Abstract
Emerging complementary metal oxide semiconductor (CMOS)-based, high-density microelectrode array (HD-MEA) devices provide high spatial resolution at subcellular level and a large number of readout channels. These devices allow for simultaneous recording of extracellular activity of a large number of neurons with every neuron being detected by multiple electrodes. To analyze the recorded signals, spiking events have to be assigned to individual neurons, a process referred to as “spike sorting.” For a set of observed signals, which constitute a linear mixture of a set of source signals, independent component (IC) analysis (ICA) can be used to demix blindly the data and extract the individual source signals. This technique offers great potential to alleviate the problem of spike sorting in HD-MEA recordings, as it represents an unsupervised method to separate the neuronal sources. The separated sources or ICs then constitute estimates of single-neuron signals, and threshold detection on the ICs yields the sorted spike times. However, it is unknown to what extent extracellular neuronal recordings meet the requirements of ICA. In this paper, we evaluate the applicability of ICA to spike sorting of HD-MEA recordings. The analysis of extracellular neuronal signals, recorded at high spatiotemporal resolution, reveals that the recorded data cannot be modeled as a purely linear mixture. As a consequence, ICA fails to separate completely the neuronal signals and cannot be used as a stand-alone method for spike sorting in HD-MEA recordings. We assessed the demixing performance of ICA using simulated data sets and found that the performance strongly depends on neuronal density and spike amplitude. Furthermore, we show how postprocessing techniques can be used to overcome the most severe limitations of ICA. In combination with these postprocessing techniques, ICA represents a viable method to facilitate rapid spike sorting of multi-dimensional neuronal recordings.
Keywords: spike sorting, multiunit, multielectrode
in the field of neurophysiology research, extracellular recordings of neural activity have become an important means to study intercell interaction and firing patterns to understand better the physiology and the information processing of neuronal networks. In multiunit recordings, the electrodes monitor the simultaneous activity of a large number of individual neurons. For the analysis, the spike trains of the individual neurons then have to be extracted from the recorded data, a process usually referred to as “spike sorting” (Lewicki 1998). Generally, the spike-sorting tasks consists of two fundamental steps: to 1) detect action potential (AP) events in the data; and 2) classify them into groups. Whereas a main problem for the spike detection task is to deal with data recorded under low signal-to-noise-ratio (SNR) conditions, the most severe challenge to the classification problem is the presence of overlapping spikes from different neurons.
The sorting performance can be drastically increased by using multielectrode devices such as tetrodes (Gray et al. 1995). In these systems, which feature several closely spaced electrodes, an AP is simultaneously measured on more than one electrode. In addition to the temporal cues of the waveform, these multielectrode devices reveal information about the spatial cues of the spike shape distribution. This additional information can be efficiently used to separate units.
Planar microelectrode arrays (MEAs) are arrangements of electrodes for extracellular measurements of multiple cells on a chip surface. They are widely used to study the dynamics of the neuronal networks, as they enable simultaneous access to a large number of neurons. Traditional MEA systems incorporate 60–200 passive metal electrodes on a silicon or glass surface, which are connected to external circuitry and typically feature interelectrode distances of 100–200 μm (Stett et al. 2003). Since the signal of a neuron is detected by at most one electrode on such MEAs, the spike-sorting problem is the same as for single-electrode recordings (Shoham et al. 2003; Zhang et al. 2003).
Recently, “active” MEAs based on complementary metal oxide semiconductor (CMOS) technology have been developed (Berdondini et al. 2005; Eversmann et al. 2003; Frey et al. 2010; Hutzler et al. 2006). These devices feature signal-conditioning circuitry on-chip and provide much larger electrode densities and, thereby, enable to conduct electrophysiological experiments at cellular or subcellular level. The MEA used for our experiments (Frey et al. 2010) features 11,011 electrodes (3,161 electrodes/mm2) as well as 126 read-out channels. The possibility to select arbitrarily a subset of electrodes for recording or stimulation entails the possibility to use different electrode configurations, such as high-density (HD) or sparse arrangements.
The HD of the electrodes on the array enables recordings at subcellular resolution, with the activity of every neuron being measured by multiple electrodes. Although this feature improves sorting capabilities, the large number of channels and the highly redundant nature of HD-MEA data pose challenges to the strategies for event detection and classification. Particularly, two issues arise when applying standard analysis techniques to HD-MEA data.
How to Perform Event Detection in Redundant Data?
A neuronal AP will produce spikes on several electrodes, leading to threshold crossing events (TCEs) on these electrodes. Ideally, these events should be grouped and considered as one single spiking event, since their origin is the same AP of the same cell. This could be performed by merging TCEs, which are spatially and temporally closely aligned. However, this task gets challenging for large numbers of spatially highly overlapping neurons.
Which Features Should be Used for Unit Separation?
For recordings with hundreds to thousands of electrodes, the feature space needs to be reduced in a way that only the electrodes are used that prominently contribute to unit separation.
Only a few methods have been published that specifically target spike sorting of HD, redundant recordings. A sequential approach that targets one electrode at a time is proposed in Litke et al. (2004). This simple and robust method lacks efficiency by repeating the clustering of the same cells many times and does not handle the overlap problem. Another approach based on template matching has been presented in Segev et al. (2004) using data from MEA recordings with up to 30 electrodes. The overlap problem is addressed by this approach; this method, however, is formulated for a limited number of electrodes and templates and requires prior knowledge of the neurons and their waveforms. The challenges of efficient, automatic spike-sorting and validation techniques for multielectrode systems have been discussed in Einevoll et al. (2012).
Independent component (IC) analysis (ICA) (Hyvärinen and Oja 2000; Stone 2002) is a blind source separation technique that can be used to demix a set of independent source signals that were linearly mixed across a number of observed signals.
The high potential of ICA to be applied to spike-sorting problems has been discussed in Brown et al. (2001). Neurons have been regarded as independent signal sources, which are linearly mixed across the recording electrodes. ICA has the following requirements. 1) The source signals are non-Gaussian and statistically independent. 2) A linear, instantaneous mixture of the source signals is assumed to produce the observed signals. 3) The number of observed signals needs to be equal to or larger than the number of source signals.
Although neurons are not independent from each other, as they can be synaptically connected or may receive common input, the individual spike trains can be regarded as statistically independent, since dependence in this context refers to instantaneous overlaps rather than time-delayed dependence (Brown et al. 2001). Therefore, concerning the first requirement, ICA will only fail to separate two neurons if they fire always precisely at the same time.
Recently, some studies attempted to combine closely spaced electrode recording techniques with computationally efficient ICA algorithms such as FastICA (Hyvärinen 1999). In Hermle et al. (2004) and Snellings et al. (2006), ICA is applied as a preprocessing step on recorded data to reduce cross talk and increase data quality. The major obstacle for an efficient use of ICA has been, however, requirement 3. In standard in vivo experiments with tetrodes, the number of neurons is likely to be larger than the number of recording sites. One attempt to overcome this restriction for ICA included to perform k-means clustering of detected waveforms in a preprocessing step and to decompose the waveforms of each cluster individually with ICA (Takahashi et al. 2003), since the number of neurons in the clusters is expected to be lower than the number of recording sites. A second approach included to increase the number of recording sites to 12 (Takahashi and Sakurai 2005).
The large number of electrodes of HD-MEAs is suited to meet requirement 3, which renders ICA a good candidate to separate HD-MEA data. Ideally, the independent source signals, found by ICA, correspond to individual activity signals of neurons. Applying threshold detection on each source signal then yields the sorted spike times of the neurons that have been recorded on the array. Making use of the redundancy in the data, ICA should also provide an increase in the SNR of the demixed signal compared with the recorded signals and, therefore, improve the spike detection performance. By separating the neuronal sources, it also holds promise to solve the problem of overlapping spikes.
In this study, we explore the applicability of ICA for blind and rapid spike sorting of HD-MEA recordings.
Methods
Data Acquisition System
Recordings were made with the HD-MEA recording system described by Frey et al. (2009, 2010). The array is integrated into a microsystem chip, fabricated in a 0.6-μm CMOS process. It accommodates a total of 11,011 electrodes of 7-μm diameter on an area of 1.99 × 1.75 mm2 (19-μm hexagonal center-to-center pitch, density of 3,161 electrodes/mm2). One hundred twenty-six bidirectional channels are implemented on-chip, featuring recording and stimulation electronics. The channels are connected to the electrodes via a flexible switch matrix lying underneath the array. This system provides routing flexibility to select almost arbitrary electrode configurations, which can be changed within milliseconds.
The programmable gain amplifiers (0–80 dB) allow for recording neuronal signals throughout a wide range of amplitudes, which depend on the respective cell type. Offset and fluctuations resulting from the electrode-saline interface are removed by first-order high-pass filtering the analog signals (tunable cutoff frequency 0.3–100 Hz). The frequency range is limited toward the high-frequency end by means of a tunable second-order low-pass filter (3.5–14 kHz). The signals are multiplexed and digitalized with 8-bit analog-to-digital converters with a sampling rate of 20 kHz.
The data were stored on a standard PC, and the analysis was conducted using MATLAB. Before postprocessing, all the data were digitally band-pass filtered (500–3,000 Hz).
Extracellular Recordings from Retinal Ganglion Cells
We used the rd1 mouse retina to record spontaneous activity (Stasheff 2008) from retinal ganglion cells (RGCs). All animal experiments and procedures were approved by the Swiss Federal Veterinary Office. The retinae were isolated at ambient light from the C3H/HeNCrl (rd1) mouse strain at P80 in Ringer medium (in mM: 110 NaCl, 2.5 KCl, 1 CaCl2, 1.6 MgCl2, 10 d-glucose, 22 NaHCO3, bubbled with 5% CO2-95% O2). Once a piece of the retina was isolated, it was placed with the RGC layer adjacent to the MEA. The retina was fixed on the array by a permeable membrane (polyester, 10-μm thickness, 0.4-μm pore size) and superfused with Ringer medium at 36°C.
All recordings shown in this study were done with electrode configurations of blocks at highest possible spatial resolution. The largest HD block that can be simultaneously read in the configurable array is 6 × 17 electrodes and covers an area of approximately 80 × 320 μm2.
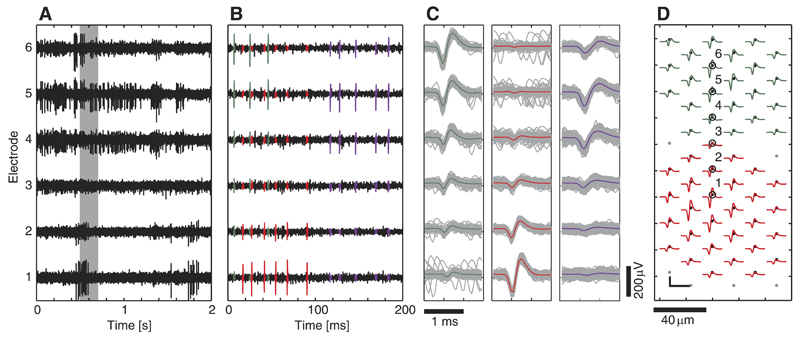
An example of recorded RGC activity is shown in Fig. 1. Three neurons were identified in an HD block, using a manually supervised custom-designed spike-sorting method based on principal component analysis (PCA) and expectation-maximization (EM) clustering (KlustaKwik; Harris et al. 2000). The characteristic multichannel signature of the neurons is obtained by spike-triggered averaging (STA) of the individual aligned traces (Fig. 1C). This cell-specific footprint will be referred to as template, namely the distribution of the average spike shape across the electrodes. Figure 1D shows the templates of two cells that have significant energy on a large number of electrodes.
Fig. 1.
Example of raw data and neuronal templates. A: 2 s o frecorded data from 6 selected electrodes, labeled in D. B: data segment indicated by a gray rectangle in A. The spikes of 3 retinal ganglion cells (RGCs) that were identified by supervised spike sorting were colored (red, green, and violet). C: superposition of all detected spike waveforms within 50 s of data (green neuron: 187 spikes; red neuron: 191 spikes; violet neuron: 1,046 spikes). The colored line shows the averaged waveform [spike-triggered averaging (STA)]. D: cell-specific templates (only green and red neuron shown for visualization purposes). Black dots indicate electrode positions on the microelectrode array (MEA). Colored waveforms correspond to the STA of the neuron on the respective electrode. The numbered electrodes refer to those in A–C. Scale bar: 100 μV/1.8 ms.
Templates and firing characteristics of recorded neurons were found to be very similar across several recording experiments (n = 10).
Assumption of Linear Dependence
ICA requires a linear and instantaneous mixture of the source signals across the electrodes. This requirement implies that the signals of the sources (neurons) on the different electrodes are linearly dependent and do not contain phase shifts. The assumption of linear dependence can be validated by testing the degree of linearity within the waveforms of the neuron template.
We measure the linearity between two vectors, a and b, using the normalized cross-correlation coefficient (CC):
| (1) |
The CC can range from −1 to 1, where two vectors with a CC of 1 are perfectly linearly dependent and two vectors with a CC of −1 are inversely linearly dependent. A CC of 0 indicates that the vectors are orthogonal. Therefore, the linearity assumption is met if the absolute CC values of the spike waveforms of the individual neuron templates are close to 1.
ICA
Let the recorded time-series signals on M electrodes be X = [x1(t),…, xM(t)]T and be the intrinsic signals generated by N single neurons. Given the assumption of linearity, stated in the previous section, we can model the recordings as a linear mixture of the neuronal signals:
| (2) |
Under the additional assumptions that the individual signals are non-Gaussian and statistically independent and that there are more recording sites than neurons, the mixing matrix A can be estimated blindly by applying ICA (FastICA; Hyvärinen 1999) directly to the recorded data. The ICs, which ideally represent individual-neuron signals, are obtained by:
| (3) |
The ICs are the rows in S = [s1(t), …, sM(t)]T, and W = A−1 is the demixing matrix. In the following, we will refer to the columns of A as the mixing coefficient vectors (MCVs), whereas the rows of W will be called demixing coefficient vectors (DCVs). Note, that the estimated mixing matrix has a dimension of M × M.
If the individual templates are known, we can evaluate the separation by directly demixing the templates with W. For a given neuron, i, consider the template matrix for which the jth row is the STA waveform at the electrode j. Then the demixed template is:
| (4) |
In the case of perfect separation, Hi contains the intrinsic neuron waveform in the ith row and zeros in all the other rows. ICA offers three main features if all the assumptions are met. 1) The redundancy is reduced so that only ICs 1, …, N contain significant signals (spikes), whereas ICs N + 1,…, M contain only noise. This allows for extracting the number of neurons from the number of ICs containing spikes. 2) The recordings are demixed in a way that every IC only contains the spikes of one corresponding neuronal source. Consequently, applying threshold detection to the ICs yields the sorted spike times. 3) ICA achieves an increase in SNR compared with single-channel signals by accumulating signals of several electrodes in the ICs.
The use of ICA, however, typically entails two major problems if the linearity assumption is not totally fulfilled. 1) One IC can contain signals from more than one neuron. In that case, demixing does not achieve perfect separation. 2) A neuron can contribute signals to two or more ICs. In that case, more than N ICs contain neuronal signals, and ICA does not completely reduce the redundancy.
Evaluation Criteria
In the following, we formulate several evaluation metrics to characterize the performance of ICA with regard to the points elaborated in the previous section.
SNR
We define the SNR of neuron i in the recorded signal X as
| (5) |
where j is the electrode on which the template Fi has its highest peak value, and where σj is the standard deviation of the noise signal on that electrode. The SNREL thus denotes the peak value in the template divided by the noise standard deviation.
Next, we define the SNR of the neurons in the ICs. FastICA normalizes the DCVs so that every IC signal has unit variance. Instead, we want the noise on the ICs to have unit variance. Therefore, we first normalize all the DCVs:
| (6) |
Here, wk is the kth row of W, and C is the instantaneous noise covariance matrix between the electrodes. is the new demixing matrix with normalized DCVs, having on its kth row. This normalization ensures equivalent noise levels on all the ICs and allows for comparison of their signals. The SNR of neuron i in the ICs is the peak value of the demixed template
| (7) |
Redundancy
Another important ICA performance measure is how well redundancy in the ICs is reduced compared with the recorded signals. Therefore, the redundancies and are defined, where the first denotes the number of electrodes and the latter denotes the number of ICs, on which the signal of neuron i exceeds the threshold value of five times the noise standard deviation.
Separability
We also introduce a measure for the separability of the neurons on the ICs. A neuron i with high separability must have a high peak signal on an IC k, on which all other neurons have only low peak signals. Thus its separability is the difference between its peak on IC k and the highest peak of any other neuron on IC k. We determine k by maximizing the separability, taking into account that the relevant peak can feature either positive- or negative-sign amplitude.
| (8) |
Analogously, we define the separability of a neuron in the recorded signals as the difference between the peak of its template and the highest peak of any other neuron template on the same electrode, divided by the noise standard deviation on that electrode.
| (9) |
Simulation of Recorded Neuronal Activity
For evaluation purposes, simulated data were generated. In this study, we used two types of simulated data sets. Data set A contained simulated activity of three neurons that had spatially overlapping templates that were extracted from digitally unfiltered, recorded data. This data set is mainly used for visualization purposes. Spike sorting to extract the templates for data simulation was performed using manually supervised PCA and EM clustering.
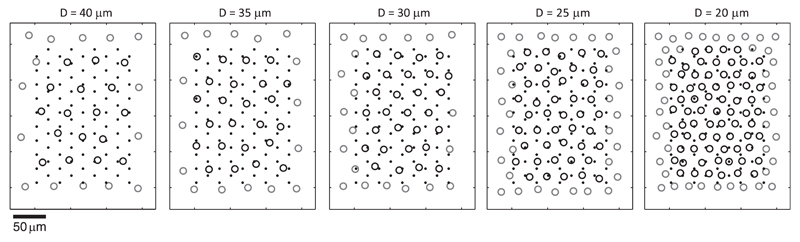
For a systematic analysis, we simulated RGC activity at different cell densities in data set B. Therefore, we used eight well-isolated, manually selected neuronal templates extracted from recorded unfiltered data as model templates. Higher spatial resolution was obtained by interpolating the model templates on a grid (5-μm pitch). By modifying position, orientation, amplitude, and spatial extension of the model templates, individual neuron templates were simulated. The modified templates were positioned on a gridlike structure with equidistant points. Peak-to-peak amplitudes were set randomly (uniformly distributed between 50 and 300 μV). The grid of neurons covered an HD block of 90 electrodes on an area of 130 × 185 μm2.
We simulated 5 different configurations with average neuron distances (D) between 40 and 20 μm (average distance to the 6 neighboring neurons in a hexagonal arrangement). Out of 20 simulations of 30 s each, which were generated for each configuration, a subset of 358 neurons per configuration was considered for the analysis. Surrounding neurons outside the electrode block were simulated but not included in the analysis. An overview of the different configurations represented in data set B is given in Table 1 and illustrated in Fig. 2.
Table 1. Simulation overview for configurations in data set B.
| Configuration | Average Neuron Distance, μm | Cell Density, Neurons/mm2 | Approximate Number of Neurons per Simulation (Included in Analysis/Simulated) | Number of Simulations Used | Neurons Considered in Analysis |
|---|---|---|---|---|---|
| 1 | 40 | 722 | 18/32 | 20 | 358 |
| 2 | 35 | 943 | 24/40 | 15 | 358 |
| 3 | 30 | 1,283 | 30/50 | 12 | 358 |
| 4 | 25 | 1,848 | 46/77 | 8 | 358 |
| 5 | 20 | 2,887 | 71/111 | 5 | 358 |
Different numbers of simulations were considered for different configurations to have equal sample sizes in the analysis. For comparison of these values, please keep in mind that the average electrode distance is 19 μm, the electrode density is 3,161 mm−2, and the number of electrodes used in the simulations is 90 (high-density arrangement of 9 × 10 electrodes).
Fig. 2.
Sample arrangements of neurons (circles) and electrodes (black dots) for the 5 configurations in data set B. Neurons lying outside the electrode block (gray) were also simulated, but only neurons inside the electrode block (black circles) were considered for the evaluation. D, average neuron distance.
Individual, uncorrelated spike trains were simulated for both data sets using sets of γ-distributed interspike intervals (ISI), which proved to be a good model for the spiking behavior of RGCs (Levine 1991). The mean firing rates ranged between 30 and 50 Hz for data set A and between 5 and 50 Hz for data set B. A refractory period of 2 ms was introduced. We upsampled the template waveforms to 160 kHz and then randomly downsampled them to the respective sampling frequency for every simulated AP, this way imitating recording conditions, under which the spikes are not always digitalized at the exact same position (Pouzat 2002; Quiroga et al. 2004). Spike shape variability (Fee et al. 1996b) was induced by multiplying the template waveforms of every spiking event on all electrodes with a random factor (normally distributed μ = 1, σ = 0.1).
A digitally unfiltered noise signal, which was recorded under experimental conditions with a retina preparation on the array that had no visible spiking activity, was added to the simulated spike data. The resulting signal was quantified to a least significant bit, similar to the one used in the measurements (5 μV). This way, the simulated data had similar characteristics as recorded data, and the same data handling and processing steps could be used.
Results
Spatiotemporal Template Analysis
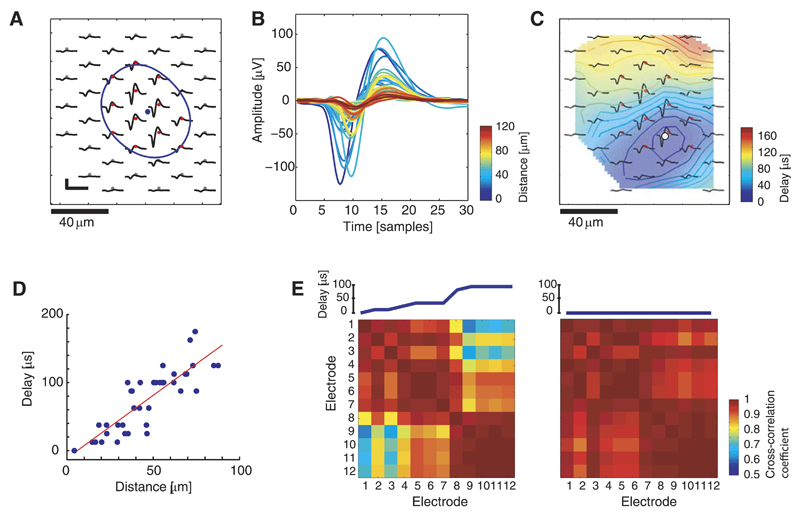
A main requirement for ICA is that the independent sources (i.e., the neuron templates) are linearly mixed over all recording electrodes. This requirement implies that the waveforms of a neuron on the different recording electrodes are linearly dependent. Therefore, the degree of linear dependence between the waveforms of a RGC template was analyzed.
A single neuron template was reconstructed from two overlapping blocks, recorded at highest spatial resolution (Fig. 3A). Because of the chip architecture, the electrodes are not sampled at the same point in time. We corrected for this by upsampling (160 kHz) and resampling the recorded data at defined time points (20 kHz). After spike detection, the multichannel spike traces of the identified neuron were again upsampled by a factor of 4 to allow a more precise spike alignment and averaging (upsampled resolution: 12.5 μs).
Fig. 3.
Ganglion cell template analysis at high spatiotemporal resolution. A: RGC template; the blue point indicates the center of gravity, and the blue line shows a Gaussian-fit equipotential line; scale bar: 100 μV/1.6 ms. B: superimposed averaged waveforms with the color code indicating the distance to the reference point marked in C. The phase shift grows with increasing distance. C: for every averaged waveform, the action potential (AP) timing was determined based on the occurrence of the respective negative peak value. The position of the earliest AP occurrence (AP reference) is indicated by the white dot, and the background color shows AP delay with respect to this reference point. D: distance to reference point vs. AP delay for all recorded waveforms (blue points) of the RGC. The red line shows a linear fit with a slope of 1.8, corresponding to a velocity of 0.55 m/s. E, left: cross-correlation coefficient (CC) matrix for waveforms on 12 electrodes with highest amplitude (red dots in A and C), arranged according to AP delay in ascending order. The increasing delay (shown at the top) causes a decrease in the CC, with values as low as 0.61. Two main groups with high intragroup but low intergroup CCs appear, particularly electrodes 1–7 and 8–12. Right: CC matrix for the same 12 waveforms, however, with the phase shift being corrected by alignment according to the occurrence of the negative peak value. The resulting CCs range between 0.89 and 1.
Figure 3B shows the superimposed waveforms from 40 electrodes, illustrating that they are not exactly in phase but shifted by up to 3 samples (150 μs). The electrode position where the AP wave appears 1st (dark blue wave in Fig. 3B) will be referred to as AP reference (white marker in Fig. 3C). The color of each waveform indicates the distance between the corresponding electrode and the AP reference. An increase in the phase shift is observed for increasing distance. This phase shift is due to the AP propagation delay.
The spatial extent of the propagation delay is visualized in Fig. 3C. For every electrode, the precise position of the negative peak in the upsampled averaged waveform was used to determine the temporal delay compared with the AP reference. After emergence close to this reference point, the AP spreads into all directions. The timing delays vs. the travelling distances for the individual waveforms, shown in Fig. 3D, give an estimate of the propagation speed. The slope of the linear fit corresponds to a velocity of 0.55 m/s.
For better visibility, we focus in the following on the waveforms recorded from the 12 electrodes providing the highest signals (peak-to-peak voltages > 60 μV, marked with a red dot in Fig. 3, A and C). In any case, signals with low amplitude only moderately influence ICA compared with high amplitude signals.
The CC matrix (Fig. 3E, left) contains the CC values (see Eq. 1 in Methods) for the waveforms of all electrode pair combinations of the selected electrodes. The electrodes were arranged in the matrix with respect to ascending time delay, which is displayed in the inset above the matrix plot. Electrodes 1–7 (electrodes in purple, bluish area in Fig. 3C) as well as 8–12 (electrodes in green, yellowish area in Fig. 3C) form electrode groups with high CC values for electrode pairs within each group but relatively small CC values for electrode pairs between both groups. The decrease in linearity is caused by a relatively large phase shift between electrodes 7 and 8. Similar abrupt phase shifts were observed for many RGCs and are presumably a characteristic physiological feature of these neurons. For electrode pairs without significant phase shift, such as for electrodes 5 and 6 or for electrodes 10–12, the resulting CCs are very close to 1.
This example shows that somatic AP signals, recorded at high spatiotemporal resolution, contain significant propagation delays and thus cannot be regarded as an instantaneous mixture. However, even if there was no propagation delay, we could not assume a perfect linear mixture for a second reason. The complex physiological structure of the neurons has effects on the spike waveforms. To analyze this, the waveforms were temporally aligned according to the occurrence of their negative peak value, and the resulting CC matrix was determined (Fig. 3E, right). For this case, the interelectrode CC values have a mean of 0.97 and a smallest value of 0.89, compared with a mean of 0.89 and smallest CC value of 0.61 in the nonaligned case.
These characteristics of limited linear dependence between the waveforms of a neuronal unit on different recording electrodes, mainly caused by the phase shift due to the AP propagation delay, imply that the linearity assumption of ICA is not fulfilled.
Nonlinearity Effects on Linear Demixing and ICA
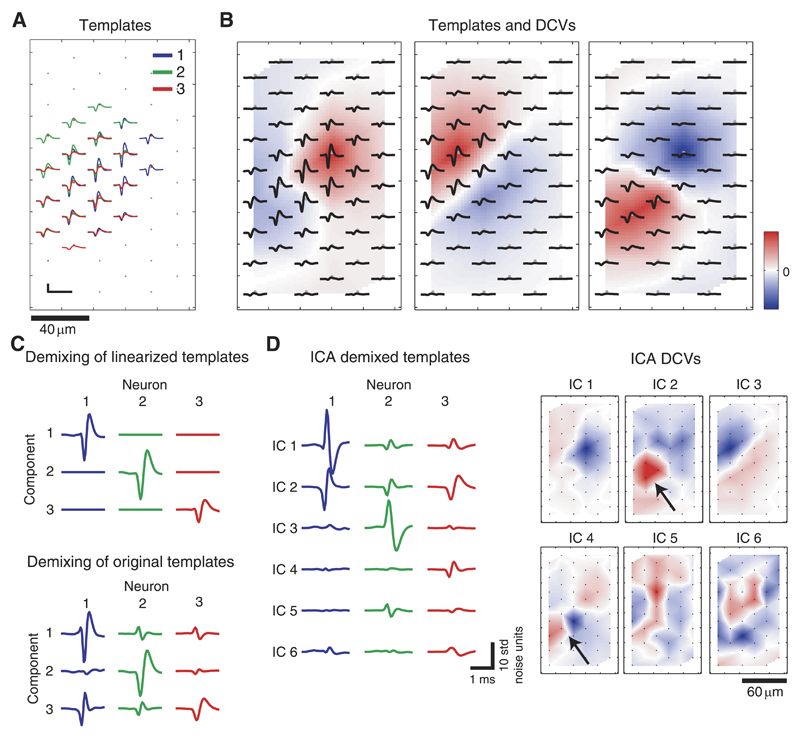
Next, we investigated how nonlinearity in the neuronal templates affects the possibilities of separating neuronal signals by means of linear demixing and ICA. For this, we used data set A, which contained simulated activity of three spatially overlapping neuronal templates (Fig. 4, A and B) extracted from a retinal recording.
Fig. 4.
A: STA templates of 3 spatially overlapping neurons identified and extracted from retinal recordings. Scale bar: 100 μ;V/1.8 ms. B: individual templates and respective demixing coefficient vectors (DCVs), directly derived from the templates (see Appendix). C: demixed templates using the DCVs, which were directly derived from the templates. In this representation, the rows refer to the demixing components, and the columns refer to the neuron templates. Top: the manipulated, idealized case where linearity across each template is given. Bottom: the realistic case where linearity is not given. The DCVs used here are shown in B. D, left: template responses for the 1st 6 independent components (ICs) obtained by applying IC analysis (ICA) on the simulated data set. Right: corresponding DCV maps. The arrows indicate an example where significant weightings for demixing 1 source (neuron 3) are found on 2 ICs (ICs 2 and 4).
In a first trial, the template of every neuron was manipulated to achieve linearity across the electrodes. This was realized by replacing the template waveform in each channel by scaled versions of the waveform observed on the electrode with maximum signal so that the peak-to-peak amplitude value at each electrode was preserved. The DCVs, directly derived from the manipulated templates (see Eqs. 10–11 in Appendix) led to perfect separation in the demixed templates (Fig. 4C, top), which means that there is only one high peak signal in each component (row). This shows that neuronal templates can be completely separated by means of linear demixing, given the assumption of perfect linear mixtures under the boundary condition that there are more electrodes than neurons.
The DCVs were extracted in the same way from the realistic templates, and the demixed templates were computed (Fig. 4C, bottom). The signals along the diagonal of both plots have similar magnitudes, suggesting that the accumulation of the template energies in the components leads to a similar SNR increase for the linearized and the real case. However, compared with the linearized ideal case, there is significant cross talk. As a result, e.g., neuron 3, having the smallest spike amplitudes and being therefore the most challenging to demix, fails to be separated.
In a next step, ICA was applied to the simulated data set A, which contained spikes from these three neurons. The demixed templates and corresponding DCVs maps (Fig. 4D) show that some of the responses (i.e., for ICs 1, 3, 5, and 6) feature a reversed sign, which is due to the fact that ICA cannot derive the correct sign of the source signals. Besides this, ICA leads to comparable SNR and separability on the first three ICs as the demixing using directly derived DCVs from the templates.
If the linearity criterion of ICA would be met, we would not only achieve perfect separation, but also redundancy reduction so that the number of ICs with significant signals would be equal to the number of neurons. This would imply that we find only one high peak signal per column of the demixed templates. Since there are still signals in the demixed templates for ICs 4–6, ICA does not completely reveal the right number of sources here.
We observe that ICs 2 and 4 have significant DCV weightings in the area of neuron 3 (black arrows in Fig. 4D, right) while having nearly orthogonal MCVs (CC = 0.11). However, the positive weightings (red) in the DCV of IC 4 are centered around a subset of three electrodes, whereas one electrode has large negative weight (blue). The CC values of spike waveforms of the three electrodes with positive weights are >0.995, however, significantly lower for combinations with the waveform of the electrode with negative weight (CC values 0.79, 0.81, and 0.83). The effect of splitting the source between different ICs was also consistently found on simulated data sets containing spikes from only single neurons (data not shown). This suggests that ICA splits the source due to the phase shift between the waveforms across the electrodes described in the previous section. As a consequence, the similarity of MCVs, as used in Takahashi and Sakurai (2005), might be a poor indicator for determining if the sources underlying two ICs are coming from a single or two separate neurons.
Interestingly, there are cases where a neuron does not exhibit the best separation performance on the first IC on which its signal is visible. Whereas neuron 3 has a strong signal but no separability on IC 2, better separability is achieved on IC 4 At the same time, the DCV for IC 4 has large weights on a smaller area (Fig. 4D, right), and therefore it features less template energy and a smaller SNR increase compared with IC 2
Advantages of ICA for Resolving Overlapping Spikes
We have shown in the previous section that the violated linearity criterion complicates clean separation of the neuronal sources using ICA-based linear demixing. However, in the following, we show that ICA has substantial advantages compared with traditional PCA methods for dealing with overlapping spikes in the case of sufficient separability on the ICs.
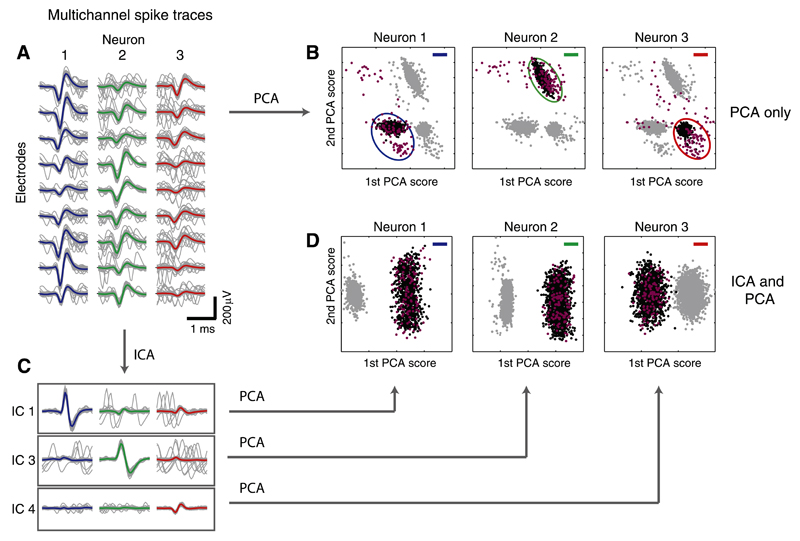
Figure 5A shows individual spike traces and average waveforms of the neurons in Fig. 4 on nine selected electrodes. The high firing rates of the simulated neurons caused many overlapping spikes, which evidently challenge alignment and classification of the spikes. To demonstrate this challenge, multichannel PCA (i.e., PCA performed on the concatenated single-channel waveform traces; see Litke et al. 2004) was applied to the spike traces of the three neurons on the selected electrodes. Figure 5B shows the resulting PCA scores of the first two principal components for each individual neuron; the black dots indicate spikes of the neuron of interest, and the gray dots mark spikes of other neurons. Violet dots indicate spikes that overlapped with another neuron within a time frame of 10 samples (0.5 ms). The nonoverlapping events are located in different, defined regions of the PCA space (ellipses) and can thus be separated using the representation of the PCA scores. However, the overlapping events are distributed all over the space and cannot be correctly identified using standard clustering methods.
Fig. 5.
Using ICA to resolve overlaps. A: template waveforms (colored) and individual spike traces (gray, only 80 traces are shown per neuron for better visualization) on 9 selected electrodes for the neurons shown in Fig. 4. Many overlapping spikes are visible in the gray traces. B: principal component analysis (PCA) applied to the multichannel spike traces on the 9 selected electrodes in A. Black dots indicate spikes from the neuron of interest, and gray dots denote spikes from other neurons. Spikes, which temporally overlapped with a spike from another neuron (timing difference of 10 samples or less), are colored violet (the respective overlapping spikes from the other neurons are not shown). Note that overlaps mostly lie outside of the main cluster. C: spike traces on ICs 1, 3, and 4 after applying ICA. The clean waveforms along the diagonal indicate that separability is given and that overlaps have been resolved. D: PCA applied individually to the IC spike traces on the 3 selected ICs.
In the following, ICs 1, 3, and 4 (from Fig. 4D) were considered for separating neurons 1–3 (Fig. 4, A and B). The spike traces projected on the corresponding ICs (traces along the diagonal in Fig. 5C) exhibit clean waveforms, which indicate that the overlaps shown in Fig. 5A have been resolved. Therefore, the ICA separation for this example is sufficient to enable proper spike assignment based only on threshold detection on the ICs.
The capability to resolve overlaps is also demonstrated by applying PCA to the IC spike traces of the three neurons for each IC independently. The representation of the scores (Fig. 5D) shows clear separation of the spikes of the respective neuron of interest (black dots) from the spikes of other neurons (gray dots). The overlapping spikes (violet dots) also lie within the cluster, which enables a correct assignment.
An important advantage of ICA usage for dealing with overlapping spikes appears here. Since the ICs can be treated independently, event detection is applied to the individual ICs. Thus two neurons that fire simultaneously evoke signals on two ICs and can also be detected as two independent spike sources.
It is important to mention in this context that, as shown in the previous section, ICA yields different separabilities on different ICs. However, it is not known which of the IC achieves the best separability for every neuron. Therefore, the best IC cannot be selected blindly.
ICA Performance Evaluation
To use ICA, followed by simple threshold detection for spike sorting, the neuronal sources need to be well-separated, and each neuron needs to be prominent and detectable on exactly one IC. To evaluate the applicability of such an ICA-based spike-sorting method, we analyzed the performance of ICA with regard to achieving three goals: 1) SNR increase; 2) separation of the neuronal sources; and 3) redundancy reduction. For this purpose, we used the basic evaluation metrics, which were defined in Evaluation Criteria.
ICA was applied to simulated RGC activity at different cell densities (data set B; see Methods). For calculation of the evaluation criteria, the noise signals and the templates that were used in the simulations have been considered.
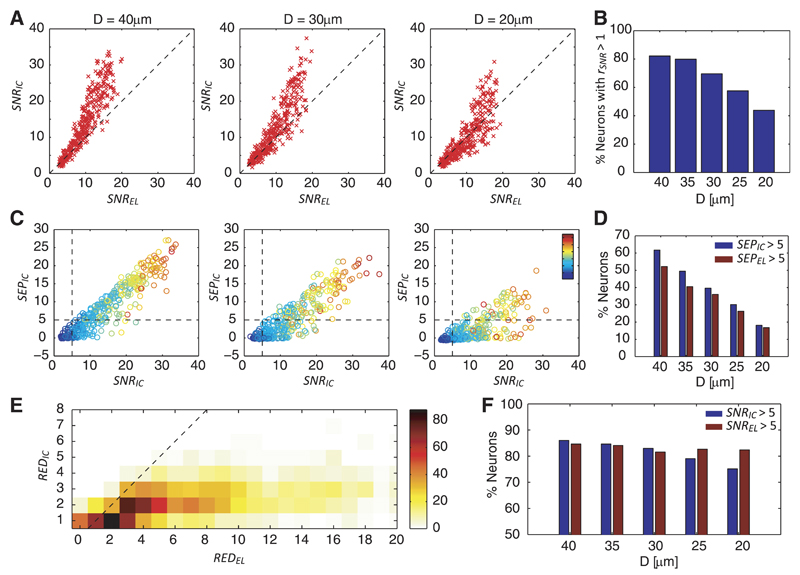
First, the SNR of the neurons on the raw data (SNREL) was compared with that on the ICs (SNRIC; Fig. 6A), whereat an overall increase in the SNRIC was observed. The SNR ratio rSNR = SNRIC/SNREL was found to be particularly large for neurons with high SNREL, which suggests that ICA is more effective in demixing signals of high-SNR neurons. However, the SNR increase drops for configurations with higher cell densities. Whereas for D = 40 μm, 82% of the neurons have rSNR > 1, for D = 20 μm this is only the case for 44% of the neurons (Fig. 6B).
Fig. 6.
A: signal-to-noise-ratio (SNR) conditions for electrodes (SNREL) vs. ICs (SNRIC) for simulated neurons (red crosses) of 3 different configurations. The dashed line shows SNREL = SNRIC. B: percentage of neurons, for which SNRIC > SNREL for the 5 configurations. C: SNR vs. separability for neurons of 3 different configurations. The dashed lines indicate threshold values for separability of neurons (SEPIC = 5 and SNRIC = 5). The color code indicates the template energies. D: percentage of neurons with SEPIC > 5 (blue) and SEPEL > 5 (red). E: histogram showing the counts of redundancies REDEL and REDIC for a total of 1,130 neurons (330 neurons having REDIC = 0 were excluded from the graphic representation). The dashed line indicates REDEL = REDIC. F: percentage of neurons that can be detected according to the criteria SNRIC > 5 (blue) and SNREL > 5 (red) for the 5 configurations.
Next, we addressed the question of how well the neuronal sources are separated in the representation of ICs. Separability SEPIC denotes the difference between the peak of a neuron on an IC and the next highest peak of another neuron. If a neuron has a large enough SEPIC, the sorted spikes can be obtained by simply applying threshold detection to the respective IC. We define the condition for a neuron to be separable if it has an SEPIC above a threshold of 5. Note that the SEPIC is given in units of standard deviations of the noise in the IC, like the SNRIC, and, therefore, every neuron with an SEPIC > 5 also has an SNRIC > 5.
Figure 6C shows the separability and SNR values of the neurons for different configurations; the color code indicates the template energy. The dashed lines confine the thresholds with regard to detectability, and therefore all neurons positioned in the upper right area of the plot are detectable and separable according to the defined criteria. For largely spaced neurons, separability increases approximately linearly with the SNR. The overall separability drops for larger neuron densities, and several large-SNR neurons feature low separability for the most tightly spaced configuration D = 20 μm. ICA yields an increase in separability compared with the raw data (Fig. 6D). However, a substantial decrease in separability is observed for tightly spaced neuron configurations, reflected by the low percentage of neurons featuring SEPIC > 5 or SEPEL > 5.
The third performance criterion is the reduction of redundancy in the IC space. The redundancies in the IC and electrode space were computed for a total of 1,460 simulated neurons in all configurations. Figure 6E shows the histogram counts for REDEL and REDIC. A majority of the neurons yields REDIC values between 1 and 2, and thus ICA performs well in reducing the dimensionality of the data.
Note that the zero value in the y-axis is not shown, and thus neurons with REDIC = 0 were excluded in this graphic. Whereas low REDIC values are desired, as they mean that the individual neurons are not detected many times, REDIC = 0 means that the neuron cannot be detected at any of the ICs.
Finally, the effect of an increased SNR (Fig. 6, A and B) on the detection of the neurons was evaluated. We found that for the configurations with D ≥ 30 μm, more neurons are detectable on the ICs (Fig. 6F) due to the increase in SNR obtained by using ICA. One has to note, however, that the SNR increase, as shown in Fig. 6A, is relatively small for low-SNR neurons. Therefore, ICA only slightly increases the percentage of detectable neurons. For configurations with D < 30 μm, the percentage of detectable neurons based on the ICs drops below the percentage of detectable neurons based on the raw signals, which is approximately constant across all configurations. We conclude that ICA fails to improve signal quality for very dense neuron populations but is beneficial for lower density populations.
The presented analyses revealed that ICA, applied to HD-MEA recordings, yields only limited separation performance and is, therefore, not suited to be used as a stand-alone spike-sorting tool in combination with threshold detection.
ICA Applied to Recorded Data
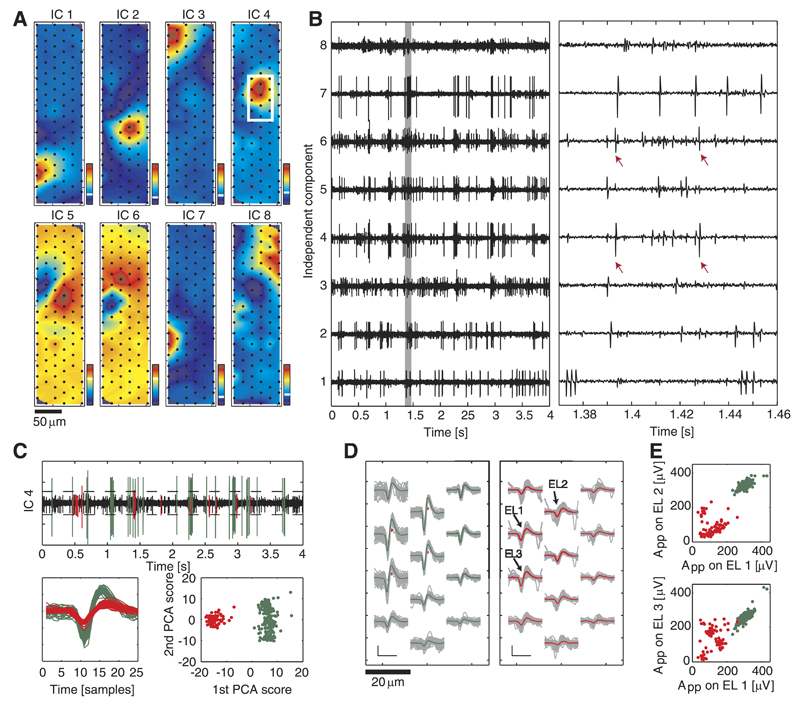
An example of ICA, applied to block recordings with spontaneous RGC activity is illustrated in Fig. 7. The DCVs, depicted in Fig. 7A, yield spatially localized high weightings. Note that the input to ICA is the multielectrode signal without any information on the electrode positions and that the neuronal templates are localized in space. Therefore, spatially localized high weighting values in the DCVs are a good indicator that the signals, underlying the ICs, originate from neuronal units.
Fig. 7.
Example of ICA applied to retinal recordings. A: spatial map of DCVs for the 1st 8 components. The coefficients are normalized with regard to the resulting IC signal having a standard deviation of 1; therefore, no absolute values are shown here. The white line in the corresponding color bars represents the 0 value. B: corresponding IC signals (left) and close-up (right, data segment is indicated by a gray rectangle on left plot). Except for the large spikes in ICs 4 and 6, which originate from the same neuron (red arrows), the spikes on different components belong to different neurons, as they are not correlated in their timing. C: threshold detection and clustering for IC 4. Top: dashed line representing the threshold level; detected events are colored. Bottom left: superimposed IC traces for all detected events above threshold within 20 s. Bottom right: 1st and 2nd PCA scores for detected events, which can be clustered (colors). D: spatial spike distributions for both clusters over selected electrodes marked by the white rectangle in A, showing the average spike shape (colored) and the individual traces (gray). Scale bar: 100 µV/1 ms. E: peak-to-peak amplitude values of the 2 clusters on pairs of electrodes, EL1–EL2 and EL1–EL3; the electrodes are indicated in D.
In looking at the spike waveforms, some ICs (e.g., ICs 1 and 7 in Fig. 7B) feature high separability and, practically, represent single-unit spike trains of neurons. However, the problems of limited separability and dimensionality reduction that were discussed in the previous sections are also visible in the IC signals. Several ICs (e.g., ICs 3–6 and 8) presumably contain spikes from multiple neurons as indicated by spike waveforms of different amplitudes on the IC. On the other hand, in some cases, spikes from a single neuron were observed on multiple components (red arrows in Fig. 7B).
Approaches for ICA-Based Spike Sorting
Since most ICs do not represent single-unit spike trains (Fig. 7B), the spikes cannot be sorted by just applying event detection to the ICs. The reduced redundancy in the data as well as the increased SNR and separability are, however, still valuable features of the IC representation. In this section, we propose to use postprocessing techniques to overcome the most severe limitations arising from the nonlinearity of the templates.
For the case that an IC contains spikes of multiple units, the spikes can be separated by means of PCA-based clustering of the IC spike waveforms. This is exemplarily shown for IC 4 in Fig. 7, C–E. The green cluster, which exhibits large spike signals in this IC, corresponds most likely to the neuron that can be associated with this component. The smaller spikes, grouped into the red cluster, can be discarded and may be detected on other ICs. Note that the peak-to-peak amplitudes in the red cluster (Fig. 7E) show large variability, suggesting that these spikes arise from multiple neurons.
The problem of multiple detection of neurons can be addressed by using an aggregation method, which, e.g., compares the identified spike times of the questionable neurons and merges them if they have a specific number of spike times in common (Litke et al. 2004). Additionally, waveform similarity and ISI statistics can be included as aggregation criteria (Fee et al. 1996a).
Furthermore, ICA-based spike sorting can be performed in an iterative procedure: ICA is applied to the data, and spikes are identified by applying threshold detection to the ICs. Following clustering and merging, the STA waveforms of identified neurons are subtracted from the raw data. In a next iteration, ICA is applied to the residual signals. Similar, subtractive methods were proposed for spike sorting using optimal filters (Gozani and Miller 1994) and template matching (Vargas-Irwin and Donoghue 2007). This iterative scheme, which adds a nonlinear feature to the linear ICA approach, is motivated by two reasons. On the one hand, neurons featuring large signal amplitudes render the separation of neighboring, spatially overlapping neurons with smaller amplitudes difficult. Therefore, the identification and subtraction of dominant neuronal sources allows for identifying less dominant signals in the subsequent ICA iteration. On the other hand, ICA facilitates the detection and classification of a spike A even though it temporally overlaps with another spike B. The proper subtraction of spike A will improve detection and classification of spike B in the proximate iteration in case that it cannot be identified on another IC.
An algorithm based on the described approaches was implemented. ICA was applied to the band-pass filtered recordings, decomposing the data into ICs. AP events, identified by applying threshold detection to the IC signals, were clustered (KlustaKwik), based on the principal components of the IC spike traces. Clusters with high standard deviation on the multichannel spike traces were believed to be erroneous and discarded. After an intermediate merging step, during which clusters of multiple-detected neurons were aggregated, the STA spikes were subtracted from the raw data. In the next iteration, ICA was applied to the residual signal. This iterative scheme was repeated for a defined number of iterations. A detailed description of the individual algorithm steps is given in ICA-Based Spike-Sorting Algorithm in Appendix.
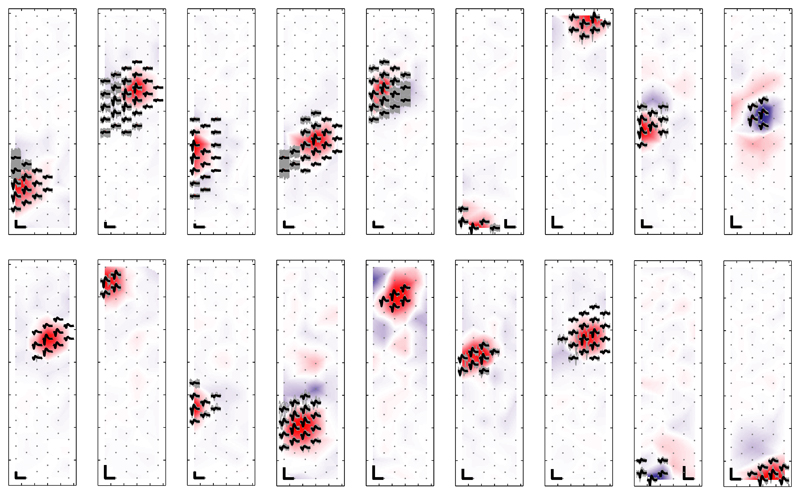
Figure 8 shows the templates (black) of 18 sorted cells from recorded RGC activity on an HD block. The DCV weightings (background colors) largely overlap with the active electrodes of the neuron templates. The individual traces are depicted in gray. Concurrent high-amplitude spiking activity of neighboring neurons results in visible gray traces near the neuron template, which can be observed for several neurons (e.g., for templates 1, 2, 4, 5, and 6). These traces, which lie mostly outside the DCV active area, indicate that ICA allowed to classify correctly the spikes despite overlaps.
Fig. 8.
STA templates (black) and individual traces (gray) of neurons as identified using the iterative ICA-based approach. The DCVs are indicated through the color code in the background. Scale bars: 100 μV/1 ms. For visualization purposes, the templates were individually scaled, which results in variable-size scale bars.
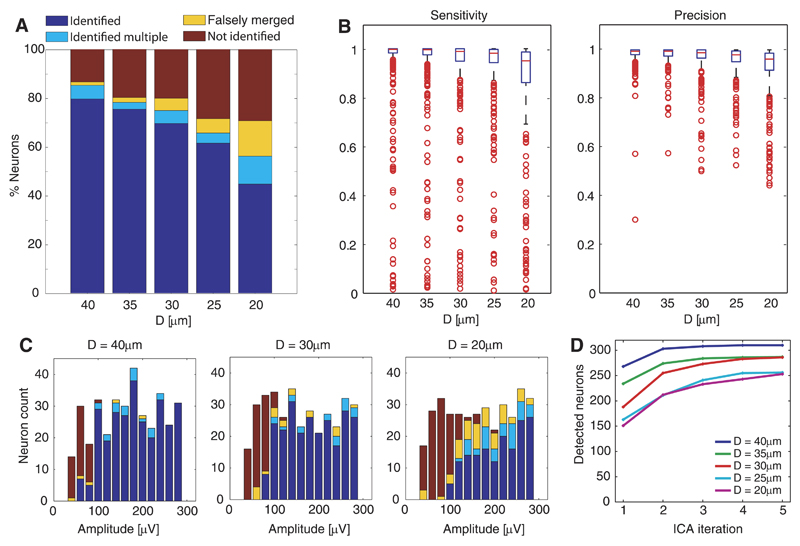
Finally, an unsupervised version of the spike-sorting algorithm was applied to the simulated data set B, and the sorting output was matched with the simulated data. A sorted neuron was assigned to a simulated neuron, if the number of matching spikes exceeded 10% of the total number of spikes of the simulated neuron. We classified the simulated neurons as identified (if detected as 1 neuron), identified multiple (if detected as 2 or more separate neurons), falsely merged (if detected but merged with 1 or more simulated neurons), or as not found (if not detected at all). The classification percentages of the simulated neurons are depicted in Fig. 9A. As for ICA alone, the sorting performance was found to depend on the cell density. Additionally, the neuronal signal amplitude played a dominant role. As shown in Fig. 9C for the different configuarations, most detected neurons featured a certain amplitude range, whereas neurons with lower amplitudes passed constantly undetected. This correlates to the observation that the separability of a neuron on the IC strongly depends on its SNR.
Fig. 9.
Evaluation of spike sorting as applied to simulated data sets. A: percentage of simulated neurons classified as identified, identified multiple, falsely merged, and not identified for the different configurations. B: box plots of the sensitivity and precision values for all sorted neurons, showing the median values (red horizontal line), the interquartile ranges (IQR; blue boxes), highest and lowest data values that are within 1.5 times the IQR (black whiskers), and outliers outside 1.5 times the IQR (red circles). C: histogram counts of neuron classification vs. neuronal-signal peak-to-peak amplitude for 3 configurations. D: cumulative sum of detected neurons vs. ICA iterations.
To quantify the sorting quality, each sorted neuron was assigned to a simulated neuron (based on the number of matching spikes), and the numbers of true-positive (TP), false-positive (FP), and false-negative (FN) events were computed. We used the performance measures “sensitivity” [TP/(TP+FN)] and “precision” [TP/(TP+FP)]; a sensitivity value of 1 means that all spikes were detected (no FNs), and a precision value of 1 signifies that only correct spikes were detected (no FPs). Figure 9B shows box plots of the performance results. The blue boxes indicate the interquartile ranges (IQR), and the black whiskers the highest and lowest data values that are within 1.5 times the IQR. Therefore, the lower performance bounds are given by the lower border of the blue boxes (for 75% of the neurons) and by the lower whiskers (for 87.5% of the neurons). Although there were numerous neurons with poor performance (outliers: red circles), the majority of the sorted neurons (indicated by the medians: red lines) yielded sensitivity and precision values >0.95 throughout all configurations. The performance was again found to depend on the cell density. For configurations D ≥ 35 μm, 87.5% of the neurons yield in performance values >0.93.
The iterative approach allowed to increase the number of detected neurons (i.e., substantial increases for iterations 2 and 3). This is evident from the number of detected neurons after each ICA iteration shown in Fig. 9D.
We also compared the detection performance for overlapping and nonoverlapping spikes (Table 2). This analysis revealed that the ICA approach performs well in classifying spikes despite the fact that they are temporally overlapping with spikes from nearby neurons. The error probability associated with overlaps (right column in Table 2) was determined using Eq. 16 in Appendix. These error probabilities varied between 1.4 and 6% for the different configurations and are on the same order as the probabilities for nonoverlap errors. Accordingly, even for the highest-density case, only 1 out of 16 overlapping spikes was missed due to spatiotemporal interference with a spike from another neuron.
Table 2. Classification performance (false-negative events) of nonoverlapping vs. overlapping spikes.
| Nonoverlaps |
Overlaps |
||||
|---|---|---|---|---|---|
| D, µm | Total | Missed (pE) | Total | Missed (pOE) | Overlap-Specific Error Probability (pO) |
| 40 | 75,824 | 1,332 (1.8%) | 2,507 | 81 (3.2%) | 1.4% |
| 35 | 63,425 | 1,282 (2.0%) | 2,214 | 97 (4.4%) | 2.4% |
| 30 | 70,797 | 1,860 (2.6%) | 3,158 | 204 (6.4%) | 3.9% |
| 25 | 57,908 | 2,220 (3.8%) | 4,873 | 426 (8.7%) | 5.1% |
| 20 | 49,803 | 2,510 (5.0%) | 6,311 | 694 (11%) | 6.3% |
For this particular analysis, all sorted neurons with sensitivities >0.6 were considered. Overlaps were defined as spikes featuring a time difference of 10 samples or less to spikes of other neurons that were closer than 50 μm. The observed error probabilities (pO) for nonoverlapping spikes (pE) and for overlapping spikes (pOE) were used to compute the probability associated with overlapping spikes pO shown in the last column, using Eq. 16 in Appendix. D, distance.
Discussion
Recently, ICA has received increasing attention as a tool to analyze biomedical signals, such as EEG or functional MRI, as well as for spike sorting of optical brain recordings (Hill et al. 2010; Mukamel et al. 2009; Reidl et al. 2007; Stone 2002). Being an automatic tool for source separation of redundant data sets, ICA represents a promising candidate to facilitate rapid spike sorting of HD-MEA data.
In this paper, the suitability of ICA for demixing HD-MEA recordings was evaluated for the first time. We analyzed neuronal activity, recorded at high spatiotemporal resolution, and found that the fundamental requirement for ICA, a linear mixture of the source signals, is not fully satisfied by the characteristics of the data. Instead, the linearity between the waveforms of a neuron on different electrodes is decreased, which is mainly due to AP propagation delays.1 The compromised linearity was found to be a limiting factor already in sparse neuron arrangements, which impeded perfect source separation. As a consequence, ICA, followed by threshold detection, cannot be used as a stand-alone method for spike sorting of HD-MEA data. These findings presumably also hold for other devices and planar microelectrode systems (Csicsvari et al. 2003; Du et al. 2009; Takahashi and Sakurai 2005). The limitations of the applicability of ICA as a consequence of the nonlinear characteristic of the neuronal signals, as shown for tetrode recordings in Shiraishi et al. (2009), cannot be completely compensated by spatial oversampling using high electrode density.
The analysis of ICA applicability using simulated data sets of RGC activity as presented here revealed that the ICA performance strongly depends on the neuronal density in the preparation. Particularly, when the neuronal density approached the electrode density (3,161 electrodes/mm2) and the number of simulated neurons (111) exceeded the number of electrodes (90), the separation performance clearly decreased. Neurons that produced high-SNR signals entailed superior separation performance.
Except for very dense neuronal populations, ICA led to a significant overall SNR increase, which allowed for detecting more neurons. Moreover, the redundancy was clearly reduced in the ICs, which helps to overcome the problem of detecting the same APs multiple times on several electrode signals. For a limited number of neuronal sources, ICA automatically provided separation, which could serve to resolve efficiently overlapping spikes from these sources.
We showed that limitations arising from the nonlinearity of the sources could be addressed by combining the ICA output with postprocessing techniques. In particular, we proposed an algorithm, based on applying PCA and clustering, to the detected IC traces. For densities up to 1,300 neurons/mm2, >80% of the neurons were detected (>70% correctly identified as single neurons) using the unsupervised, ICA-based algorithm. The detection of the majority of the neurons for these densities was highly accurate (87.5% of the neurons had sensitivity and precision values above 0.86 and 0.91). Additionally, the algorithm performed well in resolving overlapping spikes. The percentage of misclassified overlaps (FNs), compared with nonoverlaps, was increased by factors of up to 2, and the particular error probability associated with the overlaps was between 1.4 and 6.3%.
The method of combining ICA with the proposed postprocessing techniques was not efficient for sorting the complete neuronal population but yielded good results in sorting large fractions of the cells with high accuracy. By iteratively subtracting identified spike waveforms from the data and applying ICA, the number of detected neurons could be increased by 15–68% for the different simulated neuron densities.
In conclusion, our results suggest that ICA applied to HD-MEA data does not yield complete separation of the neuronal signals. However, the IC-representation of the data has some valuable features, e.g., the reduced redundancy, which entails that spikes from a neuron are only prominent on one or a few ICs. This allows for treating the individual ICs as separate signals, which facilitates spike detection in redundant MEA data. In addition, the increased separability contributes to resolving overlaps. These features make ICA a valuable tool to serve as a preprocessing step to spike sorting.
Supplementary Material
Acknowledgments
We thank Jan Müller, Ian Jones, Ralph Streichan, Branca Roscic, Douglas Bakkum, and Alexander Stettler for help with the measurements and the experimental setup. U. Wahlen is acknowledged for contributions to data analysis. Three anonymous reviewers are expressly thanked for providing very helpful comments on an earlier version of this paper.
Grants
This work was financially supported by the European Community through the European Research Council (ERC) Advanced Grant 267351, NeuroCMOS. F. Franke acknowledges individual support through Marie Curie Training Network Grant CT 2006-035854, CELLCHECK.
Footnotes
Disclosures
No conflicts of interest, financial or otherwise, are declared by the author(s).
Author Contributions
D.J. and U.F. conception and design of research; D.J. analyzed data; D.J., U.F., and F.F. interpreted results of experiments; D.J. prepared figures; D.J. drafted manuscript; U.F., F.F., and A.H. edited and revised manuscript; M.F. performed experiments; A.H. approved final version of manuscript.
Additional analyses (data not shown) revealed that the most relevant frequency bands for the phase differences between the waveforms on different channels are in the range between 1 and 4 kHz (for digitally unfiltered data). Consequently, low-pass filtering the data before ICA with a cutoff frequency ~1 kHz reduced the nonlinearity effect and led to an increased ICA performance with respect to finding efficient MCVs and reducing redundancy. However, the achieved source separation was still limited. Furthermore, by removing the frequencies >1 kHz, where spikes have a significant part of their energy, important information for discrimination gets lost, and the SNR decreases.
References
- Berdondini L, van der Wal PD, Guenat O, de Rooij NF, Koudelka-Hep M, Seitz P, Kaufmann R, Metzler P, Blanc N, Rohr S. High-density electrode array for imaging in vitro electrophysiological activity. Biosens Bioelectron. 2005;21:167–174. doi: 10.1016/j.bios.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Brown GD, Yamada S, Sejnowski TJ. Independent component analysis at the neural cocktail party. Trends Neurosci. 2001;24:54–63. doi: 10.1016/s0166-2236(00)01683-0. [DOI] [PubMed] [Google Scholar]
- Csicsvari J, Henze DA, Jamieson B, Harris KD, Sirota A, Bartho P, Wise KD, Buzsaki G. Massively parallel recording of unit and local field potentials with silicon-based electrodes. J Neurophysiol. 2003;90:1314–1323. doi: 10.1152/jn.00116.2003. [DOI] [PubMed] [Google Scholar]
- Donoho DL, Johnstone JM. Ideal spatial adaptation by wavelet shrinkage. Biometrika. 1994;81:425–455. [Google Scholar]
- Du JG, Riedel-Kruse IH, Nawroth JC, Roukes ML, Laurent G, Masmanidis SC. High-resolution three-dimensional extracellular recording of neuronal activity with microfabricated electrode arrays. J Neurophysiol. 2009;101:1671–1678. doi: 10.1152/jn.90992.2008. [DOI] [PubMed] [Google Scholar]
- Einevoll GT, Franke F, Hagen E, Pouzat C, Harris KD. Towards reliable spike-train recordings from thousands of neurons with multielectrodes. Curr Opin Neurobiol. 2012;22:11–17. doi: 10.1016/j.conb.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eversmann B, Jenkner M, Hofmann F, Paulus C, Brederlow R, Holzapfl B, Fromherz P, Merz M, Brenner M, Schreiter M, Gabl R, et al. A 128 × 128 CMOS biosensor array for extracellular recording of neural activity. IEEE J Solid-State Circuits. 2003;38:2306–2317. [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Automatic sorting of multiple unit neuronal signals in the presence of anisotropic and non-Gaussian variability. J Neurosci Methods. 1996a;69:175–188. doi: 10.1016/S0165-0270(96)00050-7. [DOI] [PubMed] [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Variability of extracellular spike waveforms of cortical neurons. J Neurophysiol. 1996b;76:3823–3833. doi: 10.1152/jn.1996.76.6.3823. [DOI] [PubMed] [Google Scholar]
- Frey U, Egert U, Heer F, Hafizovic S, Hierlemann A. Microelectronic system for high-resolution mapping of extracellular electric fields applied to brain slices. Biosens Bioelectron. 2009;24:2191–2198. doi: 10.1016/j.bios.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Frey U, Sedivy J, Heer F, Pedron R, Ballini M, Mueller J, Bakkum D, Hafizovic S, Faraci FD, Greve F, Kirstein KU, et al. Switch-matrix-based high-density microelectrode array in CMOS technology. IEEE J Solid-State Circuits. 2010;45:467–482. [Google Scholar]
- Gozani SN, Miller JP. Optimal discrimination and classification of neuronal action potential waveforms from multiunit, multichannel recordings using software-based linear filters. IEEE Trans Biomed Eng. 1994;41:358–372. doi: 10.1109/10.284964. [DOI] [PubMed] [Google Scholar]
- Gray CM, Maldonado PE, Wilson M, McNaughton B. Tetrodes markedly improve the reliability and yield of multiple single-unit isolation from multi-unit recordings in cat striate cortex. J Neurosci Methods. 1995;63:43–54. doi: 10.1016/0165-0270(95)00085-2. [DOI] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsaki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Hermle T, Schwarz C, Bogdan M. Employing ICA and SOM for spike sorting of multielectrode recordings from CNS. J Physiol (Paris) 2004;98:349–356. doi: 10.1016/j.jphysparis.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Hill ES, Moore-Kochlacs C, Vasireddi SK, Sejnowski TJ, Frost WN. Validation of independent component analysis for rapid spike sorting of optical recording data. J Neurophysiol. 2010;104:3721–3731. doi: 10.1152/jn.00691.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzler M, Lambacher A, Eversmann B, Jenkner M, Thewes R, Fromherz P. High-resolution multitransistor array recording of electrical field potentials in cultured brain slices. J Neurophysiol. 2006;96:1638–1645. doi: 10.1152/jn.00347.2006. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans Neural Netw. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Levine MW. The distribution of the intervals between neural impulses in the maintained discharges of retinal ganglion cells. Biol Cybern. 1991;65:459–467. doi: 10.1007/BF00204659. [DOI] [PubMed] [Google Scholar]
- Lewicki MS. A review of methods for spike sorting: the detection and classification of neural action potentials. Network. 1998;9:R53–R78. [PubMed] [Google Scholar]
- Litke AM, Bezayiff N, Chichilnisky EJ, Cunningham W, Dabrowski W, Grillo AA, Grivich M, Grybos P, Hottowy P, Kachiguine S, Kalmar RS, et al. What does the eye tell the brain?: Development of a system for the large-scale recording of retinal output activity. IEEE Trans Nucl Sci. 2004;51:1434–1440. [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouzat C. Using noise signature to optimize spike-sorting and to assess neuronal classification quality. J Neurosci Methods. 2002;122:43–57. doi: 10.1016/s0165-0270(02)00276-5. [DOI] [PubMed] [Google Scholar]
- Quiroga RQ, Nadasdy Z, Ben-Shaul Y. Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput. 2004;16:1661–1687. doi: 10.1162/089976604774201631. [DOI] [PubMed] [Google Scholar]
- Reidl J, Starke J, Omer DB, Grinvald A, Spors H. Independent component analysis of high-resolution imaging data identifies distinct functional domains. Neuroimage. 2007;34:94–108. doi: 10.1016/j.neuroimage.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Segev R, Goodhouse J, Puchalla J, Berry MJ. Recording spikes from a large fraction of the ganglion cells in a retinal patch. Nat Neurosci. 2004;7:1154–1161. doi: 10.1038/nn1323. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Katayama N, Takahashi T, Karashima A, Nakao M. Multi-neuron action potentials recorded with tetrode are not instantaneous mixtures of single neuronal action potentials. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:4019–4022. doi: 10.1109/IEMBS.2009.5333505. [DOI] [PubMed] [Google Scholar]
- Shoham S, Fellows MR, Normann RA. Robust, automatic spike sorting using mixtures of multivariate t-distributions. J Neurosci Methods. 2003;127:111–122. doi: 10.1016/s0165-0270(03)00120-1. [DOI] [PubMed] [Google Scholar]
- Snellings A, Anderson DJ, Aldridge JW. Improved signal and reduced noise in neural recordings from close-spaced electrode arrays using independent component analysis as a preprocessor. J Neurosci Methods. 2006;150:254–264. doi: 10.1016/j.jneumeth.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Stasheff SF. Emergence of sustained spontaneous hyperactivity and temporary preservation of OFF responses in ganglion cells of the retinal degeneration (rd1) mouse. J Neurophysiol. 2008;99:1408–1421. doi: 10.1152/jn.00144.2007. [DOI] [PubMed] [Google Scholar]
- Stett A, Egert U, Guenther E, Hofmann F, Meyer T, Nisch W, Haemmerle H. Biological application of microelectrode arrays in drug discovery and basic research. Anal Bioanal Chem. 2003;377:486–495. doi: 10.1007/s00216-003-2149-x. [DOI] [PubMed] [Google Scholar]
- Stone JV. Independent component analysis: an introduction. Trends Cogn Sci. 2002;6:59–64. doi: 10.1016/s1364-6613(00)01813-1. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Anzai Y, Sakurai Y. Automatic sorting for multi-neuronal activity recorded with tetrodes in the presence of overlapping spikes. J Neurophysiol. 2003;89:2245–2258. doi: 10.1152/jn.00827.2002. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Sakurai Y. Real-time and automatic sorting of multi-neuronal activity for sub-millisecond interactions in vivo. Neuroscience. 2005;134:301–315. doi: 10.1016/j.neuroscience.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Vargas-Irwin C, Donoghue JP. Automated spike sorting using density grid contour clustering and subtractive waveform decomposition. J Neurosci Methods. 2007;164:1–18. doi: 10.1016/j.jneumeth.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PM, Wu JY, Zhou Y, Liang PJ, Yuan JQ. Spike sorting in multi-channel extracellular recordings of retinas. Neural Networks and Signal Processing, 2003. Proceedings of the 2003 International Conference on; 2003. pp. 712–715. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.