Summary
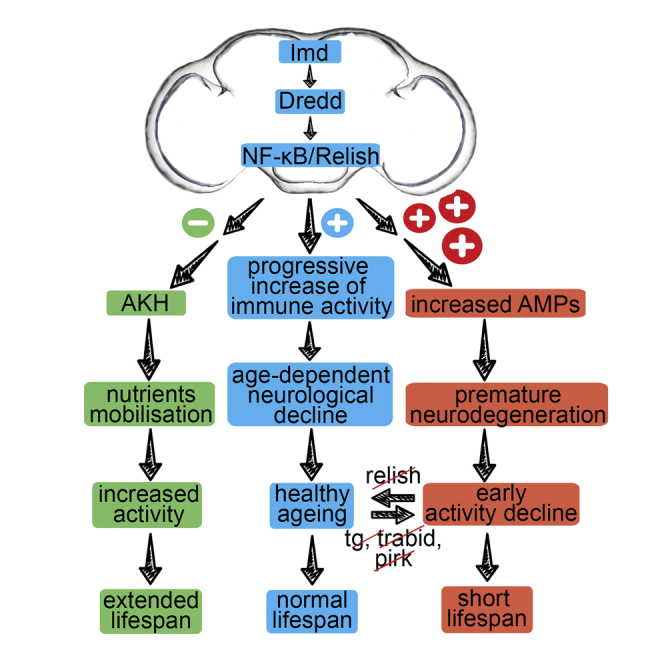
During aging, innate immunity progresses to a chronically active state. However, what distinguishes those that “age well” from those developing age-related neurological conditions is unclear. We used Drosophila to explore the cost of immunity in the aging brain. We show that mutations in intracellular negative regulators of the IMD/NF-κB pathway predisposed flies to toxic levels of antimicrobial peptides, resulting in early locomotor defects, extensive neurodegeneration, and reduced lifespan. These phenotypes were rescued when immunity was suppressed in glia. In healthy flies, suppressing immunity in glial cells resulted in increased adipokinetic hormonal signaling with high nutrient levels in later life and an extension of active lifespan. Thus, when levels of IMD/NF-κB deviate from normal, two mechanisms are at play: lower levels derepress an immune-endocrine axis, which mobilizes nutrients, leading to lifespan extension, whereas higher levels increase antimicrobial peptides, causing neurodegeneration. Immunity in the fly brain is therefore a key lifespan determinant.
Keywords: Drosophila, NF-κB, Imd, Relish, brain, innate immunity, lifespan
Graphical Abstract
Highlights
-
•
Constitutive immunity predisposes to short lifespan with severe neurodegeneration
-
•
Blocking constitutive immunity in glia rescues predisposed flies
-
•
Suppression of immunity in glia of healthy flies triggers adipokinetic signaling
-
•
This immune-endocrine axis mobilizes nutrients and extends active lifespan
In humans, both healthy aging and age-dependent neurodegeneration are accompanied by an upregulation of innate immunity. What is the cause and what is the consequence remain unclear. Kounatidis et al. show that, in flies, NF-κB immune signaling controls lifespan in healthy flies, as well as in those predisposed to early neurodegeneration.
Introduction
In the later stages of human life, there is a sustained increase in innate immune activity that has been associated with aging (Franceschi et al., 2007). This is in contrast to the induction of an immune response by infection and its rapid termination when the pathogen is cleared. This heightened age-associated pro-inflammatory activity has been termed “inflammageing” (Franceschi et al., 2007). It is thought to be partly balancing the exhaustion of T cell-mediated immunity caused by a decrease in the number of naive T cells produced by the thymus because of organism-wide cell senescence (Franceschi et al., 2007, Deeks, 2011). Other causes include the accumulation of tissue damage during the removal of senescent cells in various tissues and a defective autophagy response (Salminen et al., 2012). One of the most important transcriptional signatures in inflammageing of both humans and mice is the activation of the pro-inflammatory nuclear factor κB (NF-κB) pathway (de Magalhães et al., 2009, Zhang et al., 2013).
It has been hypothesized that, under certain conditions of heightened innate immunity, inflammageing may lead to age-associated brain neurodegeneration (Giunta et al., 2008). Although it is not known what predisposes individuals to disease, central to this immune-to-brain signaling are microglia. In the aging brain, the microglia have a primed phenotype as a consequence of changes in their local environment (Moreno et al., 2011). Nevertheless, whether the process involves the age-dependent activity of the NF-κB pathway is still an open question.
Taken together, the data mentioned above indicate the important influence of innate immunity on both healthy aging and age-related neurodegeneration. However, because cause and effect are so intricately linked (neurodegeneration itself causes inflammation), it is unclear how common themes in signaling may end up with such distinct phenotypes (healthy aging versus neurological disease). In this context, we used Drosophila to explore the potential influence of NF-κB-controlled immune signaling in predisposition to age-related neurological disease as well as healthy aging.
In Drosophila, systemic infection triggers two NF-κB signaling pathways; namely, Toll and immune deficiency (IMD). The latter pathway has extensive similarities with the tumor necrosis factor receptor 1 (TNFR1) signaling cascade (Kounatidis and Ligoxygakis 2012). Upon infection, activation of IMD is triggered when fragments of peptidoglycan released by Gram-negative bacteria or Gram-positive bacilli bind the transmembrane peptidoglycan recognition protein (PGRP)-LC or the intracellular PGRP-LE (Kaneko et al., 2006). The signal is then transduced through a receptor-adaptor complex to the NF-κB homolog Relish (Rel). Specifically, IMD (a RIP1 homolog) associates with the Fas-associated death domain protein (FADD), which then recruits the caspase-8 homolog death related ced-3/Nedd2-like caspase (DREDD), which is activated by ubiquitylation (Meinander et al., 2012). DREDD cleaves IMD, thus unmasking a domain of interaction with the Drosophila inhibitor of apoptosis-2 (dIAP-2), which ubiquitinates and stabilizes IMD (Paquette et al., 2010). This creates a transient signaling platform for the recruitment of transforming growth factor β (TGF-β)-activating kinase 1 (TAK1) and its binding adaptor TAB2 (Fernando et al., 2014). The TAK1/TAB2 complex mediates phosphorylation of the IκB kinase (IKK) on one hand and Jun nuclear kinase (JNK) on the other (Silverman et al., 2003). In turn, IKK phosphorylates the N-terminal domain of Rel, whereas DREDD cleaves the C-terminal. N-terminal Rel is then free to move to the nucleus and regulate transcriptional targets, including induction of antimicrobial peptide (AMP) genes (Stoven et al., 2003). As the signal is transmitted from the cell surface to the nucleus, there is negative regulation at every step. There is inhibition of PGRP-LC signaling by the transmembrane PGRP-LF (Basbous et al., 2011), inhibition of the receptor-adaptor complex through Rudra/Pirk (Aggarwal et al., 2008), and blocking of the signaling flow by successive de-ubiquitination enzymes targeting IMD (dUSP36) (Thevenon et al., 2009), TAK1 (the A20 homolog Trabid) (Fernando et al., 2014), or IKK (the cylidromatosis disease homolog cylindromatosis [CYLD]) (Tsichritzis et al., 2007). Moreover, ubiquitin-mediated proteolysis depletes the pathway from DREDD (via Dnr1) (Guntermann et al., 2009), TAK1 (via plenty of SH3 [POSH]) (Tsuda et al., 2005), and Relish (via ring and YY1 binding protein [dRYBP]) (Aparicio et al., 2013), whereas transglutaminase (TG)-catalyzed protein-protein cross-linking prevents Relish from entering the nucleus (Shibata et al., 2013). Finally, Caspar inhibits DREDD-dependent cleavage of Relish (Kim et al., 2006). In addition, there are extracellular negative regulators represented by secreted catalytic PGRP proteins (PGRP-LB and PGRP-SC), which reduce the epithelial and/or systemic response by scavenging peptidoglycan (Paredes et al., 2011). The safeguarding of the IMD pathway at all levels and with multiple means underlines the notion of an important cost paid if these safeguards were to decrease or collapse. Indeed, lack of Trabid, Pirk, PGRP-SC, or PGRP-LB compromise lifespan (Fernando et al., 2014, Paredes et al., 2011), whereas mutations in dnr-1 or overexpression of AMPs in the brain result in neurodegeneration (Cao et al., 2013). Similarly, continuous overexpression of PGRP-LE, leading to chronic upregulation of AMPs, compromised lifespan in a Relish-dependent manner, linking immunity, inflammation, and longevity in flies (Libert et al., 2006). Moreover, TG has been associated with neurotoxicity in a spinocerebellar ataxia model (Lin et al., 2015),whereas mutations in relish suppress neurodegeneration in an ataxia-telangiectasia model (Petersen et al., 2013). Nevertheless, innate immune genes are upregulated in fly models of neurodegeneration, raising the possibility that this upregulation may be protective (Cantera and Barrio, 2015). In this context also, cause and consequence might be intimately linked.
In addition to the link between components of the immune system and neurodegeneration, there is an intimate connection between immunity and metabolism. In mammals, adipose tissues and infiltrating immune cells produce numerous bioactive factors that have pro-inflammatory or anti-inflammatory activities. Dysregulated production of so-called adipokines can contribute to the pathogenesis of obesity-linked metabolic disease (for a review, see Ouchi et al., 2011). These players have been shown to drive type 2 diabetes, whereas cytokines regulate lipid stores (for a review, see Donath and Shoelson, 2011). In flies, prolonged immune activation in the context of bacterial or viral infections has also been associated with deregulation of metabolism, mainly through the insulin signaling pathway (Dionne et al., 2006; DiAngelo et al., 2009). More recently, a switch between immunity and metabolism has been identified in the transcription factor Mef2. There, sustained immune activity removed Mef2 from metabolic regulation, whereas, in the absence of infection, Mef2 associated primarily with metabolic transcriptional signatures (Clark et al., 2013).
The results presented here show that, with age, NF-κB-dependent AMP gene expression increased, and this was accompanied by progressive neurodegeneration and locomotion decline. Constitutive NF-κB immune signaling (in pirk, trbd, or tg mutants) resulted in high head and brain AMPs. Flies had a short lifespan, severe neurodegeneration, and locomotor defects. Conversely, reducing the normal levels of NF-κB in the brain of healthy flies resulted in an extended lifespan with improved activity in old age, accompanied by increased hormonal signaling and elevated glucose, trehalose, and triglycerides. Our results demonstrate that IMD/NF-κB/Relish immune signaling in glia determines lifespan.
Results
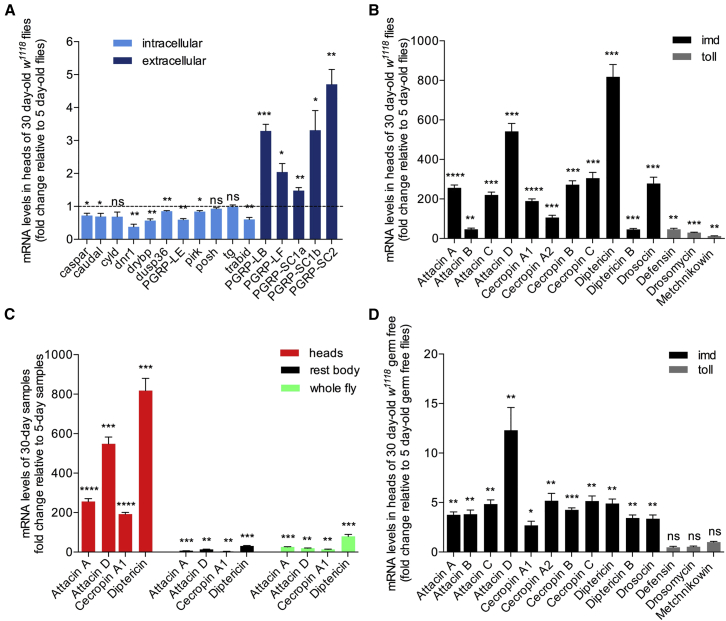
Age-Dependent Immune Regulation in Flies
We monitored the age-related expression of negative regulators of IMD, Toll, and JNK by comparing healthy whole flies, heads, and the rest of the body in the absence of infection. Our results indicated a consistent pattern: age-dependent gene expression of IMD intracellular negative regulators was significantly reduced in heads (Figure 1A) compared with the rest of the body (where expression of some was increased, compare Figure S1A with Figure 1A) or to whole flies (where expression appeared unchanged, compare Figure S1B with Figure 1A). In contrast, expression of IMD extracellular negative regulators was increased with age in heads (Figure 1A), the rest of the body (Figure S1A), and whole flies (Figure S1B). The decrease of intracellular negative regulation in the heads of aging flies was paralleled by a dramatic increase in AMP gene expression in the heads of 30- and 50-day-old flies (compare Figure 1B with Figures S1C and S1D for the rest of the body and whole flies respectively; see Figure 1C for a combined graph for a 30-day-old dataset; see Figure S1E for a 50 day-old dataset). We also observed a statistically significant increase in expression levels of rel in heads and whole bodies of 30-day-old flies compared with young 5-day-old controls (Figure S1F). Dif expression was also increased in the heads of 30-day-old flies (Figure S1F). However, this increase was not to the same extent as rel (3.2-fold versus 1.9-fold induction for rel and dif, respectively; Figure S1F). No significant change in expression of other components of the Toll or JNK pathway was observed with age (Figure S1F).
Figure 1.
Healthy Aging in Drosophila Is Characterized by Age-Dependent Upregulation of Immunity
(A–D) In the heads of 30-day old flies, gene expression of intracellular negative regulators of IMD was reduced, whereas gene expression of extracellular negative regulators was increased (A). This pattern was accompanied by a significant increase in AMP gene expression in the head (B), the rest of the body, and whole flies (C, comparative graph). IMD-dependent (but not TOLL-dependent) AMP gene expression was also increased with age in the heads of 30-day-old germ-free flies (D). Values shown are mean ± SEM.
Comparison of AMP gene expression levels in the heads of conventionally reared versus germ-free flies indicated the importance of the microbial environment in the age-dependent increase observed (compare Figure 1D with Figure 1C). The largest part of this increase (>90%) was microbiota-dependent (Figures S1G and S1H). Nevertheless, this still indicated that there was a microbially independent, intrinsic propensity for age-dependent deregulation of the IMD pathway (Figures S1G and S1H). (Of note, that measurements under axenic conditions showed that the AMP gene expression increase concerned only IMD-regulated AMPs and not TOLL-dependent ones; Figure 1D).
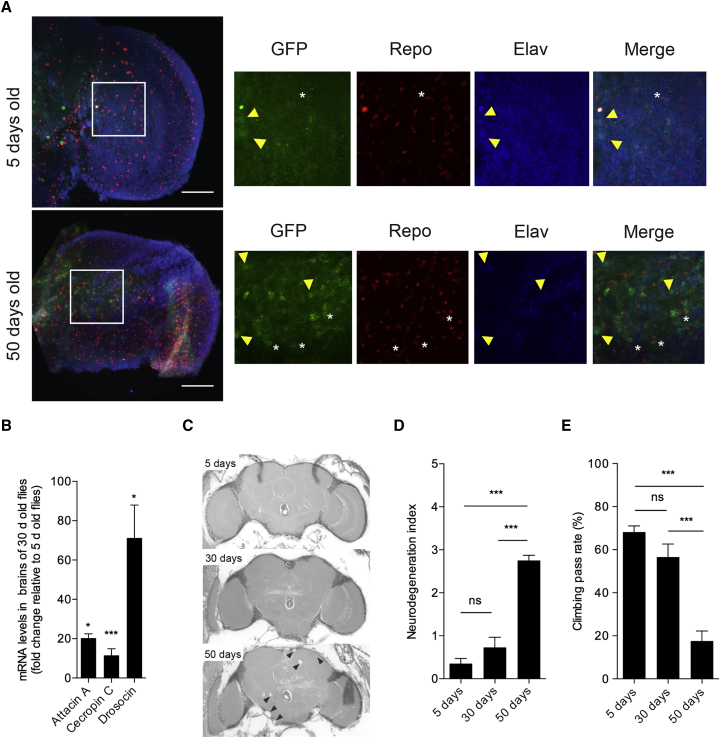
We also observed an age-dependent increase in GFP expression in the brains of flies carrying a reporter gene in which GFP expression was under the control of the attacinA promoter (Tzou et al., 2000) (Attacin-GFP; Figure 2A). In addition, measurements by qPCR revealed an increase in AMP expression in brains of 30-day-old flies compared with their 5-day-old counterparts (Figure 2B). The results from these experiments indicated that the age-dependent increase in IMD-related AMP gene expression was concerning the brain itself and not just the surrounding tissues.
Figure 2.
Increase of AMP Levels in the Brain Correlates with Neurological Decline
(A) Confocal stacks of 5- and 50-day-old (top and bottom panels, respectively) Attacin-GFP optic lobes of brains immunostained for GFP (green), the glial marker Repo (red), and the neuronal marker Elav (blue). All three markers are shown in the merged image. An increase in GFP staining is observed in the brains of older flies, where GFP co-localizes with both Repo (asterisks) and Elav (arrowheads). The boxed regions show an enlarged image of the optic lobes. Representative images are shown. n = 19 (5 days old) and n = 12 (50 days old). Scale bars, 50 μm.
(B–E) Significant age-dependent upregulation of AMPs in brains was also observed by qPCR (B). This increase was accompanied by neurodegeneration (arrowheads) in 50-day-old flies as observed (C) and quantified (D) in midbrain sections. This was coupled to a reduction in locomotor activity in 50-day-old flies (E).
In all experiments, the fly strain used was w1118. Values shown are mean ± SEM. Asterisks denote statistically significant differences (∗p ≤ 0.05, ∗∗∗p ≤ 0.001. ns, non-significant.
This age-dependent shift in IMD-related AMP transcription was the result of a specific pattern: a combined decrease in some negative intracellular regulators and an increase in extracellular negative regulators with a sum total culminating in an increase in rel mRNA levels and a rise in AMPs controlled by Rel. Furthermore, the age-dependent increase of AMPs was accompanied by an increase in brain neurodegeneration (see Figure 2C for sections, Figure 2D for index, and Table S1 for statistics) and locomotion defects (see Figure 2E and Table S2 for statistics) in 50-day-old flies. Taken together, the data above indicated a phenotype where an age-related increase in IMD/NF-κB-driven immunity was combined with a neurological decline. Whether this was a cause-and-effect relationship was our next question.
Loss of NF-κB Intracellular Negative Regulation Predisposes to Early Neurodegeneration and Short Lifespan
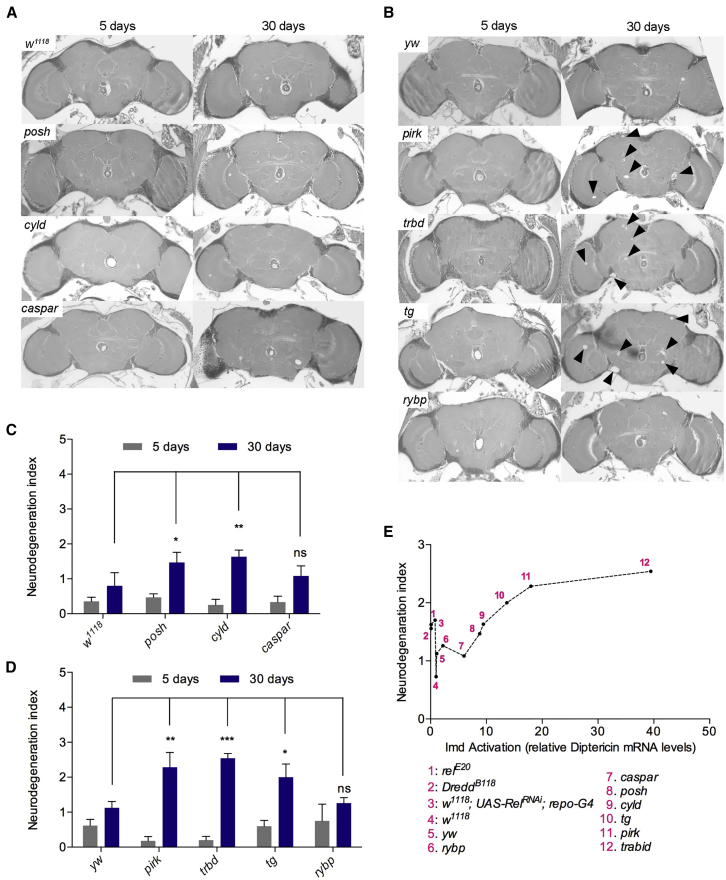
We reasoned that one way to explore this question would be to manipulate IMD/NF-κB signaling and measure any changes in the age-dependent neurological state of our flies. The causative relationship between increased IMD/NF-κB signaling levels and age-dependent neurological deterioration was identified previously for loss of Dnr1 (Cao et al., 2013). Therefore, we asked whether disabling other negative regulators and, thus, increasing IMD/NF-κB signaling levels could predispose flies to early onset of neurological decline, brain neurodegeneration, and, finally, reduced lifespan. Brain sections of 5- and 20- or 30-day-old loss-of-function mutants of the intracellular negative regulators (regardless whether their age-dependent expression declined or stayed the same) posh, cyld, and caspar (with their w1118 genetic background as a control) and tg, trbd, pirk, and drybp (with their yw genetic background as a control) as well as of the extracellular negative regulator pgrp-lb (with its w1118 genetic background as a control) were examined at both 25°C (see Figures 3A and 3B for brain sections and Figures 3C and 3D and Table S1 for index) and 29°C (accelerated aging; see Figures S2A, S2B, and S2E for brain sections, Figures S2C, S2D, and S2F for index, and Table S1 for statistical analysis). Early studies examining Drosophila lifespan at different temperatures have established a positive relationship between mortality and increase in temperature, therefore suggesting that flies are living and aging faster at 29°C than at 25°C (Alpatov and Pearl, 1929, Miquel et al., 1976). At both temperatures, our results revealed a statistically significant increase in the neurodegeneration index for three mutants in intracellular regulators of the IMD pathway; namely, tg, trbd, and pirk. In addition, plotting the neurodegeneration index against Imd transcriptional output (measured by gene expression levels of the AMP diptericin) indicated that the dipt levels in these three mutants correlated with the highest index (Figure 3E).
Figure 3.
Loss of Intracellular Negative Regulators of NF-κB/Relish Triggers Early Onset of Neurodegeneration
5- and 30-day-old mutants for intracellular negative regulators of the IMD pathway, aged at 25°C, were examined for age-dependent neurodegeneration by histology and then quantified.
(A) Midbrain sections of caspar mutant flies did not show any differences compared with their genetic background (age-matched w1118 controls). However, some sections showed an increase in cyld and posh mutants (data not shown).
(B) Mutants for the intracellular negative regulators pirk, trbd, and tg, but not rybp, showed increased age-dependent neurodegeneration in midbrain sections.
(C) When quantified, caspar mutant flies had a neurodegeneration index indistinguishable to w1118 controls, whereas posh and cyld had a significant increase in neurodegeneration.
(D) Quantification of pirk, trbd, and tg, but not rybp, revealed a neurodegeneration index that was significantly different from yw controls.
(E) Neurodegeneration index in correlation with Imd pathway output (as measured by dipt gene expression). The increase in neurodegeneration correlated with an increase in mRNA levels of dipt.
Values shown are mean ± SEM. Asterisks denote statistically significant differences (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
Of note, we found that the neurodegeneration index for conventionally reared mutants of PGRP-LB was comparable with the wild-type. This indicated that, despite the reduction in lifespan reported in the literature (Paredes et al., 2011), neurodegeneration was not the cause of early death in this mutant. Moreover, in contrast to trbd and tg (see below), under germ-free conditions, the lifespan of flies lacking PGRP-LB was restored to normal (Paredes et al., 2011 and data not shown). This result confirmed previous studies that indicated that the reduction in lifespan in PGRP-LB mutants was due to commensal dysbiosis (Paredes et al., 2011).
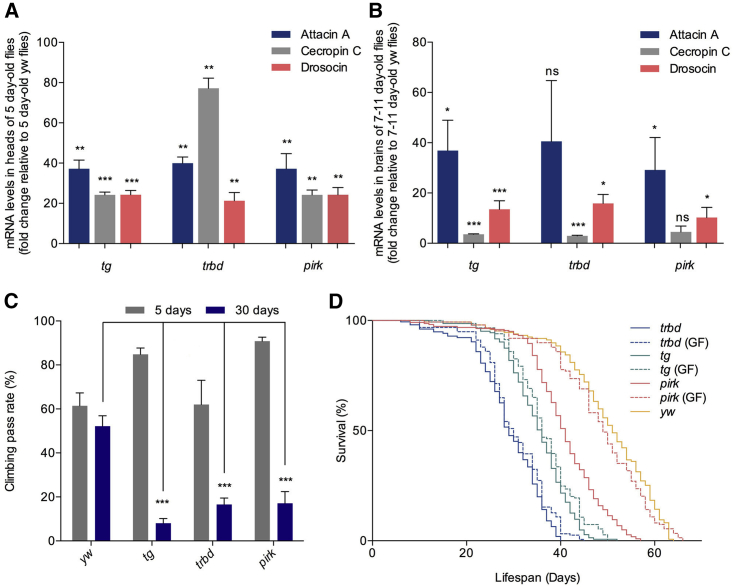
Along with the age-dependent neurodegeneration in tg, trbd, and pirk, these mutants showed a statistically significant increase in AMP gene expression in heads (Figure 4A), dissected brains (Figure 4B), and the rest of the body (Figure S3A) compared with the control. They were therefore exposed to “increased immune signaling from a young age” (predisposition) and were studied further. Tg, trbd, and pirk mutant flies exhibited an early reduction in locomotor activity (see Figure 4C and Table S2 for statistics) and showed a 24.2%, 37.1%, and 9.1% reduction in maximum lifespan, respectively, compared with wild-type controls (see Figure 4D and Table S3 for statistics). In the case of trbd and tg, lifespan patterns were not influenced by the absence of bacterial populations because they exhibited a statistically non-distinguishable reduction in lifespan under germ-free conditions (Figure 4D; Table S3). However, the reduction in lifespan of pirk mutants was rescued in germ-free flies (Figure 4D; Table S3), as has been reported elsewhere (Paredes et al., 2011). This result was in accordance with the published observation that pirk expression was regulated by the presence of the gut microflora (Lhocine et al., 2008) and underscored the differences between microbe-independent and microbe-dependent intracellular negative regulation of the IMD pathway (see Discussion).
Figure 4.
Loss of Intracellular Negative Regulators of NF-κB/Relish Leads to a Brain-Specific Increase in Expression of AMPs, Locomotor Defects, and Shorter Lifespan
(A–D) A significant increase in AMP gene expression in comparison with yw controls was observed in heads (A) and dissected brains (B) of pirk, trbd, and tg mutants that correlated with a decrease in locomotor activity (C). The shorter lifespan of pirk, but not trbd or tg mutants, was rescued under germ-free (GF) conditions (D). Values shown are mean ± SEM. Asterisks denote statistically significant differences (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
In contrast to pirk, trbd, and tg, negative regulator mutants that did not show age-dependent neurodegeneration, such as drybp (Figure 3A) and caspar (Figure 3B), had levels of AMPs in their brains similar to controls (Figure S3B) and a lifespan that was not reduced compared with controls (the lifespan for caspar was actually increased; see Figure S3C and Table S3 for statistics). These results indicated that loss of different intracellular negative regulators had different effects on brain-specific AMP expression and, as a consequence, different effects on brain tissue, locomotion, and, ultimately, lifespan. This result also suggested possible tissue specificity (see Discussion). These differences notwithstanding, our results indicated that an increase in AMP gene expression did correlate with an increase in neurological decline and reduced lifespan. Indeed, Cao et al. (2013) have shown that overexpression of single AMPs increased neurodegeneration). We explored further the underlying neurological and lifespan phenotype and found that individual overexpression of drosocin, attacinC, or cecropinA1 in neurons resulted in reduced lifespan and age-dependent climbing ability (Figures S3D and S3E, respectively). Similarly, overexpression of drosocin, attacinC, or cecropinA1 in glia resulted in reduced lifespan and age-dependent climbing ability (Figures S3F and S3G, respectively).
Silencing NF-κB in the Brain of Predisposed Flies Restores Lifespan and Suppresses Neurodegeneration
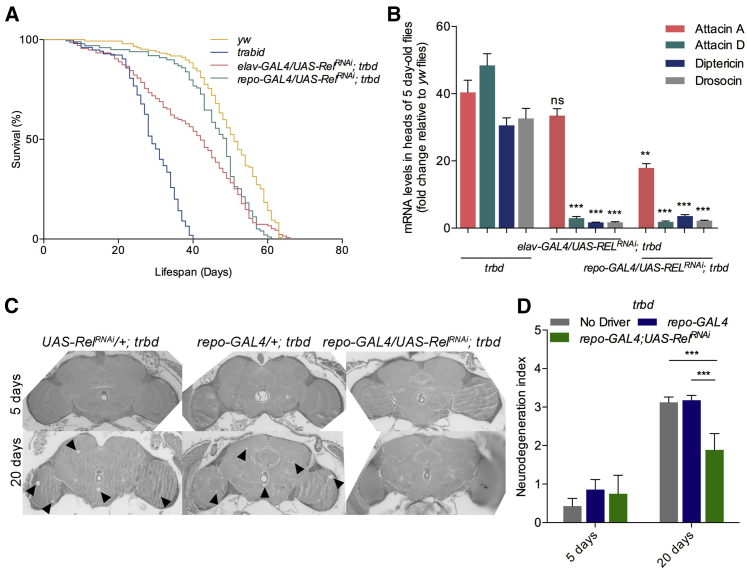
In trbd mutants, the median lifespan (LT50) was 29 days (Figure 5A; statistics in Table S3; see also Fernando et al., 2014). This phenotype was suppressed when rel was silenced in neurons (w1118; elav-GAL4/UAS-RelRNAi; trbd, LT50 = 42) and even more when rel was suppressed in glia (w1118; repo-GAL4/UAS-RelRNAi; trbd), with an LT50 of 48 days that was in line with the LT50 (52 days) of its genetic background (Figure 6A; Table S3). Mating rates were statistically indistinguishable in all different RNAi interventions compared with controls and trbd mutants (Table S6). Finally, the lifespan of trbd flies with only either of the GAL4 drivers or just the UAS-RelRNAi transgene was statistically indistinguishable from trbd alone (data not shown).
Figure 5.
Predisposition to Neurodegeneration Is Suppressed by Reducing NF-κB/Relish in the Brain
(A and B) In trbd mutants, reduction of lifespan (A) and upregulation of AMPs (B) were suppressed by silencing rel in neurons (elav-GAL4) or glia (repo-GAL4).
(C and D) Neurodegeneration in brain sections (C) and quantification (D) showed that lifespan extension was accompanied by fully rescued neurodegeneration only when rel was silenced in glia (compare with Figure S4B for sections when rel was silenced in neurons).
Values shown are mean ± SEM. Asterisks denote statistically significant differences (∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
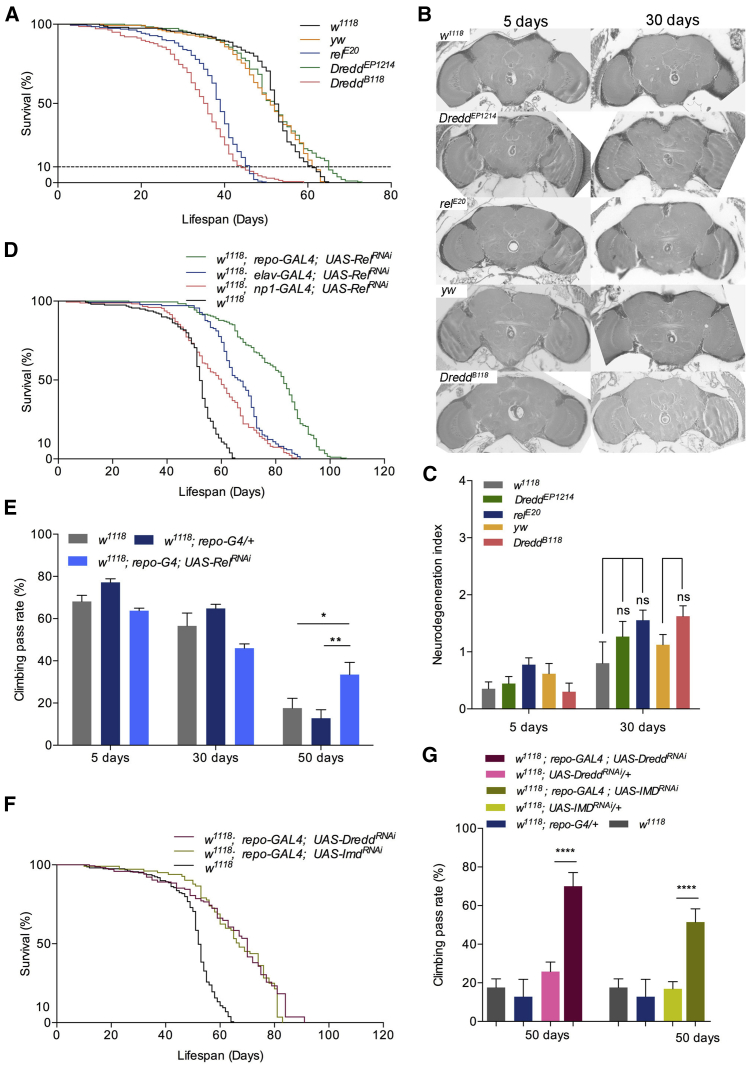
Figure 6.
Glial Suppression of IMD/NF-κB Signaling in Healthy Flies Extends Active Lifespan
(A–C) The maximum lifespan of a hypomorphic allele of Dredd (DreddEP1214) showed a modest extension compared with w1118 controls. Nevertheless, midbrain sections (B) and quantification (C) of yw; dreddB118, w1118; relE20-null mutants and a w1118; dreddEP1412 hypomorphic mutant showed no difference in neurodegeneration compared with controls.
(D–G) RNAi-dependent silencing of rel in glia resulted in a very substantial extension of lifespan (D) and amelioration of the age-dependent decline in locomotor activity (E). Similarly, glial knockdown of Dredd and Imd resulted in lifespan extension (F) and amelioration of the age-dependent decline in locomotor activity (G).
Values shown are mean ± SEM. Asterisks denote statistically significant differences (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001).
Compared with 5-day-old and 30-day-old controls, rel-silenced flies exhibited reduced AMPs in heads (Figure 5B). Neurodegeneration was reduced, and locomotor activity increased when rel was silenced in glia (see Figure 5C for brain sections, Figure 5D and Table S1 for index, and Figure S4A and Table S2 for locomotor assays), but not when it was silenced in neurons (see Figure S4B for brain sections, Figure S4C and Table S1 for index, and Figure S4D and Table S2 for locomotor assays). This result underlined the role of glia in the immunity-to-brain axis whereby suppressing immunity in glia rescued both neurodegeneration and locomotion and, thus, restored lifespan to wild-type levels. The data also supported the notion that predisposition to early neurodegeneration was determined by IMD/NF-κB pathway levels.
Suppressing NF-κB Immune Signaling in Healthy Flies Extends Lifespan
If increased levels of IMD/NF-κB signaling led to the reduction of lifespan with early onset of age-related neurological decline, we wanted to test whether a decrease in normal IMD signaling levels in healthy flies from the beginning of their life could have the opposite effect. To this end, we initially assayed the effects on lifespan of two IMD-null mutants (yw; dreddB118 and w1118; relE20), one hypomorphic mutant (w1118; dreddEP1412), and their genetic backgrounds to which the mutant strains were backcrossed for ten generations as recommended (He and Jasper, 2014). Dredd- or rel-null flies had a statistically significant reduction in lifespan compared with their genetic backgrounds (Figure 6A; Table S3). However, the hypomorphic w1118; dreddEP1412, although it showed the same significant reduction in LT50 as the null IMD pathway mutants compared with controls, also displayed a statistically significant increase in less than 90% (LT90) and maximum lifespan (Figure 6A; Table S3). However, no difference was observed in neurodegeneration in both brain sections (Figure 6B; Table S1) and index (Figure 6C; Table S4).
Nevertheless, this result suggested the possibility that a lower (but not null) IMD pathway activity could increase the maximum lifespan in some flies by reducing the cost of the age-dependent increase in levels of immune activity. To test this idea, we used the galactose responsive transcription factor 4/upstream activation sequence (GAL4/UAS) system to suppress rel via RNAi in either neurons (elav-GAL4) or glia (repo-GAL4) or the intestine (np1-GAL4). The latter GAL4 driver was used because the gut has been suggested to be the tissue where upregulation of rel causes age-related commensal dysbiosis (Guo et al., 2014). Previous studies have demonstrated that induction of the RNAi mechanism itself in the nervous system or gut does not influence adult lifespan (Alic et al., 2012). In addition, the mating rates were statistically indistinguishable in all different RNAi interventions compared with controls (Table S6).
Our results indicated a significant extension of lifespan when rel was suppressed in either neurons or glia (Figure 6D). In our hands (see Experimental Procedures), the genetic background w1118 had an LT50 of 52 days and an LT90 of 62 days, with a maximum lifespan of 65 days (see Figure 6D for a graph and Table S3 for statistics). In comparison, the w1118; elav-GAL4; UAS-RelRNAi LT50 was 67 days (an increase of 29%) and an LT90 of 79 days with a maximum lifespan of 88 days (35% increase). For w1118; repo-GAL4; UAS-RelRNAi, the LT50 was 84 days (an increase of 61%), the LT90 was 95 days, and the maximum lifespan was 106 days (an increase of 63%; Figure 6D and Table S3). We also observed a comparatively modest (but statistically significant) increase in lifespan in w1118; np1-GAL4; UAS-RelRNAi, as recently documented (Guo et al., 2014). In w1118; np1-GAL4; UAS-RelRNAi flies, the LT50 was 61 days and the maximum lifespan 70 days. However, w1118; np1-GAL4 alone had a slightly increased LT50 compared with w1118, which was not the case for w1118; elav-GAL4 or w1118; repo-GAL4 (Figure 6D; Table S3). Interestingly, flies with glia- or neuron-specific suppression of immunity signaling did not show a reduction in brain neurodegeneration levels compared with controls (see Figure S5A for brain sections, Figure S5B for glia index, and Figure S5C for neuron index) even though AMPs in the heads were significantly reduced as expected (Figure S5D). This indicated that a reduction in the basal level of neurodegeneration was not the reason for lifespan extension. Nevertheless, glia-specific (but not neuron-specific) suppression resulted in increased locomotion in older flies (Figure 6E). This indicated that the lifespan extension in w1118; repo-GAL4; UAS-RelRNAi flies was not just an extension of the moribund phase but corresponded to more activity in old age. An extension of lifespan statistically comparable with w1118; repo-GAL4; UAS-RelRNAi flies was also observed in w1118; repo-GAL4; UAS-dreddRNAi flies and w1118; repo-GAL4; UAS-imdRNAi flies, two IMD pathway components upstream of Relish (see Figure 6F and Table S3 for statistics). To verify whether mating had an effect, we reproduced the same result in mated females only (see Figure S5E and Table S6 for statistics). Crucially, this extension was also correlated with a greater locomotor activity in later life (see Figure 6G and Table S5 for statistics). These results reinforced the notion that IMD/NF-κB signaling in the brain was linked to life expectancy in flies.
An Immune-Neuroendocrine Axis Mediates Lifespan Extension in Flies
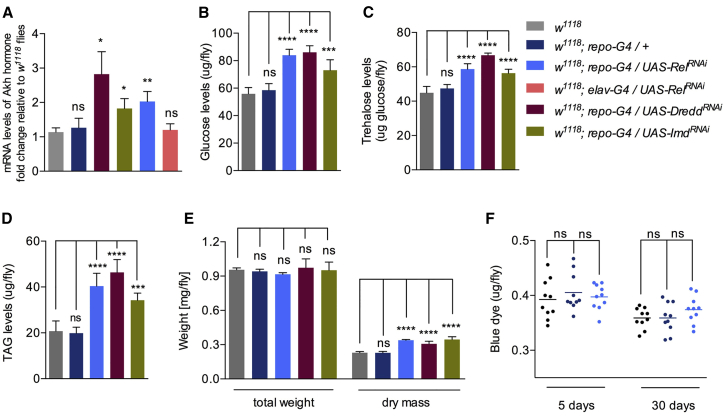
The observed increase in physical performance in old age in flies with suppressed Imd signaling in glial cells was accompanied by doubling of the transcriptional levels of the adipokinetic hormone (Akh) hormone (Figure 7A). In contrast, this was not the case for mutants of negative regulators (trbd, tg, and pirk), where the pathway was constitutively active (Figure S6A).
Figure 7.
Lifespan Extension following Glial Suppression of IMD/NF-κB Signaling Is Accompanied by Metabolic Changes
(A) The RNAi-dependent silencing of rel, dredd, and imd resulted in an increase in Akh transcription.
(B–F) Increased AKH signaling triggered rises in (B) glucose, (C) trehalose, and (D) TAG levels that built up an increase in dry mass (E). Nevertheless, feeding rates were comparable with controls when rel was silenced in glia (F).
Values shown are mean ± SEM.
Akh, which has glucagon-like functions, has been shown to increase the levels of blood glucose (Kim and Rulifson, 2004) as well as to extend lifespan and modulate climbing activity in Drosophila (Waterson et al., 2014). It has also been shown to sustain flight in locusts, tobacco hornworm moths, and certain beetles (reviewed in Gäde and Auerswald, 2003). In addition, its homolog in mice decelerates aging (Zhang et al., 2013). Consistent with the above, injections of a locust Akh in 30-day-old (see Figure 7B for a 5-cm climb in 10 s) and 50-day-old wild-type Drosophila increased locomotion (see Figure 7C for a 5-cm climb in 10 s and Figure 7D for a 2.5-cm climb in 10 s).
To corroborate the role of AKH as the effector of lifespan extension, we measured metabolic markers that have been associated with AKH signaling in Drosophila (Lee and Park, 2004, Bharucha et al., 2008). 30-day-old w1118; repo-GAL4; UAS-RelRNAi flies showed a statistically significant increase in glucose levels (free glucose levels in whole flies; Figure 7B) as well as in circulating trehalose (representing glucose dimers; Figure 7C). Of note is that levels of glycogen, which represented stored glucose, were unchanged relative to controls (Figure S6E). Consistent with the action of AKH signaling on mobilization of lipid stores, triglycerides (TAGs) as well as total dry mass (but not total weight) were increased in long-lived flies (Figure 7E). In contrast, total protein levels were indistinguishable from w1118 controls and w1118; repo-GAL4 flies (Figure S6F). Finally, the metabolic changes observed were not due to calorie restriction because the feeding rates between long-lived and control flies were statistically indistinguishable (Figure 7F). Taken together, the above results indicated that, during healthy aging, suppression of NF-κB activity delayed neurological decline through a mechanism involving an immune-endocrine axis that seems to be evolutionary conserved in mice (Zhang et al., 2013).
Discussion
Loss of IMD Homeostasis Predispose to Age-Dependent Neurodegeneration
We have shown that, in Drosophila, an age-related pattern of increased NF-κB-controlled immune activity is caused by a parallel reduction in intracellular negative regulation. As seen in germ-free flies, this age-dependent rise in immune levels was observed downstream of IMD but not TOLL. Moreover, axenic conditions revealed that this increase was, to a large extent, due to the microbial environment. However, the trend of this increase (albeit at a reduced scale) remained in germ-free flies, indicating a mechanism independent of the microbiota.
This phenomenon was also accompanied by neurological and locomotor decline, but it was unclear whether this was a consequence of increased immune activation. If this were the case, we hypothesized that loss-of-function mutations in intracellular negative regulators of IMD would predispose flies to high AMP levels and, thus, to immune-dependent early neurodegeneration. Indeed, overexpression of AMPs with repo- or elav-GAL4 can cause lesions in the brain at 25 days of age (Cao et al., 2013). Moreover, overexpression of the AMPs drosocin, attacinC, and cecropinA1 in neurons or glia was accompanied by a reduction in lifespan and early appearance of locomotor defects in comparison with controls. Taken together, these results indicate a causative relationship between high levels of AMPs and neurodegeneration. Indeed, plotting the neurodegeneration index against the levels of the AMP gene diptericin in various mutant and control genetic backgrounds showed that increased neurodegeneration correlated with increased dipt levels (Figure 3E).
Nevertheless, Petersen et al. (2013) have suggested that it shows correlation, not causation, because imd alleles still show neurodegeneration. This view correlates with our results showing that both loss of function of IMD signaling components as well as silencing by RNAi in the brain of these same components did not reduce basal neurodegeneration. Therefore, the basal levels of neurodegeneration in healthy aging are primarily NF-κB-independent. Alternatively, the histological method we have used may not be sensitive enough to detect any potential small changes. Nevertheless, downregulation of NF-κB extends the active lifespan despite levels of neurodegeneration remaining constant. This shows that, when IMD is under normal regulatory conditions, longevity is determined by control of metabolism rather than neurodegeneration (see below). Conversely, when IMD control is lost, and AMPs increase to toxic levels, causing early neurodegeneration (Cao et al., 2013; this study), NF-κB activity is the dominant lifespan determinant over and above any metabolic process.
Loss of four intracellular IMD negative regulators, Trbd, Pirk, Tg (this study), and Dnr-1 (Cao et al., 2013), resulted in early neurodegeneration in combination with a reduction in lifespan. Nevertheless, the IMD pathway is negatively regulated at most steps. Therefore, the question arises of why loss of these particular four intracellular negative regulators produced age-dependent neurodegeneration.
Our view is that the data underscore the context and/or tissue-dependent nature of IMD signaling and the difference between intracellular control exerted when interacting with the gut microbiota and/or upon infection (dusp36, pgrp-le, pirk, and drybp) (see Thevenon et al., 2009, Bosco-Drayon et al., 2012, Lhocine et al., 2008; and Aparicio et al., 2013; respectively) versus infection-independent homeostatic control (dnr-1, trbd, and tg) (see Cao et al., 2013, Fernando et al., 2014; and Shibata et al., 2013; respectively). For example, Pirk is the upstream-most flora-dependent intracellular negative regulator that is a target of the pathway, and, therefore, its loss will result in the absence of an important negative feedback loop (Lhocine et al., 2008). Nevertheless, this feedback loop is not important for lifespan in the absence of the gut microflora (Paredes et al., 2011). In contrast, flies reared under germ-free conditions reveal that intracellular regulators such as Trbd are important for IMD pathway homeostasis independent of the microbial environment (Fernando et al., 2014). A non-canonical or tissue-specific IMD pathway may also be at play in the brain, as has been suggested for the fly retina (Chinchore et al., 2012) or in an ataxia telangiectasia model (Petersen et al., 2013).
Loss of negative regulation in IMD signaling in the brain led to tissue disruption and early neurodegeneration. This phenotype was accompanied by a severe reduction in lifespan and locomotor defects. However, when Relish was silenced in glia of trbd mutants, brain lesions, lifespan, and locomotion were restored and comparable with healthy control flies. This showed that all effects were due to the derepression of NF-kB signaling.
Our results indicate that reduction of lifespan in tg and trbd is microbe-independent. We suggest that the main reason for this lifespan reduction is the microbiota-independent part of the early increase in AMPs in those mutants. Interestingly however, the lifespan reduction in pirk mutant flies was rescued under germ-free conditions. Because pirk mutants display early neurodegeneration, this raises the possibility of a gut-brain axis that may include a microflora-dependent component, which, in turn, influences brain neurodegeneration. Indeed, it has been shown previously that, as flies age, their gut becomes leaky, resulting in aging decline with a strong microbiota-driven component (Rera et al., 2012). More work is needed to understand this interaction.
Finally, our work indicates that the age-dependent regulation of the Imd pathway is at a state of allostasis (when homeostasis is achieved through change). Taken at specific time points, signaling output is defined by the microbial environment, and, in the absence of infection, it is tightly regulated to a basal level. However, this basal level increases in an age-dependent manner. By culturing germ-free Drosophila, we were able to pinpoint the component of this signaling that is microbe-independent and intrinsic to the host.
Suppression of IMD Extends Lifespan
Lowering the levels of IMD signaling by RNAi in glia of healthy flies extended the lifespan by more than 60% in LT50 and increased physical activity in old age. Our results agree with a previous observation that pharmacological inhibition of NF-κB modestly extends the Drosophila lifespan (20% of LT50; Moskalev and Shaposhnikov, 2011). However, we go further to indicate mechanism and tissue specificity. Lifespan extension was not achieved by lower levels of neurodegeneration but by an increase in AKH signaling. This indicated that the basal level of neurodegeneration in healthy flies was independent of IMD/NF-κB.
Extension of lifespan was accompanied by an altered metabolic profile, including statistically significant increases in free glucose, trehalose, and TAGs, whereas there was no significant difference in glycogen and protein levels. This suggested a shift toward catabolism, leading to the immediate use of carbohydrates and fats (as opposed to storing them) to fulfill the energetic requirements of long-lived flies. Increases in glucose, trehalose, and TAGs were consistent with the upregulation of akh expression and confirmed IMD signaling as a potent modulator of lifespan upstream of AKH.
More work is needed to identify the NF-κB targets in glia that enable AKH activation and establishment of this immune-neuroendocrine axis. We speculate that this is not a direct transcriptional relationship because akh cis-regulatory elements do not have NF-κB binding sites (unpublished data). One possible connection however, could be through Mef2 (Clark et al., 2013). Mef2 is an in vivo immune-metabolic switch controlled by NF-κB that can alternate between immune and metabolic gene expression. Upon infection, NF-κB activity recruits Mef2 as an immune regulator. A tempting hypothesis is that, by lowering NF-κB signaling under homeostatic conditions, Mef2 is freer to increase its involvement in metabolic activity.
Our results point to IMD/NF-κB signaling in the brain as a major determinant of Drosophila lifespan. The LT50 (84 days) of w1118; repo-GAL4; UAS-RelRNAi is a statistically indistinguishable percent extension of LT50 compared with w1118; methuselah (Lin et al., 1998), lifespan extension by loss of insulin signaling (Clancy et al., 2001), or calorie restriction (Mair et al., 2003). Our data put under an evolutionary perspective studies in mice where a similar NF-κB-controlled immune-neuroendocrine integration led to modulation of hormone levels and lifespan extension (Zhang et al., 2013). However, our results go further to indicate that NF-κB signaling levels regulate both healthy aging as well as age-related neurodegenerative disease. Given the evolutionary conservation of NF-κB innate immune signaling in flies, mice, and humans, opposing the inflammatory effects of NF-κB may represent a common strategy to increase active lifespan in both the context of healthy aging as well as in cases of predisposition to age-related neurological disease.
Experimental Procedures
Drosophila Stocks and Genetics
Flies were maintained on cornmeal-molasses medium at 25°C unless otherwise stated. For a complete list of the strains used, check the Supplemental Experimental Procedures.
Lifespan Studies and Production and Maintenance of Germ-free Flies
For lifespan studies, all fly strains were backcrossed for ten generations to their respective genetic background as recommended previously (He and Jasper 2014). When these “lifespan-ready” strains were established, cohorts of 20 flies (10 males and 10 females) were put in vials and monitored for their survival in 12 biological replicates (n = 240/strain). The flies were transferred to fresh food every 2 days. Germ-free flies were produced as described previously (Fernando et al., 2014) and checked every 5 days for bacterial contamination by performing PCR analysis on fly homogenates using 16S eubacterial primers (63F//1387R) as well as by culturing the homogenates on LB plates.
Histology and Neurodegeneration Score
Histological analysis and determination of the neurodegeneration index were done as described previously (Cao et al., 2013). For a detailed description, check the Supplemental Experimental Procedures.
Gene Expression in Brains
Quantitative real-time PCR was used to measure mRNA expression. 30–40 fly brains were dissected at the indicated time points, and RNA was isolated using TrizolRT (Molecular Research Center) according to the manufacturer’s instructions. cDNA was prepared from 0.5 μg total RNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was carried out using iQ SYBR Green Supermix (Bio-Rad). Primer sequences are presented in the Supplemental Experimental Procedures.
Gene Expression in Heads, the Rest of the Body, and Whole Flies
Total RNA was extracted from whole flies (6 females), the rest of the body (8 females), and heads (30 females) using the Total RNA Purification Plus kit (Norgen Biotek), and cDNA was prepared from 0.5 μg total RNA using the Maxima First Strand cDNA synthesis kit (Thermo Scientific). Triplicate cDNA samples were amplified with the SensiFASAT SYBR No-ROX kit (Bioline) in a Corbet Rotor-Gene 6000 qPCR machine (QIAGEN) according to the manufacturer’s protocols. Primer sequences are presented in the Supplemental Experimental Procedures.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism software (GraphPad). All qPCR data were analyzed using nonparametric unpaired Student’s t test corrected for multiple comparisons (Bonferroni correction). Significant differences between the survival data of different genotypes were identified using log rank and Wilcoxon tests (chi-square and p values). The neurodegeneration index and climbing assay data were analyzed using one- or two-way ANOVA with Tukey and Bonferroni post tests, respectively. In all tests, p < 0.05 was considered significant.
Author Contributions
P.L., I.K., and S.C. designed the experiments. I.K. and S.C. performed the experiments with input from Y.C., M.H., and D.J. I.K., S.C., and P.L. analyzed the data with input from B.G. P.L. wrote the paper with input from I.K., S.C., and B.G.
Acknowledgments
We thank many colleagues for providing Drosophila stocks. We thank Aki Ikeda for use of his microtome facility, Ling Ling Ho for excellent technical help, Peter Burns for technical assistance, Maria Iossifidou for her help, members of the Ganetzky and Ligoxygakis labs for helpful discussions, and Daniel Babcock and David Wassarman for critical reading of the manuscript. We thank John Yoder (University of Alabama) for the use of his microscope facility and Janna Fierst (University of Alabama) for help with statistical tests. P.L. would like to dedicate this paper to the loving memory of our dear friend and colleague Thanasis Loukeris, the most sincere of scientists. He will forever keep a place in our hearts and minds. Work in Oxford was supported by the European Research Council (Consolidator Grant 310912 to P.L.). Work in Madison was supported by the NIH (grant R01 AG033620) and the Steenbock professorship (to B.G.).
Published: April 25, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.04.007.
Supplemental Information
References
- Aggarwal K., Rus F., Vriesema-Magnuson C., Ertürk-Hasdemir D., Paquette N., Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N., Hoddinott M.P., Foley A., Slack C., Piper M.D., Partridge L. Detrimental effects of RNAi: a cautionary note on its use in Drosophila ageing studies. PLoS ONE. 2012;7:e45367. doi: 10.1371/journal.pone.0045367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpatov W.W., Pearl R. Experimental studies on the duration of life. XXI. Influence of temperature during the larval period and adult life on the duration of life of the image of Drosophila melanogaster. American Naturalist. 1929;63:33–67. [Google Scholar]
- Aparicio R., Neyen C., Lemaitre B., Busturia A. dRYBP contributes to the negative regulation of the Drosophila Imd pathway. PLoS ONE. 2013;8:e62052. doi: 10.1371/journal.pone.0062052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbous N., Coste F., Leone P., Vincentelli R., Royet J., Kellenberger C., Roussel A. The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 2011;12:327–333. doi: 10.1038/embor.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha K.N., Tarr P., Zipursky S.L. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J. Exp. Biol. 2008;211:3103–3110. doi: 10.1242/jeb.016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Drayon V., Poidevin M., Boneca I.G., Narbonne-Reveau K., Royet J., Charroux B. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. 2012;12:153–165. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Cantera R., Barrio R. Do the genes of the innate immune response contribute to neuroprotection in Drosophila? J. Innate Immun. 2015;7:3–10. doi: 10.1159/000365195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Chtarbanova S., Petersen A.J., Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. USA. 2013;110:E1752–E1760. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchore Y., Gerber G.F., Dolph P.J. Alternative pathway of cell death in Drosophila mediated by NF-κB transcription factor Relish. Proc. Natl. Acad. Sci. USA. 2012;109:E605–E612. doi: 10.1073/pnas.1110666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D.J., Gems D., Harshman L.G., Oldham S., Stocker H., Hafen E., Leevers S.J., Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Clark R.I., Tan S.W., Péan C.B., Roostalu U., Vivancos V., Bronda K., Pilátová M., Fu J., Walker D.W., Berdeaux R. MEF2 is an in vivo immune-metabolic switch. Cell. 2013;155:435–447. doi: 10.1016/j.cell.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães J.P., Curado J., Church G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G. HIV infection, inflammation, immunosenescence, and aging. Annu. Rev. Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAngelo J.R., Bland M.L., Bambina S., Chery S., Birnbaum M.J. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc. Natl. Acad. Sci. USA. 2009;106:20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne M.S., Pham L.N., Shirasu-Hiza M., Schneider D.S. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Donath M.Y., Shoelson S.E. Type-2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 2011;11:98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- Fernando M.D., Kounatidis I., Ligoxygakis P. Loss of Trabid, a new negative regulator of the Drosophila immune-deficiency pathway at the level of TAK1, reduces life span. PLoS Genet. 2014;10:e1004117. doi: 10.1371/journal.pgen.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Capri M., Monti D., Giunta S., Olivieri F., Sevini F., Panourgia M.P., Invidia L., Celani L., Scurti M. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Gäde G., Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen. Comp. Endocrinol. 2003;132:10–20. doi: 10.1016/s0016-6480(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Giunta B., Fernandez F., Nikolic W.V., Obregon D., Rrapo E., Town T., Tan J. Inflammaging as a prodrome to Alzheimer’s disease. J. Neuroinflammation. 2008;5:51–66. doi: 10.1186/1742-2094-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntermann S., Primrose D.A., Foley E. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev. Comp. Immunol. 2009;33:127–134. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Guo L., Karpac J., Tran S.L., Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Jasper H. Studying aging in Drosophila. Methods. 2014;68:129–133. doi: 10.1016/j.ymeth.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Yano T., Aggarwal K., Lim J.H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W.E., Oh B.H. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Rulifson E.J. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Kim M., Lee J.H., Lee S.-Y., Kim E., Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc. Natl. Acad. Sci. USA. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidis I., Ligoxygakis P. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2012;2:120075–120089. doi: 10.1098/rsob.120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Park J.H. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhocine N., Ribeiro P.S., Buchon N., Wepf A., Wilson R., Tenev T., Lemaitre B., Gstaiger M., Meier P., Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Libert S., Chao Y., Chu X., Pletcher S.D. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell. 2006;5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- Lin Y.-J., Seroude L., Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- Lin Y., He H., Luo Y., Zhu T., Duan R. Inhibition of transglutaminase exacerbates polyglutamine-induced neurotoxicity by increasing the aggregation of mutant ataxin-3 in an SCA3 Drosophila model. Neurotox. Res. 2015;27:259–267. doi: 10.1007/s12640-014-9506-8. [DOI] [PubMed] [Google Scholar]
- Mair W., Goymer P., Pletcher S.D., Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Meinander A., Runchel C., Tenev T., Chen L., Kim C.H., Ribeiro P.S., Broemer M., Leulier F., Zvelebil M., Silverman N., Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel J., Lundgren P.R., Bensch K.G., Atlan H. Effects of temperature on the life span, vitality and fine structure of Drosophila melanogaster. Mech. Ageing Dev. 1976;5:347–370. doi: 10.1016/0047-6374(76)90034-8. [DOI] [PubMed] [Google Scholar]
- Moreno B., Jukes J.P., Vergara-Irigaray N., Errea O., Villoslada P., Perry V.H., Newman T.A. Systemic inflammation induces axon injury during brain inflammation. Ann. Neurol. 2011;70:932–942. doi: 10.1002/ana.22550. [DOI] [PubMed] [Google Scholar]
- Moskalev A., Shaposhnikov M. Pharmacological inhibition of NF-κB prolongs lifespan of Drosophila melanogaster. Aging (Albany NY) 2011;3:391–394. doi: 10.18632/aging.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi N., Parker J.L., Lugus J.J., Walsh K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011;11:85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N., Broemer M., Aggarwal K., Chen L., Husson M., Ertürk-Hasdemir D., Reichhart J.M., Meier P., Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol. Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes J.C., Welchman D.P., Poidevin M., Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Petersen A.J., Katzenberger R.J., Wassarman D.A. The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics. 2013;194:133–142. doi: 10.1534/genetics.113.150854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M., Clark R.I., Walker D.W. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K., Kauppinen A. Inflammaging: disturbed interplay between autophagy and inflammasomes. Aging (Albany NY) 2012;4:166–175. doi: 10.18632/aging.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Sekihara S., Fujikawa T., Miyaji R., Maki K., Ishihara T., Koshiba T., Kawabata S. Transglutaminase-catalyzed protein-protein cross-linking suppresses the activity of the NF-κB-like transcription factor relish. Sci. Signal. 2013;6:ra61. doi: 10.1126/scisignal.2003970. [DOI] [PubMed] [Google Scholar]
- Silverman N., Zhou R., Erlich R.L., Hunter M., Bernstein E., Schneider D., Maniatis T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- Stoven S., Silverman N., Junell A., Hedengren-Olcott M., Erturk D., Engstrom Y., Maniatis T., Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc. Natl. Acad. Sci. USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon D., Engel E., Avet-Rochex A., Gottar M., Bergeret E., Tricoire H., Benaud C., Baudier J., Taillebourg E., Fauvarque M.O. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6:309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Tsichritzis T., Gaentzsch P.C., Kosmidis S., Brown A.E., Skoulakis E.M., Ligoxygakis P., Mosialos G. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134:2605–2614. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Langmann C., Harden N., Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou O., Ohresser S., Ferrandon D., Capovilla M., Reichhart J.M., Lemaitre B., Hoffmann J.A., Imler J.L. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- Waterson M.J., Chung B.Y., Harvanek Z.M., Ostojic I., Alcedo J., Pletcher S.D. Water sensor ppk28 modulates Drosophila lifespan and physiology through AKH signaling. Proc. Natl. Acad. Sci. USA. 2014;111:8137–8142. doi: 10.1073/pnas.1315461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li J., Purkayastha S., Tang Y., Zhang H., Yin Y., Li B., Liu G., Cai D. Hypothalamic programming of systemic ageing involving IKK-β, NF-κB and GnRH. Nature. 2013;497:211–216. doi: 10.1038/nature12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.