Abstract
RNA editing is an essential post-transcriptional process that creates functional mitochondrial mRNAs in Kinetoplastids. Multiprotein editosomes catalyze pre-mRNA cleavage, uridine (U) insertion or deletion, and ligation as specified by guide RNAs. Three functionally and compositionally distinct editosomes differ by the mutually exclusive presence of the KREN1, KREN2 or KREN3 endonuclease and their associated partner proteins. Because endonuclease cleavage is a likely point of regulation for RNA editing, we elucidated endonuclease specificity in vivo. We used a mutant gamma ATP synthase allele (MGA) to circumvent the normal essentiality of the editing endonucleases, and created cell lines in which both alleles of one, two or all three of the endonucleases were deleted. Cells lacking multiple endonucleases had altered editosome sedimentation on glycerol gradients and substantial defects in overall editing. Deep sequencing analysis of RNAs from such cells revealed clear discrimination by editosomes between sites of deletion versus insertion editing and preferential but overlapping specificity for sites of insertion editing. Thus, endonuclease specificities in vivo are distinct but with some functional overlap. The overlapping specificities likely accommodate the more numerous sites of insertion versus deletion editing as editosomes collaborate to accurately edit thousands of distinct editing sites in vivo.
INTRODUCTION
RNA editing is an essential process that inserts or deletes uridine nucleotides (Us) to recode most mitochondrial mRNAs using information provided by guide RNAs (gRNAs) in Trypanosoma brucei (1–3). Editing is limited in some mRNAs, such as COII, which has three editing sites (ESs) that undergo four U insertions that are specified by a single gRNA. Other mRNAs are extensively edited, such as ND7, which has 291 ESs that undergo 553 U insertions and 89 U deletions that are specified by numerous gRNAs. RNA editing is developmentally regulated by unknown mechanisms that alter the extent of editing in specific mRNAs as the parasite cycles between the procyclic form (PF) in the tsetse fly and the bloodstream form (BF) in mammals (4). RNA editing is performed by multiprotein complexes, variously called ∼20S editosomes or RNA Editing Core Complexes (RECCs), which contain several catalytic activities: endonucleases KREN1, KREN2, or KREN3 that cleave mRNA; terminal uridylyl transferase (TUTase) KRET2 that adds Us at insertion sites; exoribonucleases (exoUases) KREX1 or KREX2 that remove Us at deletion sites; and ligases KREL1 or KREL2 that ligate mRNA fragments after U addition or removal (5–13). Three distinct ∼20S editosomes contain mutually exclusive sets of proteins: KREN1/KREPB8/KREX1, KREN2/KREPB7, or KREN3/KREPB6, in addition to the 12 proteins that they share in common (14–16). Each endonuclease imparts functional differences to their respective editosome, resulting in distinct ES specificities in vitro. Isolated KREN1 editosomes cleave deletion ESs, KREN2 editosomes cleave insertion ESs, and KREN3 editosomes cleave an ES modelled on COII (5,6,12,14).
The editing endonucleases belong to the RNase III family, which in all characterized members have dimeric active sites (17,18). The archetypal Escherichia coli RNase III is a homodimer, while two RNase III domains in human Dicer form an intramolecular heterodimer to create the catalytic fold. In contrast, editosomes have a single KREN1, KREN2, or KREN3, each with only one RNase III domain, suggesting that they may form an intermolecular heterodimeric active site with other editosome proteins (16). Editosome proteins KREPB4 and KREPB5 have degenerate RNase III motifs and have been suggested to fulfil this role (16). However, divergent RNase III motifs were recently identified in KREPB6, KREPB7, and KREPB8, as were the interactions of these proteins, respectively, with KREN3, KREN2 and KREN1 (19). Thus, these RNase III-like domains of KREPB6, KREPB7, and KREPB8 may also, or alternatively, form heterodimeric active sites with their partner endonuclease, i.e. KREN3, KREN2 and KREN1 respectively. Such diversity might provide for the recognition and cleavage of thousands of similar, albeit distinct, sites that are accurately edited in vivo.
Analysis of cleavage activity in vitro is complicated by the complex nature of the editing endonucleases and by the limitations of the available assays. Recombinant editosome endonucleases perform poorly in vitro (20), and thus investigation of endonuclease function often employs editosomes that have been isolated, typically by affinity tagging the endonuclease (14). However, isolated editosomes inefficiently cleave the RNA substrate in vitro, and progression from one ES to another in vitro is even more inefficient and essentially impractical for in vitro study (21). Furthermore, these assays are low throughput, which prevents investigation of more than a handful of ESs, which may not accurately represent the multitude that exists in vivo. In vivo experimentation is complicated by the simultaneous presence of three endonucleases, all of which are normally essential. The discovery that a mutant gamma-ATP synthase (MGA) can circumvent the necessity for mitochondrial gene expression in BF provided an opportunity to study the editing endonucleases in isolation in vivo (22). We therefore created transgenic BF cells that express the MGA and which contain all, one or no editing endonucleases, and used them to investigate ES recognition by each endonuclease in vivo.
Examination of in vivo editing products has until recently been limited to a small number of cloned and sequenced products (23–25). These studies have shown that editing progresses generally 3΄ to 5΄ relative to mRNA and many products are partially edited and are presumptive intermediates. The 3΄ regions of these partially edited mRNAs contain sequences that match that of fully edited mRNA, i.e. represent canonical editing. These regions also frequently contain edited sequences that do not match that of fully edited mRNA, i.e. are non-canonical, and are located in the junction between the 3΄ fully edited and 5΄ unedited sequence. Non-canonical editing can either represent a difference in U content at sites that are edited in fully edited mRNA or editing at sites that are not edited in mature mRNA. The advent of deep sequencing technologies provides the means to characterize in vivo RNA editing products in much greater detail (26–28).
We report here the analysis of editosomes and deep sequencing of in vivo RNA editing in cell lines in which one, two or all three of the editing endonucleases were eliminated. In order to represent the diversity of insertion and deletion editing and transcript-specific differences, we analysed five edited RNAs: MURF2, ND7-5΄ domain, A6, CYb and COII. Cells lacking multiple endonucleases had altered editosome sedimentation and defects in overall editing. Extremely limited ‘editing’ was observed in the absence of all three editing endonucleases, perhaps due to an unknown endonuclease. We found that editing of some ESs employs a particular endonuclease, e.g. KREN1 for U deletion at all deletion ESs and KREN3 for U insertion in COII. However, we showed that other insertion ES can employ either KREN2 or KREN3, albeit with an apparent ES preference by each. Thus KREN1 is specific for deletion ESs, but KREN2 and KREN3 have overlapping insertion ES specificities with distinctly different relative frequencies. Together, these results reveal insights into how editosomes that differ in ES recognition by their endonucleases collaborate to and edit numerous distinct ESs in vivo.
MATERIALS AND METHODS
Transfection constructs
Plasmids for the knockout of endonuclease alleles have been previously published (5,6,12). For sequential elimination of endonucleases, the alleles of the first endonuclease were eliminated using previously published plasmid constructs derived from pLEW13 and pLEW90 (6,12), which use selection with G418 (neoR) or hygromycin (hygR), respectively. Prior to elimination of the second allele of the first endonuclease, cells were transfected with pEnT6+ATPaseGammaWT+3UTR plasmid (a gift from Matthew K. Gould and Achim Schnaufer) that contains the L262P mutation to introduce Mutant Gamma ATP synthase allele using selection with blasticidin (bsdR) (22). For elimination of subsequent endonucleases, knockout constructs were generated by fusion PCR, which use selection with phleomycin (bleR) or puromycin (pacR). These PCR constructs included loxP sites flanking the drug resistance cassettes to permit recycling of drug markers and the Herpes Simplex Virus Thymidine Kinase (HSVTK) gene from either pyr-FEKO-BLE (Addgene plasmid 24023; George Cross) or pSM07 (29). Oligos used to create these knockout constructs are in Supplemental Materials. For these PCR constructs, distinct homologous targeting sequences to eliminate the first and second alleles were used as previously described (29,30). Cell lines were subsequently transfected with pLEW100Cre_del_tetO (Addgene plasmid 24019; a gift from George Cross) to transiently express Cre recombinase and eliminate loxP-flanked sequences, and thereby make cells sensitive to both phleomycin and puromycin again (31,32). Resistance to ganciclovir (GCV) (Invivogen) selected for Cre-mediated loss of the drug cassette that includes the HSVTK gene.
Cell lines
All cell lines examined are derived from BF 427 wild type. The MGA cell line was made by transfecting 427 cells with pEnT6+ATPaseGammaWT+3UTR, followed by blasticidin selection. The KREN2 null cell line was made by transfecting the SKO-KREN2 cell line that has the first KREN2 allele replaced by the neoR cassette (6) with pEnT6+ATPaseGammaWT+3UTR, and selecting with blasticidin. The second KREN2 allele was then eliminated by transfection with hygR cassette as previously described (6). The KREN3 null cell line was made by transfecting the SKO-KREN3 cell line that has the first KREN3 allele replaced by the neoR cassette (12) with pEnT6+ATPaseGammaWT+3UTR, and selecting with blasticidin. The second KREN3 allele was then eliminated by transfection with hygR cassette as previously described (12). The KREN1 only cell line was made by transfecting the KREN2 null cell line with KREN3-BLE-KO PCR DNA, and selecting with phleomycin. The second KREN3 allele was then eliminated by transfection with KREN3-PAC-KO PCR DNA, and selection with puromycin. The KREN2 only cell line was made by transfecting the KREN3 null cell line with KREN1-BLE-KO PCR DNA, and selecting with phleomycin. The second KREN1 allele was then eliminated by transfection with KREN1-PAC-KO PCR DNA, and selection with puromycin. The KREN3 only cell line was made by transfecting the KREN2 null cell line with KREN1-BLE-KO PCR DNA, and selecting with phleomycin. The second KREN1 allele was then eliminated by transfection with KREN1-PAC-KO PCR DNA, and selection with puromycin. The Triple null cell line was made by transient transfection of the Cre-expressing pLEW100Cre_del_tetO plasmid into the KREN3 only cell line to remove bleR and pacR cassettes in the KREN1 locus, and selecting with ganciclovir. The resulting cell line was then transfected with KREN3-BLE-KO PCR DNA, and selected with phleomycin. The second KREN3 allele was then eliminated by transfection with KREN3-PAC-KO PCR DNA, and selection with puromycin. Cells were grown in HMI-9 media with 10% fetal bovine serum at 37°C, with appropriate drug selection at the following concentrations: 2.5 μg/ml G418, 2 μg /ml blasticidin, 5 μg/ml hygromycin, 1 μg /ml phleomycin, 0.1 μg /ml puromycin and/or 25 μg/ml ganciclovir. Transfections were performed with the Amaxa Nucleofector (Lonza) as previously described (29,30). All cell lines were screened by analyzing genomic DNA using PCR with specific primers to either demonstrate loss of the knocked out endonuclease, replacement with transfection construct, altered size of the entire locus, or sequence of introduced allele in the case of MGA (data not shown).
RNA preparation from cells
Total RNA was extracted from cells using TRIzol (Life Technologies) following manufacturer's instructions. RNA samples were DNase I treated using the TURBO DNA-free kit (Ambion) following manufacturer's instructions.
Real-time PCR analysis
DNase I treated total RNA was converted into cDNA using random hexamer priming as previously described (6). Briefly, 4.5 μg of RNA was converted to cDNA using random hexamers and Taqman Reverse Transcription Reagents (Applied Biosystems) in a 30 μl reaction. Each experiment had a reaction without reverse transcriptase as a control. The 30 μl reaction was diluted by adding 170 μl of water. Fluidigm BioMark Real-time PCR was performed as previously described, with values normalized to TERT as an internal control (30). For each RNA measured, an average threshold cycle (Ct) value from four measurements was used for calculations. Relative changes in target amplicons were determined by using the ΔΔCt method (33), with undetected transcripts given a Ct value of 40. Results were confirmed by independent experiments.
Glycerol gradients
Fractionation of BF whole cell lysates on 10–30% glycerol gradients was performed as previously described (6) with the following differences: ∼1.2 × 109 cells were lysed and fractionated on each gradient, and the gradients were centrifuged at 38 000 rpm in a Beckman SW40 Ti rotor for 5 h at 4°C. Briefly, cells were resuspended in lysis buffer (10 mM Tris pH 7.2, 10 mM MgCl2, 100 mM KCl, 1 mM Pefabloc, 2 μg/ml leupeptin, 1 μg/ml pepstatin, cOmplete protease inhibitors (Roche, as directed by manufacturer), and 1 mM DTT) to final volume of 700 μl, and 70 μl 10% Triton X-100 was added. Samples were mixed by inversion for 10 min at 4°C and cleared by centrifugation at 17 000 × g for 9 min at 4°C. After fractionation, glycerol gradients were divided into 0.5 ml fractions from the top, flash frozen on liquid nitrogen, and stored at –80°C.
Western analysis
For each cell line, equivalent cell numbers were lysed and loaded onto glycerol gradients. 42 μl of a gradient fraction was resolved in each lane of a Criterion 10% SDS-PAGE gel (BioRad), transferred to Immobilon-P membrane (Millipore), and blocked overnight at 4°C in 1× PBS-T with 5% milk. Blots were probed with monoclonal antibodies against KREPA1, KREPA2, KREL1 and KREPA3 as previously described (34), mitochondrial Heat Shock Protein 70 (HSP70) as previously described (34), or 1:2000 KRET2 rabbit polyclonal primary antibody, followed by 1:2000 goat anti-rabbit HRP (BioRad) secondary antibody and washed with 1× PBS-T. Blots were developed with ECL kit (Pierce) per manufacturer's instructions. Positive control ∼20S samples from purified PF mitochondria (IsTaR 1.7a strain) were generated as previously described (6,35).
Adenylation assays
Auto-adenylation KREL1 and KREL2 with [α-32P]-ATP was performed as previously described (36). Proteins were resolved on 10% Criterion (BioRad) SDS-PAGE gels that were then fixed in 50% methanol/10% acetic acid, equilibrated in 10% methanol/4% glycerol, dried, and analyzed by PhosphorImager (Molecular Dynamics).
Endonuclease cleavage assays
Triple-site substrate was assayed as previously described (12,16). Reaction products were detected by polyacrylamide-urea gel electrophoresis and analyzed by PhosphorImager (Molecular Dynamics). Assays were performed in the presence or absence of 1 mM ADP, which stimulates deletion cleavage and inhibits insertion cleavage (37). Each cleavage assay used 15 μl of glycerol gradient fraction.
Library construction
Editing amplicon libraries for Illumina MiSeq sequencing were prepared as follows. cDNA was made by random hexamer priming (as for real-time PCR). cDNAs were PCR amplified with Primestar polymerase (Takara) using oligos that contained both ∼20 nucleotides annealing sequence targeting specific mitochondrial mRNAs as well as adapter sequences for subsequent rounds of PCR to add Nextera Index Kit indexing primers (Illumina). Oligo sequences used for PCR amplification are in Supplemental Materials. The specific targeting primer sequences were designed to anneal to either never-edited or pre-edited sequences flanking the 3΄ section of an editing domain within each mRNA. Control PCR reactions using minus reverse transcriptase mock cDNA preparations were performed to ensure amplification products were bona fide measures of RNA content (data not shown). To ascertain the range of linear amplification for these cDNAs and specific primer sets, real-time PCR using SsoAdvanced Universal SYBR Green Supermix (BioRad) was performed (data not shown). Based on these results, 17 cycles of PCR amplification were performed using mRNA specific primers in 150 μl total volume, with three 50 μl technical replicates pooled after amplification. These PCR reactions were ethanol precipitated with 15 μg of glycogen, and pellets resuspended in 10 μl water. MiSeq adapter oligos that include specific indices for each sample source were added in a second PCR reaction, using 2 μl of the resuspended PCR products in a 100 μl reaction, with 15 cycles of amplification, which was determined to be within the linear range of amplification as above. These PCR reactions were ethanol precipitated with 10 μg of glycogen, and pellets resuspended in 8 μl 1× GelPilot Loading Dye (Qiagen). All 8 μl was then purified on 1.5% agarose 1× TAE gels, stained with ethidum bromide, and the region corresponding to the expected range of PCR products was excised. PCR amplicon DNA was isolated from gel slices using NuceloSpin Gel and PCR Clean-up spin columns, eluting in 30 μl volume (Macherey-Nagel). PCR amplicon concentrations were determined using Qubit Fluorometer (Thermo Fisher Scientific), and molar concentrations calculated using pre-edited amplicon size. Equimolar amounts of all indexed amplicons were then combined into a single library for analysis on MiSeq sequencer (Illumina). All amplicons were sequenced together on a single flow cell to eliminate the possibility of uncontrolled variability between sequencing runs. A single biological sample was analyzed to ensure sufficient sequencing depth for multiple mRNA targets from each cell line.
Illumina MiSeq operation
Amplicons were sequenced through one single end sequencing reads of 50 nucleotides. Libraries were denatured and loaded onto Illumina 50-cycle V2 cartridges, according to the manufacturer's suggested workflow. Briefly, libraries were combined at a concentration 4 nM and denatured for 5 min at room temperature with freshly prepared 0.2N NaOH. After incubation, the reaction was neutralized with ice-cold, HT1 Hybridization Buffer (Illumina) diluted to 12 pM and loaded onto the cassette. Illumina PhiX Control was used as internal quality control. PhiX was denatured as mentioned above and diluted in buffer HT1 to a concentration of 12.5 pM. PhiX was added to denatured, diluted editing amplicon libraries to a concentration final concentration of 1% of the combined library volume.
Read sequence analysis
Read data have been deposited at the NCBI Sequence Read Archive under accession numbers SRR4450419, SRR4450418, SRR4450417, SRR4450416, SRR4450382, SRR4450381, SRR4450383 and SRR4450317. Indexed reads from Illumina sequencing were processed using custom made Pipeline Analyzing RNA Editing RNA Sequencing (PARERS) programming package. PARERS starts by identifying reads that a) contain exact string match for 12–15 nucleotides of primer sequence for each targeted transcript, and b) contain an exact string match between the reverse complemented read with T nucleotides removed and the entire maxicircle reference sequence (GenBank M94286.1) with T nucleotides removed. Respective primer sequences for string matches are ND7-5΄: GCATCGTGGTACAGA; CYb: GTCTTTTAATGTCAG; COII: CCTGGTAGGTGTAATG; MURF2: GTCGTGTTTTTGATTTG; A6: GGAGTTATAGAATAA. Once these reads have been identified and mapped to a maxicircle target, the original read sequences (with T nucleotides) are categorized using exact string matches to find either pre-edited or fully-edited sequences, which leaves partially edited sequences in the unmatched set.
RESULTS
Cells with restricted endonuclease expression
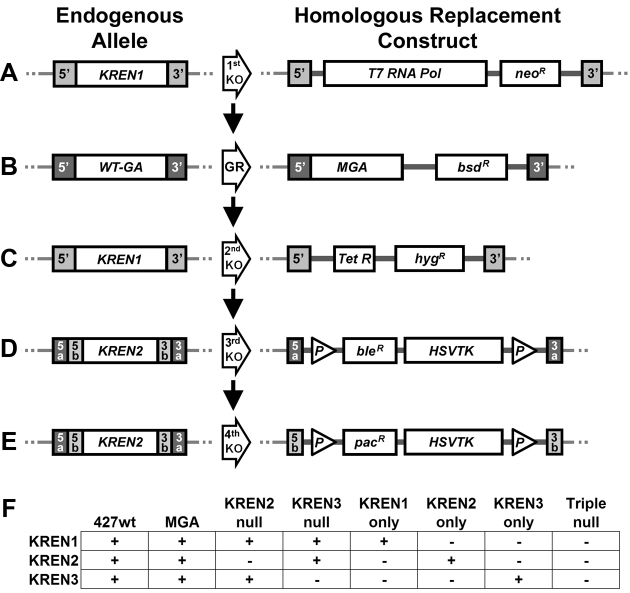
In order to determine the in vivo cleavage capabilities of each editing endonuclease, we used homologous recombination to generate cell lines that express only a single endonuclease, or no endonuclease at all as a control. Because the editing endonucleases are normally essential genes, we introduced the Mutant Gamma ATP synthase (MGA) allele into the endogenous locus, which permits survival in the absence of kDNA expression (22). The transfection constructs required to make cells that express only one of the characterized editing endonuclease (only KREN3 in this example) are diagrammed in Figure 1, and follow established protocols for construction (6,29). Briefly, after elimination of the first allele of the first endonuclease, the MGA allele replaced the wild-type Gamma ATP synthase, which allowed cells to grow in the absence of kDNA expression. The second endonuclease allele was then eliminated, followed by the homologous knockout of the first and then second alleles of a second endonuclease. The final two transfection constructs have lox P sites flanking the selectable marker and HSTVK, which permitted the subsequent elimination of the intervening sequences by transient transfection with a Cre recombinase expressing plasmid, followed by ganciclovir counter-selection for the loss of HSVTK. This Cre-mediated elimination permitted reuse of the selectable markers, which had been eliminated, in order to knockout the alleles of the third endonuclease and generate the Triple null cell line. As a control, the parental 427 wild-type strain was transfected with the MGA construct alone, so that any effect of MGA expression could be assessed. After transfections, genomic DNA from cell lines was screened by PCR to examine the targeted locus to confirm the intended homologous replacement had occurred (data not shown). Growth of these cell lines with restricted endonuclease repertoires was similar to the parental 427 wild-type cell line or a cell line expressing MGA alone (Supplementary Figure S1). The endonucleases that are present in the cell lines thus generated are depicted in Figure 1F.
Figure 1.
Schematic of homologous recombination knockout constructs. The homologous recombination constructs used to create the N3 only cell line are depicted as an example of the methods used to generate all cell lines, with endogenous allele depicted on the left and homologous replacement on the right. (A) Knockout of the first KREN1 allele (first KO) by replacement with T7 RNA polymerase and neomycin resistance gene (neoR) using KREN1 5΄ and 3΄ UTR targeting sequences (light gray). (B) Gene Replacement (GR) of the Wild-Type Gamma ATP synthase (WT-GA) with the Mutant Gamma ATP synthase (MGA) and blasticidin resistance (bsdR) gene using Gamma ATP synthase 5΄ and 3΄ UTR targeting sequences (dark gray). (C) Knockout of the second KREN1 allele (second KO) by replacement with tetracycline repressor (Tet R) and hygromycin resistance gene (hygR) using the same KREN1 5΄ and 3΄ UTR targeting sequences (light gray) used for first KO. (D) Knockout of the first KREN2 allele (third KO) by replacement with a cassette containing phleomycin resistance gene (bleR) fused to Herpes Simplex Virus Thymidine Kinase (HSVTK) flanked by lox P sites (P) using the outer KREN2 5΄ and 3΄ UTR targeting sequences (dark gray; 5a and 3a respectively). (E) Knockout of the second KREN2 allele (fourth KO) by replacement with a cassette containing puromycin resistance gene (pacR) fused to Herpes Simplex Virus Thymidine Kinase (HSVTK) flanked by lox P sites (P) using the inner KREN2 5΄ and 3΄ UTR targeting sequences (light gray; 5b and 3b respectively). (F) Presence or absence of editing endonuclease genes in examined cell lines. The presence (+) or absence (–) of KREN1, KREN2, and KREN3 is shown for the eight cell lines examined in this work. Absence reflects elimination of both alleles of the indicated gene by homologous recombination.
Loss of editing in vivo is associated with loss of endonuclease expression
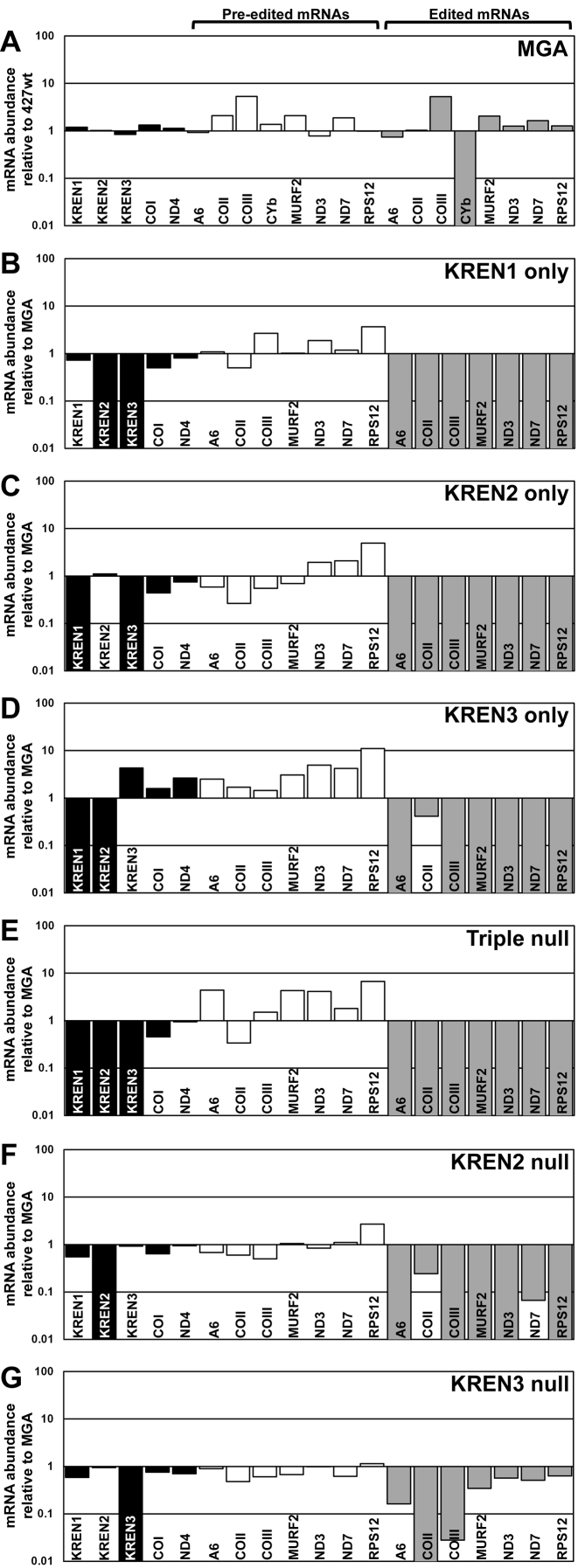
Real-time PCR analysis reveals that elimination of editing endonuclease expression causes a dramatic reduction in the editing of numerous mRNAs in vivo (Figure 2). Total RNA was isolated from cells, treated with DNase I, reverse transcribed into cDNA, and then analysed by real-time PCR. Relative amounts of each target RNA were calculated using TERT internal control. The average standard deviation of measured Ct values was 0.12, reflecting consistent similarity between measured values. For MGA cells, 427wt was used as a reference control for relative mRNA expression (Figure 2A), while the remaining analyses used MGA as the reference control (Figure 2B–G). Comparison of mRNA abundance in MGA relative to 427wt revealed either minor or no differences in all transcripts, with the notable exception of edited CYb, which was undetected in MGA. Because edited CYb was undetected in MGA, subsequent relative comparisons to MGA do not include examination of CYb transcript levels. As expected, mRNAs for editing endonucleases KREN1, KREN2, and KREN3 were only detected in cell lines in which their coding sequence had not been eliminated by homologous recombination. This result experimentally verifies that the intended genetic modifications were successful. The abundance of maxicircle transcripts that do not get edited (COI and ND4) were not noticeably altered in any tested cell line. The amounts of edited mRNAs in endonuclease-restricted cell lines were either severely reduced or not detected, with some notable exceptions. Edited COII appeared only slightly reduced in KREN3 only and KREN2 null cells, both of which contain KREN3. Small amounts of edited ND7 were also detected in KREN2 null cells, in which editing is detected at the 5΄ end of the 5΄ ND7 domain. Editing of COII is lost and editing of COIII is noticeably decreased in KREN3 null cells, while other edited transcripts have variable amounts of diminishment compared to MGA (Figure 2G). This result shows the distinct consequences of KREN3 versus KREN2 loss on editing. In contrast to edited transcripts, the relative amounts of pre-edited RNAs were unchanged or slightly increased after loss of endonuclease expression.
Figure 2.
Real-time PCR analysis shows loss of endonuclease expression and associated loss of editing. Relative RNA abundance is shown for KREN1, KREN2, KREN3 and never-edited mRNAs COI and ND4 (black bars), pre-edited mRNAs (white bars), and edited mRNAs (gray bars). For each target amplicon, the relative change in RNA abundance was determined by using telomerase reverse transcriptase (TERT) mRNA as an internal control, with each cell line compared relative to either 427wt or MGA control as indicated. (A) MGA cells have no large changes in mRNA abundance relative to 427wt, with the exception of the complete loss of CYb editing. As the remaining cell lines are compared to MGA, CYb is excluded from those analyses. (B) KREN1 only cells have no detectable KREN2 or KREN3 mRNA, and a broad loss of RNA editing. (C) KREN2 only cells have no detectable KREN1 or KREN3 mRNA, and a broad loss of RNA editing. (D) KREN3 only cells have no detectable KREN1 or KREN2 mRNA, and a broad loss of RNA editing, with COII editing notably retained. (E) Triple null cells have no detectable KREN1, KREN2, or KREN3 mRNA, and a broad loss of RNA editing. (F) KREN2 null cells have no detectable KREN2 mRNA, and a broad loss of RNA editing with some amount of COII and ND7 editing retained. (G) KREN3 null cells have no detectable KREN3 mRNA, and a loss of COII RNA editing with variable amounts of other edited mRNAs retained.
Loss of multiple endonucleases alters editosome sedimentation
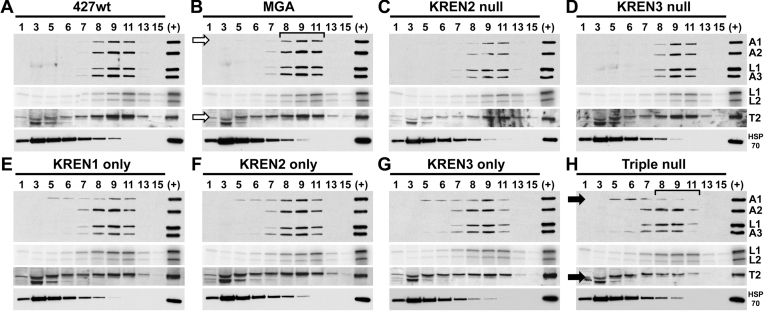
Editosomes from cells lacking multiple endonucleases are shifted to lower S values on glycerol gradients compared to 427wt and MGA control cells (Figure 3). Western analysis with a mixture of monoclonal antibodies against KREPA1, KREPA2, KREL1 and KREPA3 and a rabbit polyclonal antibody against KRET2 shows a ∼20S peak of editosome proteins in fractions 8–11 from lysates from 427wt and MGA cell lines (Figure 3A and B). Sedimentation of editosomes from KREN2 null and KREN3 null cell lines appears indistinguishable from these controls (Figure 3C and D). In contrast, analysis of cells expressing only one endonuclease or those without any endonuclease reveals an obvious shift of KREPA1 and KRET2 towards the top (lower numbered fractions; smaller S values) of the gradient (Figure 3E–H). While the majority of KREPA2, KREL1, KREPA3 and KRET2 signals are retained in fractions 8–11 in cells expressing a single endonuclease, the amount of signal in fraction 7 appears to increase slightly compared to controls. Notably, in Triple null cells the peak of the western signal for KREPA1 and KRET2 is shifted to fractions 5–7 compared to the peak signals for KREPA2, KREL1 and KREPA3 that are in fractions 7–9. The peak of KREPA1 and KRET2 sedimentation in fractions 5–7 in Triple null sample is in stark contrast with MGA control, where these peak in fractions 8–11. Editosomes that lack editing endonuclease are therefore substantially altered, with an apparent dissociation of insertion heterotrimeric subcomplex proteins (KREPA1 and KRET2) from other editosome proteins. Adenylation assays, performed on the same gradient fractions analysed by western blot, radiolabel ligases KREL1 and KREL2 that are capable of autoadenylation. Although KREL2 is a component of the insertion heterotrimeric subcomplex like KREPA1 and KRET2, any apparent shift in KREL2 signal to the top of the gradient is subtle. As adenylation assays require catalytic activity of the ligase, this assay may disproportionately detect ligases in functional complexes. Conversely, KREL2 may be more tightly associated with the remainder of the ∼20S editosome in the absence of endonuclease in comparison to KREPA1 and KRET2.
Figure 3.
Western and adenylation analyses of glycerol gradient fractionated editosomes. Gradient fractions from 427wt (A), MGA (B), KREN2 null (C), KREN3 null (D), KREN1 only (E), KREN2 only (F), KREN3 only (G) and triple null (H) were analysed using antibodies recognizing editosome proteins KREPA1, KREPA2, KREL1 and KREPA3 (first panels), by adenylation of ligases KREL1 and KREL2 (second panels), using antibody against KRET2 (third panels), or using antibody against mitochondrial HSP70 as a non-editosome control. Typical ∼20S editosome peak signal is centered on fraction 9, as highlighted by the bracket in the MGA control. In KREN1 only, KREN2 only, KREN3 only, and Triple null cells the sedimentation of KREPA1 is notably shifted toward upper part of the gradient (i.e. smaller in size) relative to 427wt and MGA controls, as is KRET2 in Triple null cells (indicated by solid arrows in Triple null compared to open arrows in MGA). Bracket in Triple null cells highlights difference in the ∼20S region compared to MGA, A control sample of ∼20S fraction from purified mitochondria (+) is included in each analysis.
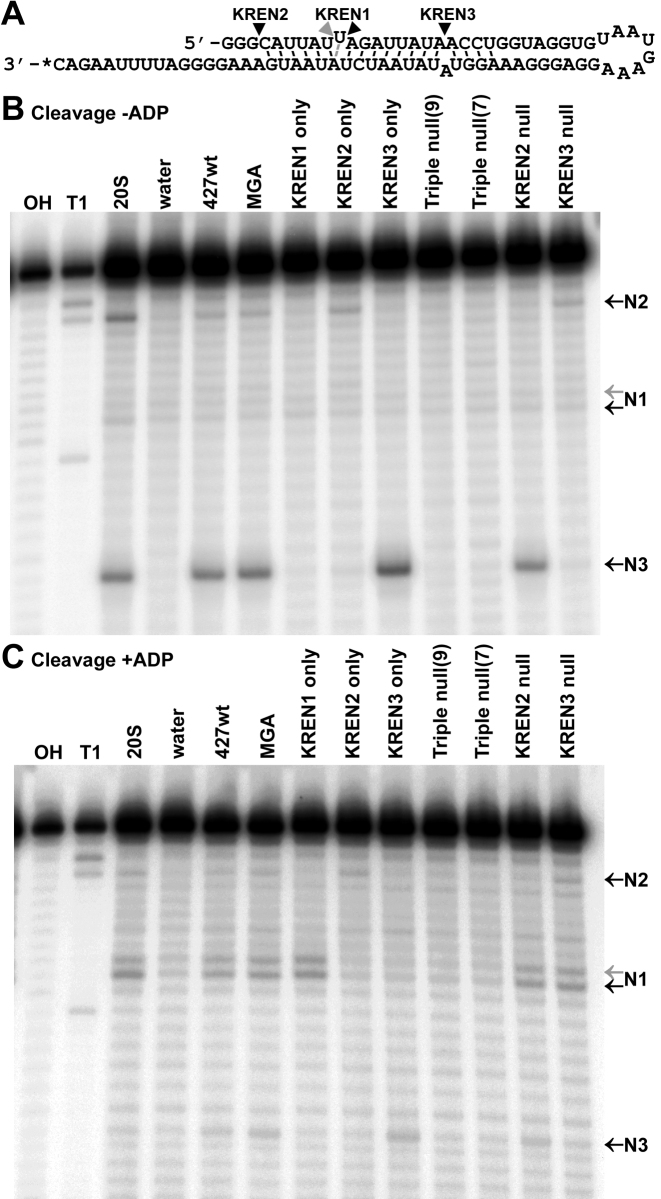
Cells lacking endonuclease lose specific cleavage activities in vitro
The in vitro cleavage activities of each cell line were examined using a ‘Triple-site’ substrate (16) that contains distinct sites specifically cleaved by each type of editosome (Figure 4). For all cell lines, the ∼20S peak of the glycerol gradient (fraction 9) was assayed; because the editosome signal shifts considerably in Triple null cells, fraction 7 from the Triple null gradient was also assayed. In order to detect both insertion and deletion cleavage activities, assays were performed either in the absence of ADP, which favors insertion cleavage (Figure 4B), or in the presence of ADP, which favors deletion cleavage (Figure 4C) (37). KREN1, KREN2 and KREN3 cleavage activities are all observed in positive control ∼20S fraction from purified mitochondria, as well as fractions from 427wt and MGA cell lines. In contrast, specific cleavage products are lost when the different endonucleases are eliminated: KREN1 only has only KREN1 cleavage; KREN2 only has only KREN2 cleavage; KREN3 only has only KREN3 cleavage; KREN2 null has only KREN1 and KREN3 cleavage; KREN3 null has only KREN1 and KREN2 cleavage; and Triple null have no detectable cleavage activity above the background represented by negative control reactions using water. Interestingly, the KREN3 only sample has noticeably greater cleavage activity at the KREN3 site compared to samples either containing all three endonucleases (20S mitochondrial positive control, 427wt, and MGA) or two endonucleases including KREN3 (KREN2 null). KREN1 only and KREN2 only also appear to have slightly more cleavage product at the KREN1 and KREN2 sites, respectively, compared to 427wt and MGA controls. The observed in vitro cleavage activities are consistent with the genetic modifications intended for each transgenic cell line, and reflect the distinct cleavage preferences for each editosome endonuclease in vitro.
Figure 4.
Editing endonuclease cleavage assays using ‘Triple-site’ substrate RNA. (A) Schematic of ‘Triple-site’ substrate RNA, with wedges indicating distinct cleavage sites for KREN1, KREN2 and KREN3 endonucleases. Asterisk denotes location of the radiolabel. Black wedges denote typical cleavage sites corresponding to activity of KREN1 (N1), KREN2 (N2) or KREN3 (N3). Note that the N1 cleavage site can shift +1 nt closer to the 5΄ end (gray wedge) in alternate conformers of the ‘Triple-site’ substrate. (B) Cleavage of ‘Triple-site’ substrate RNA in reactions lacking ADP to favor insertion cleavage by either ∼20S glycerol gradient fraction from purified mitochondria, or ∼20S peak of glycerol gradients (fraction 9) of indicated cell lines. Fraction 7 from Triple null cell line is also included. Water is used as a negative control for background degradation of the substrate. Reference ladders were generated by cleavage using alkaline hydrolysis (OH) or RNase T1 (T1). (C) Cleavage of ‘Triple-site’ substrate RNA in reactions with ADP to favor deletion cleavage, otherwise similar to (B).
RNAseq analysis of in vivo editing activities
RNAseq of RT-PCR products from cells with restricted endonuclease expression revealed a comprehensive view of endonuclease capabilities and substrate preferences, providing detailed insight into the editing process in vivo. Using RNAseq, we examined the sequences that represent the initial editing sites, i.e. 3΄ regions. The initial RT-PCR amplifications were performed in the linear range to reduce PCR artifacts and which match never-edited or pre-edited sequences in MURF2, ND7 5΄ domain, CYb, A6 or COII. RNAseq primers contained adaptor sequences used in a second PCR reaction, which was also performed in the linear range. The second PCR reaction used indexing primers to barcode the product from each cell type and included sequences for annealing to the flow cell. These DNA products were resolved on agarose gels, and those with the sizes in the region predicted for pre-edited and edited sequences were isolated. Products for five targeted amplicons from all eight cell lines generated 40 indexed libraries and were combined in equimolar amounts and sequenced in a single RNAseq flow cell. The sequencing generated 14 638 584 indexed reads, with 95.2% of reads having a Q score greater than 30, indicating high quality sequence data.
To analyse the resulting RNAseq data, we developed custom software called PARERS (Pipeline Analyzing RNA Editing RNA Sequencing) that identifies and maps sequences that may contain variable numbers of U nucleotides. PARERS removes T nucleotides from each reverse complement of the read sequence and performs an exact string match to the maxicircle reference sequence that has also had T nucleotides removed. Because the T nucleotides in read sequences represent U nucleotides from the source mRNA, string matches based solely on A, C, and G sequence allow identification regardless of U insertion or deletion editing events. After PARERS identifies and maps a read to the maxicircle target, subsequent string matches identify the original T-inclusive read sequence as a pre-edited, fully-edited, or partially edited sequence. Using PARERS, 11 344 810 reads (77.5% of indexed reads) were mapped to the maxicircle, which forms the dataset used in all subsequent analyses (Supplementary Table S1). Analysis of the RNAseq data demonstrates a broad diversity of unique read sequences within each sample, with expected variations that are consistent with the distinct biological modifications in each cell line. (Supplementary Table S2). Among these reads, 7 292 652 were pre-edited, 540 852 were fully edited and 3 511 306 were partially edited sequences (Supplementary Table S3).
Loss of endonuclease alters in vivo editing products
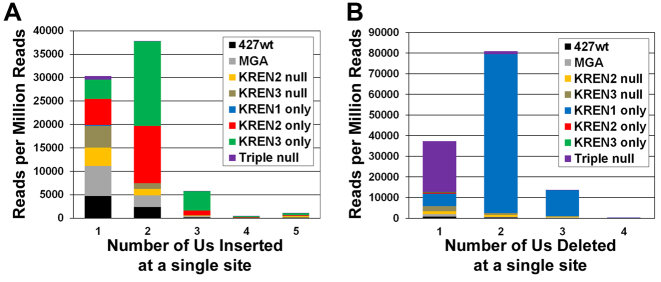
The frequency of both U insertion and deletion events reflects the genetic modifications made in the examined cell lines. They also reveal an unknown source of cleavage activity. For our initial examination of the RNAseq data, we opted to examine a simpler subset of the entire dataset: those reads that had editing at a single site, which corresponds to 154 468 insertion and 204 313 deletion events found in all cell lines (Figure 5, Supplementary Tables S4–S6). For each of these editing events, we determined how many reads from each cell line had one or more U nucleotides inserted or deleted, and normalized those read counts to the total number of mapped reads identified for each cell line. The relative frequencies of reads that correspond to the insertion of a single U are similar for 427wt, MGA, KREN2 null, KREN3 null, KREN2 only and KREN3 only; however, KREN1 only and Triple null samples are starkly smaller in comparison (Figure 5A). Both KREN1 only and Triple null samples contain extremely few reads that have any number of Us inserted, consistent with the loss of insertion endonuclease activity. The frequencies of reads that have either two or three Us inserted are dominated by two samples: KREN2 only and KREN3 only, cell lines that exclusively have insertion endonucleases. Examination of reads that correspond to the deletion of a single U reveals that KREN1 only and particularly Triple null predominate (Figure 5B). While the relatively higher frequency of the KREN1 only sample is consistent with this cell line exclusively having deletion endonuclease activity, the large number of reads containing the deletion of a single U in the Triple null sample was unexpected, because all of the characterized editing endonucleases have been eliminated in this cell line. Triple null cells must therefore contain an unknown activity that primarily deletes a single U. The frequencies of reads that have either two or three Us deleted is dominated by the KREN1 only sample, with comparatively few reads observed from any other samples, including Triple null. In addition to revealing an unknown deletion activity, these results indicate that the underlying sequence data are consistent with the characterized biological activities of the editing endonucleases on a global level, validating the computational analysis. The position of the single editing events within the read sequence also provides insight into the initiation of editing in each mRNA. While ES1 is frequently the site edited in reads containing a single edited site, editing at other sites is also observed (Supplementary Tables S5 and S6). This result suggests that editing is not strictly locked into using the 3΄ most site in the mRNA for initiation.
Figure 5.
Frequency of various numbers of Us inserted or deleted in reads edited at a single site. Reads with editing at a single site were counted based on the number of Us either (A) inserted or (B) deleted, and these read counts were normalized to the total number of mapped reads for each sample. Reads from each cell line are indicated by colors noted in legend.
RNAseq of in vivo editing products reveals distinct and overlapping endonuclease activities
Detailed examination of read sequences isolated from each cell line reveals complex interactions between editing sites and endonucleases, because few sites are exclusively recognized by particular endonucleases, and the majority of sites have overlapping specificities that differ in frequency of endonuclease recognition. Close inspection of reads from each cell line showed that many sequences were observed repeatedly, and the frequency of particular read sequences was altered after genetic modification to restrict endonuclease expression.
MURF2
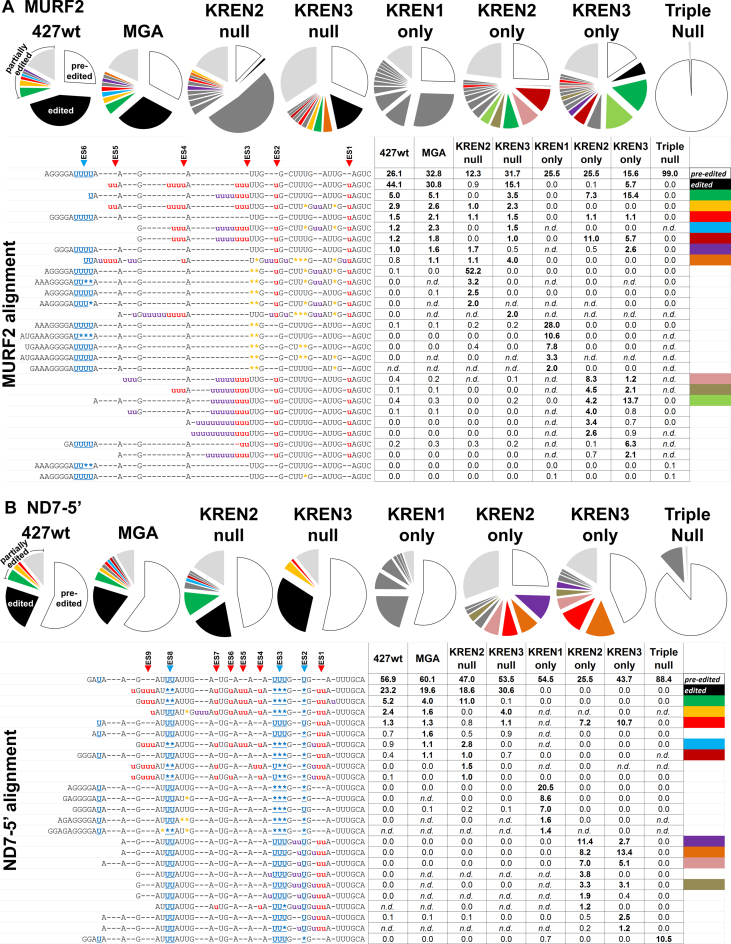
MURF2, which is edited robustly in both life cycle stages, has a relatively small editing domain with a single deletion site in the middle of 10 insertion sites (4,38). Examination of MURF2 reads reveals editing profiles that vary considerably depending upon the endonucleases present. RNAseq of MURF2 generated 805 748 reads corresponding to pre-edited sequence, 240 716 reads corresponding to edited sequence and 2 187 796 reads corresponding to partially edited sequence when all samples are combined (Figure 6A, Supplementary Table S2). The length of the read covers the first four canonical ES in the fully edited sequence, all of which are insertion sites. The profiles for 427wt and MGA appeared largely similar, with pre-edited and edited sequences representing a majority of the reads. Comparison of the most abundant partially edited reads between 427wt and MGA also appears similar, with the four most abundant sequences present in both cases. The profile for KREN3 null cells differs only slightly from these controls, with the majority of reads being partially edited sequences, in a very similar pattern to 427wt and MGA. This probably reflects the absence of deletion editing sites in MURF2. The profile for KREN2 null, by contrast, is noticeably different from those of 427wt and MGA, with only 0.9% of the reads fully edited and 52.2% of the reads representing a single partially edited sequence with both insertion and deletion events, many of which are non-canonical. Many of the most frequently observed partially edited sequences found in KREN2 null cells are vanishingly infrequent in other samples, and they notably contain non-canonical deletion events. In KREN1 only cells, only deletion editing is present in all of the frequently observed partially edited sequences and these sequences are rarely detected in other cell lines. Of the 418 238 MURF2 reads in the KREN1 only sample, only six were fully edited, indicating that KREN1 can only very rarely generate insertion editing. The most frequent partially edited MURF2 reads from both KREN2 only and KREN3 only cells exclusively contain insertion events, and although the same partially edited read sequences were often observed in both samples, their relative frequencies were distinct. For example, the most frequent partially edited read in KREN2 only, representing 11% of the sample, is the fourth most frequent in KREN3 only, where it is 5.7% of the sample. Interestingly, this read sequence differs only in the insertion of three Us at ES4 in comparison to the most frequent partially edited read in KREN3 only, where it represents 15.4% of that sample, but just 7.3% of the KREN2 only sample. This suggests that KREN2 and KREN3 have distinct recognition preferences at ES4. The second most frequent partially edited read in KREN2 only, representing 8.3% of the sample, is just 1.2% of the KREN3 only sample, in which it ranks eleventh. This sequence also differs from the most frequent partially edited read in KREN3 only by the insertion of three Us, in this case at a non-canonical site 5΄ to ES4. This result again indicates that distinct endonuclease preferences can exist at a particular ES. Thus, while both KREN2 and KREN3 can cleave the same editing sites, the differences in the relative frequency of this cleavage reflect distinct preferences for each endonuclease. At the far end of the spectrum, 99.0% of the reads from Triple null cells are pre-edited sequence, with fully edited reads not detected. The remaining 1.0% of the reads represented partially edited sequences, with the most frequent of these having one or two Us deleted, including at the single canonical deletion site in MURF2.
Figure 6.
Distinct and overlapping endonuclease specificities are revealed by sequencing of in vivo editing products. Analysis of RNAseq reads for MURF2 (A), ND7-5΄ (B), A6 (C), CYb (D) and COII (E) show activities corresponding to endonuclease repertoire in various cells lines. Pie charts show percent of mapped reads for each target that have a unique read sequence. For each sample, the proportion of reads that perfectly match pre-edited sequence are white and those that match fully edited sequence are black. Each partially edited read sequence that represents more than 1% of the samples reads for that transcript is shown in dark gray, unless this read sequence is also found in another sample, in which case it is color-coded. This color-coding is used to identify the same read sequence within other samples, and is also shown in the far right column of the alignment table below to indicate the sequence. The proportion of partially edited read sequences that individually represent less than 1% of the samples reads are pooled into a section shown in light grey. An alignment of the most frequent read sequences is shown below, with a table indicating the proportion of each sample's reads that each sequence represents. Within the alignment, a lowercase red ‘u’ indicates canonical insertion, while a lowercase purple ‘u’ indicates non-canonical insertion. A blue underlined asterisk indicates canonical deletion, while an orange asterisk indicates non-canonical deletion. Hyphens are used to maintain alignment throughout the reads. Within the table numbers are rounded to the nearest tenth of a percent, and bold numbers highlight frequencies greater than 1%. Samples that had two or fewer reads for a given read sequence are classified as not detected (n.d.).
ND7
ND7 is pan-edited, with 291 sites in two distinct domains: a 5΄ domain that is edited robustly in both life cycle stages, and a 3΄ domain that is predominantly edited in BF parasites (39). RNAseq of ND7 5΄ domain generated 1 133 209 reads corresponding to pre-edited sequence, 240 674 reads corresponding to edited sequence, and 990 361 reads corresponding to partially edited sequence when all samples are combined (Figure 6B). The length of the read covers the first nine canonical ES in the fully edited sequence, including three deletion sites. As with MURF2, the sequence profiles for 427wt and MGA appeared largely similar, with pre-edited and edited sequences representing a majority of the reads. The most abundant partially edited reads are also very similar, with the three most abundant sequences in 427wt ranked in the top four in MGA. The profiles of both KREN2 null and KREN3 null have subtle but distinct differences from these controls. In particular, the most frequent partially edited read is the same in 427wt, MGA, and KREN2 null samples, and its sequence differs from fully edited by a single U inserted at a non-canonical site 3΄ to ES1. To our knowledge, editing at this site has not been previously reported. This read represents 5.2% of 427wt, 4.0% of MGA, 11.0% of KREN2 null, but only 0.1% of KREN3 null, indicating that the non-canonical insertion present in this sequence preferentially occurs when both KREN1 and KREN3 activities are present. Conversely, the second most frequent partially edited read in 427wt and MGA is the most frequent partially edited read in KREN3 null, but virtually absent in KREN2 null. This sequence, which has edited ES1–ES7 and non-canonical insertion and deletion 5΄ of ES7, therefore preferentially occurs in the presence of KREN1 and KREN2 activities. These results mirror those with MURF2, indicating distinct ES preferences for KREN2 and KREN3. As with MURF2, only deletion editing is present in all of the frequently observed partially edited ND7-5΄ sequences in KREN1 only cells, and these sequences are rarely detected in other cell lines. Of the 289 855 ND7-5΄ reads in the KREN1 only sample, only 6 were fully edited, again demonstrating the inability of KREN1 to efficiently process multiple insertion ESs. Similar to observations with MURF2, the most frequent partially edited ND7-5΄ reads from both KREN2 only and KREN3 only cells exclusively contain insertion events. The top three most frequent partially edited reads in KREN3 only are the second, third, and fourth most frequent in KREN2 only, again indicating functional overlap between these endonuclease. However, the most frequent partially edited read in KREN2 only, representing 11.4% of the sample, is the fifth most frequent read in KREN3 only, at 2.7% of that sample. The sequence of this read includes the non-canonical insertion of two Us at ES2, a site that would have the deletion of a single U in canonical editing. Thus, while both KREN2 and KREN3 can cleave ES2, this site appears to have a stronger preference for KREN2. Surprisingly, 10.5% of the reads from Triple null cells are a single partially edited sequence, having a single U deleted at ES2. This read is relatively rare in other cell lines, with the 0.7% found in KREN1 only being the sole exception. The prevalence of this sequence makes it highly unlikely to be an artifact. 88.4% of the remaining reads in Triple null cells represented pre-edited sequence. Together these results demonstrate that the editing endonucleases possess distinct preferences, despite a significant degree of functional overlap between KREN2 and KREN3 in vivo, and although the three characterized endonucleases are responsible for the vast majority of editing, another uncharacterized endonuclease can function in editing.
A6
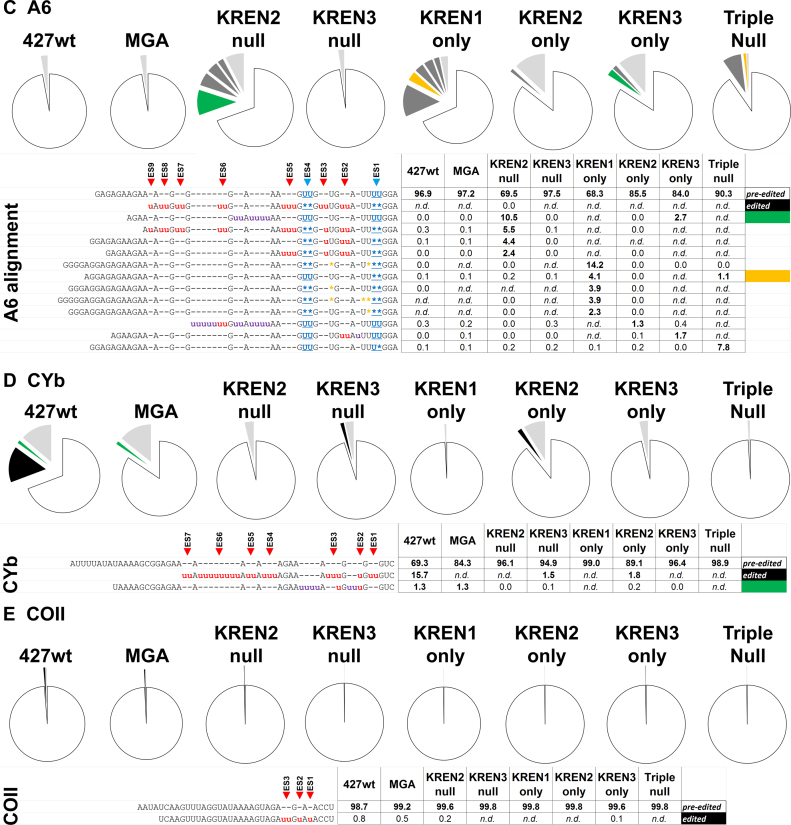
A6 is pan-edited, with 186 sites that are edited in both life cycle stages, and unique among transcripts studied here, the first editing site is a deletion site (40). RNAseq of A6 generated 699,083 reads corresponding to pre-edited sequence, 163,556 reads corresponding to partially edited sequence, and only nine reads corresponding to canonical edited sequence when all samples are combined (Figure 6C). The length of the read includes all or part of the first nine canonical ES in the fully edited sequence, including two deletion sites. The profiles for 427wt and MGA are again very similar, with roughly 97% of the reads corresponding to pre-edited sequence and no fully edited reads in both samples. All nine fully edited A6 reads were found in the KREN2 null sample, which is 0.004% of the KREN2 null reads. More than 30% of the KREN2 null reads are partially edited, with the most frequent read containing non-canonical insertion between ES5 and ES6. However, the second most common read differs from a fully edited sequence by a single U: ES3 has one U inserted instead of the canonical two. Interestingly, this nearly complete edited read sequence is also observed in 427wt, MGA and KREN3 null at a frequency between 0.1 to 0.3%. Thus, while the canonical fully edited sequence was only observed in nine reads, a nearly identical variant was observed in 12 355 reads. As editing in this region of A6 is in the 3΄ UTR, this variant sequence may represent the actual fully edited sequence in these 427-derived strains. The third and fourth most common reads in the KREN2 null sample have edited ES1–ES4 and ES1–ES5 respectively, with the caveat that ES3 again has a single U inserted, suggesting that these are precursors to the variant fully edited sequence. The KREN3 null profile is very similar to 427wt and MGA controls, with 97.5% of reads being pre-edited. The KREN1 only profile is again dominated by deletion editing, with the first, third, fourth and fifth most frequent reads having canonical deletion activity at ES1 and ES4 with additional non-canonical deletions, while the second most frequent read has canonical deletion at ES1 only. KREN2 only and KREN3 only profiles appear to be very similar, with pre-edited sequence predominant and non-canonical insertion activity present in all frequent partially edited reads. The most frequent partially edited read in KREN3 only is also the most frequent in KREN2 null, indicating that KREN1 activity is dispensable for these insertions which KREN3 alone can generate. 90.3% of the Triple null sample is pre-edited, with 7.8% of the reads having a single U deleted at ES1, and 1.1% having both canonical Us deleted at the same site. These A6 editing events in Triple null cells provide additional evidence for an unknown activity in vivo.
CYb
CYb is predominantly edited in PF, and all 13 sites are edited by insertion (41). Analysis of CYb reads indicates that complete editing can be accomplished by KREN2 activity alone. RNAseq for CYb generated 2 596 745 reads corresponding to pre-edited sequence, 56 085 reads corresponding to edited sequence and 164 884 reads corresponding to partially edited sequence when all samples are combined (Figure 6D). The predominance of pre-edited reads in this sample is expected as CYb is infrequently edited in bloodstream form cells. The length of the read includes all or part of the first seven canonical ES in the fully edited sequence, all of which are insertion sites. Unlike other transcripts, the profiles for 427wt and MGA are obviously distinct, with roughly 15.7% of the reads corresponding to fully edited sequence in 427wt, and no fully edited reads detected in MGA. This result mirrors observations from real-time PCR analysis that indicated a loss of CYb editing in MGA (Figure 2). Interestingly, the most frequent partially edited read sequence is the same in 472wt and MGA, which indicates that both cell lines still have editing similarities despite the loss of fully edited CYb reads in MGA. With the exception of MGA, fully edited CYb sequence is observed in all cell lines that contain KREN2, namely 427wt, KREN3 null and KREN2 only, but not in other samples. In both KREN2 null and KREN3 only samples, more than 96% of the reads are pre-edited with various partially edited sequences containing insertions. KREN1 only and Triple null are 99% pre-edited, indicating very little editing of CYb in either sample.
COII
COII uniquely contains its gRNA in its 3΄ UTR, has only three insertion sites, and is predominantly edited in PF (42). RNAseq data reveal that KREN3 activity alone is required to edit all three insertion sites in COII. COII analysis detected 2 057 867 reads corresponding to pre-edited sequence, 3368 reads corresponding to edited sequence and 4709 reads corresponding to partially edited sequence when all samples are combined (Figure 6E). Because COII editing is developmentally down-regulated in bloodstream form cells, the predominance of pre-edited reads is expected. Although all samples had ∼99% pre-edited reads, fully edited COII was detected in four cell lines: 427wt, MGA, KREN2 null and KREN3 only. This result is consistent with real-time PCR analyses showing COII editing only in the presence of KREN3 activity (Figure 2).
Examination of all editing events reveals distinct activity profiles for each endonuclease
A global examination of editing events throughout each dataset reveals endonuclease signatures that further support distinct editing site preferences concomitant with functional overlap between KREN2 and KREN3. While the inspection of individual partially edited read sequences from each sample yields detailed insights using the most frequently observed reads, we sought to complement this analysis with a more global approach. We analyzed all editing events independently at each potential editing site, which we define as the site between any two non-U nucleotides, and determined the proportion of all edited and partially edited reads with a given number of Us at these sites. We graphically represented the totality of editing at each site in order to enable visualization of the editing patterns in the difference cell lines (Figures 7–11; Supplementary Figure S2).
Figure 7.
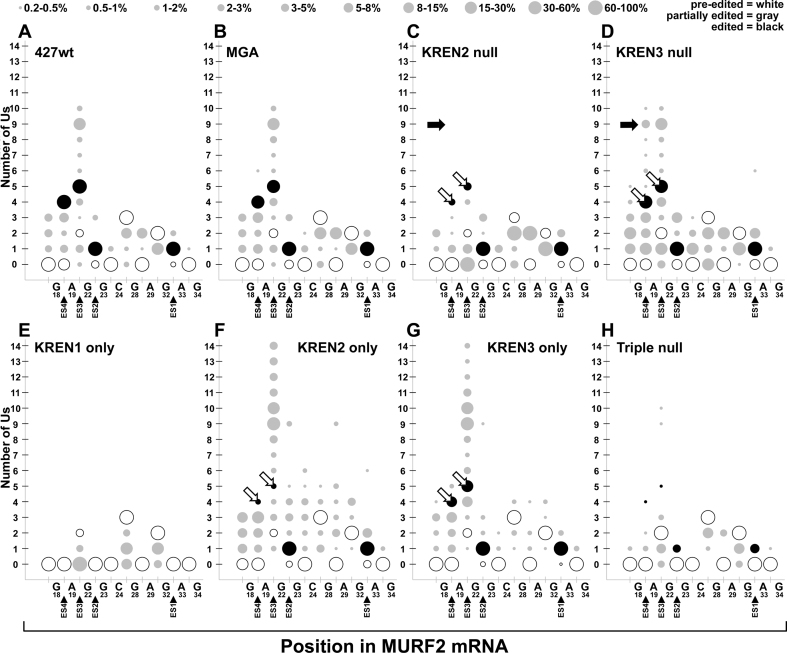
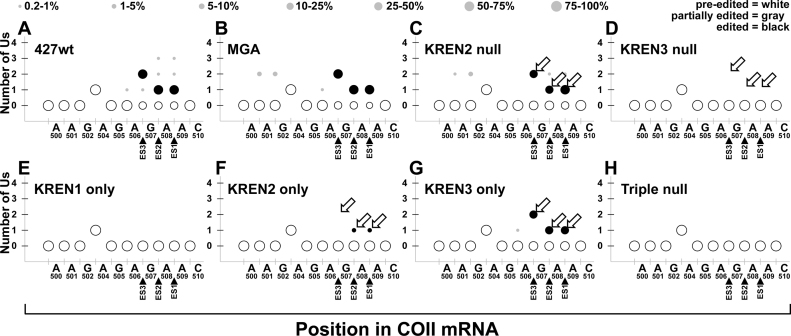
Bubble plot analyses of MURF2 edited read sequences show the frequency of site-specific insertion and deletion activities. Bubble plots show number of Us inserted at potential editing sites at the 3΄ end of MURF2 for each cell line. All mapped reads for MURF2 are analyzed to determine the amount (number of Us, y-axis) and position (location within 3΄ end of edited region, x-axis) of editing events within each cell line. The x-axis shows the MURF2 mRNA position using the non-U nucleotide sequence, with position numbering starting at the first base of the start codon in the pre-edited sequence. When the number of Us at a position matches the pre-edited number of Us, the bubble is white; when the number of Us is altered by editing, the number of Us matching a fully edited mRNA is colored black. When the number of Us at a position matches neither pre-edited nor fully edited sequence, bubbles are colored grey to denote partial editing. The size of the bubble correlates with the proportion of reads that have that number of Us at each position, with the legend at top giving the percent range for each size bubble. To decrease noise in these plots, data points that represent <0.25% of reads or fewer than five reads total are not shown. Open arrows highlight differences in fully edited ES3 and ES4 corresponding to differences in KREN2 and KREN3 activities. Closed arrows highlight predominant non-canonical insertions at ES4 that are distinctive in KREN3 null cells.
Figure 11.
Bubble plot analyses of COII edited read sequences show the frequency of site-specific insertion and deletion activities. Bubble plots of COII editing for each cell line displayed as described for Figure 8. Open arrows highlight differences in fully edited ES1, ES2 and ES3 corresponding to strong preferences for KREN3 compared to KREN2 at these sites.
MURF2
The MURF2 data reveals strikingly similar profiles for 427wt and MGA despite the diversity in the amounts of editing at nearly every site, including both insertion and deletion at both canonical and non-canonical sites (Figure 7). In addition to a large proportion of editing at the four canonical editing sites covered in this region of MURF2, both 427wt and MGA have a noticeable amount of non-canonical insertion at ES3, which is also observed in the most frequently observed partially edited read (Figure 6A). The profile for KREN2 null shows a marked decrease in canonical editing at ES4 and ES3, and a loss of the non-canonical insertions at ES3. KREN3 null by contrast appears very similar to 427wt and MGA controls, with the notable exception of increased non-canonical insertion at ES4. The KREN1 only profile displays only non-canonical deletion events. KREN2 only and KREN3 only samples have largely similar profiles with some notable differences. The most prominent feature is that both samples have increased non-canonical insertion at ES3 compared to controls. KREN2 only, however, differs from KREN3 only by having reduced canonical editing at ES4 and ES3, and generally increased non-canonical insertion between nucleotides G50 and A61. KREN2 activity therefore appears to preferentially edit ES3 and ES4, but in the absence of KREN1 activity, increased non-canonical insertions occur at the expense of canonical editing. As 99.0% of Triple null reads are pre-edited, the editing profile of this sample reflects only 1% of reads, and may therefore reflect more noise than other samples and make comparisons less tractable. Nonetheless, infrequent editing events are detected in the Triple null sample, including both insertion and deletion, and both canonical and non-canonical.
ND7
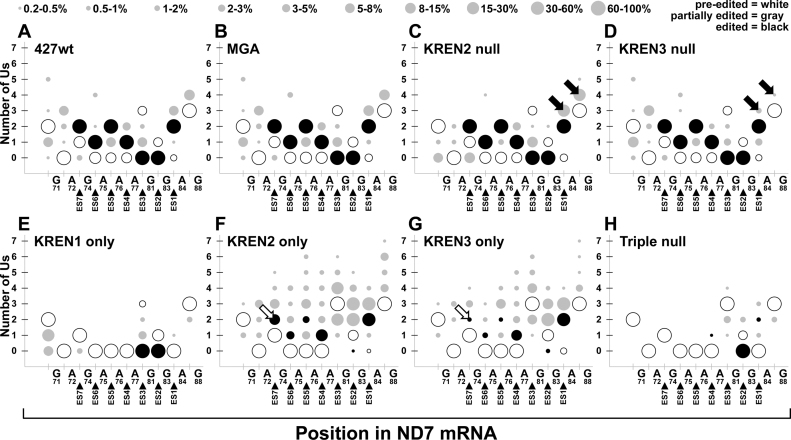
Similar analysis of editing events spanning the region that includes ES1–ES7 in ND7-5΄ also reveals distinct profiles amongst different samples (Figure 8). Again, the profiles for 427wt and MGA are overwhelmingly similar, with varying amounts of insertion and deletion editing at both canonical and non-canonical sites. KREN2 null and KREN3 null both also look similar to these controls, with notable exceptions at particular sites. KREN2 null has decreased amounts of partial editing flanking G105 that differs from the profile of 427wt, MGA and KREN3 null. KREN3 null has decreased amounts of partial editing flanking A118 that differs from the profile of 427wt, MGA and KREN2 null. The KREN1 only profile again primarily represents deletion events, including the two canonical deletion sites in ND7-5΄, ES2 and ES3. KREN1 only does have limited amounts of one U inserted at ES1. As with MURF2, analysis of ND7-5΄ editing in KREN2 only and KREN3 only samples shows mostly similar profiles with some differences. The overall KREN2 only profile has more non-canonical insertions and more canonical editing at ES5–7 compared to KREN3 only. This result is consistent with observations of MURF2, suggesting that KREN2 promotes increased non-canonical insertions relative to KREN3 in the absence of KREN1 activity. Analysis of the Triple null profile is again restricted by the predominance of pre-edited reads in this sample. With ND7-5΄, however, a partially edited single sequence represents 10.5% of the Triple null reads (Figure 6B), which translates into >90% of the reads with editing shown in this analysis. The single U deletion at ES2 that is contained in this read sequence therefore dominates the editing Triple null profile. The remaining 1.1% of the Triple null reads that contain editing thus accounts for the infrequent insertion and deletion editing restricted to the 3΄ end of the region, between A110 and G122.
Figure 8.
Bubble plot analyses of ND7-5΄ edited read sequences show the frequency of site-specific insertion and deletion activities. Bubble plots of ND7-5΄ editing for each cell line displayed as described for Figure 8. Open arrows highlight differences in fully edited ES7 corresponding to differences in KREN2 and KREN3 activities. Closed arrows highlight predominant non-canonical insertions before and at ES1 that reflect differences in KREN2 and KREN3 activities.
A6
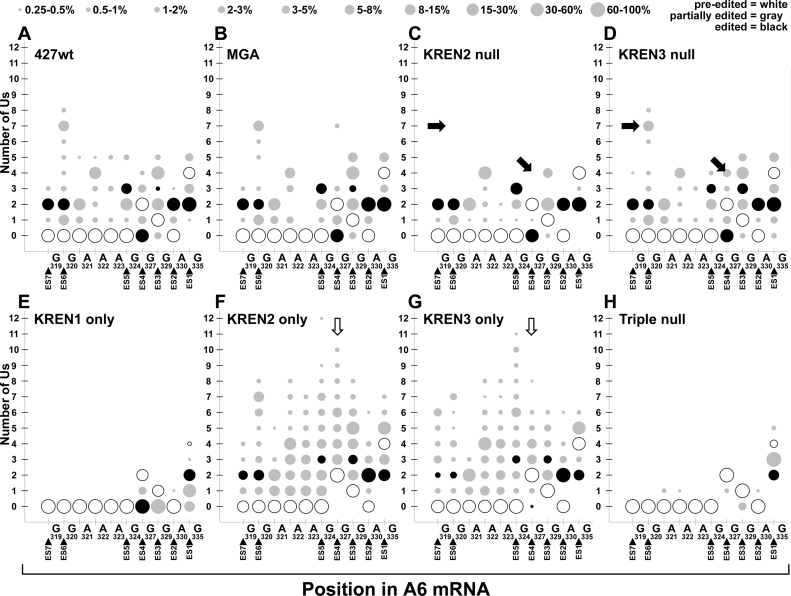
Global analysis of editing events covering ES1–ES7 in A6 also reveals insight into the endonuclease activities present in each sample (Figure 9). The profiles for 427wt, MGA, KREN2 null and KREN3 null are again broadly similar, with some exceptions. Canonical insertion editing at ES3 (+3Us) as well as non-canonical insertions at ES4 (+3 to 4 Us) and ES6 (+3 to 8 Us) are notably absent in KREN2 null. Analysis of the 427wt, MGA, and KREN3 null profiles is somewhat hampered by the predominance of pre-edited reads in these samples, and may complicate comparisons with KREN2 null. The KREN1 only profile displays nearly exclusive deletion events, including the two canonical deletion sites in A6: ES1 and ES4. Full deletion at ES4 therefore occurs in the near total absence of insertion editing at ES2 and ES3. KREN1 only does have infrequent insertion of a single U at ES2. KREN2 only and KREN3 only samples are again very similar with notable differences. Canonical deletion editing at ES1 is surprisingly frequent in both KREN2 only and KREN3 only, as is non-canonical insertion at this same site. Canonical editing at ES6 and ES7 is more frequent in KREN2 only compared to KREN3 only. While KREN2 only has more predominant non-canonical insertion events at ES4 (+3 to 10 Us), KREN3 only differs by having some canonical deletion at this site (–2 Us), and less frequent non-canonical insertion (+3 to 8 Us). In the Triple null sample, the two frequent partially edited read sequences are reflected in the deletion of one or both Us at ES1, but notably infrequent insertion and deletion events are observed, almost always corresponding to a single U change.
Figure 9.
Bubble plot analyses of A6 edited read sequences show the frequency of site-specific insertion and deletion activities. Bubble plots of A6 editing for each cell line displayed as described for Figure 8. Closed arrows highlight predominant non-canonical insertions at ES4 and ES6 that are distinctive in KREN3 null cells. Open arrows highlight non-canonical insertions at ES4 corresponding to differences in KREN2 and KREN3 activities, as well as the loss of canonical deletion editing at this site.
CYb
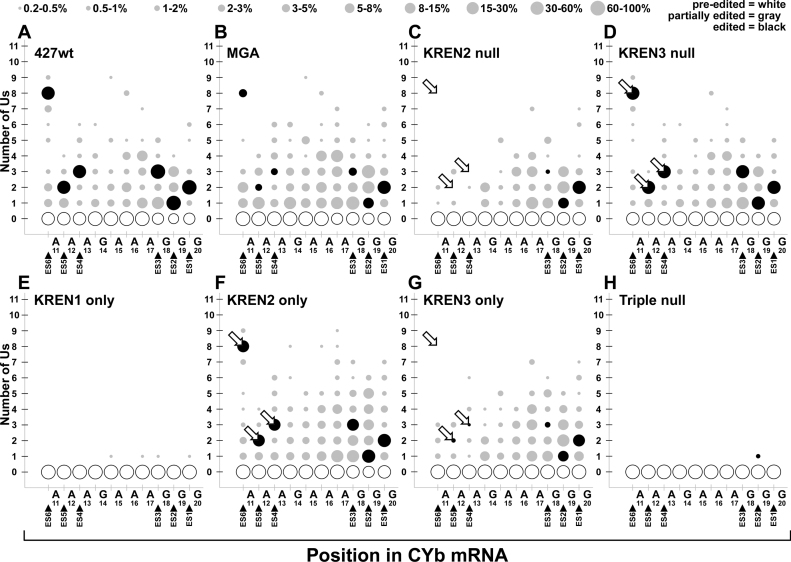
Detailed analysis of all editing events covering ES1-ES6 in CYb shows KREN2 activity correlates with full editing and that canonical editing by KREN3 is primarily restricted to ES1 and ES2 (Figure 10). Despite the complete lack of fully edited CYb reads in the MGA sample, the comparison of the 427wt and MGA editing profiles show significant similarity, with the exception being a notable decrease in canonical editing at ES3–ES6 in MGA. KREN2 null and KREN3 only have similar profiles, with the bulk of editing events found at the 3΄ end between A47 and G52, and little or no fully edited ES3-ES6. In contrast, KREN3 null and KREN2 only samples appear similar to 427wt, with a small increase in non-canonical insertions in KREN2 only at sites such as ES2. KREN1 only and Triple null samples have virtually no editing activity at all, with the infrequent insertion of a single U observed in very few reads.
Figure 10.
Bubble plot analyses of CYb edited read sequences show the frequency of site-specific insertion and deletion activities. Bubble plots of CYb editing for each cell line displayed as described for Figure 8. Open arrows highlight differences in fully edited ES4, ES5 and ES6 corresponding to strong preferences for KREN2 compared to KREN3 at these sites.
COII
Examination of all editing events for COII demonstrates that KREN3 activity is required for full editing, and although KREN2 very infrequently edits ES1 and ES2, editing of ES3 was not observed (Figure 11). The COII profiles for 427wt, MGA, KREN2 null and KREN3 only are very similar, containing full editing for all three ES. In contrast, the profiles for KREN3 null, KREN1 only, and Triple null all lack full editing for COII. The KREN2 only sample has 12 reads that contained full editing at both ES1 and ES2, but none at ES3, indicating very poor recognition of the COII insertion sites.
DISCUSSION
The data we present here show that the T. brucei editing endonucleases recognize ESs in vivo by a combination of the KREN1 endonuclease having specificity for deletion editing and KREN2 and KREN3 endonucleases having overlapping preferences for insertion editing. Previously published work suggested that KREN3 primarily edited COII sites, but this work shows much broader insertion cleavage activity for this endonuclease (12). MGA cells that we constructed with restricted endonuclease repertoires retained editing complexes, albeit with altered gradient sedimentation, even when all three endonucleases were eliminated. These complexes had substantially altered editing both in vitro and in vivo. RNAseq analysis of pre-edited, edited, and partially edited mRNAs from these cells was performed using PARERS, a custom editing analysis tool that we created to identify editing profiles that were characteristic of each cell line. These patterns reveal the relative in vivo specificity of each of the three editing complexes, and provide insight into how multiple editosomes collaborate to edit functionally distinct ESs. Analysis of these cell lines has also revealed a previously unknown structural role for the endonucleases, which provides insight into how editosomes function in vivo. Our results also indicate the presence of an unknown endonuclease activity that can result in sequence changes akin to editing in the absence of the characterized endonucleases.
To maximize the breadth of information obtained from RNAseq data, multiple transcripts were chosen based on distinct biological differences in their editing in vivo. MURF2, CYb and COII have relatively small editing domains containing 11, 13 and 3 ESs, respectively, and are almost exclusively insertion editing, with MURF2 possessing a single deletion ES. In contrast, ND7 and A6 are pan-edited, with 291 and 186 ESs respectively, and multiple deletion ES in both. The first ES in A6 is a deletion site, while all others start with an insertion ES. MURF2, ND7 5΄ domain and A6 are edited robustly in both PF and BF, while COII and CYb are preferentially edited in PF (4). Our examination of a diverse set of ESs and transcripts illustrates the non-uniformity of editing substrates in vivo, and the complex dynamics that must be at work. Although both CYb and COII are infrequently edited in BF, our approach detected editing of these mRNAs, which critically delineate ESs that have strong endonuclease specificity. Despite limited amounts of editing in vivo, the RNAseq data clearly demonstrate that CYb editing requires KREN2 and COII editing requires KREN3 in vivo. Interestingly, fully edited CYb was not detected in MGA cells by either QPCR (Figure 2) or RNAseq (Figure 10), even though partially edited species were detected (Figure 10). Because 427wt and MGA lines behave in virtually identical fashion in other assays (Figures 2–4, 6–9, and 11), the loss of fully edited CYb in MGA may be an idiosyncratic difference that arose during the cloning and isolation of this cell line. As CYb is not essential for the MGA line to survive, loss of a CYb gRNA might explain these results, although it is clear from RNAseq data that canonical editing of many CYb ESs does occur (Figure 10).
Investigation of multiple edited transcripts by RNAseq provides insight into the complex behaviors of the editing endonucleases. The substantial defects in editing in cells expressing a single endonuclease revealed by QPCR demonstrate that collaboration between endonucleases is required for complete editing of most transcripts in vivo. With the exception of COII editing, which appears to require only KREN3, expression of a single endonuclease leads to loss of editing in QPCR assays and significant alterations in editing events detected by RNAseq.
Examination of editing events by RNAseq in KREN1 only, KREN2 only and KREN3 only cells reveals that catalytic activities that follow cleavage, namely the insertion or deletion of Us, are tightly linked to the endonucleolytic event. KREN1, KREN2 and KREN3 endonucleases can cleave ESs that are canonically insertion or deletion sites, but the subsequent editing activities are highly segregated such that deletion editing occurs after KREN1 cleavage and insertion editing occurs after KREN2 or KREN3 cleavage. KREN1 cleavage only rarely results in insertion activity at canonical insertion sites (e.g. ES2 in A6, ES1 in CYb or ES1 in ND7-5΄). Similarly, while KREN2 and KREN3 do cleave canonical deletion sites, this rarely results in deletion activity, with the more frequent result being non-canonical insertion at these sites (e.g. ES1 in A6, ES4 in A6, or ES2 in ND7). Although editing generally proceeds in a 3΄ to 5΄ direction relative to the mRNA, our results demonstrate that some editing can progress in the absence of either insertion or deletion cleavage. For example, analysis of A6 editing shows that KREN2 only cells can perform insertion at ES2, ES3, ES5, ES6 and ES7 despite the total absence of deletion at ES4 (Figure 9). The converse is also true, as KREN1 only cells can perform full deletion at ES4 despite the near total absence of insertion editing. Thus, a strict dependence on editing at sites 3΄ prior to editing at sites 5΄ is not required for editing to occur in vivo. Many ESs may experience both insertion and deletion events, as editing intermediates are known to vary from the canonical sequence. ND7 5΄ and A6 RNAseq profiles clearly illustrate this point, as elimination of KREN1 leads to noticeably increased non-canonical insertion at multiple insertion ESs, for example when KREN2 only is compared to KREN3 null or when KREN3 only is compared to KREN2 null (Figures 8 and 9).
Analysis of KREN2 null and KREN3 null cells was also informative, as the interplay between the remaining insertion endonuclease and KREN1 produce editing profiles similar to 427wt and MGA controls in many cases. QPCR results from KREN3 null (Figure 2G) show that aside from strong defects in COII and COIII editing, the amounts of many other edited transcripts are barely altered. The combination of KREN1 and KREN2 activities are therefore sufficient for near normal editing of A6, MURF2, ND3, ND7 and RPS12, a conclusion that is also supported by RNAseq data. KREN3 null RNAseq profiles are very similar to both 427wt and MGA for MURF2, ND7 5΄ and A6, indicating that KREN3 activity is largely dispensable for many mRNAs. Similarly, incomplete loss of COII and ND7 editing in KREN2 null cells by QPCR (Figure 2F) is mirrored by KREN2 null RNAseq profiles that show KREN1 and KREN3 activities are sufficient for editing these two mRNAs (Figure 11). The analysed ESs in the 5΄ domain of ND7 can therefore be canonically edited by either KREN2 or KREN3 in collaboration with KREN1, but none of the endonucleases can perform this editing alone. Because the QPCR and RNAseq primers that analyse editing in A6, MURF2, and ND7 target distinct portions of each mRNA, they serve as independent assessments for the editing of these transcripts, with QPCR measuring the 5΄ most edited region and RNAseq the 3΄ most portion of the edited region. The agreement between these assays supports the conclusion that collaboration between the endonucleases is complex, as ESs can be recognized in different ways by each of the endonucleases. The mechanism of ES recognition by the endonucleases does not appear to be defined by the mRNA sequence, as analysis of the nucleotides flanking ESs preferentially cleaved by KREN2 are virtually indistinguishable from those preferentially cleaved by KREN3 (data not shown). This result is consistent with other RNase IIIs, which recognize a wide variety of dsRNA sequences with limited sequence conservation (18). Because T. brucei encodes many redundant gRNAs that can edit the same ES, the differences in KREN2 and KREN3 cleavage preferences may reflect differential gRNA characteristics that affect substrate selection.
The similarity of the A6 RNAseq profiles for 427wt, MGA, and KREN3 null samples suggests that KREN3 is dispensable for normal editing; however, the lack of canonical fully edited sequences in these samples is unexpected (Figure 9). The in vivo kinetics of the editing process is poorly understood, and the absence of canonical fully edited reads may be a limitation of the assay. In particular, the PCR amplification of cDNAs uses a pre-edited forward primer, which could prevent robust detection of the fully edited read sequence. For example, if A6 editing is either discontinuous (i.e. the primer binding site gets edited prior to all 3΄ sites being edited) or extremely rapid (i.e. mRNAs that have fully edited ES1–ES9 and pre-edited primer binding site are extremely transient) then fully edited sequences would be difficult to detect in this assay. Another possible explanation for the absence of fully edited A6 is that the examined strain has a different fully edited sequence in this region. All of the editing sites detected in the RNAseq reads are in the 3΄ UTR of the A6 mRNA, therefore editing in this region does not need to be a perfect match for the canonical sequence to be translatable. Consistent with this, an A6 read sequence that differs from the canonical sequence by a single U at ES3 was detected at a frequency of 0.3% in 427wt, 0.1% in MGA, 0.1% in KREN3 null, and 5.5% in KREN2 null. This sequence may represent a fully edited mRNA in these 427-derived strains. The predominance of this sequence in KREN2 null cells indicates that the combination of KREN1 and KREN3 activities is fully capable of editing the insertion and deletion sites at the 3΄ end of A6, but editing with just these two endonucleases differs from controls either in the order of editing events, the kinetic rate, or some other fashion.
Our RNAseq data also reveal that in the absence of all three editing endonucleases, some cleavage activity persists that originates from an unknown source. Although this activity is comparatively weak in MURF2, COII, and CYb, the frequency of single U deletion at a single ES in ND7-5΄ and A6 supports the conclusion that cells have a bona fide endonuclease activity outside of the three characterized editing endonucleases (Figure 6). Although this novel cleavage activity was not detected in vitro using the Triple-site substrate (Figure 4), this likely reflects either the limited dynamic range of the assay, or the possibility that the unknown endonuclease does not recognize this particular substrate in vitro. Indeed, this RNA substrate is derived from COII sequence, and very little editing activity was detected for this mRNA in RNAseq data from Triple null cells (Figure 11). RNAseq data in general indicate that this cleavage activity in the Triple null cells is relatively weak, as the vast majority of partially edited read sequences are typically lower abundance in Triple null cells when compared to other samples. While the source of this endonuclease activity is unknown, other editosome proteins have been suggested as endonucleases. Although KREPA3 lacks an identifiable catalytic motif, it has been proposed to act as an endonuclease, as recombinant KREPA3 cleaves atypical RNA substrates in vitro (43). Additionally, an unidentified endonuclease that sediments distinctly from ∼20S editosomes and cleaves A6 at ES1 and CYb at ES2 in a gRNA-independent manner has been previously reported (44). The biological significance of the lower frequency cleavage by the unknown endonuclease is uncertain; it may be spurious, i.e. have no function associated with editing, but we cannot exclude a role in editing.
The endonuclease activities of KREN1, KREN2 and KREN3 appear to be in competition with each other, such that loss of one activity increases activity of the other. This phenomena can be observed in vitro, as the amount of cleavage product specific for each endonuclease increases when the corresponding endonuclease is the only one expressed, particularly with KREN3 (Figure 4). The increase in specific in vitro cleavage activity in KREN1 only, KREN2 only, and KREN3 only cell lines is consistent with increased relative amount of each type of editosome. This suggests that the endonucleases are stabilized by interactions with other editosome proteins that are normally limiting. This conclusion is also supported by the RNAseq data, where non-canonical insertions increase in the absence of KREN1 activity, as noted above. This increase is manifest in two ways, both in the number of ESs with non-canonical insertions as well as the number of U insertions at each site. A potential explanation for the increased non-canonical insertions is that KREN1 activity normally re-edits ES that have more than the canonical number of Us inserted, deleting them to match gRNA template. Thus it appears that a stoichiometric balance between the endonucleases exists, and this conclusion is also supported by sedimentation analysis of isolated editosomes.
Analysis of editosome sedimentation reveals that the endonucleases also perform a structural role in the complex, as cells expressing a single endonuclease have significant dissociation of KREPA1 from the ∼20S editosome. This result is consistent with a stoichiometric balance for the three endonucleases, as the expression of two endonucleases results in normal editosome sedimentation, apparently compensating for the loss of one endonuclease. Previous experiments have shown that isolated editosomes containing a single endonuclease sediment at ∼20S, thus supporting the contention of a disrupted stoichiometric balance of the endonucleases in vivo (14,16). A stronger sedimentation defect occurs when all three endonucleases are eliminated, such that loss of KREPA1 and KRET2 from the 20S region are apparent. Interactions between the endonucleases and KREPA1 have been circumstantially indicated previously (45), and a general association between the insertion subcomplex (containing KREPA1) and the endonucleases has been shown (15,16). Recent cross-linking mass spectrometry experiments have shown that KREN1, KREN2, and KREN3 are all in distinct complexes, and each endonuclease has proximity to KREPA1 and KRET2, consistent with our results here (19). How editosomes collaborate during the editing process has largely remained unclear, as existing data did not distinguish between editosomes that could be: (i) stably processive in vivo, with multiple ∼20S editosomes remaining on a substrate from beginning to end of editing; (ii) dynamically coming on and off substrate at various ESs, with each endonuclease-specific ∼20S editosome being exchanged as needed or (iii) partially stable on substrates, with only endonucleases and associated partner proteins dynamically exchanging while the remainder of the editosome persisted on the substrate. Our discovery that the endonucleases are required for retaining components of the insertion subcomplex strongly suggests that the latter hypothesis is unlikely, as the remainder of the editosome is structurally instable in the absence of endonuclease. Distinguishing between the first two models, however, will require additional experimentation.
Because endonucleolytic cleavage is the first step in the series of catalytic events performed by editosomes, regulation of ES recognition is a logical point for controlling the entire process. This work reveals the complex nature of the distinct and overlapping specificities of these editing endonucleases, which reflects the vast diversity of ESs in vivo. Recent evidence has shown that multiple editosome proteins have distinct functional differences in PF and BF stages, suggesting the developmental regulation of editing may be a consequence of changes within the editosome (30). The same principle may developmentally alter the function of the endonucleases, with a shift in ES recognition subsequently resulting in the observed changes in editing overall. Indeed, the preferences we observe for the editing endonucleases in BF may differ in PF, and such changes could be crucial for the developmental regulation of editing. Similar developmental shifts in the function of KREPB6, KREPB7 and KREPB8 could facilitate this change as well, thereby regulating mitochondrial gene expression to result in the necessary adaptations for energy production in radically different host environments.
ACCESSION NUMBERS
SRR4450419, SRR4450418, SRR4450417, SRR4450416, SRR4450382, SRR4450381, SRR4450383, SRR4450317.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Matthew K. Gould, Achim Schnaufer, and George Cross for sharing the plasmids, Irina Kurtz for technical assistance, Bob Morrison and Isabelle Phan for bioinformatics assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [AI014102 to K.S.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K.. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005; 30:97–105. [DOI] [PubMed] [Google Scholar]
- 2. Aphasizheva I., Aphasizhev R.. U-insertion/deletion mRNA-editing holoenzyme: definition in sight. Trends Parasitol. 2016; 32:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Read L.K., Lukes J., Hashimi H.. Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley. Interdiscip. Rev. RNA. 2016; 7:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schnaufer A., Domingo G.J., Stuart K.D.. Natural and induced dyskinetoplastid trypanosomatids: how to live without mitochondrial DNA. Int. J. Parasitol. 2002; 32:1071–1084. [DOI] [PubMed] [Google Scholar]
- 5. Trotter J.R., Ernst N.L., Carnes J., Panicucci B., Stuart K.. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005; 20:403–412. [DOI] [PubMed] [Google Scholar]
- 6. Carnes J., Trotter J.R., Ernst N.L., Steinberg A.G., Stuart K.. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16614–16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ernst N.L., Panicucci B., Igo R.P. Jr, Panigrahi A.K., Salavati R., Stuart K.. TbMP57 is a 3΄ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003; 11:1525–1536. [DOI] [PubMed] [Google Scholar]
- 8. Kang X., Rogers K., Gao G., Falick A.M., Zhou S., Simpson L.. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McManus M.T., Shimamura M., Grams J., Hajduk S.L.. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001; 7:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schnaufer A., Panigrahi A.K., Panicucci B., Igo R.P. Jr, Salavati R., Stuart K.. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001; 291:2159–2162. [DOI] [PubMed] [Google Scholar]
- 11. Huang C.E., Cruz-Reyes J., Zhelonkina A.G., O’Hearn S., Wirtz E., Sollner-Webb B.. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 2001; 20:4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K.. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008; 28:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carnes J., Ernst N.L., Wickham C., Panicucci B., Stuart K.D.. KREX2 is not essential for procyclic or bloodstream form Trypanosoma brucei. PLoS ONE. 2012; 7:e33405–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Stuart K.D.. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006; 12:1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo X., Carnes J., Ernst N., Winkler M., Stuart K.D.. KREPB6, KREPB7, and KREPB8 are important for editing endonuclease function in Trypanosoma brucei. RNA. 2012; 18:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carnes J., Zelaya-Soares C., Wickham C., Stuart K.. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J. Biol. Chem. 2011; 286:19320–19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Macrae I.J., Doudna J.A.. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin Struct Biol. 2007; 17:138–145. [DOI] [PubMed] [Google Scholar]
- 18. Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley. Interdiscip. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDermott S.M., Luo J., Carnes J., Ranish J.A., Stuart K.. The architecture of Trypanosoma brucei editosomes. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6476–E6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang X., Gao G., Rogers K., Falick A.M., Zhou S., Simpson L.. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alatortsev V.S., Cruz-Reyes J., Zhelonkina A.G., Sollner-Webb B.. Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex. Mol. Cell. Biol. 2008; 28:2437–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dean S., Gould M.K., Dewar C.E., Schnaufer A.C.. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14741–14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ammerman M.L., Presnyak V., Fisk J.C., Foda B.M., Read L.K.. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010; 16:2239–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo X., Ernst N.L., Carnes J., Stuart K.D.. The Zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei. PLoS ONE. 2010; 5:e8913–e8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koslowsky D.J., Bhat G.J., Read L.K., Stuart K.. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell. 1991; 67:537–546. [DOI] [PubMed] [Google Scholar]
- 26. Bundschuh R., Altmuller J., Becker C., Nurnberg P., Gott J.M.. Complete characterization of the edited transcriptome of the mitochondrion of Physarum polycephalum using deep sequencing of RNA. Nucleic. Acids Res. 2011; 39:6044–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. David V., Flegontov P., Gerasimov E., Tanifuji G., Hashimi H., Logacheva M.D., Maruyama S., Onodera N.T., Gray M.W., Archibald J.M. et al. . Gene loss and error-prone RNA editing in the mitochondrion of perkinsela, an endosymbiotic kinetoplastid. MBio. 2015; 6, doi:10.1128/mBio.01498-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Simpson R.M., Bruno A.E., Bard J.E., Buck M.J., Read L.K.. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA. 2016; 22:677–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merritt C., Stuart K.. Identification of essential and non-essential protein kinases by a fusion PCR method for efficient production of transgenic Trypanosoma brucei. Mol Biochem. Parasitol. 2013; 190:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McDermott S.M., Guo X., Carnes J., Stuart K.. Differential editosome protein function between life cycle stages of Trypanosoma brucei. J. Biol Chem. 2015; 290:24914–24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barrett B., LaCount D.J., Donelson J.E.. Trypanosoma brucei: a first-generation CRE-loxP site-specific recombination system. Exp. Parasitol. 2004; 106:37–44. [DOI] [PubMed] [Google Scholar]
- 32. Scahill M.D., Pastar I., Cross G.A.. CRE recombinase-based positive-negative selection systems for genetic manipulation in Trypanosoma brucei. Mol Biochem. Parasitol. 2008; 157:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 34. Panigrahi A.K., Gygi S., Ernst N., Igo R.P. Jr, Palazzo S.S., Schnaufer A., Weston D., Carmean N., Salavati R., Aebersold R. et al. . Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001; 21:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stuart K., Panigrahi A.K., Schnaufer A.. Gott JM. Identification and characterization of trypanosome RNA editing complex components. Methods in Molecular Biology. 2004; 265, Totowa, NJ: Humana Press Inc; 273–291. [DOI] [PubMed] [Google Scholar]
- 36. Sabatini R., Hajduk S.L.. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 1995; 270:7233–7240. [DOI] [PubMed] [Google Scholar]
- 37. Cruz-Reyes J., Rusché L.N., Piller K.J., Sollner-Webb B.. T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell. 1998; 1:401–409. [DOI] [PubMed] [Google Scholar]
- 38. Feagin J.E., Stuart K.. Developmental aspects of uridine addition within mitochondrial transcripts of Trypanosoma brucei. Mol. Cell. Biol. 1988; 8:1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koslowsky D.J., Bhat G.J., Perrollaz A.L., Feagin J.E., Stuart K.. The MURF3 gene of T. brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell. 1990; 62:901–911. [DOI] [PubMed] [Google Scholar]
- 40. Bhat G.J., Koslowsky D.J., Feagin J.E., Smiley B.L., Stuart K.. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990; 61:885–894. [DOI] [PubMed] [Google Scholar]
- 41. Feagin J.E., Jasmer D.P., Stuart K.. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987; 49:337–345. [DOI] [PubMed] [Google Scholar]
- 42. Benne R., Van den Burg J., Brakenhoff J.P.J., Sloof P., Van Boom J.H., Tromp M.C.. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contain four nucleotides that are not encoded in the DNA. Cell. 1986; 46:819–826. [DOI] [PubMed] [Google Scholar]
- 43. Brecht M., Niemann M., Schlüter E., Müller U.F., Stuart K., Göringer H.U.. TbMP42, a protein component of the RNA editing complex in African trypanosomes has endo-exoribonuclease activity. Mol. Cell. 2005; 17:621–630. [DOI] [PubMed] [Google Scholar]
- 44. Piller K.J., Rusché L.N., Cruz-Reyes J., Sollner-Webb B.. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA. 1997; 3:279–290. [PMC free article] [PubMed] [Google Scholar]
- 45. Law J.A., Huang C.E., O’Hearn S.F., Sollner-Webb B.. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell. Biol. 2005; 25:2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.