Abstract
Discoba (Excavata) is an evolutionarily important group of eukaryotes that includes Jakobida, with the most bacterial-like mitochondrial genomes known, and Euglenozoa, many of which have extensively fragmented mitochondrial genomes. However, little is known about the mitochondrial genomes of Heterolobosea, the third main group of Discoba. Here, we studied two heteroloboseids—an undescribed amoeba “BB2” and Pharyngomonas kirbyi. Phylogenomic analysis revealed that they form a clade that is a sister group to all other Heterolobosea. We characterized the mitochondrial genomes of BB2 and P. kirbyi, which encoded 44 and 48 putative protein-coding genes respectively. Their gene contents were similar to that of Naegleria. In BB2, mitochondrially encoded RNAs were heavily edited, with ∼500 mononucleotide insertion events, mostly guanosines. These insertions always have the same identity as an adjacent nucleotide. Editing occurs in all ribosomal RNAs and protein-coding transcripts except one, and half of the transfer RNAs. Analysis of Illumina deep-sequencing data suggested that this RNA editing is very accurate and efficient, and most likely co-transcriptional. The dissimilarity of this editing process to other RNA editing phenomena in discobids, as well as its apparent absence in P. kirbyi, suggest that this remarkably extensive system of insertional editing evolved independently in the BB2 lineage, after its divergence from the P. kirbyi lineage.
Keywords: protist, phylogenomics, eukaryote, amoeba “BB2”, Pharyngomonas, transcription
Introduction
Mitochondria are eukaryotic organelles that originated from α-proteobacteria by endosymbiosis and contain their own DNA and transcription/translation machinery (Gray et al. 1999). This ancient endosymbiosis was followed by massive gene loss in the ancestral mitochondrial (mt) genome, by deletion of unnecessary genes and gene transfer from the mitochondrial genome to the nucleus (Gray et al. 1999). Mitochondrial gene content has continued to decrease to varying degrees in different eukaryote groups, giving rise to the huge variety among mitochondrial genomes we now observe (Lang et al. 1997; Kamikawa et al. 2016).
Discoba (Excavata) is a major (kingdom-level) group of protistan eukaryotes that includes species with some of the most extraordinary mitochondrial genomes known (Gray et al. 2004; Simpson et al. 2006; Hampl et al. 2009). It comprises three main subgroups: Jakobida, Euglenozoa, and Heterolobosea, plus the isolated genus Tsukubamonas (Hampl et al. 2009; Kamikawa et al. 2014). Jakobida (e.g., Reclinomonas, Andalucia) have the most bacterial-like (ancestral) and gene-rich mitochondrial genomes discovered to date (Lang et al. 1997; Burger et al. 2013). For example, the mitochondrial genome of Andalucia godoyi contains 66 protein-coding and 34 structural RNA genes. Jakobid mtDNAs encode multiple subunits (almost always four) of bacteria-type RNA polymerase, whereas all other eukaryotes possess instead a nucleus-encoded, single-subunit enzyme homologous to bacteriophage RNA polymerases (Burger et al. 2013).
The mitochondria of Euglenozoa also have unusual features, including gene fragmentation and extensive editing of mitochondrial transcripts (Flegontov et al. 2011). For example, the mitochondrial genome of the model euglenid Euglena gracilis encodes fragmented ribosomal RNAs (Spencer and Gray 2011; Dobakova et al. 2015), while those of diplonemids contain fragmented genes where each nonoverlapping piece is encoded on a separate circular chromosome (Marande et al. 2005). In diplonemids these fragments are transcribed separately and assembled into an mRNA by a unique trans-splicing mechanism, which sometimes also involves insertion of uridines and substitution of cytosine by uridine and adenosine by inosine (Marande and Burger 2007; Kiethega et al. 2013; Moreira et al. 2016, Yabuki et al. 2016). Most famously, many mitochondrial pre-mRNAs in kinetoplastids, the sister group of diplonemids, are massively edited post-transcriptionally by uridine insertion/deletion to produce the functional RNAs (Benne et al. 1986; Horton and Landweber 2002; Lukes et al. 2005). Uridine insertion/deletion in kinetoplastids is mediated by small antisense RNAs (guide RNAs), which specify editing sites, and 20S multi-subunit protein complexes called “editosomes” (Knoop 2011).
This mechanism in kinetoplastid mitochondria is one of the best studied examples of “RNA editing”, which is defined as targeted modifications to the RNAs that result in sequence differences, via nucleotide insertions/deletions or substitutions, between the transcriptome and the corresponding genomic sequences (Gray 2003; Knoop and Rudinger 2010). RNA editing is a diverse phenomenon that has evolved multiple times independently, and it occurs in the mitochondria, plastids or nuclei of a wide range of other eukaryotes, including dinoflagellates, myxomycetes (Amoebozoa), and land plants (Chaterigner-Boutin and Small 2011; Knoop 2011). Both protein-coding RNAs (mRNAs) and structural RNAs can be affected (e.g., ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs); Gott and Emeson 2000; Horton and Landweber 2002; Gray 2003).
While the mitochondrial genomes of Jakobida and Euglenozoa have been intensively studied, only three mitochondrial genomes have been characterized from Heterolobosea: those of Naegleria gruberi (Fritz-Laylin et al. 2010), Naegleria fowleri (Herman et al. 2013), and Acrasis kona (Fu et al. 2014). The N. gruberi and N. fowleri mitochondrial genomes have 42 protein-coding and 21 structural RNA genes. In contrast, the mt genome of A. kona contains only 26 protein-coding genes and 13 structural RNA genes. In addition, a small number of instances of substitutional C-to-U RNA editing were reported in mitochondrial transcripts of N. gruberi and A. kona, along with the presence of a DYW-type pentatrico-peptide repeat (PPR) protein, previously only found in land plants (Knoop and Rudinger 2010; Fu et al. 2014). In plants, the PPR protein recognizes and binds to specific C residues for RNA editing (Yagi et al. 2013); it seems plausible that heteroloboseid homologues have the same function. Interestingly, Fu et al. (2014) suggested that the DYW-type PPR proteins in N. gruberi and A. kona were acquired by multiple independent lateral gene transfer events and thus were not ancestral to the Heterolobosea.
In order to better understand mitochondrial genome diversity and evolution in Discoba, we here characterize the mitochondrial genomes of two putatively early-diverging species within Heterolobosea: the undescribed amoeba “BB2” and Pharyngomonas kirbyi (Park and Simpson 2011; Harding et al. 2013). Our phylogenomic analyses demonstrate that they form a single clade that emerges at the base of the Heterolobosea, and we describe the dynamics of mitochondrial genome evolution in this group in light of this newly resolved phylogeny. Unexpectedly, we found that an extremely efficient form of insertional RNA editing is very widespread in BB2 mitochondria, occurring at nearly 500 positions, affecting transcripts of all but one of the protein-coding genes, as well as the rRNAs, and half of the encoded tRNAs. This phenomenon is clearly different from the forms of mitochondrial RNA editing documented previously in discobid mitochondria, and, presumably, evolved independently.
Material and Methods
Transcriptomic Sequencing of BB2
Amoeba BB2 strain PRA-19 was obtained from the American Type Culture Collection (ATCC) and grown at 42 °C in ATCC medium 1034 (modified PYNFH medium: Bacto-peptone 10.0 g/l, yeast extract 10.0 g/l, yeast nucleic acid 1.0 g/l, folic acid 15.0 mg/l, hemin 1.0 mg/l, fetal bovine serum 10%, KH2PO4 0.36 g/l, Na2HPO4 0.5 g/l). Pharyngomonas kirbyi strain AS12B was grown at 12.5% salt at 37 °C as described in Harding et al. (2016). Total RNA was isolated from BB2 cells harvested using TRIzol (Rio et al. 2010) following the manufacturer’s instructions (Ambion), and treated with Turbo DNAse (Ambion) to remove residual DNA.
For BB2, a cDNA library was constructed using the TruSeq RNA sample preparation kit version 2 (Illumina) and sequenced on a MiSeq platform, generating 19.2 million 150-bp paired-end reads. Reads were trimmed to remove adapter sequences and low-quality sequences using Trimmomatic 0.32 with a PHRED33 quality threshold of 25 (Bolger et al. 2014). Reads were assembled using Trinity 2.0.2 (Grabherr et al. 2011), and open-reading frames (ORFs) were predicted using TransDecoder.
Mitochondrial ORFs missing from the assembled transcripts but present in genomic sequences were sequenced by RT-PCR. First-strand cDNA was synthesized from DNase-treated total RNA using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo) with random hexamer primers, following the manufacturer’s instructions.
Phylogenetic Analysis
To clarify the phylogenetic position of our study organisms, we modified a curated “phylogenomic” dataset containing 252 nucleus-encoded house-keeping genes from a broad range of eukaryotes, described in Harding et al. (2016). To the original dataset, we added sequences from the BB2 transcriptomic data, as well as from Pharyngomonas kirbyi transcriptome (data from Harding et al. 2016; GECH01000000), plus six other Excavates: Spironucleus vortens, Tritrichomonas foetus, Diplonema papillatum, Leishmania major, “Seculamonas ecuadoriensis”, “Jakoba bahamensis” (all available in GenBank, http://www.ncbi.nlm.nih.gov/, last accessed February 10, 2016), and Stygiella incarcerata (Leger et al. 2016).
Orthologous protein sequences were aligned by MAFFT-linsi (Katoh et al. 2005), and any sites in the alignments with more than 40% gaps were masked using BMGE v1.1 (Criscuolo and Gribaldo 2010). Single gene trees were generated using RAxML v7.2.6 with the PROTGAMMALG model, and manually examined to remove putative contaminants, paralogs or laterally transferred genes. Sequences of remaining proteins were concatenated into a super-matrix containing 67 taxa and 68,718 amino acid positions. Maximum-likelihood trees were estimated using IQ-TREE (Nguyen et al. 2014) under the LG + C20 + F + gamma model. Topological support was assessed by 1000 ultrafast bootstrap replicates and the SH-like approximate likelihood ratio test with 1000 replicates. ML trees were also estimated using RAxML under the LG4X model (from 100 starting trees), with topological support assessed by 100 bootstrap replicates.
Bayesian inference was conducted using PhyloBayes-MPI v. 1.6.5 under the CAT-Poisson model. Five independent Markov chain Monte Carlo chains were run for 5,000 generations, sampling every two generations. Five hundred generations were discarded as burn-in. Convergence was achieved for three of the chains, with the largest discrepancy observed across all bipartitions (maxdiff) less than 0.26.
Genomic DNA Sequencing of BB2 and P. kirbyi
Total DNA was extracted from BB2 and P. kirbyi using a salt-based separation method (Aljanabi and Martinez 1997). For sequencing of genomic DNA, libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina). Sequencing was done using the MiSeq platform, yielding 43.4 million 150-bp paired-end reads for BB2 and 31.6 million 250-bp paired-end reads for P. kirbyi. Reads were trimmed as described above. Reads generated from P. kirbyi were filtered to remove sequences derived from food prokaryotes as described in Harding et al. (2016).
Genomic contigs for BB2 and P. kirbyi were assembled with the de novo assemblers Ray v2.3.1 (Boisvert et al. 2010) and/or MIRA v4.9.5_2 (Chevreux et al. 2004). For P. kirbyi, a second round of decontamination was performed by using assembled contig sequences as queries in BLASTn searches against the NT database. Alignments longer than 100 bp showing more than 90% identity to a prokaryotic sequence were discarded as potential contaminants.
Mitochondrial Genome Assembly and Annotation
The genomic contigs from both species were screened for regions homologous to the mitochondrial genomes of other heteroloboseids (Naegleria gruberi, NC_002573; Naegleria fowleri, NC_021104; Acrasis kona, NC_026286), as well as the jakobid Reclinomonas americana (NC_001823) and Tsukubamonas globosa (NC_023545), using BLASTn and BLASTx. For BB2, eight contigs with sizes ranging from 6 to 18 kb were highly similar to mitochondrial-derived sequences (identities >25%). These contigs were linked together into a circular-mapping mitochondrial genome with a size of 119,312 bp after PCR-amplification of “bridging” fragments using the LongAmp Taq PCR kit (NEB) and combinations of specific primers (supplementary table S1 and fig. S1A, Supplementary Material online). These amplicons were Sanger-sequenced. For P. kirbyi, two contigs (sizes 55 and 19 kb) were linked into one linear 75,717-bp scaffold by the same approach (supplementary table S1 and fig. S1B, Supplementary Material online).
Annotation was performed using the automated gene annotation tools, MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl, last accessed April 26, 2016) and RNAweasel (http://megasun.bch. umontreal.ca/RNAweasel/, last accessed April 26, 2016), and BLASTp searches against the NR database (NCBI) with an E-value cutoff of 1 × 10−10. Transfer RNA genes were confirmed using tRNAscan-SE v1.23 (Lowe and Eddy 1997). For gene prediction in the BB2 mitochondrial genome, the assembled transcripts were aligned to the mitochondrial genome in order to compare the sequences, then subjected to MFannot to identify genes that were initially not recognized from the analysis of mtDNA alone. To identify genes potentially missed by MFannot we searched for proteins by using HMMER 3.1b2 (Eddy 1998). We created hidden Markov models with the appropriate protein sequences (aligned by MAFFT-linsi) from the mitochondrial genomes of Andalucia godoyi, Reclinomonas americana, Naegleria fowleri, Naegleria gruberi, Tsukubamonas globosa, Histiona aroides, “Jakoba bahamiensis”, Jakoba libera, and “Seculomonas ecuadoriensis” (supplementary table S2, Supplementary Material online).
The secondary structure of tRNAs were predicted using tRNAscan-SE v1.23. The secondary structure of the SSU rRNA was predicted and adjusted manually according to the secondary structure conserved among bacteria, archaea, eukaryotes, plastids, and mitochondria (as compiled in the Comparative RNA Web Site and Project; http://www.rna.icmb.utexas.edu, last accessed May 18, 2016), and generated using the xrna program (http://rna.ucsc.edu/rnacenter/xrna/, last accessed May 18, 2016).
Genome maps were drawn using GenomeVx (Conant and Wolfe 2008) followed by manual adjustment. Mitochondrial gene content was compared amongst eukaryotes from diverse lineages by extending the analyses of Kamikawa et al. (2016).
Confirmation of RNA Editing in BB2
Comparison of transcript sequences to the genome indicated the presence of nonencoded nucleotides (insertions) in the former. These insertions were confirmed by mapping RNA-seq reads onto the genome using Bowtie 2 v.2.3.1 (Langmead et al. 2009) and by assessing variant sites using FreeBayes (ploidy of 100, mapping quality > 30; Garrison and Marth 2012). DNA-derived reads were also treated the same way in order to predict genomic polymorphism and verify that predicted editing sites were not mistakenly identified due to the presence of polymorphisms at these loci. A subset of these RNA editing sites (50/475 sites) was confirmed by sequencing eight distinct PCR products using cDNA as template (supplementary table S3, Supplementary Material online).
To characterize the efficiency and fidelity of the RNA editing mechanism, transcriptomic-sequencing reads were aligned to the mature transcript sequences using BLASTn (NCBI). We used 10-nucleotide-long sliding windows along the transcript sequences to examine the frequency and types of mismatches (insertion, missing nucleotide, or substitution in RNA-derived sequencing reads compared to the transcriptomic consensus sequences) near editing sites (windows with editing sites) and remote from editing sites (windows with no editing sites). We then performed Z-tests to determine if the rates of various types of errors were significantly different between editing and nonediting sites. The Z-scores for each error type were calculated as follows:
where p1 is the frequency of mismatches near editing sites, p0 is the frequency of mismatches remote from editing sites, p is the frequency of mismatches for all sites, while n1 and n0 are the total numbers of nucleotides near editing sites and remote from editing sites, respectively. The p-value for the null hypothesis that p1 and p0 are equal was determined from the Z-score based on the standard normal distribution.
Results
Phylogenetic Positions of BB2 and P. kirbyi
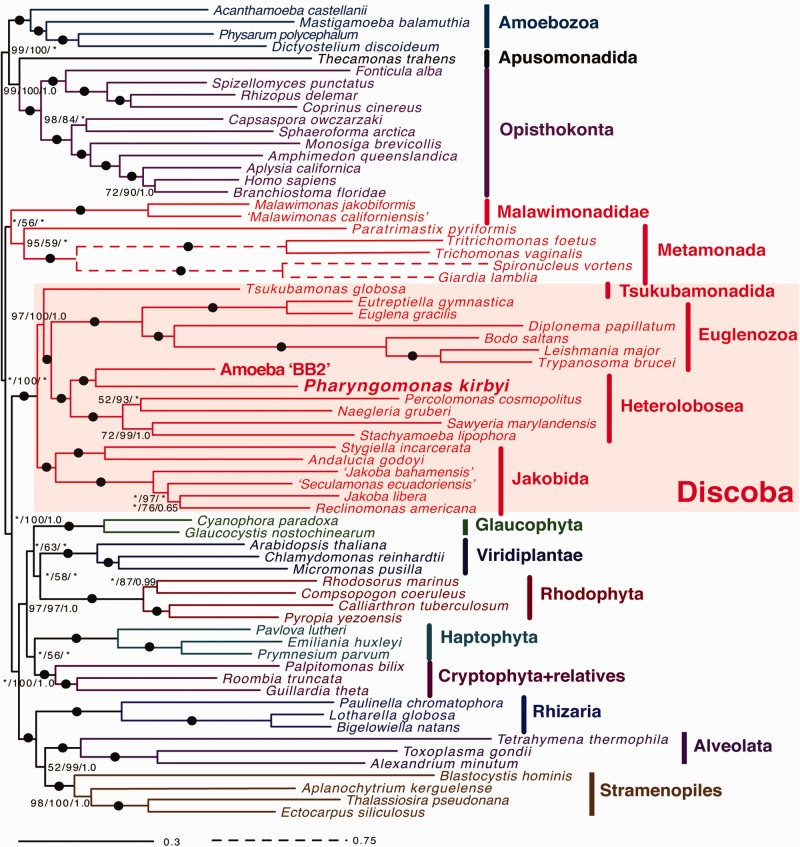
We searched the RNA-Seq data of “BB2” and P. kirbyi for orthologs of 252 conserved nucleus-encoded proteins that comprise a eukaryote-wide “phylogenomic” super-matrix. After including these orthologs within the aligned super-matrix and trimming of ambiguously aligned regions, we inferred the phylogenetic position of these two taxa within the eukaryote tree of Life. Both maximum likelihood (ML) analyses and Bayesian inference (BI) showed that BB2 and P. kirbyi robustly group with other heteroloboseids with strong statistical support (100% ML-bootstrap (MLBP), 100% ML-ultrafast bootstrap (UFBoot) and Bayesian posterior probability (BPP) of 1; fig. 1, see legend for model details). Interestingly, BB2 and P. kirbyi were inferred to be sister taxa in both ML and BI analyses, with 100% MLBP, 100% UFBoot and BPP of 1.0 (fig. 1). This BB2 + P. kirbyi clade formed the deepest branch within Heterolobosea, and the remaining heteroloboseids (Percolomonas cosmopolitus, N. gruberi, Sawyeria marylandensis, and Stachyamoeba lipophora) formed a well-supported group (100% MLBP, 100% UFBoot, and BPP of 1.0); in other words, there was maximal support for the basal placement of the BB2 + P. kirbyi clade within Heterolobosea.
Fig. 1.
—Phylogenetic tree estimated from the 252-protein dataset, inferred by IQtree under the LG + C20 + F + Gamma model with ML ultrafast bootstrap support (UFBoot). ML bootstrap support (MLBP) was also estimated by RAxML under the LG4X model, and BI posterior probabilities (BPP) were estimated by Phylobayes-MPI under the CAT-Poisson model. Support values are shown at each branch in the following order: MLBP, UFBoot and BPP. Black dots indicate 100% MLBP, 100% UFBoot and 1.0 BPP. Asterisks (*) indicate branches that were not recovered in the RAxML or Phylobayes-MPI analysis.
Mitochondrial Genome Overview
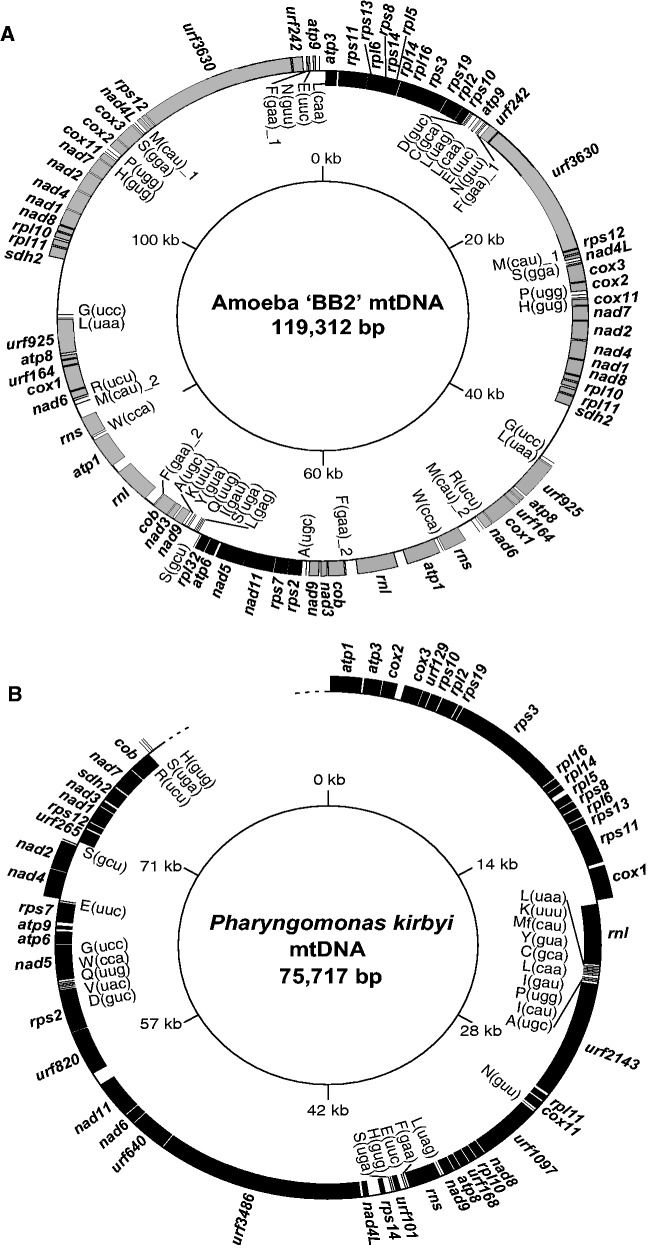
The mitochondrial genome of BB2 was assembled as a single circular-mapping molecule 119,312 bp in length (fig. 2A). It contained a 49 kbp-long repeated region (fig. 2A, shown in grey) with the two copies in an inverted orientation and situated opposite one another on the map (inverted repeat: IR). This organization, presented in figure 2A, is the simplest one among many possible organizations (involving multiple different linear or circular molecules) that are consistent with our sequencing of regions between the IR and nonrepeated regions (shown in black in fig. 2A). In addition, mapping of reads derived from genomic DNA onto the mitochondrial genome revealed higher coverage (4–5 times on average) in the IR region compared to nonrepeated regions (supplementary fig. S2, Supplementary Material online). This difference in coverage depth is consistent with the presence of multiple copies of the 49 kb-long region, although these coverage data should be treated with caution since the read depth was highly variable (not observed in the coverage analysis of Pharyngomonas, see supplementary fig. S2, Supplementary Material online). The existence of such a large IR is somewhat unusual among mt genomes of protists, although mt genomes are extremely diverse in size and organization (Gray et al. 1998; Burger et al. 2003). However, large IR are often observed in plastid genomes (Palmer and Thompson 1982; Cosner et al. 1997) and are also present in the mt genomes of Malawimonas jakobiformis (NC_002553), Proteromonas lacertae (Pérez-Brocal et al. 2010), Palpitomonas bilix (Nishimura et al. 2016), and Acavomonas peruviana (Janouškovec et al. 2013; Tikhonenkov et al. 2014). Furthermore, to determine whether this unusual genome organization resulted from mis-assembly artifacts caused by nuclear mitochondrial DNA segments (NUMTs), we searched for nuclear genomic contigs encoding sequences similar to the mt genome of BB2. We did not detect such cases, indicating that NUMTs probably do not occur in amoeba BB2 and did not affect our mt genome assembly.
Fig. 2.
—The mitochondrial genome maps of (A) amoeba “BB2” and (B) Pharyngomonas kirbyi. Genes encoding proteins and ribosomal RNAs are shown in boxes, transfer RNA genes are shown by lines. Boxes on the outside of the circle represent RNAs encoded on the positive strand, boxes inside are RNAs encoded on the negative strand. The duplicated “IR” regions are highlighted in grey. Single copy regions are shown in black.
In total, the mitochondrial genome of BB2 is 81% coding sequence and contains 40 known protein-coding genes, 2 rRNA genes, 24 tRNA genes, and 4 Unknown open Reading Frames (URFs; for these counts, the IR region is only considered once and RNA editing is taken into account—see below). Genes are tightly packed, and many of them are partially overlapping, such as rps8–rps14, rpl14–rpl16, and cox1–nad6. No introns were detected. The overall adenine + thymine (AT) content is 70.1%, the lowest among currently known mt genomes of heteroloboseids (N. gruberi: 77.8%, N. fowleri: 74.8%, and Acrasis kona: 83.3%).
The mitochondrial genome of P. kirbyi was assembled into a single linear-mapping contig with a size of 75,717 bp (fig. 2B). Attempts at closing the genome into a circular map by bridging-PCR experiments were unsuccessful (supplementary table S1 and fig. S1B, Supplementary Material online). No telomeric repeats could be detected at the ends of the sequence. Such repeats would normally be expected for a linear DNA molecule, although the P. bilix mt genome was shown recently to be linear and to lack telomeric repeats (Nishimura et al. 2016). Consequently, this P. kirbyi mitochondrial genome assembly is possibly partial, although, if so it is likely near-complete because the demonstrated gene content is very similar to that of BB2 (see below). The overall AT content is 87.5%, the highest known within Heterolobosea. The mitochondrial genome is gene-dense, with 92% of the sequence in coding regions, and it contains 39 protein-coding genes, small and large subunit rRNA genes, 23 tRNA genes and 9 URFs. There are many partially overlapping ORFs including cox3–URF129, URF129–rps10, rps19–rpl2, rps19–rps3, rpl16–rpl14, rpl5–rps8, rpl6–rps13, rps13–rps11, nad8–rpl10, rpl10–URF168, URF168–atp8, atp8–nad9, URF640–nad6, and URF820–rps2. As for BB2, no introns were detected in the P. kirbyi mitochondrial genome.
Mitochondrial Gene Content and Synteny
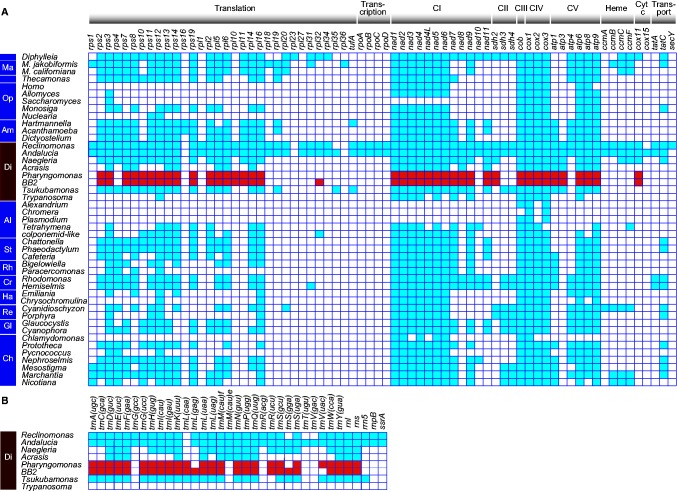
The gene contents of these mitochondrial genomes were compared to eukaryotes from diverse lineages (fig. 3). BB2 and P. kirbyi mtDNAs appear to have extremely similar gene contents, although the uncertainty regarding the completeness of the P. kirbyi mitochondrial genome prevents definitive conclusions. Their mtDNAs both encoded ribosomal proteins (RPS2, 3, 7, 8, 10–14, 19, RPL2, 5, 6, 10, 11, 14, 16), components of electron chain transport complexes I (Nad1-4, 4L, 5–9, 11), II (SDH2), III (Cob), IV (COX1-3), and V (ATP1, 3, 6, 8, 9), and a cytochrome c oxidase assembling protein (COX11). One additional ribosomal protein gene was found in the BB2 mtDNA: rpl32. On the other hand, both BB2 and P. kirbyi lack four of the genes encoded on the mtDNAs of Naegleria spp.: two cytochrome c maturase subunits (ccmC, ccmF), twin arginine translocase (tatC) and ribosomal protein S4 (rps4; fig. 3). These genes were not detected even in the (predominantly nuclear) transcriptome data from BB2 and P. kirbyi when searches were performed using hidden Markov models built with the corresponding protein sequences from other Discoba, suggesting that they were completely absent in both BB2 and P. kirbyi.
Fig. 3.
—Presence and absence of (A) protein-coding genes in the mitochondrial genomes of various eukaryotes and (B) transfer RNA genes among Discoba. Ma: Malawimonas, Op: Opisthokonta, Am: Amoebozoa, Di: Discoba, Al: Alveolata, St: Stramenopiles, Rh: Rhizaria, Cr: Cryptophyceae, Ha: Haptophyta, Re: Red algae, Gl: Glaucophyta, Ch: Chloroplastida, CI-CV: electron transport chain complex I–V (following Kamikawa et al. 2016).
Gene order comparison among representative mitochondrial genomes from Discoba (N. fowleri, BB2, P. kirbyi, Andalucia godoyi, and Tskubamonas globosa) showed vestiges of a highly conserved ribosomal gene cluster that is similar to the S10, spectinomycin, alpha ribosomal protein gene clusters of the close bacterial relatives of mitochondria (e.g., Rickettsia;supplementary fig. S3, Supplementary Material online). This ribosomal gene cluster was not detected in the mitochondrial genome of Acrasis kona. The mitochondrial genomes of BB2 and P. kirbyi also showed two other pairs of genes in the same order (nad2–nad4 and cox2–cox3).
RNA Editing in Mitochondria of BB2
Initial examinations of the mitochondrial genome of BB2 showed numerous apparent frameshifts interrupting the protein-coding regions. By comparing transcripts (or assembled RNA-seq data) and RNA-seq read sequences to genomic DNA sequences, we identified 475 unique sites where transcripts almost always contained a single nucleotide insertion relative to the mitochondrial genome sequence. In contrast, no consistent deletions or substitutions were observed. Fifty of these insertion sites were confirmed by sequencing PCR products from both genomic DNA and cDNA. Mononucleotide insertions were detected in 43 out of 44 protein-coding genes, small subunit (SSU) and large subunit (LSU) rRNAs, 12 out of 25 tRNAs, and intergenic regions (table 1, see supplementary table S4, Supplementary Material online for numbers and types of nucleotide inserted in each gene). Insertion-type RNA editing observed in protein-coding regions resolves apparent gene fragmentation and frameshifts in the mtDNA, resulting in one continuous ORF for each transcribed gene (fig. 4).
Table 1.
Number and Type of Insertions Found in Edited Mitochondrial Transcripts of Amoeba “BB2”
| Type | # Genes Edited/Total # of Genes | G | A | C | U | Total |
|---|---|---|---|---|---|---|
| Protein-coding genes | 39/40 | 265 | 20 | 17 | 7 | 309 |
| URFs | 4/4 | 90 | 12 | 4 | 7 | 113 |
| tRNA | 12/25 | 10 | 1 | 0 | 1 | 12 |
| rRNA | 2/2 | 35 | 0 | 4 | 1 | 40 |
| Intergenic regions | 0 | 1 | 0 | 0 | 1 | |
| Total | 400 | 34 | 25 | 16 | 475 | |
| (84.2%) | (7.1%) | (5.3%) | (3.4%) |
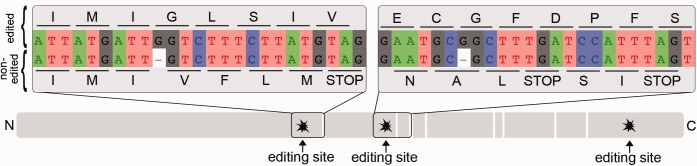
Fig. 4.
—Representation of the conceptual translation of the first 5′ → 3′ reading frame of the nad3 genomic sequence, indicating in-frame stop codons (vertical white bars) and the location of three editing sites. Partial alignments are shown for the first two editing sites to illustrate the recovery of the proper reading frame after insertional editing. The placement of the inserts before the identical encoded nucleotide, rather than after, is arbitrary (see text).
The most frequently inserted nucleotide by far is guanosine (84.2%), followed by adenosine (7.1%), cytidine (5.3%), and uridine (3.4%; table 1). Strikingly, all the inserted nucleotides are located next to one or more nucleotides with the same identity that were encoded by the mtDNA. This characteristic made it impossible to determine whether nucleotides were inserted before or after the identical nucleotide (or nucleotides) already specified by the mitochondrial genome.
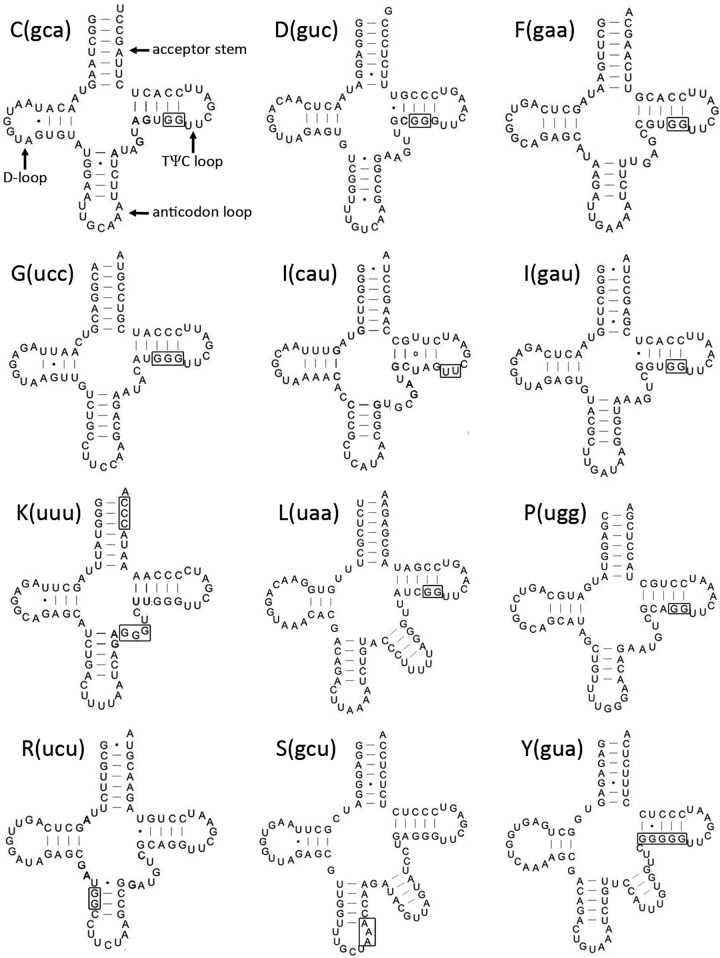
The secondary structures of tRNA predicted using mtDNA sequences showed the general cloverleaf structure but some lacked normally well-conserved features, such as the highly conserved GUUC motif, the acceptor stem or the anticodon loop. These features were recovered when edited RNA sequences were used to reconstruct secondary structures (fig. 5). Most interestingly, tRNAs for R(ucu) and S(gcu) were identified from their edited transcripts; these genes were initially unrecognizable from analysis of the mtDNA alone. The tRNA for R(ucu) has a guanosine insertion in the anticodon stem, which creates a G-C base pair and a typical 4–5 bp-long anticodon stem (an unusually short 3 bp-long stem is implied by the mtDNA sequence alone). In the tRNA for S(guc), a single adenosine is inserted at either the anticodon stem or loop, creating a typical seven nucleotide-long loop (whereas the mtDNA implies an aberrant six nucleotide-long anticodon loop).
Fig. 5.
—Secondary structures of mitochondrial tRNAs of amoeba “BB2”. Regions including an editing site are shown in boxes. For each editing site, only one nucleotide is inserted (the exact insertion sites are unknown since nucleotides are inserted next to one or more encoded nucleotides with the same identity).
The SSU rRNA of BB2 mitochondria is 1,654 nucleotides in length in mature form. Maturation requires editing at 18 sites with single nucleotide insertions (15 guanosines, 1 uridine, and 2 cytidines). Editing sites are distributed over the entire length of the rRNA. After inferring the secondary structure of the SSU rRNA, we determined that the localization of editing sites is not limited to any particular features of the predicted structure (supplementary fig. S4, Supplementary Material online). We found most insertions (11/18) in base-paired regions (stems) and 2 insertions that were unambiguously within loop structures. The location of other insertions (5/18) were uncertain, as the run of the same nucleotide (one of them being editing site) were localized in regions covering both loop and stem structures. If RNA editing is assumed to be consistently ordered (insertions always occur either before or after the identical encoded nucleotide), then either 14 edits are placed in stems and 4 in loops (edits after), or 13 in stems and 5 in loops (edits before). Either way these proportions are similar to the proportions of nucleotides in stems and loops (1,026 versus 628), and the difference is not significant (Z-tests; P = 0.17 for edits after, or 0.37 for edits before).
Sequence Conservation around Editing Sites
We also examined sequence conservation near editing sites in order to identify any motifs that may be used as a localization signal for RNA editing. For this analysis, we collected 311 60-nucleotide-long sequences near editing sites (29 nucleotides before and after two identical nucleotides including the inserted one; we excluded editing sites where two or more nucleotides identical to the inserted nucleotide are encoded on the genome), and 328 randomly selected 60-nt-long sequences as a negative control, and for each set generated a sequence logo. Apart from the enrichment of guanosine at editing sites (discussed previously), general patterns in sequence logos were similar between the control and test sets (supplementary fig. S5, Supplementary Material online). Since we only considered insertions that happened next to a single identical nucleotide, the flanking nucleotides (nucleotides right before and after the identical nucleotides at editing sites) were constrained to be low in G (and rich in less commonly inserted nucleotides) as an artifact of this site selection. No motifs were apparent when the sequence length was extended to 120 nucleotides (not shown).
Accuracy of Editing Mechanism
RNA sequence reads were compared with consensus transcript sequences to examine the accuracy of the editing mechanism. The overall “apparent error rate” (which would include any instances where editing sites have not been edited yet; see below) was 0.07% (5,397 mismatches over 7,942,887 nucleotides) for nonediting windows and 0.37% (2,416/648,826) for editing windows. Inside nonediting windows, missing nucleotide, substitution, and insertion-type errors were 0.01, 0.05, and 0.01%, respectively. At editing sites, apparent missing nucleotide-type errors were 30 times more common (0.33%), while rates of substitution-type errors (0.03%) and insertion-type errors (0.01%) were similar to those in nonediting windows.
Z-Tests indicated that only the rate of missing nucleotide-type error was different between editing and nonediting sites (P < 0.00001). The excess missing nucleotide-type errors presumably represent instances where RNA editing has not happened (rather than transcription errors) and suggest that a proportion of transcripts were not edited, or had not been edited yet (i.e., were immature pre-edited transcripts), when RNA was extracted (though see below). Nonetheless, this proportion was very small in absolute terms.
Discussion
Mitochondria Genome Evolution in Discoba
Previously reported phylogenetic analyses based on the 18S rRNA gene alone did not resolve the relative phylogenetic positions of BB2 and P. kirbyi (Harding et al. 2013). A Bayesian analysis showed BB2 as the deepest-branching member of Heterolobosea, while BB2 and P. kirbyi were sister taxa at the base of Heterolobosea in an ML analysis, but with negligible statistical support in each case (Harding et al. 2013). Our phylogenomic analyses of 252 protein-coding genes have resolved this uncertainty, clearly showing that BB2 and P. kirbyi are sister taxa at the base of Heterolobosea.
Amoeba BB2 and P. kirbyi have very similar mitochondrial gene contents that are subsets of the gene complement of the jakobid species. This suggests that BB2 and P. kirbyi do not have unusually “ancestral” (gene-rich) mitochondrial genomes, relative to other Discoba, and despite their phylogenetic position as the deepest branch within Heterolobosea, their mitochondrial genome coding capacity is relatively similar to that of Naegleria. The presence of four extra genes encoded on Naegleria mitochondrial genomes (ccmC, ccmF, tatC, and rps4), but not on the mtDNA of BB2 and P. kirbyi, or Acrasis kona, is likely the result of parallel gene losses; one set of events in the common ancestor of P. kirbyi and BB2 and a second in the lineage leading to A. kona. Interestingly, both BB2 and P. kirbyi expressed nuclear transcripts encoding holocytochrome c synthase, a key component of one of the pathways involved in the covalent attachment of heme to apocytochrome c in mitochondria that can substitute for the absence of ccmC and ccmF (Nishimura et al. 2016). In addition, both BB2 and P. kirbyi have the nucleus-encoded phage-type mitochondrial RNA polymerase (with a sequence highly similar to that of Naegleria; data not shown) instead of the bacteria-type mitochondrial RNA polymerase present in jakobids. Since Jakobida does not appear to be the earliest branching eukaryotic lineage (Derelle et al. 2015), it seems likely that the last eukaryote common ancestor had both types of RNA polymerase, and the bacterial one was lost from the Heterolobosea lineage after the split from Jakobida (Stechmann and Cavalier-Smith 2002). The absence of the bacteria-type mitochondrial RNA polymerase from the deepest branch of Heterolobosea supports the already parsimonious inference that this loss was an ancient one shared by Heterolobosea and their probable closest relatives, Euglenozoa.
Comparison of BB2 RNA Editing with Other Systems
The mitochondrial transcripts of BB2 require RNA editing to produce functional RNAs. In BB2 transcripts, mononucleotides are inserted next to one or more nucleotides of the same identity encoded by mtDNA by a very accurate and efficient RNA editing mechanism (only 0.37% apparent error rate). A somewhat similar RNA editing system occurs in paramyxoviruses; the mature P protein mRNA of paramyxoviruses is produced after insertion of one or more additional G residues next to an encoded G residue (Jacques et al. 1994). This happens by co-transcriptional polymerase “stuttering” at a homo-polymer tract (AnGn; Jacques et al. 1994). Although RNA editing in BB2 adds one additional nucleotide next to an identical encoded nucleotide, it is unlikely to use exactly the same mechanism as paramyxoviruses because BB2 pre-mRNA did not show a predominance of homo-polymer tracts around editing sites, and the error rate of RNA editing is much lower for BB2 (see below).
RNA editing in the amoebozoan Physarum shares some similar characteristics with editing in BB2 in that all four types of nucleotides are inserted, and all types of RNA (mRNA, rRNA, and tRNA) are edited (Mahendran et al. 1994; Antes et al. 1998; Bundschuh et al. 2011). However, unlike BB2 in which all nucleotides are inserted next to an identical encoded nucleotide, this is true of only some edits in Physarum. In addition, dinucleotide insertions are observed in a few editing sites in Physarum (1.73%) whereas only mononucleotide insertions occur in BB2.
In kinetoplastids, each U insertion/deletion site is specified by a guide RNA (gRNA) and editing is post-transcriptional (Horton and Landweber 2002). It is unlikely that BB2 mitochondria use the same RNA editing mechanism as kinetoplastids, based on our findings. Firstly, regarding the existence of gRNAs in BB2, the likelihood of detecting these in our transcriptomic data was low since we did not select for small RNA molecules during library construction. However, we searched for gRNA-like sequences in the nuclear and mitochondrial genome sequence datasets for BB2 using 10–30 nucleotide-long sequences around editing sites as queries, but did not detect any high-identity matches that could have suggested the presence of gRNA genes in BB2. Second, the low missing-nucleotide rate at editing sites in BB2 RNA-derived sequencing reads suggests that RNA editing is not post-transcriptional. On the contrary, editing in kinetoplastid mitochondria is relatively “error-prone”; for example, ∼50% of transcripts at a given editing sites are mis-edited or not fully edited in Perkinsela (David et al. 2015). By comparison, the co-transcriptional insertion editing of Physarum is much more “accurate”, with only 5% mis-edited transcripts in RNAs synthesized by partially purified mitochondrial transcription elongation complexes (Visomirski-Robic and Gott 1995; Byrne et al. 2002). Although approaches used to estimate error rate vary between organisms, the extremely low missing nucleotide-type error rate in BB2 (0.33%) implies that the RNA editing mechanism is unusually accurate and efficient. Also, the very low proportion of unedited transcripts suggests that RNA editing probably takes place during transcription, or very soon after.
In BB2, RNA editing is essential for generating functional tRNAs through the creation of conserved features such as the GUUC motif (which is needed for tRNA recognition), proper acceptor stems and anticodon loops. Similarly, in some myxogastrid amoeobozoans (Physarum polycephalum and Didymium nigripes), single-nucleotide insertions restore the GUUC sequence, anticodon stem, DHU stem, or acceptor stem of tRNAs (Antes et al. 1998). Editing of tRNAs in Acanthamoeba is different in that it uses base-pairing in the stems as the template for editing (Byrne et al. 2002). In BB2, the mechanism must be different since editing sites are not always in base-paired regions (either in tRNAs or in rRNA).
Phylogenetic Context of RNA Editing in the Heterolobosea
RNA editing likely arose independently in the BB2 lineage since no other heteroloboseid studied to date, including its sister lineage P. kirbyi, has this type of editing of their mitochondrial transcripts. Within Heterolobosea, A. kona and N. gruberi are known to undergo some C-to-U RNA editing of their mitochondrial transcripts (Knoop and Rudinger 2010; Fu et al. 2014), however, we found no evidence of C-to-U RNA editing, or any other type of substitution editing, in either BB2 or P. kirbyi. This type of substitution-type RNA editing is distinct from what we observed in BB2 mitochondria, where insertion-type RNA editing is extensive, and takes place in most of the protein-coding ORFs, rRNAs and many tRNAs. Just two editing sites are identified in N. gruberi and six sites in A. kona (compared to 475 sites in BB2), and these are all in protein-coding ORFs. Therefore, it is unlikely that RNA editing arose in the common ancestor of all Heterolobosea, rather these appear to be two different phenomena that have evolved independently.
Possible Mechanisms for RNA Editing in BB2 Mitochondria
RNA editing in BB2 mitochondria is the first case of insertion-type RNA editing in a heteroloboseid and, as discussed above, seems to be a novel one. The mechanism is unknown, but some speculation about the architecture of the system is possible from our data, and could be helpful in directing further research. Given that the inserted nucleotide is identical to one of its neighbors and all insertions are accurately incorporated, it is possible that the editing mechanism is a “stutter” during the transcriptional process, wherein the RNA polymerase uses the same DNA base as a template twice in a row. This simple mechanism would imply no distinct machinery for the “RNA editing” itself (though the system for recognition of editing sites could be distinct from the typical transcription elongation complex). A more complex and less likely model would invoke a separate RNA editing machinery that intervenes to reuse the template base after a stalling of the RNA polymerase near an editing site. These scenarios in which transcription and “RNA editing” are essentially co-incident are consistent with the very low frequency of unedited transcripts (which indicates that the time interval between transcription of an editing site and the RNA editing action is relatively short). On present data, it is much more challenging to propose a plausible mechanism for editing site recognition, since no sequence conservation was detected around editing sites.
Further studies will be needed to determine salient features of the RNA editing mechanism. Obvious questions include: where precisely the insertions happen (i.e., before or after the identical encoded nucleotide, noting that this is a moot point if the RNA editing mechanism is stuttering by RNA polymerase), how editing sites are specified and if RNA editing in BB2 is truly co-transcriptional. To resolve these questions, many molecular biology experiments must follow, including the development of in vitro assays using isolated mitochondria, similar to experiments which showed that RNA editing in Physarum is co-transcriptional (Visomirski-Robic and Gott 1995; Cheng et al. 2001).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Matt Brown for providing a pipeline for phylogenomic tree construction and Dr Michael Gray, Dr John Archibald and Dr Claudio Slamovits for useful discussions. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (grant number 2016-06792 to AJR) as well as the Tula Foundation and the Canadian Institute for Advanced Research (CIfAR).
Literature Cited
- Aljanabi SM, Martinez I.. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antes T, Costandy H, Mahendran R, Spottswood M, Miller D.. 1998. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol Cell Biol. 18:7521–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, et al. 1986. Major transcript of the frameshifted cox11 gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF.. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF.. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709–716. [DOI] [PubMed] [Google Scholar]
- Bundschuh R, Altmuller J, Becker C, Nurnberg P, Gott JM.. 2011. Complete characterization of the edited transcriptome of the mitochondrion of Physarum polycephalum using deep sequencing of RNA. Nucleic Acids Res. 39:6044–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J.. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comp Biol. 17:1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EM, Stout A, Gott JM.. 2002. Editing site recognition and nucleotide insertion are separable processes in Physarum mitohcondria. EMBO J. 21:6154–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YW, Visomirski-Robic LM, Gott JM.. 2001. Non-template addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 20:1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, et al. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaterigner-Boutin A, Small I.. 2011. Organellar RNA editing. WIREs RNA 2:493–506. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Jansen R, Palmer JD, Downie SR.. 1997. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet. 31:419–429. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David V, et al. 2015. Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, and endosymbiotic kinetoplastid. Mbio 6:e01498–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, et al. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci USA. 112:E693–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobakova E, Flegontov P, Skalicky T, Lukes J.. 2015. Unexpectedly streamlined mitochondrial genome of the Euglenozoan Euglena gracilis. Genome Biol Evol. 7:3358–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. [DOI] [PubMed] [Google Scholar]
- Fu C, Sheikh S, Miao W, Andersson SG, Baldauf SL.. 2014. Missing genes, multiple ORFs, and C-to-U type RNA editing in Acrasis kona (Heterolobosea, Excavata) mitochondrial DNA. Genome Biol Evol. 6:2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642. [DOI] [PubMed] [Google Scholar]
- Flegontov P, Gray MW, Burger G, Lukes J.. 2011. Gene fragmentation: a key to mitochondrial genome evolution in Euglenozoa? Curr Genet. 57:225–232. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907.
- Gott JM, Emeson RB.. 2000. Functions and mechanisms of RNA editing. Annu Rev Genet 34:499–531. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, et al. 1998. Genome structure and gene content in protest mitochondrial DNAs. Nucleic Acids Res. 26:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2003. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55:227–233. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF.. 1999. Mitochondrial evolution. Science 283:1476–1481. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G.. 2004. Mitochondria of protists. Annu Rev Genet. 38:477–524. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T, et al. 2013. Amoeba stages in the deepest branching heteroloboseans, including Pharyngomonas: evolutionary and systematic implications. Protist 164:272–286. [DOI] [PubMed] [Google Scholar]
- Harding T, Brown MW, Simpson AGB, Roger AJ.. 2016. Osmoadaptative strategy and its molecular signature in obligately halophilic heterotrophic protists. Genome Biol Evol. 8:2241–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EK, et al. 2013. The mitochondrial genome and a 60-kb nuclear DNA segment from Naegleria fowleri, the causative agent of primary amoebic meningoencephalitis. J Eukaryot Microbiol. 60:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton TL, Landweber LF.. 2002. Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr Opin Microbiol. 5:620–626. [DOI] [PubMed] [Google Scholar]
- Jacques JP, Hausmann S, Kolakofsky D.. 1994. Paramyxovirus mRNA editing leads to G deletions as well as insertions. EMBO J. 13:5496–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec, et al. 2013. Colponemids represent multiple ancient alveolate lineages. Curr Biol. 23:2546–2552. [DOI] [PubMed] [Google Scholar]
- Kamikawa R, et al. 2014. Gene content evolution in Discobid mitochondria deduced from the phylogenetic position and complete mitochondria; genome of Tsukubamonas globosa. Genome Biol Evol. 6:306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, Shiratori T, Ishida K, Miyashita H, Roger AJ.. 2016. Group II intron-mediated trans-splicing in the gene-rich mitochondrial genome of an enigmatic eukaryote, Diphylleia rotans. Genome Biol Evol. 8:458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiethega GN, Yan Y, Turcotte M, Burger G.. 2013. RNA-level unscrambling of fragmented genes in Diplonema mitochondria. RNA Biol. 10:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. 2011. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 68:567–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Rudinger M.. 2010. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer?. FEBS Lett. 584:4287–4291. [DOI] [PubMed] [Google Scholar]
- Lang BF, et al. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387:493–497. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger M, Eme L, Hug L, Roger AJ.. 2016. Novel hydrogenosomes in the microaerophilic jakobid Stygiella incarcerata. Mol Biol Evol. 33:2318–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukes J, Hashimi H, Zıkova A.. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 48:277–299. [DOI] [PubMed] [Google Scholar]
- Mahendran R, et al. 1994. Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J. 13:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marande W, Burger G.. 2007. Mitochondrial DNA as a genomic jigsaw puzzle. Science 318:415.. [DOI] [PubMed] [Google Scholar]
- Marande W, Lukes J, Burger G.. 2005. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot Cell 4:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S, Valach M, Aoulad-Aissa M, Otto C, Burger G.. 2016. Novel modes of RNA editing in mitochondria. Nucleic Acids Res. 44:4907–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ.. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, et al. 2016. Mitochondrial genome of Palpitomonas bilix: derived genome structure and ancestral system for cytochrome c maturation. Genome Biol Evol. 8:3090–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell Biol. 29:537–550. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AGB.. 2011. Characterization of Pharyngomonas kirbyi (= “Macropharyngomonas halophila” nomen nudum), a very deep-branching, obligately halophilic heterolobosean flagellate. Protist 162:691–709. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V, Shahar-Golan R, Clark CG.. 2010. A linear molecule with two large inverted repeats: the mitochondrial genome of the stramenopile Proteromonas lacertae. Genome Biol Evol. 2:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC, Ares MJ, Hannon GJ, Nilsen TW.. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 6:pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Inagaki Y, Roger AJ.. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 23:615–625. [DOI] [PubMed] [Google Scholar]
- Spencer DF, Gray MW.. 2011. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: fragmented genes in a seemingly fragmented genome. Mol Genet Genomics 285:19–31. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T.. 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297:89–91. [DOI] [PubMed] [Google Scholar]
- Tikhonenkov, et al. 2014. Description of Colponema vietnamica sp. n. and Acavomonas peruviana n. gen. n. sp., two new alveolate phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS One 9:e95467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic LM, Gott JM.. 1995. Accurate and efficient insertional RNA editing in isolated Physarum mitochondria. RNA 1:681–691. [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, Tanifuji G, Kusaka C, Takishita K, Fujikura K.. 2016. Hyper-eccentric structural genes in the mitochondrial genome of the algal parasite Hemistasia phaeocysticola. Genome Biol Evol. [Epub ahead of print]. doi: 10.1093/gbe/evw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, et al. 2013. Pentatricopeptide repeat proteins involved in plant organellar RNA editing. RNA Biol. 10:1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.