Abstract
Melanoma antigen (MAGE) genes are conserved in all eukaryotes and encode for proteins sharing a common MAGE homology domain. Although only a single MAGE gene exists in lower eukaryotes, the MAGE family rapidly expanded in eutherians and consists of more than 50 highly conserved genes in humans. A subset of MAGEs initially garnered interest as cancer biomarkers and immunotherapeutic targets due to their antigenic properties and unique expression pattern that is primary restricted to germ cells and aberrantly re-activated in various cancers. However, further investigation revealed that MAGEs not only drive tumorigenesis, but also regulate pathways essential for diverse cellular and developmental processes. Therefore, MAGEs are implicated in a broad range of diseases including neurodevelopmental, renal, and lung disorders, as well as cancer. Recent biochemical and biophysical studies indicate that MAGEs assemble with E3 RING ubiquitin ligases to form MAGE-RING ligases (MRLs) and act as regulators of ubiquitination by modulating ligase activity, substrate specification, and subcellular localization. Here, we present a comprehensive guide to MAGEs highlighting the molecular mechanisms of MRLs, their physiological roles in germ cell and neural development, oncogenic functions in cancer, and potential as therapeutic targets in disease.
Graphical abstract
OVERVIEW OF THE MAGE FAMILY
Discovery of the MAGEs
During the 1980s, researchers identified a patient, MZ-2, with stage IV amelanotic melanoma of an unknown primary tumor who had strong T cell reactivity against autologous tumor cells in culture (reviewed in [1]). Despite surgical intervention and chemotherapy, the patient never achieved a complete remission. However, following multiple vaccinations of patient-derived clones that had been mutagenized in vitro and irradiated before being injected intradermally, patient MZ-2 had a remarkable recovery. This dramatic response led researchers to undertake the monumental task of identifying the tumor-associated antigen that allowed for recognition by cytotoxic T cells (CTLs). Through the elegant application of autologous typing and transfection of a cosmid library into the patient-derived MZ2-E cell line, Boon and colleagues discovered and cloned the first human tumor antigen, melanoma antigen-1 (MAGE-1) [2]. Subsequent studies and homology searches revealed that MAGE-1, later renamed MAGE-A1, belongs to a larger family of genes that are now known as MAGEs [3].
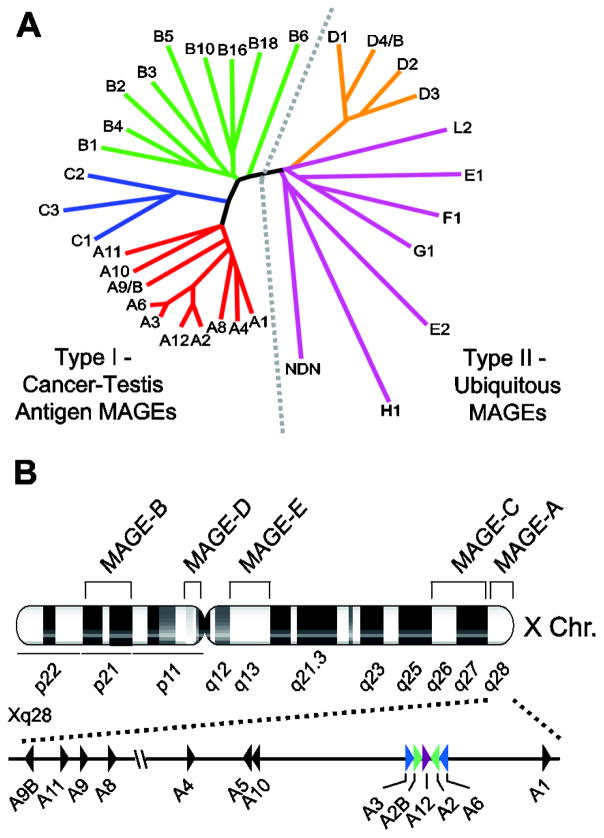
In humans, the MAGE family consists of about 60 genes (some of which are designated pseudogenes) that are categorized into two classes based on their chromosomal location and expression pattern (Figure 1A) [3, 4]. Collectively, the MAGE-A, -B, and -C subfamily members located on the X-chromosome comprise the type I MAGE cancer testis antigens (CTAs), in that the genes of all three subfamilies are primarily expressed in the testis and are aberrantly expressed in cancers [3]. Conversely, the MAGE-D, -E, -F, -G, -H, -L and Necdin genes are classified as type II MAGEs, which are not restricted to the X chromosome and are expressed in a variety of tissues [3].
Figure 1.
The MAGE family. (A) Dendrogram tree of the human MAGE protein family. Type I MAGEs include MAGE-A, -B, and -C subfamilies. Type II MAGEs include MAGE-D, -E, -F, -G, -H, -L2, and Necdin families. (B) Chromosomal locations of MAGE subfamilies on the X chromosome. The MAGE-A genes are clustered in the q28 region of the X chromosome. Triangles indicate gene orientation. Colored triangles represent the palindrome arrangement of MAGE-A genes.
Chromosome organization
Consistent with their classification as type I MAGEs, the MAGE-A genes are clustered in the q28 region of the X chromosome, the MAGE-B genes at Xp21, and the MAGE-C genes at Xq26-27 (Figure 1B) [5–11]. This distinct clustering pattern on the X chromosome is not exclusive to the type I MAGE CTAs. In fact, several CTAs, including GAGEs and NY-ESO-1, are also encoded by multi-gene families on the X chromosome [12]. Interestingly, the X chromosome contains a disproportionately high number of large, highly homologous inverted repeats that predominantly contain genes expressed in the testis [13]. It has been estimated that CTA genes constitute approximately 10% of the DNA sequence on the X chromosome, suggesting that these families are the result of gene duplications [14].
Aside from the four MAGE-D genes located at Xp11 and the three MAGE-E genes at Xq13, the remaining type II MAGEs are single-copy genes and do not exhibit clustering on the X chromosome [15]. Interestingly, MAGE-G1 (15q13.1), MAGE-L2, and Necdin (15q11.2) cluster together on chromosome 15, while MAGE-F1 is located at 3q13 and MAGE-H1 at Xp11.21.
Evolution of the MAGE genes
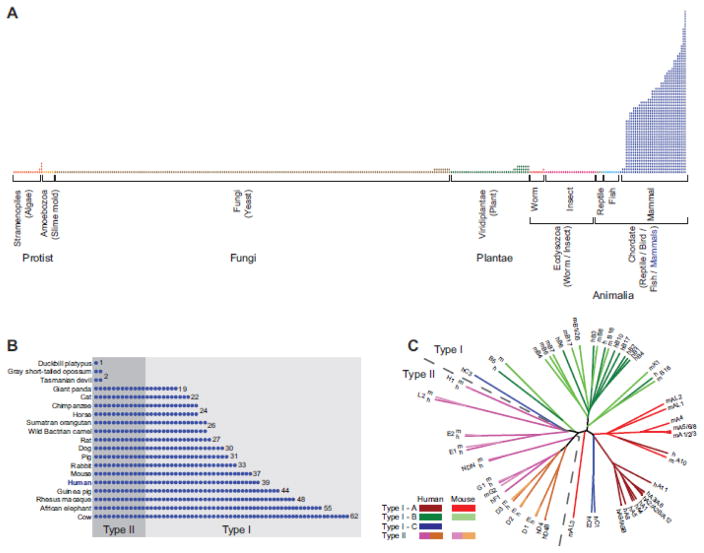
The MAGE genes are an ancient protein family that can be evolutionarily traced back to a single gene in protozoa that only recently underwent rapid expansion in placental mammals to create a multi-gene family (Figure 2) [16]. Katsura and Satta propose that the evolutionary history of the MAGE gene family can be divided into four phases [17]. In phase I, the ancestral MAGE exists as a single gene, as evidenced by the existence of only a single MAGE homolog in non-mammalian species [3, 16, 17]. Phase II is characterized by the emergence of eutherian mammals and LINE elements (Figure 2A). It is during this eutherian radiation that the subfamily ancestors were formed via retrotransposition, with the exception of MAGE-C, which was generated by gene duplication of MAGE-A [3, 17]. During phase III, gene duplications occur within the subfamilies and palindromes form in the MAGE-A subfamily (Figure 1B) [3, 17]. Finally, in phase IV, the human MAGE-A genes undergo sequence divergence, specifically in epitope-coding regions, to allow for the generation of diverse epitopes that can bind various human leukocyte antigen (HLA) class I molecules [17]. Intriguingly, this co-evolution of HLA and MAGE epitopes may have promoted functional differentiation whereby MAGEs acquired a novel human-specific role in cancer immunity in addition to their established function in germ cell development.
Figure 2.
Evolution of the MAGE genes. (A) The MAGE family is evolutionarily conserved in all eukaryotes. Following the emergence of eutherian mammals (blue), the MAGE family underwent a rapid and dramatic expansion from a single MAGE in lower eukaryotes to a large multi-gene family. Each column represents an organism with the number of circles denoting the number of MAGE proteins in each organism based on pfam annotation. (B) A detailed view of the recent expansion of MAGEs in select mammals. The type II MAGEs (designated based on the human MAGEs) are more evolutionarily ancient while the type I MAGEs appear to be the result of recent gene duplications. (C) The type II MAGEs share high homology with their mouse orthologs whereas type I MAGEs share much higher sequence conservation within their respective subfamilies compared to their mouse orthologs.
While the type I MAGEs are composed of three or four exons, with the terminal exon encoding the entire protein, many of the type II MAGEs are characterized by a single exon. The MAGE-D genes, however, have a particularly unique genomic structure. Each MAGE-D gene contains 13 exons with the open reading frame split over 11 exons, thus allowing for alternatively spliced mRNAs. Based on their unique, complex genomic structure, it has been proposed that the MAGE-D genes may be closely related to the ancestral MAGE. In addition, the gene duplication events that generated the MAGE-D genes appear to be much older than the duplication events of other subfamilies. For example, the N- and C-termini of the MAGE-D proteins are highly conserved between human and mouse orthologs, indicating that these genes evolved independently, long before the phylogenic separation of the two species [3]. However, it is important to note that the single MAGE genes found in Entamoeba histolytica and Drosophila are encoded by a single exon, and the number of introns in MAGE genes increases in the different animal phyla as they evolve [16]. Therefore, the ancestral MAGE gene was likely encoded by a single exon and it acquired introns during the course evolution.
Alternatively, others have suggested that MAGE-G1 is more functionally related to the ancestral MAGE. The yeast MAGE, Nse3, is a component of the SMC5/6 complex, which plays an essential role in homologous recombination [18–20]. Using proteomics, Taylor et al. identified MAGE-G1 as the human ortholog of yeast Nse3 and determined MAGE-G1 and its cognate RING ligase, NSE1, to be essential components of the human SMC5/6 complex [21]. In addition, the Drosophila MAGE protein shows highest sequence identity to MAGE-G1 [22]. Moreover, the chicken MAGE protein and human MAGE-G1 interact with E2F1 and the p75 neurotrophin receptor [16]. Therefore, while genomic architecture points to MAGE-D genes, functional studies suggest that MAGE-G1 may be most related to the ancestral MAGE.
Although it is not entirely clear which specific MAGE is the most evolutionarily ancient, it is clear that type II MAGEs appeared earlier than the type I MAGEs (Figure 2B). Overall, type II MAGEs share high homology with their orthologs, with at least 82% nucleotide sequence identity [23]. Conversely, the mouse Mage-a and -b genes share much higher sequence conservation within their respective subfamilies than with their human orthologs (Figure 2C) [3, 5, 24, 25]. These discrepancies, in addition to the absence of MAGE-C genes in mice, imply a more recent and rapid evolution of the type I MAGE subfamilies [3].
Zhao et al. attribute the distinct evolution of type I and type II MAGEs to differential selection acting on the two classes of genes [23]. Their statistical analyses indicate that type I MAGEs evolved under positive selection while type II MAGEs evolved under purifying or negative selection [23]. Where positive selection allows for diversification or the acquisition of additional functions for the redundant type I MAGEs, purifying selection maintains the established essential, non-redundant functions of the type II MAGEs [23].
MAGE homology domain
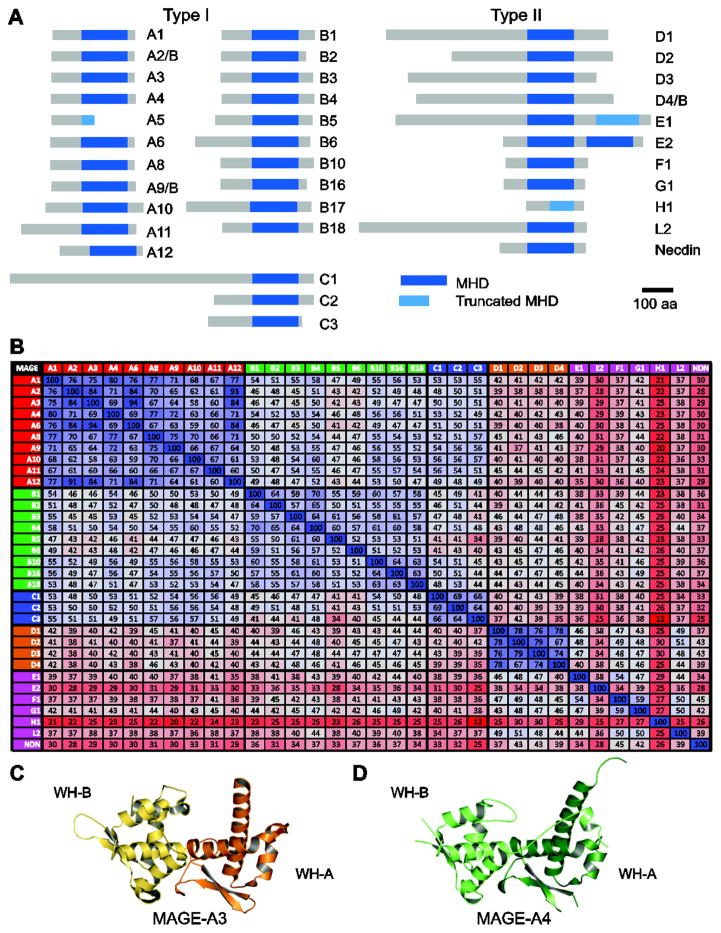
The MAGE homology domain (MHD) is a feature common to both type I and type II MAGEs (Figure 3A). The approximately 170-amino acid domain is highly conserved, such that all human MHDs share 46% protein sequence identity and most contain a conserved dileucine motif (Figure 3B) [4, 26]. The MHDs within specific subfamilies share even higher conservation; for example, the twelve MAGE-A MHDs and the four MAGE-D MHDs are 70% and 75% conserved, respectively [26].
Figure 3.
The MAGE homology domain. (A) List of human MAGE proteins and their conserved MAGE homology domain (MHD) highlighted in blue. (B) The percentage of identical amino acids between the various human MHDs is given and colored on a three-color sale (blue = high, gray = medium, red = low). Crystal structures of the MAGE-A3 (C) and MAGE-A4 (D) MHDs. The two winged-helix motifs (WH-A and WH-B) are noted.
Structural studies have revealed that the MHD consists of two tandem winged-helix (WH) motifs, referred to as WH-A and -B (Figures 3C, D) [26, 27]. Each WH features a characteristic helix-turn-helix motif packed against a three-stranded antiparallel β-sheet “wing”, however, WH-B also contains additional α-helices [26, 27]. Overall, the MAGE-A3 and -A4 MHD structures share the same relative orientation and both exhibit a peptide extension binding into the conserved cleft between the two WH motifs; however, the C-terminus of MAGE-A4 is more closely associated with the rest of the molecule and forms a longer section of α-helix [27].
Despite the sequence and structural similarities shared among MHDs, mounting evidence suggests that MHDs are more versatile and complex than one might expect. Rather than recognizing and binding a common motif, MHDs confer binding specificity to multiple unique interaction motifs [26]. In addition, biophysical interrogation of MAGE-A4 by native mass spectrometry revealed a broad charge state distribution, indicating that MAGEs are structurally dynamic proteins [28]. Therefore, the flexible MHD may undergo conformational changes that allow for interaction with distinct protein domains thereby conferring unique functions to individual MAGEs.
FUNCTION AND MECHANSIM OF MAGE-RING LIGASES
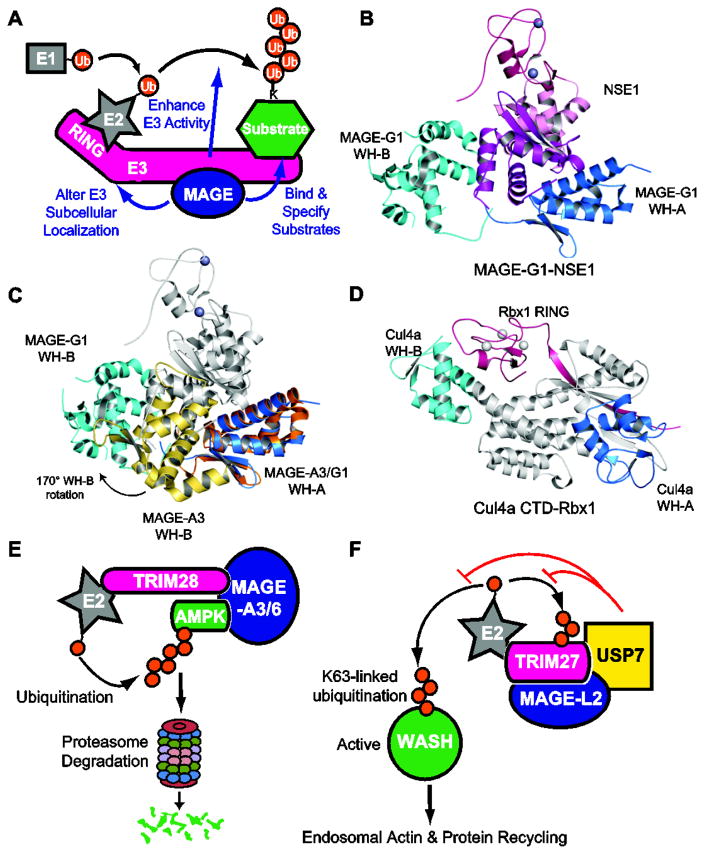
Following the initial discovery of MAGEs, the major emphasis, by far, has been on their expression in cancer. While these studies have revealed many valuable aspects regarding the prognostic and therapeutic potential of MAGEs, efforts to characterize their molecular functions in physiology and pathology are limited. However, a growing body of literature has demonstrated that MAGEs assemble with E3 RING ubiquitin ligases to form MAGE-RING ligases (MRLs) that function in a myriad of cellular processes (Figure 4A). Interaction studies including targeted and global proteomics have reported more than 50 distinct MRLs, including MAGE-A1-TRIM31, MAGE-A2-TRIM28, MAGE-A3-TRIM28, MAGE-A6-TRIM28, MAGE-B18-LNX1, MAGE-C2-TRIM28, MAGE-D1-PRAJA-1, MAGE-G1-NSE1, and MAGE-L2-TRIM27, that have been studied to various degrees. Within these complexes, MAGEs have been shown to regulate their cognate RING proteins by (1) enhancing ligase activity, (2) specifying novel substrates for ubiquitination, and (3) altering subcellular location. In the sections below, we discuss in detail specific MRL complexes and the mechanisms by which they function.
Figure 4.
Structure and function of MAGE-RING ligases. (A) Schematic representing biochemical and cellular functions of MAGE-RING ligases (MRLs). (B) Crystal structure of MAGE-G1-NSE1. (C) Alignment of MAGE-G1-NSE1 and MAGE-A3 based on WH-A. The orientation of the WH motifs differs between unbound MAGE-A3 and NSE1-bound MAGE-G1. (D) Crystal structure of Cul4A C-terminal domain (CTD)-Rbx1 (PDB: 2HYE) shares structural features similar to MAGE-G1-NSE1. (E) Model of MAGE-A3/6-TRIM28 ubiquitination and degradation of the AMPK tumor suppressor. MAGE-A3/6 directly bind AMPK and recruit it to the TRIM28 ubiquitin ligase for ubiquitination and subsequent proteasome-mediated degradation. (F) Model of MAGE-L2-TRIM27, USP7, and K63-ubiquitin regulation of endosomal protein recycling. MAGE-L2-TRIM27-USP7 form stable complex where TRIM27 mediates non-degradative K63-linked ubiquitination of WASH. This ubiquitination events leads to WASH activation, generation of F-actin accumulation, and recycling of proteins through the retromer pathway. USP7 functions to fine-tune WASH ubiquitination to allow for precise levels of endosomal F-actin and stabilize the complex through preventing TRIM27 auto-ubiquitination induced degradation.
Ubiquitination and RING ligases
Ubiquitination is the covalent post-translational modification of lysines on substrate proteins with the small 76 amino acid ubiquitin protein and regulates nearly all aspects of cell function. The most well characterized function of ubiquitination is the targeting of proteins for proteasomal degradation; however, ubiquitination also regulates proteasome-independent processes such as endocytosis and lysosomal targeting, nuclear export, DNA repair, and activation of kinases and transcription factors depending on the specific type of ubiquitin chain linkage (reviewed in [29]). In the ubiquitination cascade, an E1 ubiquitin-activating enzyme activates ubiquitin; this activated ubiquitin is transferred to an E2 ubiquitin-conjugating enzyme, and is subsequently ligated to a substrate via an E3 ubiquitin ligase (Figure 4A) [30]. Therefore, by providing substrate specificity in the ubiquitin pathway, E3 ligases play a critical regulatory role in various cellular pathways.
The majority of known E3 ubiquitin ligases include the RING domain and RING-like proteins, which mediate substrate recognition and subsequent ubiquitin ligation through activation of the E2 enzyme [29]. The RING domain contains a conserved cysteine- and histidine-rich consensus sequence that coordinates two zinc ions via a cross-brace arrangement [30]. Interestingly, many RING ligases have been implicated in cancer due to their roles in the maintenance of genomic integrity and cellular homeostasis (reviewed in [31]).
MAGE-RING ligase architecture
Upon determining that MAGE family proteins form complexes with E3 RING ubiquitin ligases, our lab sought to characterize the biochemical and biophysical properties of MRLs. By in vitro binding assays, we demonstrated that each MAGE generally binds one specific RING ligase, and highly homologous MAGEs tend to bind the same RING protein [26]. Subsequent mapping of the minimal regions required for MRL complex formation revealed that MAGEs bind their cognate RING ligases via the MHD; however, the region on which MAGEs bind RING ligases varies between different MRLs [26]. For example, MAGE-C2 binds the coiled-coil region of TRIM28, MAGE-B18 binds a basic region between the RING and first PDZ domain of LNX1, and MAGE-G1 binds the WH motifs of NSE1 [26]. These findings further validate the notion that the MHD is flexible and its conformational plasticity allows for unique and complex interactions with specific RING ligases.
In an effort to gain insights into the structural properties of MRLs, we also determined the crystal structure of MAGE-G1 in complex with its cognate NSE1 RING ligase (Figure 4B) [26]. Consistent with the in vitro domain mapping experiments, MAGE-G1 WH-A interacts with both WH motifs of NSE1 via a series of hydrogen bonds and a large hydrophobic interface that includes the conserved dileucine motif [26]. In addition, mutation of the dileucine motif disrupts binding of not only MAGE-G1 and NSE1, but also complex formation of other MRLs, suggesting that these conserved residues play an important role in the binding interface [26].
Intriguingly, while the WH-A and WH-B motifs of NSE1-bound MAGE-G1 and free MAGE-A4 are similar, with rmsd values of 1.05Å and 1.07Å, respectively, their relative orientations are very distinct in the two structures (Figure 4C) [26, 27]. The WH motifs in MAGE-G1 are distantly separated, such that WH-B rotates approximately 170° and translates about 30Å relative to WH-A [26, 27]. Moreover, the peptide extension bound between the two WH motifs in the MAGE-A3 and -A4 structures is not present in the MAGE-G1-NSE1 structure [26, 27]. Therefore, the two different conformational states demonstrate that the MHD undergoes extensive rearrangement for MRL complex formation and the structural changes required to accommodate association with RING ligases may confer binding specificity.
Structurally, MRLs share a number of key features with Cullin-RING ligases (CRLs) (Figure 4D). CRLs, the largest family of multi-component E3s, generally consist of four subunits: cullins (CUL1-7), RINGs (Rbx1-2), adaptor proteins, and substrate recognition proteins (reviewed in [32]). The cullin acts as the core molecular scaffold that binds to an adaptor protein and a substrate receptor protein at the N-terminus and a RING protein at the C-terminus [33]. Like MRLs, these modular CRLs can assemble with multiple substrate recognition proteins to recruit various unique substrates [32]. In addition, the cullin C-terminal domain (CTD) shares marked structural similarities to MAGEs, in that two WH motifs contribute to a groove where the RING domain binds [33]. Interestingly, this region of the cullin CTD, referred to as the cullin homology region, is conserved in several other proteins including the APC2 subunit of the anaphase-promoting complex/cyclosome (APC/c) E3 ligase, suggesting a conserved function that also extends to MRLs [33–36].
MAGEs enhance E3 RING ubiquitin ligase activity
One of the earliest indications that MAGEs form complexes with E3 RING ubiquitin ligases came from the discovery that MAGE-A2, -A3, -A6, and -C2 directly bind and regulate the TRIM28 (also referred to as KAP1) E3 ubiquitin ligase, a multi-functional protein implicated in transcriptional regulation, cellular differentiation, and DNA damage repair [26, 37]. Biochemical analysis of TRIM28 ubiquitin ligase activity revealed that these MAGEs stimulate both TRIM28 auto-ubiquitination and ubiquitination of its substrates, the p53 tumor suppressor and ZNF382, in vitro and in cells [26, 38, 39]. Moreover, the enhanced ubiquitin ligase activity of MAGE-TRIM28 reduced p53 and ZNF382 protein levels in a proteasome-dependent manner [26, 38, 39]. In addition to MAGEs regulating the TRIM28 ligase, MAGE-A1 has been reported to stimulate the ligase activity of TRIM31 [40]. Furthermore, this ability of MAGEs to enhance E3 ubiquitin ligase activity is not limited to the type I MAGEs. NSE1 normally has weak in vitro ubiquitin ligase activity in the presence of the E2 ubiquitin-conjugating enzyme, UbcH13/Mms2; however, upon addition of MAGE-G1, NSE1 activity is significantly enhanced [18, 26]. Therefore, the ability to enhance E3 RING ubiquitin ligases is a feature common to both type I and type II MAGEs.
To gain a better understanding of how MAGEs enhance the ligase activity, we tested four possible mechanisms whereby MAGEs (1) induce a conformational change in the E3 RING ligase, thus promoting increased activity, (2) promote substrate binding to the E2-E3 ubiquitin ligase machinery, (3) stimulate charging of the E2 ubiquitin-conjugating enzyme by the ubiquitin E1, or (4) bind and recruit E2 ubiquitin-conjugating enzymes to the E3 substrate complex. However, we found that MAGE-G1 did not alter NSE1 conformation, neither MAGE-A2 nor -C2 enhanced p53 binding to TRIM28, and MAGE-C2 did not affect UbcH2 charging [26]. Interestingly, we demonstrated that MAGE-A2/C2 specifically bind the UbcH2 E2 ubiquitin-conjugating enzyme, suggesting that MAGEs may enhance ubiquitin ligase activity by recruiting and/or stabilizing the E2 enzyme at the E3-substrate complex [26].
Depending on whether MAGE-A2/C2 can bind UbcH2 and TRIM28 simultaneously, or if the two binding interactions are mutually exclusive, Feng and colleagues propose two mechanistic models by which these MAGEs facilitate TRIM28 E3 ligase activity [41]. In the first model, MAGE-A2/C2 binding to TRIM28 and UbcH2 are mutually exclusive. After transferring one ubiquitin to the substrate, UbcH2 is recharged by an E1 ubiquitin-activating enzyme while remaining in close proximity to the TRIM28 machinery via interactions with MAGE-A2/C2. In this way, MAGEs promote the on-site recharging of the E2 enzyme. In the second model, MAGE-A2/C2 binds to TRIM28 and UbcH2 at the same time. Here, two UbcH2 molecules are recruited to the TRIM28 machinery—with one UbcH2 interacting with the TRIM28 RING domain and the other with MAGE-A2/C2—to promote the sequential assembly of a polyubiquitin chain on the substrate [41]. However, whether MAGE-A2/C2 can bind TRIM28 and UbcH2 simultaneously remains unclear and further work must be done in order to validate these proposed models, as well as determine whether additional MAGEs bind to their cognate E2 enzymes.
Regulation of AMPK by MAGE-A3/6-TRIM28
MAGE-A3 and the highly similar protein MAGE-A6 (referred to as MAGE-A3/6) also bind TRIM28 [26]. However, expression of MAGE-A3/6 does not inversely correlate with p53 mutational status, suggesting this MRL may have additional targets relevant to its function in cancer cells [42]. In an in vitro screen to identify direct substrates of MAGE-A3/6-TRIM28, we found that expression of MAGE-A3/6 enhances ubiquitination of AMPKα1 (Figure 4D) [42]. Importantly, MAGE-A3/6 not only promotes ubiquitination and subsequent proteasomal degradation of AMPKα1, but also directly interacts with and specifies AMPKα1 as a substrate for TRIM28 [42]. Thus, unlike p53, which can be targeted by TRIM28 in the absence of MAGEs, AMPKα1 is only targeted by TRIM28 in the presence of MAGE-A3/6. Therefore, expression of MAGE-A3/6 in cancer cells reprograms the ubiquitous TRIM28 ubiquitin ligase to degrade a key metabolic regulator and tumor suppressor to enhance tumorigenesis.
AMPKα1 is the catalytic subunit of AMP-activated protein kinase (AMPK) heterotrimer, a crucial energy sensor in cells [43]. In response to even modest decreases in ATP production, AMPK is activated and promotes catabolic ATP-generating pathways while also inhibiting anabolic ATP-consuming pathways, such as mTOR signaling to maintain energy homeostasis [43]. In this way, AMPK and mTOR signaling play opposing functions in the regulation of autophagy, a degradative process important for balancing energy sources during development and in response to nutrient stress [43]. Due to its role as a master regulator of cellular energy, AMPK functions as a critical tumor suppressor to stop cell growth and its activity is often perturbed in various diseases such as cancer.
Further investigation into the functional consequences of AMPK regulation by MAGE-A3/6TRIM28 revealed increased glucose consumption and lactate production upon TRIM28 knockdown, suggesting that the inhibition of AMPK by the MRL affects cell metabolism [42]. In addition, we found that MAGE-A3/6-TRIM28 is critical for the maintenance of mTOR activity [42]. Consistent with the role of MAGE-A3/6 in regulating AMPK and mTOR signaling, we demonstrated that the MAGE-A3/6-TRIM28 MRL inhibits autophagy [42, 44].
Taken together, these results suggest that the oncogenic MAGE-A3/6-TRIM28 MRL regulates several cellular metabolic regulatory pathways via ubiquitination and degradation of AMPKα1 [42]. Interestingly, breast invasive carcinoma, colon adenocarcinoma, and lung squamous cell carcinoma tumors expressing MAGE-A3/6 have significantly reduced total and active AMPKα protein levels and reduced downstream AMPK signaling, indicating that this regulation of AMPK by MAGE-A3/A6-TRIM28 is relevant in human tumors [42]. However, further work will be necessary to examine whether MAGE-A3/6-TRIM28 suppression of autophagy is important for its oncogenic activity and whether MAGE-A3/6-TRIM28 regulates additional AMPK cellular responses. In addition, how this function relates to the physiological role of MAGE-A3/6 in the testis remains unclear. Given that germ cells in the testis change carbon energy sources as they differentiate from spermatogonial stem cells to mature spermatids, it will be intriguing to determine if MAGE-A3/6-mediated AMPK regulation contributes to this metabolic switch during spermatogenesis [45].
Regulation of WASH-mediated endosomal protein trafficking by MAGE-L2-TRIM27
Endosomal protein trafficking is an essential process that allows for the delivery of membrane components, receptor-associated ligands, and solute molecules to various intracellular destinations such as the lysosome for degradation, the cell surface, or the trans-Golgi network (TGN) [46]. The primary function of the retromer complex is to select cargo proteins for endosome-to-Golgi transport, or retrograde transport. In addition to cargo recognition, the retromer also recruits the WASH complex, which promotes actin filament (F-actin) nucleation by the Arp2/3 complex [47, 48]. The formation of actin patches plays a critical role in endosomal sorting by generating discrete domains into which specific proteins are sorted for transport to their respective destinations [46, 49].
Our lab found that MAGE-L2 interacts with the TRIM27 E3 RING ubiquitin ligase and localizes to retromer-positive endosomes through interactions between MAGE-L2 and the VPS35 component of the retromer complex (Figure 4E) [50]. Importantly, the ubiquitin ligase activity of MAGE-L2-TRIM27 is required for proper recycling of endosomal proteins through the retromer pathway to the TGN or plasma membrane [50]. Detailed analysis of the retromer pathway demonstrated that MAGE-L2-TRIM27 is required for WASH-mediated F-actin assembly on endosomes. Additionally, more in-depth studies revealed that MAGE-L2-TRIM27, in conjunction with the Ube2O E2 ubiquitin-conjugating enzyme, facilitates the non-degradative K63-linked ubiquitination of WASH [50]. Interestingly, additional mechanistic studies revealed that this ubiquitination of WASH on lysine 220 (K220) by MAGE-L2-TRIM27 destabilizes auto-inhibitory contacts in the WASH complex, thus allowing for its activation and F-actin assembly on endosomes and recycling through the retromer pathway [50]
In a subsequent study, we made the surprising discovery that the USP7 deubiquitinating enzyme (DUB) is an integral component of the MAGE-L2-TRIM27 MRL complex (Figure 4E) [51]. Through in vitro binding experiments, we found that USP7 directly binds both MAGE-L2 and TRIM27 to form an intricate and stable protein complex. In the trimeric complex, MAGE-L2 binds the USP7 N-terminal TRAF domain as well as the C-terminal HUBL1-3 regulatory domains, whereas the C-terminal domains of TRIM27 interact with the catalytic domain of USP7 [51]. Although previous examples of DUBs regulating ligases have been described, the intricate and obligate nature of USP7 for stable complex formation of MAGE-L2-TRIM27 suggests an important linkage between conjugating (TRIM27) and deconjugating (USP7) enzymes for proper cellular function.
Functional interrogation of the complex demonstrated that USP7 knockdown or disruption of the MAGE-L2-USP7 interaction impaired endosomal actin accumulation and protein recycling, indicating that USP7 acts in concert with MAGE-L2-TRIM27 and is required for regulation of WASH-mediated protein trafficking [51]. Interestingly, USP7 performs dual functions in the endosomal protein-recycling pathway: (1) deubiquitination of TRIM27 to protect TRIM27 from auto-ubiquitination-induced degradation and (2) deubiquitination of WASH to precisely regulate WASH activity [51]. Through these seemingly opposing activities, USP7 serves as molecular rheostat to fine tune endosomal F-actin levels [51]. This buffering capacity of USP7 is critical given that too little or too much F-actin on endosomes can both be detrimental to retromer-mediated recycling [51]. Thus, it is no surprise that nature has elegantly linked the conjugation and deconjugation machinery in a single complex to allow precise control of WASH ubiquitination and activity. Additional studies will be necessary to determine if MAGE-L2 regulates TRIM27 and USP7 enzymatic functions in addition to acting as a molecular scaffold in the complex and mediating localization to endosomes. Importantly, these findings open up the exciting possibility that the functional cooperativity observed between USP7 and MAGE-L2-TRIM27 may be conserved among other MRLs and DUBs.
Regulation of cyclins by MAGEs and SCF
In contrast to the majority of reports on MRLs, recently two MAGEs have been shown to associate with and regulate CRLs. In the first case, MAGE-C2 was identified as a component of the SCF CRL through interactions with Rbx1 E3 RING ubiquitin ligase [52]. Interestingly, unlike many of the MRL examples described previously, MAGE-C2 stabilized cyclin E by inhibiting SCF-dependent ubiquitination and subsequent proteasomal degradation [52]. This MAGE-C2-mediated stabilization of cyclin E, an essential regulator of cell cycle transition from G1 to S phase, promotes cell cycle progression and cell proliferation [52]. Importantly, cyclin E expression positively correlates with MAGE-C2 expression in melanoma tumor samples, suggesting that this newly identified function of MAGE-C2 may be relevant in tumorigenesis [52].
A subsequent study found that MAGE-A11 interacts with Skp2, an F-box domain substrate recognition protein of the SCF CRL [53]. In this case, MAGE-A11 regulates substrate specificity of Skp2, such that MAGE-A11 enhances Skp2-mediated degradation of cyclin A and the retinoblastoma-related protein p130, but inhibits Skp2-mediated degradation of the E2F1 transcription factor [53]. The authors account the differential effects of MAGE-A11 on Skp2 by proposing a competitive relationship between MAGE-A11 and Skp2 in binding cyclin A [53]. Collectively these data suggest that MAGEs may regulate cell cycle progression by modulating SCF ubiquitin ligase activity and substrate recognition.
Regulation of transcription
Intriguingly, a number of MAGEs have been implicated in the regulation of various transcription factors. Although some of the means by which MRLs regulate p53 and E2F1 are described in earlier sections, here we highlight additional mechanisms of MAGE-mediated transcriptional regulation.
p53 is a transcription factor that responds to a variety of stress signals and coordinates a gene expression program that contributes to tumor suppression. In addition to modulating p53 stability through MAGE-TRIM28-induced ubiquitination and proteasome-dependent degradation, several other roles for MAGEs in regulating p53 have been reported. For example, MAGE-A2 may sterically occlude the p53 DNA-binding domain, recruit histone deacetylases (HDACs), or inhibit the MDM2 E3 ligase to repress p53 transcriptional activity [54–56].
E2F transcription factors are key regulators of cell cycle progression and E2F1 is essential for the transactivation of target genes involved in the G1/S transition [57]. The retinoblastoma (Rb) tumor suppressor is a critical regulator of E2F1 transcriptional activity [57]. During G1 phase, Rb is in a hypophosphorylated state and binds to the transactivating domain of E2F1, thereby repressing E2F1-depedent transcription [57]. As cells progress toward S phase, cyclin-dependent kinases phosphorylate Rb and E2F1 is released [57]. E2F1 is then free to transactivate cell cycle progression genes [57]. In the same way, Necdin and MAGE-G1 bind to the transactivation domain of E2F1 and repress E2F1 transcriptional activity [58].
In contrast, investigation into the mechanisms of MAGE-A11 function demonstrated that MAGE-A11 stabilizes the retinoblastoma-related protein p107 and promotes p107 binding to E2F1, thus activating E2F1 transcriptional activity [59]. Recent work by Peche et al. also showed that MAGE-B2 interacts with HDAC1, an E2F1 repressor, to enhance E2F1 transactivation [60]. Therefore, whereas some type II MAGEs target and inhibit E2F function, other type I MAGEs may stimulate E2F activity to promote tumor cell proliferation.
The androgen receptor (AR), a member of the nuclear receptor superfamily, is a transcriptional regulator that responds to androgens and is particularly important for the growth and progression of prostate cancer [61]. Therefore, androgen deprivation therapy is often the first-line of treatment for patients with prostate cancer [61]. However, over time, prostate cancer cells can develop resistance to androgen deprivation, this type of relapse is referred to as castration-recurrent prostate cancer and is associated with high mortality [61].
MAGE-A11 was first identified as an AR coactivator by a yeast two-hybrid screen of a human testis library [62]. Additional studies revealed that MAGE-A11 binds the AR N-terminal FXXLF motif, thereby recruiting the steroid receptor coactivator (SRC)/p160 coactivators and promoting AR transcriptional activity [63]. However, by directly interacting with the p160 coactivator transcriptional mediator protein (TIF2) and the transcriptional regulator p300, MAGE-A11 is also able to enhance AR-mediated gene activation [64]. Subsequent studies demonstrated that epidermal growth factor (EGF), in the presence of dihydrotestosterone, stabilizes the MAGE-A11-AR complex through phosphorylation of MAGE-A11 at threonine 360 and ubiquitination of lysine residues 240 and 245 [65]. Interestingly, during androgen deprivation therapy, MAGE-A11 levels increase in prostate cancer, suggesting that MAGE-A11 plays a key role in the progression of castration-recurrent prostate cancer by enhancing AR transcriptional activity [66].
Summary of MRL function
Since the discovery that MAGEs function in complex with E3 RING ubiquitin ligases, they have been shown to regulate not only E3 enzymatic activity, but also substrate recognition and cellular localization. These MRLs act on a diverse array of cellular pathways that have clear implications in various pathologies associated with MAGEs, including transcription, metabolism, protein trafficking, and cell proliferation. However, these mechanistic studies are small in number and further work to determine the functions of additional MAGE members as well as their direct contributions to disease will be invaluable in our understanding of the MAGE family of proteins. In addition, important challenges for the future include identifying the E3 RING ubiquitin ligases that function with orphan MAGEs and discovering the substrates of novel and known MRLs, such as MAGE-A1-TRIM31, MAGE-B18-LNX1, and MAGEG1-NSE1.
PHYSIOLOGICAL EXPRESSION & FUNCTION
Type I MAGEs and germ cell development
Many of the type I MAGEs, consistent with their classification as CTAs, are normally expressed only in germ cells and/or placenta. However, due to the high homology between subfamily members and lack of specific antibodies, determination of the exact spatiotemporal expression profiles of these proteins is not trivial. Initial characterization by RT-PCR revealed that MAGE-A1-A4, -A6, and -A12 are expressed in testis; and MAGE-A4 as well as MAGE-A8-A11 are also expressed in placenta [5]. Like the MAGE-A subfamily, the MAGE-B and MAGE-C genes are also expressed only in testis, with MAGE-B2 expressed in both testis and placenta [3, 8, 11, 67]. This restricted expression to testis suggests a functional role for type I MAGEs in germ cell development.
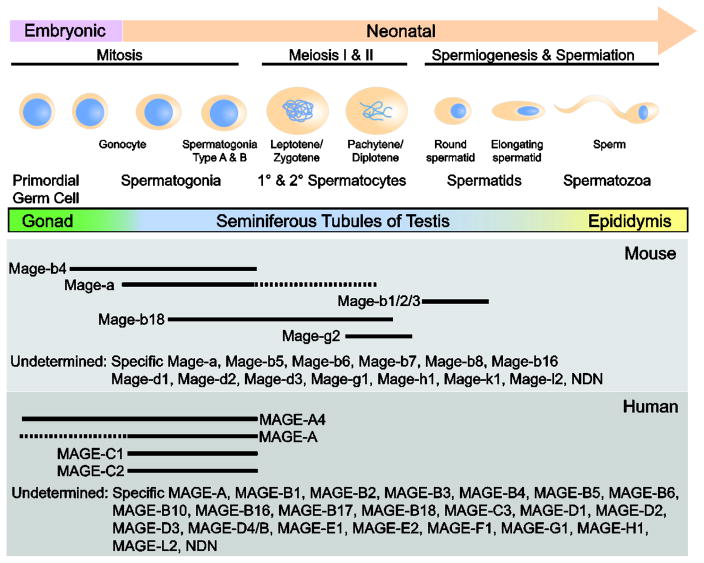
In males, the primordial germ cells (PGCs), the progenitor cells of gametogenesis, are surrounded by somatic Sertoli cells and become prospermatogonia, which proliferate for a few days and then arrest at G0/G1 until birth (Figure 5) [68]. At puberty, proliferation resumes to initiate spermatogenesis. Spermatogonia are the germ cells of spermatogenesis that remain proliferative throughout life to maintain the pool of stem cells, or undergo differentiation to produce spermatozoa. This process occurs through two meiotic divisions, in which tetraploid primary spermatocytes undergo meiosis I to form diploid secondary spermatocytes, which then undergo meiosis II to form haploid spermatids that develop into spermatozoa.
Figure 5.
MAGEs are differentially expressed during spermatogenesis. Schematic diagram of germ cell development and where specific MAGE genes are expressed. The expression profiles of most MAGE genes are unknown.
Early immunohistochemical staining demonstrated MAGE-A1 and -A4 expression in spermatogonia and primary spermatocytes, but not in spermatids or Sertoli cells of adult testes [69]. Subsequent studies using a different antibody that reacts with MAGE-A1, -A3, -A4, -A6, and -A12 detected MAGE-A expression in migrating primordial germ cells in 5-week-old human embryo as well as in the nuclei and cytoplasm of spermatogonia and spermatocytes in adult testes [70, 71]. Like the human MAGE genes, the murine Mage-a genes are expressed in spermatogonia undergoing maturation toward the spermatocyte stage [72, 73]. These findings indicate that the expression of these proteins is highly regulated and could play an active role in spermatogenesis. Consistent with this notion, a recent mouse model with deletion of Mage-a1, -a2, -a3, -a5, -a6, and -a8 exhibited reduced size of the testes and diameter of seminiferous tubules [74]. Furthermore, deletion of the Mage-a cluster led to an increase in apoptotic germ cells, primarily in the first wave of testicular apoptosis, as well as activation of p53 and induction of Bax in response to genotoxic stress [74]. Moreover, mutations in MAGE-A9B, -C1, and -C3 were identified in a cohort of infertile men [75, 76]. Collectively, these results support the hypothesis that these proteins play a critical role in germ cell development.
Interestingly, Mage-b4, like Mage-a genes, is preferentially expressed in spermatogonia, whereas Mage-b1 and -b2 are found in postmeiotic spermatids [25, 72, 73]. In male germ cells, Mage-b4 is preferentially expressed during cell cycle arrest. When cells resume mitosis and enter meiosis, Mage-b4 protein levels decrease and are hardly detectable in pachytene cells, suggesting that Mage-b4 may be important for cell cycle arrest of male germ cells. This differential expression of MAGEs led investigators to hypothesize that Mage-a and Mage-b4 might be involved in germ cell differentiation while Mage-b1 and -b2 regulate spermiogenesis.
In female germ cells, MAGE-A1 is expressed in the human oogonia prenatally and MAGE-A4 is expressed in some migrating PGCs and early oogonia in female human embryos [77, 78].
In addition, Mage-b4 is expressed in premeiotic germ cells and during the pachytene and telophase portions of meiosis, suggesting that MAGEs might also function in developing oocytes [25]. In addition to their role in germ cells, MAGE-A proteins may also be involved in neuronal development. By immunohistochemistry, MAGE-A reactivity was detected in the spinal cord and brain stem of the early developing CNS as well as in peripheral nerves. Investigators also report MAGE-A-positive PGCs in the adrenal cortex of early fetuses [71]. Therefore, MAGE-A might also function during embryonic development. However, additional mouse models and mechanistic studies are required to demonstrate the functional relevance of MAGEs in germ cells and to determine how their physiological roles may be co-opted in the context of cancer.
Type II MAGEs and neural development
Type II MAGEs include the MAGE-D, -E, -F, -G, -H, -L and Necdin genes. In contrast to the type I MAGEs, type II MAGE genes are ubiquitously expressed at various levels in many tissues. Intriguingly, a number of these type II MAGEs are enriched in the brain and have been implicated in various neural processes.
MAGE-D1, also referred to as NRAGE or Dlxin, is a type II MAGE that has been implicated in multiple pathways including apoptosis, cell cycle progression, and differentiation [4]. Initial analysis showed that MAGE-D1 is highly expressed in the brain, but is also detected in most embryonic and adult tissues [3]. Additional investigation into MAGE-D expression showed that members of this subfamily are widely expressed throughout the human adult brain, with strongest signals in the cerebral cortex and medulla [79]. Based on the widespread distribution of MAGE-D genes in the brain, it is likely that this MAGE subfamily plays a general role in neural differentiation and maintenance. Interestingly, the MAGE-D genes are located in a chromosomal region associated with many monogenic X-linked neurodevelopmental disorders [80]. Therefore, the enrichment of MAGE-D in the cerebral cortex and hippocampus—structures involved in higher function—suggests that these genes can be candidates for such disorders.
Consistent with this, Mage-d1 knockout mice demonstrate symptoms of depression such as decreased locomotor activity, social interaction, and reward responsiveness, as well as increased anxiety and immobility time; and treatment with antidepressants attenuated some of these behavioral changes related to depression [81]. Notably, Mage-d1-null mice display decreased extracellular serotonin levels and increased serotonin transporter (SERT) protein levels, suggesting that deficiency in MAGE-D1 induces both behavioral and neurological phenotypes of depression [81]. Further investigation revealed that MAGE-D1 regulates ubiquitination and proteasomal degradation of SERT, consistent with previous reports associating MAGE-D1 with PRAJA-1 E3 ligase and modulation of Msx2- and Dlx5-dependent transcription as well as neuronal differentiation [81–83]. In an alternative Mage-d1-deficient (hemizygous) mouse model, loss of Mage-d1 results in reduced social interactions, decreased sexual activity leading to infertility in males, reduced motor activity, late-onset obesity associated with hyperphagia, and increased anxiety-like behaviors [84]. Several of these phenotypes can be explained by significantly reduced levels of mature oxytocin in the brain of Mage-d1-deficient mice; and suggest that the combined effects of reduced SERT and oxytocin can contribute to an altered serotonergic system as well as the observed phenotypes in the Mage-d1 knockout model [81, 84].
Intriguingly, several of the phenotypes reported in the Mage-d1-deficient mice, including hyperphagia and reduced sociability, mimic symptoms of individuals with Prader-Willi Syndrome (PWS) and autism spectrum disorder (ASD). In fact, one of the earliest insights into the physiological function for MAGE genes came from the genetic analysis of PWS, which showed that individuals with PWS bear deletions or mutations within a specific chromosomal region containing NECDIN and MAGEL2 and with MAGE-G1 in close proximity [85, 86].
Necdin mRNA is ubiquitously expressed and is detected in all developing neurons in the central and peripheral nervous systems in early development [87, 88]. After E13, Necdin is enriched in discrete regions of the nervous system, such as the hypothalamus, thalamus, and pons; suggesting a specific spatial and temporal function therein [89]. Like MAGE-D1, various studies have implicated Necdin in neuronal differentiation and survival [90–92].
In two independent Necdin-deficient mouse models, mutant mice exhibited respiratory distress and death [87, 93]. Strikingly, individuals with PWS exhibit similar breathing defects with irregular rhythm and often manifest sleep apneas [94]. Investigation into the observed respiratory failure in mice demonstrated that Necdin is expressed in medullary serotonergic neurons and Necdin deficiency alters the serotonergic metabolism, thereby contributing to abnormal respiratory rhythmogenesis [94, 95].
Although the surviving Necdin-deficient mice showed a normal growth pattern, the mutant mice exhibited altered behavioral phenotypes reminiscent of PWS patients, such as increased skin scraping and enhanced spatial learning and memory [87]. In addition, immunohistochemistry revealed a significant decrease in the number of oxytocin- and luteinizing hormone-releasing hormone (LHRH)-producing neurons in the hypothalamus—the primary region of the brain involved in PWS.
MAGE-L2 is widely expressed in fetal tissues and is enriched in various parts of the brain [96]. Unlike its human counterpart, mouse Magel2 is almost exclusively expressed in the hypothalamus and peaks during neurogenesis during E15–17; however, it is also detected in non-neuronal tissues such as the genital tubercle, midgut region, and placenta [51, 96].
Loss of MAGE-L2 is also implicated in neurodevelopmental disorders and multiple mouse models have been generated to determine its physiological function. Magel2-null mice, generated by lacZ knock-in allele, exhibited 10% postnatal lethality, normal birth weights, and slightly decreased food intake [97]. However, close observation of their growth pattern revealed that the Magel2-deficient mice displayed a two-phase weight curve as seen in PWS patients; such that mutant mice showed delayed growth and decreased weight gain in the first four weeks of life, followed by increased weight gain, higher fat mass, and elevated leptin, insulin, and cholesterol levels [98]. It was later discovered that Magel2-null mice have reduced muscle mass and increased expression of atrophy genes, indicating that loss of Magel2 contributes to hypotonia and related musculoskeletal abnormalities such as scoliosis and digital contractures [99].
Consistent with PWS phenotypes as well as cellular evidence that MAGE-L2 modulates the activity of circadian rhythm proteins, Magel2-null mice exhibit reduced daytime activity and disrupted circadian regulation [97, 100, 101]. In addition, the amounts of orexins, the neuropeptides that regulate wakefulness, orexin-positive neurons, and orexin-2 receptors were all reduced in the hypothalamus of Magel2-deficient mice [97]. Taken together, these data suggest that Magel2 is required for proper hypothalamic function and maintains circadian rhythm potentially though the regulation of orexin levels.
Additional behavioral assays reported increased anxiety-like behavior in Magel2-null mice. MRI analysis found that regions of the brain with moderate to high Magel2, such as the amygdala, hippocampus, and the nucleus accumbens, but not the hypothalamus, were significantly smaller in Magel2-deficient mice [102]. In addition to reduced brain volume, the Magel2-mutant mice have reduced serotonin and its metabolite 5-HIAA as well as reduced dopamine. Both serotonergic and dopaminergic pathways are implicated in various neurobehavioral disorders typically seen in PWS patients, including anxiety, depression, and obsessive behavior. Administration of selective serotonin reuptake inhibitors (SSRIs) in PWS mitigates aggressive and compulsive behaviors; further substantiating the notion that altered serotonergic pathway contributes to some of the behavioral aspects of PWS [103, 104].
Investigation into the reproductive functions of MAGE-L2 showed early-onset reproductive decline in Magel2-null mice [105]. In males, testosterone, but not luteinizing hormone or follicle-stimulating hormone levels, were reduced; however, despite reduced testosterone, the male reproductive organs and sperm show no overt differences. Interestingly, the male Magel2-deficient mice had altered olfactory preference. Female mice had delayed and lengthened puberty and were infertile by 24 weeks. Although there were no differences in the gross anatomy of ovaries and uteri collected from the infertile 26-week old Magel2-null females, the ovarian histology of these mice showed an absence of corpora lutea, suggestive of normal folliculogenesis with missed ovulations. In fact, Magel2-null females exhibited abnormal estrous cycles, similar to other circadian mutant female mice and women with PWS.
In a more recent Magel2 mouse model, Magel2-deficient mice exhibited 50% postnatal mortality and the impaired suckling and subsequent feeding deficits seen in PWS newborns [106]. In the hypothalamus, Magel2 mutant neonates had reductions in the hypothalamic neuropeptides oxytocin, orexin-A, and arginine-vasopressin. Interestingly, injection of oxytocin just after birth rescued the suckling initiation defects and the neonatal lethality.
These observations from multiple mouse models, as well as complementary cellular studies, draw striking parallels to neurological disorders. Taken together, the data suggest that many type II MAGEs play critical roles in differentiation and neural development, such that loss of function leads to a spectrum of cognitive, behavioral, and developmental deficits.
MAGEs in DISEASE
MAGE-L2 in Prader-Willi and Schaaf-Yang syndromes
Prader-Willi syndrome (PWS, OMIM 176270) is a complex neurodevelopmental disorder that was first described in the medical literature in 1956 [107]. It is characterized by infantile hypotonia with poor suck and failure to thrive, often necessitating assisted feeding [108]. Beginning in early childhood and over time, individuals with PWS exhibit hyperphagia, rapid weight gain, developmental delay, intellectual disability, hypogonadism, and short stature [108]. In addition to these cardinal features of PWS, a characteristic behavior and cognitive profile, including reduced activity, obsessive-compulsive traits, and temper outbursts (often associated with food and eating), has also been ascribed to this multi-system syndrome [108, 109].
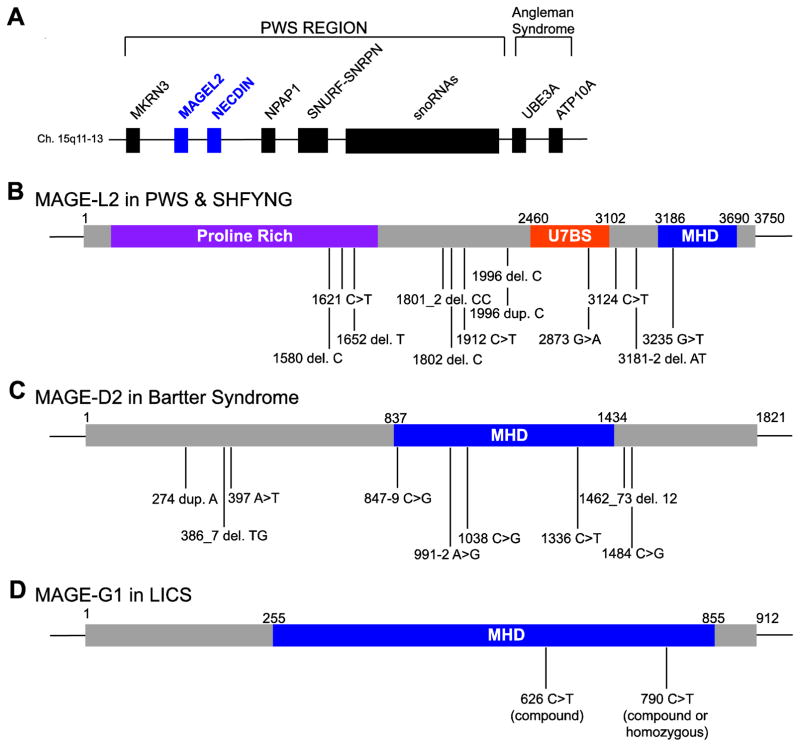
The genetic causes of PWS include deletion of paternal 15q11–q13 (65–75% of cases), maternal uniparental disomy (20–30%), and imprinting defects (1–3%) [85]. Although the deletion sizes can be variable, most individuals with PWS lose expression of genes in the PWS locus (15q11–q13), which comprises MKRN, MAGEL2, NDN, NPAP1, SNURF-SNRPN, a family of six small nucleolar RNA (snoRNA) genes, and several lncRNAs (Figure 6A) [85]. This locus has also been associated with general neuropsychiatric illness and autism spectrum disorder (ASD) [110, 111]. In fact, 10–40% of individuals with PWS meet the criteria for ASD [112]. Genotypic and phenotypic correlations have revealed that individuals with PWS caused by maternal uniparental disomy are more commonly affected with ASD (38%) than those with PWS caused by microdeletion (18%) [113].
Figure 6.
Type II MAGEs are associated with various diseases. (A) Schematic representation of the PWS locus. Identified mutations in MAGE-L2 (B), MAGE-D2 (C), and MAGE-G1 (D) associated with PWS and SHFYNG, Bartter syndrome, and LICS, respectively.
Despite the evident importance of genes in the PWS locus, the individual phenotypic contribution of each gene is not entirely clear. Previous reports have shown that individuals with deletions of the SNORD116 snoRNA cluster presented with key characteristics of PWS; however, an individual with paternal deletion of MKRN3, MAGEL2, and NDN exhibited obesity and intellectual disability but not the typical PWS phenotype [114–117]. This indicates that although loss of multiple genes may be required to produce the syndrome, the loss of individual genes could contribute to the various distinct phenotypes of this complex disorder.
Schaaf et al. initially identified four individuals with truncating mutations on the paternal allele of MAGEL2—the first individuals reported as having point mutations in a protein-coding gene within the PWS locus (Figure 6B) [118]. All four individuals presented with ASD, intellectual disability, and varying degrees of PWS phenotype. For example, while subject 2 exhibited classic PWS according to the diagnostic criteria established by Holm et al.; the other three subjects did not meet the full clinical criteria [108, 118]. Due to the phenotypic overlap with PWS, the condition was initially considered a Prader-Willilike syndrome. However, as the cohort of individuals with truncating MAGEL2 mutations grew, it became more apparent that the clinical condition caused by these mutations manifests specific phenotypes distinct from PWS. For example, hyperphagia and subsequent obesity—hallmarks of PWS—were either absent or only mildly present in individuals with MAGEL2 mutations [118, 119]. In addition, ASD was over-represented among individuals with molecularly confirmed mutations in MAGEL2 [118, 119]. Moreover, joint contractures, a phenotype rarely reported in PWS, were found in 23 of 28 cases with MAGEL2 mutations [118–120]. To highlight these phenotypic differences, the clinical condition caused by truncating MAGEL2 mutations was renamed Schaaf-Yang syndrome (SHFYNG, OMIM 615547).
In determining the molecular function of MAGE-L2, our lab demonstrated that MAGE-L2, in complex with the TRIM27 E3 RING ubiquitin ligase and USP7 deubiquitinating enzyme, regulates WASH-dependent actin polymerization and protein trafficking (refer to Regulation of WASH-mediated endosomal protein trafficking by MAGE-L2-TRIM27, Figure 4F) [50, 51]. These findings, taken together with the characterization of MAGE-L2 truncating mutations in individuals with SHFYNG, led us to hypothesize that similar phenotypes may also present in individuals with USP7 mutations. We identified seven cases with either heterozygous deletion or mutation of USP7 that resulted in phenotypes similar to those seen in SHFYNG and PWS, including intellectual disability, ASD, hypotonia, and hypogonadism [51]. Interestingly, these individuals with USP7 haploinsufficiency did not present with the typical PWS phenotypes of infantile feeding difficulties, hyperphagia, excessive weight gain, and characteristic craniofacial features [51]. They did however exhibit phenotypes specific to USP7 mutation or deletion, such as seizures and aggressive behavior, suggesting that specific genes in the PWS locus contribute to a spectrum of shared and independent phenotypes [51].
Intriguingly, individuals with mutations in WASH complex components also exhibit neurological pathologies. For example, autosomal dominant mutations in spastic paraplegia (SPG8), the gene encoding Strumpellin, result in hereditary spastic paraplegia [121, 122]. Likewise, recessive mutations in KIAA1033, the gene encoding SWIP, are associated with autosomal recessive intellectual disability and late-onset Alzheimer disease [123, 124]. In addition, TRIM27 has been implicated in ASD and Parkinson disease, suggesting that alterations in protein recycling may contribute to neurological disorders [125, 126].
MAGE-D2 in Bartter syndrome
Polyhydramnios is the excessive accumulation of amniotic fluid caused by an imbalance between production and removal the fluid [127]. While most cases of polyhydramnios are mild and result from the gradual increase of amniotic fluid, severe polyhydramnios may result in preterm birth and an increased risk of other perinatal complications [127]. Some of the known causes of polyhydramnios include fetal esophageal atresia and maternal diabetes; however, in 30 to 60% of cases, the cause remains unknown [127–129]. Antenatal Bartter syndrome (OMIM 300971), one of the few Mendelian diseases associated with polyhydramnios, is a rare, often life-threatening autosomal recessive renal tubular disorder characterized by fetal and postnatal polyuria, renal salt wasting, hypercalciuria, hypokalemia [130].
Previously it was known that mutations in SLC12A1 (encoding NKCC2), KCNJ1, CLCNKA, CLCNKB, or BSND impair the kidneys’ ability to reabsorb salt and can cause Bartter syndrome [131]. Recently, Laghmani et al. identified mutations in MAGE-D2 that cause X-linked polyhydramnios with prematurity and a transient but severe form of antenatal Bartter syndrome (Figure 6C) [129]. These patients initially exhibited a more severe presentation of antenatal Bartter syndrome with earlier onset of polyhydramnios and preterm labor for male offspring [132]. Immediately after birth, the infants developed progressive polyuria and severe hypercalciuria; however, within weeks, clinical symptoms spontaneously resolved [132]. Through genetic sequencing of nine families with transient antenatal Bartter syndrome and idiopathic polyhydramnios, the authors identified seven truncating mutations (two nonsense, two frameshift, and three splice-site mutations) and two nontruncating mutations (one missense and one in-frame deletion) in MAGE-D2 [129].
Further investigation revealed that MAGE-D2 promotes the expression and activity of the two crucial sodium chloride cotransporters (NKCC2 and NCC) necessary for proper ion reabsorption in the thick ascending limb of the loop of Henle and distal tubules [129]. Additionally, Laghmani and colleagues demonstrate that MAGE-D2 interacts with HSP40, a cytoplasmic chaperone that interacts with both NKCC2 and NCC and has been shown previously to regulate the biogenesis of NCC [129, 133]. Collectively, these findings indicate that MAGE-D2 plays a key role in fetal renal salt absorption, amniotic fluid homeostasis, and maintenance of normal pregnancy. However, further work will be important to determine how MAGE-D2 regulates NKCC2 and NCC, and if this regulation is dependent on ubiquitin-mediated trafficking.
MAGE-G1 in lung disease immunodeficiency and chromosome breakage syndrome
Members of the structural maintenance of the chromosome complex (SMC) family of proteins form three highly conserved heterodimeric complexes that regulate mitotic proliferation, meiosis, and DNA repair to support genomic stability [19]. One such complex, the SMC5/6 complex, consists of SMC5 and SMC6 in addition to non-SMC elements including the MAGE-G1-NSE1 MRL [18, 21]. The SMC5/6 complex promotes homologous recombination-mediated DNA repair and is essential for DNA damage response and telomere lengthening by recombination [19, 20, 134].
Recently, a report associated missense mutations in MAGE-G1 with an autosomal recessive chromosome breakage syndrome that leads to severe lung disease in early childhood (referred to as lung disease immunodeficiency and chromosome breakage syndrome, LICS) (Figure 6D) [135]. Two sisters (subjects A and B) with homozygous MAGE-G1 mutations (c.790C>T) exhibited B and T cell abnormalities, increased infection susceptibility, and eczema. In addition, they experienced feeding difficulties, failure to thrive, weight loss, psychomotor retardation, and axial hypotonia—phenotypes reminiscent of PWS and SHFYNG. After the onset of pediatric acute respiratory distress syndrome (PARDS), these individuals also manifested respiratory complications such as pneumomediastinum, pneumothorax, and subcutaneous emphysema. Following multiple episodes of virus-induced pneumonia, the affected individuals experienced severe progressive irreversible lung damage and died at 12 and 14 months. An affected brother and sister (subjects C and D, respectively) from a second family also developed a similar clinical history of progressive, severe PARDS and infectious pneumonia. Exome sequencing of the second family revealed compound heterozygous mutations in MAGE-G1 with a maternally inherited c.790C>T mutation (identical to the first family), and a paternally inherited c.626C>T mutation [135].
In the SMC5/6 complex, MAGE-G1-NSE1 and NSMCE4 form a molecular bridge between SMC5 and SMC6 and are essential for complex formation [136, 137]. Interestingly, the two identified variants of MAGE-G1 result in p.Leu264Phe (L264F) and p.Pro209Leu (P209L), and were both shown to disrupt interactions with NSMCE4 and destabilize the SMC5/6 complex [135]. Consistent with this, SMC5 and SMC6 protein levels were significantly reduced while MAGE-G1 protein was not detectable in fibroblasts from affected individuals [135]. Moreover, cells from subject B exhibited increased numbers of micronuclei, a hallmark of genome instability [135]. These cells also demonstrated hypersensitivity to various DNA damaging agents and defective homologous recombination. In addition, cells from subject B displayed defects in recovery from replication stress, similar to MMS21/NSMCE2-defective cells, and could be rescued by expression of wildtype MAGE-G1 [135].
Therefore, the identified mutations in affected individuals alters the stability of the SMC5/6 complex and result in faulty homologous recombination and impaired recovery from replication stress. Although the affected individuals manifest clinical features similar to those seen in Nijmegen breakage and AT chromosomal breakage syndromes, the severe and ultimately fatal pulmonary disease is unique to this novel chromosome breakage syndrome [135].
Type I MAGEs in Cancer
In addition to their physiological expression in reproductive tissues, the type I MAGE CTAs are re-activated in a wide variety of tumors (Table 1). This aberrant expression of type I MAGEs as well as their prognostic value in various cancers has been extensively documented (Table 2).
Table 1.
Summary of MAGE expression in select cancer types
| Percent of MAGE-positive patient tumors | ||||||
|---|---|---|---|---|---|---|
| MAGE a | Melanoma | Lung | Breast | Ovarian | HNSCC | References |
| A1 | 16–61% | 24–49% | 6% | 11–54% | 16–23% | [138, 139, 143, 146, 147, 149, 207] |
| A2 | 44–84% | 40% | 20% | 3% | 20–31% | [138, 149] |
| A3 | 48–80% | 38–55% | 10–26% | 3–37% | 29–41% | [42, 138, 141, 143, 147–150, 207] |
| A4 | 9–34% | 19–65% | 13–19% | 32–47% | 23–47% | [138, 139, 143, 147, 149, 207] |
| A6 | 48–82% | 26–50% | 28% | 5% | 31–43% | [42, 141, 148–150] |
| A8 | 4–9% | 7% | 4% | 3% | 1–3% | [208] |
| A9 | 8–23% | 69% | 16–45% | 25–37% | 25–33% | [144, 207] |
| A10 | 13–61% | 43% | 8% | 19–52% | 13–19% | [147, 207] |
| A11 | 15–41% | 61% | 8–67% | 33% | 18–38% | [208] |
| A12 | 45–77% | 27% | 9–15% | 5% | 13–28% | [208] |
| B1 | 4–23% | 9% | 25% | 8% | 0–7% | [208] |
| B2 | 10–27% | 41% | 3% | 8% | 19–35% | [151] |
| B3 | 0% | 0% | 1% | 5% | 1% | [208] |
| B4 | 0% | 0% | 3% | 0% | 0% | [208] |
| B6 | 18–25% | 55% | 1% | 5% | 20–27% | [150] |
| B16 | 6–21% | 6% | 21% | 1% | 1–4% | [208] |
| B18 | 9–20% | 9% | 4% | 18% | 1–5% | [208] |
| C1 | 24–62% | 28% | 5–38% | 16–35% | 9–13% | [138, 140, 143, 153, 207, 209] |
| C2 | 33–67% | 21% | 8–58% | 13% | 4–13% | [142, 209] |
MAGE expression correlates with DNA methylation status (highlight)
Table 2.
Correlation between MAGE expression and overall survival in select cancer types
| Overall survival
|
|||||
|---|---|---|---|---|---|
| Cancer type | MAGE | HR | CI (95%) | P-value | References |
| Lung | A3 | 3.226 | 1.446–7.918 | 0.004 | [42, 146, 210] |
| A9 | 2.334 | 1.664–3.274 | 0.001 | [147] | |
| C2 | 1.909 | 0.954–3.821 | 0.068 | [210] | |
| Breast | A1 | 2.285 | 0.910–5.732 | 0.078 | [211] |
| A3 | 3.446 | 1.00–11.77 | 0.049 | [211] | |
| A9 | 3.702 | 1.392–9.845 | 0.009 | [212] | |
| C2 | 3.07 | 1.47–12.01 | 0.003 | [142] | |
| Ovarian | A9 | 2.271 | 1.372–3.761 | 0.001 | [144] |
| Colorectal | A9 | 2.376 | 1.380–4.089 | 0.002 | [213] |
| LSCC | A9 | 3.57 | 1.457–8.762 | 0.005 | [214] |
| HCC | A9 | 2.17 | 1.121–4.205 | 0.022 | [215] |
| ESCC | A11 | 2.689 | 1.434–5.040 | 0.002 | [216] |
| Gastric | A12 | 1.78 | 1.23–2.58 | 0.002 | [217] |
HR, hazard ratio; CI, confidence interval
Given that the MAGE CTAs were originally identified in melanoma cells, it is not surprising that a number of MAGEs are expressed at high frequencies in melanoma. However, it is interesting that MAGE-A1-4, and -C1 expression changes over the course of cancer progression, such that these MAGEs express at higher frequencies in metastases compared to primary melanoma samples [138–140]. In addition, expression of these MAGE-A genes associates with thicker tumors and ulcerated melanomas, supporting the notion that high MAGE CTA expression correlates with advanced tumor grade [138, 139].
This trend is not limited to melanoma. For example, MAGE-A3/6 and -C2 expression in breast cancer associates with tumor estrogen receptor- and progesterone receptor-negative status, high histologic grade, as well as worse survival [141, 142]. In ovarian cancer, MAGE-A1, -A9, and -A10 expression also correlate with poor survival [143, 144]. Likewise, MAGE-A1, -A2, -A3/6, -A12, -B2, and -C1 are expressed in non-small-cell lung cancer where MAGE-A3/6 and -A9 expression is associated with advanced tumor type and decreased survival [145–147]. Similarly, expression of MAGE-A1-6, -B2 and -B6 in patients with head and neck squamous cell carcinoma (HNSCC) correlates with advanced clinical stage of cancer and poor oncologic outcomes [148–151]. Furthermore, MAGE-A1, -A6, -A8, -A9, and -A11 are expressed at significantly higher levels at the tumor front of advanced stages of HNSCC, suggesting that these MAGEs contribute to malignancy [152].
Indeed, a growing body of evidence supports the notion that MAGEs function as oncogenic drivers, rather than simple biomarkers or passengers of global genomic dysregulation. Consistent with this idea, some studies have reported that MAGEs can be turned on early during the process of tumorigenesis even before clinical signs of the disease [145]. Furthermore, investigation into the oncogenic potential of these genes has demonstrated that MAGE-A3/6 is required for the viability of patient-derived breast, colon, lung cancer and multiple myeloma cells; whereas MAGE-A3/6 does not significantly alter the viability of MAGE-A3/6-negative cells [42, 153]. This specificity suggests that upon expression of MAGE-A3/6, these cancer cells become dependent on or addicted to MAGE-A3/6 expression for viability [42]. Consistent with these findings, MAGE-A3/6 expression drives several hallmarks of cancer such as cell proliferation, cell migration, invasion, and anchorage-independent growth (Figure 7A) [42, 154]. Remarkably, expression of MAGE-A3/6 is sufficient to stimulate foci formation in fibroblasts and promote anchorage-independent growth in non-transformed human colonic epithelial cells [42]. In an orthotopic xenograft mouse model for thyroid cancer, MAGE-A3/6 expression results in significantly increased tumor growth and larger, more numerous lung metastases [154]. These results are consistent with a model wherein MAGE-A3/6 functions as a potent driver of tumorigenesis.
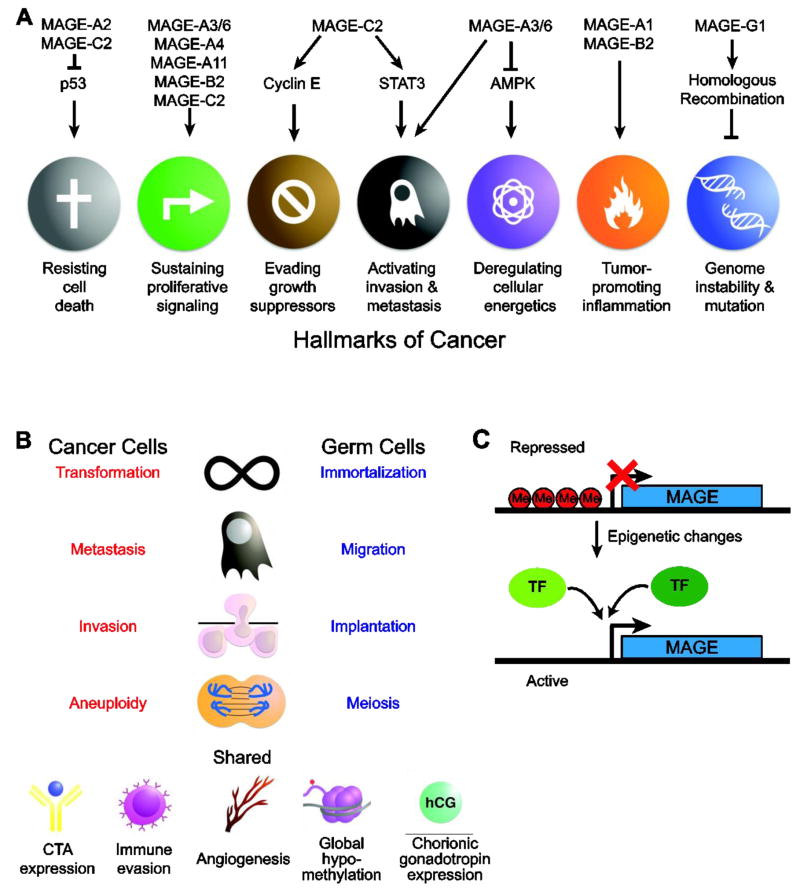
Figure 7.
Type I MAGEs promote tumorigenesis. (A). Specific functions of several type I MAGEs in driving the hallmarks of cancer is shown. (B) The shared phenotypes between cancer cells and germ cells suggest that activation of the gametogenic program might contribute to tumorigenesis. (C) Transcriptional activation of MAGEs. Type I MAGEs are typically silenced but can be activated by epigenetic changes (including DNA CpG demethylation) and specific transcription factors.
While most MAGE-related studies focus on the MAGE-A subfamily and their involvement in cancer, the MAGE-B and -C subfamilies are also associated with tumor growth and progression (Figure 7A). For example, knockdown of mouse Mage-b genes reduces cell viability in melanoma and mast cell lines [37, 155]. Similarly, MAGE-B2 promotes cell proliferation in transformed oral keratinocytes, osteosarcoma, and colon cancer cell lines [60, 151]. Moreover, Mage-b knockdown suppresses growth in a syngeneic mouse model, whereas over-expression of MAGE-B2 enhances tumor growth in mice [37, 60]. Likewise, MAGE-C2 has also been shown to promote cell proliferation in malignant melanoma cells and tumor metastasis in vivo by enhancing STAT3 signaling [52, 156]. These findings indicate that MAGE CTAs have oncogenic functions. Additional studies are needed, especially with regard to the MAGE-B and -C subfamilies, to determine the specific activities and mechanisms by which MAGE CTAs promote cancer development.
Interestingly, MAGE CTAs exhibit preferential expression in cancer stem cells (CSCs), a small population of cancer cells that have the ability to self-renew, differentiate, and initiate tumor growth [157]. Analysis of the MAGE CTA expression profiles in cancer stem-like cells demonstrated that MAGE-A2 and -A3/6 are enriched in the CSC-like side population cells derived from lung and colon adenocarcinoma cells while MAGE-A4 and -B2 are preferentially expressed in the CSC-like side population derived from colon adenocarcinoma cells [158]. Additionally, MAGE-A3 exhibits enriched expression in the CSC-like side population in bladder cancer [159]. Consistent with these findings, MAGE-C1 expresses in stem cells as well as immature B cells [160]. This preferential expression of MAGE CTAs in CSCs is reminiscent of their physiological enrichment in spermatogonial stem cells. The marked similarities between these cells types, such as immortalization, immune evasion, and induction of specific differentiation pathways has led investigators to speculate that the expression of MAGEs and activation of their physiological functions in cancer initiates a gametogenic program that might contribute to the tumor formation, progression and CSC maintenance (Figure 7B).
TRANSCRIPTIONAL REGULATION
Although there is an abundance of evidence for the deregulated expression of type I MAGE CTAs in cancer, the precise mechanisms regulating their aberrant expression is only beginning to be elucidated. While some cancers such as melanoma and lung cancers frequently express MAGEs, others including leukemia and uveal melanoma rarely do. In addition, some tumors tend to express a single or few MAGEs, while others express multiple MAGEs simultaneously [161]. Although the details of how specific MAGE genes get turned on in cancer are not fully understood, it is apparent that epigenetic events, including DNA methylation and histone modifications, contribute to the regulation of MAGE expression in both normal and neoplastic cells (Figure 7C).
DNA methylation
DNA methylation of promoter CpG dinucleotides by DNA methyltransferases (DNMTs) exerts a repressive effect on transcription by preventing transcription factor binding and by recruiting methyl-CpG-binding domain proteins (MBDs), which in turn bind chromatin remodeling co-repressor complexes to repress gene expression. A growing body of literature has shown that DNA methylation is the primary regulation of CTA expression in normal and cancer cells. In fact, Weber and colleagues provided the first insights into the transcriptional regulation of MAGEs by demonstrating that 5-aza-2’-deoxycytidine (decitabine), a DNMT inhibitor, induces MAGE-A1 expression in cultured melanoma cells [162]. Subsequent analysis revealed that expression of several MAGE-A genes correlates with methylation status of its promoter in various types of neoplastic cells [163, 164]. Importantly, an unmethylated MAGE-A1 promoter was shown to drive expression of a reporter gene in MAGE-A1 nonexpressing cells, indicating that the methylation status of the MAGE CTA promoter is the leading mechanism for regulating expression [165, 166]. Likewise, downregulation of DNMT1 results in the activation and stable hypomethylation of a methylated MAGE-A1 transgene in melanoma cells [167]. Interestingly, Sigalotti et al. reported an association between DNA hypomethylation of CTA promoters and CTA expression in populations of putative melanoma stem cells, suggesting that this form of epigenetic regulation is potentially an important mechanism of CTA gene regulation in cancer stem cells [168]. Therefore, methylation-specific PCR can be used to evaluate the hypomethylation status of CpG sites in the promoter regions of MAGEs as a method for early cancer detection [145]. However, while DNA methylation clearly plays an important role in regulating MAGE expression, there is mounting evidence that demethylation is not sufficient to drive expression of all MAGEs and additional nonepigenetic mechanisms are required [162, 169, 170].
Histone modifications
Histone modifications have also been shown to function in the regulation of CTA expression. Histones assemble with DNA into nucleosomes, the basic unit of chromatin. Their flexible N-terminal tails protrude from the nucleosomes and are targeted for post-translation modification, including acetylation and methylation (reviewed in [171]). Histone acetylation by histone acetyltransferases (HATs) results in chromatin decompaction and gene transcription. Conversely, histone deacetylases (HDACs) serve the opposite function and promote the compaction of chromatin to prevent the accessibility of DNA to transcription factors and RNA polymerase. In the case of MAGE-A2 and MAGE-A12, treatment with HDAC inhibitors results in up-regulation of transcriptional activity [172].
Depending on the lysine residue that is modified, histone methylation is associated with both transcriptional activation and repression. For example, methylation of histone H3 lysine 9 or lysine 27 (H3K9 or H3K27, respectively) are repressive marks whereas methylation of histone H3 lysine 4 (H3K4) is an active mark. In a study from Tachibana et al., knockout of the histone methyltransferases that catalyze H3K9me2, G9a and/or GLP, induced the expression of Mage-a genes in mouse embryonic stem cells [173]. In addition, in G9a- or GLP-null mouse ES cells, Mage-a genes are hypomethylated [173]. These results suggest that histone modifications are also involved in the repression of MAGE CTAs.
Transcription factors
Although less is known about the nonepigenetic mechanisms of MAGE gene regulation, the ETS transcription factor sites have been show to function in the regulation of MAGE-A genes. The ETS protein family is one of the largest families of transcription factors and members of this family are implicated in the development of various tissues as well as cancer progression. Work from de Smet and colleagues revealed that two inverted ETS motifs near the transcriptional start site of MAGE-A1 drive transcriptional activity of the unmethylated promoter [165, 174]. In cells where the endogenous MAGE-A1 promoter is methylated and inactive, MAGE-A1 promoter transgenes are highly active and dependent on ETS sequences [165, 174]. Further studies have shown that methylation of a CpG in the ETS consensus sequence inhibits ETS binding [165, 174]. Although the identity of the exact ETS factor that drives MAGE-A expression remains unclear, one potential candidate is ETS1, which was shown to activate MAGE-A genes when overexpressed.
BORIS, a paralog of the imprinting regulator and chromatin insulator protein CTCF, has also been implicated in promoting the expression of MAGE-A1. In fact several MAGE-A genes contain BORIS binding sites and the proteins have been shown to coexpress in head and neck cancer [175]. Moreover, BORIS overexpression in immortalized oral keratinocytes led to MAGE-A induction and DNA hypomethylation [175].
Signal transduction pathways
In addition to transcription factors, signal transduction pathways such as activated tyrosine kinases, have been implicated in MAGE CTA expression. The KIT receptor tyrosine kinase is a proto-oncogene that binds to c-kit ligand, also known as steel factor or stem cell factor, to activate its tyrosine kinase activity and signal transduction pathway. Yang et al. reported that treatment of mast cells with the tyrosine kinase inhibitor imatinib results in downregulation of MAGE-A, -B, and -C and potentially alters DNA methylation levels [176]. In addition to KIT, others have reported that fibroblast growth factor receptor 2 (FGFR2) activation downregulates MAGE-A3 expression [177]. In addition, FGFR2 and MAGE-A3 promoters show reciprocal DNA methylation patterns [178]. Therefore, FGFR2 and KIT appear to have opposing effects of MAGE-A expression. Interestingly, FGFRs are known to play a critical role in a variety of processes including tissue repair and angiogenesis [179]. Given that MAGE-A1 is expressed during wound healing as well as in the joints of patients with juvenile arthritis, MAGE-A1 may have a role in inflammation [180, 181]. Furthermore, MAGE-B2 was originally identified as being expressed in patients with systemic lupus erythematosus where MAGE-B2 auto-antibodies can be found [182, 183]. Thus, additional investigation of roles for type I MAGE CTAs in other disease contexts besides cancer is warranted in the future.
THERAPY
Cancer immunotherapy
Since the identification of MAGEs, significant effort has gone into the development of CTA-based immunotherapeutic strategies (Table 3). Compared to chemotherapy, which often has limited efficacy in patients with relapsed cancer or advanced disease, immunotherapy has the potential to provide long-lasting responses by modulating the immune response against specific cancer proteins.
Table 3.
Clinical trials with MAGEs
| MAGE | Tumor type | Phase | Intervention | Identifier | Status |
|---|---|---|---|---|---|
| A3 | Melanoma | I/II | MAGE-A3.A1 peptide and CpG7909 | NCT00145145 | Terminated |
| A3 | Melanoma | II | GSK1203486A recombinant MAGE-A3 ASCI |
NCT00706238 | Terminated |
| A3 | Melanoma | II | GSK2132231A, dacarbazine | NCT00849875 | Terminated |
| A3 | Melanoma | III | GSK 2132231A | NCT00796445 | Terminated |
| A3 | Lung | I | GSK1572932A, cisplatin, vinorelbine, radiotherapy | NCT00455572 | Terminated |
| A3 | Lung | III | GSK1572932A | NCT00480025 | Terminated |
| A3, A12 | Metastatic | I/II | anti-MAGE-A3/12 TCR, aldesleukin, cyclophosphamide, fludarabine | NCT01273181 | Terminated |
| A3 | Melanoma | I/II | Enhanced TCR Transduced Autologous T Cells | NCT01350401 | Ongoing |
| A3 | Multiple myeloma | I/II | Enhanced TCR Transduced Autologous T Cells | NCT01352286 | Ongoing |
| A3, C2 | Melanoma | II | TriMix-DC and ipilimumab | NCT01302496 | Ongoing |
| A3, C1 | Multiple myeloma | I | CT7, MAGE-A3, and WT1 mRNA- electroporated Langerhans cells (LCs) | NCT01995708 | Recruiting |
| A3 | Metastatic | I/II | Anti-MAGE-A3-DP4 TCR, cyclophosphamide, fludarabine, aldesleukin | NCT02111850 | Recruiting |
| A3 | Metastatic | I/II | MG1MA3 (MG1 Maraba/MAGE-A3 adenovirus vaccine) | NCT02285816 | Recruiting |
| A10 | Urinary bladder, head and neck, melanoma | I | Autologous genetically modified MAGE A10C796T cells | NCT02989064 | Recruiting |
| A10 | Lung | I/II | Autologous genetically modified MAGE A10C796T cells | NCT02592577 | Recruiting |
| A3 | Lung | I/II | Ad-MAGEA3, MG1-MAGEA3, pembrolizumab | NCT02879760 | Recruiting |
From an immunological perspective, MAGE CTAs are ideal target molecules for cancer immunotherapies due to their widespread, prominent expression in various cancers, but restricted normal expression to the immune-privileged testis, thus limiting the possibility of an autoimmune response but maximizing their potential for broad application to large cohorts of patients [184–186]. MAGE-A3, which codes for an antigenic nonapeptide that is recognized by CTLs on the HLA-A1 molecule, garnered particular interest as an immunotherapeutic. In a pivotal clinical trial where tumor-bearing HLA-A1-positive patients with metastatic melanoma were treated with subcutaneous injections of MAGE-A3 peptide, seven out of 25 patients displayed significant tumor regression, including three complete responses [187]. However, MAGE-A3-specific CTL responses were not detected during the course of vaccinations, even in patients with positive clinical responses [187]. Subsequent studies confirmed these immunologic and clinical responses in melanoma patients treated with MAGE-A3-pulsed DCs [188, 189]
Additional MAGE CTA-based vaccines have utilized recombinant proteins to (1) induce both CD8+ and CD4+ immune responses thereby magnifying the CTL response, (2) generate responses against multiple epitopes, and (3) avoid specific HLA-type requirements for patients [190]. In a phase II clinical trial where 36 patients with stage III or IV M1a melanoma were treated with MAGE-A3 combined with AS15 immunostimulant, three patients exhibited complete responses [191].
Based on these preliminary data, two large clinical trials were set forth—PREDICT and DERMA. PREDICT was a phase II study with the goal evaluating the clinical activity of MAGE-A3 antigen-specific cancer immunotherapeutic (ASCI) in patients with MAGE-A3-positive unresectable metastatic melanoma [192]. The goal of DERMA (phase III) was to evaluate the benefit MAGE-A3 antigen-specific cancer immunotherapeutic (ASCI) in melanoma patients after surgical tumor removal. Unfortunately, the objective response rate was lower than in previous studies [192]. Consistent with these findings, DERMA was terminated early following assessment showing lack of efficacy of the treatment.
Similarly, MAGE-A3 ASCI showed promising results in phase II clinical trials of patients with NSCLC and inspired the largest phase III therapeutic trial in lung cancer—MAGRIT [193–196]. In the case of patients with MAGE-A3-positive surgically resected NSCLC, MAGE-A3 ASCI failed to increase disease-free survival compared with placebo and further development of the MAGE-A3 ASCI was stopped [197].
More alarming than lack of efficacy, however, are the unexpected deaths associated with MAGE-based immunotherapies. In order to overcome the complications associated with low frequency of cancer antigen-specific cells and low avidity of expanded effector T cells, T cells can be genetically engineered to expressed T cell receptors (TCRs) that have high affinity and specificity. Unfortunately, in a study utilizing anti-MAGE-A3 TCR gene therapy as a treatment for metastatic cancers, one patient developed transient Parkinson-like symptoms, while two patients lapsed into comas and died [198]. Further investigation revealed that T cells transduced with the generated TCR also recognized MAGE-A12, which is expressed in the brain [198]. Therefore, treatment resulted in neuronal cell destruction that manifested as necrotizing leukoencephalopathy [198]. In a separate study using anti-MAGE-A3 TCR therapy, two patients developed progressive cardiogenic shock and died within a week of infusion. In this case, the generated TCR recognized another peptide derived from titin, a component of striated muscle, and the off-target reactivity resulted in severe myocardial damage with T-cell infiltration [199]. These studies demonstrate the need for more rigorous studies of MAGE expression and stringent analysis of engineered TCRs.
Combination therapy
In an effort to develop alternative methods to target MAGE-expressing cancers and improve clinical outcomes, investigators are utilizing combinatorial approaches including conventional therapy, immunotherapy, and molecular targeting.
One obstacle to overcome in the development of immunotherapies is T cell recognition of cancer cells expressing MAGE CTAs. In cancer cells, promoter hypermethylation suppresses MHC expression, thereby impeding antigen presentation [200]. Therefore, several groups have indicated that treatment with demethylating agents such as decitabine, can potentially enhance antigen presentation and cancer cell recognition by upregulating both MHC and MAGE expression [162, 201–203]. In a phase I clinical trial combining dectabine and DC vaccine targeting MAGE-A1 and -A3 for patients with relapsed neuroblastoma demonstrated a response in six of nine patients with complete response in one patient [204]. Although the preliminary data of this combination therapy looks promising, future trials are needed to better determine the efficacy of this treatment.
A major obstacle to the practical application of cancer immunotherapy is the ability of tumor cells to evade the immune system by promoting an immunosuppressive tumor microenvironment. Therefore, investigators have turned to immune checkpoint therapies, which target regulatory pathways in T cells to enhance the antitumor immune response. Rather than activating the immune system to directly target tumor cells, immune checkpoint therapies remove inhibitor pathways that block effective anti-tumor T cell responses (reviewed in [205]). Therefore, combination approaches with immune checkpoint therapies may be beneficial in the treatment of MAGE-expression cancers and are currently on going (Table 3).
In addition to these proposed treatments, molecular targeting of specific protein-protein interactions may prove to be a powerful therapeutic strategy. A prime candidate is targeting specific MRLs. For example, Bhatia and colleagues screened a library of compounds and identified three potential compounds that interfere with MAGE-C2-TRIM28 binding, promote death in MAGE-positive cells, and activate p53 [206]. This data supports the therapeutic value of inhibiting MAGE binding to cognate E3 ligases to block oncogenic functions of MAGE proteins. Furthermore, by knowing the precise cellular targets and functions of MRLS, novel drug susceptibilities can be predicted based on MAGE expression status in individual tumors. For example, considering the interplay between MAGE-A proteins, p53, and HDAC, the use of HDAC inhibitors in combination with other therapeutic approaches could help restore p53 tumor suppressor activity. Furthermore, utilization of AMPK agonists (such as metformin) or mTOR inhibitors may be an effective future treatment for MAGE-A3/6 positive cancers. Future studies validating these approaches in cell and animal models will be important.
CONCLUSIONS AND PERSPECTIVES
Since their initial discovery, the MAGEs have gained much attention and interest as cancer biomarkers and targets of cancer immunotherapies. Despite the fact that initial attempts to develop MAGE-based immunotherapies have not been entirely successful, recent studies highlighting a role for MAGEs in the regulation of E3 RING ubiquitin ligases have greatly expanded our understanding of the diverse functions of MAGE family members and their impact on various molecular processes (such as p53 signaling, cell metabolism, and protein trafficking), and have opened up novel avenues for personalized cancer-specific therapies targeting MRLs. However, many unanswered questions remain.
How do the cellular functions attributed to MAGEs contribute to disease progression? How do MAGEs regulate pathways that effect tissue-specific outcomes? What transcriptional programs lead to the aberrant re-expression of type I MAGEs in cancer? Do type I MAGEs have similar functions in their physiological context of germ cells as they do in cancer cells? What are the cellular and physiological functions of the relatively unexplored MAGEs? What are the substrates of many of the uncharacterized MRLs?
Although our understanding of the molecular mechanisms by which MRLs function is still developing, continued identification and characterization of the functional interplay between MAGEs, RING ligases, and deubiquitinating enzymes will provide valuable insights into the role of MAGEs. In addition, structural studies may help us not only to understand the biophysical underpinnings of MRLs, but also design specific MAGE inhibitors for therapeutic purposes. The continued study of MAGEs clearly holds great promise for answering fundamental biological questions and may reveal new ways to target them for the treatment of disease.
HIGHLIGHTS.
The MAGE family is evolutionarily conserved and consists of >50 genes in humans
MAGE proteins function in MAGE-RING ligase (MRL) complexes to direct ubiquitination
Type I MAGEs are expressed in reproductive tissues and function in germ cell development
When aberrantly expressed, type I MAGEs function as oncogenes to drive tumorigenesis
Type II MAGEs are broadly expressed and disruption results in neurodevelopmental defects
Acknowledgments
We would like to thank members of the Potts lab for advice and critical discussions. We apologize for our inability to discuss and reference all work in the field due to space limitations. This work was supported by NIGMS grant R01GM111332, Worldwide Cancer Research grant 15-0177, and Foundation for Prader-Willi Syndrome Research grant 342604 to P.R.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jager E, Knuth A. The discovery of cancer/testis antigens by autologous typing with T cell clones and the evolution of cancer vaccines. Cancer Immun. 2012;12:6. [PMC free article] [PubMed] [Google Scholar]
- 2.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 3.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–51. [PubMed] [Google Scholar]
- 4.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–12. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 5.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–9. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 6.Rogner UC, Wilke K, Steck E, Korn B, Poustka A. The melanoma antigen gene (MAGE) family is clustered in the chromosomal band Xq28. Genomics. 1995;29:725–31. doi: 10.1006/geno.1995.9945. [DOI] [PubMed] [Google Scholar]
- 7.Dabovic B, Zanaria E, Bardoni B, Lisa A, Bordignon C, Russo V, et al. A family of rapidly evolving genes from the sex reversal critical region in Xp21. Mamm Genome. 1995;6:571–80. doi: 10.1007/BF00352360. [DOI] [PubMed] [Google Scholar]
- 8.Lurquin C, De Smet C, Brasseur F, Muscatelli F, Martelange V, De Plaen E, et al. Two members of the human MAGEB gene family located in Xp21.3 are expressed in tumors of various histological origins. Genomics. 1997;46:397–408. doi: 10.1006/geno.1997.5052. [DOI] [PubMed] [Google Scholar]
- 9.Muscatelli F, Walker AP, De Plaen E, Stafford AN, Monaco AP. Isolation and characterization of a MAGE gene family in the Xp21.3 region. Proc Natl Acad Sci U S A. 1995;92:4987–91. doi: 10.1073/pnas.92.11.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas S, De Smet C, Arden KC, Viars CS, Lethe B, Lurquin C, et al. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58:743–52. [PubMed] [Google Scholar]
- 11.Lucas S, De Plaen E, Boon T. MAGE-B5, MAGE-B6, MAGE-C2, and MAGE-C3: four new members of the MAGE family with tumor-specific expression. Int J Cancer. 2000;87:55–60. [PubMed] [Google Scholar]
- 12.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–21. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–9. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas S, Brasseur F, Boon T. A new MAGE gene with ubiquitous expression does not code for known MAGE antigens recognized by T cells. Cancer Res. 1999;59:4100–3. [PubMed] [Google Scholar]
- 16.Lopez-Sanchez N, Gonzalez-Fernandez Z, Niinobe M, Yoshikawa K, Frade JM. Single mage gene in the chicken genome encodes CMage, a protein with functional similarities to mammalian type II Mage proteins. Physiol Genomics. 2007;30:156–71. doi: 10.1152/physiolgenomics.00249.2006. [DOI] [PubMed] [Google Scholar]
- 17.Katsura Y, Satta Y. Evolutionary history of the cancer immunity antigen MAGE gene family. PLoS One. 2011;6:e20365. doi: 10.1371/journal.pone.0020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pebernard S, Perry JJ, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol Biol Cell. 2008;19:4099–109. doi: 10.1091/mbc.E08-02-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potts PR. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 2009;8:499–506. doi: 10.1016/j.dnarep.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–88. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. Identification of the proteins, including MAGEG1, that make up the human SMC5–6 protein complex. Mol Cell Biol. 2008;28:1197–206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura I, Shimizu S, Sakoda JY, Yoshikawa K. Expression of Drosophila MAGE gene encoding a necdin homologous protein in postembryonic neurogenesis. Gene Expr Patterns. 2007;7:244–51. doi: 10.1016/j.modgep.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Q, Caballero OL, Simpson AJ, Strausberg RL. Differential evolution of MAGE genes based on expression pattern and selection pressure. PLoS One. 2012;7:e48240. doi: 10.1371/journal.pone.0048240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Backer O, Verheyden AM, Martin B, Godelaine D, De Plaen E, Brasseur R, et al. Structure, chromosomal location, and expression pattern of three mouse genes homologous to the human MAGE genes. Genomics. 1995;28:74–83. doi: 10.1006/geno.1995.1108. [DOI] [PubMed] [Google Scholar]
- 25.Osterlund C, Tohonen V, Forslund KO, Nordqvist K. Mage-b4, a novel melanoma antigen (MAGE) gene specifically expressed during germ cell differentiation. Cancer Res. 2000;60:1054–61. [PubMed] [Google Scholar]
- 26.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–74. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman JA, Cooper CD, Roos AK, Aitkenhead H, Oppermann UC, Cho HJ, et al. Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding. PLoS One. 2016;11:e0148762. doi: 10.1371/journal.pone.0148762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagiwara Y, Sieverling L, Hanif F, Anton J, Dickinson ER, Bui TT, et al. Consequences of point mutations in melanoma-associated antigen 4 (MAGE-A4) protein: Insights from structural and biophysical studies. Sci Rep. 2016;6:25182. doi: 10.1038/srep25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–12. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 31.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 33.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 34.Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, et al. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–9. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–22. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 36.Grossberger R, Gieffers C, Zachariae W, Podtelejnikov AV, Schleiffer A, Nasmyth K, et al. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J Biol Chem. 1999;274:14500–7. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- 37.Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer research. 2007;67:9954–62. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 38.Xiao TZ, Suh Y, Longley BJ. MAGE proteins regulate KRAB zinc finger transcription factors and KAP1 E3 ligase activity. Arch Biochem Biophys. 2014;563:136–44. doi: 10.1016/j.abb.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao TZ, Bhatia N, Urrutia R, Lomberk GA, Simpson A, Longley BJ. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS One. 2011;6:e23747. doi: 10.1371/journal.pone.0023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozakova L, Vondrova L, Stejskal K, Charalabous P, Kolesar P, Lehmann AR, et al. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle. 2015;14:920–30. doi: 10.1080/15384101.2014.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Gao J, Yang M. When MAGE meets RING: insights into biological functions of MAGE proteins. Protein Cell. 2011;2:7–12. doi: 10.1007/s13238-011-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–28. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pineda CT, Potts PR. Oncogenic MAGEA-TRIM28 ubiquitin ligase downregulates autophagy by ubiquitinating and degrading AMPK in cancer. Autophagy. 2015;11:844–6. doi: 10.1080/15548627.2015.1034420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura M, Okinaga S, Arai K. Metabolism of round spermatids: evidence that lactate is preferred substrate. Am J Physiol. 1984;247:E234–42. doi: 10.1152/ajpendo.1984.247.2.E234. [DOI] [PubMed] [Google Scholar]
- 46.Seaman MN, Gautreau A, Billadeau DD. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 2013;23:522–8. doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–23. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, et al. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143:761–73. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, et al. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell. 2013;152:1051–64. doi: 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao YH, Fountain MD, Jr, Fon Tacer K, Xia F, Bi W, Kang SH, et al. USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol Cell. 2015;59:956–69. doi: 10.1016/j.molcel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao J, Song X, Wang J, Guo C, Li Y, Li B, et al. Cancer-testis antigen MAGE-C2 binds Rbx1 and inhibits ubiquitin ligase-mediated turnover of cyclin E. Oncotarget. 2015;6:42028–39. doi: 10.18632/oncotarget.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su S, Chen X, Geng J, Minges JT, Grossman G, Wilson EM. Melanoma antigen-A11 regulates substrate-specificity of Skp2-mediated protein degradation. Mol Cell Endocrinol. 2017;439:1–9. doi: 10.1016/j.mce.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcar L, Maclaine NJ, Hupp TR, Meek DW. Mage-A cancer/testis antigens inhibit p53 function by blocking its interaction with chromatin. Cancer Res. 2010;70:10362–70. doi: 10.1158/0008-5472.CAN-10-1341. [DOI] [PubMed] [Google Scholar]
- 55.Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, et al. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A. 2006;103:11160–5. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcar L, Ihrig B, Hourihan J, Bray SE, Quinlan PR, Jordan LB, et al. MAGE-A Cancer/Testis Antigens Inhibit MDM2 Ubiquitylation Function and Promote Increased Levels of MDM4. PLoS One. 2015;10:e0127713. doi: 10.1371/journal.pone.0127713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuwako K, Taniura H, Yoshikawa K. Necdin-related MAGE proteins differentially interact with the E2F1 transcription factor and the p75 neurotrophin receptor. J Biol Chem. 2004;279:1703–12. doi: 10.1074/jbc.M308454200. [DOI] [PubMed] [Google Scholar]
- 59.Su S, Minges JT, Grossman G, Blackwelder AJ, Mohler JL, Wilson EM. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J Biol Chem. 2013;288:24809–24. doi: 10.1074/jbc.M113.468579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peche LY, Ladelfa MF, Toledo MF, Mano M, Laiseca JE, Schneider C, et al. Human MageB2 Protein Expression Enhances E2F Transcriptional Activity, Cell Proliferation, and Resistance to Ribotoxic Stress. J Biol Chem. 2015;290:29652–62. doi: 10.1074/jbc.M115.671982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson EM. Androgen receptor molecular biology and potential targets in prostate cancer. Ther Adv Urol. 2010;2:105–17. doi: 10.1177/1756287210372380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol Cell Biol. 2005;25:1238–57. doi: 10.1128/MCB.25.4.1238-1257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Askew EB, Bai S, Hnat AT, Minges JT, Wilson EM. Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators. J Biol Chem. 2009;284:34793–808. doi: 10.1074/jbc.M109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Askew EB, Bai S, Blackwelder AJ, Wilson EM. Transcriptional synergy between melanoma antigen gene protein-A11 (MAGE-11) and p300 in androgen receptor signaling. J Biol Chem. 2010;285:21824–36. doi: 10.1074/jbc.M110.120600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai S, Wilson EM. Epidermal-growth-factor-dependent phosphorylation and ubiquitinylation of MAGE-11 regulates its interaction with the androgen receptor. Mol Cell Biol. 2008;28:1947–63. doi: 10.1128/MCB.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karpf AR, Bai S, James SR, Mohler JL, Wilson EM. Increased expression of androgen receptor coregulator MAGE-11 in prostate cancer by DNA hypomethylation and cyclic AMP. Mol Cancer Res. 2009;7:523–35. doi: 10.1158/1541-7786.MCR-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li B, Qian XP, Pang XW, Zou WZ, Wang YP, Wu HY, et al. HCA587 antigen expression in normal tissues and cancers: correlation with tumor differentiation in hepatocellular carcinoma. Lab Invest. 2003;83:1185–92. doi: 10.1097/01.lab.0000080605.73839.96. [DOI] [PubMed] [Google Scholar]
- 68.Gilbert SF. Spermatogenesis. 6. Sunderland MA: Sinauer Associates; 2000. Developmental Biology. [Google Scholar]
- 69.Takahashi K, Shichijo S, Noguchi M, Hirohata M, Itoh K. Identification of MAGE-1 and MAGE-4 proteins in spermatogonia and primary spermatocytes of testis. Cancer Res. 1995;55:3478–82. [PubMed] [Google Scholar]
- 70.Jungbluth AA, Busam KJ, Kolb D, Iversen K, Coplan K, Chen YT, et al. Expression of MAGE-antigens in normal tissues and cancer. Int J Cancer. 2000;85:460–5. [PubMed] [Google Scholar]
- 71.Gjerstorff MF, Harkness L, Kassem M, Frandsen U, Nielsen O, Lutterodt M, et al. Distinct GAGE and MAGE-A expression during early human development indicate specific roles in lineage differentiation. Hum Reprod. 2008;23:2194–201. doi: 10.1093/humrep/den262. [DOI] [PubMed] [Google Scholar]
- 72.Chomez P, Williams R, De Backer O, Boon T, Vennstrom B. The SMAGE gene family is expressed in post-meiotic spermatids during mouse germ cell differentiation. Immunogenetics. 1996;43:97–100. doi: 10.1007/BF00186613. [DOI] [PubMed] [Google Scholar]
- 73.Clotman F, De Backer O, De Plaen E, Boon T, Picard J. Cell- and stage-specific expression of mage genes during mouse spermatogenesis. Mamm Genome. 2000;11:696–9. doi: 10.1007/s003350010116. [DOI] [PubMed] [Google Scholar]
- 74.Hou S, Xian L, Shi P, Li C, Lin Z, Gao X. The Magea gene cluster regulates male germ cell apoptosis without affecting the fertility in mice. Sci Rep. 2016;6:26735. doi: 10.1038/srep26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krausz C, Giachini C, Lo Giacco D, Daguin F, Chianese C, Ars E, et al. High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PLoS One. 2012;7:e44887. doi: 10.1371/journal.pone.0044887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pastuszak AWCC, Bekheirniac M, Lamb DJ. Melanoma antigen protein MAGEC1 mutation identified in familial non-obstructive azoospermia 71st Annual Meeting of the American Society for Reproductive Medicine; 2015; p. e80. [Google Scholar]
- 77.Gjerstorff MF, Kock K, Nielsen O, Ditzel HJ. MAGE-A1, GAGE and NY-ESO-1 cancer/testis antigen expression during human gonadal development. Hum Reprod. 2007;22:953–60. doi: 10.1093/humrep/del494. [DOI] [PubMed] [Google Scholar]
- 78.Mollgard K, Jespersen A, Lutterodt MC, Yding Andersen C, Hoyer PE, Byskov AG. Human primordial germ cells migrate along nerve fibers and Schwann cells from the dorsal hind gut mesentery to the gonadal ridge. Mol Hum Reprod. 2010;16:621–31. doi: 10.1093/molehr/gaq052. [DOI] [PubMed] [Google Scholar]
- 79.Bertrand M, Huijbers I, Chomez P, De Backer O. Comparative expression analysis of the MAGED genes during embryogenesis and brain development. Dev Dyn. 2004;230:325–34. doi: 10.1002/dvdy.20026. [DOI] [PubMed] [Google Scholar]
- 80.Chelly J, Mandel JL. Monogenic causes of X-linked mental retardation. Nat Rev Genet. 2001;2:669–80. doi: 10.1038/35088558. [DOI] [PubMed] [Google Scholar]
- 81.Mouri A, Sasaki A, Watanabe K, Sogawa C, Kitayama S, Mamiya T, et al. MAGE-D1 regulates expression of depression-like behavior through serotonin transporter ubiquitylation. J Neurosci. 2012;32:4562–80. doi: 10.1523/JNEUROSCI.6458-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J Biol Chem. 2002;277:22541–6. doi: 10.1074/jbc.M109728200. [DOI] [PubMed] [Google Scholar]
- 83.Teuber J, Mueller B, Fukabori R, Lang D, Albrecht A, Stork O. The ubiquitin ligase Praja1 reduces NRAGE expression and inhibits neuronal differentiation of PC12 cells. PLoS One. 2013;8:e63067. doi: 10.1371/journal.pone.0063067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dombret C, Nguyen T, Schakman O, Michaud JL, Hardin-Pouzet H, Bertrand MJ, et al. Loss of Maged1 results in obesity, deficits of social interactions, impaired sexual behavior and severe alteration of mature oxytocin production in the hypothalamus. Hum Mol Genet. 2012;21:4703–17. doi: 10.1093/hmg/dds310. [DOI] [PubMed] [Google Scholar]
- 85.Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med. 2012;14:10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- 86.Chibuk TK, Bischof JM, Wevrick R. A necdin/MAGE-like gene in the chromosome 15 autism susceptibility region: expression, imprinting, and mapping of the human and mouse orthologues. BMC Genet. 2001;2:22. doi: 10.1186/1471-2156-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le Moal M, Cau P, et al. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet. 2000;9:3101–10. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- 88.Niinobe M, Koyama K, Yoshikawa K. Cellular and subcellular localization of necdin in fetal and adult mouse brain. Dev Neurosci. 2000;22:310–9. doi: 10.1159/000017455. [DOI] [PubMed] [Google Scholar]
- 89.Andrieu D, Watrin F, Niinobe M, Yoshikawa K, Muscatelli F, Fernandez PA. Expression of the Prader-Willi gene Necdin during mouse nervous system development correlates with neuronal differentiation and p75NTR expression. Gene Expr Patterns. 2003;3:761–5. doi: 10.1016/s1567-133x(03)00138-8. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi Y, Matsuyama K, Takagi K, Sugiura H, Yoshikawa K. Arrest of cell growth by necdin, a nuclear protein expressed in postmitotic neurons. Biochem Biophys Res Commun. 1995;213:317–24. doi: 10.1006/bbrc.1995.2132. [DOI] [PubMed] [Google Scholar]
- 91.Maruyama K, Usami M, Aizawa T, Yoshikawa K. A novel brain-specific mRNA encoding nuclear protein (necdin) expressed in neurally differentiated embryonal carcinoma cells. Biochem Biophys Res Commun. 1991;178:291–6. doi: 10.1016/0006-291x(91)91812-q. [DOI] [PubMed] [Google Scholar]
- 92.Matsumoto K, Taniura H, Uetsuki T, Yoshikawa K. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001;272:173–9. doi: 10.1016/s0378-1119(01)00544-3. [DOI] [PubMed] [Google Scholar]
- 93.Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- 94.Zanella S, Watrin F, Mebarek S, Marly F, Roussel M, Gire C, et al. Necdin plays a role in the serotonergic modulation of the mouse respiratory network: implication for Prader-Willi syndrome. J Neurosci. 2008;28:1745–55. doi: 10.1523/JNEUROSCI.4334-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ren J, Lee S, Pagliardini S, Gerard M, Stewart CL, Greer JJ, et al. Absence of Ndn, encoding the Prader-Willi syndrome-deleted gene necdin, results in congenital deficiency of central respiratory drive in neonatal mice. J Neurosci. 2003;23:1569–73. doi: 10.1523/JNEUROSCI.23-05-01569.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, et al. Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet. 2000;9:1813–9. doi: 10.1093/hmg/9.12.1813. [DOI] [PubMed] [Google Scholar]
- 97.Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–72. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- 98.Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–9. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- 99.Kamaludin AA, Smolarchuk C, Bischof JM, Eggert R, Greer JJ, Ren J, et al. Muscle dysfunction caused by loss of Magel2 in a mouse model of Prader-Willi and Schaaf-Yang syndromes. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw225. [DOI] [PubMed] [Google Scholar]
- 100.Camfferman D, McEvoy RD, O'Donoghue F, Lushington K. Prader Willi Syndrome and excessive daytime sleepiness. Sleep Med Rev. 2008;12:65–75. doi: 10.1016/j.smrv.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 101.Devos J, Weselake SV, Wevrick R. Magel2, a Prader-Willi syndrome candidate gene, modulates the activities of circadian rhythm proteins in cultured cells. J Circadian Rhythms. 2011;9:12. doi: 10.1186/1740-3391-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mercer RE, Kwolek EM, Bischof JM, van Eede M, Henkelman RM, Wevrick R. Regionally reduced brain volume, altered serotonin neurochemistry, and abnormal behavior in mice null for the circadian rhythm output gene Magel2. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1085–99. doi: 10.1002/ajmg.b.30934. [DOI] [PubMed] [Google Scholar]
- 103.Hellings JA, Warnock JK. Self-injurious behavior and serotonin in Prader-Willi syndrome. Psychopharmacol Bull. 1994;30:245–50. [PubMed] [Google Scholar]
- 104.Warnock JK, Kestenbaum T. Pharmacologic treatment of severe skin-picking behaviors in Prader-Willi syndrome. Two case reports. Arch Dermatol. 1992;128:1623–5. [PubMed] [Google Scholar]
- 105.Mercer RE, Wevrick R. Loss of magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLoS One. 2009;4:e4291. doi: 10.1371/journal.pone.0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schaller F, Watrin F, Sturny R, Massacrier A, Szepetowski P, Muscatelli F. A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum Mol Genet. 2010;19:4895–905. doi: 10.1093/hmg/ddq424. [DOI] [PubMed] [Google Scholar]
- 107.Prader ALA, Willi H. Ein Syndrom von Adipositas, Kleinwuchs Kryptorchismus und Oligophrenie nach myatonieartigem Zustand im Neugeborenenalter. Schweiz Med Wochenschr. 1956;86:1260–1. [Google Scholar]
- 108.Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics. 1993;91:398–402. [PMC free article] [PubMed] [Google Scholar]
- 109.Verhoeven WM, Tuinier S. Prader-Willi syndrome: atypical psychoses and motor dysfunctions. Int Rev Neurobiol. 2006;72:119–30. doi: 10.1016/S0074-7742(05)72007-9. [DOI] [PubMed] [Google Scholar]
- 110.Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–34. [PMC free article] [PubMed] [Google Scholar]
- 111.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 112.Dykens EM, Lee E, Roof E. Prader-Willi syndrome and autism spectrum disorders: an evolving story. J Neurodev Disord. 2011;3:225–37. doi: 10.1007/s11689-011-9092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fountain MD, Jr, Schaaf CP. MAGEL2 and Oxytocin-Implications in Prader-Willi Syndrome and Beyond. Biol Psychiatry. 2015;78:78–80. doi: 10.1016/j.biopsych.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 114.de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18:3257–65. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–90. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013;45:1405–8. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fountain MD, Aten E, Cho MT, Juusola J, Walkiewicz MA, Ray JW, et al. The phenotypic spectrum of Schaaf-Yang syndrome: 18 new affected individuals from 14 families. Genet Med. 2016 doi: 10.1038/gim.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mejlachowicz D, Nolent F, Maluenda J, Ranjatoelina-Randrianaivo H, Giuliano F, Gut I, et al. Truncating Mutations of MAGEL2, a Gene within the Prader-Willi Locus, Are Responsible for Severe Arthrogryposis. Am J Hum Genet. 2015;97:616–20. doi: 10.1016/j.ajhg.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.de Bot ST, Vermeer S, Buijsman W, Heister A, Voorendt M, Verrips A, et al. Pure adult-onset spastic paraplegia caused by a novel mutation in the KIAA0196 (SPG8) gene. J Neurol. 2013;260:1765–9. doi: 10.1007/s00415-013-6870-x. [DOI] [PubMed] [Google Scholar]
- 122.Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152–61. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ropers F, Derivery E, Hu H, Garshasbi M, Karbasiyan M, Herold M, et al. Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum Mol Genet. 2011;20:2585–90. doi: 10.1093/hmg/ddr158. [DOI] [PubMed] [Google Scholar]
- 124.Vardarajan BN, Bruesegem SY, Harbour ME, Inzelberg R, Friedland R, St George-Hyslop P, et al. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33:2231, e15–e30. doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Y, Zhu M, Lin L, Fan X, Piao Z, Jiang X. Deficiency of Trim27 protects dopaminergic neurons from apoptosis in the neurotoxin model of Parkinson's disease. Brain Res. 2014;1588:17–24. doi: 10.1016/j.brainres.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 126.St Pourcain B, Whitehouse AJ, Ang WQ, Warrington NM, Glessner JT, Wang K, et al. Common variation contributes to the genetic architecture of social communication traits. Mol Autism. 2013;4:34. doi: 10.1186/2040-2392-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Magann EF, Chauhan SP, Doherty DA, Lutgendorf MA, Magann MI, Morrison JC. A review of idiopathic hydramnios and pregnancy outcomes. Obstet Gynecol Surv. 2007;62:795–802. doi: 10.1097/01.ogx.0000290349.58707.e0. [DOI] [PubMed] [Google Scholar]
- 128.Dorleijn DM, Cohen-Overbeek TE, Groenendaal F, Bruinse HW, Stoutenbeek P. Idiopathic polyhydramnios and postnatal findings. J Matern Fetal Neonatal Med. 2009;22:315–20. doi: 10.1080/14767050802531870. [DOI] [PubMed] [Google Scholar]
- 129.Laghmani K, Beck BB, Yang SS, Seaayfan E, Wenzel A, Reusch B, et al. Polyhydramnios, Transient Antenatal Bartter's Syndrome, and MAGED2 Mutations. N Engl J Med. 2016;374:1853–63. doi: 10.1056/NEJMoa1507629. [DOI] [PubMed] [Google Scholar]
- 130.Seyberth HW, Rascher W, Schweer H, Kuhl PG, Mehls O, Scharer K. Congenital hypokalemia with hypercalciuria in preterm infants: a hyperprostaglandinuric tubular syndrome different from Bartter syndrome. J Pediatr. 1985;107:694–701. doi: 10.1016/s0022-3476(85)80395-4. [DOI] [PubMed] [Google Scholar]
- 131.Jeck N, Schlingmann KP, Reinalter SC, Komhoff M, Peters M, Waldegger S, et al. Salt handling in the distal nephron: lessons learned from inherited human disorders. Am J Physiol Regul Integr Comp Physiol. 2005;288:R782–95. doi: 10.1152/ajpregu.00600.2004. [DOI] [PubMed] [Google Scholar]
- 132.Reinalter S, Devlieger H, Proesmans W. Neonatal Bartter syndrome: spontaneous resolution of all signs and symptoms. Pediatr Nephrol. 1998;12:186–8. doi: 10.1007/s004670050433. [DOI] [PubMed] [Google Scholar]
- 133.Donnelly BF, Needham PG, Snyder AC, Roy A, Khadem S, Brodsky JL, et al. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem. 2013;288:13124–35. doi: 10.1074/jbc.M113.455394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–90. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 135.van der Crabben SN, Hennus MP, McGregor GA, Ritter DI, Nagamani SC, Wells OS, et al. Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J Clin Invest. 2016;126:2881–92. doi: 10.1172/JCI82890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hudson JJ, Bednarova K, Kozakova L, Liao C, Guerineau M, Colnaghi R, et al. Interactions between the Nse3 and Nse4 components of the SMC5–6 complex identify evolutionarily conserved interactions between MAGE and EID Families. PLoS One. 2011;6:e17270. doi: 10.1371/journal.pone.0017270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guerineau M, Kriz Z, Kozakova L, Bednarova K, Janos P, Palecek J. Analysis of the Nse3/MAGE-binding domain of the Nse4/EID family proteins. PLoS One. 2012;7:e35813. doi: 10.1371/journal.pone.0035813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brasseur F, Rimoldi D, Liénard D, Lethé B, Carrel S, Arienti F, et al. Expression of MAGE genes in primary and metastatic cutaneous melanoma. International journal of cancer Journal international du cancer. 1995;63:375–80. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- 139.Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, et al. Tumor antigen expression in melanoma varies according to antigen and stage. Clin Cancer Res. 2006;12:764–71. doi: 10.1158/1078-0432.CCR-05-1544. [DOI] [PubMed] [Google Scholar]
- 140.Jungbluth AA, Chen YT, Busam KJ, Coplan K, Kolb D, Iversen K, et al. CT7 (MAGE-C1) antigen expression in normal and neoplastic tissues. Int J Cancer. 2002;99:839–45. doi: 10.1002/ijc.10416. [DOI] [PubMed] [Google Scholar]
- 141.Ayyoub M, Scarlata CM, Hamai A, Pignon P, Valmori D. Expression of MAGE-A3/6 in primary breast cancer is associated with hormone receptor negative status, high histologic grade, and poor survival. J Immunother. 2014;37:73–6. doi: 10.1097/CJI.0000000000000013. [DOI] [PubMed] [Google Scholar]
- 142.Yang F, Zhou X, Miao X, Zhang T, Hang X, Tie R, et al. MAGEC2, an epithelial-mesenchymal transition inducer, is associated with breast cancer metastasis. Breast Cancer Res Treat. 2014;145:23–32. doi: 10.1007/s10549-014-2915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daudi S, Eng KH, Mhawech-Fauceglia P, Morrison C, Miliotto A, Beck A, et al. Expression and immune responses to MAGE antigens predict survival in epithelial ovarian cancer. PLoS One. 2014;9:e104099. doi: 10.1371/journal.pone.0104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu Y, Wang C, Zhang Y, Jia L, Huang J. Overexpression of MAGE-A9 Is Predictive of Poor Prognosis in Epithelial Ovarian Cancer. Sci Rep. 2015;5:12104. doi: 10.1038/srep12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jang SJ, Soria JC, Wang L, Hassan KA, Morice RC, Walsh GL, et al. Activation of melanoma antigen tumor antigens occurs early in lung carcinogenesis. Cancer Res. 2001;61:7959–63. [PubMed] [Google Scholar]
- 146.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–62. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 147.Zhang S, Zhai X, Wang G, Feng J, Zhu H, Xu L, et al. High expression of MAGE-A9 in tumor and stromal cells of non-small cell lung cancer was correlated with patient poor survival. Int J Clin Exp Pathol. 2015;8:541–50. [PMC free article] [PubMed] [Google Scholar]
- 148.Filho PA, Lopez-Albaitero A, Xi L, Gooding W, Godfrey T, Ferris RL. Quantitative expression and immunogenicity of MAGE-3 and -6 in upper aerodigestive tract cancer. Int J Cancer. 2009;125:1912–20. doi: 10.1002/ijc.24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Noh ST, Lee HS, Lim SJ, Kim SW, Chang HK, Oh J, et al. MAGE-A1–6 expression in patients with head and neck squamous cell carcinoma: impact on clinical patterns and oncologic outcomes. Int J Clin Oncol. 2016;21:875–82. doi: 10.1007/s10147-016-0989-6. [DOI] [PubMed] [Google Scholar]
- 150.Zamuner FT, Karia BT, de Oliveira CZ, Santos CR, Carvalho AL, Vettore AL. A Comprehensive Expression Analysis of Cancer Testis Antigens in Head and Neck Squamous Cell Carcinoma Revels MAGEA3/6 as a Marker for Recurrence. Mol Cancer Ther. 2015;14:828–34. doi: 10.1158/1535-7163.MCT-14-0796. [DOI] [PubMed] [Google Scholar]
- 151.Pattani KM, Soudry E, Glazer CA, Ochs MF, Wang H, Schussel J, et al. MAGEB2 is activated by promoter demethylation in head and neck squamous cell carcinoma. PLoS One. 2012;7:e45534. doi: 10.1371/journal.pone.0045534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hartmann S, Brisam M, Rauthe S, Driemel O, Brands RC, Rosenwald A, et al. Contrary melanoma-associated antigen-A expression at the tumor front and center: A comparative analysis of stage I and IV head and neck squamous cell carcinoma. Oncol Lett. 2016;12:2942–7. doi: 10.3892/ol.2016.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Atanackovic D, Hildebrandt Y, Jadczak A, Cao Y, Luetkens T, Meyer S, et al. Cancer-testis antigens MAGE-C1/CT7 and MAGE-A3 promote the survival of multiple myeloma cells. Haematologica. 2010;95:785–93. doi: 10.3324/haematol.2009.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68:8104–12. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 155.Yang B, O'Herrin S, Wu J, Reagan-Shaw S, Ma Y, Nihal M, et al. Select cancer testes antigens of the MAGE-A, -B, and -C families are expressed in mast cell lines and promote cell viability in vitro and in vivo. J Invest Dermatol. 2007;127:267–75. doi: 10.1038/sj.jid.5700548. [DOI] [PubMed] [Google Scholar]
- 156.Song X, Hao J, Wang J, Guo C, Wang Y, He Q, et al. The cancer/testis antigen MAGEC2 promotes amoeboid invasion of tumor cells by enhancing STAT3 signaling. Oncogene. 2016 doi: 10.1038/onc.2016.314. [DOI] [PubMed] [Google Scholar]
- 157.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 158.Yamada R, Takahashi A, Torigoe T, Morita R, Tamura Y, Tsukahara T, et al. Preferential expression of cancer/testis genes in cancer stem-like cells: proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens. 2013;81:428–34. doi: 10.1111/tan.12113. [DOI] [PubMed] [Google Scholar]
- 159.Yin B, Zeng Y, Liu G, Wang X, Wang P, Song Y. MAGE-A3 is highly expressed in a cancer stem cell-like side population of bladder cancer cells. Int J Clin Exp Pathol. 2014;7:2934–41. [PMC free article] [PubMed] [Google Scholar]
- 160.Wienand K, Shires K. The use of MAGE C1 and flow cytometry to determine the malignant cell type in multiple myeloma. PLoS One. 2015;10:e0120734. doi: 10.1371/journal.pone.0120734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, et al. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105:20422–7. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2'-deoxycytidine. Cancer Res. 1994;54:1766–71. [PubMed] [Google Scholar]
- 163.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93:7149–53. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Honda T, Tamura G, Waki T, Kawata S, Terashima M, Nishizuka S, et al. Demethylation of MAGE promoters during gastric cancer progression. Br J Cancer. 2004;90:838–43. doi: 10.1038/sj.bjc.6601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.De Smet C, Courtois SJ, Faraoni I, Lurquin C, Szikora JP, De Backer O, et al. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics. 1995;42:282–90. doi: 10.1007/BF00176446. [DOI] [PubMed] [Google Scholar]
- 166.Sigalotti L, Coral S, Nardi G, Spessotto A, Cortini E, Cattarossi I, et al. Promoter methylation controls the expression of MAGE2, 3 and 4 genes in human cutaneous melanoma. J Immunother. 2002;25:16–26. doi: 10.1097/00002371-200201000-00002. [DOI] [PubMed] [Google Scholar]
- 167.Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem. 2006;281:10118–26. doi: 10.1074/jbc.M510469200. [DOI] [PubMed] [Google Scholar]
- 168.Sigalotti L, Covre A, Zabierowski S, Himes B, Colizzi F, Natali PG, et al. Cancer testis antigens in human melanoma stem cells: expression, distribution, and methylation status. J Cell Physiol. 2008;215:287–91. doi: 10.1002/jcp.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suyama T, Ohashi H, Nagai H, Hatano S, Asano H, Murate T, et al. The MAGE-A1 gene expression is not determined solely by methylation status of the promoter region in hematological malignancies. Leuk Res. 2002;26:1113–8. doi: 10.1016/s0145-2126(02)00048-6. [DOI] [PubMed] [Google Scholar]
- 170.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 171.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–49. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 173.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Serrano A, Garcia A, Abril E, Garrido F, Ruiz-Cabello F. Methylated CpG points identified within MAGE-1 promoter are involved in gene repression. Int J Cancer. 1996;68:464–70. doi: 10.1002/(SICI)1097-0215(19961115)68:4<464::AID-IJC11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 175.Smith IM, Glazer CA, Mithani SK, Ochs MF, Sun W, Bhan S, et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS One. 2009;4:e4961. doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang B, Wu J, Maddodi N, Ma Y, Setaluri V, Longley BJ. Epigenetic control of MAGE gene expression by the KIT tyrosine kinase. J Invest Dermatol. 2007;127:2123–8. doi: 10.1038/sj.jid.5700836. [DOI] [PubMed] [Google Scholar]
- 177.Kondo T, Zhu X, Asa SL, Ezzat S. The cancer/testis antigen melanoma-associated antigen-A3/A6 is a novel target of fibroblast growth factor receptor 2-IIIb through histone H3 modifications in thyroid cancer. Clin Cancer Res. 2007;13:4713–20. doi: 10.1158/1078-0432.CCR-07-0618. [DOI] [PubMed] [Google Scholar]
- 178.Zhu X, Asa SL, Ezzat S. Fibroblast growth factor 2 and estrogen control the balance of histone 3 modifications targeting MAGE-A3 in pituitary neoplasia. Clin Cancer Res. 2008;14:1984–96. doi: 10.1158/1078-0432.CCR-07-2003. [DOI] [PubMed] [Google Scholar]
- 179.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–97. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 180.Becker JC, Gillitzer R, Brocker EB. A member of the melanoma antigen-encoding gene (MAGE) family is expressed in human skin during wound healing. Int J Cancer. 1994;58:346–8. doi: 10.1002/ijc.2910580306. [DOI] [PubMed] [Google Scholar]
- 181.McCurdy DK, Tai LQ, Imfeld KL, Schwartz M, Zaldivar F, Berman MA. Expression of melanoma antigen gene by cells from inflamed joints in juvenile rheumatoid arthritis. J Rheumatol. 2002;29:2219–24. [PubMed] [Google Scholar]
- 182.Hoftman AD, Tai LQ, Tze S, Seligson D, Gatti RA, McCurdy DK. MAGE-B2 autoantibody: a new biomarker for pediatric systemic lupus erythematosus. J Rheumatol. 2008;35:2430–8. doi: 10.3899/jrheum.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McCurdy DK, Tai LQ, Nguyen J, Wang Z, Yang HM, Udar N, et al. MAGE Xp-2: a member of the MAGE gene family isolated from an expression library using systemic lupus erythematosus sera. Mol Genet Metab. 1998;63:3–13. doi: 10.1006/mgme.1997.2639. [DOI] [PubMed] [Google Scholar]
- 184.Bart J, Groen HJ, van der Graaf WT, Hollema H, Hendrikse NH, Vaalburg W, et al. An oncological view on the blood-testis barrier. Lancet Oncol. 2002;3:357–63. doi: 10.1016/s1470-2045(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 185.Fiszer D, Kurpisz M. Major histocompatibility complex expression on human, male germ cells: a review. Am J Reprod Immunol. 1998;40:172–6. doi: 10.1111/j.1600-0897.1998.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 186.Kalejs M, Erenpreisa J. Cancer/testis antigens and gametogenesis: a review and "brain-storming" session. Cancer Cell Int. 2005;5:4. doi: 10.1186/1475-2867-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marchand M, van Baren N, Weynants P, Brichard V, Dreno B, Tessier MH, et al. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int J Cancer. 1999;80:219–30. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 188.Nestle FO, Alijagic S, Gilliet M, Sun Y, Grabbe S, Dummer R, et al. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat Med. 1998;4:328–32. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 189.Thurner B, Haendle I, Roder C, Dieckmann D, Keikavoussi P, Jonuleit H, et al. Vaccination with mage-3A1 peptide-pulsed mature, monocyte-derived dendritic cells expands specific cytotoxic T cells and induces regression of some metastases in advanced stage IV melanoma. J Exp Med. 1999;190:1669–78. doi: 10.1084/jem.190.11.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Marchand M, Punt CJ, Aamdal S, Escudier B, Kruit WH, Keilholz U, et al. Immunisation of metastatic cancer patients with MAGE-3 protein combined with adjuvant SBAS-2: a clinical report. Eur J Cancer. 2003;39:70–7. doi: 10.1016/s0959-8049(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 191.Kruit WH, Suciu S, Dreno B, Mortier L, Robert C, Chiarion-Sileni V, et al. Selection of immunostimulant AS15 for active immunization with MAGE-A3 protein: results of a randomized phase II study of the European Organisation for Research and Treatment of Cancer Melanoma Group in Metastatic Melanoma. J Clin Oncol. 2013;31:2413–20. doi: 10.1200/JCO.2012.43.7111. [DOI] [PubMed] [Google Scholar]
- 192.Saiag P, Gutzmer R, Ascierto PA, Maio M, Grob JJ, Murawa P, et al. Prospective assessment of a gene signature potentially predictive of clinical benefit in metastatic melanoma patients following MAGE-A3 immunotherapeutic (PREDICT) Ann Oncol. 2016;27:1947–53. doi: 10.1093/annonc/mdw291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Atanackovic D, Altorki NK, Stockert E, Williamson B, Jungbluth AA, Ritter E, et al. Vaccine-induced CD4+ T cell responses to MAGE-3 protein in lung cancer patients. J Immunol. 2004;172:3289–96. doi: 10.4049/jimmunol.172.5.3289. [DOI] [PubMed] [Google Scholar]
- 194.Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25(Suppl 2):B61–71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 195.Tyagi P, Mirakhur B. MAGRIT: the largest-ever phase III lung cancer trial aims to establish a novel tumor-specific approach to therapy. Clin Lung Cancer. 2009;10:371–4. doi: 10.3816/CLC.2009.n.052. [DOI] [PubMed] [Google Scholar]
- 196.Vansteenkiste J, Zielinski M, Linder A, Dahabreh J, Gonzalez EE, Malinowski W, et al. Adjuvant MAGE-A3 immunotherapy in resected non-small-cell lung cancer: phase II randomized study results. J Clin Oncol. 2013;31:2396–403. doi: 10.1200/JCO.2012.43.7103. [DOI] [PubMed] [Google Scholar]
- 197.Vansteenkiste JF, Cho BC, Vanakesa T, De Pas T, Zielinski M, Kim MS, et al. Efficacy of the MAGE-A3 cancer immunotherapeutic as adjuvant therapy in patients with resected MAGE-A3-positive non-small-cell lung cancer (MAGRIT): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2016;17:822–35. doi: 10.1016/S1470-2045(16)00099-1. [DOI] [PubMed] [Google Scholar]
- 198.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–9. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–71. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ye Q, Shen Y, Wang X, Yang J, Miao F, Shen C, et al. Hypermethylation of HLA class I gene is associated with HLA class I down-regulation in human gastric cancer. Tissue Antigens. 2010;75:30–9. doi: 10.1111/j.1399-0039.2009.01390.x. [DOI] [PubMed] [Google Scholar]
- 201.Serrano A, Tanzarella S, Lionello I, Mendez R, Traversari C, Ruiz-Cabello F, et al. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2'-deoxycytidine treatment. Int J Cancer. 2001;94:243–51. doi: 10.1002/ijc.1452. [DOI] [PubMed] [Google Scholar]
- 202.Sigalotti L, Altomonte M, Colizzi F, Degan M, Rupolo M, Zagonel V, et al. 5-Aza-2'-deoxycytidine (decitabine) treatment of hematopoietic malignancies: a multimechanism therapeutic approach? Blood. 2003;101:4644–6. doi: 10.1182/blood-2002-11-3458. discussion 5–6. [DOI] [PubMed] [Google Scholar]
- 203.Bao L, Dunham K, Lucas K. MAGE-A1, MAGE-A3, and NY-ESO-1 can be upregulated on neuroblastoma cells to facilitate cytotoxic T lymphocyte-mediated tumor cell killing. Cancer Immunol Immunother. 2011;60:1299–307. doi: 10.1007/s00262-011-1037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Krishnadas DK, Shusterman S, Bai F, Diller L, Sullivan JE, Cheerva AC, et al. A phase I trial combining decitabine/dendritic cell vaccine targeting MAGE-A1, MAGE-A3 and NY-ESO-1 for children with relapsed or therapy-refractory neuroblastoma and sarcoma. Cancer Immunol Immunother. 2015;64:1251–60. doi: 10.1007/s00262-015-1731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 206.Bhatia N, Yang B, Xiao TZ, Peters N, Hoffmann MF, Longley BJ. Identification of novel small molecules that inhibit protein-protein interactions between MAGE and KAP-1. Arch Biochem Biophys. 2011;508:217–21. doi: 10.1016/j.abb.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tajima K, Obata Y, Tamaki H, Yoshida M, Chen YT, Scanlan MJ, et al. Expression of cancer/testis (CT) antigens in lung cancer. Lung Cancer. 2003;42:23–33. doi: 10.1016/s0169-5002(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 208.Yao J, Caballero OL, Yung WK, Weinstein JN, Riggins GJ, Strausberg RL, et al. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res. 2014;2:371–9. doi: 10.1158/2326-6066.CIR-13-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hou S, Sang M, Zhao L, Hou R, Shan B. The expression of MAGE-C1 and MAGE-C2 in breast cancer and their clinical significance. Am J Surg. 2016;211:142–51. doi: 10.1016/j.amjsurg.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 210.Chen X, Wang L, Liu J, Huang L, Yang L, Gao Q, Zhang Y. Expression and prognostic relevance of MAGE-A3 and MAGE-C2 in non-small cell lung cancer. Oncology Letters. 2017;13:1609–28. doi: 10.3892/ol.2017.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Balafoutas D, zur Hausen A, Mayer S, Hirschfeld M, Jaeger M, Denschlag D, et al. Cancer testis antigens and NY-BR-1 expression in primary breast cancer: prognostic and therapeutic implications. BMC cancer. 2013;13:271. doi: 10.1186/1471-2407-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu X, Tang X, Lu M, Tang Q, Zhang H, Zhu H, et al. Overexpression of MAGE-A9 predicts unfavorable outcome in breast cancer. Exp Mol Pathol. 2014;97:579–84. doi: 10.1016/j.yexmp.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 213.Zhan W, Zhang Z, Zhang Y, Ma J, Wu T, Gu Y, et al. Prognostic value of MAGE-A9 expression in patients with colorectal cancer. Clinics and research in hepatology and gastroenterology. 2016;40:239–45. doi: 10.1016/j.clinre.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 214.Han L, Jiang B, Wu H, Zhang S, Lu X. Expression and prognostic value of MAGE-A9 in laryngeal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:6734–42. [PMC free article] [PubMed] [Google Scholar]
- 215.Gu X, Fu M, Ge Z, Zhan F, Ding Y, Ni H, et al. High expression of MAGE-A9 correlates with unfavorable survival in hepatocellular carcinoma. Sci Rep. 2014;4:6625. doi: 10.1038/srep06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sang M, Gu L, Liu F, Lian Y, Yin D, Fan X, et al. Prognostic Significance of MAGE-A11 in Esophageal Squamous Cell Carcinoma and Identification of Related Genes Based on DNA Microarray. Archives of medical research. 2016;47:151–61. doi: 10.1016/j.arcmed.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 217.Wu J, Wang J, Shen W. Identification of MAGEA12 as a prognostic outlier gene in gastric cancers. Neoplasma. 2017:64. doi: 10.4149/neo_2017_210. [DOI] [PubMed] [Google Scholar]