Abstract
BACKGROUND. Melanoma is a heterogeneous malignancy. We set out to identify the molecular underpinnings of high-risk melanomas, those that are likely to progress rapidly, metastasize, and result in poor outcomes.
METHODS. We examined transcriptome changes from benign states to early-, intermediate-, and late-stage tumors using a set of 78 treatment-naive melanocytic tumors consisting of primary melanomas of the skin and benign melanocytic lesions. We utilized a next-generation sequencing platform that enabled a comprehensive analysis of protein-coding and -noncoding RNA transcripts.
RESULTS. Gene expression changes unequivocally discriminated between benign and malignant states, and a dual epigenetic and immune signature emerged defining this transition. To our knowledge, we discovered previously unrecognized melanoma subtypes. A high-risk primary melanoma subset was distinguished by a 122-epigenetic gene signature (“epigenetic” cluster) and TP53 family gene deregulation (TP53, TP63, and TP73). This subtype associated with poor overall survival and showed enrichment of cell cycle genes. Noncoding repetitive element transcripts (LINEs, SINEs, and ERVs) that can result in immunostimulatory signals recapitulating a state of “viral mimicry” were significantly repressed. The high-risk subtype and its poor predictive characteristics were validated in several independent cohorts. Additionally, primary melanomas distinguished by specific immune signatures (“immune” clusters) were identified.
CONCLUSION. The TP53 family of genes and genes regulating the epigenetic machinery demonstrate strong prognostic and biological relevance during progression of early disease. Gene expression profiling of protein-coding and -noncoding RNA transcripts may be a better predictor for disease course in melanoma. This study outlines the transcriptional interplay of the cancer cell’s epigenome with the immune milieu with potential for future therapeutic targeting.
FUNDING. National Institutes of Health (CA154683, CA158557, CA177940, CA087497-13), Tisch Cancer Institute, Melanoma Research Foundation, the Dow Family Charitable Foundation, and the Icahn School of Medicine at Mount Sinai.
Keywords: Dermatology, Oncology
The TP53 family of genes and genes regulating the epigenetic machinery demonstrate strong prognostic and biological relevance during progression of early melanoma.
Introduction
Melanomas of the skin are aggressive malignancies. They can arise from preexisting benign melanocytic nevi (moles), advance through local stages of progression, and, ultimately, result in distant metastasis (1). Approximately 85% of melanoma patients present with localized disease (stages I and II, American Joint Committee on Cancer [AJCC]), 10% with regional lymph node metastasis (stage III), and 5% with distant metastases (stage IV) (2, 3). Even for stage I disease, in which the 5-year survival rate is greater than 90%, a small subset of these patients develop metastasis (4). The risk prediction of stage II and III disease is more challenging, as the 5-year survival rates vary widely, from 53%–82% and 22%–68%, respectively (2). The management of early localized and regional metastatic disease is confined to a “watchful waiting” strategy after surgical removal of the primary tumor (5). Limited genetic, genomic, or transcriptional information is routinely used and widely accepted in clinical practice for risk stratification of cutaneous melanoma patients. Adjuvant treatment strategies for stage III patients consist of a few immunotherapeutic agents, such as interferon-α2b, pegylated interferon-α2b, and ipilimumab, while others are being actively tested in clinical trials (6–8). Adjuvant therapy with interferon is not widely accepted due to marginal effect on survival (9).
Oncogenic mutations in BRAF (50%) or NRAS (25%–30%), leading to mitogen-activated protein kinase pathway activation, are common early events that are also present in melanocytic nevi (10–12). BRAFV600E in melanocytes, after an initial phase of cell proliferation, induces a senescence-like state by engaging regulatory circuits of the p16INK4A/pRb pathway, chromatin alterations, and a tight transcriptional control of cell proliferation (13, 14). Malignant transformation occurs only in the presence of additional genetic alterations, such as inactivation of tumor suppressors PTEN, CDKN2A, or TP53 (1, 15–17). Although genetic mechanisms of melanoma development are studied in depth and subtypes are defined based on mutations and copy number changes (1, 18–20), these efforts have led to limited information on the identification of aggressive, high-risk tumors with poor outcomes.
The cancer cell’s genetic and epigenetic interplay with the immune system is complex, and regulatory functions of noncoding regions of the transcriptome add another layer of modulation. Yet, beyond traditional genetic mechanisms, in melanoma these processes remain vastly unknown. Epigenetic-driven aberrant transcription has been shown to regulate immunoregulatory signals, which in turn contributes to the make up and behavior of the tumor’s immune microenvironment (21). For example, DNA methylation together with p53 can silence noncoding repetitive element transcription for maintenance of genetic stability, a condition recognized in normal cells (22). However, in cancer, “unleashed” or “high levels” of repetitive element expression can result in a state of “viral mimicry” (a response of the cell to an exogenous viral infection) and a cytotoxic interferon response (23, 24). Despite these emerging themes, this interplay between melanoma and its premalignant benign precursors, in particular, during tumor initiation and early stages of progression (stages I and II) is largely unexplored. Major drawbacks, including those of The Cancer Genome Atlas (TCGA) (18), have been in part due to small tissue size of precursor lesions and primary tumors, difficulty in sparing fresh tissue for research biasing studies toward advanced primary tumors and metastatic disease, and sequencing protocols focusing only on protein-coding regions of the transcriptome, thereby neglecting, for example, noncoding repetitive element analysis. Thus, TCGA data does not include primary melanomas in early phases or nevi. The existing limited information on the melanoma cell–transcriptome and immune cell phenotypes of nevi and early primary tumors represents data and literature prior to the next-generation sequencing era.
We set out to examine transcriptional landscape changes of primary melanomas of early-, intermediate-, and late-stage tumors (AJCC tumor thickness categories T1–4, stages I–III) and benign precursor lesions. Additionally, interrogation of the transcriptome beyond protein-coding regions led to a deeper understanding of transcript deregulation during early disease and its progression. A distinct, previously unrecognized subset representing high-risk melanomas with poor survival characteristics was discovered. This subtype exhibited TP53 family and epigenome deregulation and repressed noncoding endogenous retrotransposons and viral defense signaling that are mechanistically tied together. We validated the high-risk subtype and their prognostic significance in different melanoma cohorts, thereby providing a molecular subtyping framework that can be used in clinical and translational studies for melanoma in its early stages.
Results
Patient characteristics and study design.
The discovery cohort consisted of 78 melanocytic neoplasms, including 27 cases of common acquired nevi and 51 cases of primary melanomas of the skin, which were subjected to RNA sequencing (see Methods). The study design is depicted in Figure 1A. Tumors were collected from treatment-naive patients; none had chemotherapy, targeted therapy, or immune-based treatments. All samples examined were from the primary tumor site; metastatic sites were not included. Nevus cases were histologically either compound or intradermal melanocytic nevi, and those with high melanocytic cell content were selected for the study. The AJCC staging system was used for clinical classification (2). Primary melanomas were comprised of invasive melanomas with tumor thicknesses of 4.25 mm (mean) and 2.41 mm (median); they represented early- (T1, ≤1.0 mm, n = 10), intermediate- (T2 and T3, 1.01–4.0 mm, n = 27), and late-stage or advanced (T4, >4.0 mm, n = 14) tumors. The distribution of tumor thickness within the cohort represented the spectrum of T1 through T4 as compared with the primary melanomas of TCGA that consist mostly of thicker tumors (Figure 1B). Overall survival of the patient cohort was 90 months (mean) and 93.5 months (median). The details of the patient cohort, clinical data, and histological characteristics are described in the Supplementary Appendix (Supplemental Table 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92102DS1).
Figure 1. Gene expression–based classification of primary melanomas and benign melanocytic nevi.
(A) Schematic diagram summarizing the study design. (B) Tumor thickness distribution of the primary melanoma cases of the discovery cohort (blue line) versus the primary melanomas of TCGA (yellow line). (C) The top bar indicates subtypes: nevus (light blue) and primary melanoma clusters (dark blue). In the heatmap, expression scores are median centered and clustered by complete linkage hierarchical clustering. Each row indicates a differentially expressed gene and each column represents a tumor sample. The heatmap is color coded on the basis of median-centered log2 gene expression levels. The bars at the bottom represent molecular parameters (nonsynonymous mutational load and BRAFV600E genotype) and clinical characteristics of the patient cohort using AJCC staging and classification for melanoma (age, gender, tumor thickness, ulceration, stage at diagnosis, and patient outcome at last follow-up). The variables are color coded as indicated. (D) Principal component analysis of the melanocytic tumors (nevus, light blue; primary melanoma, dark blue). (E) Gene expression classifier distinguishing nevus and primary melanoma groups.
Nevus versus melanoma — functional categorization underlying melanoma initiation.
Transcriptional profiling of 78 melanocytic tumors using unsupervised consensus-based hierarchical clustering identified three major clusters that distinguished benign nevi and identified two groups of melanomas in an unbiased manner (Figure 1C). Principal component analysis supported the distinction of these groups. In particular, the separation of melanoma and nevus samples was quite evident (Figure 1D). We constructed a machine learning–based gene expression classifier for melanoma versus nevus samples, capturing a minimal set of differentially expressed genes that best distinguished the two groups, information that could be utilized for delineating benign and malignant states within melanocytic tumors (Figure 1E and Supplemental Tables 2 and 3).
Benign melanocytic nevi exemplify an oncogene-induced senescence-like state driven by BRAFV600E (13). All nevus cases in our cohort, except one with an unknown genotype, harbored the BRAFV600E mutation. We reasoned that RNA transcripts and pathways deregulated between the nevus and melanoma groups represented processes during malignant transformation and transition from a senescence-like state. Overall, we found 4,639 genes to be significantly differentially expressed between melanoma and nevus samples (Figure 2A and Supplemental Table 2). Functional annotation analysis showed marked enrichment of genes regulating immune response and inflammation, cell cycle and proliferation, epigenetic modifiers, apoptosis, extracellular matrix, and cell adhesion (Figure 2, B and C). MITF, which encodes the master melanocyte transcription regulator and a melanoma-lineage survival oncogene (25), was uniformly upregulated in nevi, and melanomas with MITF “high” and “low” subgroups, as previously described (18), were notable. Other well-characterized oncogenic signaling pathways, such as loss of PTEN gene expression (12), loss of tumor suppressor FBXW7 and activation of NOTCH signaling (26), and its target genes, were highly deregulated in melanoma versus nevi (Figure 2C). We examined the molecular estimates measuring the influence of transcriptomic signals from tumor or stromal cell compartments computationally using a normal epidermal melanocyte expression signature (Supplemental Table 4 and Supplemental Figure 2). Additionally, tumor-infiltrating lymphocyte (TIL) scores were generated across the data set by histologic examination (Figure 2D and Supplemental Table 1). High TIL scores were evident in a subset of primary melanomas, while the precursor nevi uniformly had low scores or scores of 0 (nevus vs. melanoma, P < 0.0001). Together, these data illustrate robust transcriptional profiling, discriminating between benign nevi and primary melanomas using a next-generation RNA sequencing strategy.
Figure 2. Transcriptomic profiling identifies dysregulated epigenetic and immune response processes underlying melanoma initiation.
(A) Volcano plot showing differentially expressed genes between primary melanoma (n = 51) and nevus (n = 27) samples using cutoffs of Benjamini-Hochberg–corrected P ≤ 0.005 and a linear fold change ±1.5. Protein-coding mRNA with counts greater than 10 per million in at least 2 samples were considered. Downregulated (n = 2,350) and upregulated genes (n = 2,289) are indicated. (B) Top functional pathways disrupted between primary melanoma and nevus cases using the Database for Annotation, Visualization and Integrated Discovery (DAVID). (C) Examples of known oncogenic signaling pathways deregulated during malignant transformation. Heatmaps represent supervised hierarchical clustering of gene signatures that were differentially expressed between primary melanoma and nevus groups. Tumors are ordered based on the clusters identified and shown in Figure 1C (x axis), and genes are ordered according to functional categories (y axis). Examples are color coded as upregulated (pink) or downregulated (blue) in primary melanomas. (D) Tumor-infiltrating lymphocyte (TIL) scores on H&E examination of nevus versus melanoma samples (P < 0.0001, Fisher’s exact test). N.A., not available. (E) Heatmaps represent immune and inflammatory processes deregulated between primary melanomas and nevi. Immune cell lineage–specific transcript signatures, in particular TIL and myeloid cell populations involved in innate (dendritic cells, macrophages, NK cells, and neutrophils) and adaptive (B and T, Th1, Th2, CD8+ T cells) arms of the immune system, differentially expressed between benign and malignant categories. Several upregulated immune checkpoint molecules are highlighted. (F) The epigenetic gene signature that is differentially expressed between the primary melanoma and nevus groups is depicted. Genes encoding proteins that regulate chromatin structure are ordered on the y axis according to the functional groups: “writers” (histone acetyltransferase, histone methyltransferase, or protein arginine methyltransferase), “erasers” (histone deacetylase and histone demethylase), and “readers” (bromodomain, chromodomain, and Tudor domain–containing molecules), as well as DNA modifiers. For more detailed information, see the Methods.
Melanoma is known to be an immunogenic tumor. By examining immune cell lineage–specific transcript signatures, we determined a wide repertoire of TIL and myeloid cell populations involved in innate and adaptive arms of the immune system to be significantly enriched in melanoma tumors compared with nevi (Figure 2E and Supplemental Table 2). The presence or lack of T cell immune infiltrate (CD8+ T cells) correlates with patient outcomes and response to immunotherapy (27). We found T cell–associated genes, specifically, inhibitory checkpoint molecules (CTLA4, PDCD1 or PD1, TIGIT, HAVCR2 or TIM3, and BTLA) and their ligands, which can be expressed on antigen-presenting cells and/or tumor cells (CD86, PDL1, PDL2, TNFRSF14 or HVEM, and GAL9), as well as Toll-like receptors (TLR1–2, TLR6–8, and TLR10), a proinflammatory milieu of chemokine, cytokine, and complement cascade molecules, to be significantly enriched in the majority, but not all, of melanoma tumors (Figure 2E, top). Importantly, most of these molecules were coexpressed in the same melanomas without mutual exclusivity, suggesting possible coordinated immune suppression due to an immune response. Striking enrichment of genes associated with inflammasome activation or their regulation (IL-18 and NLRP10), IL-1 family members (IL-33 and IL-37), and lymphocyte cell survival, activation, and homing molecules (IL-2R2, IL-7, and CD44) was observed in the benign tumors (Figure 2E, bottom). These transcripts were significantly downregulated in melanoma. Due to a close link of endogenous retrovirus (ERV) sequences triggering double-stranded RNA-mediated viral defense gene responses, we also examined and found endogenous element transcription to be significantly downregulated in melanomas (Supplemental Figure 3). Together, these data reveal highly enriched immune and inflammatory signal output and transcriptional silencing (or derepression) of endogenous elements in primary melanomas as compared with benign nevi.
An emerging theme in melanoma biology is the disruption of the epigenetic machinery; however, key players are largely unknown. By examining transcripts of the epigenetic machinery, we identified a gene signature (n = 108) with discrete expression profiles between melanoma and nevus groups involving DNA methylation and chromatin modification processes (Figure 2F and Supplemental Table 2). We found significant downregulation of TET family enzymes, TET1 and TET2, a well-characterized mechanism for global loss of 5-hydroxymethylcytosine marks, leading to gene dysregulation (28). When chromatin-modifying enzymes were examined, we noted significant enrichment of BRD4, a member of the BET subfamily (29), as well as histone variant H2AZ.2 (30), both of which enhance cell proliferation in melanoma. Notably, we found transcriptional dysregulation of several genes regulating the epigenome that have not been previously linked to melanoma or any other cancer, thus representing novel discriminatory events.
Primary melanoma subgroups — high-risk melanomas underlie epigenome and p53 family deregulation.
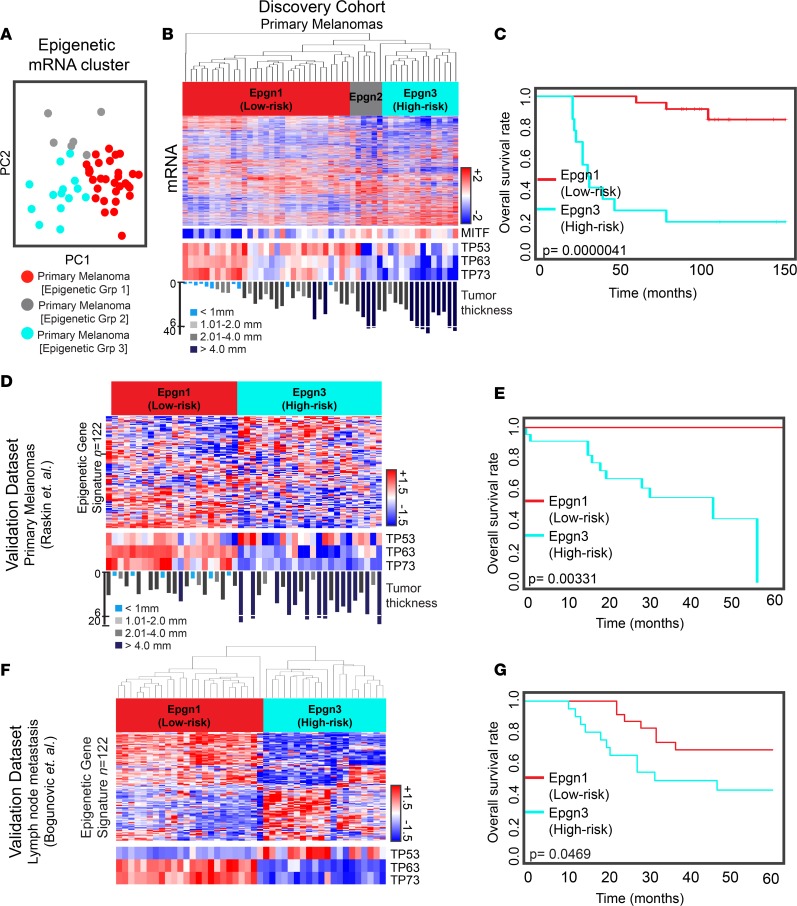
An unbiased approach as well as a focused examination of the epigenome and immune-related genes led to three distinct clusters of melanomas. We determined whether these unbiased groups were best reflected by differential expression of immune or epigenetic genes. To our surprise, our unbiased clusters were best recapitulated by focusing on epigenetic genes and not immune gene categories. We therefore referred to our melanoma clusters as “epigenetic” clusters (referred to as Epgn1, Epgn2, and Epgn3 subtypes) (Figure 3A). Epgn2 was an outlier group that contained fewer tumors (n = 6); thus, we focused on the Epgn1 and Epgn3 clusters.
Figure 3. “Epigenetic” transcriptomic clusters and TP53 family members identify a high-risk melanoma subtype.
(A) Principal component analysis of the discovery cohort. Colored circles indicate “epigenetic” (Epgn) primary melanoma subtypes (Epgn1, red; Epgn2, gray; and Epgn3, turquoise). (B) Unsupervised clustering of primary melanoma subtypes: Epgn1, Epgn2, and Epgn3. Heatmaps show differential expression of mRNA (top) and MITF, TP53, TP63, and TP73 (bottom). (C) Survival curves for Epgn1 and Epgn3 subgroups of the discovery cohort (P = 4.1e-06). (D) Validation of the Epgn1 and Epgn3 clusters utilizing the epigenetic gene signature (n = 122 genes) in an external data set of primary melanomas (n = 46) (Raskin et al., ref. 31). Expression levels of TP53, TP63, and TP73 (bottom) are indicated. (E) Survival curves for the Epgn1 and Epgn3 clusters of this data set (P = 3.31e-03) are depicted. (F) Validation of the Epgn1 and Epgn3 clusters in an external data set of melanoma metastases to the lymph nodes (n = 44) (Bogunovic et al., ref. 33). (G) Survival curves for the Epgn1 and Epgn3 clusters of this data set (P = 0.0469). Log-rank test was used for statistical analysis.
When interrogated, a 122-epigenetic gene signature was found to discriminate between Epgn1 and Epgn3 (Supplemental Table 5). The Epgn3 subclass represented a group of thicker melanomas (geometric mean = 4.3 mm, median = 6.1 mm) with a significantly poorer overall survival rate than the Epgn1 group (P = 4.1e-06), suggesting Epgn3 as an aggressive or a high-risk melanoma subtype (Figure 3, B and C). This high-risk subset involved 27% of the melanomas in our cohort. Enrichment of genes involved in cell proliferation such as cell cycle checkpoint molecules and E2F-target genes in the Epgn3 cluster was noted. When oncogenic signaling pathways were examined, we found TP53 family gene members, TP53 (–1.53 linear fold change), TP63 (–6.11), and TP73 (–4.59), to be significantly repressed in this cluster (Figure 3B). The presence of Epgn1 (low-risk) and Epgn3 (high-risk) groups and their correlation with better or worse patient outcomes, respectively, were found in another independent data set of primary melanomas (Figure 3, D and E), validating our findings (31, 32). In a separate cohort of melanomas metastatic to the lymph nodes (stage III), we found the low- versus high-risk clusters in the lymph nodes for which the high-risk tumors showed poor survival characteristics (Figure 3, F and G), thus further validating the signature and its persistence in the lymph node metastasis (33). Additionally, this high-risk cluster was verified in TCGA melanoma data set (Supplemental Figure 4) (18). Enrichment of mutations in TP53 in the high-risk tumors was not found in our discovery cohort or TCGA.
We next tested the Epgn1/3 classification on the prognostic effect. When patient outcomes were stratified by the Epgn1/3 classification after correcting for tumor thickness in our cohort and in an external data set of primary melanomas (31), we found that the Epgn3 (high-risk) signature was an independent prognostic indicator for shorter survival (Supplemental Figure 5). In summary, these findings illustrate the identification of a high-risk melanoma cluster distinguished by an epigenetic gene signature, a prognostic value on patient outcome, and a link to TP53 family of genes.
High-risk melanomas and associated functional groups.
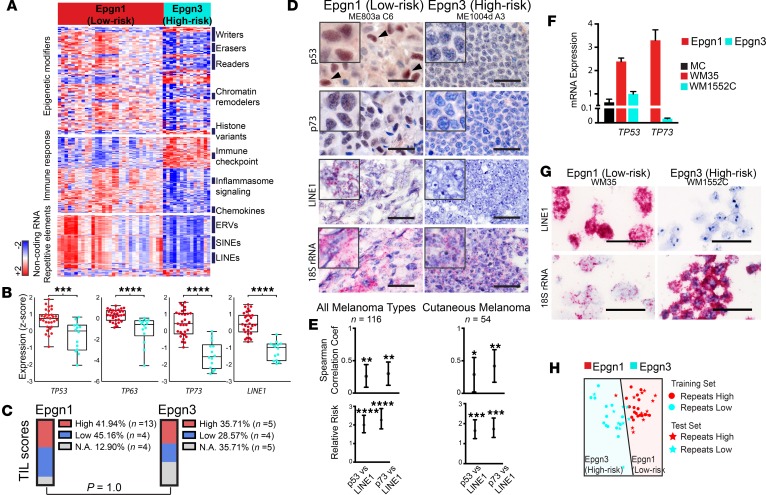
When further examined in the context of functional groups, these clusters displayed a discrete signature of epigenetic modifiers (n = 122), immune response genes, and noncoding repetitive elements (Figure 4, A and B, and Supplemental Table 5). Epigenetic regulators frequently mutated in human cancer, including melanoma, which function as tumor suppressors (1, 34) were significantly downregulated in Epgn3, such as components of the SWI/SNF chromatin remodeling complexes (ARID1A/B, SMARCA2, SMARCA4, and SMARCC2). Several chemokines essential for T-lymphocyte recruitment to tumor sites (CCL22, its receptor CCR4, and CCL21) were significantly suppressed. We found significant downregulation of various NOD-like receptor family pyrin domain (NLR) genes (NLRP1 and NLRP10), IL-1R family members (IL18R, IL1RL1, and IL1RL2), and pattern recognition receptors (NOD-like receptors; NOD2, NLRX1, and NLRC5) in Epgn3 as compared with the prognostically better Epgn1 group. We next examined the noncoding regions of the transcriptome, in particular, noncoding repetitive elements that are regulated by epigenetic mechanisms (i.e., DNA methylation), and likely by the protein product of TP53 (p53) (22). Indeed, differential repetitive element expression alone across our tumor cohort distinguished the Epgn1 and Epgn3 groups. The Epgn3 group showed significant transcriptional repression of long- and short-interspersed nuclear elements (LINE and SINE) and ERV transcription at these particular repetitive sequences as compared with Epgn1 and with benign nevi, in which expression was increased (Figure 4, A and B, and Supplemental Figure 3). Importantly, TIL scores did not show significant differences between Epgn1 and Epgn3 clusters (P = 1.0, Figure 4C). TP53, TP63, and TP73 correlated with LINE1 and the 122-epigenetic signature (Supplemental Figure 6). These findings demonstrate the associated functional groups and noncoding transcripts with the high-risk melanoma cluster.
Figure 4. Noncoding repetitive element transcripts are repressed in high-risk melanomas.
(A) Heatmaps indicating differentially expressed protein-coding transcripts involved in epigenetic regulation and immune response between the Epgn1 and Epgn3 subtypes as well as noncoding repetitive element transcripts (retrotransposons). Tumors are ordered based on the clusters identified and shown in Figure 3B (x axis, Epgn1 and Epgn3), and genes are ordered according to functional categories or sequence homology in accordance with retrotransposon classification (y axis). ERV, endogenous retrovirus; SINE, short-interspersed nuclear element; LINE, long-interspersed nuclear element. (B) Box plots represent expression of TP53 family genes and LINE1 between the Epgn1 and Epgn3 groups (2-tailed Mann-Whitney test). (C) TIL scores on H&E examination of Epgn1 versus Epgn3 clusters (statistically not significant, 2-tailed Fisher’s exact test). N.A., not available. (D and E) Melanoma tissue arrays (n = 116) and those limited to primary cutaneous melanomas (n = 54) were assayed for p53 and p73 protein expression by immunohistochemistry and for LINE1 and 18S rRNA (housekeeping gene) transcripts by RNA in situ hybridization. Representative examples are depicted. Scale bar: 25 μm. Inset: 18 μm x 18 μm. Graphs show correlation coefficient and relative risk values for p53 versus LINE1 or p73 versus LINE1 expression (Spearman correlation). (F) Melanoma cell lines established from primary melanoma tumors (WM35, stage I and WM1552C, stage III) were obtained. TP53 and TP73 transcripts were examined by qPCR. Normal human primary melanocytes (MC) are shown as a reference (1-tailed Student’s t test). (G) Cell pellets were formalin-fixed and paraffin-embedded and assayed for endogenous elements (LINE1) by RNA in situ hybridization. 18S rRNA was employed as a housekeeping transcript. Representative examples are shown. Scale bar: 25 μm. (H) Gene expression classifier distinguishing low-risk Epgn1 and high-risk Epgn3 groups based on repetitive element expression.
Noncoding endogenous element transcription is repressed in high-risk melanomas and co-occurs with loss of p53 and p73.
To validate the signature and dissect the signal output derived from the tumor cells, we utilized melanoma tissue microarrays (n = 116) of independent tumor sets and cell lines. We used immunohistochemistry to examine p53 and p73 expression and RNA in situ hybridization for LINE1. We found a subset of melanomas with co-occurring high levels of p53, p73, and LINE1 and another subset with loss of p53 and p73 protein expression and repression of LINE1 transcription (Figure 4, D and E). We next identified primary melanoma cell lines and designated them as Epgn1 or Epgn3 by qPCR analysis based on several genes within our signature. Cell lines defined as Epgn1 or Epgn3 corresponded to lines derived from patients with stage I or stage III disease, respectively. Epgn1 and Epgn3 cell lines showed coexisting expression patterns or loss of TP53, TP73, and LINE1 (Figure 4, F and G). We constructed a machine learning–based classifier for repeat “high” versus repeat “low” tumor types that could be utilized for prognosis determination (Figure 4H and Supplemental Table 3). Together, these findings verify the co-occurrence of p53 family and LINE1 expression or loss in melanoma cells.
“Immune” subtypes.
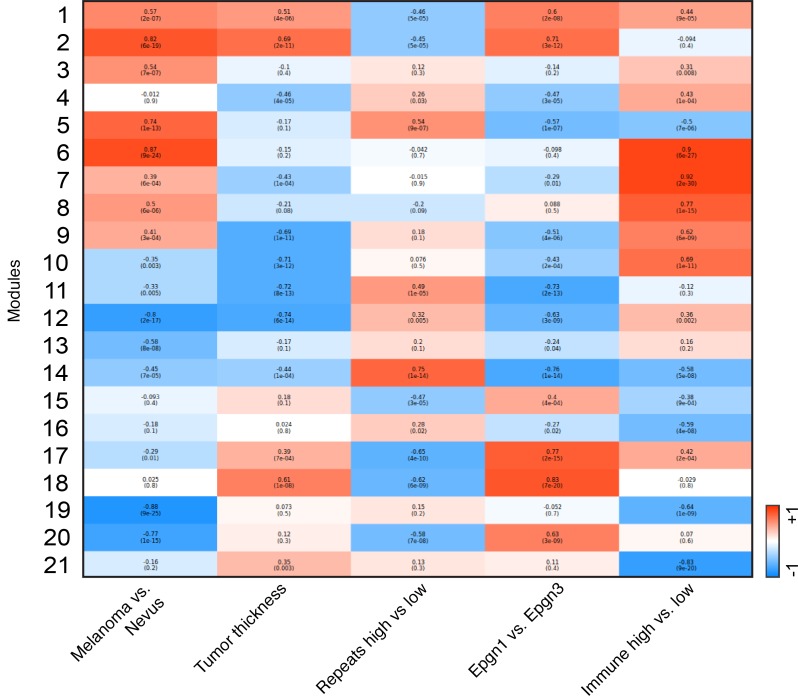
Transcriptional subtypes of melanoma with “low” or “high” levels of immune response genes are well characterized and have recently been underscored by TCGA using next-generation RNA sequencing (18, 35). Similarly, during our analyses, we noted subsets of immune “low,” “medium,” and “high” melanomas, with expression levels of “low” and “high” clusters being quite distinct (Figure 5A and Supplemental Table 6). TIL scores verified immune-based clustering into “low” and “high” groups (Figure 5B). We found TEAD transcription factors and several chromatin modifiers as differentially expressed between the two clusters (Figure 5, C and D). Lower expression levels of TEAD1 and LCMT1 in immune “high” melanoma tumor samples as compared with immune “low” cases were noted in an independent set of primary melanomas by qPCR (Figure 5E). Discriminatory transcripts (immune response and epigenetic modifiers) in immune “low” versus “high” tumors were distinct from those expressed between Epgn1 and Epgn3 subtypes. Together, our data verify the presence of immune “low” and “high” clusters among primary melanomas and identify several candidates linked to these clusters. Finally, we summarize the different tumor clusters identified in our study (Epgn1 vs. Epgn3, immune “low” vs. “high,” benign vs. malignant) and biologically relevant variables using coexpression network modeling (Figure 6).
Figure 5. Defining “immune” transcriptomic subclasses.
(A) Principal component analysis of two distinct primary melanoma subsets: immune “low” (n = 9) and immune “high” (n = 12). (B) TIL scores by H&E examination of immune “low” and “high” clusters (P = 0.035, Fisher’s exact test). N.A., not available. (C) Genes involved in Yap signaling (TEAD1, TEAD2, TEAD3, and TEAD4) and those involved in epigenetic processes are found to be differentially expressed between the immune “low” and “high” subgroups (2-tailed Mann-Whitney test). (D) Correlation matrix of differentially expressed genes with representative upregulated and downregulated immune genes across the primary melanoma samples of the discovery cohort (n = 51). Red indicates a positive correlation while blue indicates an anticorrelation. (E) Validation of the candidates (TEAD1 and LCMT1) was performed on an extension cohort of primary melanoma samples (n = 49) using qPCR. *P < 0.05, 2-tailed Student’s t test. The bar graph shows samples that were classified as either immune “low” or “high.”
Figure 6. Summary of the findings via coexpression network modeling.
Coexpression network analysis was built across tumor subtypes and biologically relevant variables (tumor thickness).
Discussion
Molecular signatures associated with distinct clinical outcomes have been delineated in various solid tumors, thereby laying the groundwork for improved clinical management through the development of personalized risk stratification and therapeutic approaches. RNA sequencing analysis of TCGA has identified several subtypes, such as “immune,” “MITF,” and “keratin” groups, that linked immune “high” subtypes to better prognosis, an observation noted previously (18). Here, we report transcriptional classification of melanoma in treatment-naive tumors that links the p53 family and the epigenome together with a specific immune milieu to clinical prognosis and patient outcomes. We discovered a molecular signature depicting high-risk and aggressive biologic behavior by examining primary melanoma tumor samples (not metastases). The signature showed its persistence during progression in the regional lymph nodes (stage III) and distant metastasis (stage IV). This information is expected to shed light onto risk stratification and development of therapeutic strategies accordingly.
p53 acts as a tumor suppressor by transcriptionally regulating a wide variety of target genes, thus dictating various cellular fates (36). While the TP53 family genes, TP53, TP63, and TP73, have independent functions and target genes, they can be interdependent on each other in the suppression of tumorigenesis, and their loss can cooperate for an aggressive phenotype (37, 38). For example, when p53 is lost, p73 plays an important role in preventing further deterioration of polyploidy and aneuploidy by acting as a stabilizing factor on the genome (37). In the mouse model system, while TP53+/– TP73+/– and TP53+/– TP63+/– mice develop the same tumor types as TP53+/– mice, the phenotype is highly aggressive and metastatic in the former scenario (39). Collectively, based on previous literature on p53 family members and their coregulated function, our data in this study suggest that intact p53, p63, and p73 associate with low-risk disease, may be indicative of genomic integrity, and serve a protective role. Conversely, loss or inactivation of these family members is associated with high-risk tumorigenesis. TP53 is mutated in approximately 15% of melanomas (18), and its inactivation leads to oncogenic transformation in the mouse system driven by BRAFV600E (15). Here, our study focused solely on the transcriptome (mRNA and noncoding sequences, including repetitive elements). TP53 mutations from RNA sequencing analysis did not show enrichment of mutations in high- or low-risk clusters. Examining somatic mutations in TP53 within the TCGA data set led to similar results, though TCGA melanomas are not representative of our cohort or the goals set for this study. Our work opens new avenues for tumor progression and risk stratification; however, the cooperative function of p53 family members in the context of p53 mutations or other mechanisms of p53 deregulation in the melanocytic lineage requires further studies.
An emerging role of p53 together with DNA methylation is to silence noncoding repetitive element transcription for maintenance of genetic stability, a condition recognized in normal cells (22). However, in cancer, “unleashed” or “high level” repetitive element expression can result in a state of “viral mimicry” and a cytotoxic interferon response (23, 24). Our study displayed “high” levels of endogenous element transcription in low-risk tumors (Epgn 1) and, in particular, in benign nevi and a downregulated (derepressed) state in high-risk tumors. While additional work is needed to decipher the understudied noncoding regions of the transcriptome, our study showed co-occurrence of p53 family deregulation together with repression of LINE1, SINE1, and inflammasome components (“viral mimicry” response).
“Immune” subtypes, previously described by others, were evident in our cohort, verifying previous observations (18, 35), and served as biological controls. Tumors upregulate inhibitory checkpoint receptors and repress antitumor immune responses. Therapies directed at these molecules (CTLA4, PD-1, and its ligand PD-L1) have recently shown significant and, in some cases, durable responses in melanoma patients (40, 41), thus shifting the treatment paradigm not only of melanoma, but cancer in general toward immunotherapy. Importantly, our data showed that there was no correlation between immune “low” or “high” subtypes and AJCC disease stage or tumor thickness categories. In treatment-naive tumors, we showed that the association of innate immune activation with better outcome is more complex that previously thought, where some patients in the Epgn3 (high-risk) category, with exclusively poor survival characteristics, had the paradoxical finding of immune “high” phenotypes. Exhausted T cell states within the tumor microenvironment (42) may, in part, explain these observations. However, the high-risk subset mostly correlated with thicker tumors, and the 122-epigenetic signature was an independent prognostic variable in predicting patient outcomes within the T3 and T4 tumor categories. Further studies with large independent cohorts validating these findings are needed.
Our work underscores a need for future clinical trials not only after metastasis, but also early in the course of the disease. The concept of delivering mechanism-based treatments for high-risk patients (i.e., those with high-risk molecular signatures) in an adjuvant setting followed by standard surgical management rather than “watchful waiting” requires further attention. Given the effective immune checkpoint inhibitors that are currently available, identifying patients that may respond to these therapies in the adjuvant setting represents a logical future direction for this highly aggressive lethal cancer.
Methods
Patient tissue samples.
Patients were treatment naive; none had received chemotherapy, targeted therapy, or immune-based treatments prior to study entry. Common acquired nevi, including compound and intradermal nevi (JTC) and primary melanomas of the skin (GB), were collected fresh and either snap-frozen or placed in RNAlater RNA stabilization reagent (Qiagen). Clinical annotations were entered, and pathology review of the H&E-stained sections of the tumor samples was performed. Additionally, two dermatopathologists (CC and RP) reviewed the tumor samples of the study cohort to confirm the diagnosis and generated the TIL scores. RNA sequencing was carried out on the discovery cohort that consisted of common acquired nevi (n = 27) and primary melanoma (n = 51) cases, whereas the extension cohort of primary melanomas (n = 49) was utilized for validating several candidates using qPCR. Sequencing data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE98394).
RNA sequencing.
Total RNA was extracted from samples using the miRNeasy Mini Kit (Qiagen). The RNA integrity number of each sample was determined using the 2100 Bioanalyzer instrument (Agilent Technologies). Samples were selected with stringent criteria based on an RNA integrity number of 7.0 or greater and a minimum of 500 ng of total RNA. Total RNA was subjected to ribosomal RNA depletion using the Epicentre Ribo-Zero Gold kit (Epicentre). The resulting RNA samples were used as input for library construction using the Illumina TruSeq Total RNA library preparation kit (Illumina). The libraries were then sequenced on the Illumina HiSeq 2500 system (Illumina) using 2 × 100-bp paired-end protocol to a minimum mean coverage of ×50. The sequencing was performed in the Genomics Core Facility of the Icahn School of Medicine at Mount Sinai.
Expression quantification, differential expression, and functional pathway analysis.
Raw reads were quality filtered by trimming the read ends; bases with quality <20 were removed. We then computed the quality at the 20th percentile. The entire read was discarded if the quality at the 20th percentile was less than 15. Reads shorter than 40 bases after trimming were discarded as well. If one or more reads in the pair failed the quality check both reads were discarded. Paired-end RNA sequencing reads were mapped to the human reference genome (Ensembl annotation, build 37) (43) and to sequences from the Repbase database of human repetitive elements (release 19, http://www.girinst.org/repbase) (44) using the STAR aligner (45). Aligned reads were assigned to genes using the featureCounts function of the Rsubread package via the external Ensembl annotation (46). This procedure generated the raw read counts for each gene. Gene expression in the form of log2 counts per million reads was computed and normalized across the samples using the TMM method as implemented in the calcNormFactors function of edgeR package (47, 48).
Differential expression analysis was performed using the limma software package (49, 50). Expression data were used in conjunction with the weights computed by the voom transformation based on the mean-variance relationship of log read counts to calculate moderated t-statistics using empirical Bayes (51). For protein-coding RNA (mRNA), we identified the gene signature differentiating each pair of subtypes. For this analysis, we considered only genes with counts per million reads greater than 10 in at least 2 samples. To create a comprehensive global heatmap depicting the gene signature of different subtypes, we considered genes to be differentially expressed with a Benjamini-Hochberg–corrected P value of P < 0.005 and a fold change of >1.5. Principal component analysis of the resulting sets of differentially expressed genes confirmed effective grouping of subtypes. To determine the biological modules that are affected in our differentially expressed gene set, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) (52), Ingenuity Pathway Analysis, and the Gene Ontology database (53). We examined pathways with a Benjamini-Hochberg–adjusted P value of P < 0.05.
Mutation detection and mutational load on RNA sequencing data.
Mutations were called using the exactSNP function of Rsubread package (46); only SNPs were considered. All detected mutations present in dbSNP (downloaded from Ensembl) (43) were removed. SNPs with a quality of 12 or higher only were included (Rsubread default). The effect of mutations was computed using SnpEff software with the same annotation that was used for building the genome index for mapping (54). If multiple features overlapped a SNP (i.e., sequences belonging to different isoforms of two overlapping genes on the opposite strands), the effect of the SNP was averaged across them. In this case, instead of the characterization of a SNP as synonymous/nonsynonymous, we used a fraction of nonsynonymous isoforms overlapping it. We added these numbers for all SNPs and rounded the result to estimate the number of nonsynonymous mutations.
Incorporation of normal epidermal melanocytic signature into differential gene and functional pathway analyses.
For details, see Supplemental Figure 2.
Validation of transcript signatures in tissues and cell lines.
Validation of the candidates was performed on an extension cohort of primary melanoma (n = 49) samples as well as in primary melanoma cell lines. Total RNA was extracted from the tumor samples and cell lines using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. One μg total RNA was reverse transcribed using the RNA to cDNA EcoDry Premix (Oligo dT) (Clontech). Quantitative PCR (qPCR) was performed using the QuantiNova SYBR Green PCR Kit (Qiagen) on the Applied Biosystems 7500 Real-Time PCR System (Agilent Technologies). The GAPDH gene was used as a housekeeping gene for normalization.
Immunohistochemistry and tissue microarrays.
Slides were stained for p53 (sc-126, clone DO-1, Santa Cruz Biotechnology) and p73 (ab40658, Abcam). Immunostaining was performed using Dako reagents (Target Retrieval Solution [S2367 and S2369], Serum-Free Protein Block [X909], Antibody Diluent with Background Reducing Components [S3022], Streptavidin/HRP [P0397]) and the Vector AEC peroxidase substrate kit detection system (SK-4200). Images were taken at ×40 on a Nikon Eclipse Ci microscope and analyzed using NIS-Elements BR Analysis software. ME803a and ME1004d Tissue Microarrays (US Biomax Inc.) were utilized for staining. ME803 contained 40 malignant melanoma cases, while ME1004d had 62 malignant and 20 metastatic malignant melanoma cases (n = 122 total melanomas). Six cases were excluded due to missing tissue after staining for a final count of 116.
Constructing classifiers.
We constructed gene expression classifiers for two distinctions: primary melanomas versus nevi and repetitive element transcriptional activity “high” versus “low.” Classifiers were constructed for predicting these features from the coding gene expression data. In order to train the classifier, we computed the top N differentially expressed genes with at least a 2-fold change between the two phenotypes based on statistical significance. We then used the gene expression data and performed dimensionality reduction to 2 dimensions using the partial least squares method, as implemented in the scikit-learn machine learning suite (55). We trained the Linear Support Vector Machine as implemented in scikit-learn in these 2 dimensions. For several values of N, we evaluated the performance of the classifier using the cross-validation technique. It was done by randomly selecting 60% of the samples as a training set and leaving out the remaining 40% as a test set. This partitioning into the training and test subsets was performed so that the proportion of representatives of the two phenotypes in the training data set was unchanged. The procedure was repeated multiple times, and the sensitivity and specificity were computed.
Coexpression network analysis of protein-coding genes.
Weighted gene coexpression network analysis was performed using the WGCNA software package (56, 57). Analysis was performed on the discovery cohort, which consisted of nevi (n = 27) and primary melanoma (n = 51) samples. For this analysis, we set the value of the parameter soft-thresholding power as equal to 10.
Statistics.
A moderated 2-tailed t test, as implemented in the limma package, was utilized for computing the significance of differential gene expression. The R survival package was utilized to perform survival analyses (https://www.r-project.org/). The log-rank test was used for the differential survival analysis. In order to evaluate the significance of association between the categorical data, Fisher’s exact test was performed.
Study approval.
The institutional review boards of Icahn School of Medicine at Mount Sinai and Fachklinik Hornheide approved this study, and written informed consent was received from participants prior to study inclusion.
Author contributions
JTC conceptualized and designed the project and supervised the entire study. BDG designed and supervised the bioinformatics analysis. BB, AS, SDC, JMC, LC, FA, RI, KSA, AT, EB, NB, DTT, EES, BDG, and JTC performed the data interpretation. BB, AS, JMC, BDG, and JTC wrote the manuscript. BB, SDC, ITA, and GSR performed the laboratory-based experiments. AS, JMC, LC, and RI performed the bioinformatics analysis. CC and RP performed the histopathologic analysis. RP supervised the histopathologic analysis. KSA performed and DTT supervised the RNA in situ hybridization experiments. SBG, TMJ, DRF, and LR provided survival data of one validation cohort. GB and JTC coordinated sample collection and processing of the discovery cohort.
Supplementary Material
Acknowledgments
We respectfully acknowledge our patients who participated in the study. We thank Yumi Kasai and the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai; Vanessa Angel in the Department of Dermatology at the Icahn School of Medicine at Mount Sinai; Ilaria Laface for immunohistochemistry experimentation at the Icahn School of Medicine at Mount Sinai; the Biostatistics Shared Resource Facility and National Cancer Institute Cancer Center support grant (P30 CA196521-01), and the Icahn School of Medicine at Mount Sinai for the statistical analysis of tissue microarray data. This work has been supported in part by the Icahn School of Medicine at Mount Sinai (to EES, JTC, and BDG), a pilot developmental research grant from the Tisch Cancer Institute (to JTC and EB), a gift from the Dow Family Charitable Fund (to JTC), research grants from the Melanoma Research Foundation (to EES and JTC), and the National Institutes of Health (CA154683 to EB; CA158557 and CA177940 to JTC; CA087497-13 to BDG).
Version 1. 05/02/2017
Electronic publication
Footnotes
GB’s present address is: NeraCare GmbH, Bönen, Germany.
Role of funding source: The funding sources played no role in the study design; data collection, analysis, or interpretation; writing of the report; or the decision to submit the paper for publication.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(8):e92102. https://doi.org/10.1172/jci.insight.92102.
Contributor Information
Brateil Badal, Email: brateil.badal@icahn.mssm.edu.
Alexander Solovyov, Email: alexander.solovyov@mssm.edu.
Serena Di Cecilia, Email: serenadicecilia@gmail.com.
Joseph Minhow Chan, Email: jminchan@gmail.com.
Li-Wei Chang, Email: lc2973@cumc.columbia.edu.
Ramiz Iqbal, Email: ramiz.iqbal@mssm.edu.
Chen Chen, Email: marychenchen@gmail.com.
Franco Abbate, Email: franco.abbate@mssm.edu.
Kshitij S. Arora, Email: KARORA@PARTNERS.ORG.
Antoine Tanne, Email: a.tanne14@gmail.com.
Stephen B. Gruber, Email: sgruber@usc.edu.
Leon Raskin, Email: leonid.raskin@vanderbilt.edu.
Robert Phelps, Email: rpdermpath@aol.com.
Nina Bhardwaj, Email: Nina.Bhardwaj@mssm.edu.
Emily Bernstein, Email: Emily.bernstein@mssm.edu.
Georg Brunner, Email: brunner@neracare.de.
References
- 1.Shain AH, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015;373(20):1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Han D, et al. Clinicopathologic predictors of sentinel lymph node metastasis in thin melanoma. J Clin Oncol. 2013;31(35):4387–4393. doi: 10.1200/JCO.2013.50.1114. [DOI] [PubMed] [Google Scholar]
- 5.Dickson PV, Gershenwald JE. Staging and prognosis of cutaneous melanoma. Surg Oncol Clin N Am. 2011;20(1):1–17. doi: 10.1016/j.soc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggermont AM, et al. Adjuvant ipilimumab versus placebo after complete resection of high-risk stage III melanoma (EORTC 18071): a randomised, double-blind, phase 3 trial. Lancet Oncol. 2015;16(5):522–530. doi: 10.1016/S1470-2045(15)70122-1. [DOI] [PubMed] [Google Scholar]
- 7.Kirkwood JM, Strawderman MH, Ernstoff MS, Smith TJ, Borden EC, Blum RH. Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol. 1996;14(1):7–17. doi: 10.1200/JCO.1996.14.1.7. [DOI] [PubMed] [Google Scholar]
- 8.Eggermont AM, et al. Adjuvant therapy with pegylated interferon alfa-2b versus observation alone in resected stage III melanoma: final results of EORTC 18991, a randomised phase III trial. Lancet. 2008;372(9633):117–126. doi: 10.1016/S0140-6736(08)61033-8. [DOI] [PubMed] [Google Scholar]
- 9.Mocellin S, Pasquali S, Rossi CR, Nitti D. Interferon alpha adjuvant therapy in patients with high-risk melanoma: a systematic review and meta-analysis. J Natl Cancer Inst. 2010;102(7):493–501. doi: 10.1093/jnci/djq009. [DOI] [PubMed] [Google Scholar]
- 10.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 12.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12(5):349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 13.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 14.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viros A, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature. 2014;511(7510):478–482. doi: 10.1038/nature13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin L, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11(21):2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44(9):1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanne A, et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc Natl Acad Sci USA. 2015;112(49):15154–15159. doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110(1):E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 26.Aydin IT, et al. FBXW7 mutations in melanoma and a new therapeutic paradigm. J Natl Cancer Inst. 2014;106(6):dju107. doi: 10.1093/jnci/dju107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji RR, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61(7):1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150(6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segura MF, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73(20):6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vardabasso C, et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell. 2015;59(1):75–88. doi: 10.1016/j.molcel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin L, et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J Invest Dermatol. 2013;133(11):2585–2592. doi: 10.1038/jid.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nsengimana J, et al. Independent replication of a melanoma subtype gene signature and evaluation of its prognostic value and biological correlates in a population cohort. Oncotarget. 2015;6(13):11683–11693. doi: 10.18632/oncotarget.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogunovic D, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106(48):20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy DM, Walsh LA, Chan TA. Driver mutations of cancer epigenomes. Protein Cell. 2014;5(4):265–296. doi: 10.1007/s13238-014-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 36.Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet. 2009;5(10):e1000680. doi: 10.1371/journal.pgen.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Talos F, Nemajerova A, Flores ER, Petrenko O, Moll UM. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol Cell. 2007;27(4):647–659. doi: 10.1016/j.molcel.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 38.Tan EH, et al. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene. 2014;33(25):3325–3333. doi: 10.1038/onc.2013.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalian SL, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32(10):1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43(Database issue):D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 45.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 51.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 53.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. doi: 10.4161/fly.19695. Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham A, et al. Machine learning for neuroimaging with scikit-learn. Front Neuroinform. 2014;8:14 [Google Scholar]
- 56.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46(11):i11. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.