Abstract
Ionizing radiation produces clustered damage to DNA which is difficult to repair and thus more harmful than single lesions. Clustered lesions have only been investigated in dsDNA models. Introducing the term ‘clustered damage to G-quadruplexes’ we report here on the structural effects of multiple tetrahydrofuranyl abasic sites replacing loop adenines (A/AP) and tetrad guanines (G/AP) in quadruplexes formed by the human telomere d[AG3(TTAG3)3] (htel-22) and d[TAG3(TTAG3)3TT] (htel-25) in K+ solutions. Single to triple A/APs increased the population of parallel strands in their structures by stabilizing propeller type loops, shifting the antiparallel htel-22 into hybrid or parallel quadruplexes. In htel-25, the G/APs inhibited the formation of parallel strands and these adopted antiparallel topologies. Clustered G/AP and A/APs reduced the thermal stability of the wild-type htel-25. Depending on position, A/APs diminished or intensified the damaging effect of the G/APs. Taken together, clustered lesions can disrupt the topology and stability of the htel quadruplexes and restrict their conformational space. These in vitro results suggest that formation of clustered lesions in the chromosome capping structure can result in the unfolding of existing G-quadruplexes which can lead to telomere shortening.
INTRODUCTION
Endogenous and exogenous chemicals and ionizing radiation damage genomic DNA by causing the formation of various base lesions, abasic sites, strand breaks, and cross-linkages (1). The biological effects of such damage is not only associated with the type and site of lesion but also with their distribution in the DNA (2). Reactive chemicals create mainly single or isolated damage sites. Ionizing radiation and radiomimetic anticancer agents, however, also induce clusters of damage in the DNA, both in solution and in cells. Although the chemical structure of lesions produced by chemicals and ionizing radiations are similar, the unique feature of the latter is the creation of clustered lesions. Ionizing radiation damages the target molecules along its track, and thus clusters are induced at very low radiation dose. Single radiation hits can also induce damage clusters. The type of radiation determines the damage spectrum (3).
By definition, clustered DNA damage or multiple DNA damage sites form when two or more lesions appear in the DNA strands within one to two helical turns in the double-stranded DNA. These are 10–20 base pairs. The notion of clustered DNA lesions first appeared in the 1990s. Since then, these types of lesions have been found and characterized in vitro in various cell types, for both irradiated and non-irradiated samples (2,4). Cells maintain genome integrity by repairing isolated DNA lesions using base excision repair (BER) and strand break repair pathways. Clustered lesions are, however, difficult to repair and are therefore very detrimental to cells and they can lead to mutagenesis, cancer, aging, and neurological disorders in humans. In repair-capable cells, the probability of non-repairing or misrepairing a single damaged site is 1 in 109 to 1010 bp. In a cluster this could be orders of magnitude greater (5).
Lesion types include abasic (AP) sites, both apurinic and apyrimidinic, oxidized sugar derivatives, a plethora of base modifications that include oxidized bases, such as 8-oxoguanine (8-oxoG), 8-oxoadenine (8-oxoA), thymine glycol, cytosine glycol, dihydrothymidine and single- and double-strand breaks. The most frequently occurring lesions are the purine AP sites formed either spontaneously by hydrolysis of the C1’-N9 glycosidic linkage of purine nucleotides or enzymatically as intermediates in the BER pathway. Esposito et al. (6) were the first to study the effects of abasic sites on quadruplex structures. The most common oxidation product is 8-oxoG and its further oxidized derivatives which are generated by hydroxyl free radical attack at the C8 position of guanine. Both the AP sites and 8-oxos can also form in clusters (7–11). A double-strand break (DSB) is thought to consist of two nearby single-stranded breaks (SSB) on the opposite DNA strands. Before the concept of clustered damage was formulated, DSBs were considered typical radiation injuries, although in human cells DSBs constitute only about 30% of the total damages. DSB is malignant damage due to its mutagenicity and inhibition of DNA replication. The clusters containing base lesions, which remained unrecognized for a long time, can however produce even more complex biological outcomes (11,12). Non‐DSB clusters compromise the efficiency of eukaryotic DNA damage repair processes by reducing the ability of glycosylases to excise base lesions and AP endonucleases to incise the AP sites within a cluster, probably due to reduced steric accessibility (13).
Most human cell lines do not accumulate clusters. HF19, TK6 and WIL2-NS alone are known to do so, currently. These clusters contain mainly oxidized bases. Normal human tissues and cells however, do accumulate multiple damage sites (14). While endogenous clusters were found only at low frequencies in non-irradiated normal human cells and tissues, in cells of smokers large numbers of clusters containing oxidized bases and AP sites were detected, suggesting that the normal DNA repair capacity can be overwhelmed (15). Furthermore, it is not only ionizing radiation, X-rays too can induce abasic and oxidized base clusters in human cells (16). UV radiation is generally believed to produce only clusters of cyclobutane pyrimidine dimers but long-wavelength UV (290–400 nm) radiation also creates oxidized bases, AP sites, and strand breaks, albeit in low yield. The number of these clusters due to UVB and UVA is negligible compared to the levels of pyrimidine dimers but they cannot be ignored (17).
Clustered natural damage to DNA has been mostly investigated with double-stranded DNA models in vitro (3,4,11,17). Less is known about clusters of natural lesions in non-canonical, unusual DNA and RNA structures, such as the G-quadruplex which is currently the most comprehensively studied non-B model (18). Regarding G-quadruplex modifications that can be defined retroactively as clustered modifications, multiple mutations by natural and synthetic bases of the guanines in the G-tetrads as well as of the bases in loops should be mentioned (19–24). In this study, we only deal with clustered natural lesions in quadruplexes.
Quadruplexes functioning as regulatory elements in many gene promoters and as the capping structures of telomeres of linear eukaryotic chromosomes are potential targets of radiation and various chemical reactants that can create clustered damage. For this reason, it is important to study how the patterns of these clusters affect the stability, folding and biochemistry of wild-type quadruplexes formed from various DNA sequences. For instance, how many lesions and in what distribution (positions) can the formation of a quadruplex be prevented? Alternatively, what cluster arrangements can unfold an existing stable quadruplex in solution? Here we report on the first answers to these questions with stabilized AP sites that mimic one of the most frequent natural DNA lesions incorporated site-specifically into potentially quadruplex-forming deoxyoligonucleotides. More than forty such sequences were prepared and the annealed structures were investigated by CD, UV absorption and NMR spectroscopies, and gel electrophoresis. The results shed light on crucial damage positions that yielded dramatic effects on the stability and folding changes of quadruplexes arising from human telomere sequences.
MATERIALS AND METHODS
The oligonucleotides were purchased from Sigma-Aldrich (Haverhill, UK) and dissolved in 1 mM sodium phosphate + 0.3 mM EDTA, pH 7.2 to provide stock solutions of ∼10 mM. Precise concentrations were determined on the basis of absorption at 260 nm, measured in a UNICAM 5625 UV/Vis spectrophotometer (Cambridge, UK) at 90°C using the following molar strand absorption coefficients determined as described in (25): 236 900 M−1cm−1 for wild-type htel-22, 222 500, 208 100 and 193 700 for the one, two and tree adenine AP site, resp., containing htel-22s, and 205 700 for the TTA loop triple abasic htel-22 sequence (see Supplementary Table S1); then 261 900 for wild-type htel-25, 247 500 and 218 700 for loop one and triple adenine AP site, resp., containing htel-25, 251 200 for the one guanine AP site and 236 800 M−1cm−1 and 240 500 for the guanine + adenine or guanine + guanine double AP site containing htel-25s. Melting curves were measured in a UV–vis spectrophotometer (Varian Cary 4000, Mulgrave, Victoria, Australia) from 1°C to 95°C and back. The temperature was increased/decreased in 1°C steps and the samples were equilibrated for 2 min before each measurement.
Purity, as the length homogeneity of the desalted samples was checked by denaturing electrophoresis (20% gel with 6.2 M urea running at 50°C for 1 h at the power output of 25 W). Native polyacrylamide gel electrophoresis (PAGE) was run in a temperature-controlled electrophoretic instrument (SE-600, Hoefer Scientific, San Francisco, CA, USA). Gel concentration was 16% (29:1 monomer to bis ratio, Applichem, Darmstadt, Germany). About two micrograms of DNA were loaded on the 14 cm × 16 cm × 0.15 cm gel. Samples were electrophoresed at 21°C for 19 h at 30 V. The gel was stained with Stains All (Sigma, St. Louis, MO, USA) after the electrophoresis and scanned using a Personal Densitometer SI, model 375-A (Molecular Dynamics, Sunnyvale, CA, USA).
CD measurements were carried out in a Jasco 815 (Tokyo, Japan) dichrograph in 1-cm path-length quartz Hellma cells placed in a thermostated cell holder at 23°C. The DNA strand concentration was 3.4 μM. A set of four scans was averaged for each sample with a data pitch of 0.5 nm and 100 nm min−1 scan speed. The CD signal was expressed as the difference in molar absorption, Δε of the left- and right-handed circularly polarized light, molarity being related to DNA base.
The 1D 1H spectra were measured at 700 MHz using a Bruker Avance III NMR spectrometer equipped with a triple resonance room temperature probe using the WATERGATE pulse sequence (26), including 1-ms rectangular selective pulses to suppress the residual signal of water. The spectra were processed using TopSpin 3.2 (Bruker, USA). Samples were prepared by dissolving the oligonucleotides in 20 mM potassium phosphate pH 7. The solutions were annealed by heating to 90°C and cooling at room temperature overnight. The resulting strand concentration was approximately 0.8 mM for the htel-22s, and 0.2 mM for the htel-25s.
RESULTS
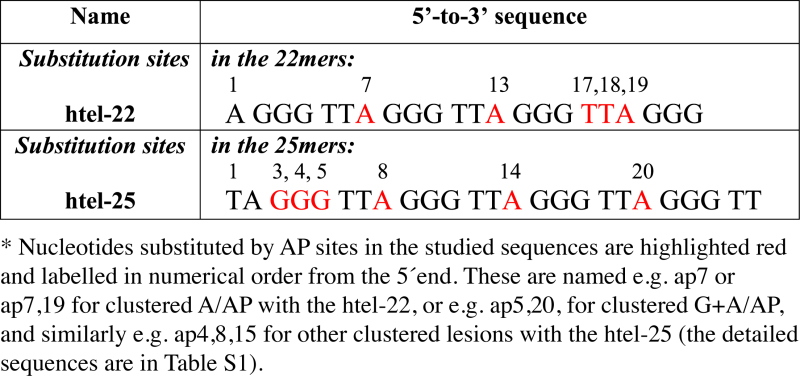
Human telomere (htel) DNA forms various quadruplex types depending on the primary structure and experimental conditions (Supplementary Figure S1). To determine the effect of clustered damage on the properties of htel quadruplexes, we used two sets of sequences modified by tetrahydrofuranyl AP sites (dSpacer) in selected positions: the 22 nucleotide long d[AG3(TTAG3)3], abbreviated as htel-22, and the d[TAG3(TTAG3)3TT], abbreviated as htel-25 (Table 1). The modified sequences denominated as api or api,j contained an AP site instead of the i- or j-th nucleotide. The full sequences are listed in Supplementary Table S1.
Table 1. Oligonucleotides used in this study*.
|
Quadruplexes arising from the htel-22 d[AG3(TTAG3)3]
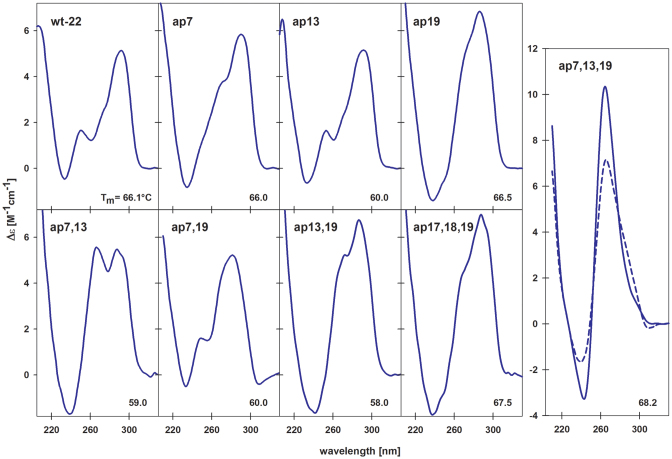
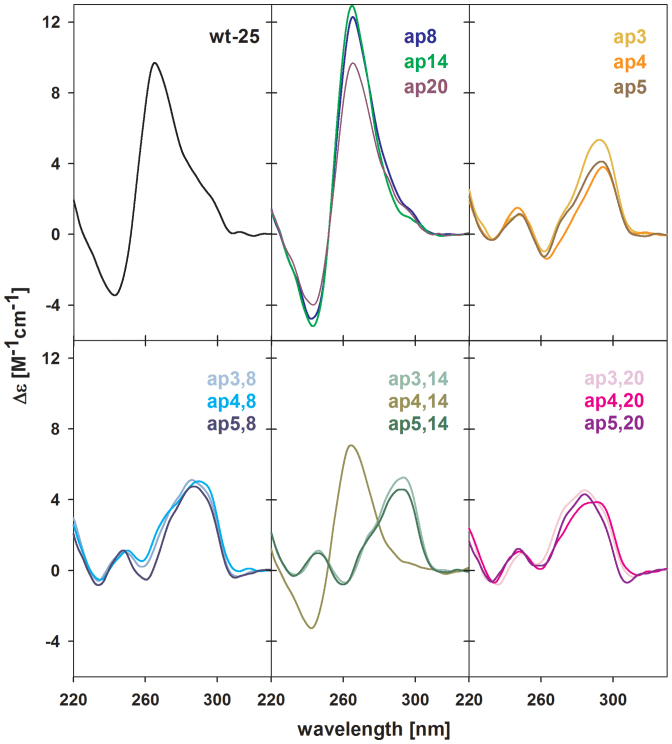
Abasic quadruplexes of d[AG3(TTAG3)3] were used in our previous NMR work to explore the effects on the structure of single adenine AP (A/AP) sites in the loops (25). To determine the impact of clustered (double and triple) loop AP lesions, the same htel-22 model was used here. The wild-type (wt) htel-22 formed a mixture of antiparallel and hybrid (h) parallel/antiparallel, so called (3+1) folds (Supplementary Figure S1) in 10–165 mM K+ at micromolar strand concentrations used for CD and UV absorption measurements (27–29). The CD spectrum of the one, two and three loop A/AP containing d[AG3(TTAG3)3] analogs, indicated formation of quadruplex-type folded structures in K+ solutions (Figure 1). In a previous study (25) we showed that the ap19, containing the AP site in the third loop (Supplementary Table S1) formed a (3+1) hybrid-2 type (h-2) quadruplex (Supplementary Figure S1d) characterized by a dominant positive CD band at 290 nm with a shoulder on its short wavelength site (Figure 1). The AP site thus stabilized a double chain reversal (propeller) type loop. The ap13 formed an even more heterogeneous mixture of conformations with only modest CD spectral changes compared to wt htel-22 but with ∼6°C lower Tm (Figure 1, Supplementary Table S2). Finally, we observed a (3+1) hybrid-1 type (h-1) for ap7 (Supplementary Figure S1c), but only at high, millimolar DNA concentrations. In low DNA concentrations, close to those used for CD, the ap7 formed a mixture of quadruplex species as shown by 1H NMR (25). These provided a CD spectrum with slightly higher CD shoulder around 260 nm than the htel-22 wt (Figure 1) indicating an increased proportion of parallel strands in its quadruplex structure. Quadruplexes of ap7 and ap19 were similarly thermostable as the wt htel-22 (Supplementary Table S2). In contrast, they were clearly destabilized in 100 mM Na+ (Supplementary Table S3).
Figure 1.
CD spectra of htel-22 wt, and of its single (upper row), double (bottom row) and triple (side panel) A/AP site containing analogs measured in the presence of 100 mM K+ (10 mM K-phosphate buffer, pH 7 + 85 mM KCl) at 23°C. The dashed blue spectrum of ap7,13,19 was measured at 23°C and the solid line spectrum at 45°C.
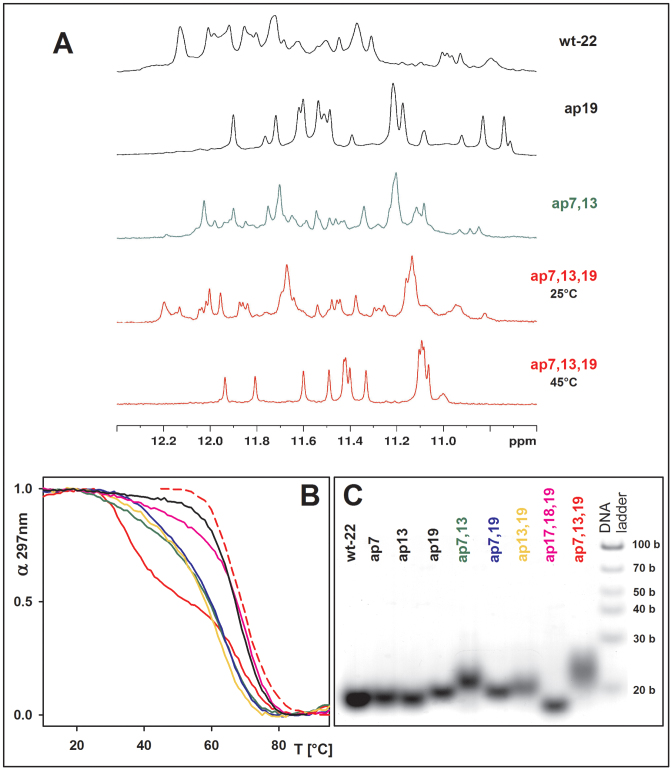
The CD spectral shapes of all multiple A/AP containing htel-22s differed from the wt (Figure 1, bottom panel): the CD spectrum of ap7,13 displayed two positive peaks of almost equal height at 270 and 290 nm. This indicated a mixture of quadruplex folds, as proven by 1H NMR spectroscopy (Figure 2A), with even greater number of parallel strands than in the single A/AP quadruplexes. The spectrum of ap7,19 with a positive peak around 286 nm also refers to a mixture of structures. The CD spectrum of ap13,19 that contained one of the two AP sites in the third loop (position 19) was similar to the spectrum of ap19 quadruplex, identified as an h-2 topology (25). The spectrum of ap17,18,19 with three AP sites substituting for the whole third TTA loop, was virtually identical to that of the ap19 (Figure 1, bottom panel) and its Tm value was close to both ap19 and wt (Supplementary Table S2, panel A), while the thermal stabilities of the double A/AP containing htel-22s were much lower (Figure 2B, and Supplementary Table S2). The two loop A/AP sites were thus detrimental to quadruplex stability. The ap7,13,19, which contained an A/AP site in each of the three TTA loops displayed a CD spectrum with a dominating positive band close to 260 nm (Figure 1, right panel) characteristic of parallel quadruplexes (28). This quadruplex unfolded in two steps (Figure 2B). Keeping the solution at temperatures higher than the temperature at which the first step was completed close to 50°C, the 260 nm CD band distinctly increased (Figure 1 and Supplementary Figure S2) indicating formation of a stable parallel fold. The 1H NMR spectrum revealed (Figure 2A) that the low-temperature mixture of ap7,13,19 transformed into a nearly single quadruplex fold at a higher temperature. In this way we observed a parallel quadruplex of the htel DNA in aqueous solution. The parallel quadruplex then melted in one step (Figure 2B) with a Tm value ∼68°C (Figure 1 and Supplementary Table S2A), which was higher than the Tm of the wild-type. Cooling the denatured ap7,13,19 back to 50°C immediately restored its parallel form. Its high 260 nm CD band with a slow kinetics diminished on further cooling and returned to the form observed at room temperature (Supplementary Figure S2). The parallel quadruplex of ap7,13,19 was only formed in K+ solution. In 100 mM Na+ the quadruplex was by nearly 19°C destabilized as compared with the wt htel-22 (Supplementary Table S3).
Figure 2.
(A) Imino proton region of 1H NMR spectra, (B) normalized UV absorption melting curves monitored at 297 nm (1 stands for native, 0 for denatured state) and (C) native PAGE of the wt-22 and of its A/AP containing analogs. The same colors are used for the oligonucleotides in particular subfigures. Red dashed line in (B) corresponds to melting of ap7,13,19 monitored at 265 nm by CD. All measurements were done in 10 mM K-phosphate buffer, pH 7 + 85 mM KCl, at 25°C in A and C, and the last NMR spectrum at 45°C. NMR spectra were measured at ∼0.8 mM DNA strand concentration.
All clustered A/AP sites containing htel-22 quadruplexes were intramolecular (Figure 2C). Propeller type loops generally inhibit the migration of quadruplexes in gel (the parallel quadruplexes run more slowly than antiparallel quadruplexes (30)). This was observed here with all sequences containing AP sites in two or three loops, compared with the wt or with ap17,18,19. The mixture of ap7,13 yielded a fuzzy band. The slow mobility in gel of quadruplexes with AP sites in two loops suggests that these may form quadruplexes containing two propeller loops, while ap17,18,19 had only one such loop. The ap7,13,19 that contained an AP site in all three loops, migrated at the lowest rate of all and was extremely fuzzy (Figure 2C). The band became narrower on PAGE running at 35°C (Supplementary Figure S3A). In Na+ containing gel the ap7,13,19 ran like the other A/AP containing htel-22s as it also remained in antiparallel quadruplex (Supplementary Figure S3B).
Quadruplexes of the htel-25 d[TAG3(TTAG3)3TT] analogs
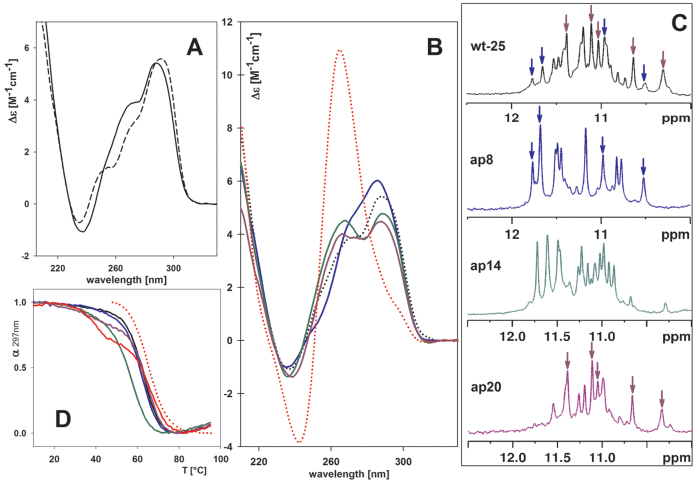
The quadruplex arising from the htel-25 d[TAG3(TTAG3)3TT] sequence in K+ solution was reported by Phan et al. (31) as adopting the (3+1)-type h-2 topology as the predominant fold at 0.5–5 mM strand concentrations based on NMR measurement. Its CD spectrum (taken at micromolar concentration) contained a dominant positive peak at 290 nm followed by a shoulder around 265 nm and a negative band close to 240 nm (Figure 3A). The shoulder was higher than that of the htel-22 quadruplex, which is in accord with the increased proportion of parallel strands in the htel-25 quadruplex structure. This sequence was chosen for further experiments as it forms a stable hybrid structure. We wanted to elucidate the effect of loop A/AP and tetrad G/AP sites occurring in the hybrid quadruplex form.
Figure 3.
(A) CD spectra of htel-22 (black dashed line) and htel-25 (black line) wt΄s quadruplexes; (B) CD, (C) 1H NMR spectra and (D) normalized UV absorption melting curves monitored at 297 nm of htel-25 wt (black, dotted line in B) and of its A/AP containing analogs ap8 (blue), ap14 (green), ap20 (violet) and ap8,14,20 (red). The red dotted line represents normalized CD melting curve monitored at 265 nm preequilibrated at 45°C. All spectra were measured in 100 mM K+ (10 mM K-phosphate buffer, pH 7 + 85 mM KCl) at 23°C or at 45°C for ap8,14,20 in (B). NMR spectra were measured at ∼0.2 mM DNA strand concentration.
Single A/AP sites in the loops of htel-25 analogs
Figure 3B illustrates the CD spectra of the three single A/AP site containing analogs of the wt htel-25, the ap8, ap14 and ap20, which are analogous to ap7, ap13 and ap19 of htel-22 (Table 1). The CD spectrum of ap8 differed most from the other spectra. On the basis of the close identity with the CD spectrum of htel-22's ap19 (Figure 1) it can be concluded that ap8 also forms a pure (3+1) hybrid quadruplex. Indeed, the 1H NMR spectrum of ap8 (Figure 3C) confirmed a single quadruplex structure. In contrast, the NMR spectrum of the wt indicated that htel-25 was not a single structure but a mixture of quadruplexes. The ap14 sequence with the A/AP site in the middle loop, displaying two positive CD bands around 260 and 290 nm (Figure 3B), folded into an even more heterogeneous mixture of structures than the wt, based on the NMR spectra (Figure 3C), while the structural heterogeneity was again reduced with ap20. The two 1H NMR spectra of ap8 and ap20 differed from each other but their sum closely matched the spectrum of the wild-type htel-25. Consequently, htel-25 wt is a mixture of quadruplex structures predominantly formed by the sequences containing a single A/AP site in the 5΄ and 3΄end loops. This finding is similar to the recent results of Burrows et al. (32) who claimed that the wt htel-25 was a mixture of h-1 and h-2 structures under their conditions.
As stated above, the CD spectrum of ap8 with the A/AP site in the first loop corresponds to a pure (3+1) hybrid (h) quadruplex structure. It is not possible to assign the type of the h structure based on the 1H NMR spectrum. Neither CD can distinguish between the two h forms. According to theoretical studies (33,34) both pure h forms should yield the same CD spectrum as a consequence of the same head to head and tail to head orientation of their tetrads and the same anti/syn disposition of guanines in particular tetrads (31) (see Supplementary Figure S4). Indeed, the CD spectrum of htel-23 d[TAG3(TTAG3)3] (Supplementary Figure S4, 35) identified as an h – 1 form (31) is of the same type as the spectrum of the htel-22 ap19 identified as an h – 2 form (25). In our previous paper, we demonstrated that the absence of adenine in the third loop of the ap19 stabilizes its propeller fold, in which the AP site is out of the quadruplex core, so that the negative enthalpic effect of adenine depurination is minimized (25). In accord with this conclusion, we assume the ap8 adopts an h – 1 quadruplex structure.
The ap8,14,20 containing an AP site in all three loops provided a CD spectrum that corresponded to a parallel quadruplex (Figure 3B). The quadruplex melted in two steps, as did the htel-22's ap7,13,19, and the pure parallel quadruplex formed only at around 50°C. It then melted at higher temperature than the wt (Figure 3D, Supplementary Table S2B). The ap14 with the A/AP site in the middle loop formed the least stable quadruplex while the Tm values of ap8 and ap20 quadruplexes were close to that of the wt htel-25. In a PAGE experiment, all the single A/AP containing htel-25s showed electrophoretic motilities similar to the wt (Supplementary Figure S5A) (all are expected to contain one parallel loop). Migration only of the ap8,14,20, containing three parallel loops, was slowed down (Supplementary Figure S5A). Formation of parallel loops was conditioned only in the presence of K+, in Na+ the ap8,14,20 quadruplex ran with the same velocity as the other htel-25s (Supplementary Figure S5B) and the quadruplex was clearly destabilized (Supplementary Table S3B).
Single guanine AP site containing htel-25 analogs
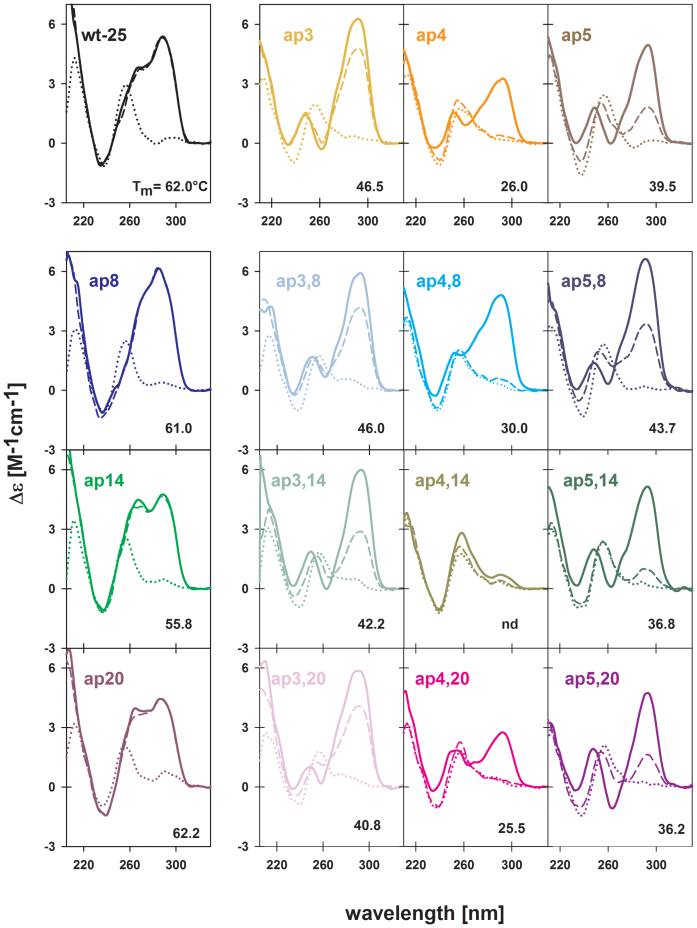
Effect of single AP sites substituting for tetrad guanines (G/AP) in htel DNA quadruplexes was earlier studied with htel-21 d[G3(TTAG3)3] (36) and htel-23 d[TAG3(TTAG3)3] (37). Both papers demonstrated site specific destabilization of the quadruplex structures, while the largest changes were induced by substitution of the middle tetrad guanine. The change in thermodynamic parameters was much more pronounced with the htel-23 than with the htel-21 but the destabilization observed with the htel-25 quadruplex d[TAG3(TTAG3)3TT] was still more drastic: the CD spectra of the wt distinctly changed (Figure 4, the upper row): spectra of ap3 and ap5 displayed a large positive peak at 290 and minima at 260 and 240 nm, which are characteristics of the antiparallel quadruplex as observed for htel-22 and htel-21 (27,28). Thus, the absence of the guanines, completely annulled the hybrid type of wt. The ap4 with G/AP site in the middle tetrad formed the least stable quadruplex (Supplementary Table S2B, Figure S6A), which provided a unique CD spectrum with distinctly reduced 290 nm band amplitude and a flat minimum around 260 nm (Figure 4, the upper row). Hyperchromicity at 297 nm, typical for quadruplex structures, was, however, close to those for ap3 and ap5 (Supplementary Figure S6B), so that the unusual spectrum corresponds to a fully formed quadruplex. All three sequences, including the least stable ap4 formed intramolecular quadruplexes (Supplementary Figure S5A).
Figure 4.
CD spectra of htel-25 wt and of its G/AP (upper horizontal panel), A/AP (left vertical panel), and (G+A)/AP (combination of the panels) containing analogs measured at their unstructured forms in 1 mM sodium phosphate with 0.3 mM EDTA, pH 7.2 (dotted lines), in 10 mM K-phosphate buffer, pH 7 (dashed lines), and in K+ added up to 100 mM (solid lines).
Clustered loop A/AP and core G/AP sites containing htel-25 analogs
We also studied the combined effect of the loop A/AP and tetrad G/AP lesions of htel-25: according to the CD spectra (Figure 4) and the sigmoid shape of their melting curves (Supplementary Figure S6) each double A/AP and G/AP containing oligonucleotide, except for ap4,14, folded into quadruplexes in 100 mM K+. In addition to CD spectra measured in 100 mM K+, Figure 4 contains CD spectra of unstructured forms of all sequences taken in the low salt and the absence of K+, and in 10 mM K+ phosphate, which enables to estimate how readily the full quadruplexes are formed. All quadruplexes were radically destabilized, by –16 to –36°C, relative to the wt htel-25 (Figure 4, Supplementary Table S2B). The ap4,20 was the least thermostable structure, while the ap4,14 did not form a quadruplex at all (Supplementary Figure S6B). The electrophoretic migration of the ap4,14 provided a footprint between single strand and quadruplex (Supplementary Figure S5A) as is usual with unstructured DNAs. Also the bands of ap4,8 and ap4,20 displayed traces of slower running species indicating less ordered quadruplex structure (Supplementary Figure S5A). This is more obvious on PAGE in Na+ (Supplementary Figure S5B). The other A/AP + G/AP oligonucleotides formed intramolecular structures (Supplementary Figure S5). The CD spectra of all G3/AP + loop A/APs (ap3,8; ap3,14 and ap3,20) and G5/AP + loop A/APs (ap5,8; ap5,14 and ap5,20) double AP containing quadruplexes (Figure 4) provided a large positive peak at 290, minima at 260 and 240 nm and lacked the positive ∼270 nm shoulder that corresponds to K+-stabilized hybrid quadruplexes (27,28). Though the wt htel-25 adopts hybrid structure and the presence of loop AP sites stabilizes propeller loop folding, thus the parallel orientation of quadruplex strands, the spectra of these clustered A/AP + G/AP quadruplexes reveal antiparallel quadruplex foldings. The clustered AP oligonucleotides thus adopt the same type of quadruplexes as their single G/AP containing analogs (Figure 4). The thermostability of the clustered quadruplexes decreased from ap3,8 to ap3,20 and from ap5,8 to ap5,20. The temperature dependences and the Tm values are shown in Supplementary Figure S6 and in Table S2B, respectively.
CD spectrum of ap4,8 became more similar to those of ap3,8 and ap5,8 as a result of combination of G4/AP with the loop A8/AP (Figure 4). The destabilization effect of the two AP sites was less negative than with the single G/AP containing ap4 (Supplementary Table S2B). In contrast, the ap4,20 provided a CD spectrum even more deformed than that of the ap4 with a single missing G4. Its spectrum with two small positive bands in 100 mM K+ appears to be intermediate in the process of quadruplex formation but its hyperchromicity at 297 nm again was the same as with the other clustered G/AP and A/AP quadruplexes (Supplementary Figure S6B).
Comparing the effect of particular loop AP sites on the stability of the clustered A/AP and G/AP quadruplexes, we can see that A8/AP slightly destabilized the quadruplex of ap3,8 but stabilized those of ap4,8 and ap5,8 compared to their respective single G/AP containing structures (Supplementary Figure S7A). The presence of A14/AP and A20/AP further increased the severe destabilizing effect of the missing guanines (Supplementary Figure S7B and C). The largest destabilization took place with ap4,20, the ΔTm was –36.5°C, and mainly with ap4,14, which did not form quadruplex at all.
The presence of G/AP in the ap3, ap4 and ap5 transformed the wt htel-25 into antiparallel quadruplexes with Tm values (Supplementary Table S2B) close to those observed for the respective quadruplexes stabilized by Na+. In combination with A/AP΄s the Tm values of all quadruplexes were, however, higher (by 10–16°C with A8/AP) in K+ than in Na+ (Supplementary Tables S2B and S3B). In conclusion, none of the G/AP containing sequences was able to form a hybrid type quadruplex in water/K+ solution (Figure 4). No change in CD spectrum was either observed on addition of the parallel quadruplex stabilizing ligand N-methyl mesoporphyrin IX (38). Moreover, none of these oligonucleotides was able to adopt intramolecular parallel quadruplex in ethanol (Figure 5) or PEG solutions (not shown) though their single A/AP analogs formed parallel quadruplexes much more easily than the wt htel-25. Only ap4,14, which does not form quadruplex in aqueous solution, displayed a CD spectrum with positive 260 nm band in ethanol reflecting a partial formation of parallel but probably intermolecular parallel quadruplex.
Figure 5.
CD spectra of (upper panel) htel-25 wt (left), A/AP (middle) and G/AP (right) and of (bottom panel) clustered A/AP + G/AP containing analogs as indicated in figures. All spectra were measured in 60% ethanol and 2 mM K+ at 23°C.
Clustered core G/AP sites containing htel-25 analogs
Substitution of AP sites for two or three guanines resulted in a significant deterioration of quadruplex structure, especially if the substitution included a G of the middle tetrad as e.g. ap3,4; ap3,4,5; ap3,22; ap4,15 or ap4,22 (Supplementary Table S2C, and selected examples are shown in Supplementary Figure S8). Of these only ap3,22 and ap4,15 formed a quadruplex, displaying a sigmoidal temperature dependence with a Tm value above room temperature (25 and 23.5°C, respectively). Interestingly, a loop adenine A8/AP site in ap4,8,15 increased the Tm of ap4,15 up to 25°C (Supplementary Table S2C).
The smallest destabilizing effect was brought about by the substitution of two G΄s in the same external tetrad (e.g. in ap3,23 or ap5,21) or by the replacement of the most distant guanines within the quadruplex (e.g. in ap3,21 or ap5,23). Their Tm values were around 34–36°C in 100 mM K+ (Supplementary Figure S8). The quadruplexes formed were antiparallel and intramolecular. The band movement of ap3,22; ap3,23 and ap5,23 but not of ap3,21, was slightly slower on the PAGE than that of wt-25. This may be a result of quadruplex deformation and loop extension due to shifts of G blocks to compensate for the lost G΄s. Oligonucleotides ap3,4; ap3,4,5; and ap4,22 remained unstructured at room temperature (Supplementary Figure S8) and inclined to form intermolecular quadruplexes at low temperatures (not shown). This is the reason of the smear of their bands observed at room temperatures (Supplementary Figure S8).
DISCUSSION
Clustered DNA damage has been defined as two or more lesions in 1–2 helical turns of a double-stranded DNA, i.e. in 10–20 base pairs. As natural base lesions also form in non-canonical nucleic acids, such as the G-quadruplexes (39–41) the original definition needs to be formulated to fit quadruplexes as well. We define it here as two or more lesions within a four-stranded core quadruplex unit assembling from 21 to 25 nucleotides.
Using this definition, few modified quadruplex structures can be qualified retrospectively as having clustered natural base mutations and damage (42–46), let alone synthetic nucleotide analogs (19). Here we investigated the structural effects of one of the most frequent natural base lesions, abasic sites when these are clustered in quadruplex structures.
According to the known multiple telomere quadruplex structures detected in K+ and the fast conformational transitions observed by single-molecule methods (32,47–49), we can consider the results found by in-solution methods, such as, CD, UV absorption and NMR spectroscopies as a description of the dynamic equilibrium between various conformations. In this article, we show how this equilibrium is shifted by clustered alterations in the human telomere DNA quadruplexes as a result of abasic damage modelled by the insertions of synthetic AP sites in place of selected loop adenines (A/AP) and tetrad guanines (G/AP).
Two types of sequences, htel-22 and htel-25, were used in these studies (Table 1 and Supplementary Table S1). Depending on solution conditions, the htel-22 d[AG3(TTAG3)3] interconverts between antiparallel, possibly a 2-tetrad basket (50,51), and the hybrid forms (31), while the antiparallel forms are predominantly formed at DNA concentrations used for optical measurements (29). In contrast, longer htel sequences with flanking nucleotides preferentially adopt hybrid (3+1) parallel/antiparallel quadruplex structures (31). The htel-25 d[TAG3(TTAG3)3TT], selected as the second sequence for study, was reported (31) to form a stable hybrid h-2 structure.
The results presented here confirm our earlier observation (25) that any single A/AP site in the predominantly antiparallel quadruplex of htel-22, causes a shift in conformational equilibrium towards a hybrid form by facilitating the formation of a propeller fold of the respective loop. Each propeller loop caused a small decrease in migration speed of htel-22 in native polyacrylamide gel (Figure 2C). The type of hybrid was determined by the position of the A/AP mutation. The most stable quadruplexes with the A/AP in the third loop, corresponded to hybrid-2 (25). The presence of A/AP in the middle loop, which could theoretically induce a (2+2) structure similar to that of d[(TTAG3)4TTA] observed for sodium (52) (Supplementary Figure S1e) has no such effect on the htel-22. This could indicate that formation of this type of (2+2) structure is not favoured in potassium ions.
The clustering of multiple A/AP lesions caused qualitative changes in quadruplex properties. The simultaneous presence of A/AP lesions in all three loops resulted in a conformational shift to the stable parallel quadruplex form (Figures 1, 2 and 3B). Thus, this result again confirms the conclusion that the A/AP facilitates the propeller loop. The quadruplexes, ap7,13,19 of the htel-22 series or the ap8,14, 20 of the htel-25 gave rather complex melting profiles (Figures 2B and 3D). The parallel structure was only stable at increased temperatures (Figure 2A). The undefined structure formed at room temperature is probably an assemble of quadruplexes as can be deduced from the long kinetics of its formation. Our results show that the quadruplexes containing a single A/AP site in the first or the third loop have the thermostability comparable to the wt, and the quadruplex with A/APs in all three loops is still more stable than the wt. In contrast, all three double A/AP htel-22 quadruplexes ap7,13; ap7,19; and ap13,19 are significantly less stable than the wt (Figure 1, Supplementary Table S2). These double A/AP containing quadruplexes provide CD spectra characterized by a clear shoulder or even separated a new signal around 260 nm indicating increased population of parallel strands, probably as a consequence of a fraction of species with more than one propeller loop. This is most obvious with ap7,13 (Figure 1), which, however, is a mixture of structures as revealed by 1H NMR (Figure 2A) rather than a single quadruplex structure containing two parallel loops. The fuzzy retarded band of ap7,13 on PAGE also indicates an additional propeller loop in a species of structural mix. The ap13,19 provides a CD spectrum that is very similar to that of ap17,18,19 and ap19, characteristic of the pure (3+1) hybrid like the ap19. This quadruplex is a candidate for adopting the form with propeller foldings in the middle and the third loop as proposed by Phan (31). It has the same mutual orientation of quadruplex tetrads and the same distribution of anti/syn glycosidic angles of the tetrad guanines as the two h-1 and h-2 forms with single propeller loops (Supplementary Figure S4).
It could be speculated that introduction of A/AP sites, instead of, in particular, the A8, into the htel-25, which already adopts a hybrid, predominantly h-2 type structure (31), might stabilize the parallel quadruplex folding (specifically when the simultaneous presence of two propeller loops is not thermodynamically favourable). However, we made the following discovery: the 1H NMR spectra showed that the A/AP sites in the first and in the third loop stabilized nearly single, mutually different quadruplexes while the wt was a mixture of quadruplexes encompassing the two quadruplex species. The first (ap8) provided a CD spectrum characteristic of a pure hybrid (3+1) structure (Figure 3B, see Supplementary Figure S4). The CD spectrum of the second one (ap20) displayed a peak at 290 nm followed by a distinct shoulder close to 260 nm (Figure 3B). Its structure is unknown. It may be similar to the h-2 hybrid as observed by An et al. (32).
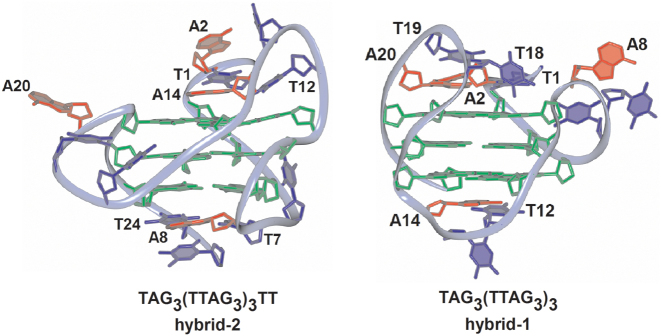
As stated above, the 1H-NMR method is not able to assign the type of hybrid structure of the ap8 and neither does the CD spectroscopy. Based on theoretical studies (33,34), both pure h-1 and h-2 forms, and all hybrid forms with the same orientation of quadruplex tetrads and anti/syn guanine glycosidic angles (53) should yield the same CD spectrum (see Supplementary Figure S4). However, in line with this and our earlier publication (25), the ap8 contains a propeller fold in the first loop, so that it is expected to form the hybrid-1 quadruplex. Figure 6 outlines the advantage of the switch of htel-25 h-2 to the h-1 structure on loss of A8: according to the molecular structures of htel-25 deposited in the database (code 2jsl) (Figure 6, left), its h-2 structure may be stabilized by A8-T24-T7 triplet on the bottom (3΄ molecule end) and by T1-A14-T12 hydrogen bonded bases on the top of the quadruplex core. However, the h-2 stabilizing A8 is missing in ap8. In the h-1 structure, according to the model of d[TAG3(TTAG3)3] (code 2jsm) (Figure 6, right), the A8 is located in the propeller loop. This minimizes the negative enthalpic effect of the adenine depurination, and its previous stabilizing role may be substituted by A14-T12 pair on the 3΄and T1-A20-A2-T19 tetrad on the 5΄side of the quadruplex (Figure 6).
Figure 6.
Molecular structures of the h-2 type d[TAG3(TTAG3)3TT] (left) and h – 1 type of d[TAG3(TTAG3)3] (right) quadruplexes deposited in database under codes (2jsl) and (2jsm), respectively. The h-2 structure is capped by T1-A14-T12 on the top and by A8-T24-T7 triplet on the bottom of the quadruplex core. In the h – 1 structure the A8 is situated in the propeller loop and the quadruplex may be stabilized by A2-T1-A20 and A2-T19 on the top site and T12-A14 on the bottom of the quadruplex.
The hybrid quadruplexes of htel-25 analogs, namely their A/AP-stabilized single hybrid structures were ideal sequences to test the effect of the clustered A/AP and G/AP lesions. Our previous spectroscopic data on htel-21 and htel-22 (36,54) and that of of Virgilio et al. on htel-23 (37) showed that any single G lesion caused strong destabilization of these quadruplexes. The destabilizing effect of the G/AP lesions were still stronger with the htel-25 quadruplexes resulting in ΔTm values of –15.5, –36 and –22.5°C for ap3, ap4, and ap5, compared to the wt (Supplementary Table S2B). The AP site in the middle tetrad was the most destructive. In spite of the hybrid structure of htel-25 and the presence of propeller loops stabilized by the A/AP sites, none of the clustered A/AP and G/AP containing sequences remained in the hybrid form. All structures were transformed into antiparallel quadruplexes. The main destabilization came from the single G/AP sites, while the additional, rather opposite effect of A/AP sites was milder and position dependent (Figure 4, Supplementary Figure S7, Table S2B). The combination of G/AP with A8/AP rather stabilized the quadruplex. The other clustered lesions destabilized the wt, and the combination of the G4/AP of the middle tetrad with the A14/AP of the middle loop, prevented quadruplex formation.
The quadruplex of ap4,20 yielded a CD spectrum very similar to that of the so called (2+2) quadruplex, which is formed by the htel 27-mer d[(TTAGGG)3TTA(BrG)GGTTA] in Na+ solution (52). The possibility that ap4,20 adopts the same type of quadruplex structure is not excluded as A/AP in the third loop of the htel-22 (analogous to A20) provides an identical spectrum to that of the htel 27-mer in Na+ solutions (unpublished), the Br atom stabilizing the (2+2) structure of the htel-27 belongs to G neighbouring the A20/AP, and missing guanines stabilize quadruplex conformational properties characteristic of Na+ solutions.
In quadruplexes with clustered G/AP and A/AP lesions, the effect of G/AP inducing the antiparallel fold always prevails over the effect of A/AP stabilizing a hybrid/parallel fold. The structural changes induced by clustered lesions should thus be considered within the antiparallel model, whereas there are only two with atomic structure reported in K+: the 2-tetrad basket of the Phan group (50) (Supplementary Figure S1f), which, in fact, is a slipped Na+-stabilized three tetrad quadruplex of Patel et al. (55). This structural change probably requires small energy input as both structures d[(G3(TTAG3)3T] in K+ and d[AG3(TTAG3)3] in Na+ have equally accessible N7 residues for interaction with dimethylsulphate (unpublished results). The second antiparallel K+-stabilized quadruplex is the 2-tetrad basket type structure of Plavec group (56) (Supplementary Figure S1g), in which one tetrad G is really missing as it was in our case.
The presence of G/AP distinctly destabilized the tetramolecular quadruplex of [d(TGGGGGT)]4 (6) but preserved its parallel type. In contrast, single G/AP in htel quadruplexes transformed their hybrid quadruplexes into antiparallel ones and their stability in K+ approached that observed in Na+ (Supplementary Tables S2B and S3B).
None of the G/AP-substituted htel-25 quadruplexes was able to convert into parallel form under crowding/dehydrating conditions (Figure 5), which is a property shared by all wt quadruplexes independent of their structure (27,28). This indicates that the hybrid or parallel htel quadruplex scaffolds are more sensitive to guanine damage than antiparallel forms. This follows from Supplementary Figure S9: changes in thermostability caused by G/AP are more extensive in K+ than in Na+ and this holds, with the exception of ap3,8 and ap5,8, also for the clustered A/AP + G/AP sites. The need for three perfect tetrads compared to two, sufficient for an antiparallel form (55,56) may be an explanation. Also, the proper stacking of tetrads, is probably inevitable for propeller type loop formation which is disturbed by deformation of tetrad planes caused by a missing G.
The changes in thermal stability caused by A/AP are much less extensive than those caused by G/AP. Nevertheless, it can be recognized that predominantly antiparallel quadruplex of htel-22 is destabilized by A7/AP and A19/AP in Na+ more than in K+ (Supplementary Figure S9). Thus, the parallel loops forming in K+ prevent the quadruplex from destabilization observed in Na+. This effect is clearly evident with both ap7,13,19 and ap8,14,20 (Supplementary Figure S9): their parallel quadruplexes formed in K+ are more stable than their wt forms, while both quadruplexes are distinctly destabilized in Na+.
The present study highlights the fact that clustered AP lesions in the human telomere quadruplex of d[TAG3(TTAG3)3TT] can change the folding and stability and can even prevent its formation. Even if the chromosomes’ capping telomere DNA–protein complexes (Shelterin) render the telomere DNA inaccessible to recognition and attack by various cellular enzymes, spontaneous depurination as well as other oxidative base damage of the DNA do occur in these tight complexes too (39–41). As the clustered damages are relatively inaccessible for repair even in double-stranded DNAs (4,57,58), these can be fateful for the telomere quadruplex. Extensive destabilization by AP sites can result in the unfolding of the quadruplex core, which in turn, can destabilize the Shelterin complex to the extent that the cellular nuclease can access and degrade the telomere DNA. The result may be telomere shortening. Conformational change of the quadruplex in vivo (59) due to the formation of clustered loop adenine and core guanine AP lesions can reduce the ability of telomerase to recognize the quadruplex as a substrate (60) or the interaction of Shelterin proteins POT1 and TPP1 complex with the telomere DNA (61).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [P205/12/0466, 13–28310S, 15–06785S]; SYMBIT [CZ.02.1.01/0.0/0.0/15_003/0000477] financed by the ERDF; Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 [LQ1601]. Funding for open access charge: SYMBIT [CZ.02.1.01/0.0/0.0/15_003/0000477].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang Z. Molecular and Biochemical Toxicology. 2007; John Wiley & Sons, Inc; 441–491. [Google Scholar]
- 2. Jenner T.J., Fulford J., O’Neill P.. Contribution of base lesions to radiation-induced clustered DNA damage: Implication for models of radiation response. Radiation Res. 2001; 156:590–593. [DOI] [PubMed] [Google Scholar]
- 3. Tokuyama Y., Furusawa Y., Ide H., Yasui A., Terato H.. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Radiat. Res. 2015; 56:446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sage E., Harrison L.. Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat. Res. 2011; 711:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gulston M., de Lara C., Jenner T., Davis E., O’Neill P.. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004; 32:1602–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esposito V., Martino L., Citarella G., Virgilio A., Mayol L., Giancola C., Galeone A.. Effects of abasic sites on structural, thermodynamic and kinetic properties of quadruplex structures. Nucleic Acids Res. 2010; 38:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Sonntag C. The Chemical Basis of Radiation Biology. 1987; London: Taylor & Francis. [Google Scholar]
- 8. O΄Neill P., Fielden E.M.. Primary free-radical processes in DNA. Adv. Radiat. Biol. 1993; 17:53–120. [Google Scholar]
- 9. Harrison L., Hatahet Z., Wallace S.S.. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999; 290:667–684. [DOI] [PubMed] [Google Scholar]
- 10. David-Cordonnier M.H., Boiteux S., O’Neill P.. Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res. 2001; 29:1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gulston M., Fulford J., Jenner T., de Lara C., O’Neill P.. Clustered DNA damage induced by γ radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002; 30:3464–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutherland B.M., Bennett P.V., Cintron-Torres N., Hada M., Trunk J., Monteleone D., Sutherland J.C., Laval J., Stanislaus M., Gewirtz A.. Clustered DNA damages induced in human hematopoietic cells by low doses of ionizing radiation. J. Radiat. Res. 2002; 43:S149–152. [DOI] [PubMed] [Google Scholar]
- 13. David-Cordonnier M.H., Laval J., O’Neill P.. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000; 275:11865–11873. [DOI] [PubMed] [Google Scholar]
- 14. Bennett P.V., Cuomo N.L., Paul S., Tafrov S.T., Sutherland B.M.. Endogenous DNA damage clusters in human skin, 3-D model, and cultured skin cells. Free Radic. Biol. Med. 2005; 39:832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bennett P., Ishchenko A.A., Laval J., Paap B., Sutherland B.M.. Endogenous DNA damage clusters in human hematopoietic stem and progenitor cells. Free Radic. Biol. Med. 2008; 45:1352–1359. [DOI] [PubMed] [Google Scholar]
- 16. Sutherland B.M., Bennett P.V., Sutherland J.C., Laval J.. Clustered DNA damages induced by X rays in human cells. Radiat. Res. 2002; 157:611–616. [DOI] [PubMed] [Google Scholar]
- 17. Song J.M., Milligan J.R., Sutherland B.M.. Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290-400 nm) radiation. Biochemistry. 2002; 41:8683–8688. [DOI] [PubMed] [Google Scholar]
- 18. Neidle S., Balasubramanian S.. Quadruplex Nucleic Acids. 2006; London: Royal Society of Chemistry. [Google Scholar]
- 19. Sagi J. G-quadruplexes incorporating modified constituents: a review. J. Biomol. Struct. Dyn. 2014; 32:477–511. [DOI] [PubMed] [Google Scholar]
- 20. Pedroso I.M., Hayward W., Fletcher T.M.. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 2009; 37:1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu Y., Sato H., Sannohe Y., Shinohara K., Sugiyama H.. Stable lariat formation based on a G-quadruplex scaffold. J. Am. Chem. Soc. 2008; 130:16470–16471. [DOI] [PubMed] [Google Scholar]
- 22. Baird D.M., Jeffreys A.J., Royle N.J.. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 1995; 14:5433–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sattin G., Artese A., Nadai M., Costa G., Parrotta L., Alcaro S., Palumbo M., Richter S.N.. Conformation and stability of intramolecular telomeric G-quadruplexes: Sequence effects in the loops. PLoS One. 2013; 8:e84113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aviňo A., Portella G., Ferreira R., Gargallo R., Mazzini S., Gabelica V., Orozco M., Eritja R.. Specific loop modifications of the thrombin-binding aptamer trigger the formation of parallel structures. FEBS J. 2014; 281:1085–1099. [DOI] [PubMed] [Google Scholar]
- 25. Babinsky M., Fiala R., Kejnovska I., Bednarova K., Marek R., Sagi J., Sklenar V., Vorlickova M.. Loss of loop adenines alters human telomere d[AG3(TTAG3)3] quadruplex folding. Nucleic Acids Res. 2014; 42:14031–14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piotto M., Saudek V., Sklenar V.. Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J. Biomol. NMR. 1992; 6:661–665. [DOI] [PubMed] [Google Scholar]
- 27. Renciuk D., Kejnovska I., Skolakova P., Bednarova K., Motlova J., Vorlickova M.. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009; 37:6625–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vorlíckova M., Kejnovska I., Sagi J., Renciuk D., Bednarova K., Motlova J., Kypr J.. Circular dichroism and guanine quadruplexes. Methods. 2012; 57:64–75. [DOI] [PubMed] [Google Scholar]
- 29. Palacký J., Vorlíčková M., Kejnovská I., Mojzeš P.. Polymorphism of human telomeric quadruplex structure controlled by DNA concentration: a Raman study. Nucleic Acids Res. 2013; 41:1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vorlickova M., Chladkova J., Kejnovska I., Fialova M., Kypr J.. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005; 33:5851–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phan A.T., Luu K.N., Patel D.J.. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006; 34:5715–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. An N., Fleming A.M., Burrows C.J.. Interactions of the human telomere sequence with the nanocavity of the α-hemolysin ion channel reveal structure-dependent electrical signatures for hybrid folds. J. Am. Chem. Soc. 2013; 135:8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Masiero S., Trotta R., Pieraccini S., De Tito S., Perone R., Randazzo A., Spada G.P.. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010; 8:2653–2872. [DOI] [PubMed] [Google Scholar]
- 34. Gray D.M., Wen J., Gray C.V., Repges R., Repges C., Raabe G., Fleischhauer J.. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008; 20:431–440. [DOI] [PubMed] [Google Scholar]
- 35. Wang Z.-F., Li M.-H., Hsu S.-T.D., Chang T.-C.. Structural basis of sodium–potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014; 42:4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skolakova P., Bednarova K., Vorlickova M., Sagi J.. Quadruplexes of human telomere dG(3)(TTAG(3))(3) sequences containing guanine abasic sites. Biochem. Biophys. Res. Commun. 2010; 399:203–208. [DOI] [PubMed] [Google Scholar]
- 37. Virgilio A., Petraccone L., Esposito V., Citarella G., Giancola C., Galeone A.. The abasic site lesions in the human telomeric sequence d[TA(G3T2A)3G3]: A thermodynamic point of view. Biochim. Biophys. Acta (BBA) - Gen. Subj. 2012; 1820:2037–2043. [DOI] [PubMed] [Google Scholar]
- 38. Sabharwal N.C., Savikhin V., Turek-Herman J.R., Nicoludis J.M., Szalai V.A., Yatsunyk L.A.. N-methylmesoporphyrin IX fluorescence as a reporter of strand orientation in guanine quadruplexes. FEBS J. 2014; 281:1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stebbeds W.J.D., Lunec J., Larcombe L.D.. An in silico study of the differential effect of oxidation on two biologically relevant G-quadruplexes: Possible implications in oncogene expression. PLoS One. 2012; 7:e43735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou C., Greenberg M.M.. DNA damage by histone radicals in nucleosome core particles. J. Am. Chem. Soc. 2014; 136:6562–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun L., Tan R., Xu J., LaFace J., Gao Y., Xiao Y., Attar M., Neumann C., Li G.M., Su B. et al. Targeted DNA damage at individual telomeres disrupts their integrity and triggers cell death. Nucleic Acids Res. 2015; 43:6334–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sagi J., Renciuk D., Tomasko M., Vorlickova M.. Quadruplexes of human telomere DNA analogs designed to contain G:A:G:A, G:G:A:A, and A:A:A:A tetrads. Biopolymers. 2010; 93:880–886. [DOI] [PubMed] [Google Scholar]
- 43. Benz A., Hartig J.S.. Redesigned tetrads with altered hydrogen bonding patterns enable programming of quadruplex topologies. Chem. Commun. 2008; 4010–4012. [DOI] [PubMed] [Google Scholar]
- 44. Singh V., Benz A., Hartig J.S.. G quadruplexes stabilised by 8-oxo-2΄-deoxyguanosine. Chemistry. 2011; 17:10838–10843. [DOI] [PubMed] [Google Scholar]
- 45. Cheong V.V., Heddi B., Lech C.J., Phan A.T.. Xanthine and 8-oxoguanine in G-quadruplexes: formation of a G·G·X·O tetrad. Nucleic Acids Res. 2015; 43:10506–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheong V.V., Lech C.J., Heddi B., Phan A.T.. Inverting the G-tetrad polarity of a G-quadruplex by using xanthine and 8-oxoguanine. Angew. Chem. 2016; 55:160–163. [DOI] [PubMed] [Google Scholar]
- 47. Ying L.M., Green J.J., Li H.T., Klenerman D., Balasubramanian S.. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14629–14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee J.Y., Okumus B., Kim D.S., Ha T.. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:18938–18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Noer S.L., Preus S., Gudnason D., Aznauryan M., Mergny J.-L., Birkedal V.. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016; 44:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T.. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009; 131:4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Z., Dai J., Veliath E., Jones R.A., Yang D.. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010; 38:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lim K.W., Ng V.C.M., Martin-Pintado N., Heddi B., Phan A.T.. Structure of the human telomere in Na+ solution: an antiparallel (2+2) G-quadruplex scaffold reveals additional diversity. Nucleic Acids Res. 2013; 41:10556–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Konvalinova H., Dvorakova Z., Renciuk D., Bednarova K., Kejnovska I., Trantirek L., Vorlickova M., Sagi J.. Diverse effects of naturally occurring base lesions on the structure and stability of the human telomere DNA quadruplex. Biochimie. 2015; 118:15–25. [DOI] [PubMed] [Google Scholar]
- 55. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 56. Galer P., Wang B., Šket P., Plavec J.. Reversible pH switch of two‐quartet G‐quadruplexes formed by human telomere. Angewandte Chemie International Edition. 2016; 55:1993–1997. [DOI] [PubMed] [Google Scholar]
- 57. Sedletska Y., Radicella J.P., Sage E.. Replication fork collapse is a major cause of the high mutation frequency at three-base lesion clusters. Nucleic Acids Res. 2013; 41:9339–9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eccles L.J., O’Neill P., Lomax M.E.. Delayed repair of radiation induced clustered DNA damage: friend or foe?. Mutat. Res. 2011; 711:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hänsel R., Löhr F., Trantirek L., Dötsch V.. High-resolution insight into G-overhang architecture. J. Am. Chem. Soc. 2013; 135:2816–2824. [DOI] [PubMed] [Google Scholar]
- 60. Moye A.L., Porter K.C., Cohen S.B., Phan T., Zyner K.G., Sasaki N., Lovrecz G.O., Beck J.L., Bryan T.M.. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015; 6:7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mullins M.R., Rajavel M., Hernandez-Sanchez W., de la Fuente M., Biendarra S.M., Harris M.E., Taylor D.J.. POT1-TPP1 binding and unfolding of telomere DNA discriminates against structural polymorphism. J. Mol. Biol. 2016; 428:2695–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.