The RQC complex involved in protein quality control mechanisms also exists as a ribosome-unbound complex during the escort of aberrant peptides to the proteasome. The E3 ubiquitin ligase Tom1 is a newly identified partner of this light version of the RQC complex and is required for aggregate prevention.
Abstract
Protein quality control mechanisms eliminate defective polypeptides to ensure proteostasis and to avoid the toxicity of protein aggregates. In eukaryotes, the ribosome-bound quality control (RQC) complex detects aberrant nascent peptides that remain stalled in 60S ribosomal particles due to a dysfunction in translation termination. The RQC complex polyubiquitylates aberrant polypeptides and recruits a Cdc48 hexamer to extract them from 60S particles in order to escort them to the proteasome for degradation. Whereas the steps from stalled 60S recognition to aberrant peptide polyubiquitylation by the RQC complex have been described, the mechanism leading to proteasomal degradation of these defective translation products remains unknown. We show here that the RQC complex also exists as a ribosome-unbound complex during the escort of aberrant peptides to the proteasome. In addition, we identify a new partner of this light version of the RQC complex, the E3 ubiquitin ligase Tom1. Tom1 interacts with aberrant nascent peptides and is essential to limit their accumulation and aggregation in the absence of Rqc1; however, its E3 ubiquitin ligase activity is not required. Taken together, these results reveal new roles for Tom1 in protein quality control, aggregate prevention, and, therefore, proteostasis maintenance.
INTRODUCTION
Protein homeostasis is an essential feature of eukaryotic cells because it prevents the accumulation of defective peptides and potentially toxic protein aggregates (for a review, see Richter et al., 2010). In the cytosol and the nucleus, most of the misfolded or defective proteins are selectively degraded by the ubiquitin-proteasome system (Bhattacharyya et al., 2014). This process requires selective recognition of substrates by E3 ubiquitin ligases, which perform substrate polyubiquitylation in concert with ubiquitin conjugating enzymes (E2) and the ubiquitin activating enzyme Uba1 (E1) (Finley et al., 2012). In Saccharomyces cerevisiae, two of these E3 ligases, Not4 and Ltn1 (listerin in mammals), have been characterized for their role in cotranslational quality control by recognition and polyubiquitylation of nascent peptides synthesized from aberrant transcripts (Dimitrova et al., 2009; Bengtson and Joazeiro, 2010). Whereas Not4 specifically targets polypeptides generated from aberrant polyadenylated mRNAs lacking a STOP codon (nonstop) or carrying an internal poly(A) tail (no-go), Ltn1 polyubiquitylates nascent peptides translated from “nonstop” mRNAs independently of their polyadenylation status (Matsuda et al., 2014), thus preventing the cellular accumulation of aberrant peptides that are potentially defective or even toxic. Polyubiquitylation requires prior dissociation of the stalled ribosomal subunits (Shao and Hegde, 2014). The dissociation process implicates Dom34 (pelota in mammals), Hbs1, and Rli1 (ABCE1 in mammals) (Shoemaker et al., 2010; Shoemaker and Green, 2011). To prevent continuous translation of defective transcripts that would lead to repeated stalling of ribosomal particles, aberrant transcripts associated with stalled ribosomes are recognized by Ski7 and the SKI complex in parallel to nascent peptide degradation, enabling mRNA degradation by the cytosolic exosome through the exonucleolytic activity of Dis3 (Frischmeyer et al., 2002; van Hoof et al., 2002; Doma and Parker, 2006; Tsuboi et al., 2012).
Ltn1 has been described as the E3 ubiquitin ligase of a multiprotein quality control complex called the ribosome-bound quality control (RQC) complex, composed of Ltn1, Rqc1, Rqc2, a hexamer of Cdc48, and its cofactors Ufd1 and Npl4 (Brandman et al., 2012; Defenouillère et al., 2013). The RQC complex detects stalled 60S particles after their dissociation from unconventional translation termination events by the presence of a peptidyl-tRNA located in their P-site and exit tunnel (Bengtson and Joazeiro, 2010; Lyumkis et al., 2014; Shao et al., 2015; Doamekpor et al., 2016). Rqc2 (NEMF in mammals) binds stalled 60S subunits at the exposed 40S interface (Lyumkis et al., 2014; Shao et al., 2015; Shen et al., 2015), recognizes the presence of a tRNA in the P-site, and recruits alanyl- and threonyl-tRNAs to perform the addition of C-terminal alanine-threonine tails (CAT-tails) to the aberrant nascent peptide (Shen et al., 2015). In parallel, the docking of Rqc2 stabilizes the interaction of Ltn1 with the 60S (Defenouillère et al., 2013; Lyumkis et al., 2014; Shao et al., 2015), thus stimulating aberrant nascent peptide polyubiquitylation by Ltn1 (Bengtson and Joazeiro, 2010; for a review, see Brandman and Hegde, 2016). This triggers the recruitment of the ubiquitin-selective AAA-ATPase Cdc48 and its cofactors Ufd1-Npl4 (Brandman et al., 2012; Defenouillère et al., 2013; Verma et al., 2013), which converts ATP hydrolysis into mechanical energy to extract the nascent peptide from the 60S exit tunnel and escorts it to the proteasome for degradation (Richly et al., 2005).
Both Rqc1 (TCF25 in mammals) and the polyubiquitylation process by Ltn1 are essential for Cdc48 recruitment to stalled 60S particles (Brandman et al., 2012; Defenouillère et al., 2013). Functional studies focused on the RQC complex revealed that Rqc1 and Ltn1 are both essential to prevent cytosolic aberrant protein aggregation (Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016), which results from a failure in recruitment of the ubiquitin-selective chaperone Cdc48 to stalled 60S and therefore inefficient proteasomal targeting of these aberrant substrates (Defenouillère et al., 2016). These aggregates contain polyubiquitin chains and are also degraded by the proteasome. However, their formation strictly relies on the addition of CAT-tails to aberrant nascent peptides by Rqc2 (Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016). In addition to its role in aberrant protein quality control, the RQC complex is also important to trigger the Hsf1 response to translational stress (Brandman et al., 2012), a process that also requires CAT-tail addition by Rqc2 (Shen et al., 2015). Finally, Ltn1 has also been shown to mediate the degradation of translation-stalled polypeptides at the endoplasmic reticulum membrane (Crowder et al., 2015), and, closely related, the RQC complex can access stalled polypeptides at the Sec 61 translocon (von der Malsburg et al., 2015), underlining the various cellular processes in which this quality control complex is involved.
Although the mechanistic steps between stalled 80S dissociation and aberrant peptide extraction from the 60S have been described, the downstream events leading to proteasomal delivery and degradation of the aberrant peptide remain to be elucidated. After its extraction from the 60S, it is not clear whether the aberrant nascent peptide is escorted to the proteasome only by Cdc48 or whether other factors, such as Ltn1, Rqc1, and Rqc2, also stay associated during proteasomal targeting. In addition, whether additional factors are recruited and also participate in this quality control process remains unknown. Here we show that the RQC complex also exists as a light-sedimenting, nonribosomal complex in vivo and interacts with the E3 ubiquitin ligase Tom1, which participates in limiting further protein aggregation. We describe mechanistic features of proteasomal targeting by the RQC complex and show that several quality control pathways participate in aberrant protein elimination and aggregate prevention.
RESULTS
The RQC complex remains bound to aberrant nascent peptides after their extraction from stalled 60S particles
Recent studies focused on the RQC complex and its mechanism of action, describing the 60S-bound steps of this quality control pathway leading to aberrant nascent peptide polyubiquitylation by Ltn1 and extraction from the stalled 60S particle by Cdc48. However, the downstream events leading to proteasomal degradation of these aberrant peptides remain unknown. To better understand the fate of 60S-extracted aberrant proteins, we performed affinity purifications using a protein reporter containing a TAP-tag (Rigaut et al., 1999) synthesized from an aberrant nonstop mRNA called TAP-NonStop (Defenouillère et al., 2016), coupled with sucrose gradient fractionation to separate the different complexes associated with this aberrant reporter by their sedimentation coefficient. To enrich ribonucleoprotein complexes (RNCs) associated with factors of the RQC complex, we performed affinity purifications in a yeast strain lacking SKI2 to increase the abundance of the aberrant mRNA reporter and depleted of Cdc48 to limit the extraction of our aberrant reporter protein from the 60S and slow down its proteasomal escort (Figure 1). The absorbance profile at 260 nm of this fractionated TEV eluate and the gel staining of corresponding fractions revealed an enrichment of proteins cosedimenting with 80S and 60S ribosomal particles, as well as proteins located in the light, nonribosomal fractions of the sucrose gradient and, to a lesser extent, polysome fractions (Figure 1A). This pattern was in agreement with the sedimentation profile of the TAP-NonStop accumulation in the absence of SKI2 and upon Cdc48 depletion that we previously observed by Western blot (Defenouillère et al., 2013). To characterize the proteins associated with this TAP-NonStop reporter within these fractionated complexes, we analyzed each fraction of the gradient by label-free quantitative mass spectrometry and observed a specific enrichment of the RQC complex proteins (Rqc2, Rqc1, Ltn1, and Cdc48) in the 60S fraction compared with the 80S fraction (Figure 1B and Supplemental Dataset S1), in agreement with the current model that the RQC complex interacts with 60S ribosomal particles only after stalled 80S dissociation (Shao et al., 2013; Lyumkis et al., 2014). As expected, proteins of the 40S subunit (RpS) were enriched in the 80S fractions compared with the 60S (Figure 1B and Supplemental Dataset S1). In parallel, we detected an enrichment of ubiquitin in the 60S-purified fractions compared with the 80S, which can be explained by the presence of polyubiquitylated aberrant peptides specifically at the 60S level (Defenouillère et al., 2013; Shao and Hegde, 2014). Moreover, analysis of the light-sedimenting, nonribosomal fractions (mainly fractions 4 and 5) revealed an enrichment of components of the proteasome and of the RQC factors Rqc1, Rqc2, Ltn1, and Cdc48 associated with nonribosomal forms of the TAP-NonStop reporter compared with stalled 80S fractions (Figure 1C and Supplemental Dataset S1), whereas the RQC complex has so far been identified as 60S-bound (Brandman et al., 2012; Defenouillère et al., 2013; Shao et al., 2015; Shen et al., 2015). This revealed that the RQC complex associated with aberrant nascent peptides can also exist as non–ribosome bound in vivo after their extraction from the stalled 60S and possibly during the escort of these substrates to the proteasome for degradation, an intermediate that could be accumulating upon Cdc48 depletion. Fractions 1–3 were devoid of RQC factors (unpublished data) and were therefore not analyzed further.
FIGURE 1:
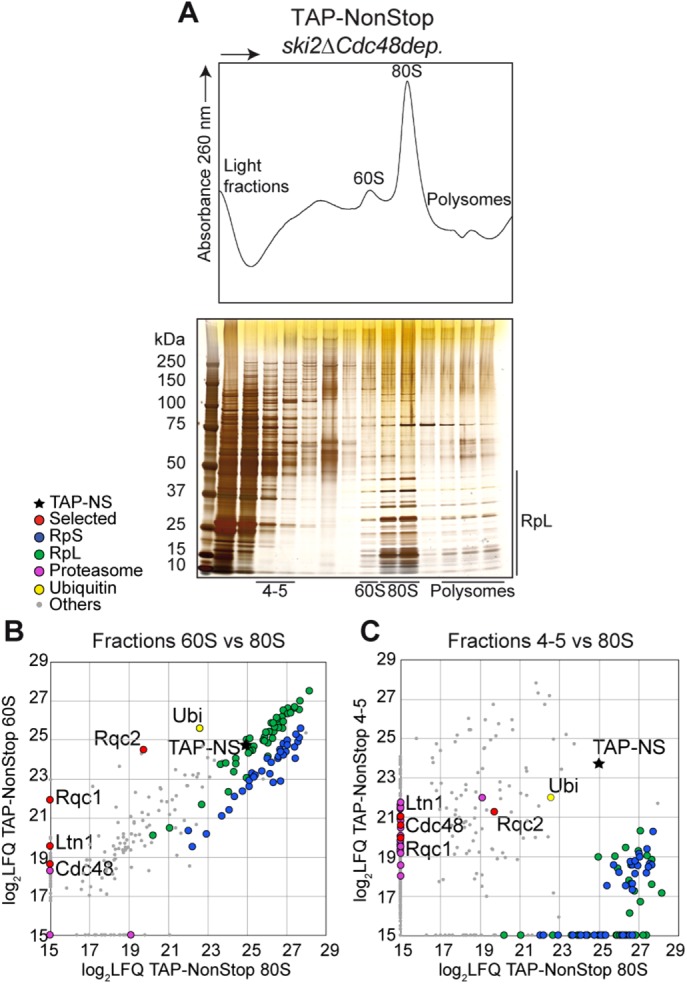
The RQC complex binds aberrant peptides in both 60S cosedimenting fractions and light fractions. (A) Sedimentation profile (absorbance at 260 nm) of a 10–50% sucrose gradient fractionation of TAP-NonStop affinity-purified complexes in ski2Δ cells depleted of Cdc48. The proteins contained in each fraction were monitored by acrylamide gel migration and silver nitrate staining. (B) The proteins associated to the TAP-NonStop complexes cosedimenting with the 60S, the 80S, and fractions 4 and 5 of the gradient were identified and quantified by LC-MS/MS. Comparison of the LFQ intensities of each protein between the 60S and 80S fractions. Each dot indicates an identified and quantified protein. The black star refers to the TAP-NonStop bait. (C) Comparison between proteins identified in fractions 4and 5 with 80S fractions. This purification coupled with gradient fractionation was repeated six times, and all LC-MS/MS analyses yielded comparable results. The list of proteins identified is given in Supplemental Dataset S1.
To verify that this “light” RQC complex detected in light-sedimenting fractions of the gradient specifically interacts with aberrant proteins, we performed a similar purification of the TAP-NonStop in Cdc48-depleted cells and, as a negative control, purified a TAP reporter containing a STOP codon (referred to as TAP-Stop). As expected, gel silver staining (Figure 2A) and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses (Figure 2B and Supplemental Dataset S1) of fractions 4 and 5 from TAP-NonStop and TAP-Stop pullouts revealed an enrichment of the RQC complex only for the TAP-NonStop purification (whereas the two TAP reporters were identified and quantified in a similar manner), which confirms that the RQC complex specifically interacts with aberrant nascent peptides. In addition, this proteomics analysis of fractions 4 and 5 revealed a specific enrichment of ubiquitin, as well as components of the proteasome, strengthening the possibility that this nonribosomal interaction between the RQC complex and its aberrant substrates occurs during their escort to the proteasome for degradation.
FIGURE 2:
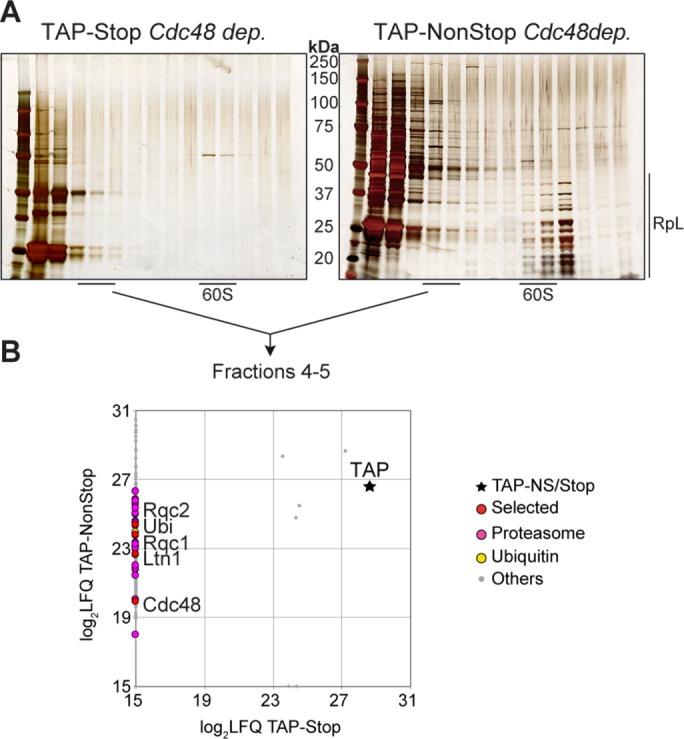
The RQC complex specifically interacts with aberrant nascent peptides in light-sedimenting fractions. (A) Silver-staining analysis of the proteins purified and fractionated from TAP-Stop (left) vs. TAP-NonStop (right) purifications upon Cdc48 depletion. (B) The proteins sedimenting in fractions 4 and 5 for both purifications were analyzed and quantified by LC-MS/MS. Comparison of LFQ intensities of all proteins identified in fractions 4 and 5 between the two purifications. The dots correspond to identified proteins, and the black star refers to the TAP-Stop or the TAP-NonStop baits (the four gray dots in the vicinity of the black star correspond to the contaminant proteins Pgk1, Act1, Pdb1, and Ssa1). The purifications using the TAP-Stop and TAP-NonStop reporters were repeated twice. The list of proteins identified is given in Supplemental Dataset S1.
Rqc1 accumulates in light-sedimenting fractions in the absence of Rqc2, Ltn1, or Cdc48
We then checked that the accumulation of the RQC complex in light fractions did not depend on the overexpression of our aberrant “nonstop” substrate. To this end, we looked at the sedimentation profile of a genomic TAP-tagged fusion of Rqc1 in various RQC complex mutants (Figure 3). As previously described, Rqc1-TAP cosediments with 60S particles in wild-type cells (Defenouillère et al., 2013). However, in all mutants of other RQC factors (rqc2Δ, ltn1Δ, and Cdc48 depletion), an additional fraction of Rqc1 accumulating in light fractions of the gradient was observed (Figure 3). Of interest, the sedimentation profile of Rqc1-TAP in the absence of Rqc2 or Ltn1 resembled the pattern previously observed for the Ltn1–3HA fusion in the absence of Rqc2 (Defenouillère et al., 2013), suggesting that in rqc2Δ cells, Ltn1 and Rqc1 cosediment and may interact both with the 60S ribosomal subunit and nonribosomal complexes. Of note, shifted species of Rqc1-TAP were visible upon Cdc48 depletion, which might correspond to ubiquitylated forms of Rqc1, a phenomenon that may be due to the autoregulation process of Rqc1 by the RQC complex itself, which has been demonstrated to be a consequence of the N-terminal polybasic stretch carried by Rqc1 (Brandman et al., 2012).
FIGURE 3:
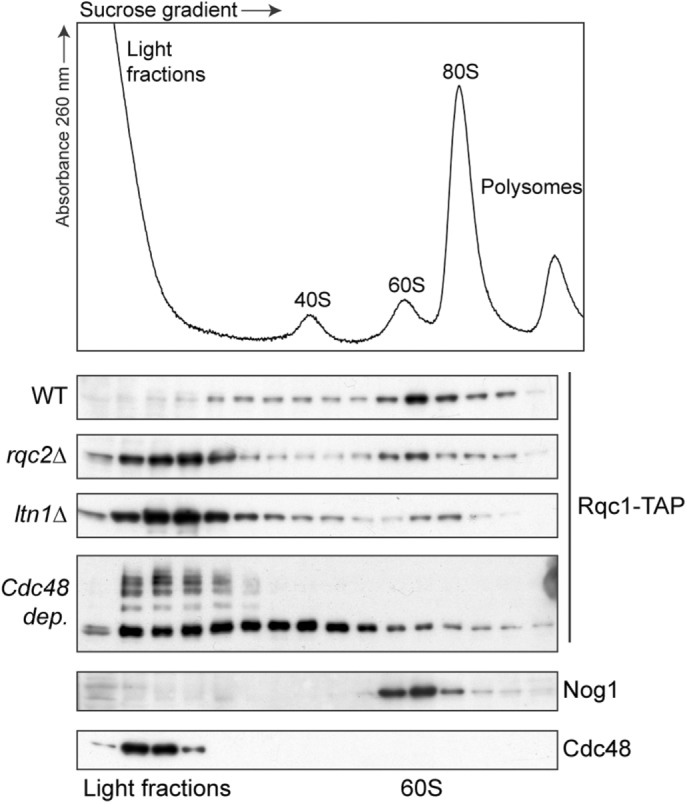
Rqc1 cosediments in light-sedimenting fractions in the absence of Rqc2, Ltn1, or Cdc48. Sedimentation profile (absorbance 260 nm) and Western blots performed with polysome extracts prepared from cells expressing Rqc1-TAP with a wild-type background, deleted for LTN1 or RQC2 or depleted for Cdc48, separated on 10–30% sucrose gradients. The localization of Rqc1-TAP was assessed using PAP antibodies; the 60S fractions and the Cdc48-sedimenting fractions were determined with anti-Nog1 and anti-Cdc48 antibodies, respectively.
The light RQC complex interacts with the E3 ubiquitin ligase Tom1
To characterize the proteins associated with the light-sedimenting form of Rqc1-TAP and its 60S-bound version, respectively, we performed purifications coupled with gradient fractionation using Rqc1-TAP as bait, with an untagged strain in parallel as a negative control. Similarly to the TAP-NonStop pullouts (Figure 2), Rqc1-TAP pullouts were performed in Cdc48-depleted cells to stabilize intermediates of the RQC quality control pathway. Fractionated samples were analyzed by gel silver staining (Figure 4A) and mass spectrometry: statistical t tests between Rqc1-TAP and the negative control were performed for the proteins quantified in fractions 4 and 5 and 60S, respectively, and were represented as volcano plots (Figure 4B and Supplemental Dataset S1). The 60S fractions purified with Rqc1-TAP showed a significant enrichment of Rqc1, Rqc2, Ltn1, ubiquitin, and ribosomal proteins of the large 60S subunit (RpL), and in this regard, they resembled the results obtained for the 60S fractions using the TAP-NonStop as bait (Figure 1B). The light-sedimenting fractions revealed an enrichment of RQC complex factors (Rqc1, Rqc2, and Ltn1) and ubiquitin (Figure 4B and Supplemental Dataset S1), as observed for the TAP-NonStop in the same fractions (Figures 1C and 2B). Cdc48 was also identified in fractions 4 and 5, although this factor was depleted, which may be due to a high binding affinity between Rqc1-TAP and Cdc48 despite the low cellular abundance of this protein after depletion. In contrast to the results obtained with TAP-NonStop purifications (Figure 2B), we did not observe any enrichment of proteasomal subunits in light-sedimenting fractions purified with Rqc1-TAP. However, analysis of these fractions revealed a significant enrichment of another partner, the E3 ubiquitin ligase Tom1, associated with the RQC complex in its light form and not with the 60S-bound version (Figure 4B). This high–molecular weight protein of 374 kDa was also visible on the silver-stained gel for the Rqc1-TAP purification but not on the negative control gel (Figure 4A) and specifically sedimented with fractions 4 and 5, consistent with Tom1 only binding the non-60S form of the RQC complex. Most of the previously described roles for Tom1 are in the nucleus (Duncan et al., 2000; Kim and Koepp, 2012; Kim et al., 2012). The fact that Tom1 might participate in aberrant protein degradation suggests that it could also be localized into the cytosol. To address this question, we used a Tom1–green fluorescent protein (GFP) fusion protein to monitor the cellular localization of Tom1 in wild-type cells and cells deleted for RQC1 or SKI2 or depleted for Cdc48 (Supplemental Figure S1). Our results show that, although most of the Tom1-GFP fluorescence signal is nuclear, a signal could be observed in nonnuclear regions of the cells, mainly corresponding to the cytosol (although Tom1 is excluded from the vacuoles), revealing that Tom1 is not strictly nuclear and can therefore also function in other cellular compartments, such as the cytosol. It is also interesting that the localization of Tom1-GFP does not significantly change in the absence of Rqc1, Ski2, or Cdc48.
FIGURE 4:
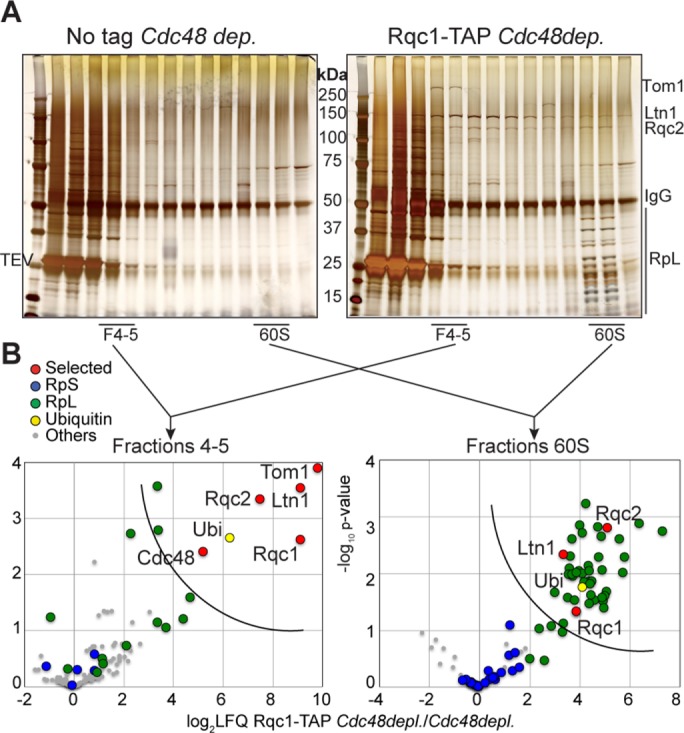
Tom1 interacts significantly with the light-sedimenting RQC complex. (A) Silver-staining analysis of the proteins fractionated from Rqc1-TAP (right) vs. negative control (left) purifications upon Cdc48 depletion. (B) Proteins associated with Rqc1-TAP complexes cosedimenting with fractions 4 and 5 (left) or the 60S (right) were identified by LC-MS/MS, and t tests of LFQ intensities for each protein were performed in comparison with the negative control. Statistical results are given in the form of volcano plots. The x-axis shows the log2 of LFQ ratios between the samples, and the y-axis shows the –log10 of the p value of each protein enrichment among statistical groups. The semicircular curve defines candidates that are significantly enriched according to a statistical t test. These purifications coupled with gradient fractionation were repeated four times. The list of proteins identified is given in Supplemental Dataset S1.
The interaction between Tom1 and the RQC complex requires the polyubiquitylation activity of Ltn1
We thus identified an additional E3 ubiquitin ligase, Tom1, able to interact with the RQC complex during an intermediate step of the quality control pathway that was likely stabilized by the Cdc48 depletion (Figure 4). We then performed Rqc1-TAP pullouts in the absence of Rqc2 and Ltn1; however, we did not observe the band corresponding to Tom1 on a silver-stained gel of the eluates in these mutant conditions and did not identify Tom1 associated to Rqc1-TAP by LC-MS/MS in these conditions (Figure 5, A–D, and Supplemental Dataset S1). In contrast, a pullout performed upon Cdc48 depletion revealed the presence of Tom1 on the gel, even without gradient fractionation. The significant enrichment of Tom1 in this gradient-free Rqc1-TAP purification was verified by performing Rqc1-TAP pullouts upon Cdc48 depletion with an untagged strain as negative control in triplicate and with mass spectrometry and statistical analysis of the identified proteins (Figure 5B and Supplemental Dataset S1). We next checked whether the presence or activity of Rqc2 and Ltn1 was required for Tom1 binding to the light RQC complex, and for this purpose, we purified Rqc1-TAP upon Cdc48 depletion in strains deleted for RQC2 and LTN1, respectively. We analyzed samples by mass spectrometry and compared them with an Rqc1-TAP purification upon Cdc48 depletion (Figure 5, C and D). The RQC2 deletion had no effect on Tom1 binding to the RQC complex (Figure 5, A and D), showing that Tom1 binding to the RQC complex does not require Rqc2 or the presence of CAT-tails on aberrant nascent peptides (Shen et al., 2015).
FIGURE 5:
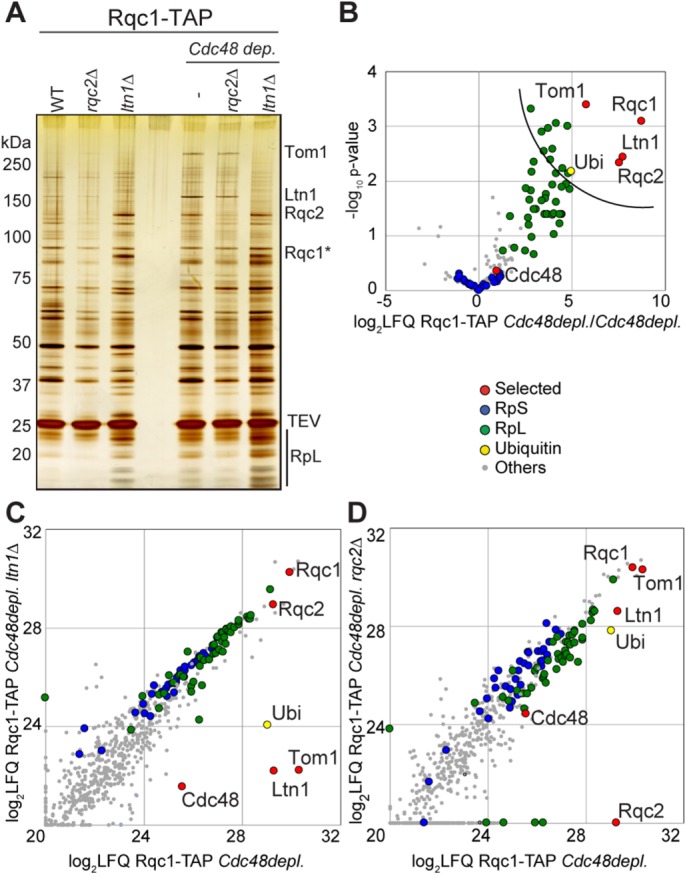
The interaction between Tom1 and the light RQC complex is Rqc2 independent but requires Ltn1. (A) Silver-staining analysis of TEV eluates purified from an Rqc1-TAP genomic fusion in various mutant strains (wild-type, rqc2Δ, and ltn1Δ) with or without Cdc48 depletion. (B) Proteins enriched with Rqc1-TAP purifications upon Cdc48 depletion were identified by LC-MS/MS, and t tests of LFQ intensities for each protein were performed in comparison with an untagged strain (negative control). Statistical results are presented in the form of a volcano plot. The x-axis shows the log2 of LFQ ratios between Rqc1-TAP and the control, and the y-axis shows the –log10 of the p value of each protein enrichment among statistical groups. The semicircular curve defines the candidates that are significantly enriched according to a statistical t test. (C) Mass spectrometry analyses of proteins identified in Rqc1-TAP purifications upon Cdc48 depletion, with or without a deletion of LTN1. Comparison of LFQ intensities obtained for each protein in the two samples; each dot represents an identified protein. (D) Comparison of Rqc1-TAP purification upon Cdc48 depletion, with or without a deletion of RQC2. The purifications presented in B were performed four times each, and those in C and D were performed once. The list of proteins identified is given in in Supplemental Dataset S1.
Strikingly, the deletion of LTN1 completely abolished the enrichment of Tom1 in the Rqc1-TAP pullout (Figure 5, A and C), revealing that Tom1 can interact with the light RQC complex only when Ltn1 is present. Because the Tom1-RQC interaction depends on the presence of Ltn1 and the mammalian orthologue of Tom1, Huwe1, contains a UBA domain that is capable of binding polyubiquitin chains (Chen et al., 2005), we tested whether the polyubiquitylation process by Ltn1, a crucial step of RQC-associated protein degradation, was essential for Tom1 binding to the RQC complex. Rqc1-TAP pullouts performed upon Cdc48 depletion in mutant strains lacking either LTN1 or its Really Interesting New Gene (RING) domain, which is essential for the interaction with its E2 ubiquitin ligases and therefore for the polyubiquitylation process, proved that Tom1 required prior polyubiquitylation of aberrant substrates by Ltn1 to interact with the light RQC complex, both with gel silver staining (Figure 6A) and mass spectrometry analysis (Figure 6B and Supplemental Dataset S1). As expected, silver-stained gel analysis of Rqc1-TAP purification upon Cdc48 depletion and in the absence of Tom1 caused a disappearance of the high–molecular weight band, confirming that this band corresponded to Tom1; however, no other difference in protein abundance was observed compared with a negative control, even by mass spectrometry (unpublished data). Together these results suggest that Tom1 interacts with the light RQC complex after polyubiquitylation of aberrant peptides by Ltn1 but independently from Rqc2, an intermediate that can be visualized in vivo in the context of Cdc48 depletion.
FIGURE 6:
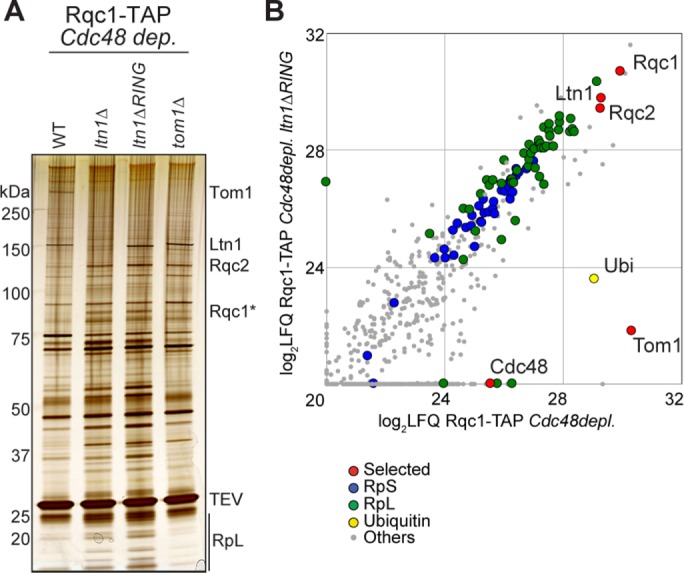
The RQC-Tom1 interaction relies on the polyubiquitylation activity of Ltn1. (A) Silver-staining analysis of TEV eluates purified from Rqc1-TAP–expressing cells in various mutant strains (wild-type, ltn1Δ, ltn1ΔRING, and tom1Δ) upon Cdc48 depletion. (B) LC-MS/MS analyses of proteins identified in Rqc1-TAP purifications upon Cdc48 depletion, with and without a deletion of the RING domain of LTN1. Comparison of LFQ intensities obtained for each protein in the two samples; each dot represents an identified protein. These purifications were performed once, except for the ltn1Δ mutant purification, which was performed twice. The list of proteins identified is given in Supplemental Dataset S1.
Tom1 physically interacts with aberrant nascent peptides in the absence of Ski2 and Cdc48
To determine whether Tom1 could directly interact with the aberrant peptides, we performed Tom1-TAP affinity purifications in cells deleted for the SKI2 gene, depleted of Cdc48, and expressing a 3FLAG-His3-NonStop aberrant reporter that derives from the His3-NonStop reporter (pAV188 plasmid; van Hoof et al., 2002; Figure 7A). We observed an enrichment of the aberrant nonstop reporter with Tom1-TAP as bait compared with an untagged strain in the same genetic background (absence of Ski2 and Cdc48; Figure 7B, top). Furthermore, the use of anti-ubiquitin antibodies revealed that Tom1 binds polyubiquitylated peptides (Figure 7B, bottom). These results are consistent with Tom1 physically interacting in vivo with the aberrant translation products. Using an Rpl1 antibody, we confirmed that the Tom1-TAP associated with 3FLAG-His3-NonStop aberrant peptides complex did not bind to the 60S subunit. In addition, we examined the sedimentation profile of Tom1-TAP in a sucrose gradient and detected the presence of Tom1-TAP in the light fractions of the gradient (Figure 7C). We also observed a subset of Tom1 cosedimenting with the fractions corresponding to higher–molecular weight complexes similar to the 40S fractions. Because Tom1 is also involved in nuclear processes, these higher–molecular weight fractions of Tom1 could be related to these additional processes (Duncan et al., 2000; Kim and Koepp, 2012; Kim et al., 2012).
FIGURE 7:
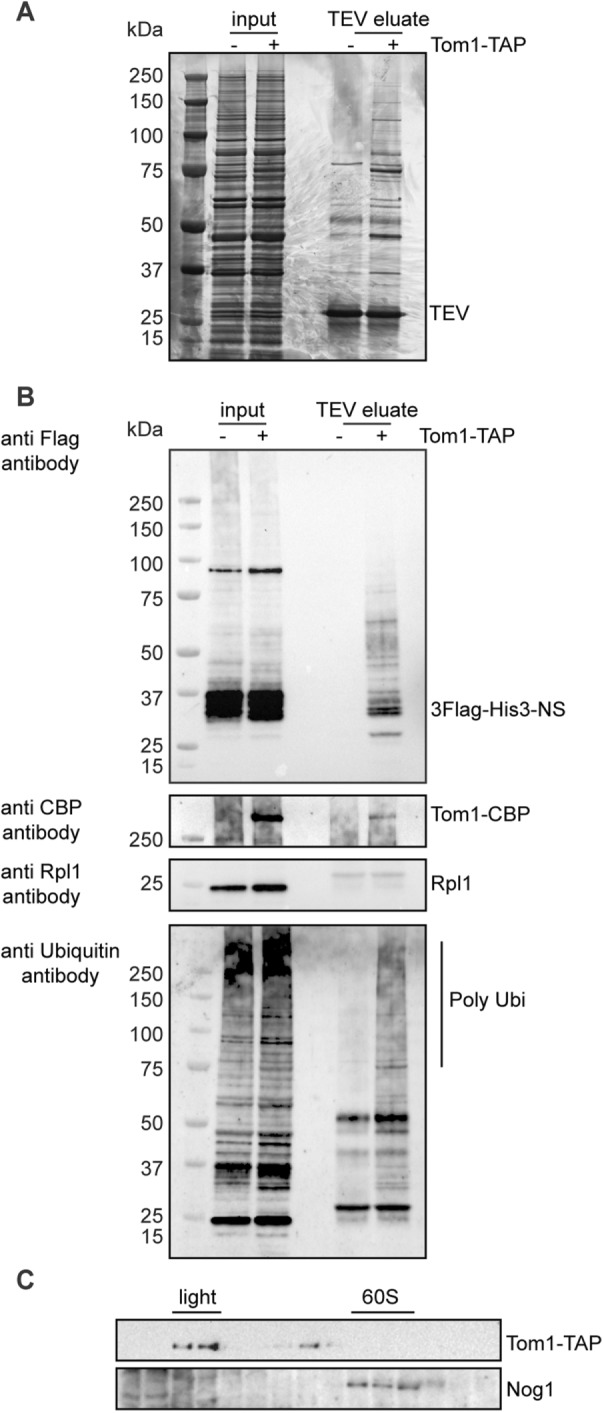
Tom1 physically interacts with aberrant nascent peptides in vivo in the absence of SKI2 upon Cdc48 depletion. (A) Proteins enriched with Tom1-TAP purifications (TEV eluate) upon Cdc48 depletion and deleted for SKI2 were separated on 4–12% gradient polyacrylamide NuPAGE gel and silver stained. For comparison, a fraction of total cellular extracts (input) was loaded. (B) Western blots were performed with input and TEV eluates fractions as in A. The presence of the 3XFlag-His3-NS aberrant peptides was assessed using anti-Flag antibodies. Tom1-CBP bait was probed with a CBP antibody. Anti-Rpl1 antibody was used to reveal Rpl1 ribosomal protein, and polyubiquitylated proteins were assessed with anti-ubiquitin antibodies. (C) Tom1 cosediments in light-sedimenting fractions. Western blots were performed with polysome extracts prepared from cells deleted for SKI2, depleted of Cdc48, and separated on 10–50% sucrose gradients. Tom1-TAP was assessed using PAP antibodies. The 40S and 60S fractions were determined with anti Rps8 and anti-Nog1 antibodies, respectively.
Tom1 limits aberrant protein aggregation in the absence of Rqc1
Tom1 is an E3 ubiquitin ligase of the homologous to E6AP C-terminus (HECT) family that has been previously described as being involved in the degradation of specific proteins, notably in the turnover of Dia2 during the cell cycle (Kim and Koepp, 2012), the stability of Cdc6 after DNA damage (Kim et al., 2012), and the degradation of excess histones (Singh et al., 2009). More recently, Tom1 has been shown to limit the excess of specific ribosomal proteins in concert with E2 enzymes Ubc4/5 (Sung et al., 2016). To investigate whether Tom1 could have a role in RQC-mediated protein quality control when bound to the light RQC complex, we assessed the effect of a TOM1 deletion on the accumulation of the TAP-NonStop reporter in a polysome gradient but did not observe any stabilization of this aberrant protein (Figure 8A). Rqc1 and Ltn1 were recently shown to prevent the aggregation of aberrant nascent peptides, a process that requires CAT-tail addition by Rqc2 (Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016). These types of aggregates can be revealed by Western blot as a protein fraction that accumulates in the stacking part of an acrylamide gel. Because Tom1 interacted with the light RQC complex in similar sedimenting fractions as the protein aggregates accumulating in rqc1Δ cells (Defenouillère et al., 2016), we checked whether Tom1 could have a role in preventing the accumulation of aggregated aberrant proteins by comparing aggregate accumulation of the TAP-NonStop reporter between rqc1Δ cells and a tom1Δrqc1Δ double mutant. Strikingly, the quantity of aggregated aberrant proteins was massively increased in the absence of both Rqc1 and Tom1 compared with the absence of Rqc1 alone (Figure 8A), which shows that Tom1 has an important role in limiting the accumulation of protein aggregates when Rqc1 is defective. However, Tom1 does not seem to participate directly in the degradation of soluble aberrant proteins, because no accumulation of the TAP-NonStop was detected in tom1Δ single-mutant cells. We then examined whether the formation of protein aggregates in the tom1Δrqc1Δ double mutant also required CAT-tail addition by Rqc2 and observed a complete disappearance of TAP-NonStop aggregates in the absence of Rqc1 and Tom1 when RQC2 was deleted (Figure 8B).
FIGURE 8:
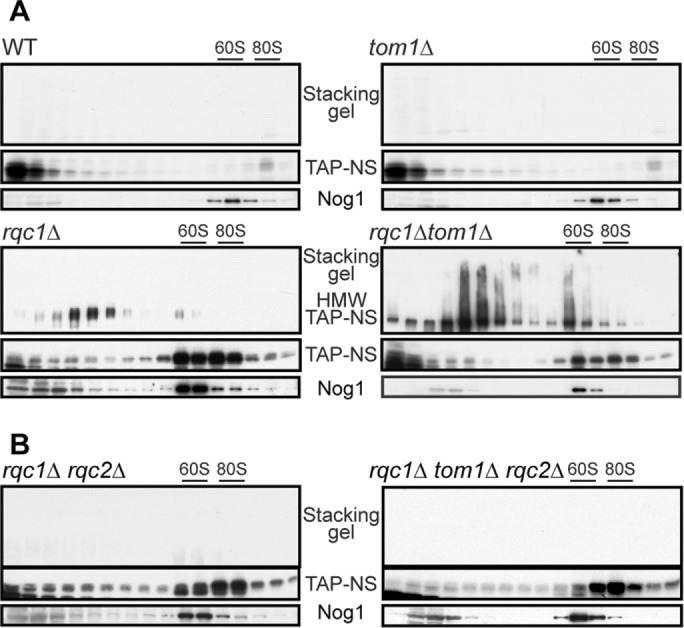
Tom1 becomes essential to limit aberrant protein aggregation when Rqc1 is impaired. (A) Western blots performed with polysome extracts prepared from TAP-NonStop–expressing cells in a wild-type background or deleted for RQC1, TOM1, or both, and separated on 10–50% sucrose gradients. The sedimentation profiles of aggregated (stacking gel) and soluble TAP-NonStop were assessed using PAP antibodies, and the 60S cosedimenting fractions were probed using anti-Nog1 antibodies. (B) As in A, using TAP-NonStop–expressing cells deleted for RQC1 and RQC2 (left) or RQC1, RQC2, and TOM1 (right).
In parallel, we used a previously described aberrant protein reporter called GFP-NonStop deriving from the TAP-NonStop reporter, except that it contains a GFP fluorescent sequence that enables the visualization of aberrant protein aggregates within yeast cells by fluorescence microscopy (Defenouillère et al., 2016). As expected, monitoring the accumulation and localization of the GFP-NonStop reporter in tom1Δ cells showed no increase in fluorescence levels compared with a wild type (Figure 9). However, the cytosolic aggregates of aberrant proteins accumulating in the absence of Rqc1 were larger and more numerous per cell in the rqc1Δtom1Δ double mutant (Figure 9), which corroborates the results obtained in these same mutants for the TAP-NonStop reporter (Figure 8A).
FIGURE 9:
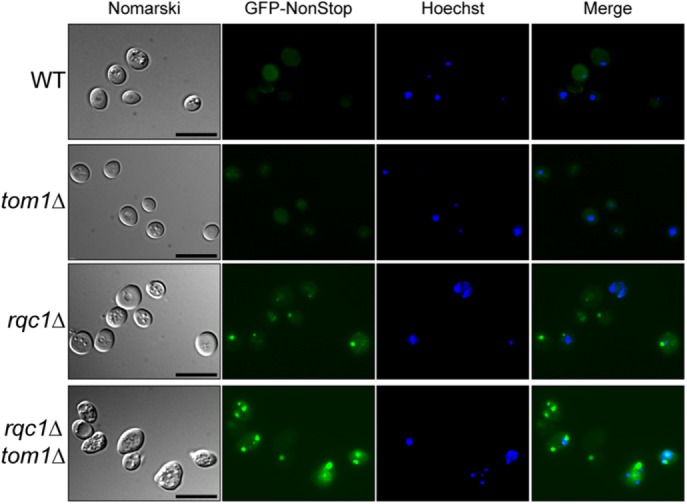
Tom1 prevents the expansion of cytosolic protein aggregates in the absence of Rqc1. WT, tom1Δ, rqc1Δ, and rqc1Δtom1Δ strains expressing the GFP-NonStop reporter protein were analyzed by fluorescence microscopy. Left, yeast cells (Nomarski); second and third sets of images, GFP and Hoechst fluorescence signals, respectively; right, their merge. Scale bar, 5 μm.
Because the aggregates accumulating in rqc1Δ cells are known to contain aberrant proteins that are polyubiquitylated in an Ltn1-dependent manner (Defenouillère et al., 2016), we assessed the level of polyubiquitylated protein aggregates in the absence of Tom1 and Rqc1 compared with the rqc1Δ single mutant in reporter-free cells. Whereas we observed no signal with an anti-ubiquitin antibody in the stacking gel of tom1Δ cells (as previously reported for a wild-type strain), we found that the quantity of aggregated polyubiquitylated proteins was higher in the rqc1Δtom1Δ double mutant than with the rqc1Δ single mutant (Figure 10A), confirming that Tom1 becomes essential to limit the accumulation of cytosolic protein aggregates that appear in the absence of Rqc1. A deletion of LTN1 in this rqc1Δtom1Δ strain completely abolished the accumulation of polyubiquitin chains in the stacking gel (Figure 10A), as described for polyubiquitylated aggregates accumulating in rqc1Δ cells (Defenouillère et al., 2016), which shows that aberrant substrates that aggregate in the absence of both Rqc1 and Tom1 are also polyubiquitylated via Ltn1.
FIGURE 10:
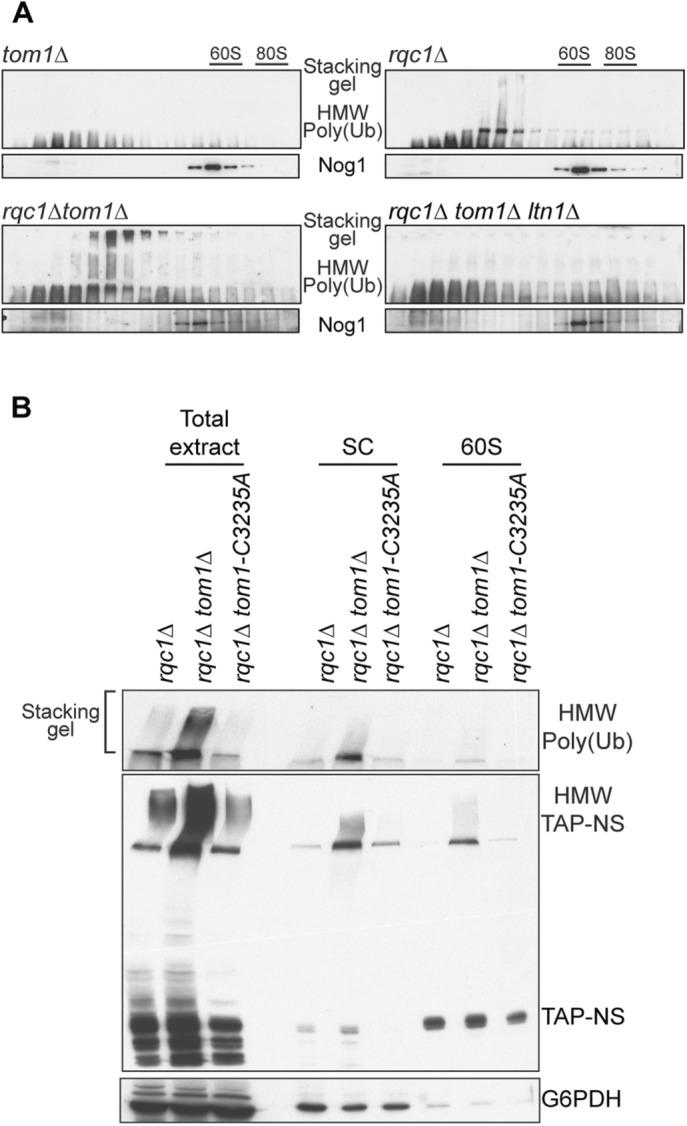
The E3 ubiquitin ligase activity of Tom1 is not required for its role in protein aggregate prevention. (A) Western blots performed with polysome extracts prepared from tom1Δ, rqc1Δ, rqc1Δtom1Δ, and rqc1Δtom1Δltn1Δ mutant cells and separated on 10–50% sucrose gradients. The sedimentation profiles of ubiquitylated proteins in their aggregated form (stacking gel) were assessed using anti-ubiquitin antibodies. The 60S cosedimenting fractions were probed using anti-Nog1 antibodies. (B) Western blots were performed using total protein extracts and polysome extracts separated on 10–50% sucrose gradients (for which the light-sedimenting fractions [SC] and the 60S-sedimenting fractions were respectively pooled), prepared from rqc1Δ, rqc1Δtom1Δ, and rqc1Δtom1-C3235A mutant yeast cells expressing the TAP-NonStop reporter. The sedimentation profiles of the aggregated and soluble versions of the TAP-NonStop reporter were probed with PAP antibodies (middle), and the levels of aggregated polyubiquitylated proteins were assessed with anti-ubiquitin antibodies (top). Anti-G6PDH antibodies were used as a loading control (bottom).
The E3 ubiquitin ligase activity of Tom1 is not required for its role in protein aggregate prevention
Finally, to determine whether the E3 ubiquitin ligase activity of Tom1 is required for its function in aberrant protein aggregate limitation, we generated a HECT-C3235A genomic point mutation within TOM1 that was previously shown to disrupt its E3 ubiquitin ligase activity (Duncan et al., 2000). In contrast to our observations upon deletion of TOM1, there was no additional accumulation of TAP-NonStop aggregates or any effect on their polyubiquitylation level when this tom1-C3235A mutant was introduced into an rqc1Δ background (Figure 10B). These results show that Tom1-mediated limitation of aggregates in the absence of Rqc1 does not rely on the E3 ubiquitin ligase activity of Tom1, suggesting that other functions carried out by this protein are required to perform this process.
Together our data show that the RQC complex remains associated with aberrant nascent peptides after their extraction from the 60S exit tunnel and thus during their escort to the proteasome for degradation and reveal the existence of an additional partner of the RQC complex, Tom1, which specifically interacts with the light, nonribosomal form of this complex. Our study reveals a new role for Tom1 in protein quality control and proteostasis by interacting with the light RQC complex and limiting the accumulation of cytosolic aberrant protein aggregates.
DISCUSSION
The RQC complex has been described as a cotranslational quality control complex that recognizes stalled 60S particles. It polyubiquitylates aberrant nascent peptides via Ltn1 (Brandman et al., 2012; Defenouillère et al., 2013), adds CAT-tails to these substrates via Rqc2 (Shen et al., 2015), and extracts these polyubiquitylated, CAT-tailed polypeptides from the 60S exit tunnel via the Cdc48 hexamer (Brandman et al., 2012; Defenouillère et al., 2013). Furthermore, functional studies focused on RQC deficiency revealed the importance of Rqc1 and Ltn1 in preventing the accumulation of cytosolic protein aggregates (Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016), a phenotype likely due to a defect in Cdc48 recruitment on stalled 60S and therefore in inefficient proteasomal targeting of aberrant translation products. The RQC complex is also known to trigger Hsf1-driven translational stress response (Brandman et al., 2012). Of interest, both protein aggregation and RQC-mediated Hsf1 activation rely on CAT-tail addition to nascent peptides, suggesting that these two systems may be linked (Shen et al., 2015; Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016). However, beyond the studies focused on the mechanism of action of the RQC complex, the respective fates of this complex and of its aberrant substrates after their dissociation from the stalled 60S remain unclear.
Here we focused on the downstream steps of this quality control pathway and showed that 1) the RQC complex remains associated to aberrant peptides independently of 60S particles, 2) the RQC complex interacts with Tom1 after 60S extraction of nascent peptides, and 3) Tom1 interacts with aberrant nascent peptides, preventing the accumulation of aberrant protein aggregates when the RQC pathway is impaired. Indeed, purification of an RQC-targeted reporter peptide showed that Rqc1, Rqc2, Ltn1, and Cdc48 remain bound to the nonstop aberrant protein reporter after its synthesis but in a form that accumulates in nonribosomal fractions of the gradient (Figures 1 and 2). This implies that this intermediate complex corresponds to a step occurring after extraction of the polyubiquitylated aberrant peptide from the stalled 60S and thus corresponds to an escorting step of the polypeptide before it is delivered to the proteasome for degradation (Richly et al., 2005). The enrichment of constituents of both the RQC complex and the proteasome specifically with our aberrant “nonstop” reporter (Figure 2B), all the more cosedimenting in the same purified fractions of a sucrose gradient, suggests that there might be an interaction between these two multiprotein complexes at the moment of aberrant substrate delivery. Our attempts to isolate such an intermediate by performing double affinity purifications using both the TAP-NonStop reporter and the proteasome itself as baits remained unfruitful (unpublished data), but this may be due to the fact that aberrant polyubiquitylated substrates are rapidly degraded when they reach the proteasome, suggesting that, if such a proteasome-RQC intermediate exists, it is likely to be very transient and unstable.
Further evidence for the existence of a “light,” nonribosomal version of a RQC-associated complex in vivo was reinforced by purifications coupled with gradient fractionation using Rqc1-TAP as bait. The fractionation experiments (Figures 3 and 4) confirmed the existence of two complexes associated with Rqc1-TAP with different sedimentation coefficients: one 60S-bound and the other corresponding to the nonribosomal RQC complex, which was physically associated with an additional partner, the E3 ubiquitin ligase Tom1 (Figure 4). Of interest, the significant enrichment of Tom1 with this nonribosomal version of Rqc1-TAP did not depend on Rqc2 (Figure 5) but strictly relied on the E3 ubiquitin ligase activity of Ltn1 (Figures 5 and 6). Because the mammalian orthologue of Tom1, Huwe1, contains a UBA domain conferring on Huwe1 the capacity to interact with polyubiquitin chains (Chen et al., 2005), one possibility is that Tom1 may interact with the light RQC complex via Ltn1-produced polyubiquitin chains. It is surprising, however, that Tom1 was identified only with Rqc1-TAP purifications and not with the TAP-NonStop reporter (Figures 1 and 2), although the purification protocols were similar and were both performed upon Cdc48 depletion. These discrepancies may reflect the fact that Rqc1-TAP pullouts are more specific and adapted to RQC complex partners because they might rely on high binding efficiencies between the different factors of the complex, whereas TAP-NonStop purifications are less selective because they enable purification of a larger variety of complexes, such as 80S particles or the proteasome (Figure 2B). However, affinity purifications using Tom1-TAP as bait in the absence of Ski2 and upon Cdc48 depletion proved to be specific enough to efficiently enrich aberrant nascent peptides (Figure 7) and confirmed that Tom1 physically binds aberrant proteins to prevent them from accumulating and therefore aggregating when the RQC complex is impaired.
In parallel, our functional study of a TOM1 deletion clearly showed the importance of Tom1 for the reduction of TAP-NonStop and GFP-NonStop aggregates when the RQC activity is compromised (Figures 8 and 9). This confirms that Tom1 indeed has a role in RQC-associated quality control because it interacts physically with the aberrant peptide associated with the light RQC complex and participates in the degradation of RQC-targeted substrates. Of note, the absence of Rqc2 did not affect Tom1 binding to the RQC complex (Figure 5D and Supplemental Dataset S1), although Rqc2 is essential for CAT-tail addition and thus aberrant aggregate formation (Figure 8B) (Shen et al., 2015, Choe et al., 2016; Defenouillère et al., 2016; Yonashiro et al., 2016), suggesting that Tom1 instead targets soluble forms of aberrant proteins. Our results also show that the E3 ligase activity of Tom1 is not required for its role in aggregate prevention, suggesting that other functional domains of Tom1 are involved in this process. Previous studies focused on the mammalian orthologue of Tom1, Huwe1, may provide some insights into this matter: indeed, in addition to its UBA domain conferring on Huwe1 the capacity to bind polyubiquitin chains (Chen et al., 2005), Huwe1 has also been described as a proteasome-associated factor (Besche et al., 2009). It is therefore possible that Tom1 recognizes RQC-bound polyubiquitylated aberrant proteins and binds the proteasome, thus acting as a physical bridge between polyubiquitylated substrates and proteasome moieties to ensure the elimination of aberrant translation products and preventing their aggregation. Further investigation of proteasomal targeting by the RQC-Tom1 complex could determine whether Tom1 can bind polyubiquitin chains and the proteasome and whether Tom1 promotes proteasomal delivery of aberrant proteins when interacting with the RQC complex.
The function of Tom1 in the degradation of excess ribosomal proteins such as Rpl26 (Sung et al., 2016) may also be relevant for the role of Tom1 in RQC-mediated quality control: indeed, optimizing the quality of the cellular ribosome pool may have an effect on translation efficiency, indirectly reducing the amount of aberrant translation products and thus the quantity of protein aggregates. However, the E3 ubiquitin ligase activity of Tom1 is required for this ribosomal protein quality control, but its impairment does not affect aberrant aggregate accumulation (Figure 10B); besides, Tom1 interacts with the RQC complex in ribosome-independent fractions (Figure 4). Therefore the data presented in our study do not support a functional link between these two roles of Tom1.
Intriguingly, the importance of Tom1 for aberrant aggregate prevention was observed only in specific mutants of the RQC complex, which raises the question of the biological relevance of this RQC-Tom1 interaction in wild-type cells. The autoregulatory loop performed by the RQC complex constantly maintains Rqc1 at a low cellular abundance (Brandman et al., 2012), and hence it is possible that during translational stress, the sudden accumulation of aberrant substrates rapidly titrates Rqc1, in which case, the Tom1-mediated quality control pathway may act as a backup mechanism to ensure the degradation of aberrant translation products and limit protein aggregation.
In conclusion, our study shows that translational quality control is not limited to the action of the RQC complex but integrates other quality control pathways, such as the role of Tom1 for aggregate limitation in case of failure of the RQC machinery. This network of quality control pathways may confer eukaryotic cells the ability to rapidly adapt and overcome stressful conditions in order to maintain proteostasis and avoid the toxicity arising from protein aggregates.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used in this study are listed in Table 1. All yeast strains were constructed from Saccharomyces cerevisiae genetic backgrounds BY4741 or BY4742 by homologous recombination. Depletions of CDC48 were performed by inserting a tetO2 promoter and incubating yeast cultures with doxycycline (10 µg/ml) for 13.5 h as described (Defenouillère et al., 2016). The pTAP-Stop and pTAP-Non-Stop plasmids were constructed by yeast homologous recombination using ProteinA-NonStop (pAV184) and ProteinA-Stop (pAV183) as host vectors as described (Defenouillère et al., 2013). The 3XFLAG-His3-NonStop reporter was constructed by yeast homologous recombination using the pAV188 as host vector (van Hoof et al., 2002) and a 3XFLAG PCR fragment. Details concerning these constructs will be provided upon request.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa, ura3Δ0, his3Δ1, leu2Δ0, met15Δ0 | Brachmann et al. (1998) |
| BY4742 | MATα, ura3Δ0, his3Δ1, leu2Δ0, lys2Δ0 | Brachmann et al. (1998) |
| LMA1951 | as BY4741, rqc1-TAP:HIS3MX6 | Ghaemmaghami et al. (2003) |
| LMA2688 | as BY4741, rqc1-TAP:HIS3MX6, rqc2Δ::KANMX4 | Defenouillère et al. (2013) |
| LMA2689 | as BY4741, rqc1-TAP:HIS3MX6, ltn1Δ::KANMX4 | Defenouillère et al. (2013) |
| LMA2648 | as BY4741, rqc1-TAP:HIS3MX6, KANMX4:PrTetO2:CDC48 | This study |
| LMA2869 | as BY4741, rqc1-TAP:HIS3MX6, KANMX4:PrTetO2:CDC48, rqc2Δ::URA3 | This study |
| LMA2870 | as BY4741, rqc1-TAP:HIS3MX6, KANMX4:PrTetO2:CDC48, ltn1Δ::URA3 | This study |
| LMA3002 | as BY4741, rqc1-TAP:HIS3MX6, KANMX4:PrTetO2:CDC48, tom1Δ::LEU2 | This study |
| LMA3058 | as BY4741, rqc1-TAP:HIS3MX6, KANMX4:PrTetO2:CDC48, ltn1ΔRING::URA3 | This study |
| LMA4747 | as BY4741, tom1-Gfp:HIS3MX6 | Ghaemmaghami et al. (2003) |
| LMA4748 | as BY4741, tom1-Gfp:HIS3MX6, ski2∆::KANMX4 | This study |
| LMA4750 | as BY4741, tom1-Gfp:HIS3MX6, rqc1∆::KANMX4 | This study |
| LMA4752 | as BY4741, tom1-Gfp:HIS3MX6, KANMX4:PrTetO2:CDC48 | This study |
| LMA2773 | as BY4741, tom1-TAP:HIS3MX6 | Ghaemmaghami et al. (2003) |
| LMA3006 | as BY4741, tom1-TAP:HIS3MX6, rqc1Δ::KANMX4, | This study |
| LMA4806 | as BY4741, tom1-TAP:HIS3MX6, ski2Δ::KANMX4, NATMX4:PrTetO2:CDC48 | This study |
| LMA1967 | as BY4742, rqc1Δ::KANMX4 | Brachmann et al. (1998) |
| LMA2070 | as BY4741, rqc1∆::KANMX4 | Brachmann et al. (1998) |
| LMA2714 | as BY4741, rqc1Δ::HIS3 | Defenouillère et al. (2013) |
| LMA2948 | as BY4742, tom1Δ::LEU2 | This study |
| LMA3123 | as BY4742, tom1Δ::LEU2, rqc1Δ::KANMX4 | This study |
| LMA3202 | as BY4742, tom1Δ::LEU2, rqc1Δ::KANMX4, ltn1Δ::NATMX4 | This study |
| LMA3193 | as BY4742, tom1-hectC3235A:LEU2, rqc1Δ::KANMX4 | This study |
| LMA2135 | as BY4742, rqc1Δ::KANMX4, rqc2Δ:: PrαNATMX4 | Defenouillère et al. (2016) |
| LMA3645 | as BY4742, rqc1Δ::KANMX4, tom1Δ::LEU2 rqc2Δ::HIS3 | This study |
| LMA2719 | as BY4742, PrαNATMX4:PrTetO2:CDC48 | Defenouillère et al. (2013) |
| LMA2746 | as BY4742, PrαNATMX4:PrTetO2:CDC48, ski2Δ::KANMX4 | Defenouillère et al. (2013) |
| LMA3121 | as BY4742, PrαNATMX4:PrTetO2:CDC48, rqc1Δ::KANMX4 | Defenouillère et al. (2016) |
Polysome gradients, proteins extraction, and Western blotting
Polysome extracts, sucrose gradients, and proteins extractions were performed as described (Defenouillère et al., 2016). For Western blot, proteins were detected by hybridization with the appropriate antibodies. Peroxidase anti-peroxidase (PAP) complex (Sigma-Aldrich) was used at 1:10,000. The rabbit antibodies against Nog1, Rpl1, and Cdc48 were used at 1:5000; the rabbit anti-CBP at 1:2000, the rabbit anti–glucose-6-phosphate dehydrogenase (G6PDH) at 1:100,000, the mouse P4D1 monoclonal antibody against ubiquitin (Covance) at 1:1000; and the monoclonal anti-FLAG M2-peroxidase antibody (Sigma-Aldrich) at 1:5000.
Affinity purifications coupled with sucrose gradient fractionation
Affinity purifications were performed as in Defenouillère et al. (2013). For Rqc1-TAP purifications, 4 l of yeast cells grown in rich medium were depleted from CDC48 for 13.5 h with 10 µg/ml doxycycline and harvested at OD600 = 1.0 by centrifugation at 4000 × g for 7 min. Cells were washed with cold water, frozen with dry ice, and lysed with a French press (twice at 1200 psi) in lysis buffer (50 mM Tris HCl, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and water with EDTA-free protease inhibitor cocktail from Roche). Lysates were centrifuged at 16,000 × g for 20 min at 4°C and incubated for 45 min at 4°C with 50 µl of magnetic beads (Dynabeads; Thermo Fisher) coupled with immunoglobulin G (IgG) antibodies, 1/2500 antifoam, and 0.075% IGEPAL. Beads were washed twice with lysis buffer with 0.075% IGEPAL and twice without. Elution was performed in lysis buffer with 10 µl of TEV protease (AcTEV; Life Technologies) for 100 min at 17°C. TEV eluates were loaded on a 10–30% sucrose gradient, and ultracentrifugation was performed at 27,000 × g for 15 h. Sucrose fractions were collected and precipitated with the methanol-chloroform method (Wessel and Flügge, 1984). Samples were divided into one-third for 4–12% gradient polyacrylamide NuPAGE gel loading and silver staining and two-thirds for analysis by LC-MS/MS. For TAP-NonStop and TAP-Stop purifications, cells transformed with one of these vectors were grown in synthetic medium containing galactose without uracil and depleted of CDC48 for 13.5 h with 10 µg/ml doxycycline. Cells were resuspended in lysis buffer and lysed with glass beads on a vortex (four times for 1 min). Lysates were clarified by centrifugation and incubated for 45 min with 50 µl of magnetic beads coupled with IgG antibodies and washed four times in lysis buffer containing 0.075% IGEPAL. TAP-associated complexes were eluted using 10 µl of AcTEV protease for 100 min. TEV eluates were loaded on 10–50% sucrose gradients, and ultracentrifugation was performed at 39,000 × g for 3 h. The rest of the sample preparation was identical to Rqc1-TAP purifications. For Tom1-TAP purifications in presence of the 3XFlag-His3-NonStop aberrant peptides, 1 l of yeast cells grown in minimal medium without uracil were depleted from CDC48 for 13.5 h with 10 µg/ml doxycycline and harvested by centrifugation at 4000 × g for 7 min. Cells were washed with cold water and lysed with Magnalyser and glass beads (three times at 3000 × g for 90 s) in lysis buffer (50 mM Tris HCl, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10 mM N-ethylmaleimide, 10 µm MG-132, 1 mM PMSF, and water with EDTA-free protease inhibitor cocktail from Roche). Lysates were centrifuged twice at 5000 × g for 5 min at 4°C and incubated for 45 min at 4°C with 50 µl of magnetic beads, followed by AcTEV protease treatment as described.
Mass spectrometry experiments and data analysis
Sample preparation was identical to that in Defenouillère et al. (2013). Briefly, proteins were precipitated with the methanol-chloroform method (Wessel and Flügge, 1984) and digested with trypsin, and peptide samples were analyzed by LC-MS/MS on an Orbitrap Velos. Protein identifications and comparative label-free quantification were performed using the MaxQuant suite (version 1.5.2.8), which includes the Andromeda search engine. Quantifications were done using the algorithm integrated into MaxQuant to calculate Label Free Quantification intensities (Cox et al., 2014).
Fluorescence microscopy
Fluorescence microscopy experiments were performed using the pGFP-Non-Stop plasmid as previously described (Defenouillère et al., 2016). Briefly, GFP-NonStop–expressing yeast cells grown in liquid synthetic medium without uracil with galactose as a carbon source were centrifuged and resuspended in MilliQ water. DNA was stained with Hoechst 33352 (5 ng/μl) for 5 min, and cells were again rinsed in water. For Tom1-GFP microscopy experiments, cells were grown overnight in rich medium until mid log phase (OD600 = 0.6), centrifuged, rinsed in water, and treated with Hoechst 33352 as described. Samples were imaged using a Leica DMRXA fluorescence microscope with fluorescence signals collected with durations of 3500 ms (GFP-NonStop), 2000 ms (Tom1-GFP), 40 ms (Hoechst 33352), and 50 ms (Nomarski). Images were collected with a Hamamatsu ORCAII-ER cooled charge-coupled device camera controlled by Openlab (version 3.5; Improvision) and processed using Adobe Photoshop CS5.
Supplementary Material
Acknowledgments
We thank the members of the Génétique des Interactions Macromoléculaires lab for discussions and criticism on the manuscript. We thank Alexander Buchberger for the anti-Cdc48 antibodies. We are grateful to the proteomics platform of the Pasteur Institute for the availability of the Orbitrap Velos. Q.D. was supported by fellowships from the Ministère de l’Enseignement Supérieur et de la Recherche and the Association pour la Recherche contre le Cancer. This work was supported by Grants ANR-2011-BSV6-011-02 and ANR-14-CE-10-0014-01 from the Agence Nationale de la Recherche, the Institut Pasteur, and the Centre National de la Recherche Scientifique.
Abbreviations used:
- CAT
C-terminal alanine-threonine
- HMW
high molecular weight
- LFQ
label-free quantification
- RQC
ribosome-bound quality control
- TAP-NonStop
tandem affinity purification-nonstop
- TEV
tobacco etch virus.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0746) on March 15, 2017.
REFERENCES
- Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Choe Y-J, Park S-H, Hassemer T, Körner R, Vincenz-Donnelly L, Hayer-Hartl M, Hartl FU. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016;531:191–195. doi: 10.1038/nature16973. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder JJ, Geigges M, Gibson RT, Fults ES, Buchanan BW, Sachs N, Schink A, Kreft SG, Rubenstein EM. Rkr1/Ltn1 ubiquitin ligase-mediated degradation of translationally stalled endoplasmic reticulum proteins. J Biol Chem. 2015;290:18454–18466. doi: 10.1074/jbc.M115.663559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci USA. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defenouillère Q, Zhang E, Namane A, Mouaikel J, Jacquier A, Fromont-Racine M. Rqc1 and Ltn1 prevent c-terminal alanine-threonine tail (CAT-tail)-induced protein aggregation by efficient recruitment of Cdc48 on stalled 60S subunits. J Biol Chem. 2016;291:12245–12253. doi: 10.1074/jbc.M116.722264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doamekpor SK, Lee J-W, Hepowit NL, Wu C, Charenton C, Leonard M, Bengtson MH, Rajashankar KR, Sachs MS, Lima CD, et al. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc Natl Acad Sci USA. 2016;113:E4151–E4160. doi: 10.1073/pnas.1605951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K, Umen JG, Guthrie C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Kim D-H, Koepp DM. Hect E3 ubiquitin ligase Tom1 controls Dia 2 degradation during the cell cycle. Mol Biol Cell. 2012;23:4203–4211. doi: 10.1091/mbc.E12-07-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Zhang W, Koepp DM. The Hect domain E3 ligase Tom1 and the F-box protein Dia 2 control Cdc6 degradation in G1 phase. J Biol Chem. 2012;287:44212–44220. doi: 10.1074/jbc.M112.401778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Oliveira Dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, Joazeiro CAP. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc Natl Acad Sci USA. 2014;111:15981–15986. doi: 10.1073/pnas.1413882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R, Ikeuchi K, Nomura S, Inada T. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells. 2014;19:1–12. doi: 10.1111/gtc.12106. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Shao S, Brown A, Santhanam B, Hegde RS. Structure and assembly pathway of the ribosome quality control complex. Mol Cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kabbaj M-HM, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M-K, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, Hess S, Deshaies RJ. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 2016;5:e19105. doi: 10.7554/eLife.19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Shao S, Hegde RS. The ribosome quality control pathway can access nascent polypeptides stalled at the Sec 61 translocon. Mol Biol Cell. 2015;26:2168–2180. doi: 10.1091/mbc.E15-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yonashiro R, Tahara EB, Bengtson MH, Khokhrina M, Lorenz H, Chen K-C, Kigoshi-Tansho Y, Savas JN, Yates JR, Kay SA, et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. Elife. 2016;5:e11794. doi: 10.7554/eLife.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


