Abstract
Nanopore technology has become a powerful tool in single molecule sensing, and protein nanopores appear to be more advantageous than synthetic counterparts with regards to channel amenability, structure homogeneity, and production reproducibility. However, the diameter of most of the well-studied protein nanopores is too small to allow the pass of protein or peptides that are typically in multiple nanometers scale. The portal channel from bacteriophage SPP1 has a large channel size that allows the translocation of peptides with higher ordered structures. Utilizing single channel conduction assay and optical single molecule imaging, we observed translocation of peptides and quantitatively analyzed the dynamics of peptide oligomeric states in real-time at single molecule level. The oxidative and the reduced states of peptides were clearly differentiated based on their characteristic electronic signatures. A similar Gibbs free energy (ΔG0) was obtained when different concentrations of substrates were applied, suggesting that the use of SPP1 nanopore for real-time quantification of peptide oligomeric states is feasible. With the intrinsic nature of size and conjugation amenability, the SPP1 nanopore has the potential for development into a tool for the quantification of peptide and protein structures in real time.
Keywords: nanobiotechnology, viral motor, bacteriophage assembly, biomotor, nanopore sensing, peptide identification
1. Introduction
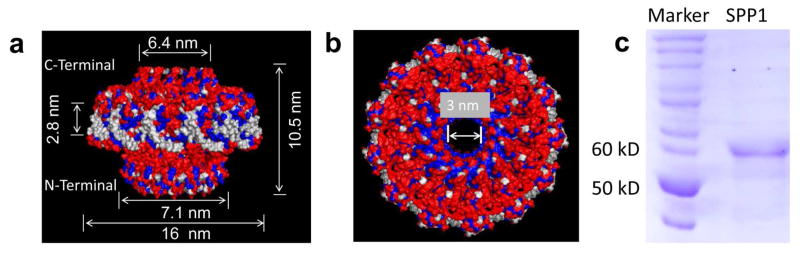
Living systems contain wide varieties of nanomachines with diverse structures and functions. The ingenious design of viral DNA packaging motors and their intriguing mechanism of action has triggered a wide range of interests among scientists in many different areas[1–3]. The portal protein is one of the essential components of the viral packaging motors with a turbine-like shape[4,5]. SPP1 is a dsDNA phage that infects Bacillus subtilis. The DNA packaging motor of SPP1 consists of a terminase composed of small (gp1) and large (gp2) subunits, portal protein gp6, and a two head completion proteins gp15 and gp16 [6,7] that power the encapsulation of 45.9 kbp genomic DNA[8]. The central core of the SPP1 motor is a portal channel, termed connector, which mainly forms a 13-subunit assembly in vitro and 12-subunit assembly inside phage particles [7,9]. Explicit engineering of the SPP1 portal protein is possible due to its available crystal structure [9,10]. The connector has an overall diameter of 16 nm and a height of 10.5 nm. The narrowest constriction of the internal channel is ~3 nm (Fig. 1)[9,10]. Portal protein is a critical component in many dsDNA bacteriophages and herpes viruses, which plays a critical role in genome packaging and ejection. Structural studies have shown that similar cone-shaped dodecameric structure is shared in portals from herpes virus and different tailed bacteriophages, such as phi29, SPP1, T4, and T3, even though their primary sequences do not show homology with varied molecular weight from 36 kDa, 56 kDa, 60 kDa and 59 kDa, respectively.
Figure 1. Structure of the SPP1 DNA packaging motor channel.
(a) Side and (b) top views showing hydrophilic (red), hydrophobic (blue) and neutral (white) amino acids; and dimensions of the channel. PDB: 2JES. This crystal structure was solved by Antson group [9]. (c) Coomasie-blue stained SDS PAGE showing the purified SPP1 channel subunits gp6.
Nanopore technology has recently emerged as a real-time and high-throughput single molecule detection method, holding great potential for sensing a wide range of analytes, molecular diagnostics and DNA sequencing applications[11–19]. Solid state nanopores generated by microfabrication generally have less reproducible pore sizes and lack chemical and location selectivity[20,21]. Protein nanopores harvested in bacteria are homogenous in size and can be functionalized with probes, but commonly used nanopores such as α-hemolysin, MspA, and aerolysin have an internal channel diameter of only ~1.3 nm[22–24]. Larger protein pores such as ClyA[25] and FhuA[26] are being developed for single molecule analysis. In search for alternative larger sized channels, we previously developed membrane-embedded phi29 motor channels[27–29] for single molecule sensing of nucleic acids[27,28,30–33], chemicals[34], peptides[35] or binding assays of antibody based on channel conformational change[30,36]. Herein, we utilized our recently developed membrane-embedded SPP1 motor channel[37,38] for quantifying the translocation and dynamics of peptide oligomeric states at single molecule level.
Elucidating the oligomeric states of proteins and peptides is critical for understanding their biological functions. A wide range of biophysical methods, such as X-ray crystallography, NMR, Circular Dichroism, Dual Polarization Interferometry, and Mass Spectrometry have been used to investigate folding and dynamic structural changes of peptides and proteins. However, these methods require expensive instrumentation and specialized labor. Nanopores offer an attractive alternative as they are intrinsically single molecule in nature requiring ultra-low sample volumes, are label free, amplification free, and function using a simple detection process requiring no specialized expertise. While the translocation of DNA and RNA have been studied extensively in biological nanopores[12,20,22,39], studies on translocation of protein or peptides are beginning to emerge[35,40–44]. The proteome can be an accurate and direct indicator of current health status of patients[45]. For example, early diagnosis and monitoring the changes of amyloid-β peptide and α-synuclein are critical for the management of Alzheimer’s and Parkinson’s disease[46].
Several studies demonstrating peptide translocation using nanopores have been published[47–60], but quantitative analysis and translocation validation remain challenging, since there is no technique available for amplifying protein substrates as in DNA or RNA qualification using PCR. The availability of only trace amounts of peptide or protein for analysis after translocation is far beyond the sensitivity threshold of classical protein detection methods. In this report, we used single molecule fluorescence microscopy to validate data obtained from resistive pulse technique to quantitatively study peptide translocation through SPP1 connectors and elucidate the structural conformations of peptides at the single molecule level. Due to the availability of crystal structure, this new nanopore can be explicitly engineered via site directed mutagenesis, and has the potential to be applied for biomarker analysis and early disease diagnosis.
2. Materials and Methods
2.1 Materials
The phospholipid 1,2-diphytanoyl-sn-glycerol-3-phosphocholine (DPhPC) was obtained from Avanti Polar Lipids, Inc. Organic solvents (n-decane and chloroform) were purchased from Fisher Scientific, Inc. and TEDIA, Inc., respectively. TAT peptide was custom-ordered from GenScript, Inc. All other reagents were purchased from Sigma or Fisher, if not specified.
2.2 Cloning and purification of the SPP1 connector
Gene gp6 encoding SPP1 portal was synthesized and cloned into PET3 vector between NhdeI and BamHI by GenScript, Inc. His-tag was inserted into the C-terminal of the connector for purification. Then the plasmid was transformed into BL21 (DE3) for expression and purification. The purification procedure has been described previously[27,29]. Briefly, the transformed bacteria were cultured in 10 mL LB medium overnight at 37°C. The bacteria were t ransferred to 1 L of fresh LB medium. When the OD600 reached ~0.5–0.6, 5 mM IPTG was added to the medium to induce protein expression. The bacteria were collected by centrifugation after 3 hours continuous culture. Bacteria was lysed by passing through French press. The cell lysate was separated by centrifugation and supernatant containing expressed protein was collected. The supernatant was purified with Nickel affinity chromatography (Novagen) [61]. His Binding Buffer (15% glycerol, 0.5 M NaCl, 5 mM Imidazole, 10 mM ATP, 50 mM Tris-HCl, pH 8.0), and the cleared lysate was loaded onto a His•Bind® Resin Column and washed with His Washing Buffer (15% glycerol, 0.5 M NaCl, 50 mM Imidazole, 50 mM Tris-HCl, pH 8.0). The His-tagged connector was eluted by His Elution Buffer (15% glycerol, 0.5 M NaCl, 0.5 M Imidazole, 50 mM Tris-HCl, pH 8.0). The purified connector from chromatography was further purified by a 15–35% glycerol gradient ultracentrifugation at 35K for 2 hrs. Before incorporating into liposomes, the purified connector was dialyzed against buffer (0.5 M NaCl, 50 mM Tris-HCl, pH 8.0) to remove excess glycerol.
2.3 Insertion of the connector into planar lipid bilayer
The protocol for the incorporation of connectors into lipid bilayer has been reported[27–30,62]. Briefly, 150 μL of 10 mg/mL DPhPC lipids in chloroform was placed in a round bottomed flask and the chloroform was evaporated under vacuum using a rotary evaporator (Buchi). The dehydrated lipid film was then rehydrated with 300 μL buffer containing 250 mM sucrose and purified connectors (0.5–1 mg/mL). The suspension was thoroughly vortexed and maintained at 45°C (above the phase transition temperature of DPhPC) followed by extrusion through 400 nm polycarbonate membranes (Avanti Polar Lipids) to generate uniform sized liposome/connector complexes.
Planar bilayer lipid membranes (BLMs) were generated in a BCH-1A horizontal BLM cell (Eastern Scientific). A Teflon partition with a 200 μm aperture was placed in the apparatus to separate the BLM cell into cis- (top) and trans- (bottom) compartments. The aperture was pre-painted with 0.5 μL of 3% (w/v) DPhPC in n-hexane. A conducting buffer (1 M KCl, 5 mM HEPES, pH 7.8) was added to both the top and bottom compartments of the BLM cell, and Ag/AgCl electrodes were placed in the buffer of each compartment. The electrode in the trans-compartment was connected to the headstage of an Axopatch 200B amplifier (Axon Instruments), and the electrode in the top compartment was grounded. A planar lipid bilayer was formed by painting the aperture with 0.5 μL of 3% (w/v) DPhPC in n-decane. 1 μL of the diluted liposome/connector complex was added to the cis-compartment directly to fuse with the planar lipid bilayer to generate membrane embedded nanopores.
2.4 Electrophysiological measurements
The headstage and Axopatch 200B patch clamp amplifier were connected to a DigiData 1440 analog-digital converter (Axon Instruments, Inc.) to monitor and record electrochemical currents through BLMs[27–29]. The current recordings were low-pass filtered at a frequency of 5 KHz. The sampling frequency was 200 KHz in all experiments, unless otherwise specified. The data were recorded a with pClamp 9.1 software (Axon Instruments, Inc.), and analyzed with the Clampfit module of pClamp 9.1 and OriginPro 8.1 (OriginLab Corporation).
2.5 Peptide translocation experiments
TAT peptide (Cys-Tyr-Gly-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg) with a final concentration of 23 μg/mL was premixed with the conducting buffer before the insertion of connector. All experiments were conducted at least three times and similar results were obtained. For quantitative validation, Cy3 fluorophore was conjugated to TAT peptide by sulfhydryl-maleimide chemistry and purified by HPLC. Multiple channels were inserted into bilayer lipid membrane and then Cy3-TAT peptide was added into cis-chamber. Samples were collected from the trans-chamber after 0, 20, 40, and 60 min. The errors represent mean ± standard deviation determined by a Gaussian fit of the data.
2.6 Single molecule fluorescence imaging
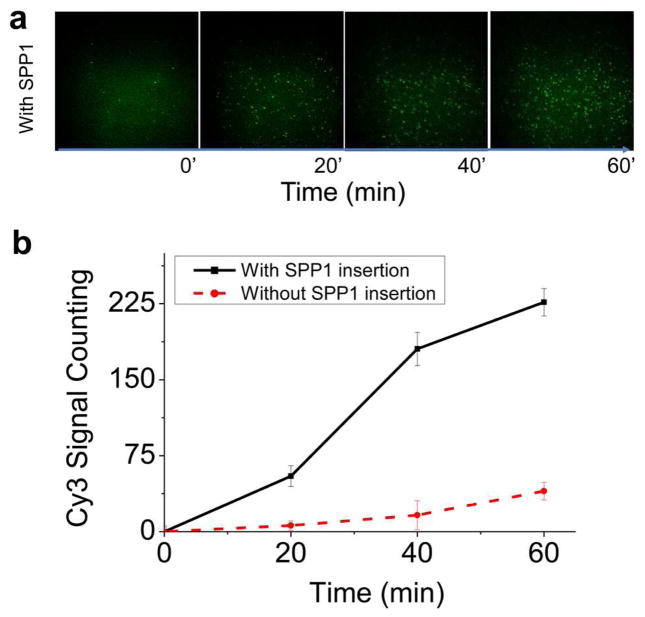
Samples collected from the patch clamp were incubated with the ozone pre-treated glass substrate for 10 min before imaging to ensure sufficient adsorption. A 532 nm green laser was used for the excitation of Cy3. A 60× objective (N.A. = 1.4, oil immersion) was used for fluorescence imaging. The signals were recorded using Andor iXon 887 V electron multiplied CCD camera. Images were taken with an exposure time of 500 ms. The number of spots in the images were counted using iSMS software[63]. These fluorescence spots in Figure 4a at 0 point may come from the impurity of the buffers or minor imperfection of the glass surfaces, which give background noise. Comparing to the signals from the translocated peptides, the number of background fluorescence spots was not significant. In the plot of Fig. 4b, the background spot numbers from the glass surface itself was subtracted for each time points. For data analysis, the number of spots at 0 min time point was subtracted from the calculation when counting the translocation peptide signals. The errors represent mean ± standard deviation from three independent imaging from within one experiment. Three independent repeats were performed and similar trend was obtained.
Fig. 4. Single molecule fluorescent images validating TAT peptide translocation.
(a) The upper row is the image showing the detection of Cy3-labeled TAT peptide from the fractions collected from patch clamp at 0, 20, 40 and 60 mins. Excitation λ: 532 nm; laser power: 5 mW; 60× objective (N.A. = 1.4, oil immersion); Exposure time: 500 ms. (b) Quantitative analysis showing the increase in Cy3-TAT peptide signal in presence of SPP1 connector compared to control without connector. The errors represent mean ± standard deviation from three independent imaging from one experiment. Three independent experiments were performed and similar trend was observed. Applied voltage: 50 mV; Buffer: 1 M KCl, 5 mM HEPES, pH 8.
3. Results
3.1 Characterization of SPP1 reengineered nanopore embedded in a lipid bilayer
Structural analysis revealed that the central region of the SPP1 connector shows slight hydrophobicity c ompared with the flanking regions at the N- and C-terminal ends which are more hydrophilic (Fig. 1a–b). We reengineered the SPP1 connector by inserting a 6×His-tag at the C-terminal end along with a 6× glycine linker for end-flexibility. The presence of His-tag enhanced the hydrophilicity of the C-terminal, t hus making the hydrophilic–hydrophobic–hydrophilic layers of the connector more distinct, which is nec essary to mimic the lipid bilayer architecture. After His-tag column purification, the purified protein was f urther purified by 15–35% glycerol gradient ultracentrifugation to further purify SPP1 portal complex fro m the single subunit (Suppl. Fig. 1). Bovine serum albumin (BSA) served as a marker to indicate the location of single subunit of SPP1 connector. BSA with molecular weight of 60 KDa, which is almost equal to a single subunit (58 KDa) of the SPP1 connector, centered at fraction 27, whereas the majority of SPP1 portal sample was at fraction 17, indicating that the single subunit and portal complex can be well separated.
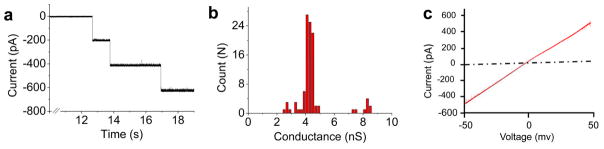
To incorporate SPP1 portal channel into planar lipid membranes, a two-step procedure was employed[27]. DPhPC lipids in chloroform were first dehydrated to remove organic solvents and then rehydrated with buffer containing 250 mM sucrose and purified connectors. The multi-lamellar lipid-connector suspension was then extruded through 400 nm polycarbonate membrane filters to generate uniform unilamellar liposomes with the connector embedded in the membrane. The resulting liposome-connector complex was fused with a planar lipid membrane to generate planar membrane-embedded SPP1 nanopore. Since the connector is not a membrane protein, direct incubation of the connector with a planar lipid bilayer did not result in connector insertion into the lipid bilayer. Single channel conductance assay was performed to measure the electrophysiological properties of membrane-embedded SPP1 connectors. The connector insertion steps were observed as distinct stepwise increase in conductance as revealed in a continuous current trace (Fig. 2a). The insertion of single portal channel results in ~200 pA in current jump under an applied potential of ±50 mV in conducting buffer (1 M KCl, 5 mM HEPES, pH 8). Occasionally ~400 pA current jumps were observed, attributed to simultaneous insertion of two connectors. The average conductance of reengineered SPP1 connectors is 4.27±0.27 nS (Fig. 2b). The conductance is uniform without displaying any voltage gating phenomena under the reported conditions of ±50 mV (Fig. 2c). At voltages greater than ±100 mV, SPP1 connector displayed discrete stepwise gating of the channel [37].
Fig. 2. Electrophysiological properties of membrane-embedded SPP1 connector.
(a) Current trace showing the insertion of SPP1 connector into the planar membrane with a characteristic step size of ~200 pA at −50 mV. (b) Conductance distribution based on 104 insertion events. (c) Current-Voltage trace acquired from −50 → +50 mV. Buffer: 1 M KCl, 5 mM HEPES, pH 8.
3.2 Characterization of peptide translocation through reengineered SPP1 connector
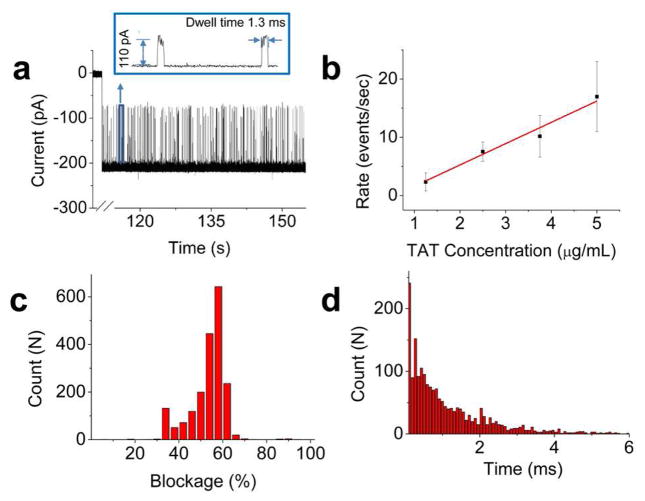
A positively charged 12 amino acid TAT peptide was used in the translocation studies with sequence Cys-Tyr-Gly-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg. Due to the presence of cysteine at the N-terminus, the peptide forms a dimer by disulfide bond under physiological conditions, which is confirmed by mass spectroscopy (data not shown). In the absence of peptide, the current trace was quiescent. In contrast, when the peptide was premixed with the conducting buffer in both cis- and trans-chambers, a burst of transient blockage events was observed immediately after the insertion of connector in the lipid membrane (Fig. 3a). As the peptide concentration was increased from 1.25, 2.5, 3.75, to 5 μg/mL, the density of current blockage events increased with a corresponding linear increase in peptide translocation rate from 2.33±1.54, 7.5±1.66, 10.17±3.58, to 16.96±6.01 events per second (Fig. 3b). One parameter used to characterize the translocation was the current blockage percentage, calculated as the ratio of current blockade resulting from peptide translocation to the open current of one portal channel, expressed as [ 1 − (Currentpeptide/Currentopen_channel) ]. The distribution of current blockage was relatively broad with a major peak centered at 55.1±3.0%, determined by Gaussian fit of the data (Fig. 3c). Another parameter was the dwell time (τ), the time taken for the peptide to traverse from one end of the connector to the other end. The dwell time distribution followed an exponential decay with a rate constant of 0.84±0.09 ms−1 (Fig. 3d).
Fig. 3. Peptide translocation through SPP1 connector.
(a) Current trace showing a burst of current blockage events with characteristic current amplitude and dwell time indicating the translocation of TAT peptides. Representative magnified events are shown in the box. [TAT peptide] =0.5 μg/mL (b) Rate of peptide translocation as a function of peptide concentration (n = 3). (c) Histogram of current blockage percentage from 1939 events. (d) Dwell time of peptide translocation events fitted with a single exponential function from 1939 events. Applied voltage: 50 mV; Buffer: 1 M KCl, 5 mM HEPES, pH 8.
3.3 Quantitative validation of peptide translocation by single molecule fluorescence imaging
To validate the translocation of peptide through SPP1 connector, we conducted single molecule fluorescence imaging of samples collected from patch clamp experiments. HPLC purified Cy3 conjugated TAT peptide at a final concentration of 2.5 ng/μL was added to the cis-chamber after stable insertion of SPP1 connectors in the lipid bilayer. Under an applied negative trans-membrane voltage, the positively charged peptide translocated through the portal channel to the trans-chamber. 50 μL samples were collected from the trans-chamber at 0, 20, 40, and 60 minutes after addition of Cy3-TAT peptide and loaded onto glass coverslips. The positively charged peptide can bind to the negatively charged glass surface through charge-charge interactions and appear as individual fluorescent spots (Fig. 4a). Fluorescence imaging revealed that the number of Cy3 spots in the field of view increased over the time course of 60 mins (Fig. 4b). In contrast, in a control experiment under the same conditions but in the absence of portal channel, very few Cy3 spots were observed, compared to the sample containing SPP1 connector (Fig. 4b). Since TAT is a membrane penetrating peptide, it is conceivable that a small fraction could potentially pass through the lipid bilayer, contributing to the small increase in the fluorescence background signal.
3.4 Kinetic analysis of TAT conformational status in real time
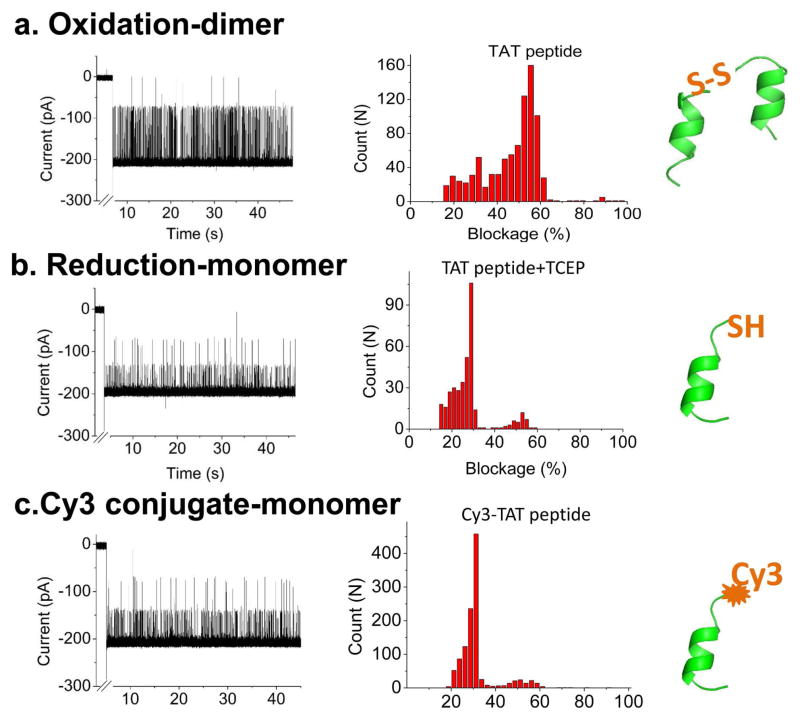
The focus of this study was to investigate whether the SPP1 motor channel can be used for kinetic analysis of peptides in real time. TAT peptide with and without a cysteine was used as a model system to study oxidation states. Structural analysis of TAT sequence using the computer program PEP-FOLD [64] revealed that without forming a disulfide bond, the TAT only adopts one α-helical conformation (Fig. 5). However, in the presence of a cysteine at the N-terminus, the peptide can be oxidized into a dimer by forming a disulfide bond between two N-terminal cysteines. The presence of two states, the oxidized and the reduced conformations, were confirmed by translocation studies with the SPP1 portal channel. Under an oxidized state, the current blockage distribution by TAT displayed a major peak, centered at 55.1±3.0% (Fig. 5a), with a minor peak centered at 28.5±1.9%. However, after adding the reducing agent TCEP [(tris(2-carboxyethyl)phosphine)] to break the disulfide bond, the blockage distribution significantly changed. The peak of 55.1±3.0% decreased significantly over time, while the majority of the blockage distribution shifted to 28.5±1.9%. This indicated that most of the peptide passing through the channel is in its single α-helical conformation (Fig. 5b). Current blockage distribution of the reduced TAT peptide was similar to Cy3-TAT signature with TCEP treatment, since the conjugation of Cy3 prevented the disulfide bond formation and resulted in similar blockage, representing the single TAT helix (Fig. 5c). In the Cy3-TAT current profile, some 55% current blockade events are observed. This can be attributed to a few unreacted TAT dimers in solution or two Cy3-TAT monomers passing through the channel at the same time.
Figure 5. Determining the conformational states of TAT peptide.
Current trace (left), current blockage distribution (middle) and conformation (right) for (a) Dimer state of TAT peptide; (b) Monomer state of TAT peptide; and (c) Cy3-conjugated TAT monomer. Applied voltage: 50 mV; Buffer: 1 M KCl, 5 mM HEPES, pH 8. Total number of events: 858 in A; 367 in B and 1128 in C.
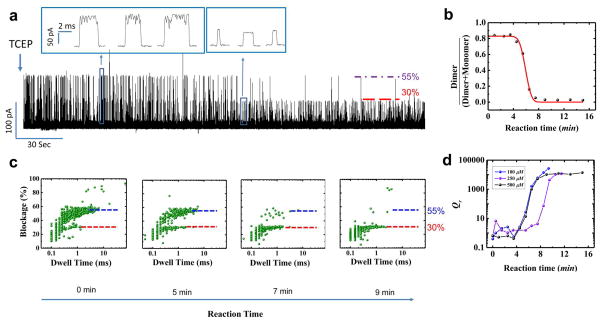
To quantify the kinetics parameter, we further examined the conformational changes of TAT peptide upon addition of TCEP and recorded the change of the current blockage profile in real time (Fig. 6). It was found that current blockade signature progressively shifted from predominantly 55.1±3% (oxidized dimer) to 28.5±1.9% (reduced monomer), representing the real-time kinetic process of disulfide bond reduction (500 μM TCEP, Fig. 6A). The TCEP reduction process can be described as a pseudo-first order reaction due to the nearly infinite TCEP amount and the reaction equation is described as follows:
| (1) |
Fig. 6. Real-time assessment of the conformational states of TAT peptide.
(a) Continuous current trace showing transition of oxidized dimer states to reduce monomer states after addition of reducing agent TCEP. (b) Quantitative analysis showing the fraction (γ) of dimer and monomer states as a function of reaction time. (c) Current blockage vs. dwell time distribution over the course of reaction time. Applied voltage: 50 mV; Buffer: 1 M KCl, 5 mM HEPES, pH 8. (d) Quantitative analysis showing the reaction quotient Qr as a function of reaction time.
C0 and εC0 are TAT dimer and monomer concentration prior to adding TCEP; Ct is TAT dimer concentration at any giving reaction time t; k is the rate constant. C(t) represents the TCEP concentration at the pore entrance. Due to the limited capture radius of nanopores, only chemicals at the pore entrance can be effectively captured and detected [65,66]. After adding TCEP into cis-chamber, the reductant gradually diffused to the pore entrance and reduced TAT dimer to monomer.
According to three dimensional diffusion equation, C(t) has the following equation: , where D is the diffusion constant of TCEP and r is the distance from the pore entrance to TCEP injection location. To simplify the representation of this reaction kinetics, two parameters γ (defined as the ratio of the number of dimer events versus the sum of dimer and monomer events, and reaction quotient Qr ( , unit is normalized) were investigated. Solving the above equations yields:
| (2) |
| (3) |
Here, CT is the final TCEP concentration at diffusion equilibrium state. Experimentally, γ and Qr can be derived from the concentration of TAT dimer and monomer, which is represented by the number of the dimer (with 40% ~ 70% blockade, 5σ) and monomer (with 19% ~ 38% blockade, 5σ) translocation events[66]. By calculating those two parameters at each reaction time, the transition of γ and Qr as shown in Fig. 6B and Fig. 6D represents the temporal evolution of the reduction reaction. The red line in Fig. 6B is the fitting curve with equation (2) (500 μM TCEP as that in Fig. 6A). γ decreased over the course of reaction time, indicating a decrease in oxidized dimer conformation and concurrent increase in reduced monomeric state. Fitting the data revealed a rate constant k = 0.989 ± 0.096 min−1. The trend is also demonstrated in current blockage percentage vs. dwell time distribution of events over the course of the reaction time (Fig. 6C). Qr (500 μM TCEP) in Fig. 6D, derived from γ with equation (3), reached a plateau when the reaction approached equilibrium. The plateau Qr is equal to standard equilibrium constant K0, estimated to be 1.4×104 and corresponding to −23.3 kJ/mol Gibbs free energy (ΔG0 = −RT lnK). All the reduction data from different concentration of TCEP show the same trend to approach equilibrium status (Fig. 6D). When the concentration of TCEP was increased from 100, 250, to 500 μM, the ΔG0 were −24.5, −23.1, and −23.3 kJ/mol (Table 1), respectively. The ΔG0 derived from our assay is very close to the Δ;G0 of −27 kJ/mol for protein disulfide bond reduction by TCEP obtained by conventional Raman and computational methods [67].
Table 1.
Parameters for the quantification of the oligomeric states of TAT peptide in real time
| [TAT] (nM) | [TCEP] (μM) | Qr (103) | K0(103) | ΔG0 (kJ/mol) |
|---|---|---|---|---|
| 300 | 100 | 26.6 | 26.6 | −24.5 |
| 300 | 250 | 11.8 | 11.8 | −23.1 |
| 300 | 500 | 13.9 | 13.9 | −23.3 |
4. Discussion
With a conductance of 4.27 nS, SPP1 portal channel is one of the larger channels and only the second viral protein channel (apart from phi29 connector)[27] to be incorporated into a lipid membrane. In the presence of TAT peptide, a burst of current blockage events with characteristic current amplitude and dwell time were observed indicating the translocation of the peptides through the SPP1 connector. In this study, we further demonstrated that this new protein nanopore is capable of detecting kinetics of TCEP mediated reduction of disulfide bond and demonstrating peptide conformational changes in real time. From Fig. 6, the frequency of dimer forms (current blockage of ~55%) decreased gradually to monomer forms (current blockage of 30%), upon addition of TCEP, as the reaction time increased. The oligomerization states of peptides was determined in real-time at single molecule level, which is not possible by ensemble methods like NMR, Circular Dichroism, and optical second harmonic generation.
Fingerprints of various peptides translocation through phi29 nanopore was reported [35]. In this study, we further analyzed the kinetic of the peptide translocation. A similar Gibbs free energy (ΔG0) was obtained when different concentrations of substrates were applied, suggesting that the use of SPP1 nanopore for real-time quantification of peptide oligomeric states is feasible. Although there are many similarities among phi29 and SPP1 nanopore, such as similar shape and conductance, several different properties have been discovered. First, DNA translocation through SPP1 nanopore has not been observed (data not shown), whereas phi29 nanopore allow dsDNA or ssDNA translocation. Second, the orientation preference of these two nanopores in lipid bilayer are found very different in lipid bilayer[38].
Direct evidence of DNA translocation through α-hemolysin was demonstrated twenty years ago by quantitative PCR[68]. Although several studies utilizing peptide and protein translocation through nanopores, have been published over the last several years [47–49,54–58], direct method for validating peptide translocations are lacking. A recent publication tried to solve the problem by conjugating a single-stranded DNA to unfolded protein and then amplify the DNA using PCR after translocation[69]. Another study demonstrated the translocation of hyaluronic acid oligosaccharides using high-resolution mass spectrometry[70]. Here, we developed a very simple method to provide evidence of peptide translocation using single molecule fluorescence imaging of samples obtained from the nanopore setup. This method can be easily adapted and employed by other laboratories to validate protein translocation and quantitatively study peptide dynamics through nanopores.
The current blockage signature based on the translocation profile can potentially be used to investigate the length, charge, hydrophobicity, secondary structures and ultimately the amino acid sequences of the peptides. The kinetics of protein folding and unfolding as well as entropic and energetic contributions can be further dissected in the future at the single molecule level.
5. Conclusion
The reengineered membrane-embedded portal channel of bacteriophage SPP1 allows translocation of peptides with higher ordered structure to produce clear and reproducible electronic signatures. The translocation of peptides observed by single channel conduction assays were verified by optical single molecule fluorescence microscopy assays. The oligomer states of peptides were clearly differentiated in real-time at single molecule level. When different concentrations of substrates were applied, a similar ΔG0 was obtained, suggesting that the use of SPP1 nanopore for real-time dynamic quantification of peptide folding is feasible.
Supplementary Material
Assembly of SPP1 gp6 subunits into 12 subunit connector complexes as shown by 15–35% glycerol gradient ultracentrifugation. BSA was used for size control.
Acknowledgments
The research was supported by NIH grant R01 EB012135 to P.G. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is a consultant of Oxford Nanopore and RNA Nanobio, Ltd. The companies licensed his patents from the University of Kentucky, University of Cincinnati and Purdue University. He is a confounder of P&Z Biological Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pi F, Vieweger M, Zhao Z, Wang S, Guo P. Discovery of a new method for potent drug development using power function of stoichiometry of homomeric biocomplexes or biological nanomotors. Expert Opin Drug Deliv. 2015;13:23–36. doi: 10.1517/17425247.2015.1082544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo P, Zhao Z, Haak J, Wang S, Wu D, Meng B, et al. Common Mechanisms of DNA translocation motors in Bacteria and Viruses Using One-way Revolution Mechanism without Rotation. Biotechnology Advances. 2014;32:853–72. doi: 10.1016/j.biotechadv.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo P, Noji H, Yengo CM, Zhao Z, Grainge I. Biological nanomotors with revolution, linear, or rotation motion mechanism. Microbiology and Molecular Biology Reviews. 2016;80:161–86. doi: 10.1128/MMBR.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuervo A, Carrascosa JL. Viral connectors for DNA encapsulation. Curr Opin Biotechnol. 2011;23:529–36. doi: 10.1016/j.copbio.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 5.Hu B, Margolin W, Molineux IJ, Liu J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A. 2015;112(35):E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, et al. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–62. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, et al. Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol. 2001;310(5):1027–37. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- 8.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–9. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 9.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26(7):1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci U S A. 2009;106(21):8507–12. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nature Nanotechnology. 2011;6:615–24. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 13.Healy K. Nanopore-based single-molecule DNA analysis. Nanomedicine. 2007;2(4):459–81. doi: 10.2217/17435889.2.4.459. [DOI] [PubMed] [Google Scholar]
- 14.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Current Opinion in Biotechnology. 2010;21(4):439–76. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasianowicz JJ, Robertson JW, Chan ER, Reiner JE, Stanford VM. Nanoscopic porous sensors. Annu Rev Anal Chem (Palo Alto Calif ) 2008;1:737–66. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 16.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38(8):2360–84. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 17.Reiner JE, Balijepalli A, Robertson JW, Campbell J, Suehle J, Kasianowicz JJ. Disease detection and management via single nanopore-based sensors. Chem Rev. 2012;112(12):6431–51. doi: 10.1021/cr300381m. [DOI] [PubMed] [Google Scholar]
- 18.Geng J, Kim K, Zhang J, Escalada A, Tunuguntla R, Comolli LR, et al. Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature. 2014;514(7524):612–5. doi: 10.1038/nature13817. [DOI] [PubMed] [Google Scholar]
- 19.Chang H, Venkatesan BM, Iqbal SM, Andreadakis G, Kosari F, Vasmatzis G, et al. DNA counterion current and saturation examined by a MEMS-based solid state nanopore sensor. Biomed Microdevices. 2006;8:263–9. doi: 10.1007/s10544-006-9144-x. [DOI] [PubMed] [Google Scholar]
- 20.Haque F, Li J, Wu HC, Liang XJ, Guo P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den HM, Hall AR, Wu MY, Zandbergen HW, Dekker C, Dekker NH. Controlling nanopore size, shape and stability. nanotechnology. 2010;21(11):115304. doi: 10.1088/0957-4484/21/11/115304. [DOI] [PubMed] [Google Scholar]
- 22.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–53. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, et al. Dynamics of unfolded protein transport through an aerolysin pore. J Am Chem Soc. 2011;133(9):2923–31. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, Hirano N, Kaneko J, Kamio Y, Yao M, Tanaka I. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure. Protein Sci. 2011;20(2):448–56. doi: 10.1002/pro.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soskine M, Biesemans A, De Maeyer M, Maglia G. Tuning the Size and Properties of ClyA Nanopores Assisted by Directed Evolution. J Am Chem Soc. 2013;135:13456–63. doi: 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J Am Chem Soc. 2012;134:9521–31. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–72. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing P, Haque F, Vonderheide A, Montemagno C, Guo P. Robust Properties of Membrane-Embedded Connector Channel of Bacterial Virus Phi29 DNA Packaging Motor. Mol Biosyst. 2010;6:1844–52. doi: 10.1039/c003010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haque F, Geng J, Montemagno C, Guo P. Incorporation of Viral DNA Packaging Motor Channel in Lipid Bilayers for Real-Time, Single-Molecule Sensing of Chemicals and Double-Stranded DNA. Nat Protoc. 2013;8:373–92. doi: 10.1038/nprot.2013.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-Way Traffic of a Viral Motor Channel for Double-Stranded DNA Translocation. Nano Lett. 2010;10:3620–7. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang H, Jing P, Haque F, Guo P. Role of channel Lysines and “Push Through a One-way Valve” Mechanism of Viral DNA packaging Motor. Biophysical Journal. 2012;102:127–35. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng J, Wang S, Fang H, Guo P. Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids. ACS Nano. 2013;7(4):3315–23. doi: 10.1021/nn400020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque F, Wang S, Stites C, Chen L, Wang C, Guo P. Single pore translocation of folded, double-stranded, and tetra-stranded DNA through channel of bacteriophage Phi29 DNA packaging motor. Biomaterials. 2015;53:744–52. doi: 10.1016/j.biomaterials.2015.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque F, Lunn J, Fang H, Smithrud D, Guo P. Real-Time Sensing and Discrimination of Single Chemicals Using the Channel of Phi29 DNA Packaging Nanomotor. ACS Nano. 2012;6:3251–61. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Z, Wang S, Zhao Z, Zhou Z, Haque F, Guo P. Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor. Small. 2016;12:4572–8. doi: 10.1002/smll.201601157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Haque F, Rychahou PG, Evers BM, Guo P. Engineered Nanopore of Phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano. 2013;7:9814–22. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Ji Z, Yan E, Haque F, Guo P. Three-step Channel Conformational Changes Common to DNA Packaging Motors of Bacterial Viruses T3, T4, SPP1, and Phi29. Virology. 2017;500:285–91. doi: 10.1016/j.virol.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Ji Z, Wang S, Haque F, Guo P. Oriented single directional insertion of nanochannel of bacteriophage SPP1 DNA packaging motor into lipid bilayer via polar hydrophobicity. Biomaterials. 2016;105:222–7. doi: 10.1016/j.biomaterials.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen S, Zeng T, Liu L, Zhao K, Zhao Y, Liu X, et al. Highly sensitive and selective DNA-based detection of mercury(II) with alpha-hemolysin nanopore. J Am Chem Soc. 2011;133(45):18312–7. doi: 10.1021/ja206983z. [DOI] [PubMed] [Google Scholar]
- 40.Wang HY, Gu Z, Cao C, Wang J, Long YT. Analysis of a single alpha-synuclein fibrillation by the interaction with a protein nanopore. Anal Chem. 2013;85(17):8254–61. doi: 10.1021/ac401496x. [DOI] [PubMed] [Google Scholar]
- 41.Mereuta L, Roy M, Asandei A, Lee JK, Park Y, Andricioaei I, et al. Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Scientific reports. 2014;4:3885. doi: 10.1038/srep03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying YL, Li DW, Liu Y, Dey SK, Kraatz HB, Long YT. Recognizing the translocation signals of individual peptide-oligonucleotide conjugates using an a-hemolysin nanopore. Chemical Communications. 2012;48:8784–6. doi: 10.1039/c2cc32636a. [DOI] [PubMed] [Google Scholar]
- 43.Mereuta L, Asandei A, Seo CH, Park Y, Luchian T. Quantitative Understanding of pH-and Salt-Mediated Conformational Folding of Histidine-Containing, b-Hairpin-like Peptides, Through Single-Molecule Probing with Protein Nanopores. ACS applied materials & interfaces. 2014;6(15):13242–56. doi: 10.1021/am5031177. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland TC, Long YT, Stefureac RI, Bediako-Amoa I, Kraatz HB, Lee JS. Structure of peptides investigated by nanopore analysis. Nano Letters. 2004;4(7):1273–7. [Google Scholar]
- 45.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4(4):384–93. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Barnham KJ, Kenche VB, Ciccotosto GD, Smith DP, Tew DJ, Liu X, et al. Platinum-based inhibitors of amyloid-beta as therapeutic agents for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2008;105(19):6813–8. doi: 10.1073/pnas.0800712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat Nanotechnol. 2012;7(4):257–63. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- 48.Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C. Fast translocation of proteins through solid state nanopores. Nano Lett. 2013;13(2):658–63. doi: 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fologea D, Ledden B, McNabb DS, Li J. Electrical characterization of protein molecules by a solid-state nanopore. Appl Phys Lett. 2007;91(5):539011–3. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang HY, Ying YL, Li Y, Kraatz HB, Long YT. Nanopore analysis of beta-amyloid peptide aggregation transition induced by small molecules. Anal Chem. 2011;83(5):1746–52. doi: 10.1021/ac1029874. [DOI] [PubMed] [Google Scholar]
- 51.Stefureac R, Long YT, Kraatz HB, Howard P, Lee JS. Transport of alpha-helical peptides through alpha-hemolysin and aerolysin pores. Biochemistry. 2006;45(30):9172–9. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- 52.Movileanu L. Squeezing a single polypeptide through a nanopore. Soft Matter. 2008;4(5):925–31. doi: 10.1039/b719850g. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Montana V, Grubisic V, Stout RF, Jr, Parpura V, Gu LQ. Nanopore sensing of botulinum toxin type B by discriminating an enzymatically cleaved Peptide from a synaptic protein synaptobrevin 2 derivative. ACS Appl Mater Interfaces. 2015;7(1):184–92. doi: 10.1021/am5056596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, Bell NA, Hernandez-Ainsa S, Thacker VV, Thackray AM, Bujdoso R, et al. Single protein molecule detection by glass nanopores. ACS Nano. 2013;7(5):4129–34. doi: 10.1021/nn4004567. [DOI] [PubMed] [Google Scholar]
- 55.Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. Protein biosensors based on biofunctionalized conical gold nanotubes. J Am Chem Soc. 2005;127(14):5000–1. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 56.Han A, Schürmann G, Mondin G, Bitterli RA, Hegelbach NG, de Rooij NF, et al. Sensing protein molecules using nanofabricated pores. Applied Physics Letters. 2006;88(9):3901. [Google Scholar]
- 57.Steinbock LJ, Krishnan S, Bulushev RD, Borgeaud S, Blokesch M, Feletti L, et al. Probing the size of proteins with glass nanopores. Nanoscale. 2014;6(23):14380–7. doi: 10.1039/c4nr05001k. [DOI] [PubMed] [Google Scholar]
- 58.Nivala J, Mulroney L, Li G, Schreiber J, Akeson M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano. 2014;8(12):12365–75. doi: 10.1021/nn5049987. [DOI] [PubMed] [Google Scholar]
- 59.Payet L, Martinho M, Pastoriza-Gallego M, Betton JM, Auvray L, Pelta J, et al. Thermal unfolding of proteins probed at the single molecule level using nanopores. Anal Chem. 2012;84(9):4071–6. doi: 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- 60.Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 2012;12(9):4895–900. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, et al. Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucleic Acids Res. 2006;34:2698–709. doi: 10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geng J, Fang H, Haque F, Zhang L, Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011;32:8234–42. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Preus S, Noer SL, Hildebrandt LL, Gudnason D, Birkedal V. iSMS: single-molecule FRET microscopy software. Nature Methods. 2015;12(7):593–4. doi: 10.1038/nmeth.3435. [DOI] [PubMed] [Google Scholar]
- 64.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37(Web Server issue):W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotechnology. 2010;5:160–5. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Z, Hu Y, Shan X, Li W, Bai X, Wang P, et al. Revealing Three Stages of DNA-Cisplatin Reaction by a Solid-State Nanopore. Sci Rep. 2015;5:11868. doi: 10.1038/srep11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.David C, Foley S, Enescu M. Protein S-S bridge reduction: a Raman and computational study of lysozyme interaction with TCEP. Phys Chem Chem Phys. 2009;11(14):2532–42. doi: 10.1039/b815492a. [DOI] [PubMed] [Google Scholar]
- 68.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93(24):13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pastoriza-Gallego M, Breton MF, Discala F, Auvray L, Betton JM, Pelta J. Evidence of unfolded protein translocation through a protein nanopore. ACS Nano. 2014;8(11):11350–60. doi: 10.1021/nn5042398. [DOI] [PubMed] [Google Scholar]
- 70.Fennouri A, Przybylski C, Pastoriza-Gallego M, Bacri L, Auvray L, Daniel R. Single molecule detection of glycosaminoglycan hyaluronic acid oligosaccharides and depolymerization enzyme activity using a protein nanopore. ACS Nano. 2012;6(11):9672–8. doi: 10.1021/nn3031047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assembly of SPP1 gp6 subunits into 12 subunit connector complexes as shown by 15–35% glycerol gradient ultracentrifugation. BSA was used for size control.