Abstract
Mutations in the Epidermal growth factor receptor (EGFR) kinase domain, such as the L858R missense mutation and deletions spanning the conserved sequence 747LREA750, are sensitive to tyrosine kinase inhibitors (TKIs). The gatekeeper site residue mutation, T790M accounts for around 60% of acquired resistance to EGFR TKIs. The first generation EGFR TKIs, erlotinib and gefitinib, and the second generation inhibitor, afatinib are FDA approved for initial treatment of EGFR mutated lung adenocarcinoma. The predominant biomarker of EGFR TKI responsiveness is the presence of EGFR TKI-sensitizing mutations. However, 30–40% of patients with EGFR mutations exhibit primary resistance to these TKIs, underscoring the unmet need of identifying additional biomarkers of treatment response. Here, we sought to characterize the dynamics of tyrosine phosphorylation upon EGFR TKI treatment of mutant EGFR-driven human lung adenocarcinoma cell lines with varying sensitivity to EGFR TKIs, erlotinib and afatinib. We employed stable isotope labeling with amino acids in cell culture (SILAC)-based quantitative mass spectrometry to identify and quantify tyrosine phosphorylated peptides. The proportion of tyrosine phosphorylated sites that had reduced phosphorylation upon erlotinib or afatinib treatment correlated with the degree of TKI-sensitivity. Afatinib, an irreversible EGFR TKI, more effectively inhibited tyrosine phosphorylation of a majority of the substrates. The phosphosites with phosphorylation SILAC ratios that correlated with the TKI-sensitivity of the cell lines include sites on kinases, such as EGFR-Y1197 and MAPK7-Y221, and adaptor proteins, such as SHC1-Y349/350, ERRFI1-Y394, GAB1-Y689, STAT5A-Y694, DLG3-Y705, and DAPP1-Y139, suggesting these are potential biomarkers of TKI sensitivity. DAPP1, is a novel target of mutant EGFR signaling and Y-139 is the major site of DAPP1 tyrosine phosphorylation. We also uncovered several off-target effects of these TKIs, such as MST1R-Y1238/Y1239 and MET-Y1252/1253. This study provides unique insight into the TKI-mediated modulation of mutant EGFR signaling, which can be applied to the development of biomarkers of EGFR TKI response.
Lung cancer is the leading cause of cancer-related deaths worldwide (1). Epidermal growth factor receptor (EGFR)1 is a predominant driver oncogene and therapeutic target mutated in 10–15% of NSCLC patients in the United States and 30–40% of patients in Asian countries. Mutations in the kinase domain, most commonly a point mutation in exon 21 (L858R) or deletions in exon 19 (e.g. E746-A750) are activating mutations associated with constitutive EGFR kinase activity and sensitivity to EGFR-specific tyrosine kinase inhibitors (TKIs), such as erlotinib (2–6). Unfortunately, approximately one year after treatment all patients treated with EGFR-TKIs develop drug-resistance. About 60% of acquired resistance to the first and second generation EGFR TKIs in patients can be attributed to acquisition of a secondary mutation at the gatekeeper residue (T790M) of the EGFR kinase domain (7, 8). Currently, there are limited options for circumventing acquired resistance to the first-generation EGFR-TKIs, gefitinib and erlotinib. Afatinib, an FDA approved second generation EGFR-TKI that was developed to circumvent T790M-mediated resistance, has not been very effective in clinical trials (9). Recently, the third-generation EGFR TKIs, osimertinib and rociletinib have shown promising results in clinical trials for the treatment of patients harboring the EGFR T790M mutation (10). Based on these promising results osimertinib was recently approved by the FDA for second line treatment of EGFR mutant patients who develop the T790M mutation.
Erlotinib and afatinib are both approved for first line treatment of patients with TKI-sensitizing EGFR mutations. However, 30–40% of patients have intrinsic resistance to these TKIs (11–14). Although several mechanisms of acquired resistance have been elucidated, mechanisms of intrinsic resistance are poorly understood. EGFR T790M mutation, MET amplification (15–19), and small cell lung cancer (SCLC) transformation (20, 21) are responsible for acquired resistance in a large number of patients, however, in many cases (estimated 20–25%) the mechanism is still unknown. Hence there is an unmet need to identify novel biomarkers of EGFR TKI response and resistance. EGFR gene mutations revealed by sequencing are the proven biomarkers of EGFR TKI sensitivity; however, the phenomenon of intrinsic resistance demonstrates that there are other factors modulating sensitivity to EGFR TKIs.
Quantitative shotgun proteomics is now widely used as a potent technology for discovery-based analysis of complex biological systems. The approach of immunoaffinity enrichment followed by mass spectrometry allows identification of low abundance tyrosine phosphorylated proteins (22). A global study has identified several oncogenic kinases such as EGFR, c-MET, PDGFRα, DDR1, and novel ALK and ROS fusions in non small cell lung cancer (NSCLC) cell lines (23) and tumor specimens (24). Quantitative profiling of phosphotyrosine performed on two adenocarcinoma cell lines with variable sensitivities to the EGFR TKI gefitinib, showed that an extensive downstream signaling network of mutant EGFR was inhibited upon treatment (25, 26). In addition, tyrosine phosphorylation of EGFR and other kinases was found to be downregulated in NSCLC cell lines treated with dasatinib (26), gefitinib (27), and afatinib (28). In our previous studies, we have employed a stable isotope labeling with amino acids in cell culture (SILAC) approach, phosphopeptide enrichment and quantitative MS to identify phosphorylation targets of mutant EGFRs in isogenic human bronchial epithelial cells and lung adenocarcinoma cells (29, 30).
In this study, we used SILAC and quantitative phosphoproteomics following enrichment of tyrosine phosphorylated peptides to elucidate the global dynamic changes of tyrosine phosphorylation upon treatment of TKI-sensitive and -resistant lung adenocarcinoma cells with either erlotinib or afatinib. We identified candidates with dynamic phosphorylation changes that are associated with the known sensitivity pattern of the TKIs, suggesting this approach can be used to discover potential biomarkers of EGFR TKI response and further elucidate the mechanisms of intrinsic or acquired resistance.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
H3255, PC9, and H1975 cell lines were obtained from ATCC. 11–18 was kindly provided by Koichi Hagiwara. The cells were cultured at 37 °C, 5% CO2 for at least five passages in RPMI medium 1640 (Pierce, Rockford, IL) containing l-arginine and l-lysine (light), 13C6-Arginine and D4-Lysine (medium), or 13C615N4-Arginine and 13C615N2-Lysine (heavy) (Cambridge Isotope laboratories, Tewksbury, MA) with 10% dialyzed fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin. After complete labeling, the cells were expanded to 15 cm dishes. In FBS experiments, the cells were grown in FBS containing medium, untreated (“light state”), or treated with erlotinib (100 nm) (“medium state”), or afatinib (100 nm) (“heavy state”) for 1 h. In H1975 cells, three more biological replicates were performed by altering the SILAC labeling between experiments, where the light and medium states were the cells grown in complete medium treated with erlotinib (100 nm) or afatinib (100 nm) for one hour, and the heavy state was the DMSO treated control cells. In parallel experiments, the cells were serum starved for 16 h prior to EGF and TKI treatment. The three states of SILAC were cells untreated (light), stimulated with EGF (100 ng/ml) for 3 min (medium), and treated with erlotinib or afatinib (100 nm) for 1 h before EGF stimulation (heavy).
Generation of EGFR Mutant Mouse Tumor Lysates
Doxycycline-inducible EGFRL858R transgenic mouse lung tumors were generated and followed by serial MRI as described before (31). Mice with lung tumors were either left untreated or treated with erlotinib at 25 mg/Kg daily by intraperitoneal injection. Mice were euthanized at early time-period (1 day) or late time period (25–41 days) to harvest lung tumors. Tumors were frozen in liquid nitrogen. About 10–15 mg of tumor tissue was lysed in 400 μl of urea lysis buffer supplemented with protease and phosphatase inhibitors using a tissue lyser (Qiagen, Germantown, MD). Lysates were centrifuged at 14,000 rpm at 4 °C for 10 mins and the clear supernatants were transferred to new tubes.
Mass Spectrometry Sample Preparation
Cells were lysed with urea lysis buffer (20 mm HEPES pH 8.0, 8 m urea, 1 mm sodium orthovanadate, 2.5 mm sodium pyrophosphate and 1 mm β-glycerophosphate). Protein concentrations were determined by the Modified Lowry method (BioRad, Hercules, CA). Equal amounts of protein from lysates of each SILAC state were mixed together to constitute 30 mg pooled lysate. The combined lysate was reduced with 45 mm dithriothreitol (Sigma Aldrich, St. Louis, MO), alkylated with 100 mm iodoacetamide (Sigma Aldrich), and subsequently digested with trypsin (Worthington, NJ) at 37 °C overnight. The digest was then acidified to 1% TFA and peptides were desalted using solid phase extraction C18 column (Supelco, Bellefonte, PA), lyophilized and stored at −80 °C.
Affinity Enrichment of Phosphotyrosine Peptides
Phosphotyrosine peptides were enriched prior to mass spectrometry analyses using a PhosphoScan Kit (p-Tyr-100 and p-Tyr-1000, Cell Signaling, Danvers, MA). The lyophilized peptide was dissolved in IAP buffer (50 mm MOPS, pH 7.2, 10 mm sodium phosphate, 50 mm NaCl) and incubated with 40 μl of immobilized anti-phosphotyrosine antibody for 1 h at 4 °C. The antibody beads were centrifuged for 1 min at 1500 g, and the supernatant was separated and saved. The antibody-bound beads were washed 3 times with 1 ml of IAP buffer and twice with water by inverting tube 5 times at 4 °C. The phosphotyrosine-containing peptides were eluted from antibody with 55 μl of 0.15% TFA by gently tap the bottom of the tube and incubate at room temperature for 10 min.
Capillary RPLC-MS/MS Analyses
Enriched phosphopeptides were analyzed on a LTQ Orbitrap XL ETD (Thermo Scientific, San Jose, CA) mass spectrometer interfaced with a dual nano pump (Eksigent, Dublin, CA) and an Agilent 1100 microwell plate autosampler. Phosphopeptides were loaded onto a trap column (75 μm x 2 cm, Magic C18AQ, 5 μm, 100Å, Michrom Bioresources), separated on an analytical column (75 μm x 15 cm, Magic C18AQ, 5 μm, 100Å, Michrom Bioresources,) at 300 nL/min flow rate with a running time of 100–130 min. The MS data were acquired at a resolution of 60,000 at m/z 350–1800 and MS/MS data were acquired on an ion trap. For each cycle of data dependent analysis, the 6 or 10 most abundant precursors were selected for MS/MS analysis with normalized collision energy of 35%. Multistage activation mode was enabled with neutral loss masses of 32.66, 48.99, and 97.97. Selected ions for fragmentation were excluded dynamically for 90 s.
Phosphopeptides enriched from the repeated experiments of H3255, 11–18, and H1975 cell lines in the presence of FBS were analyzed on a LTQ-Orbitrap Elite (Thermo Scientific, San Jose, CA) and SilicaTip emitter (New Objective, Woburn, MA) for electrospray ionization. An Easy-nLC 1000 (Thermo Scientific) was used for on-line RPLC separation. The enriched phosphopeptides were loaded onto a nano-trap column (Acclaim PepMap100 Nano Trap Column, C18, 5 μm, 100 Å, 100 μm i.d. x 2 cm) and separated on a nano-LC column (Acclaim PepMap100, C18, 3 μm, 100 Å, 75 μm i.d. × 25 cm, nanoViper). Mobile phases A and B consisted of 0.1% formic acid in water and 0.1% formic acid in 90% ACN, respectively. Peptides were eluted from the column at 250 nL/min using the following linear gradient: from 2 to 8% B in 5min, from 8 to 32% B in 100 min, from 32 to 100% B in 10 min, and held at 100% B for an additional 10 min. The spray voltage was 2.2 kV. Full spectra were collected from m/z 350 to 1800 in the Orbitrap analyzer at a resolution of 120,000, followed by data-dependent HCD MS/MS scans of the top 10 or 15 most abundant ions, using 32% collision energy. A dynamic exclusion time of 30 or 60 s were used to discriminate against the previously analyzed ions.
Data Analysis
Peptides and proteins were identified and quantified using the Maxquant software package (version 1.3.0.5) with the Andromeda search engine (32). MS/MS spectra were searched against the Uniprot human protein database (May 2013, 38523 entries) and quantification was performed using default parameters for 3 states SILAC in MaxQuant. Mouse data was searched using the Maxquant (version 1.5.5.1) against the Uniprot mouse protein database (May 2013, 26304 entries) that had human EGFR protein entry added because the transgenic mouse tumors express human mutant EGFRs. Maxquant was used to perform label free quantitation for the mouse data. The parameters used for data analysis include trypsin as a protease with two missed cleavage sites allowed. Carbamidomethyl cysteine was specified as a fixed modification. Phosphorylation at serine, threonine and tyrosine, deamidation of asparagine and glutamine, oxidation of methionine and protein N-terminal acetylation were specified as variable modifications. The precursor mass tolerance was set to 7 ppm and fragment mass tolerance to 20 ppm. False discovery rate was calculated using a decoy database and a 1% cut-off was applied to both peptide table and phosphosite table. Ninety-nine percent of the phosphosites are Class I and Class II sites (localization probability greater than 0.5).
Combined normalized SILAC ratio and intensities of the phosphosites from label-free mice were obtained from the MaxQuant search. Perseus (version 1.5.5.3) was used to view and further analyze the data. For label-free quantitation, imputation of missing values was performed in Perseus where missing values are replaced by random numbers that are drawn from a normal distribution of the whole data matrix. Hierarchical clustering of phosphorylation was obtained in Perseus using log2 SILAC ratios for cell line SILAC experiments and log intensities for the mouse label free experiments. Box and Whisker plots were made in Microsoft Office Excel for the selected genes by using the log intensities of the mouse label free quantitation data.
Kinase/Phosphatase Substrate Analysis
We used Fisher's exact test (FET) to identify kinases and phosphatases whose substrates were significantly hyper/hypo phosphorylated following EGFR inhibition. FET uses the hypergeometric test to calculate the significance of overlap of hyper/hypo phosphorylated substrates with the substrates of kinases/phosphatases. For this analysis, we used the substrates of various kinases obtained from a context-free network databases called STRING (33) for this analysis. To obtain high quality interactions from this database, we filtered out any interactions which were not reported to be experimentally validated, resulting in 423 interactions for 61 different tyrosine kinases and 957 interactions for 110 different phosphatases. We also downloaded DEPOD database (http://www.koehn.embl.de/depod/) to obtain substrates for phosphatases resulting in 396 interactions for 76 phosphatases.
We selected kinases, which significantly regulated substrates hyper-phosphorylated (up in M/L) following EGF stimulation and hypo-phosphorylated (down in H/M) following treatment with EGFR inhibitor (p value ≤ 0.05). For phosphatases, we selected those which significantly regulated substrates hypo-phosphorylated (down in M/L) following EGF stimulation and were hyper-phosphorylated (up in H/M) following the treatment with EGFR inhibitor.
The protein-protein interaction (PPI) maps of EGFR pathway substrates with altered phosphorylation upon drug treatment were imported from the “STRING: protein query” module of the cytoscape software (San Diego, CA, USA, version 3.4.0) (34) with the confidence cutoff of 0.80. These maps were analyzed for functional enrichment of the gene ontology biological process categories using the ClueGO 2.2.6 plugin (35) with the kappa statistic > = 0.4, a two-sided hypergeometric test for enrichment with Benferroni step down method for correction of the multiple hypothesis testing. A p value of 0.001 was used as the cut-off criterion.
Antibodies, Phospho-MAPK Array and Phospho-RTK Array
The primary antibodies against EGFR, pEGFR-Y1173, MAPK7, pMAPK7-Y220, ERK1/2, pERK1-Y204, ErbB3, pErbB3-Y1328, MET, pMET-Y1234/1235 EphA2, pEphA2-Y594, PKCδ, and pPKCδ-Y311 were obtained from Cell Signaling Technology. MAB108 hybridoma for mouse monoclonal antibody against human EGFR was obtained from ATCC and antibody purified in a core facility using standard procedures. Rabbit DAPP1 specific antibody was obtained from Protein Tech, Inc. Rabbit pY139-DAPP1 specific antibodies was prepared in collaboration with Rockland Immunochemicals Inc., Pottstown, PA. 4G10-HRP antibody was obtained from Millipore, Billerica, MA. Mouse monoclonal antibody against DAPP1 (BAM32/E10) was obtained from Santa Cruz Biotechnology, Santa Cruz, CA. Antibody against RhoGDI (Sigma) was used as loading control. Membranes were incubated with ECL (Amersham Biosciences, Piscataway, NJ) for 2–5 min prior to imaging in FluoChem HD2 Imaging System (Alpha Innotech) or Odyssey Fc Imaging System (LI-COR, Lincoln, NE). For phospho-MAPK and phosphor-RTK arrays, cells were washed with PBS and lysed with lysis buffer 6 (R&D systems, Minneapolis, MN) and centrifuged to remove cell debris. The human Phospho-MAPK array and human Phospho-Receptor Tyrosine Kinase array kit (R&D systems) were used according to the manufacturer's instructions.
siRNA Transfection and Cell Viability Assay
H3255, H1975, PC9, 11–18, HCC827, A549, H358, and H2303 cells were maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum and transfected with siRNA using the DharmaFECT transfection reagent (Darmacon, Lafayette, CO). Nontarget siRNA and siRNA death were used as negative and positive control respectively. The cell viability was measured 72 h after transfection using Cell Titer-Glo Luminescent Assay (Promega, Madison, WI) or AlamarBlue cell proliferation protocol (ThermoFisher).
Plasmids, Transfection, Lysate Preparation, Immunoprecipitation, and Immunoblot Analysis
Wild type DAPP1-YFP fusion plasmid was obtained from Dr. Lawrence E. Samelson (Center for Cancer Research, NCI, NIH). Mutation of DAPP1 plasmid to express Y139F DAPP1-YFP fusion protein was carried out at the National Cancer Institute Core Facility, Frederick, MD. Sources of wild type EGFR and mutant EGFR has been described elsewhere (36). For expression of wild type and mutant DAPP1 and EGFRs, HEK 293 cells were transfected with plasmid combinations using X-treamgene 9 DNA transfection reagent following manufacturer's recommendations. 24 h post-transfection, the medium was replaced with fresh growth medium. After an additional 48 h of growth, cell lysates were prepared. For immunoblot analysis, cells were lysed in modified RIPA lysis buffer (150 mm NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, and 50 mm Tris, pH 8.0), whereas for immunoprecipitation, cell extracts were prepared in Nonidet P-40 lysis buffer supplemented with protease and phosphatase inhibitors. For immunoprecipitation 1000 μg of lysate was incubated overnight at 4°C with 2 to 5 μg of mouse anti-EGF monoclonal antibody (MAB108) or mouse anti-DAPP1 monoclonal antibodies. The antigen-antibody complex was then captured, washed, and extracted as described somewhere else (36). Proteins were fractionated by SDS-polyacrylamide (4–15%) gel electrophoresis, transferred to nitrocellulose membrane by the semi-dry transfer method and probed with the specified antibody.
Experimental Design and Statistical Rationale
For the FBS experiments of H3255, 11–18 and H1975 cells, four, three and three biological replicates, respectively, were cultured independently, followed by enrichment of phosphotyrosine-containing peptides and MS analysis. In H1975 cells, three more biological replicates were performed by altering the SILAC labeling between experiments. For serum starved experiments, two biological replicates of H3255 and 11–18, three biological replicates of PC9 and four biological replicates of H1975 were performed. Phosphorylation changes were estimated by 3-state SILAC labeling on precursor peptide intensities in MaxQuant. Histograms of the log2 SILAC ratios (normalized M/L, H/L, H/M of each individual replicates and final combined ratios) of FBS and serum starved experiments were plotted for individual cell lines and a combined histogram obtained for all FBS SILAC ratios and all serum starved SILAC ratios (37). Phosphosites with 2-fold changes of final normalized combined ratio in either up or down regulated direction were considered as significant changes, which corresponds to the ±0.84 S.D. of mean in the serum starved experiment, - 0.7 S.D. and +1.6 S.D. of mean for the FBS experiment. The 2-fold cut-off used here has been used as a cut-off of biological significance in SILAC experiments (25, 37–42). In the FBS experiments, mean ± 1.5SD corresponds to SILAC ratios of 1.8 and 0.3, respectively.
RESULTS
Identification of Tyrosine Phosphorylated Peptides and Quantitation of Phosphorylation in Lung Adenocarcinoma Cells Upon Erlotinib or Afatinib Treatment
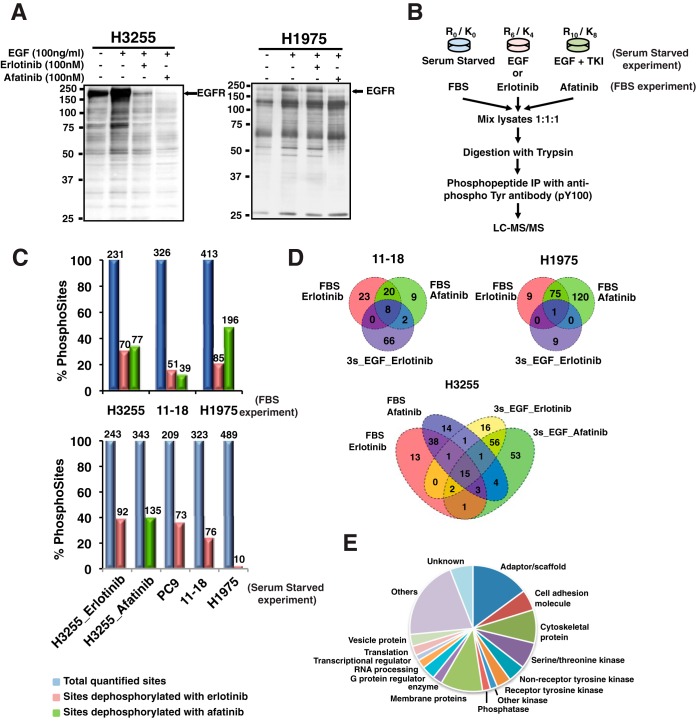
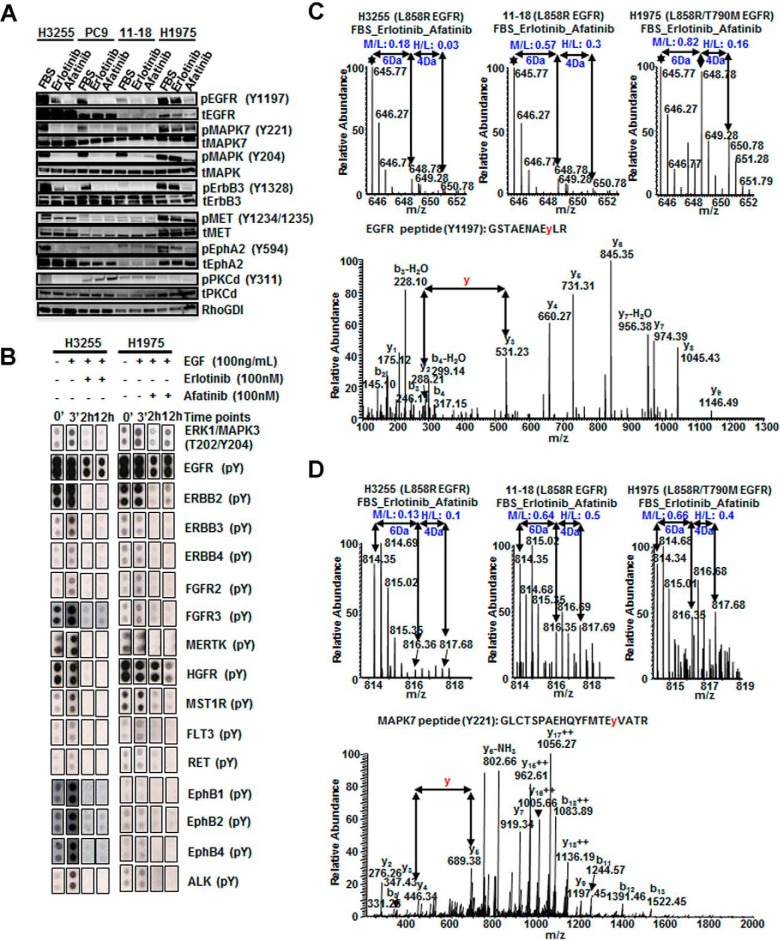
Four human lung adenocarcinoma cell lines with EGFR mutations and increasing resistance to the first-generation EGFR-TKI erlotinib were used for tyrosine phosphoproteomics analysis. H3255 (EGFRL858R) and PC9 (EGFRDel 746–750) are the most sensitive cells. 11–18 (EGFRL858R) has intermediate sensitivity, and H1975 (EGFRL858R/T790M) is erlotinib resistant. Cells were treated with either erlotinib or afatinib under the following growth conditions: (1) complete medium (FBS experiments) or (2) serum starved cells stimulated with EGF or TKI treated before EGF stimulation (serum starved experiments). Because tumor cells in vivo are always under the influence of various ligands, FBS experiments better represent the tumor cells in vivo. Because EGFR mutants exhibit constitutive activity, but may still be further stimulated with EGF, TKI treatment and EGF stimulation of serum starved cells is expected to demonstrate greater dynamic changes in phosphorylation aiding in the identification of mutant EGFR targets. Western blot analysis showed that treatment of H3255 cells with either erlotinib or afatinib followed by EGF stimulation resulted in a global decrease in tyrosine phosphorylation. Phosphorylation of a large subset of tyrosine sites was inhibited in H1975 cells upon treatment with afatinib, but not with erlotinib (Fig. 1A).
Fig. 1.
Summary of SILAC-based quantitative phosphoproteomics to identify and quantify phosphotyrosine sites in lung adenocarcinoma cells treated with erlotinib or afatinib. A, H3255 and H1975 lung adenocarcinoma cells were serum starved, treated with EGF (100 ng/ml), or pretreated with erlotinib (100 nm) or afatinib (100 nm) before EGF stimulation. Cell lysates were immunoprecipitated with anti-phosphotyrosine antibody (4G10), then IP eluates immunoblotted with 4G10-HRP. B, Experimental workflow showing treatment of SILAC-labeled cells, enrichment of phosphotyrosine peptides, and detection by tandem mass spectrometry. C, Bar graphs showing the percentage of quantified phosphotyrosine sites, and hypo-phosphorylated sites (SILAC ratio < 0.5) upon erlotinib or afatinib treatment. Top panel shows phosphosites identified in cells grown in complete medium (FBS experiments); bottom panel from the serum starved experiments. Above each bar are the actual number of phosphosites. D, Venn diagrams depicting the number of peptides dephosphorylated upon erlotinib or afatinib treatment in three lung cancer cell lines. E, GO analysis of identified phosphoproteins.
We employed quantitative mass spectrometry to identify and quantify tyrosine phosphorylation of mutant EGFR targets upon EGFR TKI treatment. Cells were cultured in medium containing light, medium, and heavy labeled amino acids to perform a “three state” SILAC experiment. Medium and heavy labeled cells were treated with erlotinib or afatinib, respectively for the experiments performed in complete growth medium (FBS experiments). In another set of experiments cells were serum starved overnight and then stimulated with EGF for 3 mins (medium state) and inhibited with erlotinib or afatinib for one hour before EGF stimulation (heavy state) (Fig. 1B). Tyrosine phosphorylated peptides were immune-enriched using a phosphotyrosine antibody (pY-100 or pY1000) prior to analysis by high-resolution liquid chromatography-tandem mass spectrometry (nLC-MS/MS). We analyzed 45 independent LC-MS/MS runs, and identified 949 distinct phosphotyrosine sites with a final FDR of less than 1%, which corresponded to 520 unique proteins (supplemental Table S1). SILAC ratios (M/L, H/L and H/M) of all identified targets followed a normal distribution (supplemental Fig. S1). Based on a 2-fold cut off (∼1.6 S.D. and -0.7 S.D. of the mean in FBS experiments), 450 tyrosine phosphorylation sites in 279 proteins were hypo-phosphorylated upon erlotinib or afatinib treatment in either FBS or serum starved experiments. SILAC labeling with heavy amino acids may affect the growth characteristics of cells, however, we have determined that this is not the case in the lung adenocarcinoma cells used in this study because these are highly proliferating cancer cell lines. We performed three additional biological replicates using H1975 cells grown in complete medium with label swapping between experimental conditions. There was good correlation in the afatinib inhibition SILAC ratios after label swapping. The correlation was less for the erlotinib inhibition SILAC ratio because H1975 cells are resistant to erlotinib and tyrosine phosphorylation at most sites remained unchanged (supplemental Fig. S2A, S2B). We also obtained good correlation between biological replicates (supplemental Fig. S2C–S2F).
The proportion of phosphotyrosine sites with reduced phosphorylation upon erlotinib inhibition was greater in the sensitive cells (H3255 and PC9), intermediate in the less sensitive 11–18 cells and least in resistant H1975 cells (Fig. 1C). Interestingly, afatinib, an irreversible second generation EGFR TKI had a greater effect on inhibiting phosphorylation in the erlotinib resistant H1975 cells, suggesting that this TKI more potently inhibits T790M mutant EGFR signaling (Fig. 1C).
Only a few hypo-phosphorylated tyrosine sites were common between both the serum starved and FBS experiments and upon erlotinib and afatinib inhibition (Fig. 1D). The 15 sites with reduced tyrosine phosphorylation in all experimental conditions in H3255 cells include ERRFI1-Y394, GAB1-Y689, EGFR-Y1110, EGFR-Y1197, MAPK1-Y187, ANXA2-Y48, DLG3-Y673, PKP3-Y84, STEAP1-Y27, ARHGAP5-Y1109, NAALADL2-Y106. The 8 tyrosine sites that were inhibited in 11–18 cells in all experimental conditions were ERRFI1-Y394, GAB1-Y406, CBL-Y406, CBL-Y674, EGFR-Y1172, EGFR-Y1110, SHC1-Y427, and TNK2-Y938. Only one phosphosite, HIPK3-Y359 showed reduced phosphorylation in all experimental conditions in the erlotinib resistant H1975 cells, suggesting that in this cell line, this is either an “off-target” or a feedback signaling effect of EGFR TKIs (Fig. 1D, Table I).
Table I. Phosphosites with decreased phosphorylation upon treatment with either of the TKIs.
| Phosphosites | FBS |
Serum starved |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H3255 Erlot | H3255 Afat | H1975 Erlot | H1975 Afat | 11–18 Erlot | 11–18 Afat | H3255 Erlot | PC9 Erlot | 11–18 Erlot | H1975 Erlot | H3255 Afat | |
| ANXA1-Y39 | 0.41 | 0.18 | 0.82 | 0.83 | 0.42 | 1.09 | |||||
| ANXA2-Y48 | 0.14 | 0.08 | 1.22 | 0.71 | 1.01 | 0.93 | 0.07 | 0.87 | 1.02 | 0.07 | |
| AP1B1-Y6 | 0.75 | 0.11 | 0.27 | 1.02 | |||||||
| ARHGAP5-Y1091 | 0.59 | 0.13 | 0.76 | 0.44 | 1.02 | 0.67 | |||||
| ARHGAP5-Y1097 | 0.91 | 0.24 | 0.28 | 2.19 | 0.25 | ||||||
| CBL-Y674 | 0.47 | 0.29 | 0.45 | 0.48 | 0.07 | 0.07 | 1.12 | ||||
| CLDN3-Y219 | 0.44 | 0.15 | 0.93 | 0.93 | 0.38 | 0.49 | |||||
| CTNND1-Y174 | 0.19 | 0.19 | 0.34 | 0.30 | 0.49 | 0.67 | 2.5 | 0.98 | |||
| CTNND1-Y248 | 0.53 | 0.26 | 0.60 | 0.79 | 0.09 | 0.67 | 1.75 | 0.12 | |||
| CRK-Y136 | 0.05 | 0.07 | 1.13 | ||||||||
| DAPP1-Y139 | 0.26 | 0.27 | 2.26 | ||||||||
| DLG3-Y705 | 0.11 | 0.04 | 0.51 | 0.17 | 0.94 | 0.6 | 0.05 | 0.11 | 0.24 | 1.92 | 0.06 |
| DSG2-Y1013 | 0.38 | 0.34 | 0.44 | 0.64 | 0.21 | 0.77 | 2.61 | ||||
| EGFR-Y998 | 0.17 | 0.22 | 0.44 | 0.17 | 0.47 | 0.13 | 0.09 | 0.92 | 0.57 | ||
| EGFR-Y1110 | 0.10 | 0.04 | 0.58 | 0.09 | 0.29 | 0.11 | 0.32 | 0.12 | 0.30 | 1.11 | 0.43 |
| EGFR-Y1172 | 0.25 | 0.07 | 0.81 | 0.15 | 0.24 | 0.16 | 0.55 | 0.15 | 0.18 | 0.92 | 0.59 |
| EGFR-Y1197 | 0.18 | 0.03 | 0.82 | 0.16 | 0.57 | 0.30 | 0.09 | 0.12 | 0.20 | 1.05 | 0.06 |
| ERBB3-Y1328 | 0.62 | 0.27 | 0.70 | 0.52 | 0.16 | 0.62 | 2.35 | 0.23 | |||
| ERRFI1-Y394 | 0.26 | 0.05 | 0.72 | 0.10 | 0.07 | 0.09 | 0.27 | 0.18 | 0.35 | 1.36 | 0.27 |
| FER-Y402 | 0.42 | 0.46 | 0.86 | 1.69 | |||||||
| GAB1-Y406 | 0.48 | 0.30 | 0.22 | 0.22 | 0.30 | 0.40 | 0.33 | 1.17 | 0.33 | ||
| GAB1-Y657 | 1.09 | 0.22 | 0.15 | 0.38 | 0.83 | 0.07 | |||||
| GAB1-Y689 | 0.11 | 0.09 | 0.40 | 0.24 | 0.18 | 0.19 | 0.03 | 0.31 | 0.36 | 0.97 | 0.06 |
| HIPK3-Y359 | 0.94 | 0.97 | 0.11 | 0.11 | 0.93 | 0.96 | 1.06 | 0.20 | |||
| ITGB1-Y783 | 0.46 | 0.31 | 0.53 | 0.36 | 0.73 | 0.82 | 0.61 | 0.79 | 0.99 | 1.93 | 0.97 |
| ITGB1-Y795 | 0.70 | 0.59 | 0.14 | 0.24 | |||||||
| LYN-Y508 | 0.45 | 0.24 | 0.52 | 0.57 | 1.32 | 1.24 | 0.93 | ||||
| MAPK1-Y187 | 0.03 | 0.03 | 0.57 | 0.16 | 0.04 | 0.03 | 0.46 | 1.04 | 0.96 | 0.94 | 0.45 |
| MAPK3-Y204 | 0.07 | 0.05 | 0.35 | 0.10 | 0.07 | 0.06 | 0.50 | 0.88 | 0.87 | 0.98 | 0.53 |
| MAPK7-Y221 | 0.13 | 0.10 | 0.66 | 0.40 | 0.64 | 0.53 | 0.78 | 0.55 | 0.45 | ||
| MET-Y1253 | 0.38 | 0.29 | 0.79 | 0.98 | 0.16 | 0.37 | 2.43 | 0.17 | |||
| MST1R-Y1238 | 0.46 | 0.51 | 0.23 | 0.18 | 0.55 | 0.72 | 0.17 | 1.37 | 2.45 | 0.15 | |
| PKP3-Y176 | 0.73 | 0.94 | 0.43 | 0.37 | 0.92 | 0.77 | 0.26 | 0.20 | 0.92 | 2.71 | 0.24 |
| PTK2-Y49 | 0.83 | 0.61 | 0.64 | 0.66 | 0.24 | 0.65 | |||||
| PTK2-Y905 | 0.78 | 0.87 | 0.34 | 0.45 | 1.00 | ||||||
| SHC1-Y427 | 0.14 | 0.04 | 0.83 | 0.22 | 0.22 | 0.19 | 0.30 | ||||
| SLITRK6-Y820 | 0.76 | 0.43 | 0.17 | 0.24 | 1.78 | 0.10 | |||||
| SPRY4-Y52 | 0.28 | 0.43 | 0.62 | 0.19 | 0.16 | 0.31 | 0.16 | 0.15 | |||
| STAT5A-Y694 | 0.03 | 0.03 | 0.52 | 0.28 | 0.77 | 0.02 | |||||
| STEAP1-Y27 | 0.28 | 0.13 | 0.64 | 0.45 | 1.30 | 0.80 | 0.11 | 0.26 | 0.22 | ||
| STX4-Y251 | 0.26 | 0.09 | 0.49 | 0.19 | 0.19 | 0.15 | |||||
| TAGLN2-Y192 | 0.57 | 0.23 | 1.01 | 1.02 | 0.58 | 0.82 | 0.46 | 1.78 | |||
| TBCB-Y98 | 0.47 | 0.27 | 0.28 | 0.95 | |||||||
| TJP2-Y1149 | 0.33 | 0.23 | 0.57 | 0.61 | 0.51 | 0.72 | 0.36 | 0.44 | 0.42 | 1.43 | 0.45 |
| TLN1-Y127 | 0.30 | 0.22 | 0.78 | 0.89 | 0.92 | 0.70 | 0.81 | 1.03 | |||
| TNS3-Y780 | 0.21 | 0.20 | 0.85 | 0.83 | 0.75 | 0.78 | 0.25 | 0.49 | 0.35 | 1.12 | 0.21 |
| TYK2-Y292 | 0.53 | 0.69 | 0.89 | 0.88 | 0.79 | 0.91 | 0.55 | 0.61 | 0.86 | 1.07 | 0.66 |
| UBASH3B-Y19 | 0.80 | 0.24 | 0.09 | 0.04 | 0.51 | ||||||
Next, we investigated the molecular functions of the identified phosphoproteins based on gene ontology (GO) annotations. Different classes of protein function were represented (Fig. 1E), including cytoskeletal, adaptor/scaffold, adhesion, and receptor and cell surface proteins. Proteins involved in transcription, translation, RNA processing and vesicle transport were also identified. Eighty-two of the phosphoproteins (16.7%) identified were kinases, including 35 serine/threonine kinases (6.7%), 21 receptor tyrosine kinase (3.7%) and 23 nonreceptor tyrosine kinase (4.5%).
Candidate Phosphotyrosine Sites Inhibited by Erlotinib or Afatinib Treatment
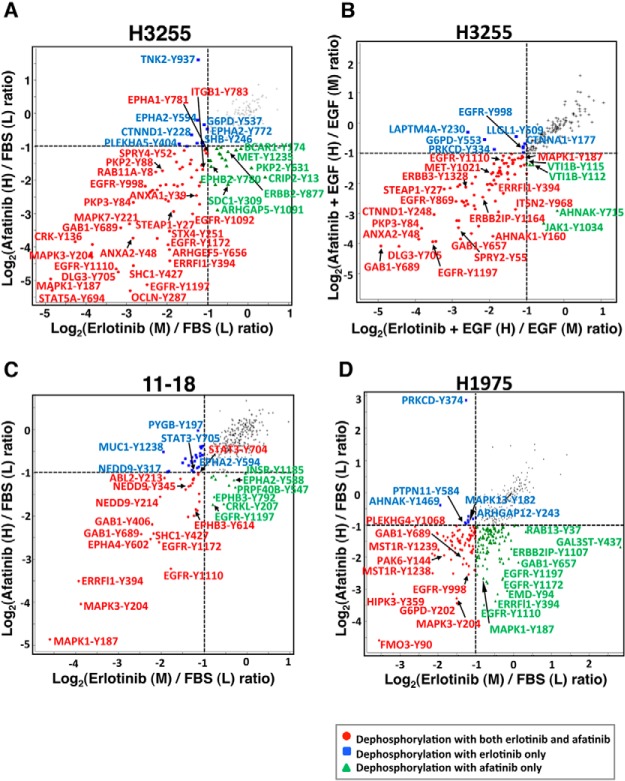
We identified phosphosites with a 2-fold change in phosphorylation upon treatment with either erlotinib or afatinib in the various lung adenocarcinoma cell lines (Fig. 2). Phosphorylation at 36 of the phosphosites identified was inhibited by both erlotinib and afatinib in H3255 cells in FBS experiments (Fig. 2A). Majority of them were also dephosphorylated in H3255 cells in serum starved condition by prior treatment with either of the TKIs (Fig. 2B). Thus, the TKI-sensitive H3255 cell line, which harbors an L858R mutation responds in a similar manner in the FBS and serum-starved conditions. The lung adenocarcinoma cell line, 11–18, also harbors an L858R mutation, but unlike H3255 cells, the mutated EGFR is not amplified and the protein is not overexpressed. This probably accounts for lower sensitivity of 11–18 cells to EGFR TKIs. Accordingly, fewer sites were hypophosphorylated upon either TKI treatment (Fig. 2C). H1975 cells harboring the T790M mutation in addition to the L858R mutation is resistant to erlotinib, but is relatively sensitive to the irreversible inhibitor, afatinib. Increased potency of afatinib in inhibiting mutant EGFR signaling in the TKI resistant cells is supported by phosphosites that are inhibited specifically by afatinib in 11–18 or H1975 cells. These include EGFR-Y1110, -Y1172, -Y1197, INSR-Y1185, EPHA2-Y588, GAB1-Y657, ERRFI1-Y394, and MAPK1-Y187. Interestingly, in complete growth medium, a large number of phosphosites were inhibited by both erlotinib and afatinib in the erlotinib resistant H1975 cells (Fig. 2D and Table I). This suggests “off-target” effects of erlotinib on potential kinases that are inhibited regardless of the presence of EGFR T790M mutation.
Fig. 2.
Scatter plot comparison of ratios of phosphorylation at phosphotyrosine sites quantified from erlotinib and afatinib treated cells. Phosphosites with significant regulation upon kinase inhibitor treatment are highlighted by color-coded dots as indicated (A, C, and D). Comparison of SILAC ratios of phosphorylation in H3255, 11–18 and H1975 cells grown in complete medium with/without erlotinib or afatinib treatment. B, Comparison of SILAC ratios of phosphorylation in H3255 cells serum starved overnight followed by EGF stimulation with/without prior erlotinib or afatinib treatment.
Cluster Analyses of SILAC Ratios of Phosphorylation of Kinases and Adaptor Proteins Upon TKI Treatment
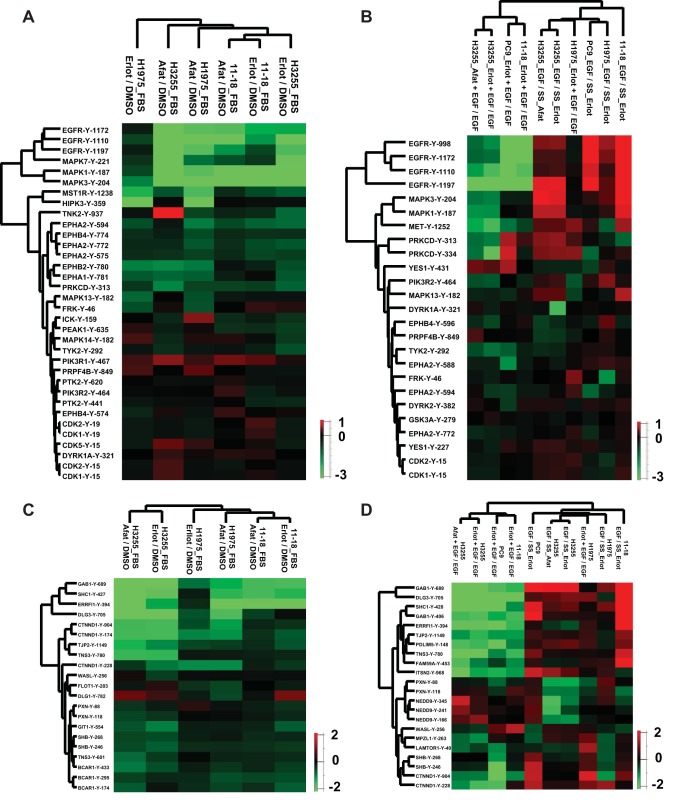
Dynamic changes in tyrosine phosphorylation of kinases and adaptor proteins regulate kinase activity and/or protein interactions, and ultimately regulate signaling pathways. We further evaluated the phosphosites on kinases and adaptor proteins identified in all cell lines from either the FBS or serum starved experiments. Hierarchical clustering of the SILAC ratios of TKI inhibition across these phosphosites showed clustering consistent with TKI sensitivity of the cell lines (Fig. 3A–3D). Different phosphosites of the same or related kinases or adaptor proteins, such as EGFR, related MAPKs, EPH receptors, CDKs, BCAR1, SHB, NEDD9 also clustered together. Three EGFR autophosphorylation sites (Y1110, Y1172, and Y1197) were hypo-phosphorylated upon treatment with both TKIs in H3255 and 11–18 cells. However, in H1975 cells these sites were hypo-phosphorylated upon afatinib, but not erlotinib inhibition. Interestingly, for both the erlotinib resistant H1975 cells and the 11–18 cells with intermediate sensitivity to erlotinib, phosphorylation of EGFR-Y1197 and MAPK7-Y221 was not inhibited upon erlotinib treatment, correlating with the extent of erlotinib sensitivity of these cells. This suggests EGFR-Y1197 and MAPK7-Y221 may be specific biomarkers of EGFR TKI sensitivity (Fig. 3A). The EGF stimulation SILAC ratios in serum starved experiments cluster together and exhibit increased tyrosine phosphorylation on kinases and adaptors (Fig 3B, 3D). This suggests that although EGFR mutants are constitutively active, they are further activated upon EGF stimulation. However, the Erlotinib + EGF/EGF SILAC ratio from H1975 cells clusters with this group because H1975 cells are resistant to erlotinib and most tyrosine phosphorylated sites are unchanged upon erlotinib treatment.
Fig. 3.
Hierarchical clustering of phosphotyrosine sites based on the SILAC ratios of phosphorylation. Columns represent different cell lines treated as indicated. Rows represent quantified phosphotyrosine sites identified in all experimental conditions. A, C, Phosphotyrosine sites in kinases (A) and adaptor proteins (C) in three lung cancer cell lines in complete medium and treated with erlotinib or afatinib. B, D, Phosphotyrosine sites in kinases (B) and adaptor proteins (D) in four lung cancer cell lines treated with erlotinib or afatinib in serum starved condition before EGF stimulation. Erlot or Afat + EGF/EGF represents the SILAC ratio of phosphorylation upon TKI inhibition. EGF/SS_Erlot or _Afat is the SILAC ratio of phosphorylation upon EGF stimulation without TKI inhibition.
The clustering analysis of adaptor proteins showed four adaptor phosphosites clustering together with the greatest inhibition of phosphorylation by TKIs in sensitive cells. These include GAB1-Y689, Y406, SHC1-Y427, DLG3-Y705, and ERRFI1-Y394. Thus, in addition to the kinase phosphosites, EGFR-Y1197 and MAPK7-Y221, these adaptor protein phosphosites are potential biomarkers of EGFR TKI sensitivity.
Functional and Pathway Analyses of EGFR TKI Regulated Phosphosites
We performed DAVID analysis to identify enriched functional categories and KEGG pathways among the proteins that were dephosphorylated upon either erlotinib or afatinib treatment in the panel of lung adenocarcinoma cells (supplemental Fig. S3). The enrichment scores are based on p value of enrichment. Receptor tyrosine kinase, cytoskeletal protein, molecular adaptor, GTPase regulator, and enzyme binding categories were enriched among proteins dephosphorylated by both TKI inhibitions in sensitive and resistant cells. The significance value of enrichment of the functional categories was less in resistant cells upon erlotinib treatment. The nonreceptor tyrosine protein kinase category was not enriched in resistant cell lines upon either TKI treatment (supplemental Fig. S3A), suggesting these proteins are indeed targets of mutant EGFRs. It is interesting to note that the serine/threonine protein kinase category was enriched but with lower significance in the sensitive compared with the resistant cell lines, suggesting some off-target effects of these TKIs in inhibiting phosphorylation of these kinases. We also identified the KEGG pathways significantly enriched among the proteins with reduced phosphorylation upon TKI inhibition (supplemental Fig. S3B). The ERBB signaling pathway was the most significantly enriched pathway. Again, the significance of enrichment of all the pathways in the resistant cell line (H1975) upon erlotinib treatment was lower than for the other groups. Several pathways, including Fc gamma R-mediated phagocytosis, JAK-STAT signaling pathway, regulation of actin cytoskeleton, insulin signaling pathway, tight junction, and non small cell lung cancer were not enriched in the resistant cell line treated with erlotinib. The greater enrichment of these diverse pathways in TKI-sensitive cells may suggest cross-talk with EGFR signaling. In summary, these bioinformatics analyses uncover functional categories of proteins and pathways that represent the potential biomarkers of TKI responsiveness.
Effect of EGFR Inhibitors on EGFR Downstream Signaling
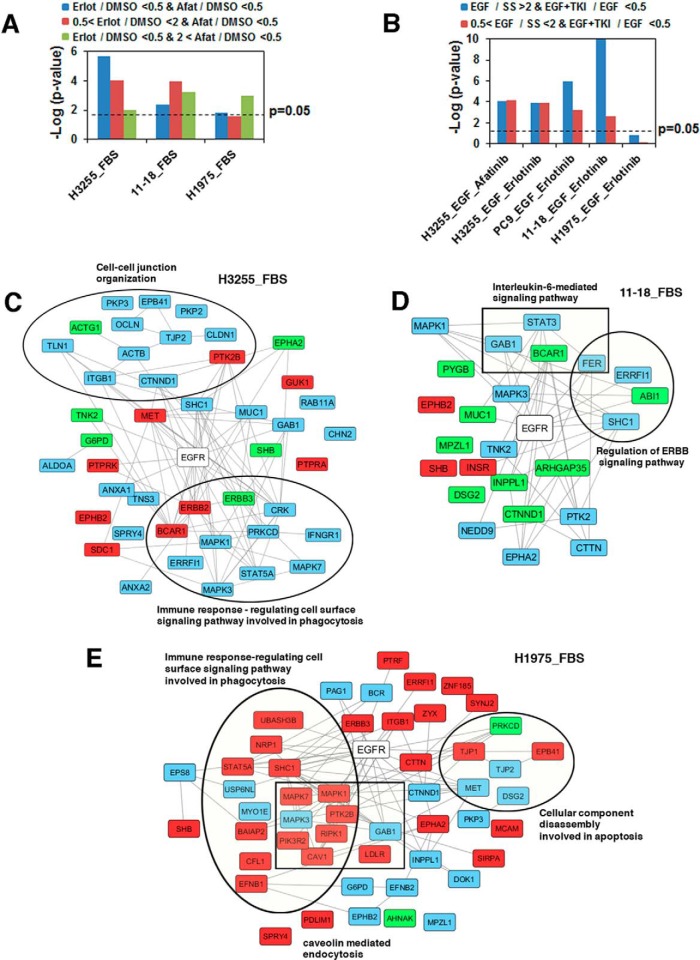
Because both erlotinib and afatinib are EGFR TKIs, we matched the proteins with reduced phosphorylation against the known, experimentally-validated EGFR signaling pathway members in the STRING database. As expected, EGFR signaling pathway proteins were indeed enriched in all our experiments in complete medium (supplemental Table S2). The enrichment scores, as represented by the p values, were lower among substrates hypo-phosphorylated with afatinib treatment only and even lower among substrates hypo-phosphorylated with erlotinib only compared with the substrates dephosphorylated by both TKIs in the sensitive cell line, H3255. The enrichment scores for EGFR signaling proteins were far lower in H1975 and 11–18, the relatively erlotinib-resistant cell lines (Fig. 4A). A similar analysis was done for the serum starved experiments both for substrates whose phosphorylation increased upon EGF stimulation and reduced upon TKI inhibition and substrates whose phosphorylation remained unchanged with EGF stimulation and reduced upon TKI inhibition. For these experiments, hypo-phosphorylation of EGFR substrates showed significance in H3255, PC9, and 11–18, but not in H1975 cells (Fig. 4B). We next identified EGFR substrates whose phosphorylation was reduced by erlotinib, afatinib, or both in the FBS experiments (Fig. 4C–4E), or the serum starved experiments (supplemental Fig. S4A–S4D). Specific gene ontology (GO) categories were significantly enriched among these EGFR interacting proteins. These include cell-cell junction organization, immune response-regulating cell surface signaling pathway involved in phagocytosis, IL6-mediated signaling pathway, regulation of ERBB signaling pathway and cellular component disassembly involved in apoptosis (Fig. 4C–4E). Interestingly, enrichment of ERBB signaling pathway and regulation of ERBB signaling pathway components were more apparent in the TKI inhibiting substrates in the serum starved experiments, suggesting more specific inhibition of these pathways among EGFR substrates inhibited in serum starved condition (supplemental Fig. S4A–S4D).
Fig. 4.
Enrichment of EGFR pathway substrates among proteins with phosphorylation modulated by erlotinib or afatinib treatment. A, Enrichment upon erlotinib or afatinib treatment of H3255, 11–18, and H1975 cells in the presence of complete medium. Colors of the bars represent specific SILAC ratio changes as indicated. B, Enrichment upon EGF stimulation and erlotinib or afatinib treatment of H3255, PC9, 11–18, and H1975 cells following serum starvation. Colors of the bars represent specific SILAC ratio changes as indicated. C–E, Networks of EGFR substrates whose phosphorylation was inhibited by erlotinib, afatinib or both in H3255 (C), 11–18 (D), and H1975 (E) cells grown in complete medium. Phosphorylation of proteins highlighted with blue was inhibited by both erlotinib and afatinib; with green by erlotinib only; and with red by afatinib only.
Validation of Select Phosphosites by Expression Analyses
We validated changes in phosphorylation on several of the key phosphosites using Western blots and protein microarrays. Phosphorylation of EGFR-Y1197, MAPK7-Y204, MET-Y1234/1235, EPHA2-Y594, ERBB3-Y1328, and PKCδ-Y1311 were analyzed on Western blots (Fig. 5A). Consistent with the MS data, all of these sites had decreased phosphorylation in H3255 cells upon erlotinib or afatinib treatment, whereas they remained unchanged in H1975 cells treated with erlotinib. In 11–18 cells, EGFR-Y1197, MAPK7-Y221, MAPK3-Y204 and ErbB3-Y1328 were dephosphorylated upon treatment with either inhibitor, whereas MET-Y1234/1235, EPHA2-Y594 and PKCδ-Y311 were unchanged.
Fig. 5.
Validation of phosphosites modulated by erlotinib and/or afatinib in lung adenocarcinoma cell lines. A, Western blots showing the effect of erlotinib or afatinib on selected phosphorylation sites relative to the protein level in H3255, PC9, 11–18 and H1975 cell lines. B, Phospho MAPK array and RTK array antibody blots showing phosphorylation changes in response to EGF stimulation upon erlotinib or afatinib treatment of H3255 and H1975 cells. Cells were serum starved overnight then treated with EGF for 3 min or with 100 nm erlotinib or afatinib for 2 h or 12 h prior to EGF stimulation. C–D, MS and MS/MS spectra of EGFR peptide with Y1197 phosphorylation (C) and MAPK7 peptide with Y221 phosphorylation (D). Phosphorylation of both sites decreased upon erlotinib or afatinib inhibition of H3255 and 11–18 cells; whereas it was only inhibited upon afatinib treatment in H1975 cells, but did not change upon erlotinib treatment.
The human phospho-Receptor Tyrosine Kinase (RTK) array and the phospho-Mitogen-activated Protein Kinase (MAPK) antibody array were used to assay for phosphorylated RTKs and MAPKs. Serum starved H3255 and H1975 cells stimulated with EGF alone or pre-treated with erlotinib or afatinib for 2 or 12 h were analyzed (Fig. 5B). There was constitutive and EGF-induced stimulation of phosphorylation for most of the kinases. The kinases tested were dephosphorylated upon erlotinib treatment of H3255 cells and afatinib treatment of H1975 cells. However, the phosphorylation of MAPK3-Y204, EGFR, and ERBB2 was restored in H1975 cells after 12 h of afatinib treatment, suggesting feedback regulation at these sites upon long term treatment with afatinib.
MS and MS/MS spectra of EGFR-Y1197 and MAPK7-Y221 phosphopeptides showed that phosphorylation of both sites decreased upon erlotinib or afatinib treatment of H3255 and 11–18 cells, whereas in H1975 cells phosphorylation at both sites was inhibited upon afatinib but not erlotinib treatment (Fig. 5C, 5D).
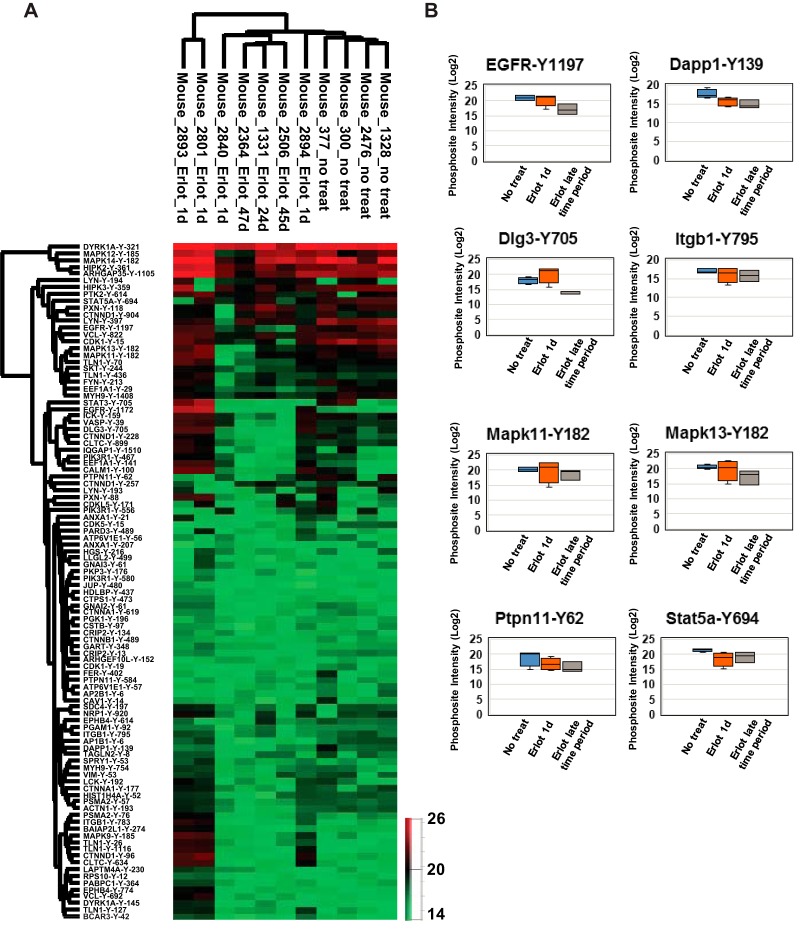
Label Free Quantitative Mass Spectrometry to Validate Specific Targets in EGFR Mutant GEM Model In Vivo
We used conditional EGFRL858R transgenic mice that develop lung adenocarcinoma upon doxycycline induction of mutant EGFR in lung type II epithelial cells (31) to treat MRI confirmed lung tumors with erlotinib. We compared tyrosine phosphosites in untreated lung tumors and tumors receiving short-term (1 day) or long-term (24–47 days) erlotinib treatment. We quantified the degree of tyrosine phosphorylation of these sites by label free quantitation algorithm of Maxquant and focused on the phosphosites identified in both the mouse tumors and human lung adenocarcinoma cell lines. Hierarchical clustering using the log intensities of the phosphopeptides common to both systems demonstrated that the untreated mouse tumors clustered together as did the tumors upon short and long-term erlotinib treatment (Fig. 6A, supplemental Fig. S5). Although there was variability in the intensities of the phosphorylated peptides, likely because of variability of the mouse stromal component, there was a pattern of reduced phosphorylation upon in vivo erlotinib treatment for 1 day and this was more pronounced upon longer treatment (Fig. 6B). Potential biomarkers of EGFR TKI response from the human lung adenocarcinoma cell lines validated with the mouse data generated in vivo. These include EGFR-Y1197, Dapp1-Y139, Dlg3-Y705, Ptpn11-Y62, and Stat5A-Y694 (Fig. 6B).
Fig. 6.
Phosphotyrosine sites validated in untreated or erlotinib-treated transgenic mice with doxycycline-inducible EGFRL858R lung tumors. A, Hierarchical clustering of phosphotyrosine sites identified in the mice based on label-free quantitation. Columns represent different mice of the same genotype (EGFRL858R) untreated or treated with erlotinib; rows represent quantified phosphotyrosine sites. Expression is based on the log2 intensity of the phosphopeptide. Only the sites identified in any of the human lung adenocarcinoma cell lines and mice are shown. B, Box plots of intensities of selected regulated phosphopeptides showing the label-free quantitation of tumor bearing mice, untreated, treated with erlotinib for 1 day, and mice receiving long term erlotinib treatment (24–47 days).
Regulation of Potential Phosphatase Targets
We identified several substrates whose phosphorylation decreased upon EGF stimulation and increased with TKI inhibition. These paradoxical dynamic changes in phosphorylation can be explained by EGFR activating a phosphatase or by feedback signaling activated upon TKI inhibition of mutant EGFR. We examined phosphatases and their substrates in our MS datasets. We used experimentally validated phosphatase-substrate and EGFR-substrate databases from STRING and DEPOD to infer potentially upstream phosphatases of this group of identified phosphosites which are also EGFR substrates. In H3255 cells, phosphorylation of BCAR1-Y174 and NEDD9-Y166, two unique substrates of PTEN, PTPN12, PTPN11, PTPRD, and INPPL1 phosphatases, decreased upon EGF stimulation and increased with TKI inhibition (supplemental Fig. S6A). EGFR activation upon EGF stimulation may have resulted in the activation of one or more of the five downstream phosphatases resulting in reduced phosphorylation of their substrates, BCAR1-Y174 and NEDD9-Y166. The same sites were hyper-phosphorylated upon erlotinib treatment resulting in EGFR inhibition and deactivation of the phosphatases.
In H1975, four phospho sites, PKP4-Y415, SDCBP-Y67, G6PD-Y552, and PPP1R18-Y230, were hypophosphorylated upon EGF stimulation and remained unchanged upon erlotinib inhibition. The upstream phosphatases of these four proteins are PTPRJ and PGAM4. In vitro phosphatase assay showed dephosphorylation of PKP4 by PTPRJ (43). PTPRJ negatively regulates EGFR signaling pathway through EGFR dephosphorylation. This further suggests EGFR activates these phosphatases, but because EGFRL858R/T790M in H1975 cells is not inhibited by erlotinib, there is no change in phosphorylation of the substrates of these phosphatases upon erlotinib treatment (supplemental Fig. S6B).
On the other hand, Cysteine-rich protein 2 (CRIP2-Y151), a unique PTPN13 substrate, had reduced phosphorylation upon EGF stimulation and increased phosphorylation with erlotinib treatment of H1975 cells (supplemental Fig. S6C). PTPN13 is a novel candidate tumor suppressor in NSCLC. Loss of PTPN13 increases EGFR and ERBB2 signaling. SRC kinase signaling inhibitor 1 (SRCIN1-Y298), a substrate of phosphatase PTPN1 and kinase CSK, and transmembrane channel-like protein 5 (TMC5-Y110), substrate of phosphatase PPM1H as well as kinase ROR1, were hypophosphorylated upon EGF stimulation and hyperphosphorylated with erlotinib inhibition in H1975 cells. This data suggests that in H1975 cells either of the phosphatases PTPN1 and PPM1H are activated upon EGF stimulation. PTPN1 dephosphorylates and inactivates EGFR (44). ROR1 has been implicated as a novel therapeutic target for EGFR-mutant nonsmall-cell lung cancer patients with the EGFR T790M mutation (45).
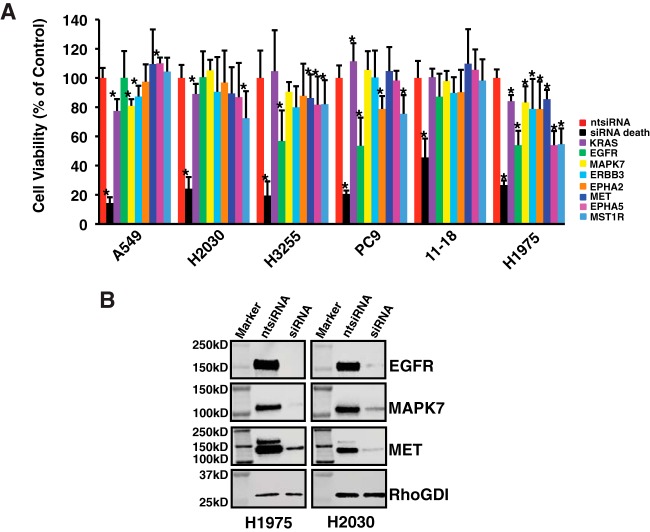
Functional Validation of Selected Mutant EGFR Targets That Are Potential EGFR TKI Biomarkers
We predicted that dynamic changes in phosphorylation of key mutant EGFR signaling pathway proteins would have functional consequences. We selected a group of 16 proteins that were differentially phosphorylated upon EGF stimulation or TKI inhibition in a manner consistent with the sensitivity pattern of the cell lines. We assayed cell viability upon siRNA-mediated knockdown of these proteins on a panel of six lung adenocarcinoma cell lines with documented EGFR or KRAS mutations. These included H3255 (EGFRL858R), 11–18 (EGFRL858R), H1975 (EGFRL858R/T790M), PC9 (EGFRDel746–750), A549 (KRASG12S), and H2030 (KRASG12C). Because the proteins we selected are mutant EGFR targets, we predicted reduced cell viability by knockdown of these proteins in EGFR mutant cells, but not the KRAS mutant cells. We identified eight genes that affected cell viability (Fig. 7A). Knockdown of MAPK7, ERBB3, EPHA5, EPHB3, MET or MST1R reduced cell viability of one or more EGFR mutant cell lines. Interestingly, knockdown of all these targets significantly decreased viability in H1975 cells but had no effect on 11–18 cells. We documented reduced protein expression of EGFR, MAPK7 and MET in H1975 and H2030 cells (Fig. 7B).
Fig. 7.
Functional characterization of lung adenocarcinoma cells upon siRNA-mediated knockdown of select target proteins with reduced tyrosine phosphorylation upon erlotinib or afatinib treatement. A, Two KRAS mutant cell lines (A549 and H2030) and four EGFR mutant cell lines (H3255, PC9, 11–18, and H1975) were transfected with 25 nm of non-targeted siRNA, siRNA death control or 8 selected siRNAs. Cell viability was measured using Cell Titer-Glo Luminescent assays. Results are shown as mean ± S.D. from three independent experiments. * p value less than 0.05 from the student t test. B, Western blots showing reduced expression of selected targets upon siRNA-mediated knockdown in H1975 and H2030.
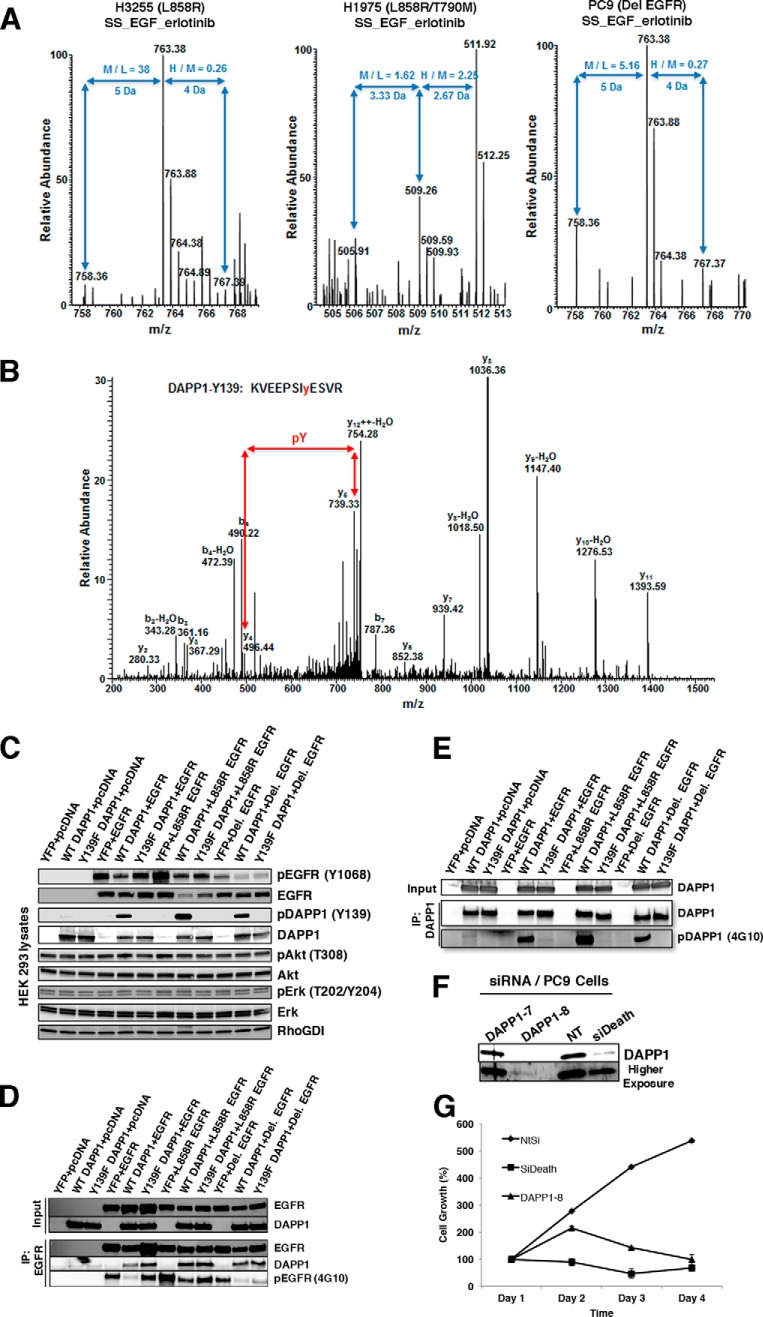
In this study, we report that DAPP1-Y139 is a novel mutant EGFR target that is also a potent EGFR TKI response biomarker. DAPP1 is an adapter protein involved in B cell receptor signaling (46, 47). Quantitative MS shows that DAPP1-Y139 phosphorylation increases upon EGF stimulation and is reduced upon TKI treatment in lung adenocarcinoma cells (Fig. 8A, 8B). To further validate Y139 phosphorylation and its functional significance we expressed wild type or Y139F mutant DAPP1 along with wild type or mutant EGFRs in HEK 293 cells (Fig. 8C). We generated a DAPP1 Y139 phospho-specific antibody that recognizes phosphorylation specifically at this site (Fig. 8C). Co-immunoprecipitation experiments show that wild type and mutant EGFRs interact with DAPP1 (Fig. 8D) and the major site of DAPP1 tyrosine phosphorylation is Y139 because tyrosine phosphorylation is abrogated in the Y139F mutant (Fig. 8E). Interestingly, coexpression of wild type DAPP1 inhibited autophosphorylation of both wild type and mutant EGFRs and Y139F mutation of DAPP1 partially rescued this effect (Fig. 8C, 8D). siRNA-mediated knockdown of DAPP1 (Fig. 8F) reduced viability of PC9 human lung adenocarcinoma cells that harbor EGFRDel mutant, suggesting the requirement of DAPP1 adapter protein in the survival of these mutant EGFR-addicted cells (Fig. 8G).
Fig. 8.
Validation of DAPP1 Y139 phosphorylation as the major site of tyrosine phosphorylation modulated by mutant EGFR signaling. A–B, MS and MS/MS spectra of DAPP1 peptide with Y139 phosphorylation. Phosphorylation increased upon EGF stimulation (M/L ratio) and decreased upon erlotinib inhibition of H3255 cells and PC9 cells, but increased in H1975 cells (H/M ratio) (A). C, Immunoblot analysis of protein lysates from HEK 293 cells expressing wild type or mutant EGFR and DAPP1 or Y139F DAPP1 mutant. Lysates prepared in modified RIPA buffer were probed with pY1068-EGFR, EGFR, pY139-DAPP1, DAPP1, pAkt, Akt, pErk, Erk, and Rho-GDI (control) specific antibodies. D, Immunoprecipitation of wild type and mutant EGFRs with EGFR specific monoclonal antibodies followed by immunoblotting with anti-EGFR, pEGFR (4G10), and anti-DAPP1 antibodies. E, Immunoprecipitation of wild type and Y139F mutant DAPP1 from HEK 293 cells expressing DAPP1 and mutant DAPP1 followed by immunoblotting with anti-DAPP1 and anti-pTyr (4G10) antibodies indicated that Y139 is the major site of DAPP1 phosphorylation. F, PC9 cells (expressing EGFRDel 746–750) were transfected with DAPP1 siRNAs, or NT siRNA (negative control) and siDeath (positive control) for 72 h followed by immunoblot analysis of cell extracts with anti-DAPP1 antibodies. G, Growth curve of PC9 cells following transfection of PC9 cells with DAPP-1 siRNA, NT-siRNA, or siDeath, showing DAPP1 knock-down significantly reduces PC9 cell growth.
DISCUSSION
We employed global, unbiased SILAC-based quantitative mass spectrometry to identify and quantify tyrosine phosphorylated sites modulated by TKI treatment of mutant EGFR-driven lung adenocarcinoma. We examined the signaling proteins affected by treatment of lung adenocarcinoma cells with the FDA approved first-generation reversible EGFR TKI, erlotinib and second-generation irreversible EGFR TKI, afatinib. To our knowledge, this is the first study comparing the tyrosine phosphorylation dynamics between erlotinib or afatinib treatment of mutant EGFR-driven lung adenocarcinoma cells. The degree of inhibition correlated with the sensitivity of each cell line to either erlotinib or afatinib. Afatinib more potently inhibited phosphorylation in both the erlotinib resistant cell line H1975 and the moderately resistant cell line, 11–18. These experiments identified novel mutant EGFR targets of phosphorylation and potential biomarkers of TKI response. These include EGFR-Y1197, MAPK7-Y221, DLG3-Y705, and DAPP1-Y139. We also identified these targets in vivo in a mouse model of mutant EGFR-induced lung tumorigenesis. Likewise, phosphorylation of these targets was inhibited by erlotinib treatment of EGFR TKI-sensitive mouse tumors. Finally, knockdown of key mutant EGFR targets decreased cell viability demonstrating the dependence on these targets for the survival of mutant EGFR cells.
H3255 cell line harboring the EGFRL858R mutant is the most EGFR TKI sensitive cell line we used in our study, whereas 11–18 cells expressing the same EGFRL858R mutant have intermediate sensitivity. Phosphorylation of many tyrosine sites did not change upon treatment of 11–18 cells with either of the inhibitors, suggesting intrinsic resistance of these cells to EGFR TKIs (Table I). It is also possible that phosphorylation at these sites in 11–18 is not dictated by EGFR alone but depends on cellular context. Hence these phosphosites may be indirect targets of mutant EGFR. 11–18 cells undergo cell cycle arrest, but not cell death upon erlotinib treatment. This has been attributed to the inability of these cells to upregulate the cell death inducer, BIM upon erlotinib treatment (48). We identified specific kinase phosphosites that were resistant to either erlotinib or afatinib in both 11–18 and H1975 cells. These include EGFR-Y1197, MAPK7-Y221, and ERBB3-Y1328. Phosphorylation at these sites was unaffected by erlotinib in both cell lines. Afatinib treatment also resulted in minimal inhibition of these sites (Fig. 5C, 5D). MAPK7 (ERK5) is the effector kinase of a canonical kinase module containing: MEKK 2/3 (MEK kinase), MEK5 (MAPK/ERK kinase), and MAPK7 itself. Clinical evidence suggests a role for dysregulated MEK5/ERK5 signaling as a driver of tumorigenesis in several cancers (49–53) and dysregulated MAPK7 was shown to drive NSCLC (54).
Aberrant EGFR signaling plays an important role in cancer by activating downstream signals essential for growth and survival. EGFR is a receptor tyrosine kinase known to drive cell growth and survival of multiple epithelial forms of cancers including NSCLC (55, 56). Various EGFR phosphorylation sites are functionally relevant. Studies using EGFR TKIs have identified sites which mediate drug effects (57–61). In this study, we identified six EGFR tyrosine sites, Y869, Y998, Y1092, Y1110, Y1172, and Y1197, which were dephosphorylated upon either erlotinib or afatinib treatment of sensitive cells (H3255, PC9) and remained unchanged upon erlotinib treatment of the TKI resistant cell line, H1975. The irreversible EGFR inhibitor, afatinib could still inhibit phosphorylation of these sites on mutant EGFRL858R/T790M -expressing H1975 cells, suggesting afatinib can inhibit the erlotinib resistant mutant EGFR, at least in cultured cells. Similarly, in the serum starved experiments, phosphorylation at EGFR Y998, Y1172, Y1110, and Y1197 increased upon EGF stimulation and decreased with erlotinib or afatinib treatment of H3255, PC9 and 11–18 cells, but not in H1975 cells. Interestingly, several phosphorylation sites on nonreceptor tyrosine kinases, such as TYK2-Y292, PTK2-Y49 and -Y905, LYN-Y508, and FER-Y402 were not inhibited by afatinib in H1975 cells. The inability to inhibit these signaling proteins along with incomplete inhibition of T790M mutant EGFR may account for the lack of in vivo response to afatinib in EGFR T790M harboring tumors in mouse models (62) or patients (63).
Our approach of employing both serum starved and cells grown in FBS containing medium in these EGFR TKI inhibition experiments sheds unique insight into the effects exerted by these TKIs in presence of ligands. Interestingly, ∼20% of phosphotyrosine sites underwent reduced phosphorylation upon erlotinib treatment of resistant H1975 cells in FBS experiments. In contrast, only 2% of sites were hypophosphorylated in the serum-starved state, underscoring the importance of growth medium in such experiments. Two MAP kinases, MAPK1-Y187 and MAPK3-Y204, were dephosphorylated upon either erlotinib or afatinib treatment in FBS experiments in all three cell lines (H3255, 11–18, and H1975). However, the degree of inhibition was lower upon erlotinib, as compared with afatinib treatment of H1975 and 11–18 cells. In the serum-starved experiments, these sites were hyper-phosphorylated upon EGF stimulation and hypo-phosphorylated with either erlotinib or afatinib inhibition in H3255. However, this was not seen upon erlotinib treatment of 11–18, PC9 or H1975 cells. This is likely because of off-target effects of these TKIs in complete growth medium containing various ligands which activate potential off-target kinases. Such off-target effects of erlotinib and afatinib on other specific kinases have been documented (64–69). In this study, we provide in vivo evidence of erlotinib or afatinib inhibiting tyrosine phosphorylation of off-target kinases or kinase substrates. For example, MST1R (RON) and MET are two members of the MET receptor tyrosine kinase family that play a role in cancer pathogenesis (70). MST1R is a prognostic marker and therapeutic target for gastroesophageal adenocarcinoma (71). Tyrosine phosphorylation at MST1R-Y1238, -Y1239 was inhibited upon either erlotinib or afatinib treatment of H1975 cells in FBS-containing medium. However, there was no inhibition of phosphorylation upon erlotinib treatment of these cells in the serum starved experiments (supplemental Fig. S7). Similarly, we observed reduced phosphorylation at MET-Y1252, -Y1253 in H1975 cells upon treatment with either erlotinib or afatinib (supplemental Fig. S8). The functional significance of these off-target effects of EGFR TKIs on MST1R and MET remains to be studied.
We have identified and quantified four tyrosine phosphosites in another RTK, EPHA2-Y575, Y588, Y594, and Y772. EPHA2 has been implicated in the regulation of a wide array of pathological conditions, including cancer (72). A recent study found that EPHA2 is over-expressed in erlotinib-resistant lung cancer cells. Loss of EPHA2 reduced the viability in vitro of erlotinib-resistant tumor cells harboring EGFRT790M mutations in vitro and inhibited tumor growth and progression in an inducible EGFRL858R+T790M-mutant lung cancer model in vivo (73). In our experiments, the EPHA2 phosphorylation sites decreased to some extent in all three cell lines (H3255, 11–18 and H1975) upon either erlotinib or afatinib treatment in FBS-containing growth medium. Phosphorylation of Y588 did not change upon erlotinib inhibition in 11–18 and H1975 cells, suggesting this phosphosite may be a potential biomarker of response to EGFR TKIs. Moreover, knockdown of EPHA2 reduced the viability of two EGFR mutant lung adenocarcinoma cell lines used in our study, PC9 and H1975 (Fig. 7), suggesting an important role for EPHA2 in mutant EGFR signaling.
We employed siRNA-mediated knockdown of specific signaling proteins identified in this study to investigate their effect on cell survival. Interestingly, knockdown of all selected targets, EGFR, ERBB3, MET, MST1R, EPHA2, EPHA5, and MAPK7, reduced survival of the TKI-resistant cell line, H1975, suggesting signaling cross-talk in survival response. This also suggests that small molecule TKI inhibitors or antibodies targeting these proteins may circumvent EGFR TKI resistance, if used in combination with EGFR TKIs. However, these knockdown experiments do not address the functional relevance of these tyrosine phosphorylation changes. As such, studies utilizing Y-F mutants of these proteins are needed.
We identified DAPP1-Y139 as a novel target of mutant EGFR signaling. Tyrosine phosphorylation at this site was inhibited by either erlotinib or afatinib treatment in TKI sensitive H3255 and PC9 cells, but not in TKI resistant H1975 cells (Fig. 8). Using DAPP1 Y139F mutant we showed that Y139 is the major site of DAPP1 tyrosine phosphorylation induced by both wild type and mutant EGFR signaling. Interestingly, DAPP1 overexpression decreases EGFR autophosphorylation, and this is partially rescued by the DAPP1 Y139F mutant (Fig. 8D). DAPP1 is also an adaptor for B cell receptor (BCR) signaling (46, 47). Hence, it is possible that mutant EGFRs cross talk with the BCR signaling through DAPP1 tyrosine phosphorylation, and phosphorylated DAPP1 in-turn executes a negative feedback to inhibit EGFR autophosphorylation. Further studies are needed to elucidate the mechanism by which mutant EGFR signals through DAPP1 and to explore the potential of EGFR regulated DAPP1 to crosstalk with BCR signaling.
This study provides in vivo evidence of tyrosine phosphorylation changes exerted on direct or indirect targets of mutant EGFR signaling in lung adenocarcinoma. These phosphosites are potential novel biomarkers of the EGFR TKI response in this dreaded disease. Further studies utilizing targeted mass spectrometry to correlate tyrosine phosphorylation of these targets with TKI sensitivity in EGFR mutant human tumors are warranted.
DATA AVAILABILITY
The MS proteomics data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (74) with the dataset identifier PXD004373. The reviewer account details are as follows: Username: reviewer70975@ebi.ac.uk Password: WD6ELzjQ.
Supplementary Material
Footnotes
Author contributions: X.Z., A.P., and U.G. designed research; X.Z., T.M., M.K.K., A.V., S.S., A.M., H.K.J., N.B., S.P., F.K., R.B., R.C., and U.G. performed research; M.B. and A.C. contributed new reagents or analytic tools; X.Z., T.M., M.K.K., M.B., A.V., S.S., S.A., A.M., H.K.J., N.B., S.P., C.M.C., S.G., F.K., R.B., R.C., A.C., and U.G. analyzed data; X.Z., T.M., M.K.K., C.M.C., and U.G. wrote the paper.
* This research was supported by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute (U.G.). The work was also supported in part by an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520) and NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036) (A.P); NCI Outstanding Investigator Award (R35CA197745–02), NCI Research Centers for Cancer Systems Biology Consortium (1U54CA209997), Leidos Biomedical Research Inc. contract 15X036 and NIH Biomedical Research Support Shared Instrumentation Grants (1S10OD021764, 1S10OD012351) (A.C). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We have declared no conflicts of interest.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EGFR
- epidermal growth factor receptor
- ACN
- Acetonitrile
- DAPP1
- Dual Adapter for Phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide
- ECL
- Enhanced Chemiluminescence
- EGF
- Epidermal Growth Factor
- FBS
- FetaL Bovine Serum
- FET
- Fisher Exact Test
- HCD
- High-energy Collision Dissociation
- HEK
- Human Embryonic Kidney
- H/L
- Heavy/Light
- H/M
- Heavy/Medium
- HEPES
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HRP
- Horseradish Peroxidase
- MAB
- Monoclonal Antibody
- M/L
- Mediam/Light
- NSCLC
- Non Small Cell Lung Cancer
- PBS
- Phosphate-buffered Saline
- PTM
- post-translational modification
- RPLC
- Reverse Phase Liquid Chromatography
- RPMI
- Roswell Park Memorial Institute
- RTK
- Receptor Tyrosine Kinase
- SILAC
- Stable Isotope labeling in Cell Culture
- SDS
- Sodium Dodecyl Sulfate
- S.D.
- Standard Deviation
- TKI
- Tyrosine Kinase Inhibitor
- TFA
- Trifluoroacetic acid
- YFP
- Yellow fluorescent protein.
REFERENCES
- 1. Siegel R., Naishadham D., and Jemal A. (2013) Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 2. Dowell J., Minna J. D., and Kirkpatrick P. (2005) Erlotinib hydrochloride. Nat. Rev. Drug Discov. 4, 13–14 [DOI] [PubMed] [Google Scholar]
- 3. Melnikova I., and Golden J. (2004) Targeting protein kinases. Nat. Rev. Drug Discov. 3, 993–994 [DOI] [PubMed] [Google Scholar]
- 4. Copeman M. (2008) Prolonged response to first-line erlotinib for advanced lung adenocarcinoma. J. Exp. Clin. Cancer Res. 27, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma S. P. (2007) New predictors of survival for early-stage NSCLC. Lancet Oncol. 8, 288. [DOI] [PubMed] [Google Scholar]
- 6. Sharma S. P. (2007) Gene signature for predicting NSCLC outcome. Lancet Oncol. 8, 105. [DOI] [PubMed] [Google Scholar]
- 7. Riely G. J., Pao W., Pham D., Li A. R., Rizvi N., Venkatraman E. S., Zakowski M. F., Kris M. G., Ladanyi M., and Miller V. A. (2006) Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 839–844 [DOI] [PubMed] [Google Scholar]
- 8. Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., and Varmus H. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller V. A., Hirsh V., Cadranel J., Chen Y. M., Park K., Kim S. W., Zhou C., Su W. C., Wang M., Sun Y., Heo D. S., Crino L., Tan E. H., Chao T. Y., Shahidi M., Cong X. J., Lorence R. M., and Yang J. C. (2012) Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 13, 528–538 [DOI] [PubMed] [Google Scholar]
- 10. Janne P. A., Yang J. C., Kim D. W., Planchard D., Ohe Y., Ramalingam S. S., Ahn M. J., Kim S. W., Su W. C., Horn L., Haggstrom D., Felip E., Kim J. H., Frewer P., Cantarini M., Brown K. H., Dickinson P. A., Ghiorghiu S., and Ranson M. (2015) AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 372, 1689–1699 [DOI] [PubMed] [Google Scholar]
- 11. Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., Fujita Y., Okinaga S., Hirano H., Yoshimori K., Harada T., Ogura T., Ando M., Miyazawa H., Tanaka T., Saijo Y., Hagiwara K., Morita S., Nukiwa T., and North-East Japan Study, G. (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 362, 2380–2388 [DOI] [PubMed] [Google Scholar]
- 12. Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., Palmero R., Garcia-Gomez R., Pallares C., Sanchez J. M., Porta R., Cobo M., Garrido P., Longo F., Moran T., Insa A., De Marinis F., Corre R., Bover I., Illiano A., Dansin E., de Castro J., Milella M., Reguart N., Altavilla G., Jimenez U., Provencio M., Moreno M. A., Terrasa J., Munoz-Langa J., Valdivia J., Isla D., Domine M., Molinier O., Mazieres J., Baize N., Garcia-Campelo R., Robinet G., Rodriguez-Abreu D., Lopez-Vivanco G., Gebbia V., Ferrera-Delgado L., Bombaron P., Bernabe R., Bearz A., Artal A., Cortesi E., Rolfo C., Sanchez-Ronco M., Drozdowskyj A., Queralt C., de Aguirre I., Ramirez J. L., Sanchez J. J., Molina M. A., Taron M., Paz-Ares L., Spanish Lung Cancer Group in collaboration with Groupe Francais de, P-C, Associazione Italiana Oncologia, T. (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13, 239–246 [DOI] [PubMed] [Google Scholar]
- 13. Wu Y. L., Zhou C., Hu C. P., Feng J., Lu S., Huang Y., Li W., Hou M., Shi J. H., Lee K. Y., Xu C. R., Massey D., Kim M., Shi Y., and Geater S. L. (2014) Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 15, 213–222 [DOI] [PubMed] [Google Scholar]
- 14. Zhou C., Wu Y. L., Chen G., Feng J., Liu X. Q., Wang C., Zhang S., Wang J., Zhou S., Ren S., Lu S., Zhang L., Hu C., Hu C., Luo Y., Chen L., Ye M., Huang J., Zhi X., Zhang Y., Xiu Q., Ma J., Zhang L., and You C. (2011) Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12, 735–742 [DOI] [PubMed] [Google Scholar]
- 15. Bean J., Brennan C., Shih J. Y., Riely G., Viale A., Wang L., Chitale D., Motoi N., Szoke J., Broderick S., Balak M., Chang W. C., Yu C. J., Gazdar A., Pass H., Rusch V., Gerald W., Huang S. F., Yang P. C., Miller V., Ladanyi M., Yang C. H., and Pao W. (2007) MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. U.S.A. 104, 20932–20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., and Janne P. A. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 17. Turke A. B., Zejnullahu K., Wu Y. L., Song Y., Dias-Santagata D., Lifshits E., Toschi L., Rogers A., Mok T., Sequist L., Lindeman N. I., Murphy C., Akhavanfard S., Yeap B. Y., Xiao Y., Capelletti M., Iafrate A. J., Lee C., Christensen J. G., Engelman J. A., and Janne P. A. (2010) Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sequist L. V., Waltman B. A., Dias-Santagata D., Digumarthy S., Turke A. B., Fidias P., Bergethon K., Shaw A. T., Gettinger S., Cosper A. K., Akhavanfard S., Heist R. S., Temel J., Christensen J. G., Wain J. C., Lynch T. J., Vernovsky K., Mark E. J., Lanuti M., Iafrate A. J., Mino-Kenudson M., and Engelman J. A. (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arcila M. E., Oxnard G. R., Nafa K., Riely G. J., Solomon S. B., Zakowski M. F., Kris M. G., Pao W., Miller V. A., and Ladanyi M. (2011) Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin. Cancer Res. 17, 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suda K., Murakami I., Sakai K., Mizuuchi H., Shimizu S., Sato K., Tomizawa K., Tomida S., Yatabe Y., Nishio K., and Mitsudomi T. (2015) Small cell lung cancer transformation and T790M mutation: complimentary roles in acquired resistance to kinase inhibitors in lung cancer. Sci. Rep. 5, 14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niederst M. J., Sequist L. V., Poirier J. T., Mermel C. H., Lockerman E. L., Garcia A. R., Katayama R., Costa C., Ross K. N., Moran T., Howe E., Fulton L. E., Mulvey H. E., Bernardo L. A., Mohamoud F., Miyoshi N., VanderLaan P. A., Costa D. B., Janne P. A., Borger D. R., Ramaswamy S., Shioda T., Iafrate A. J., Getz G., Rudin C. M., Mino-Kenudson M., and Engelman J. A. (2015) RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 6, 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salam A. A., Eyres K. S., Magides A. D., and Cleary J. (1991) Anterior dislocation of the restrained shoulder: a seat-belt injury. Arch. Emerg. Med. 8, 56–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., and Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 24. Schweppe D. K., Rigas J. R., and Gerber S. A. (2013) Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteomics 91, 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo A., Villen J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., and Comb M. J. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., Rix U., Fang B., Bai Y., Edwards A., Colinge J., Bennett K. L., Gao J., Song L., Eschrich S., Superti-Furga G., Koomen J., and Haura E. B. (2010) A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 6, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Assiddiq B. F., Tan K. Y., Toy W., Chan S. P., Chong P. K., and Lim Y. P. (2012) EGFR S1166 phosphorylation induced by a combination of EGF and gefitinib has a potentially negative impact on lung cancer cell growth. J. Proteome Res. 11, 4110–4119 [DOI] [PubMed] [Google Scholar]
- 28. Yoshida T., Zhang G., Smith M. A., Lopez A. S., Bai Y., Li J., Fang B., Koomen J., Rawal B., Fisher K. J., Chen Y. A., Kitano M., Morita Y., Yamaguchi H., Shibata K., Okabe T., Okamoto I., Nakagawa K., and Haura E. B. (2014) Tyrosine phosphoproteomics identifies both codrivers and cotargeting strategies for T790M-related EGFR-TKI resistance in non-small cell lung cancer. Clin. Cancer Res. 20, 4059–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guha U., Chaerkady R., Marimuthu A., Patterson A. S., Kashyap M. K., Harsha H. C., Sato M., Bader J. S., Lash A. E., Minna J. D., Pandey A., and Varmus H. E. (2008) Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc. Natl. Acad. Sci. U.S.A. 105, 14112–14117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X., Belkina N., Jacob H. K., Maity T., Biswas R., Venugopalan A., Shaw P. G., Kim M. S., Chaerkady R., Pandey A., and Guha U. (2015) Identifying novel targets of oncogenic EGF receptor signaling in lung cancer through global phosphoproteomics. Proteomics 15, 340–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Politi K., Zakowski M. F., Fan P. D., Schonfeld E. A., Pao W., and Varmus H. E. (2006) Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 20, 1496–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 33. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., and Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W. H., Pages F., Trajanoski Z., and Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maity T. K., Venugopalan A., Linnoila I., Cultraro C. M., Giannakou A., Nemati R., Zhang X., Webster J. D., Ritt D., Ghosal S., Hoschuetzky H., Simpson R. M., Biswas R., Politi K., Morrison D. K., Varmus H. E., and Guha U. (2015) Loss of MIG6 Accelerates Initiation and Progression of Mutant Epidermal Growth Factor Receptor-Driven Lung Adenocarcinoma. Cancer Discov. 5, 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stuart S. A., Houel S., Lee T., Wang N., Old W. M., and Ahn N. G. (2015) A Phosphoproteomic Comparison of B-RAFV600E and MKK1/2 Inhibitors in Melanoma Cells. Mol. Cell. Proteomics 14, 1599–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 39. Ong S. E., Foster L. J., and Mann M. (2003) Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 [DOI] [PubMed] [Google Scholar]
- 40. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 41. Blagoev B., Ong S. E., Kratchmarova I., and Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 42. Kim M. S., Zhong Y., Yachida S., Rajeshkumar N. V., Abel M. L., Marimuthu A., Mudgal K., Hruban R. H., Poling J. S., Tyner J. W., Maitra A., Iacobuzio-Donahue C. A., and Pandey A. (2014) Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol. Cell. Proteomics 13, 2803–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barr A. J., Ugochukwu E., Lee W. H., King O. N., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Muller S., and Knapp S. (2009) Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 136, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Flint A. J., Tiganis T., Barford D., and Tonks N. K. (1997) Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamaguchi T., Yanagisawa K., Sugiyama R., Hosono Y., Shimada Y., Arima C., Kato S., Tomida S., Suzuki M., Osada H., and Takahashi T. (2012) NKX2–1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 21, 348–361 [DOI] [PubMed] [Google Scholar]
- 46. Sommers C. L., Gurson J. M., Surana R., Barda-Saad M., Lee J., Kishor A., Li W., Gasser A. J., Barr V. A., Miyaji M., Love P. E., and Samelson L. E. (2008) Bam32: a novel mediator of Erk activation in T cells. Int. Immunol. 20, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rouquette-Jazdanian A. K., Sommers C. L., Kortum R. L., Morrison D. K., and Samelson L. E. (2012) LAT-independent Erk activation via Bam32-PLC-gamma1-Pak1 complexes: GTPase-independent Pak1 activation. Mol. Cell 48, 298–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gong Y., Somwar R., Politi K., Balak M., Chmielecki J., Jiang X., and Pao W. (2007) Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 4, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song H., Jin X., and Lin J. (2004) Stat3 upregulates MEK5 expression in human breast cancer cells. Oncogene 23, 8301–8309 [DOI] [PubMed] [Google Scholar]
- 50. Montero J. C., Ocana A., Abad M., Ortiz-Ruiz M. J., Pandiella A., and Esparis-Ogando A. (2009) Expression of Erk5 in early stage breast cancer and association with disease free survival identifies this kinase as a potential therapeutic target. PLoS ONE 4, e5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weldon C. B., Scandurro A. B., Rolfe K. W., Clayton J. L., Elliott S., Butler N. N., Melnik L. I., Alam J., McLachlan J. A., Jaffe B. M., Beckman B. S., and Burow M. E. (2002) Identification of mitogen-activated protein kinase kinase as a chemoresistant pathway in MCF-7 cells by using gene expression microarray. Surgery 132, 293–301 [DOI] [PubMed] [Google Scholar]
- 52. Esparis-Ogando A., Diaz-Rodriguez E., Montero J. C., Yuste L., Crespo P., and Pandiella A. (2002) Erk5 participates in neuregulin signal transduction and is constitutively active in breast cancer cells overexpressing ErbB2. Mol. Cell. Biol. 22, 270–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mehta P. B., Jenkins B. L., McCarthy L., Thilak L., Robson C. N., Neal D. E., and Leung H. Y. (2003) MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene 22, 1381–1389 [DOI] [PubMed] [Google Scholar]
- 54. Gavine P. R., Wang M., Yu D., Hu E., Huang C., Xia J., Su X., Fan J., Zhang T., Ye Q., Zheng L., Zhu G., Qian Z., Luo Q., Hou Y. Y., and Ji Q. (2015) Identification and validation of dysregulated MAPK7 (ERK5) as a novel oncogenic target in squamous cell lung and esophageal carcinoma. BMC Cancer 15, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hackel P. O., Zwick E., Prenzel N., and Ullrich A. (1999) Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr. Opin. Cell Biol. 11, 184–189 [DOI] [PubMed] [Google Scholar]
- 56. Zwick E., Hackel P. O., Prenzel N., and Ullrich A. (1999) The EGF receptor as central transducer of heterologous signalling systems. Trends Pharmacol. Sci. 20, 408–412 [DOI] [PubMed] [Google Scholar]
- 57. Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., and Varmus H. (2004) EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morgan M. A., Parsels L. A., Kollar L. E., Normolle D. P., Maybaum J., and Lawrence T. S. (2008) The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin. Cancer Res. 14, 5142–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tan A. R., Yang X., Hewitt S. M., Berman A., Lepper E. R., Sparreboom A., Parr A. L., Figg W. D., Chow C., Steinberg S. M., Bacharach S. L., Whatley M., Carrasquillo J. A., Brahim J. S., Ettenberg S. A., Lipkowitz S., and Swain S. M. (2004) Evaluation of biologic end points and pharmacokinetics in patients with metastatic breast cancer after treatment with erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor. J. Clin. Oncol. 22, 3080–3090 [DOI] [PubMed] [Google Scholar]
- 60. Zhang X., Zhang H., Tighiouart M., Lee J. E., Shin H. J., Khuri F. R., Yang C. S., Chen Z. G., and Shin D. M. (2008) Synergistic inhibition of head and neck tumor growth by green tea (-)-epigallocatechin-3-gallate and EGFR tyrosine kinase inhibitor. Int. J. Cancer 123, 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li T., Ling Y. H., and Perez-Soler R. (2008) Tumor dependence on the EGFR signaling pathway expressed by the p-EGFR:p-AKT ratio predicts erlotinib sensitivity in human non-small cell lung cancer (NSCLC) cells expressing wild-type EGFR gene. J. Thorac. Oncol. 3, 643–647 [DOI] [PubMed] [Google Scholar]
- 62. Regales L., Gong Y., Shen R., de Stanchina E., Vivanco I., Goel A., Koutcher J. A., Spassova M., Ouerfelli O., Mellinghoff I. K., Zakowski M. F., Politi K. A., and Pao W. (2009) Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J. Clin. Invest. 119, 3000–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Katakami N., Atagi S., Goto K., Hida T., Horai T., Inoue A., Ichinose Y., Koboyashi K., Takeda K., Kiura K., Nishio K., Seki Y., Ebisawa R., Shahidi M., and Yamamoto N. (2013) LUX-Lung 4: a phase II trial of afatinib in patients with advanced non-small-cell lung cancer who progressed during prior treatment with erlotinib, gefitinib, or both. J. Clin. Oncol. 31, 3335–3341 [DOI] [PubMed] [Google Scholar]
- 64. Fabian M. A., Biggs W. H. 3rd, Treiber D. K., Atteridge C. E., Azimioara M. D., Benedetti M. G., Carter T. A., Ciceri P., Edeen P. T., Floyd M., Ford J. M., Galvin M., Gerlach J. L., Grotzfeld R. M., Herrgard S., Insko D. E., Insko M. A., Lai A. G., Lelias J. M., Mehta S. A., Milanov Z. V., Velasco A. M., Wodicka L. M., Patel H. K., Zarrinkar P. P., and Lockhart D. J. (2005) A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23, 329–336 [DOI] [PubMed] [Google Scholar]
- 65. Conradt L., Godl K., Schaab C., Tebbe A., Eser S., Diersch S., Michalski C. W., Kleeff J., Schnieke A., Schmid R. M., Saur D., and Schneider G. (2011) Disclosure of erlotinib as a multikinase inhibitor in pancreatic ductal adenocarcinoma. Neoplasia 13, 1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weber C., Schreiber T. B., and Daub H. (2012) Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J. Proteomics 75, 1343–1356 [DOI] [PubMed] [Google Scholar]
- 67. Kitagawa D., Yokota K., Gouda M., Narumi Y., Ohmoto H., Nishiwaki E., Akita K., and Kirii Y. (2013) Activity-based kinase profiling of approved tyrosine kinase inhibitors. Genes Cells 18, 110–122 [DOI] [PubMed] [Google Scholar]
- 68. Lanning B. R., Whitby L. R., Dix M. M., Douhan J., Gilbert A. M., Hett E. C., Johnson T. O., Joslyn C., Kath J. C., Niessen S., Roberts L. R., Schnute M. E., Wang C., Hulce J. J., Wei B., Whiteley L. O., Hayward M. M., and Cravatt B. F. (2014) A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat. Chem. Biol. 10, 760–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Duong-Ly K. C., Devarajan K., Liang S., Horiuchi K. Y., Wang Y., Ma H., and Peterson J. R. (2016) Kinase Inhibitor Profiling Reveals Unexpected Opportunities to Inhibit Disease-Associated Mutant Kinases. Cell Rep. 14, 772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yao H. P., Zhou Y. Q., Zhang R., and Wang M. H. (2013) MSP-RON signalling in cancer: pathogenesis and therapeutic potential. Nat. Rev. Cancer 13, 466–481 [DOI] [PubMed] [Google Scholar]
- 71. Catenacci D. V., Cervantes G., Yala S., Nelson E. A., El-Hashani E., Kanteti R., El Dinali M., Hasina R., Bragelmann J., Seiwert T., Sanicola M., Henderson L., Grushko T. A., Olopade O., Karrison T., Bang Y. J., Kim W. H., Tretiakova M., Vokes E., Frank D. A., Kindler H. L., Huet H., and Salgia R. (2011) RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer Biol. Ther. 12, 9–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pasquale E. B. (2010) Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Amato K. R., Wang S., Tan L., Hastings A. K., Song W., Lovly C. M., Meador C. B., Ye F., Lu P., Balko J. M., Colvin D. C., Cates J. M., Pao W., Gray N. S., and Chen J. (2016) EPHA2 Blockade Overcomes Acquired Resistance to EGFR Kinase Inhibitors in Lung Cancer. Cancer Res. 76, 305–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (74) with the dataset identifier PXD004373. The reviewer account details are as follows: Username: reviewer70975@ebi.ac.uk Password: WD6ELzjQ.