Abstract
Background
In vitro studies have shown that Helicobacter pylori (H. pylori) infection induces autophagy in gastric epithelial cells. However, prolonged exposure to H. pylori reduces autophagy by preventing maturation of the autolysosome. The alterations of the autophagy-related genes in H. pylori infection are not yet fully understood.
Materials and Methods
We analyzed autophagy-related gene expression in H. pylori infected gastric mucosa compared with uninfected gastric mucosa obtained from 136 Bhutanese volunteers with mild dyspeptic symptoms. We also studied single nucleotide polymorphisms (SNPs) of autophagy-related gene in 283 Bhutanese participants to identify the influence on susceptibility to H. pylori infection.
Results
Microarray analysis of 226 autophagy-related genes showed that 16 genes were up-regulated (7%) and 9 were down-regulated (4%). We used quantitative reverse transcriptase-polymerase chain reaction to measured mRNA levels of the down-regulated genes (ATG16L1, ATG5, ATG4D and ATG9A) that were core molecules of autophagy. ATG16L1 and ATG5 mRNA levels in H. pylori positive specimens (n = 86) were significantly less than in H. pylori negative specimens (n = 50). ATG16L1 mRNA levels were inversely related to H. pylori density. We also compared SNPs of ATG16L1 (rs2241880) among 206 H. pylori-positive and 77 negative subjects. The odds ratio for the presence of H. pylori in the GG genotype was 0.40 (95% CI: 0.18-0.91) relative to the AA/AG genotypes.
Conclusions
Autophagy-related gene expression profiling using high-throughput microarray analysis indicated that down-regulation of core autophagy machinery genes may depress autophagy functions and possibly provide a better intracellular habit for H. pylori in gastric epithelial cells.
Introduction
Although Helicobacter pylori (H. pylori) is generally considered an extracellular human pathogen, H. pylori also can reside within gastric epithelial cells [1, 2]. It has been proposed that the ability of H. pylori to reside within gastric epithelial cells may be in part responsible for the difficulty in eradicating the infection with antimicrobial therapy [3]. The possibility role of intracellular expression of H. pylori genes in the development of H. pylori associated diseases remains unclear [4–6].
Macroautophagy (hereafter referred to as autophagy) is an intracellular process in which cytoplasmic material is delivered to lysosomes for degradation [7–9]. Bacterial pathogens are among the targets of selective autophagy, termed xenophagy[10]. Xenophagy is an innate immune mechanism. Autophagy can target intracellular bacteria present in either the cytosol or within vacuoles and restrict their growth. In most cases, autophagosomes form around the target bacteria and deliver them to the lysosome for degradation.
The autophagic process is regulated at both the post-translational and transcriptional level [11, 12]. In vitro studies have shown that infection of gastric epithelial cells with H. pylori can induce autophagy [13–15]. However, prolonged exposure (e.g., for 24 hours) of these cells to culture supernatants from vacuolating cytotoxin A (VacA) positive H. pylori results in prevention of autolysosome maturation resulting in an overall reduction in autophagy[16]. Tang et al.[17] showed that the expression of the key autophagy genes ATG12 and BECN1 decreased in association with up-regulation of microRNA (MIR) 30B in the gastric mucosa. They also found that conversion of LC3B-I to LC3B-II required for autophagy function was reduced in H. pylori-infected gastric mucosa compared to uninfected mucosa suggesting that the MIR30B-related reduction in autophagy function allowed intracellular H. pylori to evade autophagic clearance.
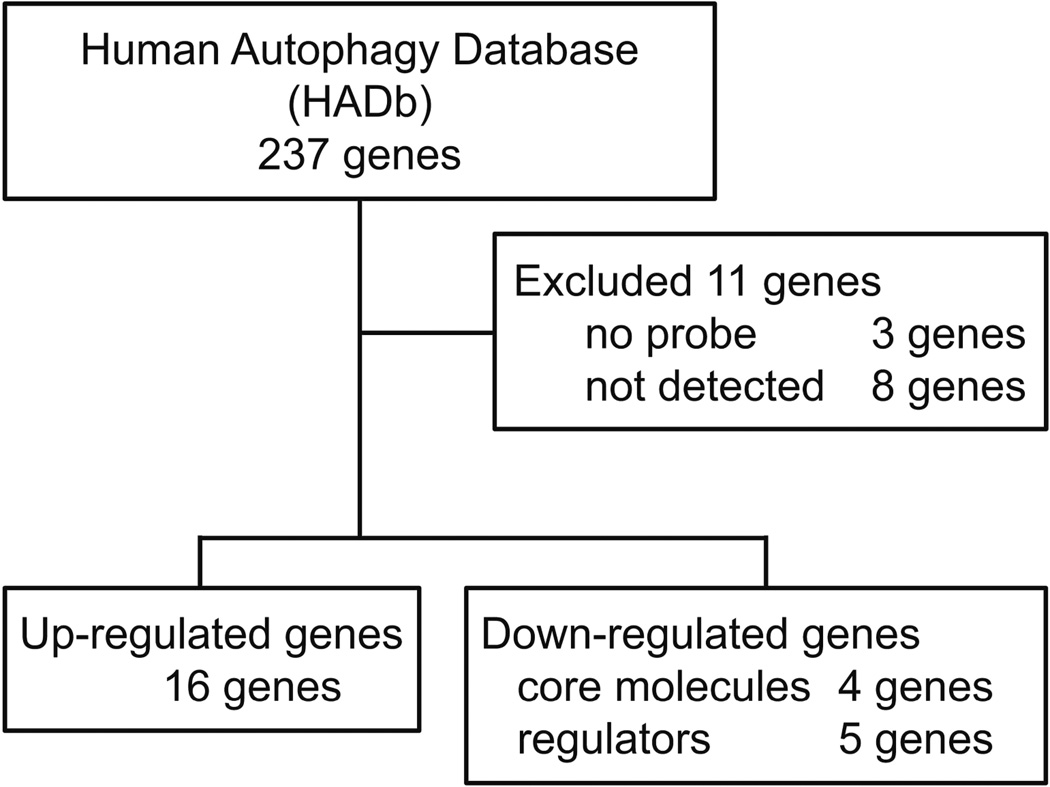
Despite recent interest changes in autophagy-related gene expression during H. pylori infection, the effect of H. pylori on autophagy function remains poorly understood. We used the Human Autophagy Database (HADb; available at www.autophagy.lu) and microarray analysis to analyze expression of autophagy-related genes in H. pylori-infected and uninfected human gastric mucosa. We also studied the relationship of single nucleotide polymorphisms (SNPs) of down-regulated autophagy-related genes in relation to susceptibility to H. pylori infection.
Methods
Subjects and gastric biopsy specimens
We used gastric mucosal biopsy specimens obtained from H. pylori infected and uninfected volunteers in Bhutan obtained as previously described [18]. In a previous study, we recruited a total of 372 volunteers with mild dyspeptic symptoms from three Bhutanese cities (Thimphu, Punaka, and Wangdue) during four days (December 6 to December 9) in 2010. During endoscopy, 4 gastric biopsy specimens were collected from healthy areas in the antrum: one each for H. pylori culture followed by DNA extraction, rapid urease test, RNA analysis, and histological examination. Written informed consent was obtained from all participants, and the protocol was approved by the ethics committee of Oita University (Japan) and Chulalongkorn University (Thailand) as well by the local hospitals where we collected the specimens[18].
All biopsy specimens for culture and RNA analyses were immediately placed in a −20°C freezer and subsequently sent on dry ice by Express Mail to Oita University Faculty of Medicine, Japan, where they were stored at −80°C until use. Biopsy specimens for histology were fixed in buffered formalin at room temperature and were sent to Oita University Faculty of Medicine for sectioning and analyses. Total RNA from the gastric specimens placed in RNA later (Ambion, Life Technologies, Carlsbad, CA) was isolated using commercially available kits (Ambion) and genomic DNA was isolated from gastric specimens following the H. pylori culture using DNeasy Blood & Tissue Kit (Qiagen).
H. pylori culture and status
H. pylori culture was performed using standard culture methods, as previously described[19]. H. pylori status was determined using the combination of rapid urease test, serology, culture, and histology. Subjects were considered to be H. pylori-negative when all four tests were negative and as H. pylori-positive when at least two of these examinations yielded positive results.
cagA and vacA genotyping of H. pylori
H. pylori DNA was extracted from H. pylori cultured on plates using the commercially available kit (DNeasy Blood & Tissue Kit; Qiagen, Valencia, CA). The cagA status was determined by polymerase chain reaction (PCR) for a conserved region of cagA and for direct sequence[20]. The cagA genotype (East-Asian type and Western type) was confirmed by sequencing the PCR products as described previously[21]. The vacA genotyping (s1, s2, m1 and m2) was also performed as described previously[22].
Histology
Biopsy specimens for histological examination were fixed in 10% (vol/vol) neutralized buffered formalin, embedded in paraffin wax and stained with hematoxylin-eosin and Giemsa stains. Specimens were evaluated by a histologist blinded to the clinical features of patients or the characteristics of the H. pylori strains. H. pylori density, the degree of mononuclear cell (MNC, inflammation) and polymorphonuclear leucocyte infiltration (PMN, activity) were determined according to the updated Sydney system. The H. pylori density was also evaluated by immunohistochemistry with polyclonal anti-H. pylori antibody, as described previously[23]. The H. pylori density was scored based on the average density on the surface and the foveolar epithelium. If areas with widely different scores were obtained on the same specimen, an average based on the general evaluation of the biopsy was considered. Only areas without metaplasia were evaluated for the presence of H. pylori.
Gene expression microarrays
Gene expression levels from the gastric specimens were analyzed by gene expression microarray. Complementary RNA was amplified, labeled, and hybridized to a 44K Agilent 60-mer oligo microarray according to the manufacturer's instructions. All hybridized microarray slides were scanned using an Agilent scanner, and relative hybridization intensities and background hybridization values were calculated using Agilent Feature Extraction Software (9.5.1.1). Differences in mRNA expressions between the two groups were considered significant if the fold change of expression values was >1.5 and the p value was <0.01 using the t test. The microarray data were registered in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/info/linking.html); the accession number is GSE47797.
Quantitative reverse transcription PCR
Expression levels of mRNAs that showed significant differences based on the microarray results were analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) from gastric biopsies. qRT-PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Beta-actin (ACTB) was used as the endogenous control for data normalization. Predesigned TaqMan Gene Expression Assays including primer set and TaqMan probe (ATG16L1: Hs01003142_m1, ATG5: Hs00169468_m1, ATG9A: Hs01036946_m1, ATG4D: Hs00262792_m1, IL1B: Hs01555410_m1, ACTB: Hs01060665_g1) were purchased from Applied Biosystems. mRNA levels of these genes in gastric specimens were quantified using ABI Prism 7300 sequence detection system (Applied Biosystems). The samples were placed in the analyzer and PCR was conducted according to the manufacturer’s instructions. The ratio change in target gene relative to the endogenous control gene (ACTB) was determined by the 2−ΔΔCT method.
SNPs genotyping analysis
TaqMan SNP Genotyping Assays (ATG16L1: rs2241880) designed with two specific probes and primers for each variant were purchased from Applied Biosystems. Genomic DNA was amplified using TaqMan Universal PCR Master Mix (Applied Biosystems) and the ABI Prism 7300 sequence detection system (Applied Biosystems) according to the manufacturer's instructions. Analysis of the results was done using the SDS Software version 1.3 (Applied Biosystems). Genotyping data were acquired by a researcher blinded to all clinical information.
Cell culture and H. pylori culture used for in vitro studies
The human gastric epithelial cancer cell line AGS was obtained from American Type Culture Collection and human gastric epithelial cancer cell lines MKN45 and MKN28 were obtained from Riken Cell Bank (Tsukuba, Japan). Cells were routinely maintained in RPMI 1640 medium (Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen, Life Technologies, Carlsbad, CA) and 1% penicillin/streptomycin at 37°C and 5% CO2.
In an attempt to investigate how H. pylori might regulate ATG16L1 and ATG5 mRNA levels in the gastric mucosa, we examined the effect of H. pylori infection on human gastric epithelial cancer cells. First, we examined three gastric epithelial cancer cell lines (AGS, MKN45 and MKN28) for the expression of ATG16L1 mRNA using qRT-PCR. In the second step, H. pylori strain 26695 was used to infect MKN45 and MKN28 for different time periods (3, 6 and 24h) and at different multiplicity of infections (MOIs) of 50 and 100. H. pylori strain 26695 was used from our stocks. H. pylori were cultured on Brain Heart Infusion (BHI; Becton, Dickinson, and Company, Sparks, MD) agar plates containing 7% defibrinated horse blood for 72 h. Before infection, bacteria were inoculated into BHI broth with 10% FBS and grown under microaerophilic conditions at 37°C overnight with shaking. Bacteria were washed with phosphate-buffered saline (PBS) (pH 7.4), resuspended in PBS for the duration of infection, and used to infect cell cultures. Cultured cells were infected with H. pylori at a multiplicity of infection (MOI) of 50 or 100. Total RNA from human cell lines was extracted using commercially available kits (Ambion).
Western blotting
Cells were washed with ice-cold PBS and then solubilized in a buffer containing 50mM Tris-HCl (pH 7.4), 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1mM EGTA, protease and phosphatase inhibitor cocktail (Roche, Mannheim, Germany) and cleared by centrifugation at 10000 g. Total protein was separated on a polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA). The membranes were blocked with 5% nonfat dry milk (Bio-Rad) for 1 h and then incubated with primary antibodies. The primary antibodies used were rabbit anti-LC3B (L7543, 1:1000, Sigma-Aldrich), and goat anti-β-actin (sc-1616, 1:500; Santa Cruz Biotechnology). After washing, the membranes were incubated in HRP-conjugated secondary antibodies. The protein signals were detected using the Clarity Western ECL Substrate (Bio-Rad).
Statistics
All statistical analyses were performed by JMP 10.0 software (SAS, Cary, NC) or Microsoft Excel for Mac 2011 (version 14.4.8, Microsoft). Clinical samples were analyzed using the χ2 test to compare discrete variables and the Mann-Whitney U test to compare continuous variables. In vitro samples were analyzed using Student's t-test. Correlation coefficients were calculated by Spearman rank correlation coefficient. For SNPs analysis, deviations of genotype frequencies from those expected under the Hardy-Weinberg equilibrium (HWE) were assessed by a goodness-of-fit χ2-test. Logistic regression adjusted by age and sex was used to analyze odds ratio (OR) and 95% confidence interval (CI). All statistical tests were two-sided, and p < 0.05 was considered statistically significant.
Results
Subjects
Samples from 136 subjects from Bhutan (median age, 40 years; age range, 16–92 years) were included in this study (Table 1). For gene expression analysis, all samples collected from participants in the capital city Thimphu on December 6th (n = 136) were investigated. For the SNP analysis, 283 of the available samples (n = 372) were investigated, the remaining samples being excluded due to lack of sufficient sample or background data. These subjects were previously reported in a survey of the prevalence of H. pylori infection[18] and the effect of H. pylori infection on gastric interleukin (IL)-8 and IL-10 mRNA levels[24]. The prevalence of H. pylori was 63% and all strains possessed cagA (Table 1).
Table 1.
Characteristics of the study subjects and H. pylori genotype
| H. pylori (−) n=50 | H. pylori (+) n=86 | p value | |
|---|---|---|---|
| Age, year (range) | 45 (18–78) | 34 (16–92) | 0.003 |
| Male (%) | 20 (40) | 40 (47) | 0.46 |
| cagA | |||
| negative (%) | 0 (0) | ||
| Western-type (%) | 2 (3) | ||
| East Asian-type (%) | 74 (97) | ||
| vacA | |||
| s1m1 (%) | 54 (71) | ||
| s1m2 (%) | 22 (29) | ||
| s2m1 (%) | 0 (0) | ||
| s2m2 (%) | 0 (0) | ||
| PMN, Grade | |||
| 0 (%) | 41 (82) | 0 (0) | |
| 1 (%) | 8 (16) | 42 (49) | |
| 2 (%) | 1 (2) | 37 (43) | |
| 3 (%) | 0 (0) | 7 (8) | |
| Mean, Median | 0.200, 0 | 1.593, 2 | <0.001 |
| MNC, Grade | |||
| 0 (%) | 16 (32) | 0 (0) | |
| 1 (%) | 33 (66) | 20 (23) | |
| 2 (%) | 1 (2) | 55 (64) | |
| 3 (%) | 0 (0) | 11 (13) | |
| Mean, Median | 0.700, 1 | 1.895, 2 | <0.001 |
| Atrophy, Grade | |||
| 0 (%) | 6 (12) | 1 (1) | |
| 1 (%) | 35 (70) | 46 (53) | |
| 2 (%) | 6 (12) | 35 (41) | |
| 3 (%) | 3 (6) | 4 (5) | |
| Mean, Median | 1.120, 1 | 1.488, 1 | <0.001 |
| H. pylori density | |||
| 0 (%) | 50 (100) | 9 (10) | |
| 1 (%) | 0 (0) | 16 (19) | |
| 2 (%) | 0 (0) | 25 (29) | |
| 3 (%) | 0 (0) | 36 (42) | |
| Mean, Median | 0, 0 | 2.023, 2 | <0.001 |
PMN, polymorphonuclear leucocyte; MNC, mononuclear cell.
Autophagy related gene expression profiles in H. pylori infection
We selected 8 samples for microarray analysis based on typical pathological findings (four samples were obtained from 50 subjects with H. pylori negative normal mucosa, and 4 from the 86 subjects with H. pylori positive gastritis mucosa). In this study, an array chipset that contained 50,599 total probe sets was used. For comprehensive expression analyses of autophagy-related genes, 226/237 genes in Human Autophagy Data base (HADb) were chosen from the microarray; 11 genes were excluded because 3 genes had no probe in this array chipset and 8 genes were not detected in all samples measured (Figure 1). Determination of the expression levels of the autophagy related genes showed 16/226 (7%) to be up-regulated (NLRC4: Fold change [FC] 11.88, p < 0.001; CXCR4: FC 7.40, p < 0.001; CCL2: FC 6.22, p < 0.001; GRID1: FC 3.49, p = 0.006; CX3CL1: FC 3.29, p =0.006; BCL2: FC 2.79, p = 0.002; RGS19: FC 2.72, p < 0.001; PRKCQ: FC 2.71, p = 0.003; FAS: FC 2.55, p = 0.007; ATG16L2: FC 2.27, p < 0.001; ARSB: FC 2.26, p = 0.006; CASP1: FC 2.09, p =0.001; ITPR1: FC 2.00, p = 0.003; BID: FC 1.90, p = 0.002; DNAJB9: FC 1.88, p = 0.009; RAB24: FC 1.64, p = 0.001) and 9/226 (4%) to be down-regulated (ATG9A: FC −1.58, p = 0.003; ITGB4: FC −1.60, p =0.001; ATG5: FC −1.71, p = 0.005; PTK6: FC −1.72, p = 0.009; ATG16L1: FC −1.73, p = 0.004; MAPK3: FC −1.80, p = 0.002; FKBP1B: FC −2.01, p < 0.001; ATG4D: FC 251 −2.18, p < 0.001; STBD1: FC −2.42, p = 0.006; respectively). Up-regulation was defined as at least a 1.5 fold increase and down regulation by at least a 1.5-fold decrease. In down-regulated genes, the core components of the autophagy machinery were included (ATG4D, ATG16L1, ATG5 and ATG9A).
Figure 1.
Flowchart of microarray analysis. Two hundred twenty-six autophagy-related genes in Human Autophagy Database (HADb; http://autophagy.lu/) were analyzed using microarray data (compared 4 H. pylori positive and 4 H. pylori negative). Differences in autophagy-related genes expression were considered significant if the fold change of expression levels was >1.5 and the p value was <0.01.
Autophagy-related genes mRNA expression levels in the antral gastric mucosa
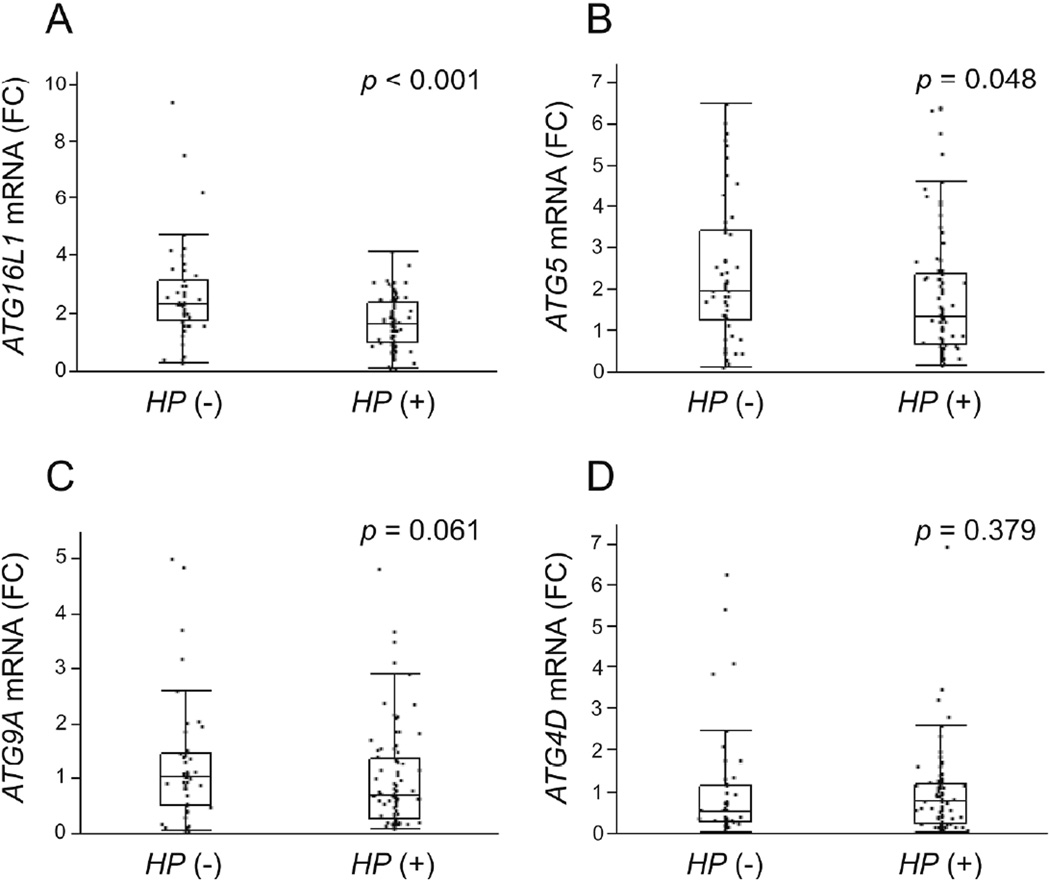
We measured the mRNA levels using qRT-PCR of the four down-regulated genes that were core molecules of autophagy: ATG16L1, ATG5, ATG4D and ATG9A. ATG16L1 mRNA levels in H. pylori positive specimens (n = 86) were significantly reduced compared to those in H. pylori negative specimens (n = 50) (H. pylori negative: median 2.34, range 0.281–9.41 and H. pylori positive: median 1.65, range 0.105–4.17, respectively; p < 0.001) (Figure 2A). ATG5 mRNA levels in H. pylori positive samples were also significantly lower than those in H. pylori negative samples (H. pylori negative: median 1.96, range 0.110–6.49 and H. pylori positive: median 1.32, range 0.171–6.42, respectively; p = 0.048) (Figure 2B). ATG9A mRNA levels in H. pylori positive samples were not statistically reduced compared to those in negative samples (H. pylori negative: median 1.04, range 0.068–5.01 and H. pylori positive: median 0.693, range 0.079–4.84, respectively; p = 0.061) (Figure 2C). ATG4D mRNA levels were not significantly different H. pylori positive and negative cases (H. pylori negative: median 0.553, range 0.050–6.26, and H. pylori positive: median 0.807, range 0.021–6.95, respectively; p = 0.379) (Figure 2D).
Figure 2.
Autophagy-related genes mRNA levels in the gastric specimens (50 H. pylori negative and 86 H. pylori positive). Down-regulated core autophagy-related genes (A, ATG16L1; B, ATG5; C, ATG9A; D, ATG4D) derived from microarray data were validated by qRT-PCR. Down-regulation of ATG16L1 and ATG5 mRNA levels were confirmed respectively. Beta-actin (ACTB) was used as the endogenous control for data normalization. Data were expressed by box plotting.
HP,Helicobacter pylori; FC, fold change.
ATG16L1 and ATG5 mRNA levels and H. pylori density
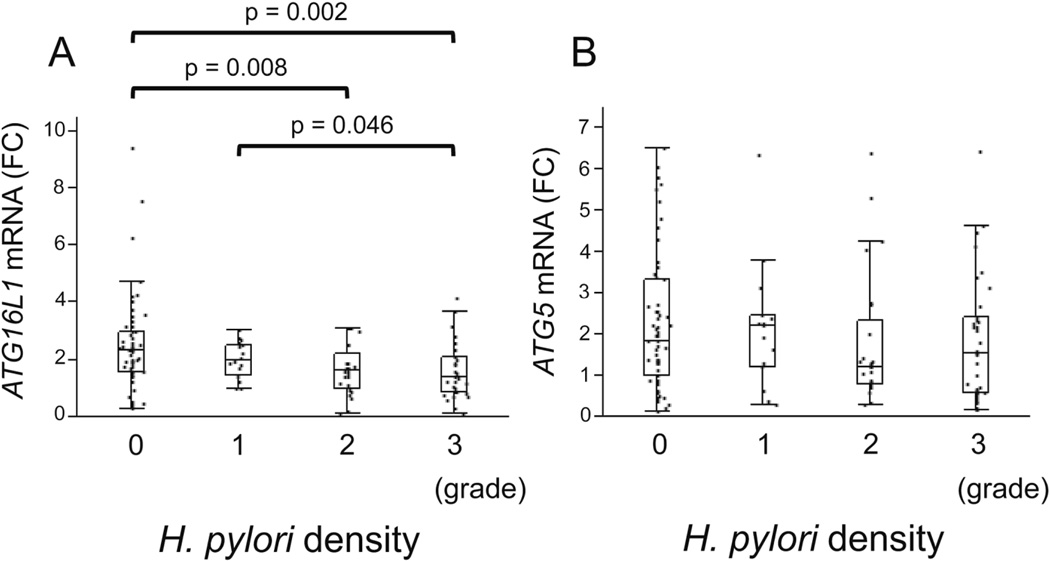
We examined the association between ATG16L1 and ATG5 mRNA levels and H. pylori density scores as determined using the updated Sydney System. There was a significant inverse relation between ATG16L1 mRNA levels and H. pylori density (score 0 vs 2, p = 0.008; score 0 vs 3, p = 0.002; score 1 vs 3, p = 0.046; respectively) (Figure 3A). On the other hand, there was no significant difference in ATG5 mRNA levels in relation to the H. pylori density score (Figure 3B).
Figure 3.
The association between ATG16L1 and ATG5 mRNA levels and H. pylori density. The ATG16L1 mRNA levels were decreased in a step-like manner by H. pylori density (Score 0, median 2.33, range 0.281–9.41; Score 1, median 1.99, range 0.997–3.03; score 2, median 1.63, range 0.135–3.12; score 3, median 1.39, range 0.105–4.17). There was no significant difference in ATG5 mRNA levels in relation to the H. pylori density score. H. pylori density were scored using the updated Sydney System. Beta-actin (ACTB) was used as the endogenous control for data normalization. Data were expressed by box plotting.
FC, fold change.
ATG16L1 and ATG5 mRNA levels and other histological findings
We examined for a possible correlation between ATG16L1 and ATG5 mRNA levels and the other histological findings (Table 2) however none of the other histological findings (PMN, MNC, atrophy) were significantly correlated with ATG16L1 or ATG5 mRNA levels (ATG16L1: PMN, p = 0.681; MNC, p = 0.291; atrophy, p = 0.406; ATG5: PMN, p = 0.155; MNC, p = 0.160; atrophy, p = 0.142; respectively).
Table 2.
Correlation between ATG16L1 and ATG5 mRNA levels and histological findings
| ATG16L1 | ATG5 | ||||
|---|---|---|---|---|---|
| Correl. Coef. | p value | Correl. Coef. | p value | ||
| PMN | 0.045 | 0.681 | PMN | −0.156 | 0.155 |
| MNC | −0.115 | 0.291 | MNC | −0.154 | 0.160 |
| atrophy | −0.091 | 0.406 | atrophy | −0.161 | 0.142 |
Correl.Coef., Correlation coefficient; PMN, polymorphonuclear leucocyte; MNC, mononuclear cell.
ATG16L1 and ATG5 mRNA levels in relation to vacA and ATG16L1 genotype
Among 86 H. pylori-infected subjects with our criteria, we were able to culture 76 H. pylori strains. We performed vacA genotyping on 76 H. pylori strains. All strains were s1 genotype (s1m1, n = 54 or s1m2, n = 22) (Table 1). There was no significant difference in ATG16L1 and ATG5 mRNA levels between vacA s1m1 and s1m2 genotypes (ATG16L1, p = 0.855; ATG5, p = 0.797) (Supplementary Figure 1). We genotyped ATG16L1 SNPs (rs2241880) using TaqMan SNP genotyping assays and analyzed whether there was an association between ATG16L1 mRNA levels and ATG16L1 genotype (Supplementary Figure 2). Twelve samples were excluded from the gene expression analysis as the genotype could not be identified (n = 124). No association was found between ATG16L1 mRNA levels and ATG16L1 genotype whether using all subjects (n = 124, p = 0.569) or specifically H. pylori positive subjects (n = 81, p = 0.930).
ATG16L1 mRNA levels and the pro-inflammatory cytokine interleukin 1β (IL-1β)
Lee et al.[25] using mouse embryonic fibroblasts reported that ATG16L1 suppressed IL-1β signaling via regulation of p62 stability and mediated ubiquitination of p62. Plantinga et al.[26] used human peripheral blood mononuclear cells to demonstrated that genetic variation in ATG16L1 was associated with higher production of IL-1β. We examined whether there was an association between ATG16L1 mRNA levels, genotype and IL-1β mRNA levels in H. pylori infected and uninfected gastric mucosa and found no correlation between ATG16L1 mRNA levels and IL-1β mRNA levels in either all subjects (n = 124; correlation coefficient, 0.092; p = 0.286) or H. pylori positive subjects (n = 81; correlation coefficient, 0.102, p = 0.350) and no association between ATG16L1 genotype and IL-1β mRNA levels in either all subjects (n = 124, p = 0.750) or H. pylori positive gastric mucosa (n = 81, p = 0.828) (Supplementary Figure 3).
ATG16L1 and ATG5 mRNA levels in gastric epithelial cells in response to H. pylori infection
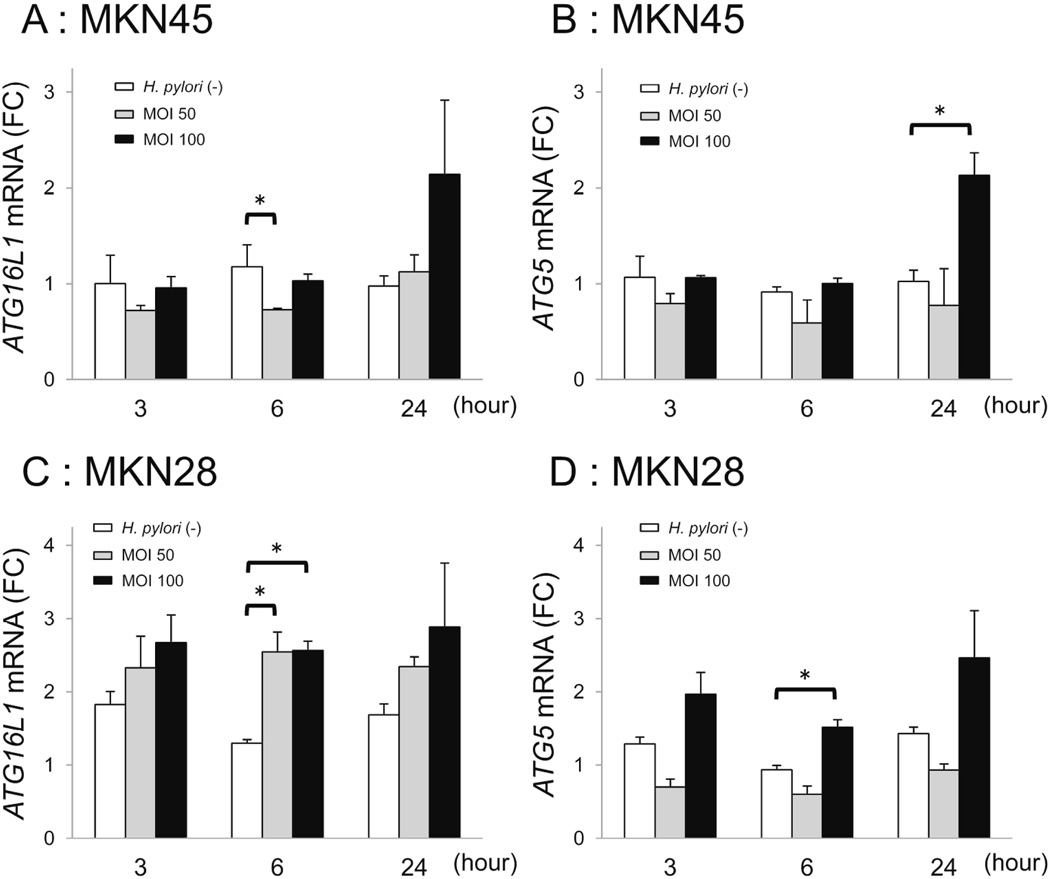
LC3B-II protein expression (Autophagy function) was increased by H. pylori infection in vitro (Supplementary Figure 4). Gastric mucosal ATG16L1 and ATG5 mRNA levels were reduced in H. pylori-infected gastric mucosa compared to uninfected mucosa (Figure 2). These results matched our hypothesis. Given this we investigated how H. pylori might regulate ATG16L1 and ATG5 mRNA levels in gastric epithelial cells. In a preliminary experiment, ATG16L1 mRNA was detected in MKN45 and MKN28, but not in AGS cells (data not shown). Based on our in vivo data showing an inverse relation between H. pylori density and ATG16L1 expression, we examined the effects on autophagy-related genes during both the early and late phases of infection and at lower and higher MOIs (Figure 4). However, we were unable to demonstrate either a time or MOI-dependency in gene expression with either cell line; MKN45 (Figure 4A, 4B) or MKN28 (Figure 4C, 4D).
Figure 4.
ATG16L1 and ATG5 mRNA levels in gastric epithelial cells in response to H. pylori infection. A, ATG16L1 mRNA levels in MKN45 cells. B, ATG5 mRNA levels in MKN45 cells. C, ATG16L1 mRNA levels in MKN28 cells. D, ATG5 mRNA levels in MKN28 cells. Cells were infected with H. pylori strain 26695. ATG16L1 and ATG5 mRNA levels in H. pylori-infected and non-infected MKN45 and MKN28 cells were measured at MOI 50 and 100 for 3, 6 and 24 h. Error bars represent the standard deviation of values obtained from three experiments. Statistical differences between treated and control samples were analyzed using Student's t-test.
* p < 0.05.
The influence on H. pylori susceptibility in relation of ATG16L1 polymorphisms
To evaluate whether there was an association between ATG16L1 polymorphisms and susceptibility to H. pylori infection, we performed a case-control study including 77 H. pylori negative and 206 H. pylori positive subjects (Supplementary Table 1). The genotype distribution for ATG16L1: rs2241880 polymorphism in this study were similar to those expected for Hardy-Weinberg Equilibrium (p = 0.172). The risk of being H. pylori infected decreased in recessive model (AA+AG vs. GG; OR, 0.40; 95% CI, 0.18-0.91; p = 0.029) (Table 3). The AG genotype was marginally associated with higher risk of H. pylori infection when compared to AA genotype (OR, 1.83; 95% CI, 1.00-3.42; p = 0.048).
Table 3.
The influence on H. pylori susceptibility in relation of ATG16L1 SNP (rs2241880)
| Genotype |
H. pylori negative |
H. pylori positive |
OR (95%CI)a | p value |
|---|---|---|---|---|
| ATG16L1, rs2241880 | (%), n=77 | (%), n=206 | ||
| AA | 43 (56) | 99 (48) | 1.00 (Ref) | |
| AG | 21 (27) | 89 (43) | 1.83 (1.00–3.42) | 0.048 |
| GG | 13 (17) | 18 (9) | 0.53 (0.23–1.22) | 0.134 |
| AA | 43 (56) | 99 (48) | 1.00 (Ref) | |
| AG/GG | 34 (44) | 107 (52) | 1.32 (0.77–2.28) | 0.311 |
| AA/AG | 64 (83) | 188 (91) | 1.00 (Ref) | |
| GG | 13 (17) | 18 (9) | 0.40 (0.18–0.91) | 0.029 |
| A allele | 107 (69) | 287 (70) | 1.00 (Ref) | |
| G allele | 47 (31) | 125 (30) | 0.99 (0.66–1.48) | 0.967 |
OR, odds ratio; CI, confidence interval; Ref, reference.
Adjusted for age and sex in logistic regression model.
Discussion
To the best of our knowledge, this is the first report of detailed expression analyses of autophagy-related genes using microarray analysis to compare H. pylori-infected and uninfected human gastric mucosa. It has previously been shown that intracellular bacterial pathogens are capable of suppressing autophagy by down-regulating autophagy-related genes[27]. The present study showed that the core component autophagy-related genes, ATG16L1 and ATG5, mRNA levels were significantly reduced in H. pylori positive human gastric mucosa (Figure 2). ATG16L1 and ATG5 encode key autophagy proteins which function as part of a complex with ATG12 responsible for the proper subcellular localization of the autophagy machinery[28]. Nguyen et al.[29] used both in vitro and a mouse model to described a reduction in ATG16L1 and ATG5 mRNA and protein levels associated with the up-regulation of MIR30C and MIR130A in intestinal epithelial cells infected with adherent invasive Escherichia coli. They also reported that ATG16L1 and ATG5 mRNA levels were reduced in ileal biopsy specimens of Crohn's disease patients compared to healthy controls. In our study, we found down-regulation of ATG16L1 and ATG5 expression in H. pylori infected gastric mucosa which is consistent with the notion that suppression of autophagy could promote H. pylori residence within gastric epithelial cells.
We also found that ATG16L1 mRNA levels in the gastric mucosa decreased in a H. pylori density-dependent manner (Figure 3), however we found no correlation between ATG16L1 mRNA levels and mucosal infiltration with acute or mononuclear inflammatory cells (Table 2). These results are similar to those of Glas et al. [30] who reported no association between ATG16L1 mRNA levels between inflamed lesions vs. non-inflamed Crohn's disease tissue biopsies. Together these results suggest that neither the presence, nor the intensity of the acute or chronic inflammatory cell infiltration of the gut mucosa significantly influence ATG16L1 mRNA levels. However, several studies have suggested that Atg16L1 may be involved in the regulation of inflammatory responses[25, 26]. For example, Saito et al. [31] reported that lipopolysaccharide stimulation of ATG16L1-deficient cells resulted in the production of large amounts of proinflammatory cytokines IL-1β and IL-18. However, we found no association between between ATG16L1 mRNA levels, genotype and IL-1β mRNA levels in H. pylori infected gastric tissue (Supplementary Figure 3). More studies are needed to examine the precise genetic and functional roles of autophagy-related genes in the pathogenesis of H. pylori–induced gastritis.
Based on the microarray analysis, ATG4D expression was found to be significantly down-regulated however, using qRT-PCR analyses up-regulation was observed. In previous studies, we have confirmed our microarray data using qRT-PCR with β-actin as the housekeeping gene. In general, our results have been similar [24, 32]. The reason why the qRT-PCR results in the current study do not match that of the microarray data is unclear. However, it is possible that this may relate to the fact we used only one house keeping gene (β-actin). Given that the use of 3−5 reference genes is currently recommended, potentially several reference genes may be required for the qRT-PCR of ATG4D. Growing evidence from in vitro studies has suggested that H. pylori virulence factors, especially VacA, are involved in the host's autophagy response to H. pylori infection[13, 14, 16]. We therefore investigated the association between vacA genotype and ATG16L1 and ATG5 mRNA expression in human gastric mucosa. We found no relation between ATG16L1 and ATG5 mRNA levels and infection with either the vacA s1m1 or s1m2 genotype (there were no s2m2 genotypes) (Supplementary Figure 1).
Our in vitro studies using H. pylori infection of human gastric cancer cells also found no relationship between the MOI or phase of infection (early vs. late) and ATG16L1 and ATG5 mRNA expression. Deen et al.[3] in their review of the interaction between H. pylori infection and host cell autophagic processes based primarily on in vitro experiments concluded that different host cell lines and bacterial strains produced different results. In contrast to in vitro experiments, H. pylori are infrequently seen within gastric mucosal cells in vivo suggesting that while invasion of epithelial cells occurs and may be a survival strategy, its role, if any, in the pathogenesis of H. pylori-related disease will be difficult to unravel. Our in vitro studies do not support a major role. In this study, we used one strain; standard H. pylori strain 26695 whose whole genome sequences had been first confirmed. Further studies using other strains will be needed to confirm our results. Autophagy is often dysregulated in a wide spectrum of human cancers[33] and most in vitro studies have used cancer cells. Possible studies using normal polarized gastric cells such as gastroids will provide a better model the in vivo interactions.
Genome-wide association studies (GWAS) have identified ATG16L1 SNP (rs2241880), encoding a Thr300Ala amino acid substitution (T300A), as a risk variant for Crohn's disease[34, 35]. Further studies also suggested that the ATG16L1 SNP was also associated with susceptibility to H. pylori infection[16, 36]. Therefore, we performed two analyses in relation to the ATG16L1 SNP. First, we analyzed whether there was an association between ATG16L1 SNP and ATG16L1 mRNA expression levels. We found no significant differences (Supplementary Figure 2). This result is similar to those of Hampe et al.[34] who demonstrated a lack of an association between ATG16L1 protein expression and ATG16L1 SNP genotype in Crohn's disease colonic mucosal biopsy specimens. Finally, we conducted a case-control study to investigate the influence of ATG16L1 SNP on susceptibility to H. pylori infection (Table 3) which suggested that the prevalence of H. pylori infection was significantly reduced in patients bearing ATG16L1 rs2241880 GG genotype compared to those bearing the AA/AG genotypes. Interestingly, our result is contrary to a report by Raju et al.[16] who found an increased risk of H. pylori infection in Scottish and German subjects bearing the GG genotype. A study in a Chinese population showed that ATG16L1 rs2241880 mutant homozygote (GG) was not found to increase the risk of gastric cancer while the heterozygote (AG) statistically increased the risk of gastric cancer. Further logistic regression analyses by these authors showed that the G allele significantly increases the risk of gastric cancer (OR: 2.38, 95% CI: 1.34-4.24) and the risk of H. pylori infection in these ethnic Chinese individuals (OR, 1.49; 95%CI, 1.02-2.16; p = 0.041). These types of association studies are easily confounded by racial or regional differences. For example, the association between ATG16L1 rs2241880 SNP and susceptibility in Crohn's disease was based on patients from Western countries[34, 35] and was not confirmed by studies in Japanese, South Korean, and Han Chinese patients[37]. Epidemiological findings from Asian countries[38] and mostly European-American countries[39] have suggested that the prevalence of H. pylori infection in inflammatory bowel disease (IBD) patients was significantly lower than in non-IBD patients. This finding is consistent with our intriguing finding that the GG genotype at rs2241880 suggested a lower risk of H. pylori infection and higher risk of Crohn's disease.
In conclusion, examination of autophagy-related gene expression profiling in human gastric mucosa with H. pylori infection showed down-regulation of core autophagy machinery genes may depress autophagy functions and possibly allow H. pylori to better reside inside gastric epithelial cells. The presence of rs2241880 GG genotype of ATG16L1 was associated with a reduced risk of H. pylori infection. New in vitro models are expected to reveal autophagy mechanism in gastric epithelial cells with H. pylori infection.
Supplementary Material
Acknowledgments
We thank Dr. Varocha Mahachai (Department of Gastroenterology, Bangkok Hospital, Bangkok, Thailand), Dr. Ratha-korn Vilaichone (Gastroenterology Unit, Department of Medicine, Thammasat University Hospital, Pathumthani, Thailand), and Dr. Lotay Tshering (Department of Surgery, Jigme Dorji Wangchuk National Referral Hospital, Thimphu, Bhutan) for participate on the endoscopy survey to obtain gastric samples with YY. This work was supported in part by grants from the National Institutes of Health [DK62813 to YY and DK56338 to DYG], Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [24406015, 24659200, 25293104, and 26640114 to YY], the Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation from the Japan Society for the Promotion of Science (JSPS), the Strategic Funds for the Promotion of Science and Technology from the Japan Science and Technology Agency (JST).
Footnotes
Disclosures
Competing interests: The authors have no financial conflicts of interest. Dr. Graham is an unpaid consultant for Novartis in relation to vaccine development for the treatment or prevention of Helicobacter pylori infection. Dr. Graham is a paid consultant for RedHill Biopharma regarding novel H. pylori therapies and he has received research support for the culture of H. pylori. He is a consultant for Otsuka Pharmaceuticals regarding diagnostic breath testing. Dr. Graham has received royalties from Baylor College of Medicine for patents covering materials related to a 13C-urea breath test.
References
- 1.Petersen AM, Krogfelt KA. Helicobacter pylori: an invading microorganism? A review. FEMS Immunology & Medical Microbiology. 2003;36:117–126. doi: 10.1016/S0928-8244(03)00020-8. [DOI] [PubMed] [Google Scholar]
- 2.Dubois A, Borén T. Helicobacter pylori is invasive and it may be a facultative intracellular organism. Cell Microbiol. 2007;9:1108–1116. doi: 10.1111/j.1462-5822.2007.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deen NS, Huang SJ, Gong L, Kwok T, Devenish RJ. The impact of autophagic processes on the intracellular fate of Helicobacter pylori: More tricks from an enigmatic pathogen? autophagy. 2013;9:639–652. doi: 10.4161/auto.23782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mora CS, Doi SQ, Marty A, Simko V, Carlstedt I, Dubois A. Intracellular and Interstitial Expression of Helicobacter pylori Virulence Genes in Gastric Precancerous Intestinal Metaplasia and Adenocarcinoma. J Infect Dis. 2003;187:1165–1177. doi: 10.1086/368133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Necchi V, Candusso ME, Tava F, Luinetti O, Ventura U, Fiocca R, et al. Intracellular, Intercellular, and Stromal Invasion of Gastric Mucosa, Preneoplastic Lesions, and Cancer by Helicobacter pylori. Gastroenterology. 2007;132:1009–1023. doi: 10.1053/j.gastro.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 6.Dubois A. Intracellular Helicobacter pylori and Gastric Carcinogenesis: An “Old” Frontier Worth Revisiting. Gastroenterology. 2007;132:1177–1180. doi: 10.1053/j.gastro.2007.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, et al. The acquisition of resistance to TNFα in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. autophagy. 2011;7:760–770. doi: 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy A, Marzec J, Clear A, Petty RD, Coutinho R, Matthews J, et al. Dysregulation of autophagy in human follicular lymphoma is independent of overexpression of BCL-2. Oncotarget. 2014;5:11653–11668. doi: 10.18632/oncotarget.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, et al. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- 14.Yahiro K, Satoh M, Nakano M, Hisatsune J, Isomoto H, Sap J, et al. Low-density Lipoprotein Receptor-related Protein-1 (LRP1) Mediates Autophagy and Apoptosis Caused by Helicobacter pylori VacA. J Biol Chem. 2012;287:31104–31115. doi: 10.1074/jbc.M112.387498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halder P, Datta C, Kumar R, Sharma AK, Basu J, Kundu M. The secreted antigen, HP0175, of Helicobacter pylori links the unfolded protein response (UPR) to autophagy in gastric epithelial cells. Cell Microbiol. 2015;17:714–729. doi: 10.1111/cmi.12396. [DOI] [PubMed] [Google Scholar]
- 16.Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo Mata E, et al. Vacuolating Cytotoxin and Variants in Atg16L1 That Disrupt Autophagy Promote Helicobacter pylori Infection in Humans. Gastroenterology. 2012;142:1160–1171. doi: 10.1053/j.gastro.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang B, Li N, Gu J, Zhuang Y, Li Q, Wang H-G, et al. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. autophagy. 2012;8:1045–1057. doi: 10.4161/auto.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilaichone R-K, Mahachai V, Shiota S, Uchida T, Ratanachu-ek T, Tshering L, et al. Extremely high prevalence of Helicobacter pylori infection in Bhutan. World J Gastroenterol. 2013;19:2806–2810. doi: 10.3748/wjg.v19.i18.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA Genotypes of Helicobacter pylori to cagA Status, Cytotoxin Production, and Clinical Outcome. Helicobacter. 1998;3:241–253. doi: 10.1046/j.1523-5378.1998.08056.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamaoka Y, Osato MS, Sepulveda AR, Gutierrez O, Figura N, Kim JG, et al. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91–96. doi: 10.1017/s0950268899003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A Comprehensive Sequence and Disease Correlation Analyses for the C-Terminal Region of CagA Protein of Helicobacter pylori. PLoS ONE. 2009;4:e7736–e7738. doi: 10.1371/journal.pone.0007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atherton JC, Cao P, Richard M, Peek J, Tummuru MKR, Blaser MJ, Cover TL. Mosaicism in Vacuolating Cytotoxin Alleles of Helicobacter pylori association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda A, Uchida T, Nguyen LT, Kawazato H, Tanigawa M, Murakami K, et al. A novel diagnostic monoclonal antibody specific for Helicobacter pylori CagA of East Asian type. APMIS. 2009;117:893–899. doi: 10.1111/j.1600-0463.2009.02548.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagashima H, Iwatani S, Cruz M, Jiménez Abreu JA, Tronilo L, Rodríguez E, et al. Differences in interleukin 8 expression in Helicobacter pylori -infected gastric mucosa tissues from patients in Bhutan and the Dominican Republic. Hum Pathol. 2015;46:129–136. doi: 10.1016/j.humpath.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Kim HR, Quinley C, Kim J, Gonzalez-Navajas J, Xavier R, et al. Autophagy Suppresses Interleukin-1β (IL-1β) Signaling by Activation of p62 Degradation via Lysosomal and Proteasomal Pathways. J Biol Chem. 2012;287:4033–4040. doi: 10.1074/jbc.M111.280065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plantinga TS, Crisan TO, Oosting M, van de Veerdonk FL, de Jong DJ, Philpott DJ, et al. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 27.Pareja M, Colombo MI. Autophagic clearance of bacterial pathogens: molecular recognition of intracellular microorganisms. Front Cell Infect Microbiol. 2013;3:54. doi: 10.3389/fcimb.2013.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen HTT, Dalmasso G, Müller S, Carrière J, Seibold F, Michaud AD. Crohn’s Disease–Associated Adherent Invasive Escherichia coli Modulate Levels of microRNAs in Intestinal Epithelial Cells to Reduce Autophagy. Gastroenterology. 2014;146:508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Glas J, Konrad A, Schmechel S, Dambacher J, Seiderer J, Schroff F, et al. The ATG16L1 Gene Variants rs2241879 and rs2241880 (T300A) Are Strongly Associated With Susceptibility to Crohn's Disease in the German Population. Am J Gastroenterol. 2008;103:682–691. doi: 10.1111/j.1572-0241.2007.01694.x. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 32.Nagashima H, Iwatani S, Cruz M, Jiménez Abreu JA, Uchida T, Mahachai V, et al. Toll-like Receptor 10 in Helicobacter pylori Infection. J Infect Dis. 2015;212:1666–1676. doi: 10.1093/infdis/jiv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu WKK, Coffelt SB, Cho CH, Wang XJ, Lee CW, Chan FKL, et al. The autophagic paradox in cancer therapy. Oncogene. 2012;31:939–953. doi: 10.1038/onc.2011.295. [DOI] [PubMed] [Google Scholar]
- 34.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2006;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 35.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castaño-Rodríguez N, Kaakoush NO, Goh K-L, Fock KM, Mitchell HM. Autophagy in Helicobacter pylori Infection and Related Gastric Cancer. Helicobacter. 2015;20:353–369. doi: 10.1111/hel.12211. [DOI] [PubMed] [Google Scholar]
- 37.Ng SC, Tsoi KKF, Kamm MA, Xia B, Wu J, Chan FKL, et al. Genetics of inflammatory bowel disease in Asia: Systematic review and meta-analysis. Inflamm Bowel Dis. 2012;18:1164–1176. doi: 10.1002/ibd.21845. [DOI] [PubMed] [Google Scholar]
- 38.Wu X-W, Ji H-Z, Yang M-F, Wu L, Wang F-Y. Helicobacter pylori infection and inflammatory bowel disease in Asians: A meta-analysis. World J Gastroenterol. 2015;21:4750–4756. doi: 10.3748/wjg.v21.i15.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luther J, Dave M, Higgins PDR, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1077–1084. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.