Abstract
While the Mre11-Rad50-Nbs1 (MRN) complex has known roles in repair processes like homologous recombination and microhomology-mediated end-joining, its role in nonhomologous end-joining (NHEJ) is unclear as Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammals have different requirements for repairing cut DNA ends. Most double-strand breaks (DSBs) require nucleolytic processing prior to DNA ligation. Therefore, we studied repair using the Hermes transposon, whose excision leaves a DSB capped by hairpin ends similar to structures generated by palindromes and trinucleotide repeats. We generated single Hermes insertions using a novel S. pombe transient transfection system, and used Hermes excision to show a requirement for MRN in the NHEJ of nonligatable ends. NHEJ repair was indicated by the >1000-fold decrease in excision in cells lacking Ku or DNA ligase 4. Most repaired excision sites had <5 bp of sequence loss or mutation, characteristic for NHEJ and similar excision events in metazoans, and in contrast to the more extensive loss seen in S. cerevisiae. S. pombe NHEJ was reduced >1000-fold in cells lacking each MRN subunit, and loss of MRN-associated Ctp1 caused a 30-fold reduction. An Mre11 dimer is thought to hold DNA ends together for repair, and Mre11 dimerization domain mutations reduced repair 300-fold. In contrast, a mre11 mutant defective in endonucleolytic activity, the same mutant lacking Ctp1, or the triple mutant also lacking the putative hairpin nuclease Pso2 showed wild-type levels of repair. Thus, MRN may act to recruit the hairpin opening activity that allows subsequent repair.
Keywords: Mre11-Rad50-Nbs1, MRN, Mre11-Rad50-Xrs2, MRX, nonhomologous end-joining, NHEJ, hairpin, hAT, transposon, Hermes
DNA double-strand breaks (DSBs) are toxic lesions that can recombine with other chromosomal DNA, and are therefore challenges to genomic integrity that must be repaired to allow cellular growth (Featherstone and Jackson 1999; Harrison and Haber 2006; Harper and Elledge 2007). DSBs in mitotic cells can be repaired by two major pathways in eukaryotes: homologous recombination (HR) and end-joining (Symington and Gautier 2011). HR is an error-free repair mechanism by which information lost at the DSB is replaced by copying the homologous sequence from another DNA molecule (Shrivastav et al. 2008; Jain and Cooper 2010; Symington and Gautier 2011). End-joining mechanisms are error-prone, and involve the direct joining of the two ends of the DSB after different amounts of DNA sequence loss and/or mutation. Nonhomologous End-Joining (NHEJ) causes only small amounts of sequence loss, while Microhomology-Mediated End-Joining (MMEJ) involves loss of larger amounts of DNA sequence. These processes are also distinct in that NHEJ requires the Ku heterodimer (Ku70 and Ku80) but MMEJ does not (Moore and Haber 1996; Daley et al. 2005; Raji and Hartsuiker 2006; McVey and Lee 2008; Chiruvella et al. 2013). The Ku heterodimer binds to DSBs and has multiple roles modulating DNA repair, dimerizing to bring DSBs together, binding to telomeres, and recruiting telomerase and/or silencing proteins (Boulton and Jackson 1996a,b, 1998; Tsukamoto et al. 1997; Mishra and Shore 1999; Walker et al. 2001; Stellwagen et al. 2003; Davis et al. 2014).
The Mre11-Rad50-Nbs1 (MRN) complex plays a central role in HR and MMEJ, as well as activation of the DNA damage checkpoint and telomerase recruitment to telomeres (Lee and Paull 2005; Viscardi et al. 2007; Rupnik et al. 2008; Lamarche et al. 2010). DNA damage checkpoints are the process of sensing the presence of DSBs, activating specific kinases, and initiating a series of events that halt the cell cycle and allow repair (Weinert 1998; Harrison and Haber 2006). MRN is one of the first complexes to bind to DSBs. Resection of a 5′ DNA strand at the DSB is required for HR and MMEJ, and Mre11 has both endo- and exonuclease activities that are important for generating the 3′ single-stranded DNA overhang (McVey and Lee 2008; Nicolette et al. 2010; Paull 2010). Rad50 is an ATPase that stimulates ATP-dependent DNA unwinding and Mre11 nuclease activation (Trujillo and Sung 2001; Chen et al. 2005). Nbs1 binds to Mre11 and allows recruitment of DNA damage checkpoint kinases ATM (in mammals) or Tel1 (in fungi) to MRN-bound DSBs (Nakada et al. 2003; Falck et al. 2005; You et al. 2005).
The role for MRN in NHEJ is much less clear. In Saccharomyces cerevisiae, where the MRN complex is called Mre11-Rad50-Xrs2 (MRX), NHEJ requires MRX in an end-joining assay that monitors recircularization of restriction enzyme-cut plasmids transformed into yeast (Zhang and Paull 2005) or one that monitors repair of a chromosomal DSB in the absence of homology for HR (Moore and Haber 1996). In contrast, Schizosaccharomyces pombe MRN has been tested in the plasmid end-joining assay and is not required (Manolis et al. 2001). Similarly, the presence or absence of MRN shows only a minor effect in a mammalian NHEJ assay that monitors chromatin cleaved in vivo by a restriction enzyme (Rass et al. 2009; Xie et al. 2009), and hypomorphic MRN mutations do not affect the NHEJ-dependent process of mammalian V(D)J joining (Carney et al. 1998; Stewart et al. 1999; Harfst et al. 2000; Bender et al. 2002; Theunissen et al. 2003). These different requirements for MRN/MRX are surprising as the nuclease functions and subunit interactions that form the complex are well-conserved (Nakada et al. 2003; Falck et al. 2005; You et al. 2009; Symington and Gautier 2011). Consequently, the biology of MRN in NHEJ is still not well-understood.
The assays that monitor restriction enzyme-cut DNA do not assess upstream processing that occurs during repair of most DNA lesions, and methods have therefore also been developed to study DNA repair by recovery from transposon excision and exposure to ionizing radiation. Transposon excision produces a lesion at a defined site, and therefore has an advantage over studying ionizing radiation that produces random breaks because the sequences of the DNA substrates and the repaired products can be examined in detail. The hAT transposon–transposase system has been valuable for this purpose, where transposon excision leaves behind covalently closed DNA hairpins in the genomic DNA (Bhasin et al. 1999; Zhou et al. 2004). Repair therefore requires nucleolytic DNA processing to generate free 5′ phosphate and 3′ hydroxyl groups for ligation. DNA hairpins can normally form in eukaryotic genomes at structures formed by palindromes, inverted repeats, trinucleotide repeats, and during vertebrate V(D)J joining (Lieber et al. 2004; Zhou et al. 2004; Lewis and Cote 2006; Lobachev et al. 2007; Sundararajan et al. 2010). The correct processing of DNA hairpins is essential for genome stability, as these structures can lead to chromosome translocations or gene amplification in tumors (Kurahashi and Emanuel 2001a,b; Kato et al. 2006; Tanaka and Yao 2009; Weil 2009). Therefore, understanding how DNA hairpins are repaired has important consequences for our understanding of genome integrity and carcinogenesis.
Work in S. cerevisiae has shown that MRX and the associated enzyme Sae2 are required for the repair of hairpins, i.e., hAT transposon events as well as palindromic genomic sequences (Paull 2010). The repair of hAT transposon excision events also requires Ku (Yu et al. 2004), suggesting a Ku-dependent NHEJ process. However, transposon excision is associated with a sequence loss (8–22 nucleotides) larger than the small loss of sequence (0–5 nucleotides) in NHEJ, which led to the conclusion that repair proceeded through an MMEJ-related process (Yu et al. 2004). The repaired excision sites (i.e., transposon footprints) in S. cerevisiae contrast with hAT transposon excision in Drosophila, Maize, and other organisms in which the majority of transposon footprints have a smaller number of base deletions, insertions, or changes (Baran et al. 1992; Atkinson et al. 1993; Scott et al. 1996; Colot et al. 1998). These comparisons suggest that S. cerevisiae may repair structures such as DNA hairpins by a combination of evolutionarily conserved processes as well as unique mechanisms not used in other eukaryotes.
The different MRN requirements of S. pombe and S. cerevisiae for NHEJ repair in the restriction enzyme-cut DNA assay, as well as the unusual repair of transposon excision events in S. cerevisiae, suggested the need for an alternative model to study end-joining repair. S. pombe has the potential to yield significant new insights into DNA repair that may be more broadly conserved, as its repair processes are more similar to mammalian cells than S. cerevisiae, e.g., repair proteins like MRN, a lower level of HR, and more similar products formed from transfected DNA [reviewed in Frank-Vaillant and Marcand (2002), Würtele et al. (2003), Bähler and Wood (2004), Decottignies (2005), and Chen et al. (2012, 2013)]. Our results on the repair of hAT transposon excision events in S. pombe revealed an NHEJ process that occurs with little or no sequence loss, and that requires MRN and the associated protein Ctp1 as well as the NHEJ factors Ku and DNA ligase 4. Mutant analysis indicated that at least one role of MRN is to form Mre11 dimers that could hold the hairpin ends together. In contrast, the nuclease activities of Mre11 were not required for efficient repair.
Materials and Methods
Hermes transposon insertion
To produce cells with only one transposon insertion, we developed a “transient transfection” assay using two plasmids that separately introduce the transposon and a transposase expression cassette. Wild-type S. pombe KRP1 or KRP201 cells (all strains are shown in Supplemental Material, Table S1 in File S1) bearing the Hermes transposase-expressing plasmid pHL2578u were grown on EMM + adenine, leucine, and histidine (EMM-ura) medium (Moreno et al. 1991), which induces and selects for the transposase. Cells were then transformed with the Hermes transposon donor plasmid pHL2577 (0.3, 1.0, or 3.0 µg) and divided between nonselective plates with either inducing (EMM + adenine, uracil, leucine, and histidine) or noninducing (yeast extract sucrose, YES) media (Moreno et al. 1991). Cells were grown for 24 hr at 30°, equivalent to two to five cell divisions. The resulting lawns of cells were then replica plated to YES medium containing supplements that both select for the transposon (200 µg/ml G418) and against both plasmids (1 mg/ml FOA). After 3 days of growth at 30°, colonies were counted and a subset was analyzed for insertions.
Mapping Hermes insertion sites
The site of insertion in several different strains was identified by inverse PCR. Genomic DNAs (5 µg) from the insertion strains (Table S1 in File S1) were digested with Sau3A I (20 units in a 50 µl reaction incubated at 37° for 16 hr, followed by inactivation at 65° for 20 min). Digested DNA (1 µg) was diluted into a 200 µl ligation reaction with 40 units of T4 DNA ligase (New England Biolabs, Beverly, MA) and incubated at 16° overnight. The resulting material was ethanol precipitated, resuspended in 10 µl 10 mM Tris–1 mM EDTA pH 8.0, and 500 ng of DNA was amplified using the outward facing primers pHL2577-3633S and pHL2577-3527AS with Expand polymerase (all primers are presented in Table S2 in File S1). The inverse PCR products were purified and sequenced with the primer Hermes_Sau3A I. The genomic DNA adjacent to Hermes was compared to the S. pombe genome using the BLAST utility at Pombase (www.Pombase.org) to map the Hermes insertion site.
Hermes transposon excision assays
General assay:
Transposon excision was induced, and the excision frequency and sequence of the excision products were examined by PCR. Two different strains, each bearing independent Hermes insertions (KRP3-3 and KRP3-4), were transformed with 1 µg of the pHL2578u plasmid expressing transposase, which mediates excision as well as integration. Transformants were plated on EMM-ura plates with 2% glucose to induce transposase expression. A small colony (1 mm diameter) was inoculated into 1 ml of EMM-ura + 2% glucose liquid medium and grown for 48 hr at 30° to 3–5 × 107cells/ml. Cells were harvested, washed with sterile milli-Q filtered water, resuspended in 100 µl Zymolyase buffer [1.2 M sorbitol, 0.1 M sodium phosphate pH 7.4, and 2.5 mg/ml Zymolyase 20T (MP Biomedicals)], and incubated at 37° for 3 hr. The cells were pelleted and incubated with 150 µl lysis buffer (0.1 M Tris, 50 mM EDTA, and 1% SDS, pH 8.0) at 65° for 20 min, followed by the addition of 50 µl of 7.5 M NH4OAc and incubation for 30 min on ice. The reaction was extracted with an equal volume of 25:24:1 phenol:chloroform:isoamyl-alcohol, precipitated with isopropanol, washed with 70% ethanol, and resuspended in 60 µl of H2O. Genomic DNA (250 ng) was used as template in a 30 µl “first-round” PCR reaction. DNA from KRP 3-4 cells was amplified with primers 3-4_S and 3-4_AS with 5 PRIME MasterMix (5 PRIME). The template DNA was denatured (94° for 2 min), followed by 45 cycles of PCR (94° for 30 sec, 52° for 30 sec, and 65° for 1 min), followed by 5 min at 65°. The first-round PCR product (0.5 µl) was used as the template in a 30 µl reaction for the “second-round” PCR using the nested primers 3-4_2S and 3-4_2AS. The DNA was denatured (at 94° for 2 min), followed by 30 cycles of PCR (94° for 30 sec, 56° for 30 sec, and 65° for 1 min). The PCR products (5 µl) were analyzed on 1% agarose gels stained with ethidium bromide. Excision events in the KPR 3-3 cells containing an independent transposon were amplified in a similar manner using the primers 3-3S and 3-3AS.
A slightly modified assay was used in the pso2∆ excision experiments. Instead of using 250 ng of genomic DNA as determined by OD260, 2 µl of genomic DNA (∼3.3% of the preparation) was used in the first PCR under the same conditions but amplified for only 35 cycles. The second PCR and gel analysis were identical to the assay described above.
To determine the sequence of the repaired excision events, PCR products were cloned into pCR2.1-TOPO using the TOPO TA kit (Life Technologies) and sequenced using the M13 forward or reverse primers. DNA was amplified from 3 to 5 colonies per experiment, and one or more clones were sequenced from each colony.
Reconstruction assay:
To quantify the frequency of Hermes excision, we generated a standard curve for excision frequency by mixing cells containing a transposon with wild-type cells that mimic transposon excision, and analyzing DNA prepared from these mixtures. KRP3-4 cells (5 × 107) bearing a Hermes insertion were mixed with 105, 104, 103, 102, or 10 wild-type KRP1 cells that lack Hermes. Genomic DNA was prepared as described for the excisions assays and then analyzed in the first and second round PCR assays. Gel images were captured on a Bio-Rad (Hercules, CA) gel scanner, and the grayscale images inverted to dark bands on a white background. Quantitation of these raw images and a comparison between the two rounds of PCR were used to determine the frequency of Hermes excision, as described in the Results. Each mixture was analyzed in triplicate to reveal the variation in amplification of rare DNA sequences, and this standard curve was then used to determine transposon excision frequency in wild-type and mutant strains. All mutant strains were analyzed in parallel with the KRP 3-4 wild-type strain, which showed median excision frequencies from 10−4 to 3 × 10−4 in all experiments.
Southern blotting to physically assay excision frequency
Southern blotting followed previously described methods (Ray and Runge 1999). Single wild-type and rad52∆ transformants bearing either the vector (pREP81) or the transposase expression plasmid (pHL2578) were grown in selective, transposase-inducing medium (EMM + adenine, uracil, and histidine + 2% glucose) for ∼10 population doublings and genomic DNA was prepared from each culture. DNA (2 µg) was digested with XbaI and analyzed by Southern blotting using the probe amplified from KRP1 genomic DNA using Hermes3-4RightProbe_S and Hermes2-4RightProbe_AS. The washed blot was exposed to a Phosphor Imager plate for 14 days, and band intensity was quantitated using the Image Quant TL software (GE Healthcare). Excision frequency was determined by the ratio of intensities of the excised band at 2.2 kb to the unexcised band at 6.6 kb.
Spot tests for drug sensitivity
The sensitivity of the mre11 mutants to hydroxyurea (HU) and camptothecin (CPT) was examined by spotting 5 µl of cells from fivefold serial dilution series on to medium containing the drug at the indicated concentration. Briefly, cells were revived from freezer stocks and streaked on nonselective YES liquid medium and allowed to form single colonies. Single colonies were used to seed 2 ml cultures in YES + G418 medium and grown overnight at 30°. Cell density was determined by optical density and 107 cells were placed in 0.5 ml of sterile water, spun down, and resuspended in 1.0 ml of sterile water. Fivefold serial dilutions were then made from the 107 cells/ml stock, and 5 µl of each dilution was spotted on plates with or without HU or CPT.
Data availability
All strains and DNA constructs described here are available upon request to the corresponding author. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
A transient transfection assay in S. pombe to maximize single-transposon insertions
To use transposon excision to investigate DNA repair, cells must bear only a single transposon; therefore, we developed an approach to efficiently generate single genomic insertions. We adapted a system in which one plasmid expresses the transposase while another contains a transposon bearing a selectable marker for G418 resistance (Evertts et al. 2007). Induction of the transposase allows the transposon to insert into the genome, and growing cells on medium with G418 can select for cells with transposon integrants. This system was efficient at generating transposon insertions, often several per cell, and has been successfully used to map S. pombe genes required for growth (Guo et al. 2013).
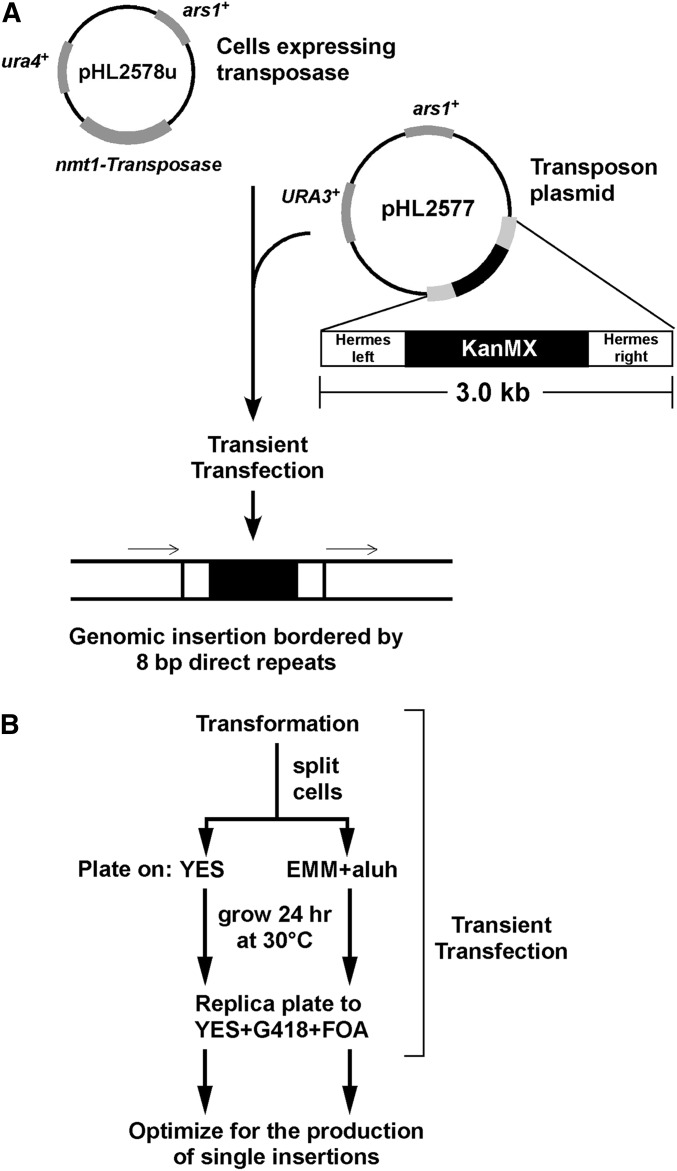
To maximize the fraction of cells with single insertions, the plasmids containing the transposon and the transposase expression cassettes were only transiently maintained prior to selection for genomic insertions, which limits the opportunity for multiple insertion events. To achieve transient expression, the plasmids (Evertts et al. 2007) were altered so that both contained markers (URA3+ and ura4+) that could be selected against with the same drug (5’-FOA, Figure 1A). S. pombe cells bearing the transposase expression plasmid were grown under inducing conditions to produce a population of cells with transposase enzyme, and then transformed with transposon plasmid. Cells were plated onto nonselective medium, allowed to grow for 24 hr, and then replica plated to medium that selects for the transposon insertion and against both plasmids (Figure 1B). This 24 hr of growth and cell division allowed cells bearing transposon insertions to lose the mitotically unstable transposon and transposase plasmids. Consequently, selection for the genomic transposon insertions and against the plasmids selected for those cells with one or more transposon insertions and no plasmids.
Figure 1.
A transient transfection assay to generate mutants with single-transposon insertions. (A) Cells grown under selection for the pHL2578u plasmid expressing the Hermes transposase from the nmt1 promoter under inducing conditions. The pHL2577 transposon donor plasmid contains the ends of the Hermes transposon flanking a kanamycin selectable marker (KanMX) that confers resistance to G418. Introducing the transposon plasmid into cells expressing the transposase allows insertion of the transposon into the genome, generating a target site duplication of 8 bp (indicated by thin arrows). (B) Cells expressing transposase were transformed with different amounts of the transposon plasmid. To monitor whether continuous expression of transposase impacted transposition, half of the transformed cells were plated onto either noninducing (YES) or inducing (EMM) nonselective media. The EMM plates contained supplements to complement cellular auxotrophies (Moreno et al. 1991). After 24 hr of growth to allow plasmid loss, the lawn of cells was replica plated onto medium that selects for the transposon (G418-resistance) and against the ura4+ and URA3+ genes (FOA) on the two plasmids. The yield of colonies bearing transposon insertions is shown in Table 1. aluh, adenine, leucine, uracil, and histidine; FOA, 5’-fluoroorotic acid; YES, yeast extract sucrose.
The growth on nonselective medium following transformation was performed using conditions where transposase was, or was not, continuously induced (Figure 1B). This approach was used because the production of cells with single-transposon insertions might require continual exposure to transposase or, alternatively, continual transposase exposure might produce multiple genomic insertions. Analysis of the number of potential transposon insertions revealed that growth for 24 hr on medium that induced the transposase produced ∼10 times more G418-resistant colonies than growth on noninducing medium (Table 1). The effect of different amounts of transposon plasmid was also tested, which showed that the number of transformants per microgram decreased as the amount of transposon DNA increased (Table 1). Thus, the transient transfection approach could produce significant numbers of cells with transposon insertions.
Table 1. Yield of transposon insertions from the different transient transfection permutations in Figure 1.
| Medium | Transforming DNA (μg) | G418R FOAR Transformantsa | |
|---|---|---|---|
| Total | #/μg | ||
| YES | 0.3 | 20 | 67 |
| 1.0 | 66 | 66 | |
| 3.0 | 95 | 32 | |
| EMM + aluh | 0.3 | 325 | 1083 |
| 1.0 | 690 | 690 | |
| 3.0 | 804 | 268 | |
FOA, 5’-fluoroorotic acid; YES, yeast extract sucrose; aluh, adenine, leucine, uracil, and histidine.
Number of colonies on the final YES + G418 + FOA plates following growth on nonselective medium in which transposase is (EMM) or is not (YES) induced.
To determine if the continuous induction of transposase produced cells containing only one transposon insertion, the transposon locations in the genome of 12 G418-resistant strains bearing insertions from two different transformations were mapped by inverse PCR (Materials and Methods) (Ochman et al. 1988). Ten of these strains had single insertions (Figure S1 in File S1). The remaining two strains had no genomic insertions but retained the transposon plasmid, possibly by mutating the URA3 negative selectable marker on the plasmid. All 10 genomic insertions showed the 8-bp target site duplication bordering the insertion site, as expected (Bhasin et al. 1999; Zhou et al. 2004). These data indicate that the transient transfection approach can efficiently produce single-transposon insertions. Two independent strains (KRP3-3 and KRP3-4) (Figure S1 in File S1) were subsequently used to examine Hermes excision and repair of the excision site.
Transposon excision footprints indicate repair by NHEJ
The Hermes transposase that catalyzes transposon insertion also mediates excision (Bhasin et al. 1999; Zhou et al. 2004), generating a DSB whose repair relies on cellular factors. Transposon excision has not been tested in S. pombe, so the nature of transposon footprints left by hAT transposon excision is unknown. An important consideration in S. pombe is that haploid cells spend most of the cell cycle in G2 and can repair DSBs by HR using the undamaged sister chromatid as a template (Ferreira and Cooper 2004). This repair will recreate the original insertion if Hermes excises from only one of the two chromosomes, because repair of the hairpin-capped DSB will copy the other chromosome that retains the Hermes transposon. Consequently, permanent removal of the transposon is expected to result only from NHEJ and MMEJ events (Figure 2A). Therefore, we expressed transposase in cells containing a transposon to determine whether the level of end-joining is sufficient to allow removal of the transposon.
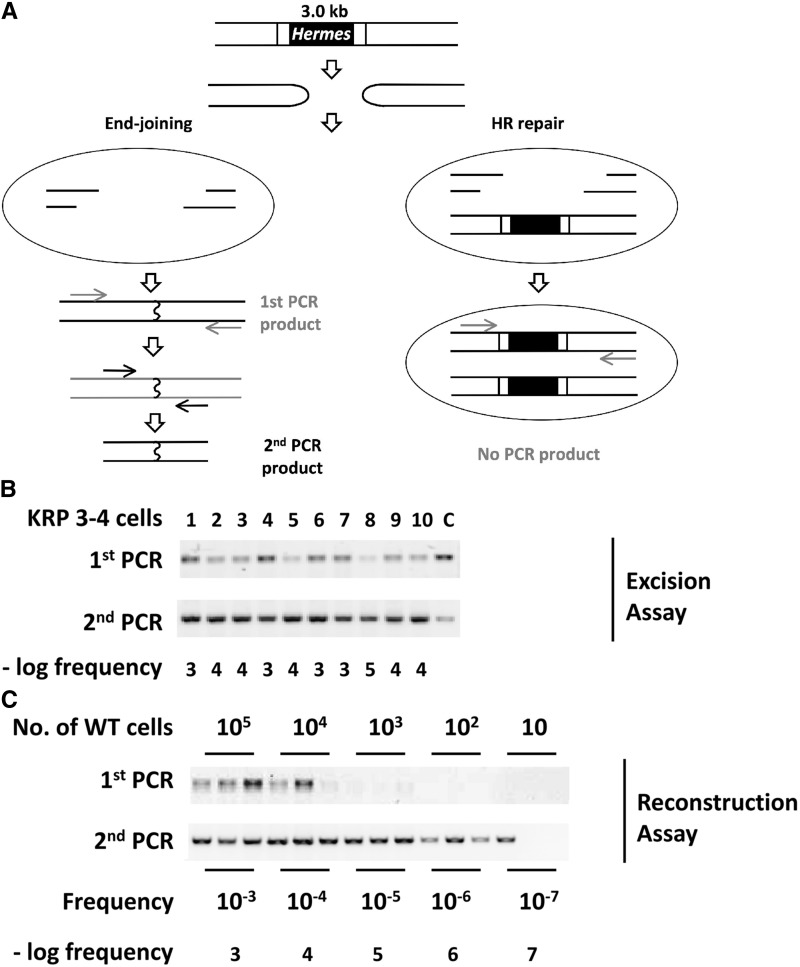
Figure 2.
Removal of a Hermes transposon requires end-joining repair. (A) Transposon excision produces hairpin-capped DSB ends that must be repaired, or cells will lose essential genetic information during mitosis and die. Repair by end-joining (NHEJ or MMEJ, left panel) involves nuclease activities and ligation of the broken ends, which can be identified by PCR. Rare events can be detected by a second PCR with a set of nested primers. Repair by HR in G2 cells that retain an unexcised transposon (right panel) will regenerate the Hermes insertion. This product will not be detected in this assay due to the use of short extension times that do not allow amplification of the Hermes insertion. The arrows indicate primers for the PCR reactions. (B) KRP 3-4 cells containing a single-transposon insertion (Figure S1 in File S1) were transformed with the transposase expression plasmid to generate colonies from single cells. The individual colonies were grown to 5 × 107 cells under conditions that induced transposase expression, and DNA was prepared and used in two rounds of PCR. Excision frequencies were determined by comparison to the standard curve shown in the next panel. The lanes labeled “C” are loaded with various amounts of a marker that show the size of the expected fragment. (C) A reconstruction test was performed to estimate the frequency of Hermes excision. The indicated number of WT, which lack a Hermes insertion, were mixed with 5 × 107 KRP 3-4 cells bearing a transposon and used to prepare DNA. PCR to detect only the excision products (as in A) was performed in triplicate for each sample. Quantitation and comparison of the first and second PCR products indicated that excision frequencies differing by 10-fold could be distinguished over the range of 10−4 to 10−6 per cell. DSB, double-strand breaks; HR, homologous recombination; MMEJ, microhomology-mediated end-joining; NHEJ, nonhomologous end-joining; wild-type, WT.
We designed a serial PCR assay to detect excision events over a wide range of frequencies. Primers were chosen at a sufficient distance from the insertion site based on the known size range of deletions generated by excision of other hAT transposons that also excise to leave hairpin-capped DSBs (i.e., S. cerevisiae, Drosophila, and Maize) (Baran et al. 1992; Scott et al. 1996; Colot et al. 1998; Yu et al. 2004)). PCR extension times were then chosen that would amplify the excision site but not the larger Hermes insertion (Figure 2A). The products of the first PCR were used in a second PCR with a set of nested primers to quantitate very rare events and to determine the frequency of transposon excision. PCR products detected in this assay (Figure 2C) indicated repair by end-joining.
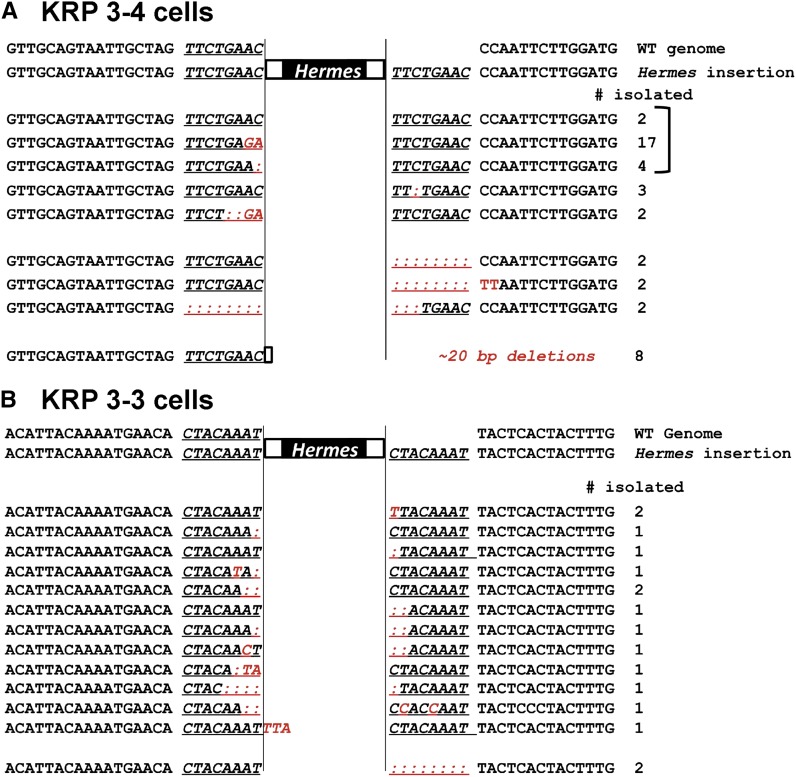
The products of the first round of PCR from individual colonies (Figure 2B) were cloned and sequenced to examine the repair of the DSB induced by Hermes excision. The majority of recovered excision events (42/58) had sequences that strongly indicated NHEJ: 0–5 bp of the flanking sequences were deleted, and the mutations were confined to sequences near the point of end-joining (Figure 3, A and B). As the same types of events were observed for the excision of two different transposons, this repair was independent of the neighboring DNA sequences. These results are inconsistent with MMEJ, which generates larger deletions (Daley et al. 2005; McVey and Lee 2008), and are quite different from excision in S. cerevisiae, where the majority of hAT transposon excision sites showed larger deletions (8–22 bp) (Yu et al. 2004). The remaining S. pombe excision events included two revertants where one of the 8-bp target site duplications had been removed, and a small number of excision events that retained a portion of the Hermes transposon accompanied by a deletion of chromosomal sequences. While the origin of these events is not due to NHEJ and is unknown, these types of products were not amplified from cells that did not express transposase (Figure S2A in File S1), indicating its requirement in generating these unusual products.
Figure 3.
Hermes excision footprint sequences indicate repair by NHEJ. Hermes excision events were monitored by cloning and sequencing the first-round PCR products from individual colonies in which transposon excision was induced. Cells bearing one of two independent transposon insertions were analyzed: KRP 3-4 cells (A) and KRP 3-3 cells (B). The PCR products for the KRP 3-4 cells are those shown in Figure 2B. The underlined sequence is the 8-bp duplication generated during transposon insertion. Base changes are indicated in red, and deletions are indicated by a colon. Most events show small deletions (0–5 nucleotides) and mutations that implicate NHEJ (Daley et al. 2005; McVey and Lee 2008). A bracket indicates the sequences used to model the mechanism of repair (Figure S4 in File S1). NHEJ, nonhomologous end-joining.
Transposon excision is rare with a large range of frequencies
The frequency of Hermes transposon excision in S. pombe is not known; therefore, we tested whether it could be determined quantitatively for the purpose of comparing wild-type and mutant strains. We first performed a reconstruction test. Different amounts of wild-type cells that lack a Hermes insertion, and can therefore give a PCR product, were mixed with cells that have an insertion that will not give a product. DNAs prepared from these mixtures were then tested by PCR. The results showed that the fraction of cells where the transposon had excised, i.e., excision frequency, could be monitored over a 104-fold range (Figure 2C). By comparing the levels of both the first- and second-PCR products, frequencies that differed by 10-fold were distinguishable in the range of 10−4 to 10−6 events/cell. This assay was then applied to the analysis of 10 independent colonies in which excision had been induced. Excision frequency varied over an ∼100-fold range, from one event in 103 to one in 105 cells (Figure 2B). The total frequency of chromosome breakage was estimated by Southern blotting of transposase-expressing cells, and was found to be ∼0.9% in wild-type cells (Figure S3 in File S1), suggesting that ∼1 in 10 to 1 in 1000 breakage events result in excision repair by NHEJ.
These data indicated the need to use fluctuation analysis to quantitate excision. Thus, the level and variation in excision frequency can be explained by the events occurring at low frequency and at random: transposon excision at early times during the growth of a culture produces a large number of cells lacking the transposon and a higher frequency in the assay, while excision in the last few population doublings before the assay is performed results in a lower frequency of cells with an excision event. The wide range in wild-type cells, and even larger range in mutant strains (described in the next section), indicated that a comparison of average excision frequencies would not be suitable because the few cultures with larger frequencies skew the average away from the majority of cultures with smaller frequencies. For example, with 10 samples giving one result of 10−3 and nine of 10−6, the most representative and frequently observed value is 10−6, but the average is 10−4, 100-fold higher. Therefore, excision frequency was determined using the “method of the median” from fluctuation analysis (Luria and Delbruck 1943; Lea and Coulson 1949). In this method, the frequencies of 10 independent cultures are obtained and ordered from highest to lowest. The median value, i.e., the average of the fifth and sixth frequencies, provides a representative frequency for a strain. This method is validated by the fact that many of the cultures have frequencies near the median, indicating that the event being measured occurs similarly in many independent cultures (Lea and Coulson 1949; Hartwell and Smith 1985; Surosky et al. 1986; Runge et al. 1991; Dershowitz and Newlon 1993; Runge and Zakian 1993; Ray et al. 2003; Dershowitz et al. 2007). Therefore, we used the median frequency of Hermes excision for each strain tested in this work. The wild-type KRP 3-4 cells showed a median excision frequency that ranged from 10−4 events/cell to 3 × 10−4 events/cell in a total of four independent assays (not shown), indicating that the median excision frequency was reproducible. These events depended upon transposase, as their frequency in the absence of this enzyme were >1000-fold less (Figure S2A in File S1).
NHEJ repair of excision events requires Ku, DNA ligase 4, and MRN
The NHEJ pathway depends upon Ku and DNA ligase 4 activities; therefore, we tested whether these factors are required for repair of Hermes excision events. Transposon excision generates hairpin ends (Figure 2A), and MRN was therefore also investigated because genetic and biochemical data implicate this complex in processing this structure (Paull and Gellert 1999; Lobachev et al. 2002; Lengsfeld et al. 2007), as well as in the processing of DNA adducts and other DNA secondary structures (Lobachev et al. 2002, 2007; Quennet et al. 2011). Mre11 can tether separated DNA ends together and facilitate base pairing of 5′ single-stranded overhangs for end-joining (Bressan et al. 1998; Lewis et al. 2004; Krogh et al. 2005; Williams et al. 2007, 2008). These properties of MRN could explain the most frequently observed KRP 3-4 transposon footprints (Figure 3A and Figure S4 in File S1).
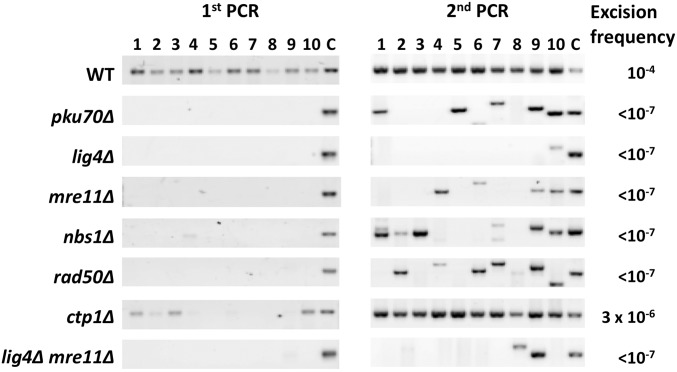
Hermes excision frequency in cells lacking a subunit of Ku (pku70∆) or DNA ligase 4 (lig4∆) was greatly reduced (Figure 4). Most of the products in the second PCR were aberrant, as indicated by the incorrect size and by sequence analysis (Figure S5 in File S1). The results from pku70∆ and lig4∆ cultures indicated that the frequency of correct excision was 10−7 events/cell or less (Figure 5). Thus, efficient repair of Hermes excision events occurs by the Ku-dependent NHEJ pathway. The aberrant products were not due to normal NHEJ, as many of these products contained portions of the Hermes transposon and were accompanied by large deletions (Figure S5 in File S1). These products were not observed in pku70∆ cells in the absence of transposase (Figure S2B in File S1), indicating that their generation requires transposase activity.
Figure 4.
Efficient NHEJ requires Ku70, DNA ligase 4, and MRN. KRP 3-4 cells bearing no mutation (WT) or the indicated mutations were tested in the excision assay. Ten individual colonies for each strain were tested by two rounds of PCR. Lane “C” contains various amounts of a PCR product from wild-type DNA loaded as a size marker for the expected NHEJ fragment. The WT gels are the same as those shown in Figure 2B. The median excision frequencies and footprint sequences are shown in Figure 5 and Figure S5 in File S1, respectively. MRN, Mre11-Rad50-Nbs1; NHEJ, nonhomologous end-joining; wild-type, WT.
Figure 5.
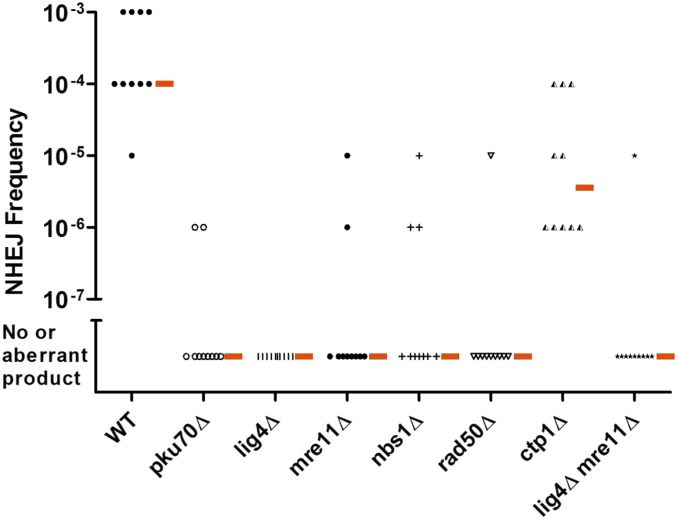
Hermes excision frequencies of different NHEJ mutants. Each data point represents the excision frequency of a single colony from the WT or mutant cells analyzed by PCR (Figure 4). The short red bars indicate the median excision frequency for the strain, which is obtained by ranking the frequencies from highest to lowest and averaging the fifth and sixth values. The differences between the WT and all strains were statistically significant (P < 0.001 for all strains by Mann–Whitney two-tailed rank sum test except for WT vs. ctp1∆, which was P = 0.0021). NHEJ, nonhomologous end-joining; wild-type, WT.
The results from cells lacking each of the MRN subunits were very similar to those in cells lacking Pku70. Products were not detected in the first PCR, and the second PCR resulted in either no product or ones that were mostly of aberrant size (Figure 4). Sequencing of products from the second PCR of the rad50∆ mutants revealed aberrant products that were similar to those identified in pku70∆ cells (Figure S5 in File S1). The frequency of correct excision in cells lacking the individual MRN subunits was 10−7 events/cell or less, similar to cells lacking Ku or DNA ligase 4 (Figure 5). A double mutant lacking DNA ligase 4 and Mre11 gave results very similar to the single mutant lacking DNA ligase 4 (Figure 4 and Figure 5), consistent with Mre11 and DNA ligase 4 acting in the same pathway. MRN is therefore required for efficient NHEJ repair of Hermes excision events in S. pombe, which contrasts with the results from end-joining assays with cut plasmids where the MRN complex is dispensable (Manolis et al. 2001).
The Mre11 dimerization domain is required for efficient NHEJ
Structural studies indicate that MRN complexes can tether separated DNA ends together, which is thought to pair chromosomal sequences for HR or MMEJ repair (Bressan et al. 1998; Lewis et al. 2004; Krogh et al. 2005; Williams et al. 2007, 2008). Mre11 is a dimer, which is thought to facilitate this process because mutations in its dimerization domain inhibit repair (Williams et al. 2008). Structural analysis of the archeal Mre11 revealed a key pair of leucine residues (Leu77 and Leu154 in S. pombe Mre11) at the dimer interface. Conversion of either of these hydrophobic residues to charged residues disrupted dimerization in two-hybrid assays and inhibited repair when placed into S. pombe Mre11 (Williams et al. 2008). Substitution of Leu77 had more of an effect on repair than that of Leu154 while maintaining interactions with Rad50 and Nbs1, and we therefore tested whether Leu77Lys affected NHEJ. We also tested the Leu77Lys Leu154Asp mutant, because simultaneous mutation of both Leu77 and Leu154 caused even greater inhibition of DNA repair and weakened interactions with Rad50 and Nbs1 (Williams et al. 2008).
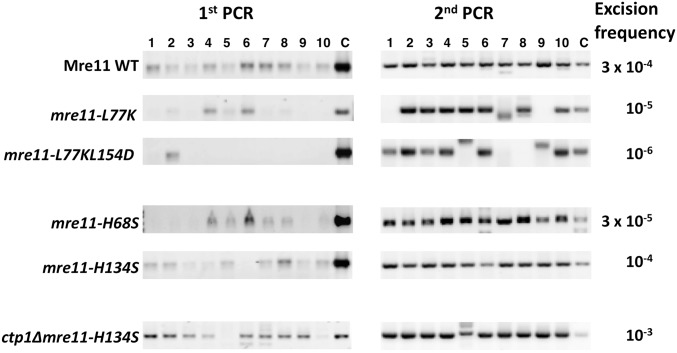
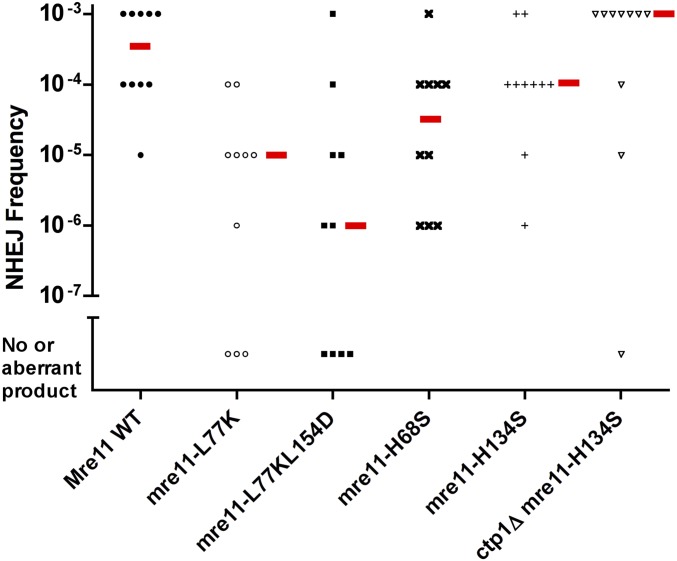
The Hermes insertion from KRP 3-4 cells was introduced into the wild-type, mre11-L77K, and mre11-L77K L154D strains, and excision frequency was monitored. The wild-type cells showed a median excision frequency similar to the original KRP 3-4 strain (i.e., between 10−3 and 10−4; Figure 6 and Figure 7). In contrast, both the mre11-L77K single mutant and mre11-L77K L154D double mutant showed reduced excision frequencies. The first PCR revealed fewer products for the single mutant and almost no products for the double mutant (Figure 6). In the second PCR, several cultures from both mutants had either no excision products or aberrant products. The median excision frequency for the mre11-L77K mutant was ∼30-fold lower than the wild-type strain, while the frequency for the double mutant was ∼300-fold lower (Figure 7). An intact dimerization domain is therefore required for efficient NHEJ.
Figure 6.
Mutations in the Mre11 dimerization domain, but not the nuclease domain, reduce NHEJ. Strains bearing the Hermes insertion and the indicated mre11 mutations were tested in the excision assay as in Figure 2 and Figure 4. The indicated median excision frequencies and excision footprint sequences are shown in Figure 7 and Figure S7 in File S1, respectively. NHEJ, nonhomologous end-joining; wild-type, WT.
Figure 7.
Hermes excision frequencies of different mre11 mutants. Each data point represents the excision frequency from a WT or mutant cell culture analyzed by PCR (Figure 6). The red bar shows the median frequency. The differences between mre11+ WT and mre11-L77K and mre11-L77KL174D were statistically significant (P < 0.001 and 0.002, respectively, by Mann–Whitney two-tailed rank sum test), the difference between mre11+ WT and mre11-H68S was borderline for significance in this nonparametric test (P = 0.023), while the differences between mre11+ and mre11-H134S or ctp1∆ mre11-H134S, and between mre11-H134S and ctp1∆ mre11-H134S, were not distinguishable (P = 0.218, 0.684, and 0.143, respectively). NHEJ, nonhomologous end-joining; wild-type, WT.
Mre11, Ctp1, and Pso2 nuclease functions are dispensable for efficient NHEJ
Nucleolytic processing of the hairpin produced by Hermes excision is required to produce a ligatable DNA end for NHEJ (Figure 2A). Mre11 in the MRN complex has both single-strand DNA endonuclease and 3′–5′ exonuclease in vitro activities, which has led to the hypothesis that, during HR, this complex plays a role in the repair of DNA hairpins (Lobachev et al. 2002; Paull 2010). The structure of the archeal Mre11-DNA complex revealed key His residues required for exonuclease activity or both nuclease activities (His68 and His134, respectively, in S. pombe Mre11) (Williams et al. 2008). Mutation of the archeal Mre11 His residue equivalent to S. pombe His68 to Ser ablated the in vitro exonuclease activity and the mutation equivalent to H134S abolished both the exo- and endonuclease activity, while both mutations maintained interactions with Rad50 and Nbs1 by two-hybrid analysis (Williams et al. 2008). S. pombe cells bearing the mre11-H134S allele deficient in both nuclease activities show increased sensitivity to DNA-damaging agents, while exonuclease-deficient mre11-H68S cells show the same sensitivity as wild-type cells (Williams et al. 2008). Therefore, we tested the effect of both nuclease-deficient alleles on NHEJ.
The excision frequency in the mre11-H68S and mre11-H134S cells was substantially higher than that of cells lacking Mre11, and only slightly less than that of wild-type cells. Products of the correct size were detected in the first round of PCR, and the excision frequencies were ∼10% or more of the wild-type strain level (Figure 6 and Figure 7). The sequences of the transposon footprints from these nuclease mutants were very similar to those from wild-type cells (Figure 3 and Figure S6 in File S1), indicating that repair occurred through NHEJ. Thus, the nuclease activities of Mre11 are largely dispensable for NHEJ repair of hairpin ends.
The results were surprising because they were strikingly different from the much larger increase in sensitivity to DNA-damaging agents seen in the S. pombe mre11-H134S mutant (Williams et al. 2008) (Figure S7 in File S1). The mre11-H134S cells that lack or have the Hermes insertion showed increased sensitivity to DNA damage, similar to that of cells with the dimerization domain mutations (mre11-L77K and mre11-L77K L154D). In contrast, the mre11-H134S cells showed a 100-fold higher level of NHEJ repair than that of the mre11-L77K L154D cells (Figure 6 and Figure 7). Endonuclease activity is required to open the hairpin, and the results with the mre11-H134S mutants that target this domain therefore indicate that another nuclease besides Mre11 must process the DNA hairpin. To date, the only MRN-associated factor implicated in hairpin cleavage is Ctp1, a homolog of mammalian CtIP and S. cerevisiae Sae2, both of which have nuclease activity in vitro (Lengsfeld et al. 2007; Limbo et al. 2007; Sartori et al. 2007; You et al. 2009; Makharashvili et al. 2014; Wang et al. 2014). Therefore, we tested whether Ctp1 is required for efficient NHEJ. The excision frequency in cells lacking Ctp1 was at least 30-fold higher than the MRN deletion mutants (Figure 4 and Figure 5). Sequencing of products from the first PCR of DNA from ctp1∆ cells revealed transposon footprints similar to those seen in wild-type cells (Figure S5 in File S1).
The small reduction in repair of the excision site in cells lacking Ctp1 compared to the other deletion mutants tested raised the question as to whether Ctp1 plays a redundant role in hairpin processing. Hermes excision in a ctp1∆ mre11-H134S mutant was therefore examined to test if cells lacking both activities would greatly reduce NHEJ. We found that the median excision frequency in the mre11 nuclease-deficient mutant lacking Ctp1 was nearly the same as in wild-type cells (Figure 7), with a clearly detectable product in the first PCR in 9 of the 10 cultures (Figure 6). Sequencing of the products from independent ctp1∆ mre11-H134S cultures showed that most of the products had small deletions and mutations (Figure S7 in File S1), similar to NHEJ repair in wild-type cells. A small number of footprints with mutations in sequences flanking the 8-bp direct repeats were also observed, revealing a more error-prone mechanism that also occurs in these cells. These results indicate that loss of the Mre11 nuclease activities and Ctp1 still allow normal NHEJ repair of Hermes excision events.
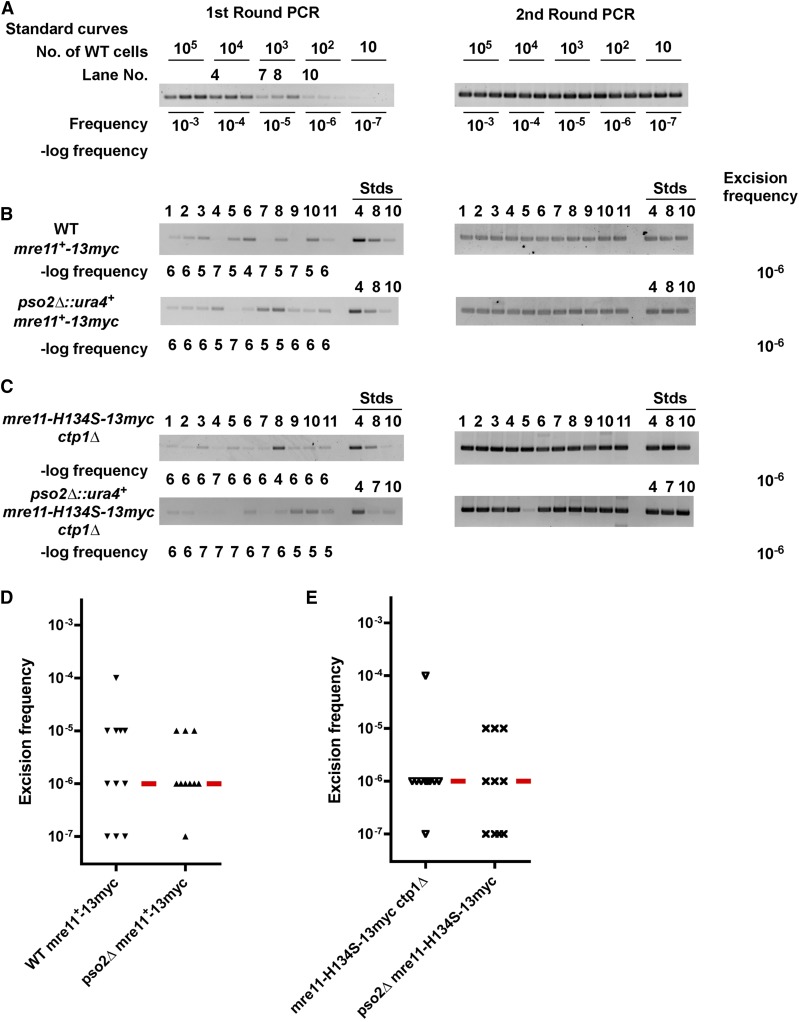
As neither Mre11 nor Ctp1 had the properties of the DNA hairpin opening activity, we tested the last known endonuclease candidate for DNA hairpin cleavage, Pso2. S. pombe Pso2 has strong sequence homology with an enzyme that can cleave covalently closed hairpin DNA substrates in vitro, S. cerevisiae Pso2 (Tiefenbach and Junop 2012). Therefore, we tested Hermes excision repair frequencies in wild-type, pso2∆, mre11-H134S ctp1∆, and mre11-H134S ctp1∆ pso2∆ cells. We found that the loss of Pso2 did not reduce excision frequencies (Figure 8). Therefore, these genetic results indicate that another process besides Mre11, Ctp1, or Pso2 opens the hairpin to allow NHEJ repair.
Figure 8.
The S. pombe homolog of in vitro hairpin nuclease Pso2 is not required for excision. S. cerevisiae Pso2 can cleave DNA hairpins in vitro (Tiefenbach and Junop 2012), suggesting that the S. pombe homolog may account for the excision healing activity in mre11 mutants that lack nuclease activity. Hermes excision was analyzed with a modified assay (see Materials and Methods) that analyzed 11 colonies. (A) A standard curve constructed using a culture of 2 × 107 cells that also contained the indicated number of wild-type cells to model the excision event as in Figure 2. The lane numbers indicate genomic DNA samples that were amplified in parallel with the colonies in (B) and (C). (B) Hermes excision frequencies in the epitope-tagged mre11-13myc strains are not affected by loss of Pso2 (P = 0.84 by Mann–Whitney test). “Stds” refers to samples from the standard curve in (A) that were amplified at the same time as each of the colonies shown. The numbers above the Stds lanes refer to the lane numbers shown in (A). (C) Excision frequencies in the epitope-tagged mre11-H134S-13myc ctp1∆ double mutant strains are not affected by loss of Pso2 (P = 0.69 by Mann–Whitney test). (D) Graphical representation of the excision frequencies in (B), where the red bar denotes the median excision frequency. (E) Graphical representation of the excision frequencies in (C). Lane No., lane number; wild-type, WT.
Discussion
The role of MRN in NHEJ is not clear, as this complex has been shown to be required for NHEJ in S. cerevisiae but not in S. pombe or mammalian cells when assayed for the repair of restriction enzyme-cut DNA (Manolis et al. 2001; Zhang and Paull 2005; Limbo et al. 2007; Rass et al. 2009; Xie et al. 2009; Li et al. 2012). Most DSBs arising from DNA damage require processing prior to ligation; therefore, we used transposon excision to study the full process of repair. We developed a novel transient transfection approach that efficiently produced Hermes transposon insertions at unique sites in the S. pombe genome, and showed that Hermes excision can be induced by transposase expression. The sequence of the excision sites combined with the requirement for Ku and DNA ligase 4 showed that these excision events occur by NHEJ (Figure 3, Figure 4, Figure 5, and Figure S5 in File S1) (Moore and Haber 1996; Daley et al. 2005; Raji and Hartsuiker 2006; McVey and Lee 2008; Chiruvella et al. 2013). However, in marked contrast to how S. pombe repairs DNA ends produced by restriction enzyme digestion (Manolis et al. 2001; Limbo et al. 2007; Li et al. 2012), this NHEJ process also required MRN. Loss of each MRN component reduced NHEJ to the same extent as loss of Ku and DNA ligase 4, and produced similar transposon footprints (Figure 4, Figure 5, and Figure S5 in File S1). A primary role for MRN is to hold DNA ends together (Bressan et al. 1998; Lewis et al. 2004; Krogh et al. 2005; Williams et al. 2007, 2008), and an intact Mre11 dimerization domain was required for repair (Figure 6 and Figure 7), indicating a role for this domain in S. pombe NHEJ. In contrast, the Mre11 nuclease functions were largely dispensable for NHEJ repair of the hairpin-capped DSB left by Hermes excision (Figure 6 and Figure 7) (Bhasin et al. 1999; Zhou et al. 2004). Hermes transposon excision in S. pombe will therefore be a valuable system for investigating the processing of DNA hairpin structures that can arise from palindromes, inverted repeats, and trinucleotide repeats, which threaten genomic stability and drive tumorigenesis in mammals (Lewis and Cote 2006; Tanaka and Yao 2009; Saini et al. 2013).
The requirement of MRN for repair of hairpins but not DNA cut by restriction enzymes (Manolis et al. 2001) indicates that MRN functions in events prior to DNA ligation, and is of interest regarding the known functions of Ku. The Ku heterodimer has the ability to bind and join the ends of restriction enzyme-cut DNA (Pang et al. 1997; Ramsden and Gellert 1998), which could allow end-joining when restriction enzyme-digested plasmids are transformed into cells (Boulton and Jackson 1996a,b; Manolis et al. 2001). Ku can also bind DNA hairpins in vitro (Arosio et al. 2002; Smider et al. 1998), and could similarly tether the hairpin-capped ends of the DSB created by Hermes excision. However, the requirement for MRN and the Mre11 dimerization domain in the NHEJ repair of Hermes excision events shows that this Ku activity is insufficient to allow the completion of end-joining in vivo. MRN-Ctp1 association with DNA breaks has been proposed to remove Ku to allow the recruitment of HR factors such as the DNA damage checkpoint kinase ATR (Langerak et al. 2011). We propose that MRN-Ctp1 also allows the recruitment of factors that facilitate NHEJ (Figure 9). One factor that needs to be recruited is the nuclease that opens the DNA hairpin to allow end ligation. Mre11 was the most obvious candidate for the end-opening activity, based on in vitro studies showing that S. cerevisiae MRX-Sae2 can cleave hairpins (Paull and Gellert 1999; Lobachev et al. 2002; Lengsfeld et al. 2007) and genetic evidence indicating that repair by HR is blocked by loss of individual MRX subunits and mre11 nuclease domain mutants (Paull and Gellert 1999; Lobachev et al. 2002; Lengsfeld et al. 2007). Based on these properties, Mre11 has been proposed to be the most likely hairpin opening activity (Lobachev et al. 2002; Dudasova et al. 2004; Yu et al. 2004; Lam et al. 2008). However, our data indicate that S. pombe Mre11 is not the hairpin-opening nuclease in NHEJ, because a mutant substituted in a residue (H134S) required for the endonuclease activity showed wild-type levels of excision and repair (Figure 6 and Figure 7). Ctp1, which associates with MRN, was also tested because it is a homolog of the nucleases Sae2 and CtIP (Paull and Gellert 1999; Lobachev et al. 2002; Lengsfeld et al. 2007; Makharashvili et al. 2014; Wang et al. 2014). While the ctp1∆ mutant showed a 30-fold decrease in excision that suggests a role in processing (Figure 4 and Figure 5), Ctp1 alone is not sufficient because the ctp1∆ mre11-H134S double mutant showed wild-type levels of excision (Figure 6 and Figure 7). We also examined the effect of removing the remaining hairpin-cleaving candidate enzyme, Pso2, but loss of this activity did not alter excision and repair (Figure 8). Consequently, Mre11, Ctp1, and Pso2 do not appear to be the major nucleases that open the hairpin for further processing.
Figure 9.
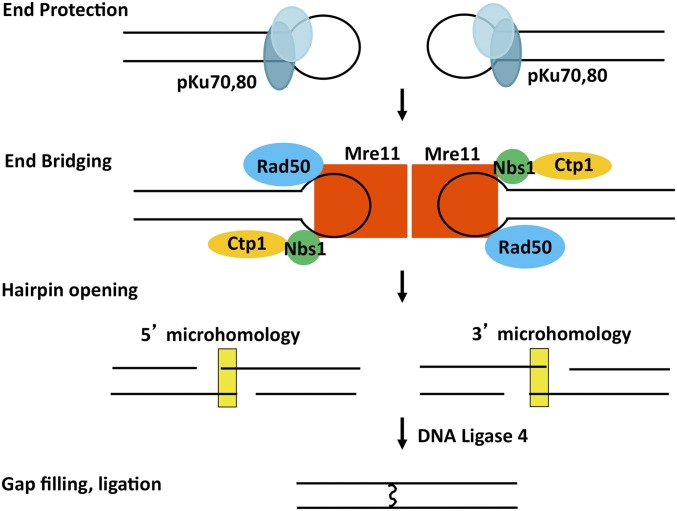
Model for the role of MRN-Ctp1 in NHEJ repair of Hermes excision sites. The initial hairpin-capped ends are bound by Ku, which has end protection functions that may prevent further processing (Ramsden and Gellert 1998; Smider et al. 1998; Arosio et al. 2002). MRN-Ctp1 is proposed to bind and replace Ku and allow synapsis to bridge the two DNA ends [as in Langerak et al. (2011) and Limbo et al. (2011)] and to recruit factors that open the hairpin. Subsequent base pairing at microhomologies (yellow box) may be potentiated by MRN or other factors prior to ligation. While MRN has a known role in strand resection (McVey and Lee 2008; Nicolette et al. 2010; Paull 2010), the most frequently observed end-joining events (Figure 3) indicated that little or no strand resection occurred to expose homologies. MRN, Mre11-Rad50-Nbs1; NHEJ, nonhomologous end-joining; wild-type, WT.
Our results showing that MRN-Ctp1 is not the hairpin-opening activity in NHEJ are surprising, but also reflect the fact that the in vivo role of this complex is not well-understood. The only relevant studies have been hAT transposon excision in S. cerevisiae, where different nuclease domain mutants gave opposing and therefore inconclusive results (Yu et al. 2004). In addition, the repair process being studied in S. cerevisiae is more like MMEJ than NHEJ (McVey and Lee 2008; Nicolette et al. 2010; Paull 2010). We found that transposon excision in S. pombe clearly occurs through NHEJ (Figure 3, Figure 4, and Figure 5). The observation that MRN deletion mutants, but not mre11 nuclease domain mutants, alter excision is therefore highly informative in showing that while MRN is required to process the hairpin, MRN-Ctp1 is not the hairpin-opening activity in NHEJ. The results are of interest in regard to the previous studies on the role of MRX in the HR-mediated repair of palindromes that can form hairpins. Loss of each MRX gene and mre11 nuclease mutants led to the accumulation of the hairpin intermediate (Paull and Gellert 1999; Lobachev et al. 2002; Lengsfeld et al. 2007), leading to the presumption that MRX cleaves hairpins (Dudasova et al. 2004; Haber 2006; Lam et al. 2008). However, those data did not distinguish whether MRX has a distinct role in cleavage vs. MRX recruiting or facilitating another hairpin-opening activity.
Currently, no other obvious candidates for the endonuclease that open the hairpin are known. Other nucleases that cleave unusual DNA structures are known, however their functions in other organisms have been implicated in more specialized types of repair (Froelich-Ammon et al. 1994; Panigrahi et al. 2005; Jonstrup et al. 2008; Hou et al. 2011; Tiefenbach and Junop 2012). Future work with the Hermes system in S. pombe could be valuable in revealing which factors resolve hairpins to repair nonligatable DSBs. Such physical analysis of DNA hairpin processing would first require improvements to increase the levels of DNA intermediates in vivo, as the level of transposase-mediated excision was relatively low (Figure 2 and Figure S3 in File S1). Potential approaches would be to increase the expression and nuclear localization of the transposase to allow the production of hairpins at much higher frequencies. Alternatively, a genetic screen for mutants that alter the frequency of Hermes excision repair may reveal factors that transiently associate with hairpin-opening factors, which would not be detectable by physical analysis [see Li (2015)].
MRN has been implicated in two other end-joining events in S. pombe, where the substrates of the reaction are more poorly defined. Decottignies (2005) described an assay where a ura4+ fragment transformed into cells spontaneously circularized to capture nonhomologous DNA, either from mitochondria or fragments cotransformed with the ura4+ DNA. Rad50 was not required for circularization but was required for the capture of exogenous DNA. Analysis of circularized plasmids revealed both large and small sequence loss, but Ku dependence was not tested. These events suggest a potential role for the MRN complex in the capture assay, which could occur by NHEJ or MMEJ.
Reis et al. (2012) studied the effect of MRN by extending the Decottignes assay, and also by studying the end-to-end joining of defective telomeres in G1-arrested cells that also lack the major double-stranded telomere-binding protein Taz1. Both events required Ku, indicating repair by NHEJ. A role for MRN and DNA ligase 4, but not Ctp1, was shown for both assays (Ferreira and Cooper 2001; Reis et al. 2012). Mutants in the Mre11 dimerization domain also blocked end-joining in both assays, similar to what we observed with transposon excision (Figure 6 and Figure 7). However, end-joining activity in both assays was defective in the mre11-H68S and mre11-H134S mutants (Reis et al. 2012), which strongly contrasts with our results showing that these two mutants were both clearly proficient in NHEJ (Figure 6 and Figure 7). These data indicate that the processing of DNA hairpins in vivo has distinct requirements compared to the events that occur during DNA transformation or the fusion of defective telomeres. Thus, the MRN complex performs related but distinct functions in these different NHEJ reactions, which can be differentiated by the mre11 nuclease-deficient alleles.
There are significant differences between the products of end-joining in S. pombe and S. cerevisiae arising from the excision of hAT transposons that leave hairpin ends. The majority of the transposon footprints we observed in S. pombe showed small deletions typical for an NHEJ reaction (Figure 3), while similar excision events in S. cerevisiae produced larger deletions more typical of MMEJ (Yu et al. 2004). This increased extent of deletion suggests that the amount of strand resection prior to end-joining is greater in S. cerevisiae compared to S. pombe. We note that the majority of NHEJ events in mammalian cells involving the ligation of DSBs created by in vivo restriction enzyme cutting also show only small amounts of DNA sequence loss (Rass et al. 2009), similar to our results in S. pombe. These considerations suggest that results on NHEJ processing in S. pombe may have broader implications for mammalian cells. For example, the repair of DNA adducts resulting from etoposide treatment of arrested human cells occurs by NHEJ in a process that requires MRN (Quennet et al. 2011), but the extent of sequence removal is unknown. An extrapolation of our S. pombe results suggests that these NHEJ events may occur with only small amounts of DNA sequence loss or mutation at each DSB, which would limit the extent of DNA damage and etoposide toxicity. Therefore, it may be possible to enhance etoposide toxicity in mammalian cells if one can devise a combination therapy that increases the level of strand resection during repair.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200972/-/DC1.
Acknowledgments
We thank Toru Nakamura and Paul Russell for S. pombe strains; Bianca Calderon, Matthew Shaughnessy, and Trihn To Tat for preliminary work and technical contributions to this project; and Julien Audry, Haitao Zhang, and Mohammed Al-Shibar for comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants RO1 GM-050752 and AG-019960 and National Science Foundation grant 1516220 to K.W.R., with funds from Case Western Reserve University-Cleveland Clinic Comprehensive Cancer Center to help purchase the Bioneer S. pombe deletion strain library. K.L.B. was supported by NIH R01 grants HL-55566 and HL-81093.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Arosio D., Cui S., Ortega C., Chovanec M., Di Marco S., et al. , 2002. Studies on the mode of Ku interaction with DNA. J. Biol. Chem. 277: 9741–9748. [DOI] [PubMed] [Google Scholar]
- Atkinson P. W., Warren W. D., O’Brochta D. A., 1993. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc. Natl. Acad. Sci. USA 90: 9693–9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wood V., 2004. The genome and beyond, pp. 13–25 in The Molecular Biology of Schizosaccharomyces Pombe, edited by Egel R. Springer-Verlag, Berlin. [Google Scholar]
- Baran G., Echt C., Bureau T., Wessler S., 1992. Molecular analysis of the maize wx-B3 allele indicates that precise excision of the transposable Ac element is rare. Genetics 130: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. F., Sikes M. L., Sullivan R., Huye L. E., Le Beau M. M., et al. , 2002. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 16: 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A., Goryshin I. Y., Reznikoff W. S., 1999. Hairpin formation in Tn5 transposition. J. Biol. Chem. 274: 37021–37029. [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1996a Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24: 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1996b Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double- strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15: 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan D. A., Olivares H. A., Nelms B. E., Petrini J. H., 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney J. P., Maser R. S., Olivares H., Davis E. M., Le Beau M., et al. , 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486. [DOI] [PubMed] [Google Scholar]
- Chen B.-R., Hale D. C., Ciolek P. J., Runge K. W., 2012. Generation and analysis of a barcode-tagged insertion mutant library in the fission yeast Schizosaccharomyces pombe. BMC Genomics 13: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.-R., Li Y., Eisenstatt J. R., Runge K. W., 2013. Identification of a lifespan extending mutation in the Schizosaccharomyces pombe cyclin gene clg1+ by direct selection of long-lived mutants. PLoS One 8: e69084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Trujillo K. M., Van Komen S., Roh D. H., Krejci L., et al. , 2005. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J. Biol. Chem. 280: 2620–2627. [DOI] [PubMed] [Google Scholar]
- Chiruvella K. K., Liang Z., Wilson T. E., 2013. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot V., Haedens V., Rossignol J. L., 1998. Extensive, nonrandom diversity of excision footprints generated by Ds-like transposon Ascot-1 suggests new parallels with V(D)J recombination. Mol. Cell. Biol. 18: 4337–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D., Wilson T. E., 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Davis A. J., Chen B. P., Chen D. J., 2014. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst.) 17: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A., 2005. Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics 171: 1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A., Newlon C. S., 1993. The effect on chromosome stability of deleting replication origins. Mol. Cell. Biol. 13: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A., Snyder M., Sbia M., Skurnick J. H., Ong L. Y., et al. , 2007. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol. Cell. Biol. 27: 4652–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudasova Z., Dudas A., Chovanec M., 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28: 581–601. [DOI] [PubMed] [Google Scholar]
- Evertts A. G., Plymire C., Craig N. L., Levin H. L., 2007. The hermes transposon of Musca domestica is an efficient tool for the mutagenesis of Schizosaccharomyces pombe. Genetics 177: 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P., 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611. [DOI] [PubMed] [Google Scholar]
- Featherstone C., Jackson S. P., 1999. DNA double-strand break repair. Curr. Biol. 9: R759–R761. [DOI] [PubMed] [Google Scholar]
- Ferreira M. G., Cooper J. P., 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7: 55–63. [DOI] [PubMed] [Google Scholar]
- Ferreira M. G., Cooper J. P., 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M., Marcand S., 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Froelich-Ammon S. J., Gale K. C., Osheroff N., 1994. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 269: 7719–7725. [PubMed] [Google Scholar]
- Guo Y., Park J. M., Cui B., Humes E., Gangadharan S., et al. , 2013. Integration profiling of gene function with dense maps of transposon integration. Genetics 195: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2006. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst.) 5: 998–1009. [DOI] [PubMed] [Google Scholar]
- Harfst E., Cooper S., Neubauer S., Distel L., Grawunder U., 2000. Normal V(D)J recombination in cells from patients with Nijmegen breakage syndrome. Mol. Immunol. 37: 915–929. [DOI] [PubMed] [Google Scholar]
- Harper J. W., Elledge S. J., 2007. The DNA damage response: ten years after. Mol. Cell 28: 739–745. [DOI] [PubMed] [Google Scholar]
- Harrison J. C., Haber J. E., 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D., 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Zhang T., Tian L., Huang J., Gu L., et al. , 2011. The role of XPG in processing (CAG)n/(CTG)n DNA hairpins. Cell Biosci. 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., Cooper J. P., 2010. Telomeric strategies: means to an end. Annu. Rev. Genet. 44: 243–269. [DOI] [PubMed] [Google Scholar]
- Jonstrup A. T., Thomsen T., Wang Y., Knudsen B. R., Koch J., et al. , 2008. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIα. Nucleic Acids Res. 36: 6165–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., et al. , 2006. Genetic variation affects de novo translocation frequency. Science 311: 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B. O., Llorente B., Lam A., Symington L. S., 2005. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B. S., 2001a Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 10: 2605–2617. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B. S., 2001b Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 29: 139–140. [DOI] [PubMed] [Google Scholar]
- Lam A. F., Krogh B. O., Symington L. S., 2008. Unique and overlapping functions of the Exo1, Mre11 and Pso2 nucleases in DNA repair. DNA Repair (Amst.) 7: 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche B. J., Orazio N. I., Weitzman M. D., 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584: 3682–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P., Mejia-Ramirez E., Limbo O., Russell P., 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–284. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T., 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554. [DOI] [PubMed] [Google Scholar]
- Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T., 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. K., Storici F., Van Komen S., Calero S., Sung P., et al. , 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166: 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M., Cote A. G., 2006. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair (Amst.) 5: 1146–1160. [DOI] [PubMed] [Google Scholar]
- Li P., Li J., Li M., Dou K., Zhang M. J., et al. , 2012. Multiple end joining mechanisms repair a chromosomal DNA break in fission yeast. DNA Repair (Amst.) 11: 120–130. [DOI] [PubMed] [Google Scholar]
- Li, Y., 2015 Construction and analysis of a genome-wide insertion library in Schizosaccharomyces pombe reveals novel aspects of DNA repair. Ph.D. Thesis, Case Western Reserve University, Cleveland, OH.
- Lieber M. R., Ma Y., Pannicke U., Schwarz K., 2004. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst.) 3: 817–826. [DOI] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., De Bruin R. A., Wittenberg C., et al. , 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O., Porter-Goff M. E., Rhind N., Russell P., 2011. Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Mol. Cell. Biol. 31: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Gordenin D. A., Resnick M. A., 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Lobachev K. S., Rattray A., Narayanan V., 2007. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front. Biosci. 12: 4208–4220. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbruck M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N., Tubbs A. T., Yang S. H., Wang H., Barton O., et al. , 2014. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54: 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolis K. G., Nimmo E. R., Hartsuiker E., Carr A. M., Jeggo P. A., et al. , 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S. E., 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K., Shore D., 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol. 9: 1123–1126. [DOI] [PubMed] [Google Scholar]
- Moore J. K., Haber J. E., 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular biology of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette M. L., Lee K., Guo Z., Rani M., Chow J. M., et al. , 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17: 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L., 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang D., Yoo S., Dynan W. S., Jung M., Dritschilo A., 1997. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 57: 1412–1415. [PubMed] [Google Scholar]
- Panigrahi G. B., Lau R., Montgomery S. E., Leonard M. R., Pearson C. E., 2005. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 12: 654–662. [DOI] [PubMed] [Google Scholar]
- Paull T. T., 2010. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst.) 9: 1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Gellert M., 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quennet V., Beucher A., Barton O., Takeda S., Lobrich M., 2011. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 39: 2144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji H., Hartsuiker E., 2006. Double-strand break repair and homologous recombination in Schizosaccharomyces pombe. Yeast 23: 963–976. [DOI] [PubMed] [Google Scholar]
- Ramsden D. A., Gellert M., 1998. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 17: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., et al. , 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16: 819–824. [DOI] [PubMed] [Google Scholar]
- Ray A., Runge K. W., 1999. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol. 19: 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Hector R. E., Roy N., Song J. H., Berkner K. L., et al. , 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33: 522–526. [DOI] [PubMed] [Google Scholar]
- Reis C. C., Batista S., Ferreira M. G., 2012. The fission yeast MRN complex tethers dysfunctional telomeres for NHEJ repair. EMBO J. 31: 4576–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge K. W., Zakian V. A., 1993. The isolation and characterization of mutants that preferentially lose linear, but not circular, chromosomes. Chromosoma 102: 207–217. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Wellinger R. J., Zakian V. A., 1991. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol. Cell. Biol. 11: 2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik A., Grenon M., Lowndes N., 2008. The MRN complex. Curr. Biol. 18: R455–R457. [DOI] [PubMed] [Google Scholar]
- Saini N., Zhang Y., Usdin K., Lobachev K. S., 2013. When secondary comes first–the importance of non-canonical DNA structures. Biochimie 95: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., et al. , 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., Lafoe D., Weil C. F., 1996. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., Nickoloff J. A., 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147. [DOI] [PubMed] [Google Scholar]
- Smider V., Rathmell W. K., Brown G., Lewis S., Chu G., 1998. Failure of hairpin-ended and nicked DNA to activate DNA-dependent protein kinase: implications for V(D)J recombination. Mol. Cell. Biol. 18: 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E., 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S., Maser R. S., Stankovic T., Bressan D. A., Kaplan M. I., et al. , 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587. [DOI] [PubMed] [Google Scholar]
- Sundararajan R., Gellon L., Zunder R. M., Freudenreich C. H., 2010. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics 184: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R. T., Newlon C. S., Tye B. K., 1986. The mitotic stability of deletion derivatives of chromosome III in yeast. Proc. Natl. Acad. Sci. USA 83: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Gautier J., 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Yao M. C., 2009. Palindromic gene amplification–an evolutionarily conserved role for DNA inverted repeats in the genome. Nat. Rev. Cancer 9: 216–224. [DOI] [PubMed] [Google Scholar]
- Theunissen J. W., Kaplan M. I., Hunt P. A., Williams B. R., Ferguson D. O., et al. , 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell 12: 1511–1523. [DOI] [PubMed] [Google Scholar]
- Tiefenbach T., Junop M., 2012. Pso2 (SNM1) is a DNA structure-specific endonuclease. Nucleic Acids Res. 40: 2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K. M., Sung P., 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276: 35458–35464. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato J., Ikeda H., 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903. [DOI] [PubMed] [Google Scholar]
- Viscardi V., Bonetti D., Cartagena-Lirola H., Lucchini G., Longhese M. P., 2007. MRX-dependent DNA damage response to short telomeres. Mol. Biol. Cell 18: 3047–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. R., Corpina R. A., Goldberg J., 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614. [DOI] [PubMed] [Google Scholar]
- Wang H., Li Y., Truong L. N., Shi L. Z., Hwang P. Y., et al. , 2014. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell 54: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C. F., 2009. Too many ends: aberrant transposition. Genes Dev. 23: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Weinert T., 1998. DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev. 8: 185–193. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Williams J. S., Tainer J. A., 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85: 509–520. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., et al. , 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele H., Little K. C., Chartrand P., 2003. Illegitimate DNA integration in mammalian cells. Gene Ther. 10: 1791–1799. [DOI] [PubMed] [Google Scholar]
- Xie A., Kwok A., Scully R., 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Chahwan C., Bailis J., Hunter T., Russell P., 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25: 5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Shi L. Z., Zhu Q., Wu P., Zhang Y. W., et al. , 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell 36: 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Marshall K., Yamaguchi M., Haber J. E., Weil C. F., 2004. Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Paull T. T., 2005. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst.) 4: 1281–1294. [DOI] [PubMed] [Google Scholar]
- Zhou L., Mitra R., Atkinson P. W., Hickman A. B., Dyda F., et al. , 2004. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432: 995–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and DNA constructs described here are available upon request to the corresponding author. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.