest. 2011;91:1181–7. doi: 10.1038/labinvest.2011.66. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Erskine PT, Coker AR, Wood SP, Cooper JB, Mares M, Baudys M. Structure of a Kunitz-type potato cathepsin D inhibitor. J Struct Biol. 2015;192:554–60. doi: 10.1016/j.jsb.2015.10.020. [DOI] [PubMed] [Google Scholar]
- Huntington JA. Serpin structure, function and dysfunction. J Thromb Haemost. 2011;9(Suppl 1):26–34. doi: 10.1111/j.1538-7836.2011.04360.x. [DOI] [PubMed] [Google Scholar]
- Jones C, Mackay A, Grigoriadis A, Cossu A, Reis-Filho JS, Fulford L, Dexter T, Davies S, Bulmer K, Ford E, Parry S, Budroni M, Palmieri G, Neville AM, O’Hare MJ, Lakhani SR. Expression profiling of purified normal human luminal and myoepithelial breast cells: identification of novel prognostic markers for breast cancer. Cancer Res. 2004;64:3037–45. doi: 10.1158/0008-5472.can-03-2028. [DOI] [PubMed] [Google Scholar]
- Khalkhali-Ellis Z, Christian AL, Kirschmann DA, Edwards EM, Rezaie-Thompson M, Vasef MA, Gruman LM, Seftor RE, Norwood LE, Hendrix MJ. Regulating the tumor suppressor gene maspin in breast cancer cells: a potential mechanism for the anticancer properties of tamoxifen. Clin Cancer Res. 2004;10:449–54. doi: 10.1158/1078-0432.ccr-1002-03. [DOI] [PubMed] [Google Scholar]
- Khalkhali-Ellis Z, Hendrix MJ. Elucidating the function of secreted maspin: inhibiting cathepsin D-mediated matrix degradation. Cancer Res. 2007;67:3535–9. doi: 10.1158/0008-5472.CAN-06-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RH, Irving JA, Buckle AM, Ruzyla K, Buzza M, Bashtannyk-Puhalovich TA, Beddoe TC, Nguyen K, Worrall DM, Bottomley SP, Bird PI, Rossjohn J, Whisstock JC. The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix. J Biol Chem. 2005;280:22356–64. doi: 10.1074/jbc.M412043200. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Suh CH, Li ZH. Clinicopathological significance of maspin expression in breast cancer. J Korean Med Sci. 2006;21:309–14. doi: 10.3346/jkms.2006.21.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yin S, Meng Y, Sakr W, Sheng S. Endogenous inhibition of histone deacetylase 1 by tumor-suppressive maspin. Cancer Res. 2006;66:9323–9. doi: 10.1158/0008-5472.CAN-06-1578. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shi HY, Nawaz Z, Zhang M. Tamoxifen induces the expression of maspin through estrogen receptor-alpha. Cancer Lett. 2004;209:55–65. doi: 10.1016/j.canlet.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Lockett J, Yin S, Li X, Meng Y, Sheng S. Tumor suppressive maspin and epithelial homeostasis. J Cell Biochem. 2006;97:651–60. doi: 10.1002/jcb.20721. [DOI] [PubMed] [Google Scholar]
- Maass N, Teffner M, Rosel F, Pawaresch R, Jonat W, Nagasaki K, Rudolph P. Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol. 2001;195:321–6. doi: 10.1002/path.948. [DOI] [PubMed] [Google Scholar]
- Machowska M, Wachowicz K, Sopel M, Rzepecki R. Nuclear location of tumor suppressor protein maspin inhibits proliferation of breast cancer cells without affecting proliferation of normal epithelial cells. BMC Cancer. 2014;14:142. doi: 10.1186/1471-2407-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen R, Biliran H, Jr, Sager R, Sheng S. The surface of prostate carcinoma DU145 cells mediates the inhibition of urokinase-type plasminogen activator by maspin. Cancer Res. 2000;60:4771–8. [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–79. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Mohsin SK, Zhang M, Clark GM, Craig Allred D. Maspin expression in invasive breast cancer: association with other prognostic factors. J Pathol. 2003;199:432–5. doi: 10.1002/path.1319. [DOI] [PubMed] [Google Scholar]
- Narayan M, Mirza SP, Twining SS. Identification of phosphorylation sites on extracellular corneal epithelial cell maspin. Proteomics. 2011;11:1382–90. doi: 10.1002/pmic.201000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawata S, Shi HY, Sugino N, Zhang M. Evidence of post-translational modification of the tumor suppressor maspin under oxidative stress. Int J Mol Med. 2011;27:249–54. doi: 10.3892/ijmm.2010.572. [DOI] [PubMed] [Google Scholar]
- Ngamkitidechakul C, Warejcka DJ, Burke JM, O’Brien WJ, Twining SS. Sufficiency of the reactive site loop of maspin for induction of cell-matrix adhesion and inhibition of cell invasion. Conversion of ovalbumin to a maspin-like molecule. J Biol Chem. 2003;278:31796–806. doi: 10.1074/jbc.M302408200. [DOI] [PubMed] [Google Scholar]
- Nicotra G, Castino R, Follo C, Peracchio C, Valente G, Isidoro C. The dilemma: does tissue expression of cathepsin D reflect tumor malignancy? The question: does the assay truly mirror cathepsin D mis-function in the tumor? Cancer Biomark. 2010;7:47–64. doi: 10.3233/CBM-2010-0143. [DOI] [PubMed] [Google Scholar]
- Odero-Marah VA, Khalkhali-Ellis Z, Schneider GB, Seftor EA, Seftor RE, Koland JG, Hendrix MJ. Tyrosine phosphorylation of maspin in normal mammary epithelia and breast cancer cells. Biochem Biophys Res Commun. 2002;295:800–5. doi: 10.1016/s0006-291x(02)00764-7. [DOI] [PubMed] [Google Scholar]
- Pandey PR, Saidou J, Watabe K. Role of myoepithelial cells in breast tumor progression. Front Biosci (Landmark Ed) 2010;15:226–36. doi: 10.2741/3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellacani D, Bilenky M, Kannan N, Heravi-Moussavi A, Knapp DJ, Gakkhar S, Moksa M, Carles A, Moore R, Mungall AJ, Marra MA, Jones SJ, Aparicio S, Hirst M, Eaves CJ. Analysis of Normal Human Mammary Epigenomes Reveals Cell-Specific Active Enhancer States and Associated Transcription Factor Networks. Cell Rep. 2016;17:2060–2074. doi: 10.1016/j.celrep.2016.10.058. [DOI] [PubMed] [Google Scholar]
- Pemberton PA, Tipton AR, Pavloff N, Smith J, Erickson JR, Mouchabeck ZM, Kiefer MC. Maspin is an intracellular serpin that partitions into secretory vesicles and is present at the cell surface. J Histochem Cytochem. 1997;45:1697–706. doi: 10.1177/002215549704501213. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010:210–3. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popnikolov NK, Ayala AG, Graves K, Gatalica Z. Benign myoepithelial tumors of the breast have immunophenotypic characteristics similar to metaplastic matrix-producing and spindle cell carcinomas. Am J Clin Pathol. 2003;120:161–7. doi: 10.1309/G6CT-R8MD-TFUW-19XV. [DOI] [PubMed] [Google Scholar]
- Ravenhill L, Wagstaff L, Edwards DR, Ellis V, Bass R. G-helix of maspin mediates effects on cell migration and adhesion. J Biol Chem. 2010;285:36285–92. doi: 10.1074/jbc.M110.177253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KB, McGowen R, Schuger L, Visscher D, Sheng S. Maspin expression inversely correlates with breast tumor progression in MMTV/TGF-alpha transgenic mouse model. Oncogene. 2001;20:6538–43. doi: 10.1038/sj.onc.1204796. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Paredes J, Silva P, Pereira EM, Maeda SA, de Carvalho LV, Schmitt FC. Novel and classic myoepithelial/stem cell markers in metaplastic carcinomas of the breast. Appl Immunohistochem Mol Morphol. 2003;11:1–8. doi: 10.1097/00129039-200303000-00001. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Schmitt FC. Maspin is expressed in the nuclei of breast myoepithelial cells. J Pathol. 2002;197:272–3. doi: 10.1002/path.1104. author reply 273–4. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Silva P, Schmitt FC. Maspin expression in myoepithelial tumors of the breast. Pathol Res Pract. 2001;197:817–21. doi: 10.1078/0344-0338-00165. [DOI] [PubMed] [Google Scholar]
- Rohilla M, Bal A, Singh G, Joshi K. Phenotypic and Functional Characterization of Ductal Carcinoma In Situ-Associated Myoepithelial Cells. Clin Breast Cancer. 2015;15:335–42. doi: 10.1016/j.clbc.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Runswick SK, O’Hare MJ, Jones L, Streuli CH, Garrod DR. Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nat Cell Biol. 2001;3:823–30. doi: 10.1038/ncb0901-823. [DOI] [PubMed] [Google Scholar]
- Shao ZM, Nguyen M, Alpaugh ML, O’Connell JT, Barsky SH. The human myoepithelial cell exerts antiproliferative effects on breast carcinoma cells characterized by p21WAF1/CIP1 induction, G2/M arrest, and apoptosis. Exp Cell Res. 1998;241:394–403. doi: 10.1006/excr.1998.4066. [DOI] [PubMed] [Google Scholar]
- Shao ZM, Radziszewski WJ, Barsky SH. Tamoxifen enhances myoepithelial cell suppression of human breast carcinoma progression in vitro by two different effector mechanisms. Cancer Lett. 2000;157:133–44. doi: 10.1016/s0304-3835(00)00466-3. [DOI] [PubMed] [Google Scholar]
- Sheng S. Maspin and the order of signaling. Cancer Biol Ther. 2003;2:404–5. doi: 10.4161/cbt.2.4.472. [DOI] [PubMed] [Google Scholar]
- Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJ, Sager R. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci U S A. 1996;93:11669–74. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S, Pemberton PA, Sager R. Production, purification, and characterization of recombinant maspin proteins. J Biol Chem. 1994;269:30988–93. [PubMed] [Google Scholar]
- Sheng S, Truong B, Fredrickson D, Wu R, Pardee AB, Sager R. Tissue-type plasminogen activator is a target of the tumor suppressor gene maspin. Proc Natl Acad Sci U S A. 1998;95:499–504. doi: 10.1073/pnas.95.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HY, Liang R, Templeton NS, Zhang M. Inhibition of breast tumor progression by systemic delivery of the maspin gene in a syngeneic tumor model. Mol Ther. 2002;5:755–61. doi: 10.1006/mthe.2002.0602. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–6. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–34. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark AM, Schem C, Maass N, Hugo HH, Jonat W, Mehdorn HM, Held-Feindt J. Expression of metastasis suppressor gene maspin is reduced in breast cancer brain metastases and correlates with the estrogen receptor status. Neurol Res. 2010;32:303–8. doi: 10.1179/016164109X12518779082192. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Kedeshian P, Shao ZM, Safarians S, Barsky SH. The human myoepithelial cell is a natural tumor suppressor. Clin Cancer Res. 1997;3:1949–58. [PubMed] [Google Scholar]
- Strien L, Joensuu K, Heikkila P, Leidenius MH. Different Expression Patterns of CXCR4, CCR7, Maspin and FOXP3 in Luminal Breast Cancers and Their Sentinel Node Metastases. Anticancer Res. 2017;37:175–182. doi: 10.21873/anticanres.11303. [DOI] [PubMed] [Google Scholar]
- Tamazato Longhi M, Cella N. Tyrosine phosphorylation plays a role in increasing maspin protein levels and its cytoplasmic accumulation. FEBS Open Bio. 2012;2:93–7. doi: 10.1016/j.fob.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamazato Longhi M, Magalhaes M, Reina J, Morais Freitas V, Cella N. EGFR Signaling Regulates Maspin/SerpinB5 Phosphorylation and Nuclear Localization in Mammary Epithelial Cells. PLoS One. 2016;11:e0159856. doi: 10.1371/journal.pone.0159856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Umekita Y, Ohi Y, Sagara Y, Yoshida H. Expression of maspin predicts poor prognosis in breast-cancer patients. Int J Cancer. 2002;100:452–5. doi: 10.1002/ijc.10500. [DOI] [PubMed] [Google Scholar]
- Umekita Y, Ohi Y, Souda M, Rai Y, Sagara Y, Sagara Y, Tamada S, Tanimoto A. Maspin expression is frequent and correlates with basal markers in triple-negative breast cancer. Diagn Pathol. 2011;6:36. doi: 10.1186/1746-1596-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umekita Y, Yoshida H. Expression of maspin is up-regulated during the progression of mammary ductal carcinoma. Histopathology. 2003;42:541–5. doi: 10.1046/j.1365-2559.2003.01620.x. [DOI] [PubMed] [Google Scholar]
- Wang KH, Kao AP, Chang CC, Lee JN, Chai CY, Hou MF, Liu CM, Tsai EM. Modulation of tumorigenesis and oestrogen receptor-alpha expression by cell culture conditions in a stem cell-derived breast epithelial cell line. Biol Cell. 2010;102:159–72. doi: 10.1042/BC20090132. [DOI] [PubMed] [Google Scholar]
- Yin S, Li X, Meng Y, Finley RL, Jr, Sakr W, Yang H, Reddy N, Sheng S. Tumor-suppressive maspin regulates cell response to oxidative stress by direct interaction with glutathione S-transferase. J Biol Chem. 2005;280:34985–96. doi: 10.1074/jbc.M503522200. [DOI] [PubMed] [Google Scholar]
- Yin S, Lockett J, Meng Y, Biliran H, Jr, Blouse GE, Li X, Reddy N, Zhao Z, Lin X, Anagli J, Cher ML, Sheng S. Maspin retards cell detachment via a novel interaction with the urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor system. Cancer Res. 2006;66:4173–81. doi: 10.1158/0008-5472.CAN-05-3514. [DOI] [PubMed] [Google Scholar]
- Zhang M, Magit D, Botteri F, Shi HY, He K, Li M, Furth P, Sager R. Maspin plays an important role in mammary gland development. Dev Biol. 1999;215:278–87. doi: 10.1006/dbio.1999.9442. [DOI] [PubMed] [Google Scholar]
- Zhang M, Sheng S, Maass N, Sager R. mMaspin: the mouse homolog of a human tumor suppressor gene inhibits mammary tumor invasion and motility. Mol Med. 1997;3:49–59. [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Shi Y, Magit D, Furth PA, Sager R. Reduced mammary tumor progression in WAP-TAg/WAP-maspin bitransgenic mice. Oncogene. 2000a;19:6053–8. doi: 10.1038/sj.onc.1204006. [DOI] [PubMed] [Google Scholar]
- Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000b;6:196–9. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor- suppressing activity in human mammary epithelial cells. Science. 1994;263:526–9. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]


