Abstract
Background
In drug-drug interactions, there are surprising cases in which the growth inhibition of bacteria by a single antibiotic decreases when a second antibiotic is added. These interactions are termed suppressive and have been argued to have the potential to limit the evolution of resistance. Nevertheless, little attention has been given to suppressive interactions because clinical studies typically search for increases in killing efficiency and because suppressive interactions are believed to be rare based on pairwise studies.
Results
Here, we quantify the effects of single-, double-, and triple-drug combinations from a set of 14 antibiotics and 3 bacteria strains, totaling 364 unique three-drug combinations per bacteria strain. We find that increasing the number of drugs can increase the prevalence of suppressive interactions: 17% of three-drug combinations are suppressive compared to 5% of two-drug combinations in this study. Most cases of suppression we find (97%) are “hidden” cases for which the triple-drug bacterial growth is less than the single-drug treatments but exceeds that of a pairwise combination.
Conclusions
We find a surprising number of suppressive interactions in higher-order drug combinations. Without examining lower-order (pairwise) bacterial growth, emergent suppressive effects would be missed, potentially affecting our understanding of evolution of resistance and treatment strategies for resistant pathogens. These findings suggest that careful examination of the full factorial of drug combinations is needed to uncover suppressive interactions in higher-order combinations.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-1017-3) contains supplementary material, which is available to authorized users.
Keywords: Higher-order interactions, Antagonism, Multiple stressors, Antibiotics, Drug resistance
Background
Antibiotic resistance is a critical public health issue [1]. With few new antibiotics in development [2] combination therapies [3, 4] have been used to overcome the problem of drug-resistant bacteria. However, it can be difficult to determine which combinations are the most rational choices because several factors must be carefully balanced in order to identify exceptional therapeutic combinations. Efficiency of killing bacteria and toxicity to the patient are critical, but just as important, yet harder to measure and seldom considered, is the likelihood to facilitate the evolution of resistance to the drugs. While there are several factors that affect the likelihood and rate of resistance evolving, including mutation rate and rate of horizontal gene transfer, the strength of selection often plays an important role. With regard to this selection pressure, there is typically a critical tradeoff: highly efficient antibiotic combinations can be good for the individual patient, but might also create stronger selection pressures for the evolution of resistance [5]. Because these effects are often at odds with each other, identifying the best drug combinations is not trivial.
Drug-drug interactions further complicate finding the best drug combinations. Depending on whether the combination effect is greater than, equal to, or less than the effect expected based on individual drugs, the interaction is termed as synergistic, additive, or antagonistic, respectively [6–8]. Oftentimes, clinicians prescribe synergistic combinations because they have maximum killing efficiency [9]. In contrast, hyper-antagonistic interactions are not used in the clinic as they yield higher bacterial growth when two drugs are combined than at least one of the drugs does on its own. This special type of interaction has been termed a “suppressive” interaction [10, 11]. In a study of pairwise antibiotic interactions, suppression accounted for a nontrivial 16 of 180 interactions (9%) [11].
These suppressive combinations have traditionally been considered a poor treatment strategy because of their decreased killing efficiency—much higher concentration of drugs would be needed in order to achieve similar bacterial growth inhibition [12]. Thus, little attention has been given to suppression because of its obvious limitations in clinical practice. However, recent studies demonstrate that suppressive or antagonistic drug combinations can be used to combat evolution of drug resistance: suppressive combinations have been shown to select against resistance [13], decrease spontaneous evolution of resistance [14], and reduce the rate of adaptation [5]. Because of the tradeoffs between killing efficiency and selection against resistance, antagonistic combinations may provide the most favorable treatment option under some conditions [15]. In sum, there is intriguing and compelling evidence (although all in vitro and in silico) that suppressive combinations might be useful to combat resistance [8], but their rare occurrence makes their identification and use more daunting.
Although suppressive interactions have been examined in pairwise combinations of both antibiotic [11] and anti-fungal [16] compounds, there have been no systematic examinations of suppressive interactions in higher-order drug combinations. Indeed, three-drug interactions themselves have not been extensively studied (but see [17]), and the classification and understanding of emergent interactions—interactions that only arise when all three drugs are present—is still developing. Indeed, we note that it is possible that a higher-order, three-drug interaction can occur even when none of the three pairs of antibiotics exhibit interactions in isolation. This kind of interaction is truly “emergent” because all three drugs must be present to observe any sort of interaction.
Here, we take advantage of new data and mathematical methods [18, 19] to classify emergent interactions and show that novel suppressive interactions occur in three-drug combinations. Using wild-type non-pathogenic Escherichia coli, a pathogenic clinical isolate of E. coli, and a non-pathogenic strain of Staphylococcus epidermidis, we measured growth of bacteria in all possible single, pairwise, and three-drug combinations in a set of 14 antibiotics to find emergent interactions. It has been suggested that synergy and antagonism occur about equally in pairwise antibiotic combinations, with suppression being more infrequent [11]. While suppression for two-antibiotic combinations ranges from 5% (this study) to 9% in a previous study on two-drug combinations, our results suggest that adding a third drug may increase the relative number of suppressive interactions. Furthermore, we find “emergent” suppressive interactions in three-drug combinations that may reduce growth as compared to the single-drug effects, but not as compared to the effect of at least one pairwise combination. Because the suppressive effect of these combinations is only revealed by comparison to constituent pairwise combinations, we term these “hidden suppressors” and discuss possible implications for their use in better understanding and potentially mitigating tradeoffs between bacterial killing efficacy and evolution of resistance.
Methods
Bacteria and antibiotics
The primary E. coli strain used in these experiments is BW25113, the wild-type strain (lacI q rrnB T14 ΔlacZ WJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78) [20] derived from the strain W1485 background [21]. BW25113 is the wild-type strain that was used to make the Keio Collection of single-gene knockouts and is a common lab strain used in a range of studies [22–24]). Additionally, drug combinations were also tested in (1) the pathogenic E. coli strain CFT073, a highly virulent pyelonephritis strain isolated from human clinical specimen (from ATCC) and (2) Staphylococcus epidermidis 14990 (from ATCC).
The 14 antibiotics in the study include: clindamycin, ciprofloxacin, tobramycin, streptomycin, cefoxitin, nitrofurantoin, ampicillin, erythromycin, gentamicin, chloramphenicol, vancomycin, fusidic acid, doxycycline, and trimethoprim (Table 1). We chose this set of antibiotics because they cover a range of mechanisms of action [25]. In addition, while some antibiotics such as vancomycin are ineffective by themselves in gram-negative bacteria, recent work has shown increased efficacy when used in combination with other drugs, making their effects in combination interesting for this study [26].
Table 1.
Summary of antibiotics used in three-drug experiments
| Drug | Drug abbreviation | Concentration rangea (xMIC) | MICa(μg/ml) | Main mechanism(s) of Action | Origin | Cidal-static |
|---|---|---|---|---|---|---|
| Clindamycin | CLI | 0.146–0.333 | 120 | Protein synthesis, 50S | Semi-synthetic | Bacteriostatic |
| Erythromycin | ERY | 0.080–0.400 | 150 | Protein synthesis, 50S | Natural | Bacteriostatic |
| Chloramphenicol | CHL | 0.203–0.374 | 187 | Protein synthesis, 50S | Natural | Bacteriostatic |
| Fusidic acid | FUS | 0.217–0.298 | 369 | Protein synthesis, 50S | Natural | Bacteriostatic |
| Gentamicin | GEN | 0.010–0.138 | 4 | Protein synthesis, 30S, aminoglycoside | Natural | Bactericidal |
| Tobramycin | TOB | 0.063–0.163 | 8 | Protein synthesis, 30S, aminoglycoside | Natural | Bactericidal |
| Streptomycin | STR | 0.056–0.278 | 18 | Protein synthesis, 30S, aminoglycoside | Natural | Bactericidal |
| Doxycycline | DOX | 0.182–0.318 | 2.2 | Protein synthesis, 30S | Semi-synthetic | Bacteriostatic |
| Cefoxitin | FOX | 0.150–0.193 | 6 | Cell wall | Semi-synthetic | Bactericidal |
| Ampicillin | AMP | 0.300–0.433 | 3 | Cell wall | Semi-synthetic | Bactericidal |
| Vancomycin | VAN | <0.350–0.750 | >100 | Cell wall | Natural | Bactericidal |
| Nitrofurantoin | NTR | 0.250–0.750 | 4 | Multiple mechanisms, DNA | Synthetic | Bactericidal |
| Ciprofloxacin | CPR | 0.200–0.375 | 0.04 | DNA gyrase | Synthetic | Bactericidal |
| Trimethoprim | TMP | 0.150–0.250 | 0.4 | Folic acid biosynthesis | Synthetic | Bacteriostatic |
ain wildtype, non-pathogenic E. coli BW25113
Three-drug suppression experiments
Single-drug concentrations were chosen to reduce growth by ~15–35% as compared to the no-drug controls. In a few cases, growth was reduced by less than 15%. For example, the ciprofloxacin concentration reduced growth by 7% in E. coli BW25113. This was because it was difficult to obtain a consistent growth reduction percentage at higher drug concentrations. Another special case was vancomycin, which increased growth to 109% compared to no-drug growth in E. coli BW25113. (Some antibiotics in some bacteria populations result in hormesis, a phenomenon where populations grow slightly better at very low levels of a stressor agent such as an antibiotic [27]. It is not thoroughly understood, although it could be an effect of antibiotics being used as communication tools rather than warfare [28–30].) We used lower concentrations of vancomycin because a higher concentration would have resulted in population death of many two-drug combinations with vancomycin. If a population had been killed with two drugs, adding a third drug would not have yielded meaningful data regarding how three-drug combinations affect bacterial growth.
Growth in three-drug combinations was compared to growth in no-drug, the three single-drug, and the three two-drug conditions. Experiments were performed as described by Tekin et al. [19]. Briefly, we started with a single colony which was used to inoculate cultures for glycerol stocks stored at −80 °C. A single colony from this glycerol culture was used to inoculate cultures in LB media (10 g/l tryptone, 5 g/l yeast extract, and 10 g/l NaCl). These cultures were resuspended in MC buffer and stored at 4 °C. We then grew bacteria for experiments in an incubator shaker by inoculating 20 μl of the MC stock into 2 ml LB for 5 h at 37 °C. 25 μl of a 10−4 dilution of this culture in LB was used to inoculate into 975 μl of media with antibiotics. From this 1 ml mix of antibiotics and bacteria in growth media, we aliquoted 100 μl per well into 4–6 wells of a 96-well plate. We grew these cultures for 18 h at 37 °C and 215 rpm. From a single experiment, we took the means of the 4–6 wells. For the primary E. coli BW25113 strain, we repeated this entire experiment for each three-drug combination at least three times, and a minimum of two times at identical concentrations between experiments. Then, we took the median of these replicates at identical concentrations and used this value as our growth measurement. Thus, for each drug condition at identical concentrations, we had at least 8 samples total. Two independent experiments, each with at least 4 replicates (or 8 total samples), were used for verification in E. coli CFT073 and S. epidermidis 14990.
Following previous studies by the authors [26] and others (e.g. [31]), we took an OD600 measurement at 18 h after bacteria encountered antibiotics and compared this to bacteria grown in no-drug environments to obtain relative growth. This measurement can be described as the relative difference in OD600 measurements at 18 h between populations with drugs and no-drug controls. Below, this is the measurement we refer to when we compare bacteria growth among populations. This growth measurement served as a reasonable proxy for other comparable measurements, including both growth rate (Additional file 1: Figure S1) and number of colony forming units (Additional file 1: Figure S2).
Measurement of suppression
Two-drug interactions
Pairwise suppressive interactions in terms of relative fitnesses were defined as described in Yeh et al. and Segre et al. [11, 32] as
where w X is the fitness (measured as growth) of the bacteria population in the presence of drug X, w Y is the fitness of the bacteria population in the presence of drug Y, and w XY is the fitness of the population in the presence of both drugs X and Y. Here, DAX , Y = w XY − w X w Y quantifies the deviation from expectation that two drugs do not interact, and the sign of this expression yields whether an interaction is synergistic (DAX , Y < 0) or antagonistic (DAX , Y > 0). The scale factor |min(w X, w Y) − w X w Y| applied to the positive DAX , Y further quantifies the degree of antagonism [11, 32–34], hence leading to a concrete criteria to determine suppressive interaction. According to this definition, suppressive interactions are those for which [DAX , Y]s > 1.15. When the maxima for both single growth and pairwise growth were greater than 90%, cases were excluded from calculations of pairwise suppression as the effect of the second drug on the growth is not obvious due to variation in growth measurements.
Three-drug interactions
Three-drug suppression was defined according to the emergent three-way interaction method [17, 18, 35] that distinguishes the pairwise interaction effects from the overall three-way interaction. According to this method, the emergent three-way interaction measure in terms of relative fitnesses is E3X , Y , Z = w XYZ − w X w YZ − w Y w XZ − w Z w XY + 2w X w Y w Z, where X, Y and Z represents the combined drugs. Note that the unscaled interaction measures defined for the two- and three-drug interactions, i.e. DAX , Y and E3X , Y , Z, are analogous to second- and third-order cumulants, which are widely used in the areas of theoretical physics [36] and statistics [37].
The interaction strength is further quantified by implementing a rescaling method introduced by Tekin et al. [19]—rescaling introduced for two-drug interactions [32] is extended and further developed for higher-order interactions, and shown to enhance the characterization of emergent properties. Accordingly, when E3 is greater than zero, corresponding to an antagonistic interaction, it is rescaled as
As shown in Tekin et al. [19], a histogram of all rescaled emergent three-way interaction ([E3]s) measures yields a multimodal distribution. Based on the location of the peaks of the distribution, suppressive interactions are those combinations for which [E3]s>1.30 [19]. The cutoff values for two- and three-drug interactions (i.e. 1.15 and 1.30, respectively) are chosen consistently based on natural breakpoints in the resulting distribution of interaction metrics. We note that the error or variability of data might be amplified because rescaling methods involve division and subtraction. The best way to avoid this is through carefully collected data with high replication.
For triple-antibiotic interaction assays in which a pairwise combination results in lethality of the two- and three-drug conditions, the [E3]s measure is non-applicable because it is impossible to determine the effect of the addition of the third drug, although we note that suppression could be found if two drugs kill a bacteria population and the addition of a third drug allowed the population to grow, “synthetic rescue.” Based on error in OD600 measurements as determined by Tekin et al. [19], growth measurements below 4.7% are defined as lethal. For E. coli BW25113, we found 37 such cases (see [19] and additional data in Additional file 1: Table S1). Thus, we examined the number of suppressive interactions out of a total 327 (rather than 364) [E3]s measures. For E. coli CFT073 and S. epidermidis 14990, the [E3]s measure was applicable in 356 and 313 cases, respectively.
Due to the full factorial design of our experiments, many more two-drug combinations were measured than three-drug combinations. In measuring the number of pairwise suppressive interactions in three-drug combinations for each bacteria strain, we first measured the interaction for each two-drug experiment. For example, in E. coli BW25113 for each two-drug combination, an average of 10.9 experiments were conducted, ranging from 6 to 12. The median interaction metric calculations (i.e. [DAX , Y]s) across these experiments was used to determine the interaction type of the two-drug combination.
Results and discussion
To identify suppressive emergent three-way interactions (E3X , Y , Z), we tested growth under the single-, double-, and triple-drug conditions in wild-type non-pathogenic E. coli (Additional file 1: Figure S3). In nearly all cases – 45 out of 46 suppressive three-drug combinations (97.8%) – the growth of the triple combination exceeded the growth of at least one of the pairwise combinations, but did not exceed growth in any of the single-drug conditions. Thus, in Fig. 1, which is a simplification of Additional file 1: Figure S3 to save space, we only show experimental data for the three-drug combination, the suppressed pairwise combination with the lowest growth, and the remaining single drug that was not part of the suppressed pair. See Additional file 1: Table S2 for all emergent interaction measures.
Fig. 1.
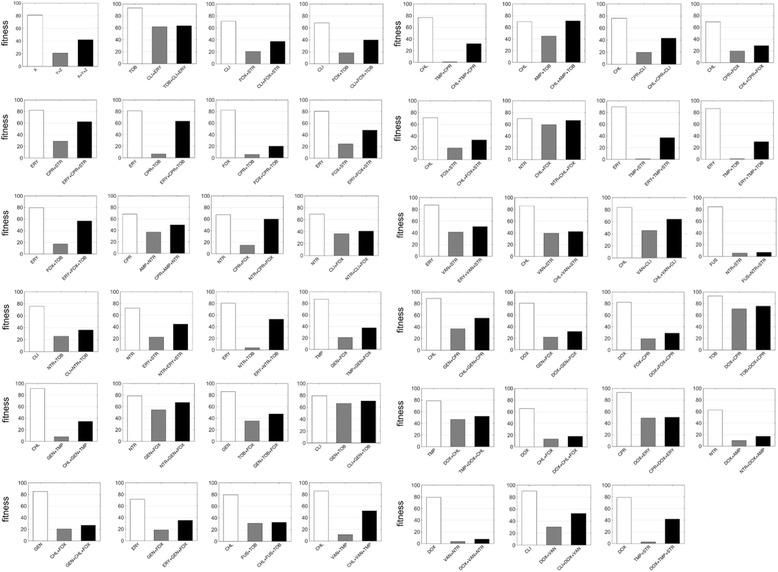
Suppressive three-drug interactions in E. coli BW25113. Growth measurements are shown for bacteria in single-drug, two-drug, and three-drug conditions relative to the no-drug control (100% growth, not shown). Emergent suppression was determined following Tekin et al. [19] (see Methods). The upper left figure is a schematic with X, Y, and Z representing three different drugs. Only the experimental data for the three-drug combination (X + Y + Z), the suppressed pairwise drug combination with the lowest growth (Y + Z), and the suppressor single drug (X) is shown. 46 triple combinations of a total 327 interaction measures were determined to be suppressive. Antibiotic abbreviations are as listed in Table 1
In three-drug combinations in the wild-type non-pathogenic E. coli strain, 14% (46 of 327 cases) of interactions were suppressive. In contrast, 2.2% (2 of 91) of two-drug combinations were suppressive (Fig. 2). There was a significant difference in the percentage of suppressive interactions in two- and three-drug combinations (z: 3.141, p-value = 0.0017). Intriguingly, we found that this pattern—antagonistic suppression is significantly more common among three-drug combinations relative to two-drug combinations—is consistent across different bacterial strains included in our study (see below for more discussion).
Fig. 2.
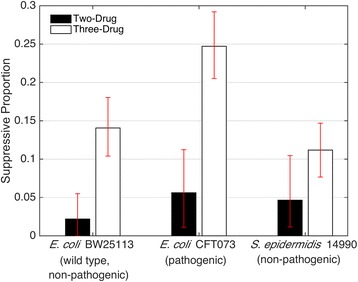
Comparison of suppressive interactions in pairwise versus emergent three-drug interactions. Bars represent the proportion of interactions that are classified as antagonistic suppressive in pairwise interactions (black) and in three-drug interactions (white) for three bacteria strains: wild-type, non-pathogenic E. coli BW25113; pathogenic E. coli CFT073; and non-pathogenic S. epidermidis 14990. For the data presented in this paper, 95% confidence intervals resulting from bootstrapping experiments are shown
For each drug, we also examined the number of times they were the suppressor (e.g. drug X in Fig. 1) compared with the number of times they were the suppressee (e.g. drugs Y or Z in Fig. 1). We did not find a significant effect, though there was a trend towards a negative correlation (Fig. 3, R = −0.375, N = 14, one-tailed p-value = 0.094).
Fig. 3.
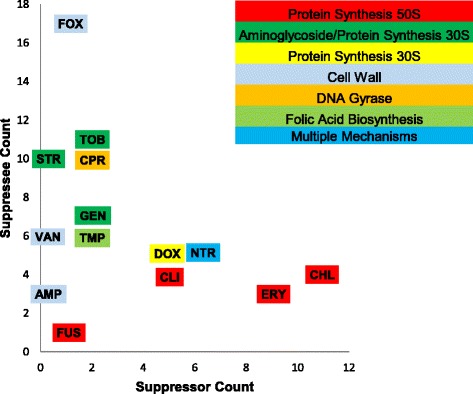
Suppressor and suppressee antibiotics. For each antibiotic, the number of suppressive interactions in E. coli BW25113 in which it acts as the suppressor (x axis) and the suppressee (y axis) are plotted. Antibiotic abbreviations are as listed in Table 1
Our finding that there was an increase in the percentage of suppressive interactions when the number of drugs used in a combination was increased is an intriguing result for several reasons. First, this suggests that clinicians should be cautious in assigning drugs for combination therapies, as one drug may have a negative impact on killing efficacy. On the other hand, there may be potential benefits for using suppressive combinations, such as slowing the evolution of resistance as previously discussed [5, 8, 13, 14].
Second, nearly all (97.8%) of the suppressive emergent interactions found in the BW25113 strain were “hidden suppressors” because they reduced growth as compared to single-drug controls, but exceeded the growth of a pairwise combination. These were “hidden” because if one did not experimentally examine two-drug growth, this suppressive effect would not be found. Suppression has been shown to be more effective at slowing the evolution of resistance both theoretically, and empirically in vitro [5, 8, 13] at least under some environments. Therefore, determining how suppression in three-drug combinations affects fitness landscapes and the evolution of resistance could be an extremely useful step (albeit one of many) towards clinical application. More generally, we need to better understand the tradeoff between slowing the evolution of resistance and increasing the bacterial killing efficiency of these combinations.
Because the effects of an antibiotic combination can vary between different bacterial strains, the results from experiments in the wild-type non-pathogenic E. coli were compared to three-drug combination results in two other bacteria strains: (1) pathogenic E. coli strain CFT073, and (2) Staphylococcus epidermidis 14990 using the same 14 antibiotics (Additional file 1: Figures. S4 and S5, Additional file 1: Tables S1 and S2). In E. coli strain CFT073, 8% of pairwise combinations were identified as suppressive, compared to 25% of triple combinations (z: 3.543, p = 0.0004). In S. epidermidis 14990, 5% of pairwise combinations were suppressive, compared to 11% of triple combinations (z: 1.806, p = 0.0709). Over three strains, 5% of pairwise were suppressive compared with 17% of triple (z: 5.016, p < 0.0001). With this data, we concluded that the general pattern that three-drugs have greater suppression cases than two-drugs still holds (Fig. 2).
Critically, as this research was conducted at sub-inhibitory concentrations following many other interaction studies [11, 13, 18, 26, 38–40], further research would be needed to determine if bacteria load would actually be reduced with suppressive drug combinations at higher drug concentrations. In addition, as shown by Chait and colleagues [13] only certain concentrations yield suppressive interactions, so interactions must also be tested across a broad range of concentrations. Therefore, before translation of these ideas to the clinic, several other substantial steps would be needed given the variance in antibiotic concentration that is expected in vivo, such as the careful consideration of concentrations, pharmacodynamics, and pharmacokinetics. Given that work on suppressive antibiotic combinations has been entirely based on in vitro and in silico studies, one critical test would be to confirm these findings using an animal infection model.
In all suppressive cases in this study, the triple-drug combination growth exceeded the growth of at least one pairwise combination, as consistent by definition. Intriguingly, we found only 5 non-hidden suppressor cases across 169 suppressive interactions in 3 bacteria strains (3%) in which the triple-drug combination growth exceeded a single-drug growth condition. It is possible that such cases would be more frequently identified by using higher antibiotic dosages, corresponding to lower growth of bacteria for which the suppressive effect would be most clearly observed. That is, because our single-drug concentrations were chosen to reduce growth only by 15–35%, it is not surprising that most triple-drug combination growth did not exceed the growth of a single-drug condition.
Our analysis shows that, within this study, there is a higher proportion of suppressive interactions in three-drug combinations across three bacteria strains (17%) than in two-drug combinations (5%). We also compared our results to Yeh and colleagues [11], which followed similar suppression criteria as in our study. These data revealed that suppressive pairwise interactions are also relatively infrequent, with only 9% showing suppressive effects.
Determining the mechanism of suppression can be very challenging. In a seminal paper on suppressive mechanisms, Bollenbach and colleagues showed that there is an optimal ratio between protein synthesis and DNA replication, and when only one of these processes is hampered, growth rates of the bacteria would be lower than when both processes are hampered [38]. Thus, many protein synthesis inhibitors and DNA synthesis inhibitors in combination could produce higher growth than just a single inhibitor by itself, leading to the observed suppressive interaction. While there are likely different mechanisms in other suppressive combinations [16, 41], there have been no studies to date that have presented alternative hypotheses and supporting data on mechanisms of suppression.
One intriguing study showed that bacteriostatic and bactericidal combinations often result in antagonism [31]. As suppression is an extreme form of antagonism, we examined whether our suppressive interactions had significantly higher rates of cidal-static combinations. Out of all three-drug combinations from 14 drugs, where eight were bactericidal and six were bacteriostatic, there were 288 (or 79%) combinations that involved at least one of each type of drug in the wildtype, non-pathogenic E. coli strain. Our data showed that 40 of 46 (or 87%) suppressive combinations involved at least one bactericidal and one bacteriostatic. This was not significantly different than expected (chi-square = 1.755, p-value = 0.1852), although statistical power was low.
It is possible that there are hubs of suppressive activity, that is, some drugs are much more likely to be involved in suppressive interactions as either suppressors or suppressees [16]. We found that, in wild-type E. coli, some drugs were much more likely to be suppressors (e.g., erythromycin and chloramphenicol) whereas other drugs were more likely to be suppressees (e.g., cefoxitin, tobramycin, and ciprofloxacin) (Fig. 3). This finding supports previous research that indicates that some drugs are much more likely to be involved in suppressive interactions [16, 38]. Interestingly, we did not find a significant correlation between these two groups, i.e. suppressor and suppressee, but patterns suggested a trend towards negative correlation, meaning that a drug was typically only a suppressor or a suppressee. This was consistent with what previous work has found, where protein synthesis inhibitors are the suppressors and DNA synthesis inhibitors the suppressees in combinations of these two classes of drugs [38]. However, this pattern is not consistent in the two other bacteria strains studied (Additional file 1: Figure S5).
In addition, in the wild-type BW25113 E. coli, the number of times a drug was a suppressee appeared to be roughly constant based on mechanism of action, and regardless of how many times it was a suppressor (and vice versa). While our sample size was small, this was suggestive that mechanism of action could be a significant predictor of suppressive interactions in three-drug combinations. This would be consistent with two-drug suppressive interactions, where certain combinations of mechanisms of action (e.g., DNA synthesis inhibitor and protein synthesis inhibitors) are more likely to result in suppression [11, 13, 38].
Exploration of mechanisms involving higher-order suppressive interactions could facilitate our understanding of how suppression translates from two-drug to higher-order combinations within drug combinations. Furthermore, other analogous forms of suppression occur in higher-order interactions in microbial communities, such as in species-species interactions or gene-gene interactions. For example, the presence of a third species can result in stable communities by affecting the interaction between the other two species (e.g. [42]), and combinations of alleles affect phenotypes such that some genes effectively suppress others in yeast populations exposed to different temperatures (e.g. [43]). Future work examining the prevalence, patterns, mechanisms, and types of suppression will increase our understanding of interactions within microbial communities.
Given the paucity of novel antibiotics in development [2] alternative strategies to overcome the problem of drug-resistant bacteria, such as combination treatments, must be considered. Another proposed strategy employs bacteriophages to either directly kill bacteria [44, 45], or to introduce genes that reverse antibiotic resistance [46]. The problem of resistance in bacteriophage therapies is further complicated by the ability of the virus to act as a vector for gene transfer, potentially arming the bacteria with antibiotic resistance or virulence genes [47]. Other strategies take advantage of virally-derived proteins that target bacteria [48] or non-viral predators of pathogenic bacteria [49]. In all cases, best practices must be considered to avoid the evolution of resistance to these new strategies. Our data highlights the need for further research in order to determine if suppressive combinations are of use in the fight against antibiotic resistance.
Acknowledgments
We thank Emily Vargas, Gabriel Ting, Tina Manzhu Kang, and Borna Shirvani for lab assistance, and Angus Buckling and three anonymous reviewers from Axios Review for comments on the manuscript.
Funding
We acknowledge an NIH Initiative to Maximize Student Development (C.B.), a James F. McDonnell Foundation Complex Systems Scholar Award (V.M.S.), and a Hellman Fellowship (P.J.Y).
Availability of data and materials
All data are made available in the Supplemental Materials.
Authors’ contributions
CB, ZM, ET, CW, VMS, and PJY performed experiments and analyzed the data. RD and JHM contributed to the acquisition of the data. CB, VMS, ET, and PJY wrote the paper. All authors revised the work for intellectual content. All authors gave a final approval of the work to be published and agree to be accountable for all aspects of the work.
Competing interests
The authors declare that that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Figure S1. Comparison of relative optical density measurement to growth rate measurement. Figure S2. Comparison of relative optical density measurement to colony forming units. Figure S3. Emergent three-way interaction measures in E. coli BW25113. Figure S4. Suppressive three-drug interactions in S. epidermidis 14990 and E. coli CFT073. Figure S5. Suppressor and suppressee antibiotics for S. epidermidis 14990 and E. coli CFT07. Table S1. Full data set for 14 drugs in E. coli BW25113, S. epidermidis 14990, and E. coli CFT073. Table S2. Emergent suppressive three-drug combinations from 14 antibiotics (see Methods, Fig. 1) for E. coli CFT073, E. coli BW25113, S. epidermidis 14990. (PDF 7668 kb)
References
- 1.World Health Organization. Antimicrobial resistance: Global report on surveillance 2014 2014.
- 2.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J, Davies D. Origins and evolution of Antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalan L, Wright G. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Reviews Mol Med. 2011;13:e5. doi: 10.1017/S1462399410001766. [DOI] [PubMed] [Google Scholar]
- 5.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci U S A. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loewe S. Die Mischiarnei. Klin Wochenschr. 1927;6:1077–1085. doi: 10.1007/BF01890305. [DOI] [Google Scholar]
- 7.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. doi: 10.1111/j.1744-7348.1939.tb06990.x. [DOI] [Google Scholar]
- 8.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klastersky J, Cappel R, Daneau D. Clinical significance of in vitro synergism between antibiotics in gram-negative infections. Antimicrob Agents Chemother. 1972;2:470–475. doi: 10.1128/AAC.2.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser TR. Lecture on the antagonism between the actions of active substances. Br Med J. 1872;2:457–459. doi: 10.1136/bmj.2.617.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 12.Jawetz E, Smith L. Combinations of antimicrobials. Calif Med. 1972;117:40-&. [PMC free article] [PubMed] [Google Scholar]
- 13.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 14.Michel JB, Yeh PJ, Chait R, Moellering RC, Kishony R. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci U S A. 2008;105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torella JP, Chait R, Kishony R. Optimal drug synergy in antimicrobial treatments. PLoS Comput Biol. 2010;6:e1000796. doi: 10.1371/journal.pcbi.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cokol M, Weinstein ZB, Yilancioglu K, Tasan M, Doak A, Cansever D, Mutlu B, Li S, Rodriguez-Esteban R, Akhmedov M, et al. Large-scale identification and analysis of suppressive drug interactions. Chem Biol. 2014;21:541–551. doi: 10.1016/j.chembiol.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood K, Nishida S, Sontag ED, Cluzel P. Mechanism-independent method for predicting response to multidrug combinations in bacteria. Proc Natl Acad Sci U S A. 2012;109:12254–12259. doi: 10.1073/pnas.1201281109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beppler C, Tekin E, Mao Z, White C, McDiarmid C, Vargas E, Miller JH, Savage VM, Yeh PJ. Uncovering emergent interactions in three-way combinations of stressors. J R Soc Interface. 2016;13:125. doi: 10.1098/rsif.2016.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tekin E, Beppler C, White C, Mao Z, Savage VM, Yeh PJ. Enhanced identification of synergistic and antagonistic emergent interactions among three or more drugs. J R Soc Interface. 2016;13:119. doi: 10.1098/rsif.2016.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko K, Wanner B. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko K, Tomita M, Wanner B, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keren I, Wu Y, Inocencio J, Mulcahy JR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 23.Cohen NR, Ross CA, Jain S, Shapiro RS, Gutierrez A, Belenky P, Li H, Collins JJ. A role for the bacterial GATC methylome in antibiotic stress survival. Nat Genet. 2016;48:581–586. doi: 10.1038/ng.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagarinova A, Stewart G, Samanfar B, Phanse S, White CA, Aoki H, Deineko V, Beloglazova N, Yakunin AF, Golshani A, et al. Systematic genetic screens reveal the dynamic global functional Organization of the Bacterial Translation Machinery. Cell Rep. 2016;17:904–916. doi: 10.1016/j.celrep.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Scott GM, Kyi MS. Handbook of essential Antibiotics. Amsterdam: Harwood Academic; 2001. [Google Scholar]
- 26.Zhou A, Kang TM, Yuan J, Beppler C, Nguyen C, Mao Z, Nguyen MQ, Yeh P, Miller JH. Synergistic interactions of vancomycin with different antibiotics against Escherichia coli: trimethoprim and nitrofurantoin display strong synergies with vancomycin against wild-type E. coli. Antimicrob Agents Chemother. 2015;59:276–281. doi: 10.1128/AAC.03502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch H. Principles and practice of antibiotic therapy. New York: Medical Encyclopedia, Inc; 1954. [Google Scholar]
- 28.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Yim G, Wang HH, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 30.Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ocampo PS, Lázár V, Papp B, Arnoldini M, Abel zur Wiesch P, Busa-Fekete R, Fekete G, Pál C, Ackermann M, Bonhoeffer S. Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob Agents Chemother. 2014;58:4573–4582. doi: 10.1128/AAC.02463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segre D, DeLuna A, Church G, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 33.Sanjuán R, Elena SF. Epistasis correlates to genomic complexity. Proc Natl Acad Sci U S A. 2006;103:14402–14405. doi: 10.1073/pnas.0604543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Qian W, Wang Z, Li Y, Zhang J. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat Genet. 2010;42:272–276. doi: 10.1038/ng.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer A, Katzir I, Dekel E, Mayo AE, Alon U. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc Natl Acad Sci U S A. 2016;113:10442–10447. doi: 10.1073/pnas.1606301113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glimm J, Jaffe A. Quantum physics: a functional integral point of view. 2. Berlin, New York: Springer-Verlag; 1987. [Google Scholar]
- 37.Kendall MG, Stuart A. The advanced theory of statistics. London: Griffin; 1969. [Google Scholar]
- 38.Bollenbach T, Quan S, Chait R, Kishony R. Non-optimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demchok J, Meletiadis J, Roilides E, Walsh T. Comparative pharmacodynamic interaction analysis of triple combinations of caspofungin and voriconazole or ravuconazole with subinhibitory concentrations of amphotericin B against Aspergillus spp. Mycoses. 2010;53:239–245. doi: 10.1111/j.1439-0507.2009.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Oliveira CE, Stamford TL, Gomes Neto NJ, de Souza EL. Inhibition of Staphylococcus aureus in broth and meat broth using synergies of phenolics and organic acids. Int J Food Microbiol. 2010;137:312–316. doi: 10.1016/j.ijfoodmicro.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 41.de Vos MG, Bollenbach T. Suppressive drug interactions between antifungals. Chem Biol. 2014;21:439–440. doi: 10.1016/j.chembiol.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JT, Taylor MB, Shen A, Ehrenreich IM. Multi-locus genotypes underlying temperature sensitivity in a Mutationally induced trait. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biswas B, Adhya S, Washart P, Paul B, Trostel AN, Powell B, Carlton R, Merril CR. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun. 2002;70:204–210. doi: 10.1128/IAI.70.1.204-210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG, Norris JS. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother. 2003;47:1301–1307. doi: 10.1128/AAC.47.4.1301-1307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R, Friedman N, Molshanski-Mor S, Qimron U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Microbiol. 2012;78:744–751. doi: 10.1128/AEM.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qbeta virion: diversity in lysis targets. Science. 2001;292:2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]
- 49.Kadouri DE, To K, Shanks RM, Doi Y. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PLoS ONE. 2013;8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are made available in the Supplemental Materials.


