Abstract
When Escherichia coli encounters stress, the endoribonuclease MazF initiates a post-transcriptional response that results in the reprogramming of protein synthesis. By removing the 3΄-terminus of the 16S rRNA, MazF generates specialized ribosomes that selectively translate mRNAs likewise processed by MazF. Given the energy required for de novo ribosome biosynthesis, we considered the existence of a repair mechanism operating upon stress relief to recycle the modified ribosomes. Here, we show that the stress-ribosomes and the 3΄-terminal 16S rRNA fragment are stable during adverse conditions. Moreover, employing in vitro and in vivo approaches we demonstrate that the RNA ligase RtcB catalyzes the re-ligation of the truncated 16S rRNA present in specialized ribosomes Thereby their ability to translate canonical mRNAs is fully restored. Together, our findings not only provide a physiological function for the RNA ligase RtcB in bacteria but highlight the reversibility of ribosome heterogeneity, a crucial but hitherto undescribed concept for translational regulation.
INTRODUCTION
Translation of mRNA-based information into proteins is one of the most intricate and fundamental processes of life. It is performed by the ribosome, a highly sophisticated ribonucleoprotein machine. The bacterial 70S ribosome is a two-subunit assembly formed by three RNA molecules and 54 proteins. Considering this complexity, protein synthesis and in particular ribosome biogenesis are of the most energy-consuming processes in the cell, which rely on a plethora of non-ribosomal factors (1,2). As every organism has to economize on energy, it is therefore tempting to envisage the existence of a repair mechanism that recognizes and mends damaged or modified ribosomes. A first study addressing this intriguing question in bacteria suggested that ribosome repair might represent an important mechanism for cell survival where the replacement of damaged ribosomal proteins can restore the translational activity of chemically inactivated ribosomes (3).
In addition to ribosomes modified or altered in their protein complement, we observed the formation of a functionally specific ribosomal subpopulation harboring a 3΄-terminally truncated 16S rRNA in Escherichia coli (4). During environmental stress the endoribonuclease MazF, the toxin component of the toxin–antitoxin (TA) module mazEF (5,6) becomes activated and specifically cleaves the 16S rRNA of 70S ribosomes at an ACA site located at positions 1500–1502 (4). Thereby, the ribosome loses a 3΄-terminal 16S rRNA fragment of 43 nucleotides (nts) in length (Figure 1A and B; henceforth referred to as RNA43) harboring helix 45 and the anti-Shine-Dalgarno (aSD) sequence (7), both vital for translation initiation of canonical mRNAs. As a consequence the translational program of the cell is modulated since the resulting specialized ribosomes (referred to as 70SΔ43 throughout the text) harboring the truncated 16SΔ43 rRNA (nts 1–1499) selectively translate specific mRNAs that are likewise processed by MazF within their 5΄-untranslated region (5΄-UTR) (4,8). Given the high number of specialized ribosomes obtained after mimicking amino acid starvation (4), this observation raises the fundamental question as to whether this 'one-step mechanism΄ of ribosome specialization might be reversible during recovery from stress. From the physiological point of view, such a mechanism would be beneficial for bacterial cells as it allows for the regeneration of the translational apparatus without de novo assembly (1,2).
Figure 1.
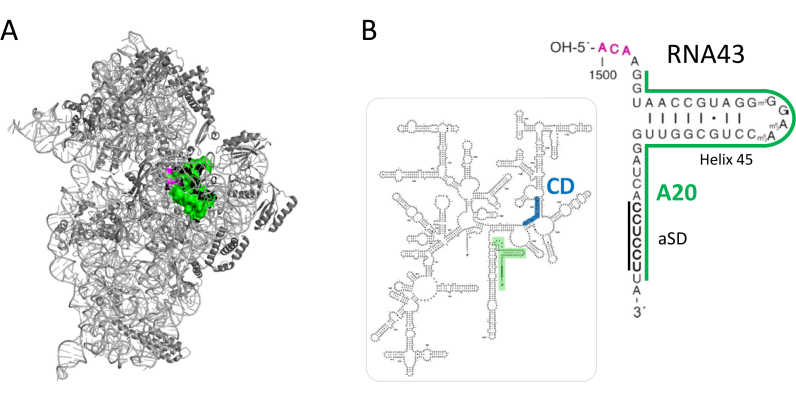
(A) The structure of the 30S subunit as seen from the solvent side. The 16S rRNA is shown in light gray, the ribosomal proteins in dark gray. The MazF cleavage site is indicated in magenta and the 3΄-terminal 43 nucleotides that are removed by the cleavage are shown in green. The structure was modeled using Polyview 3D molecular system software (42) and PDB file 2HGP (43). (B) Secondary structure of the 16S rRNA. The RNA43 fragment is highlighted and enlarged. The MazF cleavage site (magenta), the aSD sequence and the binding sites for probes A20 (green) and CD (blue) used for northern blot analysis shown in Figure 2 are given.
The E. coli RNA ligase RtcB was found to seal RNA 2΄,3΄-cyclic phosphate or 3΄-phosphate termini with 5΄-hydroxyl RNA ends (9–15). The evolutionary conserved enzyme represents the bacterial homolog of the human tRNA ligase HSPC117 (16), which joins tRNA exon halves after cleavage by the tRNA splicing endonuclease complex (17). In E. coli, the rtcB gene is genetically linked to rtcA, encoding a 3΄-terminal phosphate cyclase (11,18–20), and the expression of the rtcBA operon is regulated by the alternative sigma factor σ54 in conjunction with the transcription factor RtcR (18). The biochemical mechanism underlying the RtcB activity was extensively investigated during the last years, and RtcA and RtcB were suggested to perform a healing and sealing function, respectively, in an RNA repair pathway in response to cellular stress (10,14). A recent report indicates a link between the RtcB activity and key cellular processes like maintaining the translational machinery and chemotactic behavior (21), however, hitherto no distinct physiological function was assigned to RtcB. Since MazF cleavage generates 2΄,3΄-cyclic phosphate and 5΄-hydroxyl termini (22), and the RNA43 exhibits structural similarities in length and nucleotide modifications with 3΄-terminal exons of mammalian tRNAs, we hypothesized that RtcB might ligate the RNA43 to the truncated 16SΔ43 rRNA in the context of the stress-ribosomes, thereby regenerating the translational apparatus.
In this study, we report that despite its low abundance (18) the RNA ligase RtcB is an important factor for the recovery of E. coli upon stress release. Our data show that RtcB rescues the subpopulation of heterogeneous ribosomes by rRNA re-ligation, thereby restoring their proficiency to translate canonical mRNAs. This unprecedented mechanism suggests a pivotal function for RtcB in gene expression by modulating ribosome specificity in response to environmental conditions and challenges a paradigm in bacterial protein synthesis by extending the role of the ribosome from a mere protein synthesis machinery to a control unit with regulatory capacity.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
Escherichia coli strains and plasmids as well as oligonucleotides used in this study are summarized in Supplementary Tables S2 and S3, respectively. Escherichia coli rtcB deletion strains were constructed by ΦP1 transduction (23) using the E. coli strain BW25113ΔrtcB as donor (Keio Collection, (24)). The integrated kanamycin resistance cassette was removed employing the Flp recombinase encoded by plasmid 706-Flp (Gene Bridges GmbH; Heidelberg, Germany) following the manufacturer's instructions. Unless otherwise indicated, bacterial cultures were grown in Luria–Bertani (LB) medium (23) supplemented with ampicillin (100 μg/ml), kanamycin (40 μg/ml), chloramphenicol (20 μg/ml) or tetracyclin (30 μg/ml) when required for plasmid maintenance. Growth was monitored by measuring the optical density at 600 nm (OD600). Plasmid pSA1 is a pQE30 derivative (Qiagen, Hilden, Germany) harboring the mazF gene under control of the T5 promoter and the lac operator (25). Ectopic mazF expression was triggered by addition of 0.1 mM IPTG. Plasmid pTwin1rtcB encoding the N-terminally intein tagged RtcB protein was generated by inserting the rtcB coding sequence into the plasmid pTwin1 (New England Biolabs; MA, USA) using the ‘Phusion High-Fidelity PCR Master Mix with HF Buffer’ (Thermo Fischer Biosciences) employing primers D10 and rtcBrev. Plasmid pACA-RNA43SD was constructed as follows: The DNA fragment 5΄-aatactcgagGTGAAGTCGTAACAAGGTAACCGTAGGGGAACCTGCGGTTGGATCAGGAGGATAGCGGGCATCGTATAATGGCTATTACCTCAGCCTTCCAAGCTGATGATGCGGGTTCGATTCCCGCTGCCCGCTCCAagatctagcccgcctaatgagcgggcttttttttaagctttata-3΄ comprising an XhoI restriction site (italics), nucleotides 1488–1542 of the rrsB gene (capital letters, bold), followed by the glyT gene (capital letters, underlined), the terminator stem of the trpA gene (small letters, bold) and the HindIII restriction site (italics) was synthesized by GeneArt™ Gene Synthesis (Thermo Fisher Scientific, Regensburg, Germany). The XhoI/HindIII fragment was inserted into plasmid pBAD33 using the SalI/HindIII sites. The ACA cutting site was removed by overlap-PCR with primers IM_U12 and IM_V12 to generate plasmid pGCA-RNA43SD. The SD sequence preceding the emerald-gfp gene encoded in plasmid pUH-C_ΔACA-EmGFP (Oron-Gottesman et al., under revision) was changed to the aSD sequence by inverse PCR using the phosphorylated primers IM_I19 and IM_J19 giving rise to plasmid pUH-C_ΔACA-EmGFPaSD.
Northern blot analysis
Purified RNA was fractionated on a 8 M UREA, 8% polyacrylamide gel, transferred to Hybond membrane (Amersham, GE Healthcare, NJ, USA) using the Trans-Blot Semi-Dry Transfer Cell (Bio-Rad Laboratories, CA, USA), and hybridized to [32P]-labeled oligonucleotides exactly as described before to optimize for short RNA fragments (26). The signals were visualized using a Typhoon PhosphorImager (Molecular Dynamics) and quantified with ImageJ software (27).
Sucrose density gradient analysis
Ribosome profile analysis was essentially performed as described (28). S30 extracts were prepared as previously described (4), and separated on a 10–30% sucrose density gradient made in Tico buffer (20 mM Hepes pH 7,4 at 4°C; 6 mM MgOAc; 30 mM NH4Ac; 4 mM 2-mercapthoethanol) and analyzed using an Äkta FPLC system (GE Healthcare, NJ, USA).
Primer extension analysis
Primer extension analysis using 5 μg of total RNA was performed as previously described (5). Annealing of the 5΄-end-labeled primer Y25 was performed in 1xRT-buffer by heating for 3 min to 80°C, snap freezing in liquid nitrogen, and slowly thawing on ice. Primer extension reactions were performed in RT-buffer by using the AMV reverse transcriptase (Promega, WI, USA) by incubation at 42°C for 30 min. The samples were separated on an 8% PAA-8 M urea gel, and the extension signals were visualized by using a Molecular Dynamics PhosphorImager (GE Healthcare, NJ, USA).
RT-PCR analysis of rRNA
rRNA was isolated from sucrose density gradient fractions containing ribosomes using the acid guanidinium thiocyanate–phenol–chloroform extraction (29). 500 ng purified rRNA were used for RT-PCR employing the ‘One step-RT-PCR kit’ (Qiagen, Hilden, Germany) using primers S7, X15, Y12 or S19. RT-PCR products were analyzed on native 10% PAA gels in TBE buffer.
Purification of the RtcB protein
E. coli strain BL21(DE3) harboring plasmid pTwin1rtcB was grown in LB medium to an OD600 of 0.5. Ectopic expression of the rtcB-intein tag fusion gene was triggered by addition of 1 mM IPTG. One hour after induction, the cells were harvested and resuspended in 50 ml buffer B1 (20 mM Tris–HCl pH 8,5 at 4°C; 500 mM NaCl; 1 mM EDTA) containing 4 mg/ml lysozyme, 0.1 mM PMSF and benzamidine, 10 U RNase free DNase I. Subsequently, cells were incubated on ice for 30 min and lysed by three freeze-thaw cycles followed by sonication (70% power, 5 cycles, 30 s pulse). The cleared cell lysate was applied to equilibrated chitin beads (New England Biolabs, MA, USA) packed in Econo-Pac® Chromatography Columns (Bio-Rad Laboratories, CA, USA) and passed through the column by gravity flow. Washing was performed three times with 10x bed volumes of buffer B1. Self-cleavage of the intein tag was performed on column in buffer B2 (20 mM Tris–HCl pH 7; 500 mM KCl; 1 mM EDTA) for 48 h on 4°C. Elution fractions were checked by SDS-PAGE and subsequent Coomassie brilliant blue staining. The enzymatic activity of RtcB was determined by the tRNA maturation assay as previously described (16).
Preparation of RNA43 and RNA43AgeI
The rrsB gene encoded on plasmid pKK3535 (30) served as template for the PCR with primers W11 (introducing the T7 promoter sequence) and O18. The PCR product containing the T7 promoter followed by the rrsB sequence from position 1491 to 1542 was purified and served as template for in vitro transcription of pre-RNA43 using the ‘Ampliscribe T7, T3 and SP6 High yield transcription kit’ (Epicentre Biotechnologies, WI, USA). After purification the pre-RNA43 was cleaved in vitro using purified E. coli MazF protein in 20 mM Tris–HCl (pH 7,5) and gel purified. For the preparation of RNA43AgeI, the respective mutations were introduced by overlap PCR using plasmid pKK3535 as template and primer pairs J15/H15 and K15/I15. After purification, the PCR product generated with primers I15 and H15 was purified and served as template for the PCR with primers W11 and O18, which was used for in vitro transcription of the pre-RNA43AgeI. The pre-RNA43AgeI was further treated with MazF as described above.
Co-purification assay
Escherichia coli strain MG1655ΔrtcB harboring plasmid pPro-rtcB(+His) or pPro-rtcB(-His) were grown in LB medium at 37°C. 30 min prior to harvest at OD600 = 0.5 and 2.0 ectopic expression was induced by addition of 1 mM IPTG. Cells were resuspended in lysis buffer (20 mM Tris–HCl pH 7.4; 10 mM MgCl2, 30 mM NH4Cl, 150 mM KCl, 10 mM imidazol, 1 mM EDTA, 0.5 mg/ml lysozyme, 4 mM 2-mercapthoethanol) and disrupted by sonication at 4°C (3 pulses, 70% for 20 s; three pulses 70% for 45 s) using a Bandelin sonoplus HD70 sonicator (Berlin, Germany). The cleared lysates were treated with DNase I (Hoffmann La Roche, Basel, Swiss). Upon addition of 1 mM CaCl2, the lysates were divided and treated with or without 2 U/ml micrococcal nuclease for 5 min at 28°C. The reaction was stopped by addition of 2 mM EDTA and cooling to 4°C. Purification of the RtcB-His-tagged protein was performed using Ni-NTA agarose (Qiagen, Hilden, Germany) following the manufacturer's instructions. Fractions were analyzed by SDS-PAGE and western blot analysis employing antibodies specific for the E. coli RtcB protein. Micrococcal nuclease treated samples derived from a culture with an OD600 of 2.0 were analyzed by mass-spectrometry (Campus Science Support Facilities GmbH; Vienna, Austria).
Ribosome purification using affinity chromatography
Escherichia coli strain JE28 harboring pPro-rtcB(-His) was grown in LB medium at 37°C until OD600 of 0.5. Ectopic rtcB expression was triggered by addition of 1 mM IPTG. One hour thereafter cells were harvested and resuspended in lysis buffer (31) and lysed employing the freeze-thaw method. The purification of ribosomes harboring the His-tagged proteins L7/L12 was essentially performed as described (31) with the minor modification that affinity chromatography was performed using a Ni-NTA agarose (Qiagen, Hilden, Germany).
Generation of RtcB-specific antibodies
RtcB protein was purified as described above followed by size exclusion chromatography using a HiLoad Superdex16/60 75 PG in an Äkta FPLC system (GE Healthcare, NJ, USA). Purified RtcB was used to generate polyclonal RtcB-specific antibodies in rabbit by Pineda Antikörper-Service (Berlin, Germany), and further purified using the negative selection method (32).
Western blotting
Total protein samples were separated on denaturing polyacrylamide gels and transferred to a Protran BA83 nitrocellulose membran (GE Healthcare, NJ, USA) using a transblot SD semidry transfer cell (Bio-Rad, CA, USA) with transfer buffer (40 mM glycin, 50 mM Tris–HCl pH 8.3, 0,04% SDS (w/v), 20% methanol (v/v)). The membrane was blocked with 1% casein in 1× TBS for 1 h at RT, after washing incubated with the primary antibody α-E. coli RtcB [1:500 dilution in PBS-T (PBS containing 0,02% Tween 20)] for 1 h at RT. Anti-rabbit Ig-G conjugated with IRDye 800 (Rockland Immunochemicals, PA, USA) was used as secondary antibody in 1:15 000 dilution in PBS-T and incubated for 45 min at RT. Visualization was performed on an ODYSSEY® CLx scanner (LI-COR, NE, USA).
RNA sequencing analysis
RNA sample preparation (total RNA and polysomal RNA) from E. coli strain MC4100 F΄ without or with plasmid pSA1 15 min upon induction of mazF expression was described elsewhere (8). cDNA libraries were prepared using 50–100 ng of the rRNA-depleted RNA with NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, E7420), following the manufacturer's instructions and sequenced on Illumina HiSequ2000 (read length 100bp). Bioinformatical analysis was performed as described in (8).
Purification of 70SΔ43 ribosomes
Ribosomes were purified as described in Vesper et al. (4) with minor modifications. Escherichia coli strain MC4100 F’ harboring plasmid pSA1 was grown at 37°C in LB medium. At OD600 of 0.5 mazF expression was induced by addition of 0.1 mM IPTG. Two hours thereafter, cells were harvested and lysed in Tico buffer containing 4 mg/ml lysozyme, 0.1 mM PMSF and 0.1 mM benzamidine by three freeze and thaw cycles. S30 extract was prepared as described (4). The S30 extract was layered on a 1.1 M sucrose cushion and ribosomes were pelleted, resuspended in Tico buffer and washed with 5 ml high salt buffer (20 mM Tris–HCl, pH 7,5; 10.5 mM MgOAc; 0.4 M NH4Cl; 0.5 mM EDTA; 3 mM 2-mercapthoethanol) using Amicon Ultra columns (100 kDa cutoff; Millipore).
In vitro ribosome repair assay
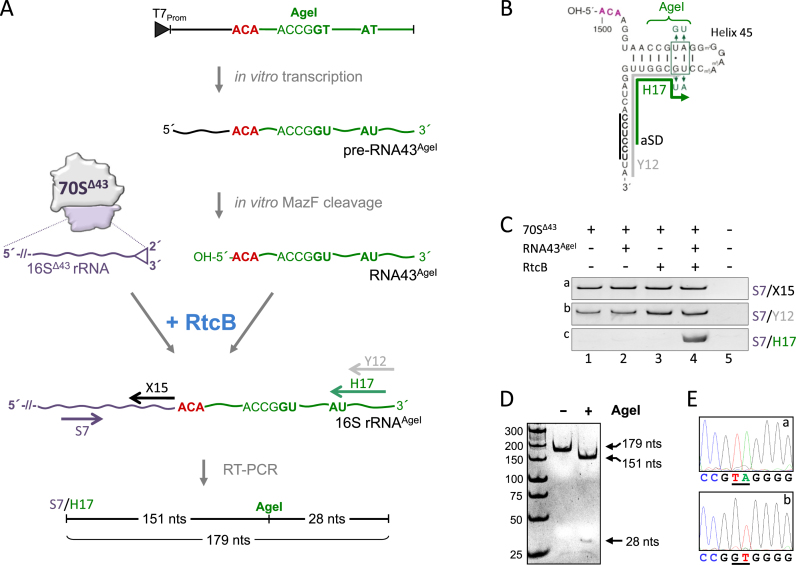
5 pmol RNA43AgeI were incubated for 10 min at 37°C in an equimolar ratio with purified E. coli 70SΔ43 ribosomes in buffer RL (20 mM Tris–HCl, pH 7.5, 6 mM MgCl2, 2 mM MnCl2, 1 mM GTP). Subsequently, 5 pmol purified RtcB were added and incubation was continued for 30 min at 37°C. Upon centrifugation of the reaction mixture over a sucrose cushion to collect ribosomes, RNA was isolated by acid guanidinium thiocyanate-phenol-chloroform extraction (33). 500 ng of the prepared RNA was used for RT-PCR employing the ‘One step-RT-PCR kit’ (Qiagen, Hilden, Germany) using primers S7, X15, Y12 and H17. The RT-PCR product was analyzed on 10% PAA gels in TBE buffer, and purified using ‘PCR clean up kit’ (Qiagen, Hilden, Germany). For AgeI restriction analysis the isolated RT-PCR product was incubated with 4 units of AgeI-HF (New England Biolabs) for 30 min at 37°C. The restriction fragments were determined using PAA gel-electrophoresis and visualized by EtBr-staining. 500 ng of the isolated RT-PCR product was sequenced by Microsynth (Switzerland).
In vivo ribosome repair assay
Escherichia coli strains MG1655 and MG1655ΔrtcB harboring plasmids pgfpaSD and either pACA-RNA43SD or pGCA-RNA43SD were grown at 37°C in M9 minimal media containing maltose (9 mM NaCl, 0.2 mM CaCl2, 2 mM MgSO4, 22 mM KH2PO4, 19 mM NH4Cl, 48 mM Na2HPO4, 10 mM maltose, 20 μg/ml casamino acids, 1 μg/ml thiamine). At OD600 of 0.15, ectopic expression of the different rrsBSD fragments and the gfpaSD gene were triggered for one hour by addition of 0.2% arabinose and 0.1 mM IPTG, respectively. Then, the cells were treated with 100 μg/ml serine hydroxamate (SHX) for 1 h to trigger the mazEF TA system. Subsequently, the cells were gently washed and resuspended in M9 minimal media containing maltose and 0.2% arabinose and 0.1 mM IPTG. Expression of the gfpaSD gene was monitored using a Synergy™ H1 multi-mode microplate reader (BioTek, VE, USA) and 96-well black flat bottom FLUOTRAC™ plates (Greiner Bio-One GmbH, Kremsmünster, A). A600 and emission at 510 nm after excitation at 480 nm were measured for 90 min. The relative fluorescence units (RFU) were calculated by normalizing the measured fluorescence using the A600 values.
RESULTS
70SΔ43 ribosomes and RNA43 are stable under stress conditions
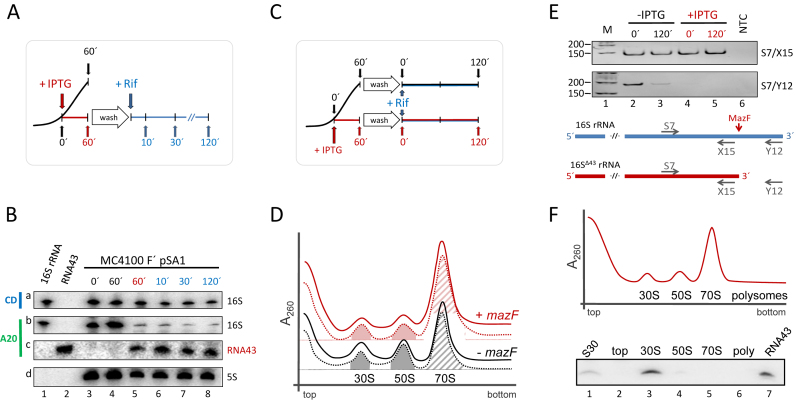
The proposed ribosome repair mechanism relies on the re-ligation of the RNA43 to the 16SΔ43 rRNA present in the specialized ribosomes. Since one prerequisite for the re-ligation reaction to occur is the persistence of the ligation substrates, we first determined the stability of the RNA43, the 16SΔ43 rRNA as well as the 70SΔ43 ribosomes. To this end, we employed E. coli strain MC4100 F΄ harboring plasmid pSA1, which encodes the IPTG-inducible mazF gene (25). As depicted in Figure 2A, 60 min after mazF induction with IPTG the cells were washed and resuspended in fresh media to remove the inductive agent. Then rifampicin (Rif) was added to stop transcription for the determination of RNA stability by northern blotting. The analysis was performed employing probes specific for the central domain of the 16S rRNA (CD, position 955–939; Figure 1B) and its 3΄-terminus (A20, position 1504–1541; Figure 1B) before mazF induction (Figure 2B, lane 3), 60 min thereafter (Figure 2B, lane 5), and 10, 30 and 120 min after addition of rifampicin (Figure 2B, lanes 6–8). A sample withdrawn from the untreated culture at 60 min served as control (Figure 2B, lane 4). Upon normalization using the 5S rRNA (Figure 2B, panel d), our analysis revealed that the amount of total 16S rRNA comprising both, full length 16S rRNA and 16SΔ43 rRNA, remains constant throughout the experiment (Figure 2B, panel a). However, after mazF induction the majority of the 16S rRNA lacks the 3΄-terminal fragment as the intensity of the signal obtained with probe A20 at the migration position of the full length 16S RNA was reduced 5- to 8-fold (Figure 2B, panel b, lanes 5–8). In addition, the amount of RNA43 did not change for 120 min upon addition of rifampicin (Figure 2B, panel c, lanes 5–8), again indicating its stability during stress conditions. Taken together, these data indicate that the relative amount of 16SΔ43 rRNA remains constant during the experiment.
Figure 2.
RNA43, 16SΔ43 rRNA and 70SΔ43 ribosomes are stable during stress conditions. (A) Schematic depiction of the experimental approach to assess the stability of RNA43 and 16SΔ43 rRNA in vivo. A schematic growth curve in the absence of IPTG is shown in black. At OD600 of 0.3 the culture was divided and IPTG was added to one half to induce mazF expression (growth is blocked as indicated in red). 60 minutes thereafter, cells were washed and resuspended in fresh medium comprising rifampicin (in blue). (B) Samples withdrawn at the time points indicated were subjected to northern blot analysis with probes specific for the central domain of the 16S rRNA (CD, panel a), the 3΄-terminus of the 16S rRNA (A20; panel b) and the RNA43 (A20; panel c). 5S rRNA was used as internal standard for quantification (panel d). In vitro transcribed 16S rRNA (lane 1) and RNA43 (lane 2) served as size markers. The experiment was performed in triplicate and one representative autoradiograph is shown. (C) Schematic of the experimental approach to assess the stability of 70SΔ43 ribosomes in vivo as described in A. (D) S30 extracts were prepared before (dotted lines) and 120 min after addition of rifampicin (solid lines) to untreated cells (black lines) or 60 min after induction of mazF expression (red lines) and subjected to sucrose density gradient analysis. Peaks representing 30S and 50S subunits and 70S ribosomes are indicated. The peak areas of the 30S and 50S subunits (filled areas) and 70S monosomes (hatched area) that were quantified to determine the subunits/monosome ratios are indicated. (E) To monitor MazF-mediated processing of the 16S rRNA, RNA was isolated from the fractions comprising the 70S monosomes. RT-PCR analysis using primers S7/X15 (upper panel), specific for both intact 16S rRNA and 16SΔ43 rRNA, and primers S7/Y12 (lower panel), yielding a product only with uncleaved 16S rRNA. NTC: no template control. Below, the binding sites of the primers are given schematically. (F) Ribosome sedimentation profiles of cell extracts 30 min after induction of mazF expression. Total RNA was purified from the indicated fractions (top, 30S, 50S, 70S and polysomes), respectively, and tested for the presence of RNA43 by northern blotting. Total RNA purified from the S30 extract withdrawn 30 min upon induction of mazF expression (lane 1), and in vitro transcribed RNA43 (lane 7) served as controls.
To assess integrity of the stress-ribosomes, we next performed ribosome profile analyses as schematically depicted in Figure 2C. Samples were withdrawn before (Figure 2D, dotted lines) and 120 min after addition of rifampicin (Figure 2D, solid lines) to untreated cells (Figure 2D, black lines) or 60 min after mazF expression was triggered by addition of IPTG (Figure 2D, red lines). Collectively, this analysis revealed no differences in the relative positions of the peaks corresponding to the 30S and 50S subunits, and the 70S monosomes before and 120 min after addition of rifampicin indicating that the stress-ribosomes are stable entities. Notably, a quantitative analysis of the respective peaks revealed that after induction of mazF expression the subunits/monosome (SU/M) ratio decreased from 0.71 ± 0.04 to 0.32 ± 0.04. This prevalence of 70SΔ43 monosomes is consistent with the 70S initiation pathway occurring on leaderless mRNAs (34) and fosters the hypothesis that selective translation of MazF-processed transcripts initiates with the recognition of the 5΄-terminus by non-dissociated 70SΔ43 ribosomes (8).
RT-PCR analyses performed on RNA purified from the respective 70S monosome fractions revealed that under the applied conditions nearly all 70S ribosomes are processed by MazF, as the RT-PCR using primers S7 and Y12, which flank the cleavage site and are thus specific for the full length 16S rRNA, did not yield a product (Figure 2E, lanes 4 and 5, lower panel), despite the presence of the processed 16SΔ43 rRNA as determined with the primers S7/X15 (Figure 2E, lanes 4 and 5, upper panel). In contrast, ribosomes purified from untreated cells containing intact 16S rRNA yield the specific products using both primer pairs (Figure 2E, lane 2). Interestingly, the weak signal specific for the full length 16S rRNA, which we obtained upon addition of rifampicin without mazF induction (Figure 2E, lane 3, lower panel) indicates that rifampicin treatment induces the formation of stress-ribosomes. Thus, our data confirm the results by Sat et al. (35), which show that rifampicin activates the TA system mazEF. In line with our previous results (4), further analysis of the protein complement of the respective subunits and monosomes did not indicate changes in protein composition of the ribosome upon induction of mazF expression (data not shown).
The RNA43 remains associated with 30S subunits during stress
We next investigated whether the RNA43 remains associated with the ribosome after MazF cleavage. We analyzed the ribosome sedimentation profile of strain MC4100 F΄ harboring plasmid pSA1 30 min after induction of mazF expression (Figure 2F). The presence of RNA43 in the respective fractions was determined by northern blotting using probe A20 specific for the 3΄ terminus of the 16S rRNA. Surprisingly, this analysis revealed that the RNA43 co-migrates exclusively with 30S subunits (Figure 2F, lane 3). We did not detect RNA43 on the top of the gradient or associated with 50S subunits, 70S monosomes, or polysomes (Figure 2F; lanes 2, 5, and 6, respectively).
The rtcB mRNA is processed by MazF and selectively translated during stress
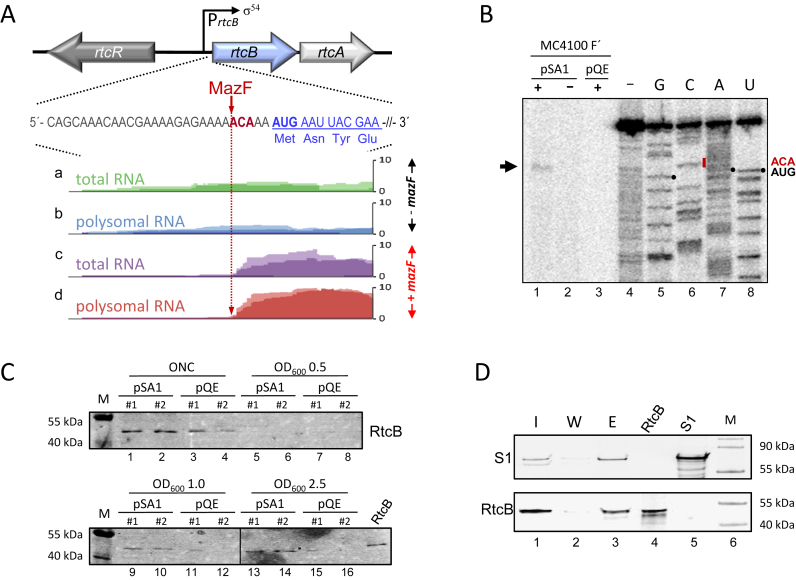
As mentioned above the expression of the rtcBA operon is controlled by the alternative sigma factor σ54 together with the transcription factor RtcR (Figure 3A), already suggesting a function in response to specific conditions. Interestingly, closely upstream of the AUG start codon of rtcB a MazF cleavage site is present, the processing of which was indicated by comparative RNA sequencing analysis performed on total RNA (Figure 3A, panels a and c) and polysomal RNA (Figure 3A, B and D) before (Figure 3A, A and B) and after ectopic overexpression of mazF (Figure 3A, panels c and d) (8). In addition, the analysis indicates that rtcB mRNA levels are increased 4–5-fold upon mazF expression (Figure 3A, panels c and d), and that the leaderless rtcB mRNA is specifically translated by the 70SΔ43 ribosomes as suggested by the presence of the leaderless rtcB mRNA in the polysomal fraction (Figure 3A, panel d). We further validated this MazF cleavage site by primer extension analysis performed on total RNA purified from strain MC4100 F΄ harboring plasmid pSA1 (Figure 3B). Without mazF induction we did not obtain an extension signal for the full length rtcB mRNA reflecting the low rtcB levels under this condition (Figure 3B, lanes 2 and 3). However, after induction of mazF expression we detected a signal, which corresponds to the 5΄-terminus processed by MazF (Figure 3B, lane 1). Further western blot analyses shown in Figure 3C likewise reveal the increased RtcB levels in response to MazF. E. coli strain MG1655 harboring either the mazF encoding plasmid pSA1 (Figure 3C; lanes 1, 2, 5, 6, 9, 10, 13 and 14) or the parental plasmid pQE30 (Figure 3C; lanes 3, 4, 7, 8, 11, 12, 15 and 16) was grown in LB at 37°C. Given the leaky expression of mazF controlled by the lac promoter in plasmid pSA1 the cells were grown in the absence of the inductive agent IPTG (see also Figure 4C). The samples were withdrawn from over night cultures (ONC), during early exponential phase (OD600 0.5), mid exponential phase (OD600 1.0), and late exponential phase (OD600 2.5) and subjected to western blot analysis using anti-RtcB antibodies. The results reveal that RtcB levels are increased when the cells harbor the mazF encoding plasmid pSA1 in contrast to cells harboring pQE. In these cells, the concentration of RtcB is below the detection level during exponential growth. However, rtcB expression increases during growth and reaches a maximum in the stationary phase as exemplified by the signal observed with the ONC samples (Figure 3C; lanes 1–4). Taken together, these results indicate that the RtcB ligase is synthesized specifically in response to the stationary phase or the presence of MazF.
Figure 3.
rtcB mRNA is processed by MazF, and RtcB interacts with the ribosome. (A) Depiction of the rtc operon comprising rtcR and rtcBA. The sequence around the AUG start codon (bold) of the rtcB mRNA is given. The 5΄ UTR (gray), the MazF cleavage site (red) and the proximal coding region (blue) are indicated. The normalized reads obtained by RNA sequencing of total (a and c) and polysomal (b and d) RNA isolated during exponential phase before (a and b; -mazF) and 15 min after induction of mazF expression encoded by plasmid pSA1 (25) (c and d; +mazF) are shown. The analysis was performed twice and the light and dark shades of the individual colors indicate the alignment of the reads in each replicate. The MazF-dependent removal of the 5΄-UTR is indicated by the lack of reads upstream of the ACA site (dotted, red arrow) after mazF induction. (B) Primer extension analysis with an rtcB specific primer using total RNA purified from strain MC4100 F΄ harboring plasmids pSA1 or pQE with (+) and without (–) addition of IPTG. The primer extension (lane 4) and sequencing reactions (lanes 5–8) of in vitro transcribed rtcB mRNA are shown. The positions of the MazF cleavage site (ACA) and the AUG start codon are given to the right. The arrow indicates the signal obtained after mazF expression. (C) Two replicates of strain MG1655 harboring either pSA1 or pQE30 were grown in LB in the absence of IPTG. Samples were withdrawn from the over night culture (ONC) and during growth at the indicated OD600 values. The same amount of total protein as determined by Coomassie staining was used to determine the RtcB levels by western blotting. The molecular weights of the marker proteins are given and purified RtcB served as control. (D) Co-elution of RtcB with affinity purified ribosomes was determined by western blot analysis using antibodies specific for ribosomal protein S1 (upper panel) and RtcB (lower panel). Samples were withdrawn during the Ni-NTA purification procedure from the input fraction (I; lane 1), the last washing fraction (W; lanes 2) and the elution fraction (E; lane 3). Purified proteins RtcB (lane 4) and bS1 (lane 5) served as controls. The molecular weights of the marker proteins are given (lane 6).
Figure 4.
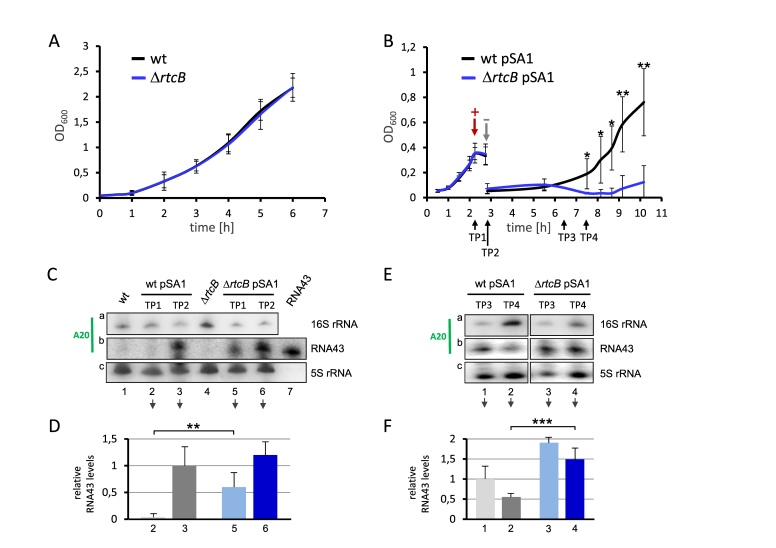
RtcB facilitates growth recovery after mazF expression and reduces the abundance of RNA43. (A) Growth of strain MC4100 F΄ (black) and its isogenic rtcB deletion mutant (blue) in LB broth at 37°C. See also Supplementary Figure S2. (B) Strain MC4100 F΄ (black) and its isogenic rtcB deletion mutant (blue) both harboring plasmid pSA1 were grown in LB broth to an OD600 of 0.35, when mazF expression was induced by addition of IPTG (+). 30 min thereafter cells were transferred in fresh medium to remove the inductive agent (–) and growth recovery was monitored for additional 7 hours. The analysis was performed in triplicate, error bars indicate the standard deviation from the mean (*P < 0.05; **P < 0.01). See also Supplementary Figure S2. (C) At time points (TP) 1 and 2 indicated in B total RNA was purified and the abundance of the 16S rRNA and RNA43 was determined by northern blotting employing specific probes for RNA43 (panel a and b), and 5S rRNA (panel c), which served as internal standard. (D) The RNA43 signals (panel b) were quantified and normalized to signals obtained for the 5S rRNA (panel c) and the relative RNA43 levels are depicted in the graph. The relative RNA43 level obtained upon mazF expression in the wild-type strain was set to 1 (lane 3). The analysis was performed in triplicate (one representative autoradiograph is shown) and error bars indicate the standard deviation from the mean (**P < 0.01). RNA samples prepared from the respective strains not harboring plasmid pSA1 (lanes 1 and 4) and in vitro transcribed RNA43 (lane 7) served as controls. (E) Likewise, during recovery from mazF overexpression at TP3 and TP4 indicated in B total RNA was prepared and northern blot analysis was performed as described in C. One representative autoradiograph of three independent experiments is shown. (F) The signals were quantified and normalized and the relative RNA43 levels are depicted in the graph. Again, the relative RNA43 level obtained upon mazF expression in the wild type strain was set to 1 (lane 1). Error bars indicate the standard deviation from the mean (***P < 0.001).
RtcB interacts weakly with ribosomes
We further studied the interaction of RtcB with the translational machinery comparing ribosome profiles of strain BW25113, its isogenic rtcB deletion strain, and strain BW25113 expressing rtcB ectopically from plasmid pBAD-rtcB. In contrast to a recent report that indicates a shift in the sedimentation of 70S ribosomes in the absence of rtcB (21), we directly analyzed the ribosome profile by separating the ribosomal subunits, 70S monosomes and polysomes from S30 extracts of the respective strains on 10–30% sucrose gradients. As shown in Supplementary Figure S1A, the absence or overexpression of rtcB did not affect the relative positions of ribosomes and subunits. To determine the co-migration of RtcB with ribosomes or subunits, we tested the respective fractions of the gradients shown in Supplementary Figure S1A as well as upon mazF expression shown in Figure 2D by western blot analysis. All experiments revealed that RtcB is present in the top fraction. We did not detect RtcB in the ribosomal fractions (data not shown), which might be attributed to the low levels of RtcB even after induction of mazF or rtcB expression from plasmids pSA1 or pBAD-rtcB, respectively. To further increase the amount of RtcB, we employed the T7 expression system. Surprisingly, these high levels of rtcB expression resulted in a strongly increased SU/M ratio (Supplementary Figure S1B), what we already observed to a lesser extent upon mild overexpression from plasmid pBAD-rtcB (Supplementary Figure S1A), suggesting that an excess of the protein might interfere with the subunit association. Subsequent western blot analysis of the gradient fractions revealed that RtcB was present in the top fractions as well as in the fractions that comprise the ribosomal subunits (Supplementary Figure S1C). However, this migration pattern might be entailed by the high amount of RtcB present in the sucrose density gradient rather than by interaction and subsequent co-migration with the ribosomal subunits. Moreover, considering the low levels of RtcB under physiological conditions that are sufficient to repair a high number of processed ribosomes, we hypothesized that the association between ribosomes and RtcB might be transient or weak, allowing the removal of RtcB during sucrose density centrifugation. Thus, we further validated the interaction by gentle co-purification with the ribosomes using E. coli strain JE28 (31). After ectopic overexpression of rtcB 70S ribosomes were purified via the His-tagged proteins L7/L12. Qualitative western blot analyses revealed that RtcB co-elutes with ribosomes (Figure 3D, lane 3).
In a complementary pull-down approach, extracts from E. coli strain MG1655ΔrtcB synthesizing either the N-terminally His-tagged RtcB variant or the untagged RtcB protein as mock control were treated with micrococcal nuclease to exclude indirect interactions mediated via nucleic acids. After affinity chromatography using Ni-NTA beads the eluted fractions were analyzed by mass spectrometry. As shown in Supplementary Table S1, the comparative analysis revealed that the majority of ribosomal proteins co-purified with His-tagged RtcB, indicating that the RNA ligase associates with the translational machinery. Besides, several stress associated proteins (e.g. the chaperonins GroEL and GroES, the universal stress protein G, the Lon protease, and others) co-eluted with RtcB.
Stress recovery is severely compromised and RNA43 levels are increased in the absence of RtcB
Next, we determined whether RtcB deficiency affects E. coli growth during stress recovery. In contrast to the marginal effect of the rtcB deletion on exponential growth (Figure 4A), the recovery of the mutant strain was severely compromised upon ectopic overexpression of mazF (Figure 4B). To exclude a polar effect on the expression of the cyclase gene rtcA, we additionally monitored growth of the isogenic rtcA deletion mutant under the same conditions (Supplementary Figure S2A and B), and found that the detrimental effect on stress recovery can be solely attributed to the deficiency of RtcB.
To test for our assumption that RtcB re-ligates RNA43 and 16SΔ43 rRNA in vivo, we performed northern blotting to determine the abundance of the RNA43 either as part of the intact 16S rRNA (Figure 4C and E, panel a) or in a free form after MazF cleavage (Figure 4C and E, panel b). Total RNA was extracted during the growth analysis shown in Figure 4B before and 30 min after induction of mazF expression (Figure 4C), and at time points indicated in Figure 4B after re-inoculation in fresh LB medium (Figure 4E). Interestingly, we did not detect RNA43 in the wild-type strain harboring plasmid pSA1 before induction of mazF expression by IPTG (Figure 4C, panel b, lane 2; Figure 4D). In striking contrast under this condition significant amounts of RNA43 were present in the rtcB deletion strain (Figure 4C, panel b, lane 5; Figure 4D). This observation is supported by the corresponding reduction of the signal obtained for the full length 16S rRNA (Figure 4C, panel a, lane 5). Taken together, our results strongly suggest that RtcB is proficient to antagonize the activity of MazF that is synthesized due to leaky expression controlled by the lac promoter in plasmid pSA1. Upon induction of mazF this difference in RNA43 abundance between wild type (Figure 4C, panel b, lane 3; Figure 4D, value was set to 1) and rtcB deletion mutant (Figure 4C, panel b, lane 6; Figure 4D) was mitigated, due to higher MazF levels that overcome RtcB activity. During outgrowth from stress RNA43 was cleared much faster in the wild-type strain (Figure 4E, panel b, lanes 1 and 2; Figure 4F) when compared to the rtcB deletion mutant (Figure 4E, panel b, lanes 3 and 4; Figure 4F) and correspondingly, the signal obtained for the intact 16S rRNA increased faster in the wild-type strain (Figure 4E, panel a, compare lane 2 and lane 4). Together, these results suggest that RtcB reduces the abundance of RNA43 in vivo.
RtcB re-ligates RNA43 to 16SΔ43 rRNA in the context of 70SΔ43 ribosomes in vitro
To directly validate that RtcB is responsible for the ligation of the MazF-processed 16SΔ43 rRNA and RNA43 in the context of 70SΔ43 ribosomes we performed an in vitro analysis using purified components (Figure 5A). To unambiguously discriminate between repaired ribosomes and accidentally co-purified intact 70S ribosomes, we examined the re-ligation of the alternative RNA43AgeI (Figure 5B). First, we in vitro transcribed a 54 nts long pre-RNA43AgeI (nts 1488–1542 of 16S rRNA) containing the ACA cleavage site and four non-canonical nucleotides, which maintain the structure of helix 45, but introduce an AgeI restriction site to the final RT-PCR product (Figure 5A and B). The pre-RNA43AgeI was further processed by MazF in vitro to generate the 5΄-hydroxyl terminus required for RtcB ligation. Then, purified 70SΔ43 ribosomes (4) were incubated with RNA43AgeI in the presence or absence of recombinant RtcB. The reaction was stopped by pelleting the ribosomes over a sucrose cushion to selectively purify the 16S rRNA incorporated in ribosomes for further analysis. The correct ligation was monitored by RT-PCR employing the forward primer S7 (Figure 5A, purple arrow) in combination with the reverse primers X15, which anneals upstream of the ACA site (Figure 5A, black arrow), Y12, which preferentially binds to the 3΄-terminus of the native 16S rRNA (Figure 5A and B, gray arrow), or H17, which is specific for the RNA43AgeI (Figure 5A and B, green arrow), respectively. The RT-PCR analysis performed with primers S7/X15 yielded the same signal intensity in all four reactions revealing that the same amount of ribosomes was present (Figure 5C, panel a). The signal obtained with primers S7/Y12 indicated the presence of accidentally co-purified intact 70S ribosomes (Figure 5C, panel b, lanes 1–4). Employing primers S7/H17 the analysis clearly showed the correct ligation of the 16SΔ43 rRNA and the RNA43AgeI in the presence of both, the RNA43AgeI and RtcB (Figure 5C, panel c, lane 4). The presence of the AgeI site in the respective RT-PCR product was further verified by subsequent restriction analysis (Figure 5D) and sequencing (Figure 5E) of the obtained RT-PCR product.
Figure 5.
RtcB re-ligates RNA43 to purified 70SΔ43 ribosomes in vitro. (A) Schematic showing the in vitro ribosome repair assay (see text for details). (B) RNA43AgeI used in the assay. Nucleotides changed in helix 45 and the binding site for primers Y12 (gray) and H17 (green) are indicated. (C) RT-PCR performed on RNA purified from 70SΔ43 ribosomes incubated in the absence of RtcB and RNA43AgeI (lane 1), in the presence of either RNA43AgeI (lane 2) or RtcB (lane 3), or both (lane 4) employing primer pairs specific for the central region of 16S rRNA (S7/X15; panel a), the 3΄end of 16S rRNA (S7/Y12; panel b), and specific for the AU nucleotide change present in 16SAgeI rRNA upon successful ligation (S7/H17; panel c). (D) AgeI restriction analysis and (E), sequencing of the RT-PCR product obtained with primer pair S7/H17 (shown in C, panel c, lane 4). Sequencing analyses from position 1509–1517 of the 16S rRNA of (a) rrsB, and (b) the RT-PCR product are shown. Nucleotides changed in RNA43AgeI are underlined.
70SΔ43 ribosomes repaired by RtcB are able to translate canonical mRNAs
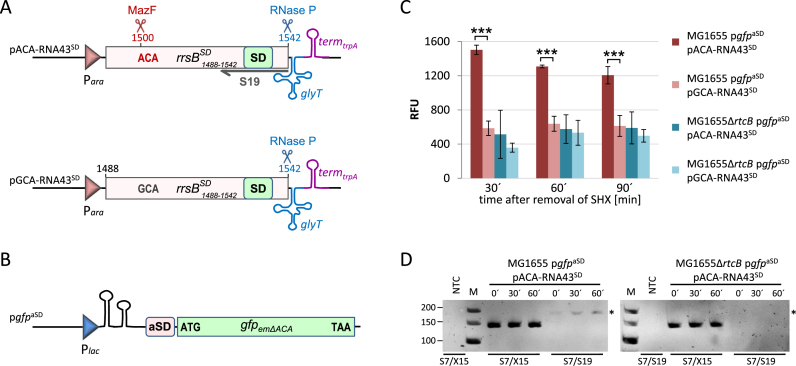
Finally, we tested whether repaired ribosomes are capable to translate canonical mRNAs in vivo employing a specialized ribosome system (36). We ectopically expressed a modified RNA43 wherein the aSD sequence was replaced by the SD sequence (RNA43SD) and confirmed its RtcB-mediated ligation to the 16SΔ43 of stress-ribosomes by monitoring the specific translation of a reporter mRNA that displays an aSD sequence upstream of the AUG start codon during recovery from stress (Figure 6). To ensure the precise 5΄- and 3΄-terminal processing of the RNA43SD, which is essential for the ligation and the functionality of the repaired ribosomes, the 3΄-terminal 54 nucleotides of the rrsBSD gene harboring the SD sequence were designed to be followed immediately by the glyT gene, encoding the tRNAGly, with transcription ending at the trpA terminator (Figure 6A). Consequently, the 3΄-end of the RNA43SD is correctly processed by RNase P, the enzyme that removes 5΄ leader sequences in pre-tRNAs (37). Upon activation of MazF the mature RNA43SD is generated, since the cleavage site at position 1500 is present in the ACA-rrsBSD fragment (Figure 6A, pACA-RNA43SD). As control, we therefore used the GCA-rrsBSD construct where we changed the ACA site to GCA to prevent MazF cleavage (Figure 6A, pGCA-RNA43SD). To determine the translational activity of the repaired ribosomes containing the modified 16SSD rRNA, we used a gfp reporter featuring the aSD sequence (Figure 6B, pgfpaSD). For the analysis strains MG1655 and MG1655ΔrtcB, both harboring plasmid pgfpaSD and either plasmid pACA-RNA43SD or pGCA-RNA43SD, respectively, were grown in minimal medium. At OD600 of 0.15, the expression of the reporter gene and the different rrsBSD fragments was induced for 60 min. Then, the cells were treated for 60 min with 100 μg/ml serine hydroxamate (SHX) to trigger the mazEF system (4). After removal of SHX the cells were resuspended in fresh medium and both, growth and GFP fluorescence were monitored during the recovery at 30, 60 and 90 min. We anticipated that besides the usual re-ligation of the endogenous RNA43 the generated RNA43SD fragments will be ligated by RtcB to the truncated 16S rRNA of a fraction of 70SΔ43 ribosomes during the recovery phase. Thereby, a subpopulation of 70SSD ribosomes that specifically translates the gfpaSD reporter mRNA will be generated. Indeed, in the wild type strain MG1655 expressing the ACA-rrsBSD construct the relative fluorescence (RFU) levels are 3-fold higher after 30 min of recovery when compared to the control strains (Figure 6C). Strain MG1655 harboring the GCA-rrsBSD fragment that cannot be processed by MazF, as well as strain MG1655ΔrtcB having either the ACA-rrsBSD or the GCA-rrsBSD gene only showed a low gfp expression, which can be attributed to unspecific translation initiation by canonical ribosomes (36). As expected, during ongoing outgrowth the specific RFU levels of strain MG1655 expressing the ACA-rrsBSD construct decreased again due to resumed growth and the concomitant de novo ribosome biosynthesis, which results in the dilution of specialized 70SSD ribosomes (Figure 6C). Concomitantly, we monitored the formation of the 16SSD rRNA as a proxy for the RtcB-mediated ribosome repair. Using primers S7 and S19, which is specific for the 3΄-terminus of the 16SSD rRNA (Figure 6A), we performed RT-PCR on total RNA extracted from strains MG1655 and MG1655ΔrtcB that express the ACA-rrsBSD construct before (time point 0΄) and 30 and 60 min after removal of SHX (Figure 6D). The analysis shows that the 16SSD rRNA is only formed in the wild type strain, whereas in the absence of rtcB we did not detect the specific product. Interestingly, we detected a small amount of the 16SSD rRNA already before removal of SHX (time point 0΄) indicating that the re-ligation by RtcB occurs already during stress treatment. However, the concentration of 16SSD rRNA is increased during the recovery phase 30 and 60 min after removal of SHX.
Figure 6.
RtcB restores the ability of the ribosomes to translate canonical mRNAs in vivo. (A) Schematic depiction of the construct encoded by plasmid pACA-RNA43SD used to ectopically express the rrsBSD fragment that can be processed by MazF at the ACA-site to generate the mature RNA43SD. Below, the control construct comprising the rrsBSD fragment without the ACA-cleavage site is shown (pACA-RNA43SD), which cannot be processed by MazF. The binding site of the primer S19 used for RT-PCR analysis shown in D is indicated. See text for details. (B) Scheme showing the corresponding gfpaSD reporter gene of plasmid pgfpaSD, which does not harbor ACA-sites not to interfere with mRNA levels due to activation of MazF during stress treatment (Oron-Gottesman et al., submitted). See text for details. (C) The relative fluorescence units (RFU, normalized to OD600) were measured for strains MG1655 and the isogenic rtcB-deletion strain harboring the gfpaSD reporter gene and either plasmid pACA-RNA43SD or plasmid pGCA-RNA43SD at the indicated time points before (0 min) and during (30 and 60 min) recovery from SHX treatment. The experiment was performed in triplicate; error bars indicate the standard deviation from the mean (***P < 0.001). (D) RT-PCR analysis to monitor the formation of the 16SSD rRNA by RtcB-mediated ligation during recovery from SHX-treatment in strains MG1655 and MG1655ΔrtcB. Primer pair S7/S19 specifically amplifies a 182 nts long product only after ligation of the 16SΔ43 rRNA and the ectopically expressed RNA43SD (indicated by an asterisk). Primers S7/X15 served as internal control. NTC: no template control. The length of the marker bands in nts is given to the left.
DISCUSSION
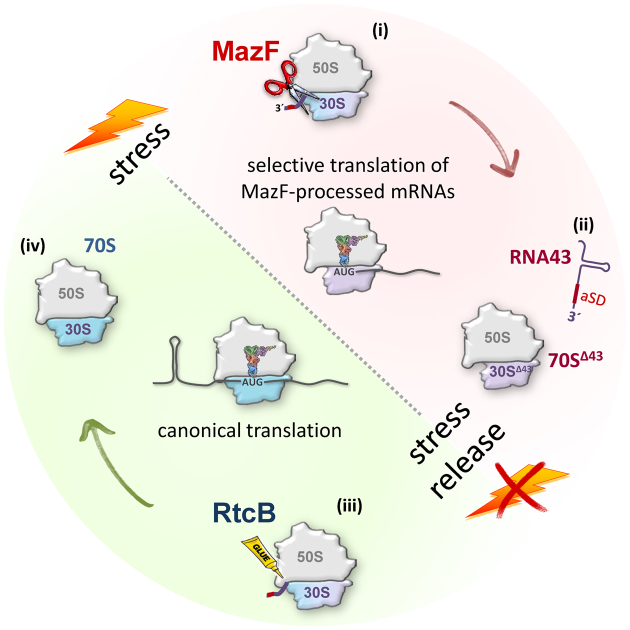
When E. coli encounters stress, several response mechanisms are triggered. In general, the bacterium adapts to the given conditions by the alteration of the transcriptome, which requires the expression of alternative sigma factors to adjust the specificity of the RNA polymerase to distinct promoters thereby ensuring the transcription of a specific set of genes. In contrast to this time- and energy-demanding transcriptional stress response we have shown that functional ribosome heterogeneity opens the possibility to instantaneously adapt protein synthesis to sudden and harsh alterations of environmental cues (4). The hallmark of this post-transcriptional stress response pathway is the modification of ribosomes by the endoribonuclease MazF, which as a result selectively translate mRNAs that are likewise processed by MazF within their 5΄-UTR and thus constitute the ‘MazF-regulon’ (Figure 7, i and ii) (4,8,38). This observation raises the fundamental question about the fate of the specialized 70SΔ43 ribosomes during the recovery from stress. When canonical mRNAs are transcribed again, these specialized ribosomes would become a burden for the cells and are expected to be discarded. Considering that disposal of the cleavage-modified ribosomes would require the energy-demanding synthesis of new ribosomes, we tested whether a repair mechanism exists that reverses the MazF-mediated ‘one-step ribosome specialization’ and restores the translational proficiency of the protein synthesis machinery to translate mRNAs harboring a canonical 5΄-UTR (Figure 7, iii and iv). Our experiments show that the RNA ligase RtcB re-ligates RNA43 and the truncated 16SΔ43 rRNA present in 70SΔ43 ribosomes. Considering the transient association of the RNA43 with 30S subunits (Figure 2E) we hypothesize that this step occurs most likely during the process of translation initiation on canonical mRNAs transcribed again during the recovery phase. Thereby, the SD–aSD interaction could contribute to the regulation of ribosome repair at two levels: first, the interaction could ensure the correct timing of the ribosome repair not before stress relief, viz. canonical mRNAs are synthesized again; second, the RNA43 would be correctly positioned on the ribosome via the interaction with the SD sequence of the canonical mRNA located in the ribosomal mRNA track to facilitate re-ligation by RtcB.
Figure 7.
Model for the reversible stress adaptation of the translational machinery in E. coli. When E. coli encounters stress (red) the endoribonuclease MazF is activated and removes the 3΄-terminal 43 nts (RNA43) from the 16S rRNA incorporated in active ribosomes (i). The resulting 70SΔ43 ribosomes selectively translate MazF-processed mRNAs to adapt protein synthesis to the adverse conditions (ii). Upon stress relief when canonical mRNAs are transcribed again, the specialized ribosomes become redundant. To regenerate these ribosomes the RNA ligase RtcB re-ligates the 16SΔ43 rRNA present in the stress-ribosomes and RNA43 (iii), thereby restoring the translational proficiency of 70S ribosomes to ensure canonical translation during relaxed conditions (green) (iv).
The abundance of the ligase RtcB is very low and its expression is controlled by the alternative σ54 factor together with the transcription factor RtcR (17) consistent with the role of RtcB during specific cellular or environmental conditions (39). In line with these results, we show that the abundance of RtcB is further regulated at multiple levels. We observed that the rtcBA transcript is processed by MazF and, as a consequence, is selectively translated by stress-ribosomes (Figure 3). Together, this feedback mechanism allows the tightly controlled synthesis of the enzyme during stress conditions that trigger the mazEF system and thus ensures its presence during stress relief when the ligase is required for ribosome repair. Collectively, our finding establishes RtcB as an important factor that reconstitutes the canonical translational apparatus, representing the first distinct physiological function of the RNA ligase RtcB in bacteria.
Notwithstanding the first indications for the generation of specialized ribosomes reported more than 40 years ago (40,41), the concept of modulating protein synthesis by reversible ribosome heterogeneity is still a matter of debate. Our results reported here provide evidence that the selectivity of the translational machinery can be constantly adapted by MazF and RtcB. These antagonizing factors provide a flexible and energy-efficient link between the number of specialized ribosomes and changing environmental conditions. This idea is supported by the observation that RtcB can effectively re-ligate the RNA43 generated by low levels of MazF (Figure 4C, TP 1). Thus, it is conceivable that even under non-stress conditions a minor active fraction of MazF might continuously generate a specialized ribosomal subpopulation responsible for translation of short-leadered and leaderless transcripts. On the other hand, the size of this subpopulation can be down-regulated to a minimum by RtcB, the expression of which is in turn controlled by MazF (Figure 3). Since the cell is constantly equipped with all factors involved in this process, this balancing mechanism provides a means for an immediate and almost reflexive response to external stimuli that trigger the mazEF TA system, what is in stark contrast to the time-consuming and concerted transcriptional stress response. Taken together, our study underscores the physiological importance of ribosome heterogeneity as a key mechanism within the bacterial stress response network and raises the translational apparatus from a cellular factory required to make proteins to a control unit with an immense regulatory capacity.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Witold Filipowicz and Stefan Weitzer for their critical reading of the manuscript and Hanna Engelberg-Kulka for providing purified protein MazF.
Author contributions: H.T., C.M., M.S., O.V., A.R., J.P., J.M. and I.M. conceived and designed the experiments; H.T., C.M. and M.S. conducted the experiments; O.V. prepared and provided ribosomes and H.T. and I.M. wrote the manuscript.
Author information: The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to I.M. (isabella.moll@univie.ac.at).
Footnotes
Present addresses:
Oliver Vesper, Center for Structural Systems Biology (CSSB), University Medical Center Eppendorf-Hamburg (UKE), Martinistrasse 52, D-20246 Hamburg, Germany, and Deutsches Elektronen-Synchrotron (DESY), Notkestrasse 85, D-22607 Hamburg, Germany, Institute of Molecular Biotechnology GmbH (IMBA), Austrian Academy of Sciences, Dr Bohr-Gasse 3–5, A-1030 Vienna, Austria, Research Institute of Molecular Pathology (IMP), Dr Bohr Gasse 7, A-1030 Vienna, Austria.
Johannes Popow, Boehringer Ingelheim—Regional Center Vienna GmbH, 1121 Vienna, Austria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Special Research Program 'RNA-REG' on ‘RNA regulation of the translatome’ by Austrian Science Fund [F4316-B09, P22249-B20, P26946-B20 to I.M.]. Funding for open access charge: Austrian Science Fund FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Keener J., Nomura M.. Curtiss R., III Neidhardt FC. Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. Regulation of ribosome biosynthesis. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 1996; 1, Washington, DC: ASM Press; 1417–1431. [Google Scholar]
- 2. Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. [DOI] [PubMed] [Google Scholar]
- 3. Pulk A., Liiv A., Peil L., Maivali U., Nierhaus K., Remme J.. Ribosome reactivation by replacement of damaged proteins. Mol. Micro. 2010; 75:801–814. [DOI] [PubMed] [Google Scholar]
- 4. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I.. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011; 147:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aizenman E., Engelberg-Kulka H., Glaser G.. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [corrected] 3΄,5΄-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R.. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Gen. 2006; 2:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shine J., Dalgarno L.. The 3΄-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sauert M., Wolfinger M.T., Vesper O., Müller C., Byrgazov K., Moll I.. The MazF-regulon: A toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakravarty A.K., Shuman S.. The sequential 2΄,3΄-cyclic phosphodiesterase and 3΄-phosphate/5΄-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 2012; 40:8558–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakravarty A.K., Subbotin R., Chait B.T., Shuman S.. RNA ligase RtcB splices 3΄-phosphate and 5΄-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3΄)pp(5΄)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Das U., Shuman S.. 2΄-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA. 2013; 19:1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka N., Chakravarty A.K., Maughan B., Shuman S.. Novel mechanism of RNA repair by RtcB via sequential 2΄,3΄-cyclic phosphodiesterase and 3΄-phosphate/5΄-hydroxyl ligation reactions. J. Biol. Chem. 2011; 286:43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka N., Meineke B., Shuman S.. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011; 286:30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka N., Shuman S.. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011; 286:7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maughan W.P., Shuman S.. Distinct contributions of enzymic functional groups to the 2΄,3΄-Cyclic phosphodiesterase, 3΄-Phosphate guanylylation, and 3΄-ppG/5΄-OH ligation steps of the Escherichia coli RtcB nucleic acid splicing pathway. J. Bacteriol. 2016; 198:1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Luhrmann R., Soll D. et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011; 331:760–764. [DOI] [PubMed] [Google Scholar]
- 17. Trotta C.R., Miao F., Arn E.A., Stevens S.W., Ho C.K., Rauhut R., Abelson J.N.. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997; 89:849–858. [DOI] [PubMed] [Google Scholar]
- 18. Genschik P., Drabikowski K., Filipowicz W.. Characterization of the Escherichia coli RNA 3΄-terminal phosphate cyclase and its sigma54-regulated operon. J. Biol. Chem. 1998; 273:25516–25526. [DOI] [PubMed] [Google Scholar]
- 19. Chakravarty A.K., Shuman S.. RNA 3΄-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5΄-monophosphate ends. J. Biol. Chem. 2011; 286:4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakravarty A.K., Smith P., Shuman S.. Structures of RNA 3΄-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:21034–21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Engl C., Schaefer J., Kotta-Loizou I., Buck M.. Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Zhang J., Hara H., Kato I., Inouye M.. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005; 280:3143–3150. [DOI] [PubMed] [Google Scholar]
- 23. Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor. 1972; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 24. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amitai S., Kolodkin-Gal I., Hananya-Meltabashi M., Sacher A., Engelberg-Kulka H.. Escherichia coli MazF leads to the simultaneous selective synthesis of both ‘death proteins’ and ‘survival proteins’. PLoS Gen. 2009; 5:e1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pall G.S., Hamilton A.J.. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008; 3:1077–1084. [DOI] [PubMed] [Google Scholar]
- 27. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaberdina A.C., Szaflarski W., Nierhaus K.H., Moll I.. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis. Mol. Cell. 2009; 33:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chomczynski P., Sacchi N.. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987; 162:156–159. [DOI] [PubMed] [Google Scholar]
- 30. Brosius J., Ullrich A., Raker M.A., Gray A., Dull T.J., Gutell R.R., Noller H.F.. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981; 6:112–118. [DOI] [PubMed] [Google Scholar]
- 31. Ederth J., Mandava C.S., Dasgupta S., Sanyal S.. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009; 37:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benet C., Van Cutsem P.. Negative purification method for the selection of specific antibodies from polyclonal antisera. BioTech. 2002; 33:1050–1054. [DOI] [PubMed] [Google Scholar]
- 33. Chomczynski P., Sacchi N.. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 2006; 1:581–585. [DOI] [PubMed] [Google Scholar]
- 34. Moll I., Hirokawa G., Kiel M.C., Kaji A., Blasi U.. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004; 32:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sat B., Hazan R., Fisher T., Khaner H., Glaser G., Engelberg-Kulka H.. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 2001; 183:2041–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hui A., de Boer H.A.. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pace N.R., Brown J.W.. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol. 1995; 177:1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sauert M., Temmel H., Moll I.. Heterogeneity of the translational machinery: variations on a common theme. Biochimie. 2014; 114:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wigneshweraraj S., Bose D., Burrows P.C., Joly N., Schumacher J., Rappas M., Pape T., Zhang X., Stockley P., Severinov K. et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Micro. 2008; 68:538–546. [DOI] [PubMed] [Google Scholar]
- 40. Deusser E. Heterogeneity of ribosomal populations in Escherichia coli cells grown in different media. MGG. 1972; 119:249–258. [DOI] [PubMed] [Google Scholar]
- 41. Bickle T.A., Howard G.A., Traut R.R.. Ribosome heterogenecity. The nonuniform distribution of specific ribosomal proteins among different functional classes of ribosomes. J. Biol. Chem. 1973; 248:4862–4864. [PubMed] [Google Scholar]
- 42. Porollo A., Meller J.. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinf. 2007; 8:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yusupova G., Jenner L., Rees B., Moras D., Yusupov M.. Structural basis for messenger RNA movement on the ribosome. Nature. 2006; 444:391–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.