Abstract
The spirochetes Brachyspira hyodysenteriae and B. pilosicoli are pig intestinal pathogens that are the causative agents of swine dysentery (SD) and porcine intestinal spirochaetosis (PIS), respectively. Although some inactivated bacterin and recombinant vaccines have been explored as prophylactic treatments against these species, no effective vaccine is yet available. Immunoproteomics approaches hold the potential for the identification of new, suitable candidates for subunit vaccines against SD and PIS. These strategies take into account the gene products actually expressed and present in the cells, and thus susceptible of being targets of immune recognition. In this context, we have analyzed the immunogenic pattern of two B. pilosicoli porcine isolates (the Spanish farm isolate OLA9 and the commercial P43/6/78 strain) and one B. hyodysenteriae isolate (the Spanish farm V1). The proteins from the Brachyspira lysates were fractionated by preparative isoelectric focusing, and the fractions were analyzed by Western blot with hyperimmune sera from challenged pigs. Of the 28 challenge-specific immunoreactive bands detected, 21 were identified as single proteins by MS, while the other 7 were shown to contain several major proteins. None of these proteins were detected in the control immunoreactive bands. The proteins identified included 11 from B. hyodysenteriae and 28 from the two B. pilosicoli strains. Eight proteins were common to the B. pilosicoli strains (i.e., elongation factor G, aspartyl-tRNA synthase, biotin lipoyl, TmpB outer membrane protein, flagellar protein FlaA, enolase, PEPCK, and VspD), and enolase and PEPCK were common to both species. Many of the identified proteins were flagellar proteins or predicted to be located on the cell surface and some of them had been previously described as antigenic or as bacterial virulence factors. Here we report on the identification and semiquantitative data of these immunoreactive proteins which constitute a unique antigen collection from these bacteria.
Keywords: Brachyspira, vaccine, immunoblot, mass spectrometry, antigen, flagellar protein, swine dysentery, spirochaetosis
Introduction
Bacteria in the Brachyspiraceae family are gram-negative and spiral-shaped Spirochaetes. Members of this phylum are characterized by their unique motility and a loosely coiled morphology caused by the existence of periplasmic flagella (Charon and Goldstein, 2002). Species of the Brachyspiraceae family are anaerobic, host-associated intestinal bacteria in pigs, humans and other species, and can cause gastrointestinal pathologies and mortality (Stanton, 2006). Currently, the Brachyspiraceae comprises 16 species. B. hyodysenteriae and B. pilosicoli are well-known Brachyspira intestinal pathogens in pigs, responsible for swine dysentery (SD, a severe mucohaemorrhagic colitis) (Alvarez-Ordóñez et al., 2013) and porcine intestinal spirochaetosis (PIS, porcine spirochaetal diarrhea, a mild, non-haemorrhagic colitis), respectively (Trott et al., 1996; Stanton, 2006).
SD is produced by B. hyodysenteriae, a restricted pig-associated species, and is a disease present worldwide that has an important economic impact on the farming business. SD mainly affects pigs in the growing-finishing periods. This contributes to the high cost of the disease, which is associated not only with mortality, which is relatively low when animals are treated (Hamdy, 1974), but also with high morbidity, growth retardation and the need for continual in-feed medication.
Unlike most Brachyspira species, B. pilosicoli has a wide host range. It is zoonotic in pigs, poultry, dogs and humans (Hampson et al., 2006), in which it can lead to intestinal spirochaetosis (IS). In humans, the prevalence of IS is very uneven, much higher in developing regions than in industrialized regions (Tsinganou and Gebbers, 2010).
The treatment for the control of a Brachyspira infection involves the use of multiple antimicrobial agents (Alvarez-Ordóñez et al., 2013). Nevertheless, several studies have highlighted the increasing occurrence of B. hyodysenteriae and B. pilosicoli isolates resistant to these antibiotics in many countries (Molnar, 1996; Karlsson et al., 2001; Rohde et al., 2004; Hidalgo et al., 2009; Ohya and Sueyoshi, 2010; Pringle et al., 2012), greatly compromising the efficacy of this global treatment.
Since Joens et al. described that pigs that recovered from SD acquired immunological protection, and therefore rarely relapse when re-exposed to the infective agent (Joens et al., 1979), several endeavors have been launched to design a vaccine. Unfortunately, these attempts have not been successful to date, and no effective vaccine against B. hyodysenteriae or B. pilosicoli is available. Early approaches in this area included immunization with an inactivated bacterin (Hampson, 2000) or attenuated strains (Hyatt et al., 1994). In addition to conferring only partial protection in the best cases, these strategies entailed a cumbersome anaerobic-culture of Brachyspira which was not appropriate for large-scale production.
In recent years, the development of subunit vaccines based on Brachyspira recombinant proteins has also been explored. Molecules studied included flagellar proteins such as FlaA and FlaB (Boyden et al., 1989; Kent et al., 1989; Gabe et al., 1995; Ge and Charon, 1997) and some structural and metabolic proteins, such as outer-membrane proteins BmpB (La et al., 2004), SmpB (Holden et al., 2008), or variable surface proteins (Vsp) (Witchell et al., 2006, 2011). Similar to an attempt to formulate a vaccine based on the ferritin protein FtnA (Davis et al., 2005), none of these antigens provided enough protection for SD. On the other hand, several surface proteins of B. pilosicoli (ClpX and two putative oligopeptide-binding proteins) have been evaluated as candidates for vaccination against IS (Movahedi and Hampson, 2007, 2010).
The recent publication of the genome sequences of B. hyodysenteriae and B. pilosicoli provided a useful tool for the exploration of new candidates for inclusion in vaccination processes. To date, the genome sequence of 20 strains of B. hyodysenteriae (including the reference strain WA1, ATCC 49526) (Black et al., 2015) and three strains of B. pilosicoli (porcine isolates P43/6/78 and 95/1,000, and an avian isolate B2904) (Wanchanthuek et al., 2010; Mappley et al., 2012; Lin et al., 2013) have been published. The availability of these data enables extensive in silico analysis to identify vaccine candidates, which can then be expressed and tested together in a subunit vaccine. The potential of this reverse vaccinology approach was demonstrated by Song et al. who explored the development of a vaccine against SD using a partial genome sequence of the B. hyodysenteriae WA1 strain (Song et al., 2009). More recently, a list of 33 ORF candidates to vaccine targets has been patented (Bellgard et al., 2015). These genes were selected on the basis of their homology with known amino acid sequences of surface proteins, secreted proteins and virulence factors from other species.
Despite these important advances on the genomic level, its translation to proteomic knowledge of B. hyodysenteriae and B. pilosicoli is still a challenge. Undoubtedly, future studies focused on the description of Brachyspira proteomes will be necessary to design an effective vaccination strategy. In this regard, we recently characterized a subset of proteins exposed on the cell surface (surfaceome) of B. hyodysenteriae and B. pilosicoli (Casas et al., 2016). This will not only contribute to select good candidates for a vaccine, but will also impart biological knowledge about invasive and pathogenic mechanisms of Brachyspira.
In this study, we extended our proteomic approach to identify potential immunogenic proteins from B. hyodysenteriae and B. pilosicoli. For this purpose we studied the immunoproteome of two B. pilosicoli strains (the isolate OLA9 and the commercial ATCC strain P43/6/78) and one B. hyodysenteriae isolate (isolate V1). Cell lysates were fractionated using preparative off-gel isoelectrofocusing and the fractions were separated by SDS-PAGE. The gels were immunoblotted using pig immune-sera, and the reactive bands were identified by mass spectrometry. Brachyspira isolates came from Spanish farms. It was reported that during 2001-2003, more than 30% of commercial pig farms in Spain had at least one positive for B. pilosicoli or B. hyodysenteriae (Carvajal et al., 2006). There is thus a major concern in relation to intestinal diseases caused by Brachyspira species in the country (Osorio et al., 2012, 2013), which is the world's fourth largest producer of pig meat and where the porcine industry has a huge socioeconomic impact. We propose the reported proteins as suitable candidates to be included in vaccines for the treatment of SD and porcine IS.
Materials and methods
Brachyspira cultures
Two isolates of Brachyspira pilosicoli and B. hyodysenteriae (strains OLA9 and V1, respectively) and a commercial B. pilosicoli (ATCC strain P43/6/78, ATCC 51139) were used for the immunoproteomics study. Three additional B. hyodysenteriae strains (the commercial ATCC strains WA1 and B-78, and the isolate INF1) were included in the study of Vsp profiles.
The isolates came from Iberian pigs that showed disease symptoms on two different farms in the Badajoz province (Spain). The medium for the isolation of Brachyspira was based on the blood agar modified medium described by Calderaro et al. (2001, 2005), supplemented with antibiotics to remove most of the fecal micropopulation (Feberwee et al., 2008). The medium was composed of blood agar base n° 2 (40 g/l) supplemented with 5% defibrinated horse blood (50 ml) (Oxoid, Thermo Scientific, Waltham, MA, USA), beef extract (3 g/l), Bacto-peptone (5 g/l), (Difco, BD, Franklin Lakes, NJ, USA) and spectinomycin (0.2 g/l), spiramycin (0.025 g/l), rifampicin (0.012 g/l) vancomycin (6.2 g/l), and colistin (6.25 mg/l) (all from Sigma-Aldrich, St. Louis, MO, USA) and 810 ml distilled water. The plates were incubated for 4–7 days at 42°C in an anaerobic jar with CO2 produced by an AnaeroGen TM 3.5 L (Oxoid, Thermo Fisher Scientific, MA, USA). The colonies were examined by phase contrast microscopy (40x). The isolates were characterized by PCR using species-specific primers for nox (B. hyodysenteriae) and 16S rRNA (B. pilosicoli) as previously described (Casas et al., 2016) and stored at −80°C.
Blood agar solid subcultures of the isolates were seeded in Brain-heart infusion (BHI) broth (Laboratorios CONDA Pronadisa, Torrejón de Ardoz, Spain) enriched with horse serum (15%) and incubated with shaking in anaerobiosis jars at 4°C for 4–7 days. The grown cultures were centrifuged at 12,900 × g for 10 min, and the pellet washed twice with TE buffer (10 mM Tris pH 8.0, 1 mM EDTA, both from Sigma-Aldrich, St. Louis, MO, USA). All bacterial growth and handling procedures were carried out under biosafety level 2 conditions.
Brachyspira cell lysate and protein quantification
Bacterial pellets (~150 mg) were resuspended in 500 μl denaturing lysis buffer containing 4% SDS, 0.1 M DTT and 100 mM Tris-HCl pH 7.5. After incubation in a Thermomixer (Eppendorf model Comfort, 600 rpm, 1 h, 95°C), the samples were homogenized in a Bullet Blender (Next Advance Storm, NY, USA) for 3 min at level 8, using 250 μl zirconium silicate beads (0.1 mm diameter, BioSpec, 11079101z). The beads were then pelleted by centrifugation at 14,000 × g for 3 min and the Brachyspira cell lysate was recovered from supernatant.
Prior to protein quantification, the excess SDS was removed from the sample using an SDS-Out ™ Precipitation Kit (Thermo Fisher Scientific). The sample was diluted with 100 mM Tris-HCl pH 7.5 (1/2 v/v) for this procedure and the SDS was precipitated with the precipitation reagent (1/20 v/v). After incubation in an ice bath for 20 min, the anionic detergent was pelleted by centrifugation (10,000 × g, 10 min) and the Brachyspira SDS-free lysate was recovered from supernatant. After clarification by an additional centrifugation (10,000 × g, 1 min) in a spin cup column, the cell lysate was aliquoted and kept at −40°C until use. A small aliquot was used to measure the protein concentration using the Bradford method (Bio-Rad Laboratories, CA, USA).
Off-gel protein fractionation
Proteins in the Brachyspira lysates were fractionated by off-gel isoelectric focusing (Ros et al., 2002; Michel et al., 2003) using an OFFGEL Fractionator 3100 (Agilent Technologies CA, USA). For this procedure, 1 mg of protein (ca. 120–140 μl of cell lysate) was diluted to a final volume of 3.6 ml with a 1.25X protein off-gel stock solution (PROSS 1.25X: 7 M urea, 2 M thiourea, 1.5 M glycerol and 0.1 M DTT) supplemented with ampholytes (GE Healthcare Life Sciences, Barcelona, Spain). Large 24-cm dry strips with a wide immobilized pH gradient of 3–10 units (GE Healthcare Life Sciences) were used for the separation. After placing the 24 well frame, the strips were rehydrated with 50 μl/well of IPG strip rehydration solution (PROSS-ampholytes 1X) for 15 min, and the sample was loaded by pipetting 150 μl/well. To have enough material for an immuno-analysis, the experiments were carried out in triplicate so a total of 3 mg of each Brachyspira isolate was processed. The samples were focused for 36 h at 64 kVh with a maximum current of 50 μA and power of 200 mW (4,500 V final voltage). Twenty-four fractions were collected per sample. Three replicates were fractionated per sample. Triplicates of each fraction were pooled in a single tube, aliquoted and stored at −40°C.
Brachyspira-challenge and control pig sera
Pig sera were provided by the company Laboratorios Larrasa S.L. (Badajoz, Spain) in the frame of a project funded by the Spanish Ministry of Economy and Competitivity (MINECO, IPT-2011-0735-010000). Laboratorios Larrasa S.L. was in compliance with Spanish legislation (R.D. 1201/2005 and Law 32/2007) and EU Council Guidelines (2003/65/CE) for the use of experimental animals. These sera were obtained from pigs from a healthy herd in Extremadura (South-Western Spain) which were inoculated at the age of 14 weeks with active viable cells of B. hyodysenteriae strain WA1 (ATCC 49526) or the B. pilosicoli porcine strain P43/6/78 (ATCC 51139). The serum was extracted from challenged animals at 12 weeks post-infection. Five different sera from each Brachyspira species challenge (B. hyodysenteriae-challenge serum 1–5 and B. pilosicoli-challenge serum 6–10) were used in this study.
SDS-PAGE separation of off-gel IEF fractions
Five (for the WB analyses) or ten (for silver staining) microliters of the off-gel fractions were separated by SDS-PAGE. A sample of 20 μl of total lysate (input) was also prepared as a control. The samples were prepared in sample loading buffer (2% w/v SDS, 10% glycerol, 0.002% w/v bromophenol blue and 25 mM Tris-HCl pH 6.8), heated at 56°C for 30 min and resolved on 12 or 7.5%-polyacrylamide gels.
Six replicates were done for each separation. The protein bands in one replicate were visualized by mass spectrometry-compatible silver staining (Shevchenko et al., 1996; Casanovas et al., 2009). The other five replicates were used for immunoblotting with the appropriate antisera.
Immunoblotting
After SDS-PAGE, the proteins in the gel were transferred to a nitrocellulose membrane using an iBlot ™ system (Life Technologies, CA, USA). Following Ponceau staining of the proteins (Sigma-Aldrich, St. Louis, MO, USA), the membrane was blocked with TBS-T (20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.2% Tween-20) containing 5% (w/v) skimmed milk (1 h, room temperature). The membrane was incubated with the appropriate pig serum diluted in blocking solution (1/3,000 for control serum and 1/5,000 for Brachyspira challenge serum) with gentle agitation (1 h, room temperature). The membranes were incubated (1 h) with a Rabbit Anti-pig IgG H&L horseradish peroxidase-conjugated secondary antibody (Abcam ab6777, 1/5,000 in blocking solution) and visualized with luminol.
Characterization of immunoreactive bands
Optical density (OD) profiles of the immunoblots and SDS-PAGE gels for all fractions were acquired using ImageJ Version 1.47 (NIH). The profiles were measured between 15 and 100 kDa, and the OD values were normalized relative to the total lane intensity. The Rf value was determined with the Quantity One 1D Analysis Software (Bio-Rad) and position of each band expressed as its molecular mass calculated from the Rf value using molecular mass markers. Calculations using bands for the same protein in a gel showed an average coefficient of variation of 0.85% (n = 14) for the calculated mass. As a consequence, bands in different lanes of the same gel with a mass difference less than 1.7% (2 × standard deviation) were considered the same band and numbered accordingly.
Bands observed in at least two OD profiles of challenge sera but absent in control sera were designated as challenge-specific, differential immunoreactive bands. Some bands were also considered differential when also potentially present in only one of the control sera but observed at a very low intensity. Other bands observed both in the control and challenge sera were considered as challenge-non-specific bands (Supplementary Tables S1, S2).
Mass spectrometry analysis and protein identification
Silver stained bands corresponding to the reactive bands detected in the immunoblots were excised and digested using an automatic device (DigestPro MS, Intavis, Cologne, Germany). The process involved reduction with dithiothreitol, derivatization with iodoacetamide, and enzymatic digestion with trypsin (37°C, 8 h) (Casanovas et al., 2009). The tryptic digests were evaporated and redissolved in 5 μl of methanol/water/trifluoroacetic acid (30/70/0.1 v/v).
Proteins in the tryptic digests (0.5 μl) were identified by MALDI-TOF peptide mass fingerprinting combined with MS/MS ion search in a 4800 TOF/TOF mass spectrometer (ABSciex, Barcelona, Spain) in the reflectron mode. The spectra were externally mass calibrated using a standard peptide mixture. Alpha-cyano-4-hydroxycinnamic acid (3 mg/ml) was used as the matrix. The five signals with the greatest intensity in each MALDI-TOF spectrum were automatically analyzed by TOF/TOF. The combined TOF and TOF/TOF spectra were interpreted by database search (Mascot, Matrix Science, MA, USA) using the following parameters: peptide mass tolerance, 50 ppm; fragment mass tolerance, 0.5 Da; fixed modification, carbamidomethyl cysteine; variable modification, oxidation of methionine; significance threshold of the MOWSE score, p < 0.05. All identifications were manually validated.
Samples which did not produce a positive identification by MALDI were reanalysed by LC-MS/MS in a Velos-LTQ or an Orbitrap-XL mass spectrometer (Thermo Fisher Scientific) equipped with a microESI ion source. Four microliters of each sample digest were diluted to 20 μl with 5% methanol and 1% formic acid, and loaded into a chromatographic system consisting of a C18 preconcentration cartridge (Agilent Technologies) connected to a 15-cm long, 150 μm i.d. (Velos-LTQ) or 100 μm i.d. (Orbitrap-XL) C18 column (Nikkyo Technos Co.). The separation was performed at 1 μl/min (Velos-LTQ) or 0.4 μl/min (Orbitrap XL) in a 30-min gradient from 3 to 40% acetonitrile (solvent A: 0.1% formic acid, solvent B: acetonitrile 0.1% formic acid). The instruments were operated in the positive ion mode with a spray voltage of 1.5 kV. The spectrometric analysis was performed in a data dependent mode. The scan range for full scans was m/z 400–1,800. The LC-MS/MS spectra were searched using SEQUEST (Proteome Discoverer v1.4, Thermo–Fisher Scientific) with the following parameters: peptide mass tolerance, 1 Da (Velos-LTQ) or 20 ppm (Orbitrap-XL); fragment tolerance, 0.6 Da; enzyme, trypsin; two missed cleavages allowed; dynamic modification, methionine oxidation (+16 Da); fixed modification, cysteine carbamidomethylation (+57 Da). The peptide identifications were filtered at 0.1% FDR and only proteins identified with two or more peptides and peptide rank 1 were considered. Relative abundance of the identified proteins in each sample was roughly estimated from the product of the total peptide sequence matches pointing to that protein and its sequence coverage. The group of more abundant proteins bearing more than 80% of the total abundance in the sample were considered for discussion (Full data is available in Supplementary Tables S3–S9).
Searches for the MALDI and LC-MS/MS methods described above were carried out against the Uniprot database (2015_11 version) restricted to Brachyspira. When results pointed to indistinguishable different accessions to the same protein in different strains, the accession for the reference ATCC strains was reported in Table 1, Supplementary Tables S1, S2.
Table 1.
Proteins identified in the immunoreactive bands from the two Brachyspira species.
| Band ida | Sample | Specificb | Observed Mass (kDa) | Protein identified | |||
|---|---|---|---|---|---|---|---|
| 12% | 7.5% | Accessionc | Name | Theor. Mass (kDa) | |||
| 2 | OLA9 | X | 86 | 76 | D8ICZ7 | Elongation factor G | 75.8 |
| 3 | OLA9 | 83 | D8IDC2 | Uncharacterized protein | 62 | ||
| 4 | ATCCBP | X | 81 | 78 | D8ICZ7 | Elongation factor G | 75.8 |
| 5* | OLA9 | X | 78 | 72 | J9UT37 | Putative polymerase | 72.3 |
| D8IEM8/A0A0G4K4U5 | Chaperone protein HtpG | 73.9 | |||||
| D8IBS0 | Chaperone protein DnaK | 67.6 | |||||
| 6 | ATCCBP | 77 | D8IDC2 | Uncharacterized protein | 62 | ||
| 7 | OLA9 | 75 | D8IDC2 | Uncharacterized protein | 62.1 | ||
| 9 | V1 | 73 | G0EJY7 | Uncharacterized protein | 65.4 | ||
| 10* | ATCCBP | X | 72 | 70 | D8IEM8 | Chaperone protein HtpG | 73.9 |
| J9UT37 | Putative polymerase | 72.3 | |||||
| D8ICG5 | Uncharacterized protein | 85 | |||||
| 12 | V1 | 72 | C0R0R7 | Putative treponemal membrane protein | 63.9 | ||
| 13 | OLA9 | X | 72 | 68 | D8IE58 | Aspartyl-tRNA synthase | 67.5 |
| 15 | ATCCBP | 69 | D8IDC2 | Uncharacterized protein | 62.1 | ||
| 16 | ATCCBP | X | 69 | D8IFS5 | Phosphoenol pyruvate carboxykinase | 67.5 | |
| 17 | ATCCBP | X | 69 | 70 | D8IE58 | Aspartyl-tRNA synthase | 67.5 |
| 18 | V1 | X | 68 | C0R0E5 | Phosphoenol pyruvate carboxykinase | 67.5 | |
| 19 | OLA9 | X | 65 | D8I9T6 | Biotin lipoyl | 65 | |
| 20 | OLA9 | 64 | 61 | D8IB78 | 60 kDa chaperonin | 58.1 | |
| 22 | ATCCBP | X | 63 | D8I9T6 | Biotin lipoyl | 65.0 | |
| 24 | ATCCBP | 62 | 61 | D8IB78 | 60 kDa chaperonin | 58.1 | |
| 25 | V1 | 61 | C0QWH4 | 60 kDa chaperonin | 58.2 | ||
| 26* | OLA9 | X | 58 | 53 | J9USS2 | 60 kDa chaperonin | 58.1 |
| D8IAM3 | Amidohydrolase 3 | 60.6 | |||||
| J9UU81 | 2-isopropylmalate synthase | 54.4 | |||||
| D8IF78 | Outer membrane efflux protein | 54.7 | |||||
| D8IDP7 | Trigger factor, C-terminal domain protein | 50.6 | |||||
| 27 | OLA9 | X | 57 | D8IBK7 | TmpB outer membrane protein | 42.5 | |
| 28 | ATCCBP | X | 56 | 55 | K0JIZ9 | Carboxyl terminal protease | 54.9 |
| 29* | ATCCBP | X | 54 | D8IBK7 | TmpB outer membrane protein | 42.4 | |
| 31 | ATCCBP | X | 51 | 50 | D8IBH9 | ATP synthase subunit beta | 45.6 |
| 32 | ATCCBP | X* | 50 | D8IET2 | Enolase | 47 | |
| 36 | V1 | X | 45 | C0QW84 | Uncharacterized protein | 38.6 | |
| 37 | OLA9 | 44 | Q9FA06 | Putative elongation factor Tu | 16.3 | ||
| D8ICZ6 | Elongation factor Tu | 44.8 | |||||
| 38 | ATCCBP | 44 | Q9FA06 | Putative elongation factor Tu | 16.3 | ||
| D8ICZ6 | Elongation factor Tu | 44.8 | |||||
| 39 | ATCCBP | X | 44 | D8ICG3 | Uncharacterized protein | 38.9 | |
| 40 | OLA9 | X | 43 | 42 | D8IBY6 | FlaA | 35.7 |
| 41 | V1 | 43 | C0QVZ4 | Elongation factor Tu | 44.7 | ||
| Q9FA06 | Putative elongation factor Tu | 16.3 | |||||
| 42 | V1 | 42 | C0QXS8 | NADH oxidase | 50.5 | ||
| C0QVZ4 | Elongation factor Tu | 44.7 | |||||
| 43 | V1 | 42 | C0R0T5 | FlaA | 24.5 | ||
| P32520 | FlaA1 | 36 | |||||
| 44 | ATCCBP | X | 41 | D8IBY6 | FlaA | 35.7 | |
| 45 | OLA9 | X | 40 | 38 | D8ICU0 | VspD | 42.7 |
| 46* | ATCCBP | X | 38 | 38 | D8ICU0 | VspD | 42.7 |
| J9TU32 | Ribonucleotide-diphosphate reductase subunit beta | 41.1 | |||||
| D8IEW7 | Mannose-1-phosphate guanylyltransferase | 40.5 | |||||
| J9URY6 | 2-oxoacid:ferredoxin oxidoreductase subunit alpha-like protein | 82.8 | |||||
| J9UBH8 | ATP-dependent 6-phosphofructokinase | 34.8 | |||||
| D8ICA2 | Toxic anion resistance family protein | 40.9 | |||||
| D8I9T4 | Uncharacterized protein | 39.9 | |||||
| D8ICR1 | Pyruvate oxireductase | 35.5 | |||||
| 47 | OLA9 | 36 | D8IAP2 | Flagellin FlaB2 | 32 | ||
| 48 | V1 | X | 35 | C0R1L9 | UDP-glucose 4 epimerase | 36 | |
| C0QYC2 | Galactose-glucose binding protein | 38.2 | |||||
| 49 | ATCCBP | 35 | D8IAP2 | Flagellin FlaB2 | 32 | ||
| 50 | V1 | 35 | Q26501 | FlaB1 | 32 | ||
| 51* | V1 | X | 35 | A0A0H0W3D6 | Enolase | 47 | |
| Q26501 | Flagellar protein FlaB1 | 32 | |||||
| A0A0HOUMF4/G0EJK5 | Fructose-bisphosphate aldolase | 35.4/35.4 | |||||
| C0QYC2 | Methyl-galactoside ABC transporter substrate-binding protein | 38.2 | |||||
| C0QZV6 | Pseudouridine-5'-phosphate glycosidase | 33.9 | |||||
| 52 | OLA9 | X* | 33 | D8ICA8 | Putative FlaA | 26.7 | |
| 53 | OLA9 | 32 | K0JHQ4 | Flagellin FlaB2 | 31.3 | ||
| 54 | V1 | 31 | P80160 | Flagellin FlaB2 | 31.1 | ||
| 55 | ATCCBP | 31 | K0JHQ4 | Flagellin FlaB2 | 31.3 | ||
| 56 | OLA9 | 29 | K0JLS4 | Flagellin FlaB3 | 30 | ||
| 57 | V1 | X* | 29 | C0QV52 | Enolase | 47 | |
| Q9F0F6 | Flagellin FlaB3 | 30.4 | |||||
| 58 | ATCCBP | 29 | D8IDG1 | Flagellin FlaB3 | 26.6 | ||
| 59 | V1 | 29 | Q9F0F6 | Flagellin FlaB3 | 30.4 | ||
| 61 | V1 | X | 26 | C0QWY9 | Putative FlaAL | 24.7 | |
| 62 | ATCCBP | X | 24 | D8ICA7 | FlaA-2 | 24 | |
Complete identification data is available in Supplementary Table S1 (MALDI TOF/TOF) and Supplementary Tables S3–S9 (LC-MS/MS). Specific, indicates immunoreactivity is only detected in challenged animals. Observed mass, correspond to the experimental mass calculated from the gel bands. ATCCBP, ATCC 51139 (P43/6/78) B. pilosicoli strain.
All identifications by MALDI TOF/TOF except those indicated with an asterisk which were identified by LC-MS/MS.
Specific bands with X* have been detected in a control serum with an intensity of one.
Mass spectrometry target analysis of Vsp proteins
LC-MS/MS analyses of Vsps were carried out in a Velos-LTQ using the configuration described above for protein identification. The equivalent to 1 μg of protein in 20 μl of 5% methanol, 1% formic acid was loaded into a system and a 120-min acetonitrile gradient from 0 to 40% was used. The spectrometric analysis was performed in the target mode, acquiring the MS/MS scans of the signals included in the corresponding mass list (Supplementary Table S10). The mass list for each species included at least two unique peptides for each Vsp protein. Potential peptide targets detected in previous shotgun analyses were preferentially selected to build the mass lists. Other targets were selected among the best proteotypic peptides predicted by the PeptideRank software (Qeli et al., 2014).
Results
Protein fractionation of B. pilosicoli and B. hyodysenteriae strains and selection of immunogenic fractions
Proteins from the cell lysates were fractionated by preparative isoelectric focusing (IEF) prior to SDS-PAGE. Via this technique, we could fractionate up to 1 mg of each Brachyspira lysate using 24-cm pH 3-10 Immobiline Drystrips. The Off-gel system provided adequate resolution and reproducibility for protein fractionation as verified in a parallel study in which replicate SDS-PAGE analyses were carried out on an arbitrary selection of 7 out of the 24 fractions collected (Supplementary Figure S1). This reproducibility allowed the pooling of fractions from the three independent IEF fractionations, thereby providing of the equivalent of a total of 3 mg of fractionated proteins for each Brachyspira strain.
To select the fractions that contained the immunogenic proteins, the 24 recovered fractions were immunoblotted with sera from pigs challenged with B. pilosicoli (serum 6–10, for OLA9 and ATCC 51139 fractionations) or B. hyodysenteriae (serum 1–5 for V1 fractionation). Most of the immunogenic bands from the three Brachyspira strains appeared in the early-middle fractions (numbers 3–13) that had isoelectric points between 3.5 and 6.5 (Figures 1, 2 and Supplementary Figure S2). Note that for all strains, an immunoblot analysis with control serum from non-challenged pigs also revealed some reactive bands (Supplementary Figure S3). This cross-reactivity was consistent with previous reports that showed that healthy pig serum detects B. hyodysenteriae surface antigens (Wannemuehler et al., 1988) or some of the recombinant proteins tested for a vaccine against SD (Song et al., 2009).
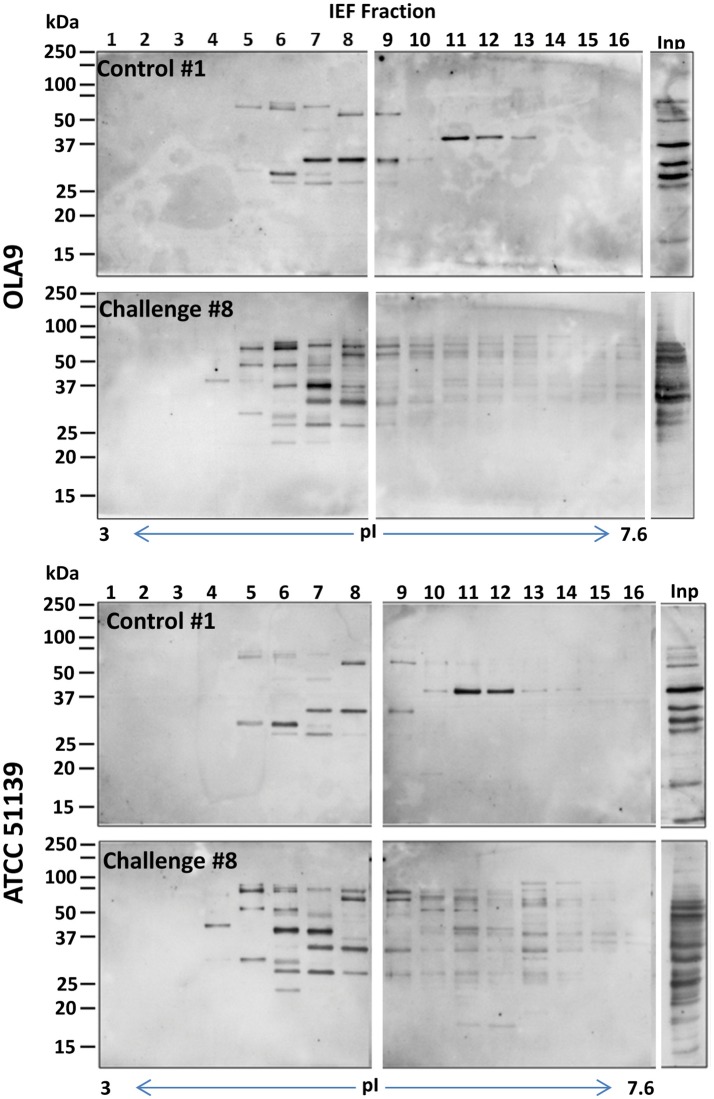
Figure 1.
General view of the immunoreactive proteins in each of the IEF fractions of the protein extracts of OLA9 (top) and ATCC 51139 (bottom) B. pilosicoli strains. The images are examples of the Western blots prepared with sera #1 and #8 (from a control and a challenged animal, respectively) (upper and lower gels for each strain). Twenty four consecutive IEF protein fractions, covering a pI range from 3 to 10, were analyzed in the corresponding lanes of three SDS-PAGE gels. Fractions presenting intense immunoreactive bands in these preliminary experiments were submitted to a detailed immunoproteomics analysis using all the individual sera available. The Figure shows only the images for the two first gels (lanes for IEF fractions 1–16), containing the more acidic fractions, and the lane corresponding to the crude proteome extract (lane Inp). IEF fractions 17–24 did not show relevant immunoreactive bands (complete images for all the fractions and individual sera tested can be found in Supplementary Figures S2A–C).
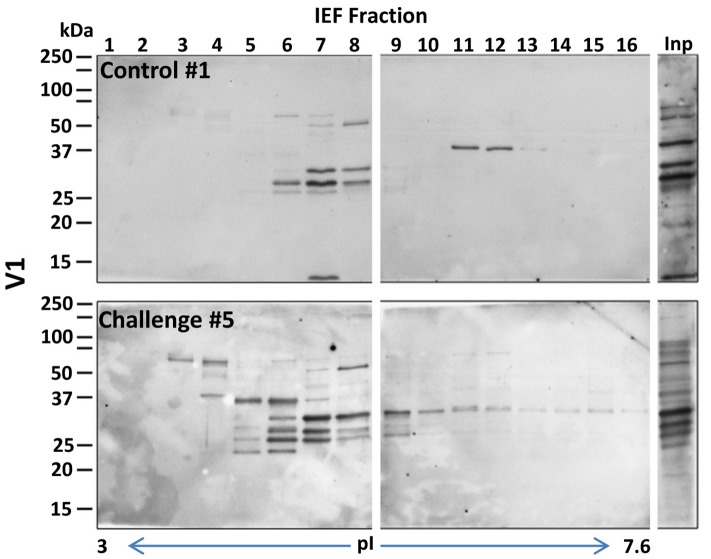
Figure 2.
General view of the immunoreactive proteins in each of the IEF fractions of the protein extracts of the B. hyodysenteriae strain V1. The images correspond to examples of the Western blots prepared with sera #1 and #5 (a control and a challenged animal, respectively) (upper and lower gels for each strain). Fractions presenting intense immunoreactive bands in these preliminary experiments were submitted to a detailed immunoproteomics analysis, see Figure 1 for details. Inp, lane corresponding to the crude extract before IEF separation.
Protein identification
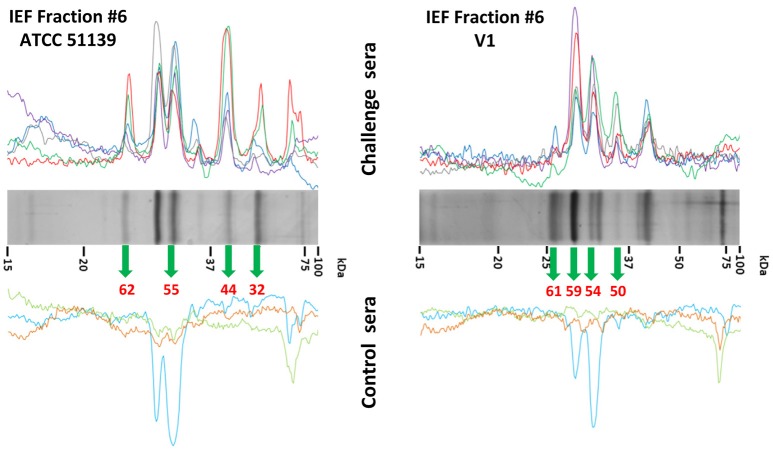
IEF fractions from the different Brachyspira strains showing the highest immunogenic response (fractions 4–9, 11, 13 and 3–9, 11–12, for B pilosicoli and hyodysenteriae respectively) (Figures 1, 2) were selected for a more detailed image analysis and band characterization. For this purpose, these fractions were reanalysed by SDS-PAGE and immunoblotted. Immunoreactive bands were identified by densitometry and the immunoblot images were matched with those obtained by silver staining on replicate SDS-PAGE separations (Figures 3, 4). The bands were then excised, trypsin digested and analyzed by mass spectrometry. Overall the components in 51 gel bands (36 from B. pilosicoli and 15 from B. hyodysenteriae strains) were identified (Table 1, Supplementary Tables S1–S9).
Figure 3.
Identification of immunoreactive proteins in the IEF fractions of the Brachyspira proteomes. The image shows the Western blot densitometry profiles (top and bottom) and the protein band profile of the corresponding silver-stained gel lane (center). Immunoreactivity traces for the 5 sera from challenged pigs (top) and the 3 sera for control pigs (bottom) are shown with different colors (see SI for color codes). Bands identified as immunoreactive were sliced from the SDS gel lane and submitted to MS analyses for identification. Code numbers for the bands analyzed from these specific lanes are indicated in red. The example given corresponds to the IEF fractions #6 from ATCC 51139 (B. pilosicoli) (left) and V1 (B. hyodysenteriae) (right) strains. The full set of images for all the fractions is given in Supplementary Figures S5A–H.
Figure 4.
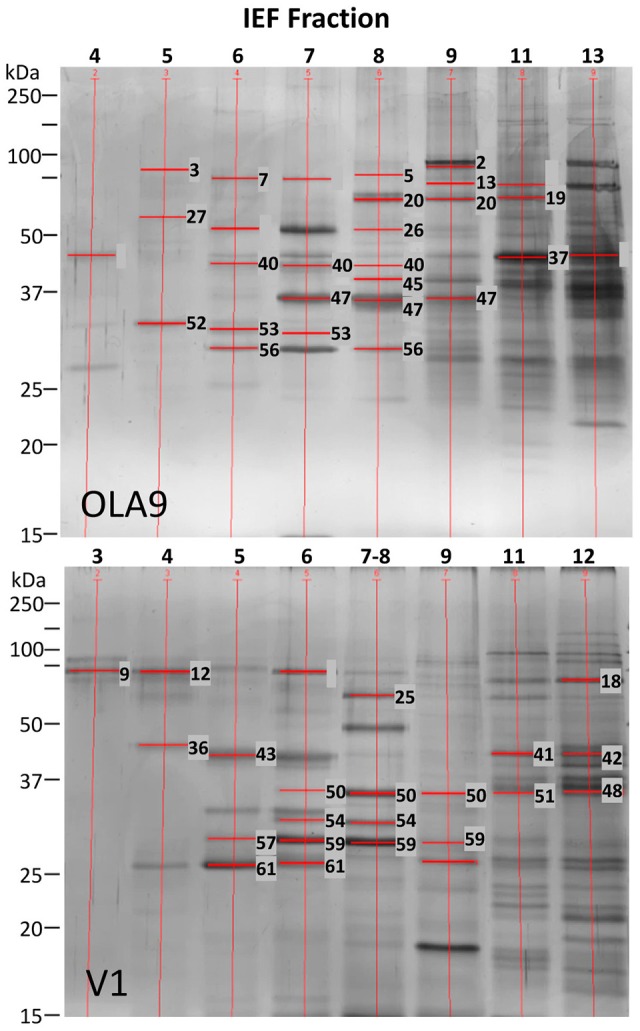
Summary of the analyzed bands from SDS-PAGE silver stained gels for the selected IEF fractions of OLA9 strain (B. pilosicoli) and V1 strain (B. hyodysenteriae).
Most of the bands (45) could be identified by MALDI, except for six of them that required an LC-MS/MS analysis. The failure of MALDI in the analysis of these bands was probably due to the presence of several major proteins in the band as confirmed from the LC-MS/MS identification data. Thus, when LC-MS/MS identifications were filtered to select the most abundant components, only one of these bands produced a single protein while the others showed the presence of 3–8 major components.
B. pilosicoli immunoreactive proteins
Sixteen and 20 immunoreactive bands were detected for the B. pilosicoli OLA9 and ATCC 51139 strains, respectively (Supplementary Figures S4, S5A–F and Table 1, Supplementary Tables S1, S2). Several of these bands were common to both strains. Some of the fractions (#8 and #9) showed a complex profile at the high mass range of the silver-stained gels. To increase the resolution of the more complex fractions, parallel SDS-PAGE and Western blot analyses were carried out using 7.5% acrylamide gels (Figure 5, Supplementary Figures S5B,E). The mass spectrometric analysis of the corresponding silver-stained SDS-PAGE bands (Figures 3, 4) produced the identification of 28 different proteins in challenge-specific immunoreactive bands (Table 1). Eighteen of the immunoreactive specific bands yield a single protein (7 for OLA9 and 11 for ATCC 51139), while 4 other bands were shown to contain more than one protein.
Figure 5.
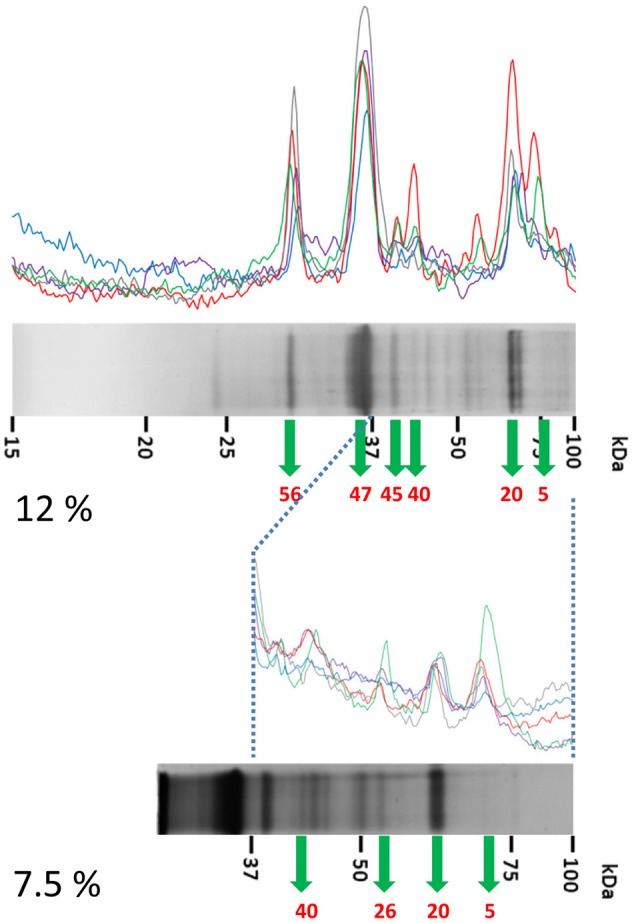
Detailed analysis of the high-mass proteins in IEF fractions #8 and #9 from ATCC 51139. IEF fractions #8 and #9 from B. pilosicoli showed unresolved, complex profiles of bands in 12% acrylamide gels (up). These profiles were resolved by a parallel analysis of these fractions in 7.5% acrylamide gels (down). Data from the 12% and 7.5% separations were later combined. The example shown corresponds to IEF fraction #8 (see Figure 3 for details).
The two B. pilosicoli strains shared eight proteins in common in the bands with specific reactivity toward the challenge sera (the outer membrane protein of the TmpB family, a flagellar filament outer layer protein FlaA, the variable surface protein VspD, the chaperone protein htpG, a putative polymerase, the aspartyl-tRNA synthase, the biotin lipoyl and the elongation factor G) (Table 1).
Seven other proteins common to both strains were found in challenge-non-specific bands including a 60 kDa chaperonin, flagellins B2 and B3 and the elongation factor Tu.
B. hyodysenteriae immunoreactive proteins
Fifteen immunoreactive bands were detected in the selected IEF fractions from the B. hyodysenteriae farm isolate V1 using sera from B. hyodysenteriae-challenged animals (Figures 3, 4, Supplementary Figures S4, S5G,H). Six of these bands showed challenge specific immunoreactivity (#18, #36, #48, #51, #57, #61) while nine others also cross-reacted with control sera from healthy pigs (#9, #12, #25, #41, #42, #43, #50, #54, and #59). Three of the challenge-specific bands, produced a single protein identification, while the other bands were shown to contain several proteins up to a total of 11 (Table 1, Supplementary Tables S1–S9). Among the challenge-specific proteins PEPCK (phosphoenolpyruvate carboxykinase) and enolase had also been identified as antigenic in the B. pilosicoli isolates.
Cross-reactivity with control sera
For all strains, the immunoblots with sera from control, non-challenged pigs also revealed several immunoreactive bands (Supplementary Figure S3).
Cross-reactivity was observed for all flagellar proteins except B. pilosicoli FlaA. The FlaB proteins were the primary targets of the control sera for B. pilosicoli and B. hyodysenteriae (Table 1, Supplementary Tables S1, S2). The three isoforms of FlaB that constitute the inner core of the spirochaetal flagella (FlaB1, FlaB2, and FlaB3) were identified in the immunoreactive bands (Table 1, Figure 4).
Other proteins immunoreactive to control sera were Elongation factor Tu, which was detected in both species and the uncharacterized protein D8IDC2, which was identified in the two B. pilosicoli strains.
Distribution of Vsp among Brachyspira species and strains
Although the variable surface protein VspD has been suggested as a potential vaccine candidate against B. hyodysenteriae we could only identify VspD in the immunoreactive bands from B. pilosicoli samples (Table 2, Supplementary Tables S11–S17). To confirm these findings, a more detailed LC-MS/MS analysis was performed targeting the family of Vsp proteins. For this, each Vsp protein was monitored by targeting two or more tryptic peptides with sequences unique to that specific protein (Supplementary Table S10). Three strains of B. hyodysenteriae (two commercial ATCC strains and one isolate) and two strains of B. pilosicoli (one commercial ATCC strain and one isolate) were analyzed.
Table 2.
Presence of Vsp proteins in Brachyspira strains.
| B. hyodysenteriae | B. pilosicoli | ||||
|---|---|---|---|---|---|
| WA1 | B-78 | INF1 | ATCCBP | OLA9 | |
| Protein | Collection | Collection | Isolate | Collection | Isolate |
| VspA | nd | nd | nd | na | na |
| VspB | *a | *a | *a | na | na |
| VspC | ** | ** | nd | na | na |
| VspD | * | nd | nd | *** | **** |
| VspE | * | ** | ** | ** | * |
| VspF | **** | nd | nd | na | na |
| VspG | nd | nd | nd | na | na |
| VspH | nd | ***a | nd | ** | nd |
| VspI | ? | ? | ** | na | na |
| VspJ | nd | nd | nd | na | na |
Asterisks indicate total number of peptide sequence matches (PSM) for the protein (
, 0–25 PSM;
, 25–100 PSM;
, 100–300 PSM;
, > 300 PSM).
PSM is correlated with protein abundance. ATCCBP, ATCC 51139 (P43/6/78) B. pilosicoli strain.
Identified from only one protein-exclusive peptide.
nd, not detected; na, not monitored (sequence not described for the species).
?, Identified from only one, non-protein-exclusive peptide (common with VspA).
The analysis showed that the profile of Vsp proteins was very variable between species and strains (Table 2). In agreement with our previous results, VspD, was highly abundant in both B. pilosicoli strains (more than 200 PSMs per strain). Contrarily, it was detected with a very low abundance (<25 PSMs) (Table 2, Supplementary Tables S13, S17) in only one of the three B. hyodysenteriae strains (WA1). Another proposed vaccine candidate in this family, VspH, was found in both species but not in all strains.
The vspC and vspF genes have not been described in B. pilosicoli. Both proteins were detected in B. hyodysenteriae although the expression was strain-dependent. VspC was detected in the commercial strains (range 25–100 PSMs), but not in the isolate. To the best of our knowledge, this result constitutes the first experimental evidence at the protein level of the expression of this protein. On the other hand, VspF was found to be greatly abundant in the B. hyodysenteriae WA1 strain but was not detected in the others. This is similar to reports by Black et al. (2015), who described the absence of the vspF gen in some strains from B. hyodysenteriae. According to Witchell et al. (2011), VspF and VspE are found in the cell in a protein complex which can include also other Vsps. Differently to the other members discussed above, VspE was detected in both species and in all strains.
We could not identify VspA, VspG, or VspJ in any of the strains analyzed. Another member, VspB, described from the genome of B. hyodysenteriae was detected in all strains but with a very low abundance.
Discussion
Challenge-specific immunoreactive proteins common to both Brachyspira species
Two proteins (PEPCK and enolase) were revealed by challenged sera from both Brachyspira species. These proteins showed a high degree of identity between species (96% identity for B. hyodysenteriae and B. pilosicoli enolases and 95% of identity for the corresponding PEPCK).
In a vaccine search, candidates are often selected from membrane-exposed proteins, because surface exposure facilitates recognition by an antibody (Boyle et al., 1997). Still, many cytosolic proteins have been described as major antigens and some of them have also been explored as vaccine components (Davis et al., 2005). In our study, many of the proteins detected were proteins annotated as cytoplasmic. This is the case for PEPCK, whose antigenic capacity had been previously reported in other species. PEPCK was identified as the antigen triggering the cellular response responsible for the hepatic granulomatous inflammation in schistosomiasis (Asahi et al., 2000). Additionally, PEPCK from Mycobacterium tuberculosis has been demonstrated to induce a strong immune response in mice and, for this reason, was proposed as a component of a subunit vaccine for tuberculosis (Liu et al., 2006).
Enolase has been reported to be immunoreactive in several pathogenic species, such as Mycobacterium tuberculosis (Rahi et al., 2017), and Borrelia burgdorferi (Barbour et al., 2008). This protein has also been described as a differential, immunogenic protein in strains of Bifidobacterium longum ssp. longum (Górska et al., 2016) and showed protective activity against colitis in mice (Srutkova et al., 2015). Enolase is a moonlighting enzyme found on the surface of some pathogens and involved in the activation of plasminogen (Rahi et al., 2017). In a previous work (Casas et al., 2016), we reported enolase to be among the ten most abundant proteins detected in the surfaceome and exoproteome compartments for B. pilosicoli and B. hyodysenteriae.
These two proteins are thus tentative candidates for vaccines against Brachyspira infections that have not been included in earlier reported vaccines.
Two other putative uncharacterized proteins (C0QW84 and D8ICG3) were identified in challenge-specific bands (bands #36 and #39, respectively) (Figure 4) from B. hyodysenteriae and B. pilosicoli, respectively. Both are predicted (PSORTb v3.0 (Yu et al., 2010), SignalP v4.1 (Petersen et al., 2011) to be extracellular or located on the outer membrane, although C0QW84 lacks the signal peptide on the N-terminal side. These proteins have the same molecular weight and isoelectric point and 62% of identity and 72% similarity between them, suggesting they are different forms of the same functional molecule in these species. In this case, a potential vaccine candidate common to both species would require identifying possible common epitopes capable of inducing an immune response.
B. pilosicoli challenge-specific immunoreactive proteins
Twenty-eight B. pilosicoli proteins were identified in the challenge-specific bands, with 8 of them common to the ATCC 51139 and environmental OLA9 strains. More than half of these proteins could be identified as proteins potentially exposed on the surface of the bacteria or secreted into the media. Thus, 9 corresponded to known or predicted (Gene Ontology Annotation (GOA), PSORTb) membrane or membrane-exposed proteins (TmpB outer membrane protein, outer membrane efflux protein, VspD, enolase, D8ICG5, D8ICG3) or flagellar proteins (FlaA, FlaA2, D8ICA8) (Supplementary Table S2). Six of these proteins (ATP-dependent 6-phosphofructokinase, 2-isopropyl malate synthase, chaperone protein DnaK, pyruvate oxidoreductase, enolase and VspD) were identified in a previous work among the most abundant proteins in the exposed proteome of these bacteria (Casas et al., 2016). Especially VspD, similar to enolase, was among the 10 most abundant proteins exclusively found on the exoproteome (i.e., proteins present in the bacterial culture media). Variable surface proteins constitute a well-known family of antigenic bacterial components. However, no evidence at the protein level had been previously reported for the expression of any Vsp protein in B. pilosicoli.
The treponemal outer membrane protein B (TmpB, D8IBK7) was identified as challenge-specific for both B. pilosicoli strains. Two putative treponemal membrane proteins (C0R0R7 and C4MGG7) with 47 and 55% homology with B. pilosicoli TmpB were included in a reverse vaccinology study against B. hyodysenteriae, but they were not immunoreactive when immunoblotted with porcine sera from challenged animals (Song et al., 2009). Contrarily, we detected C0R0R7 in immunoreactive bands of B. hyodysenteriae. However, this reactivity was observed toward both the challenge and control sera, so this protein was discarded as a specific vaccine candidate.
The flagellar protein FlaA was also found in the immunoreactive bands of both B. pilosicoli strains. FlaA constitutes the outer sheath of periplasmic flagella in spirochetes, impacting the unusual morphology and motility of this bacterial phylum (Li et al., 2000, 2008; Rosa et al., 2005; Jiang et al., 2014; Zhao et al., 2014). Flagellar proteins are major immunoreactive proteins in B. hyodysenteriae (Kent et al., 1989). It has been reported that FlaA from S. hyodysenteriae, S. innocens, and S. pilosicoli was recognized by rabbit polyclonal and murine monoclonal antibodies produced against S. hyodysenteriae lysates (Fisher et al., 1997). However, no data were previously available on the antigenicity of B. pilosicoli FlaA. In our study, both B. pilosicoli and B. hyodysenteriae FlaA were found in immunoreactive bands although only B. pilosicoli evidenced a challenge-specific response.
Two other cytoplasmic proteins, the chaperone protein HtpG and aspartyl tRNA synthase, were found to be immunogenic in both B. pilosicoli strains. HtpG has been reported to be responsible for a strong humoural response in human periodontitis caused by Porphyromonas gingivalis (Shelburne et al., 2008). On the other hand, aminoacyl tRNA synthases are the targets of many antibacterial compounds (Chopra and Reader, 2014). They play an important role in bacterial resistance as described for Mycobacterium tuberculosis strains where mutations in this protein are involved in their resistance to whole-cell inhibitors (Ioerger et al., 2013). No data were available on the antigenicity of these molecules.
B. hyodysenteriae challenge-specific immunoreactive proteins
Flagellar proteins were the most frequent class of immunoreactive proteins identified in B. hyodysenteriae (Table 1, Supplementary Tables S1, S2). However, except for a putative flagellar filament outer layer-like protein (C0QWY9), other flagellar proteins identified such as FlaA1, FlaB1, and FlaB3 were also found in bands immunoreactive toward control sera. FlaA1 had been previously described as one of the molecules that produced a highly specific immune reaction in B. hyodysenteriae (Li et al., 1993).
Other proteins detected in the challenge-specific immunoreactive bands included the galactose-glucose binding protein, a periplasmic protein, and two cytoplasmic proteins, pyridine nucleotide-disulfide oxidoreductase and fructose-bisphosphate aldolase. These proteins have been detected in the extracellular space in other species involved in processes related to the interaction/ adhesion to the host cell (Tunio et al., 2010; Roier et al., 2015; Zhe et al., 2016). The galactose-glucose binding protein was identified as the main component of the outer membrane vesicles released from 5 different strains of Haemophilus influenza (Roier et al., 2015). Extracellular nanovesicles are released by all pathogenic and non-pathogenic gram-negative bacteria (Lusta, 2015). They are composed of outer membrane components such as LPS, glycerophospholipids as well as proteins from the outer membrane and the periplasm (Kuehn and Kesty, 2005; Bai et al., 2014; Lusta, 2015). Outer membrane vesicles are considered as potent virulence factors because they provide a means for the extracellular secretion of proteins and lipids that can interact with the host tissues. The pyridine nucleotide-disulfide oxidoreductase (Zhe et al., 2016) has been identified as one of the proteins that interacts with brain microvascular endothelial cells, which may contribute to invasion by Streptococcus equi ssp. zooepidemicus through the blood-brain barrier. Finally, fructose-bisphosphate aldolase has been reported to be immunogenic in Candida albicans (Calcedo et al., 2012) and Madurella mycetomatis, in which it has been proposed as a vaccine candidate (de Klerk et al., 2012).
Vsp proteins are the most abundant outer membrane proteins of B. hyodysenteriae (Gabe et al., 1998), in which they have been postulated to have an antigenic role either as protein complexes or as individual molecules (McCaman et al., 1999; Witchell et al., 2011). Two members of the Vsp family, VspH, and VspD, have been included as components of a potential vaccine against B. hyodysenteriae (Bellgard et al., 2015). However, little is known about the expression of these proteins in Brachyspira species. The expression of VspH was reported in a B204 strain of B. hyodysenteriae (Witchell et al., 2006), although in further studies they described the absence of the gene in other strains (ATCC WA1 and X576) (Witchell et al., 2011). In the latter study, the expression of the VspD protein (together with vspF, vspE, and vspI) in a virulent Australian isolate of B. hyodysenteriae (Witchell et al., 2011) was reported. These authors suggested that Vsp proteins form complexes and that they are immunoreactive only in that form.
Interestingly, the analysis of our immunoreactive bands identified VspD only in the B. pilosicoli samples. This was in agreement with our previous work (Casas et al., 2016) on the exposed proteomes of B. pilosicoli and B. hyodysenteriae. In that study, the VspD protein was classified as exclusive from the exoproteome of B. pilosicoli because it was not found in any compartment of B. hyodysenteriae. To confirm our findings about the differential presence of VspD and to depict the distribution of the Vsp proteins in different Brachyspira strains, a more detailed targeted LC-MS/MS analysis was performed. The study confirmed the high expression of VspD in both B. pilosicoli strains and the low or no expression in B. hyodysenteriae strains.
Thus, Vsp proteins have a broad expression profile in different strains and species, a trait that could determine the efficiency of proteins such as VspH or VspD as vaccine components. Vsp proteins are components of a mechanism used by pathogenic bacteria to adapt to host conditions and optimize colonization. These proteins can show reversible on/off switching of their expression (phase variation) or antigenic changes by the expression of alternative protein phenotypes (Lysnyansky et al., 2001). Variability in the expression of variable surface proteins has been reported for several species of Mycoplasma. In in vitro experiments with Mycoplasma bovis, it was observed that exposure of the bacteria to anti-Vsp antibodies induced a change in the Vsp expression pattern (Caswell et al., 2010). Vsp proteins are the major antigenic targets in Mycoplasma bovis, however the immune response observed was not protective. In Mycoplasma mycoides, the different Vsp expression pattern observed in several outbreaks was suggested to be related to different Vsp proteins triggering the immune response in each case (Hamsten et al., 2008). These facts stress the value of determining the actual gene product levels present under different conditions as a complement of genomic-based approaches for vaccine design.
Cross-reactivity with control sera
We observed immunoreactive bands using control sera from non-challenged pigs with all strains tested (Supplementary Figure S3). This was consistent with previous reports showing that healthy pig serum can detect B. hyodysenteriae surface antigens (Wannemuehler et al., 1988). Song et al. also observed that non-vaccinated pigs from a herd with no reported history of SD developed increasing systemic lgG and lgM levels to all antigens of a vaccine being tested as the body weight of the animals increased (Song et al., 2009).
The FlaB family of proteins was the primary target of control sera in both species. This finding could be related with reports indicating that endoflagellar FlaBs, but not FlaA, from B. hyodysenteriae cross-react with the corresponding proteins from the non-pathogenic B. innocens (Li et al., 1993). In fact, it has been shown that the immunoreactivity patterns of purified flagellar proteins from different strains of B. hyodysenteriae and several non-pathogenic spirochetes have similar distributions, suggesting the existence of shared epitopes in these species (Kent et al., 1989). Thus, the observed cross-reactivity of flagellar proteins could be due to exposure to other non-pathogenic bacteria. Vaccination of pigs with the endoflagellar protein FlaB1 (recombinant or purified from B. hyodysenteriae) generated antibodies against the protein (Gabe et al., 1995). However, that response was not sufficient to protect the animals against the disease. It was suggested that the efficiency of the anti-FlaB antibodies would be reduced by the reduced accessibility to the inner part of the B. hyodysenteriae flagella (Gabe et al., 1995).
Other identified targets of the control sera were the Elongation factor Tu and the uncharacterized protein D8IDC2. Elongation factor Tu, is a protein which is conserved in different bacterial species so its detection with sera from pigs that had not been infected with SD or IS is not surprising. This protein has been detected on the surface of Leptospira interrogans (the spirochete which is the aetiological agent of leptospirosis) and it is related to the binding with the host plasminogen (Wolff et al., 2013). It is interesting to note that D8IDC2, which was identified in the two B. pilosicoli strains, has a 97% of homology with Bpmp-72, a membrane protein whose sequence was patented by Hampson and La (2009) for its use as a vaccine and a therapeutic treatment against intestinal spirochaetosis.
Conclusions
The immunoproteomics approach applied in this study has been demonstrated to be very effective for the characterization of new Brachyspira antigens. Data reported here was restricted to IgG's immunoreactivity toward these molecules. Further studies focussing on serum and secretory IgA's could potentially increase this collection of potential candidates. Most previously reported vaccine candidates were selected on the basis of previous knowledge from other species and through in silico reverse vaccinology approaches. The advantage of an immunoproteomics approach is that it intrinsically takes into account the actual expression levels, molecular characteristics and exposure to the host of the specific antigens that elicit an immune response. Consequently, the immunoreactive proteins described are unrivaled candidates to be components of vaccines for the treatment of SD and porcine IS. As discussed, we identified two abundant antigenic proteins shared by the two Brachyspira species (enolase and PEPCK) that could be considered as candidates for common vaccines for these species. In addition, 8 and 11 challenge-specific immunoreactive proteins were described for Brachyspira pilosicoli and B. hyodysenteriae, respectively. Although some of the immunoreactive bands were shown to contain more than one component and the actual antigen should be confirmed, the collection of proteins described constitutes a unique antigen collection from these bacteria.
Author contributions
JA, VC, AR, and MC conceived the project and designed the work. SV prepared the bacterial cultures and characterized the isolates by PCR. AR performed the immunoblot assays and RP the image analysis. VC collected proteomics data. VC and JA analyzed and interpreted the data. AR and VC redacted the manuscript draft and JA made the critical revision and produced the final manuscript. All the authors approved the final version of the article. VC carried out this work in the framework of the Immunology Ph.D. program of the Autonomous University of Barcelona.
Funding
This work was funded by the Spanish MINECO (IPT-2011-0735-010000). The CSIC/UAB Proteomics Laboratory of IIBB-CSIC is a member of Proteored, PRB2-ISCIII and is supported by grant PT13/0001, of the PE I+D+i 2013-2016, funded by ISCIII and FEDER.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00723/full#supplementary-material
References
- Alvarez-Ordóñez A., Martínez-Lobo F. J., Arguello H., Carvajal A., Rubio P. (2013). Swine dysentery: aetiology, pathogenicity, determinants of transmission and the fight against the disease. Int. J. Environ. Res. Public Health 10, 1927–1947. 10.3390/ijerph10051927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahi H., Osman A., Cook R. M., Loverde P. T., Stadecker M. J. (2000). Schistosoma mansoni phosphoenolpyruvate carboxykinase, a novel egg antigen: immunological properties of the recombinant protein and identification of a T-cell epitope. Infect. Immun. 68, 3385–3393. 10.1128/IAI.68.6.3385-3393.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Kim S. I., Ryu S., Yoon H. (2014). Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infect. Immun. 82, 4001–4010. 10.1128/IAI.01416-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G., Jasinskas A., Kayala M. A., Davies D. H., Steere A. C., Baldi P., et al. (2008). A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect. Immun. 76, 3374–3389. 10.1128/IAI.00048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellgard M., Hampson D. J., La T. (2015). Genes and Proteins of Brachyspira Hyodysenteriae and Uses Thereof. Patent US8992938.
- Black M., Moolhuijzen P., Barrero R., La T., Phillips N., Hampson D., et al. (2015). Analysis of multiple Brachyspira hyodysenteriae genomes confirms that the species is relatively conserved but has potentially important strain variation. PLoS ONE 10:e0131050. 10.1371/journal.pone.0131050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden D. A., Albert F. G., Robinson C. S. (1989). Cloning and characterization of Treponema hyodysenteriae antigens and protection in a CF-1 mouse model by immunization with a cloned endoflagellar antigen. Infect. Immun. 57, 3808–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J. S., Koniaras C., Lew A. M. (1997). Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int. Immunol. 9, 1897–1906. 10.1093/intimm/9.12.1897 [DOI] [PubMed] [Google Scholar]
- Calcedo R., Ramirez-Garcia A., Abad A., Rementeria A., Pontón J., Hernando F. L. (2012). Phosphoglycerate kinase and fructose bisphosphate aldolase of Candida albicans as new antigens recognized by human salivary IgA. Rev. Iberoam. Micol. 29, 172–174. 10.1016/j.riam.2011.07.004 [DOI] [PubMed] [Google Scholar]
- Calderaro A., Bommezzadri S., Piccolo G., Zuelli C., Dettori G., Chezzi C. (2005). Rapid isolation of Brachyspira hyodysenteriae and Brachyspira pilosicoli from pigs. Vet. Microbiol. 105, 229–234. 10.1016/j.vetmic.2004.10.021 [DOI] [PubMed] [Google Scholar]
- Calderaro A., Merialdi G., Perini S., Ragni P., Guégan R., Dettori G., et al. (2001). A novel method for isolation of Brachyspira (Serpulina) hyodysenteriae from pigs with swine dysentery in Italy. Vet. Microbiol. 80, 47–52. 10.1016/S0378-1135(00)00374-6 [DOI] [PubMed] [Google Scholar]
- Carvajal A., Arriba M. L. D., Rodriguez H., Vidal A. B., Duhamel G. E., Rubio P. (2006). Prevalence of Brachyspira species in pigs with diarrhoea in Spain. Vet. Rec. 158, 700–701. 10.1136/vr.158.20.700 [DOI] [PubMed] [Google Scholar]
- Casanovas A., Carrascal M., Abián J., López-Tejero M. D., Llobera M. (2009). Discovery of lipoprotein lipase pI isoforms and contributions to their characterization. J. Proteomics 72, 1031–1039. 10.1016/j.jprot.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Casas V., Vadillo S., Juan C. S., Carrascal M., Abian J. (2016). The exposed proteomes of Brachyspira hyodysenteriae and B. pilosicoli. Front. Microbiol. 7:1103. 10.3389/fmicb.2016.01103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell J. L., Bateman K. G., Cai H. Y., Castillo-Alcala F. (2010). Mycoplasma bovis in respiratory disease of feedlot cattle. Vet. Clin. North Am. Food Anim. Pract. 26, 365–379. 10.1016/j.cvfa.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Charon N. W., Goldstein S. F. (2002). Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36, 47–73. 10.1146/annurev.genet.36.041602.134359 [DOI] [PubMed] [Google Scholar]
- Chopra S., Reader J. (2014). tRNAs as antibiotic targets. Int. J. Mol. Sci. 16, 321–349. 10.3390/ijms16010321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. J., Smith S. C., Moore R. J. (2005). The Brachyspira hyodysenteriae ftnA gene: DNA vaccination and real-time PCR quantification of bacteria in a mouse model of disease. Curr. Microbiol. 50, 285–291. 10.1007/s00284-005-4472-2 [DOI] [PubMed] [Google Scholar]
- de Klerk N., de Vogel C., Fahal A., van Belkum A., van de Sande W. W. J. (2012). Fructose-bisphosphate aldolase and pyruvate kinase, two novel immunogens in Madurella mycetomatis. Med. Mycol. 50, 143–151. 10.3109/13693786.2011.593005 [DOI] [PubMed] [Google Scholar]
- Feberwee A., Hampson D. J., Phillips N. D., La T., van der Heijden H. M. J. F., Wellenberg G. J., et al. (2008). Identification of Brachyspira hyodysenteriae and other pathogenic Brachyspira species in chickens from laying flocks with diarrhea or reduced production or both. J. Clin. Microbiol. 46, 593–600. 10.1128/JCM.01829-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L. N., Duhamel G. E., Westerman R. B., Mathiesen M. R. (1997). Immunoblot reactivity of polyclonal and monoclonal antibodies with periplasmic flagellar proteins FlaA1 and FlaB of porcine Serpulina species. Clin. Diagn. Lab. Immunol. 4, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabe J. D., Chang R. J., Slomiany R., Andrews W. H., McCaman M. T. (1995). Isolation of extracytoplasmic proteins from Serpulina hyodysenteriae B204 and molecular cloning of the flaB1 gene encoding a 38-kilodalton flagellar protein. Infect. Immun. 63, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabe J., Dragon E., Chang R.-J., McCaman M. T. (1998). Identification of a linked set of genes in Serpulina hyodysenteriae (B204) predicted to encode closely related 39-kilodalton extracytoplasmic proteins. J. Bacteriol. 180, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Charon N. (1997). FlaA, a putative flagellar outer sheath protein, is not an immunodominant antigen associated with Lyme disease. Infect. Immun. 65, 2992–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska S., Dylus E., Rudawska A., Brzozowska E., Srutkova D., Schwarzer M., et al. (2016). Immunoreactive proteins of Bifidobacterium longum ssp. longum CCM 7952 and Bifidobacterium longum ssp. longum CCDM 372 Identified by gnotobiotic mono-colonized mice sera, immune rabbit sera and non-immune human sera. Front. Microbiol. 7:1537. 10.3389/fmicb.2016.01537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy A. H. (1974). Therapeutic effect of lincomycin and spectinomycin water medication on swine dysentery. Can. J. Comp. Med. Rev. Can. Med. Comp. 38, 1–6. [PMC free article] [PubMed] [Google Scholar]
- Hampson D. (2000). Influences of diet and vaccination on colonisation of pigs by the intestinal spirochaete Brachyspira (Serpulina) pilosicoli. Vet. Microbiol. 73, 75–84. 10.1016/S0378-1135(99)00200-X [DOI] [PubMed] [Google Scholar]
- Hampson D. J., La T. (2009). Brachyspira Pilosicoli 72kDa Outer-Membrane Protein and Diagnostic and Therapeutic Uses Thereof. Patent US007531176B2.
- Hampson D. J., Oxberry S. L., La T. (2006). Potential for zoonotic transmission of Brachyspira pilosicoli. Emerg. Infect. Dis. 12, 869–870. 10.3201/eid1205.051180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsten C., Westberg J., Bolske G., Ayling R., Uhlen M., Persson A. (2008). Expression and immunogenicity of six putative variable surface proteins in Mycoplasma mycoides subsp. mycoides SC. Microbiology 154, 539–549. 10.1099/mic.0.2007/010694-0 [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Carvajal A., García-Feliz C., Osorio J., Rubio P. (2009). Antimicrobial susceptibility testing of Spanish field isolates of Brachyspira hyodysenteriae. Res. Vet. Sci. 87, 7–12. 10.1016/j.rvsc.2008.10.017 [DOI] [PubMed] [Google Scholar]
- Holden J., Coloe P. J., Smooker P. M. (2008). An evaluation of the immunogenicity and protective responses to Brachyspira hyodysenteriae recombinant SmpB vaccination. Vet. Microbiol. 128, 354–363. 10.1016/j.vetmic.2007.10.026 [DOI] [PubMed] [Google Scholar]
- Hyatt D. R., ter Huurne A. A., van der Zeijst B. A., Joens L. A. (1994). Reduced virulence of Serpulina hyodysenteriae hemolysin-negative mutants in pigs and their potential to protect pigs against challenge with a virulent strain. Infect. Immun. 62, 2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T. R., O'Malley T., Liao R., Guinn K. M., Hickey M. J., Mohaideen N., et al. (2013). Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS ONE 8:e75245. 10.1371/journal.pone.0075245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Singh B., Li H. S., Kim Y. K., Kang S. K., Nah J. W., et al. (2014). Targeted oral delivery of BmpB vaccine using porous PLGA microparticles coated with M cell homing peptide-coupled chitosan. Biomaterials 35, 2365–2373. 10.1016/j.biomaterials.2013.11.073 [DOI] [PubMed] [Google Scholar]
- Joens L. A., Harris D. L., Baum D. H. (1979). Immunity to Swine dysentery in recovered pigs. Am. J. Vet. Res. 40, 1352–1354. [PubMed] [Google Scholar]
- Karlsson M., Gunnarsson A. F. A. (2001). Susceptibility to pleuromutilins in Brachyspira (Serpulina) hyodysenteriae. Anim. Heal. Res. Rev. 2, 59–65. 10.1079/AHRR200118 [DOI] [PubMed] [Google Scholar]
- Kent K. A., Sellwood R., Lemcke R. M., Burrows M. R., Lysons R. J. (1989). Analysis of the axial filaments of Treponema hyodysenteriae by SDS-PAGE and immunoblotting. J. Gen. Microbiol. 135, 1625–1632. 10.1099/00221287-135-6-1625 [DOI] [PubMed] [Google Scholar]
- Kuehn M. J., Kesty N. C. (2005). Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19, 2645–2655. 10.1101/gad.1299905 [DOI] [PubMed] [Google Scholar]
- La T., Phillips N. D., Reichel M. P., Hampson D. J. (2004). Protection of pigs from swine dysentery by vaccination with recombinant BmpB, a 29.7 kDa outer-membrane lipoprotein of Brachyspira hyodysenteriae. Vet. Microbiol. 102, 97–109. 10.1016/j.vetmic.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Li C., Corum L., Morgan D., Rosey E. L., Stanton T. B., Charon N. W. (2000). The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J. Bacteriol. 182, 6698–6706. 10.1128/JB.182.23.6698-6706.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wolgemuth C. W., Marko M., Morgan D. G., Charon N. W. (2008). Genetic analysis of spirochete flagellin proteins and their involvement in motility, filament assembly, and flagellar morphology. J. Bacteriol. 190, 5607–5615. 10.1128/JB.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Dumas F., Dubreuil D., Jacques M. (1993). A species-specific periplasmic flagellar protein of Serpulina (Treponema) hyodysenteriae. J. Bacteriol. 175, 8000–8007. 10.1128/jb.175.24.8000-8007.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., den Bakker H. C., Suzuki H., Lefebure T., Ponnala L., Sun Q., et al. (2013). Complete genome sequence of the porcine strain Brachyspira pilosicoli P43/6/78(T.). Genome Announc. 1, 6–7. 10.1128/genomeA.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Ba X., Yu J., Li J., Wei Q., Han G., et al. (2006). The phosphoenolpyruvate carboxykinase of Mycobacterium Tuberculosis induces strong cell-mediated immune responses in mice. Mol. Cell. Biochem. 288, 65–71. 10.1007/s11010-006-9119-5 [DOI] [PubMed] [Google Scholar]
- Lusta K. A. (2015). Bacterial outer membrane nanovesicles: structure, biogenesis, functions, and application in biotechnology and medicine (Review). Appl. Biochem. Microbiol. 51, 485–493. 10.1134/S0003683815040092 [DOI] [PubMed] [Google Scholar]
- Lysnyansky I., Ron Y., Sachse K., Yogev D. (2001). Intrachromosomal recombination within the vsp locus of Mycoplasma bovis generates a chimeric variable surface lipoprotein antigen. Infect. Immun. 69, 3703–3712. 10.1128/IAI.69.6.3703-3712.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mappley L. J., Black M. L., AbuOun M., Darby A. C., Woodward M. J., Parkhill J., et al. (2012). Comparative genomics of Brachyspira pilosicoli strains: genome rearrangements, reductions and correlation of genetic compliment with phenotypic diversity. BMC Genomics 13:454. 10.1186/1471-2164-13-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaman M. T., Auer K., Foley W., Gabe J. D. (1999). Sequence characterization of two new members of a multi-gene family in Serpulina hyodysenteriae (B204) with homology to a 39kDa surface exposed protein: vspC and D. Vet. Microbiol. 68, 273–283. 10.1016/S0378-1135(99)00104-2 [DOI] [PubMed] [Google Scholar]
- Michel P. E., Reymond F., Arnaud I. L., Josserand J., Girault H. H., Rossier J. S. (2003). Protein fractionation in a multicompartment device using Off-GelTM isoelectric focusing. Electrophoresis 24, 3–11. 10.1002/elps.200390030 [DOI] [PubMed] [Google Scholar]
- Molnar L. (1996). Sensitivity of strains of Serpulina hyodysenteriae isolated in Hungary to chemotherapeutic drugs. Vet. Rec. 138, 158–160. 10.1136/vr.138.7.158 [DOI] [PubMed] [Google Scholar]
- Movahedi A., Hampson D. J. (2007). Distribution of the clpX gene in Brachyspira species and reactivity of recombinant Brachyspira pilosicoli ClpX with sera from mice and humans. J. Med. Microbiol. 56, 930–936. 10.1099/jmm.0.47004-0 [DOI] [PubMed] [Google Scholar]
- Movahedi A., Hampson D. J. (2010). Evaluation of recombinant Brachyspira pilosicoli oligopeptide-binding proteins as vaccine candidates in a mouse model of intestinal spirochaetosis. J. Med. Microbiol. 59, 353–359. 10.1099/jmm.0.015842-0 [DOI] [PubMed] [Google Scholar]
- Ohya T., Sueyoshi M. (2010). In vitro antimicrobial susceptibility of Brachyspira hyodysenteriae strains isolated in Japan from 1985 to 2009. J. Vet. Med. Sci. 72, 1651–1653. 10.1292/jvms.10-0271 [DOI] [PubMed] [Google Scholar]
- Osorio J., Carvajal A., Naharro G., La T., Phillips N. D., Rubio P., et al. (2012). Dissemination of clonal groups of Brachyspira hyodysenteriae amongst pig farms in Spain, and their relationships to isolates from other countries. PLoS ONE 7:e39082. 10.1371/journal.pone.0039082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio J., Carvajal A., Naharro G., Rubio P., La T., Phillips N. D., et al. (2013). Identification of weakly haemolytic Brachyspira isolates recovered from pigs with diarrhoea in Spain and Portugal and comparison with results from other countries. Res. Vet. Sci. 95, 861–869. 10.1016/j.rvsc.2013.07.014 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pringle M., Landén A., Unnerstad H. E., Molander B., Bengtsson B. (2012). Antimicrobial susceptibility of porcine Brachyspira hyodysenteriae and Brachyspira pilosicoli isolated in Sweden between 1990 and 2010. Acta Vet. Scand. 54:54. 10.1186/1751-0147-54-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qeli E., Omasits U., Goetze S., Stekhoven D. J., Frey J. E., Basler K., et al. (2014). Improved prediction of peptide detectability for targeted proteomics using a rank-based algorithm and organism-specific data. J. Proteomics 108, 269–283. 10.1016/j.jprot.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Rahi A., Matta S. K., Dhiman A., Garhyan J., Gopalani M., Chandra S., et al. (2017). Enolase of Mycobacterium tuberculosis is a surface exposed plasminogen binding protein. Biochim. Biophys. Acta Gen. Subj. 1861, 3355–3364. 10.1016/j.bbagen.2016.08.018 [DOI] [PubMed] [Google Scholar]
- Rohde J., Kessler M., Baums C. G., Amtsberg G. (2004). Comparison of methods for antimicrobial susceptibility testing and MIC values for pleuromutilin drugs for Brachyspira hyodysenteriae isolated in Germany. Vet. Microbiol. 102, 25–32. 10.1016/j.vetmic.2004.03.023 [DOI] [PubMed] [Google Scholar]
- Roier S., Blume T., Klug L., Wagner G. E., Elhenawy W., Zangger K., et al. (2015). A basis for vaccine development: comparative characterization of Haemophilus influenzae outer membrane vesicles. Int. J. Med. Microbiol. 305, 298–309. 10.1016/j.ijmm.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Ros A., Faupel M., Mees H., Oostrum J. V., Ferrigno R., Reymond F., et al. (2002). Protein purification by Off-Gel electrophoresis. Proteomics 2, 151–156. [DOI] [PubMed] [Google Scholar]
- Rosa P. A., Tilly K., Stewart P. E. (2005). The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3, 129–143. 10.1038/nrmicro1086 [DOI] [PubMed] [Google Scholar]
- Shelburne C. E., Shelburne P. S., Dhople V. M., Sweier D. G., Giannobile W. V., Kinney J. S., et al. (2008). Serum antibodies to Porphyromonas gingivalis chaperone HtpG predict health in periodontitis susceptible patients. PLoS ONE 3:e1984. 10.1371/journal.pone.0001984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Song Y., La T., Phillips N. D., Bellgard M. I., Hampson D. J. (2009). A reverse vaccinology approach to swine dysentery vaccine development. Vet. Microbiol. 137, 111–119. 10.1016/j.vetmic.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Srutkova D., Schwarzer M., Hudcovic T., Zakostelska Z., Drab V., Spanova A., et al. (2015). Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS ONE 10:e0134050. 10.1371/journal.pone.0134050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B. (2006). The genus brachyspira, in The Prokaryotes, Vol. 7, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York, NY: Springer; ), 330–356. [Google Scholar]
- Trott D. J., Stanton T. B., Jensen N. S., Duhamel G. E., Johnson J. L., Hampson D. J. (1996). Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int. J. Syst. Bacteriol. 46, 206–215. 10.1099/00207713-46-1-206 [DOI] [PubMed] [Google Scholar]
- Tsinganou E., Gebbers J.-O. (2010). Human intestinal spirochetosis–a review. Ger. Med. Sci. 8, 1–7. 10.3205/000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunio S. A., Oldfield N. J., Berry A., Ala'Aldeen D. A., Wooldridge K. G., Turner D. P. (2010). The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol. Microbiol. 76, 605–615. 10.1111/j.1365-2958.2010.07098.x [DOI] [PubMed] [Google Scholar]
- Wanchanthuek P., Bellgard M. I., La T., Ryan K., Moolhuijzen P., Chapman B., et al. (2010). The complete genome sequence of the pathogenic intestinal spirochete Brachyspira pilosicoli and comparison with other Brachyspira genomes. PLoS ONE 5:e11455. 10.1371/journal.pone.0011455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannemuehler M. J., Hubbard R. D., Greer J. M. (1988). Characterization of the major outer membrane antigens of Treponema hyodysenteriae. Infect. Immun. 56, 3032–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchell T. D., Coutts S. A., Bulach D. M., Adler B. (2006). Differential expression of the Bhmp39 major outer membrane proteins of Brachyspira hyodysenteriae. Infect. Immun. 74, 3271–3276. 10.1128/IAI.02000-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchell T. D., Hoke D. E., Bulach D. M., Coutts S. A. J., Driesen S. J., Cordwell S. J., et al. (2011). The major surface Vsp proteins of Brachyspira hyodysenteriae form antigenic protein complexes. Vet. Microbiol. 149, 157–162. 10.1016/j.vetmic.2010.09.036 [DOI] [PubMed] [Google Scholar]
- Wolff D. G., Castiblanco-Valencia M. M., Abe C. M., Monaris D., Morais Z. M., Souza G. O., et al. (2013). Interaction of Leptospira elongation factor Tu with plasminogen and complement factor H: a metabolic leptospiral protein with moonlighting activities. PLoS ONE 8:e81818. 10.1371/journal.pone.0081818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Norris S. J., Liu J. (2014). Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53, 4323–4333. 10.1021/bi500059y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhe M., Jie P., Hui Z., Bin X., Xiaomeng P., Huixing L., et al. (2016). SILAC and LC-MS/MS identification of Streptococcus equi ssp. zooepidemicus proteins that contribute to mouse brain microvascular endothelial cell infection. Appl. Microbiol. Biotechnol. 100, 7125–7136. 10.1007/s00253-016-7579-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.