Significance
New proteins can evolve by duplication of the genes that encode them, followed by specialization of the different copies. However, how the growth rate of an organism is coupled to the changes in a protein’s structure and function occurring during this process is not known. Here we show at atomic resolution how selection for the growth of a bacterium led to the evolution of HisA proteins with either a new function or two functions (old and new). We found that a distinct protein conformation is responsible for each function, and that a better enzyme leads to faster growth only up to a certain threshold. This study provides insight into how evolution works, from atomic to whole-organism levels.
Keywords: HisA, TrpF, adaptive evolution, enzyme performance threshold
Abstract
New genes can arise by duplication and divergence, but there is a fundamental gap in our understanding of the relationship between these genes, the evolving proteins they encode, and the fitness of the organism. Here we used crystallography, NMR dynamics, kinetics, and mass spectrometry to explain the molecular innovations that arose during a previous real-time evolution experiment. In that experiment, the (βα)8 barrel enzyme HisA was under selection for two functions (HisA and TrpF), resulting in duplication and divergence of the hisA gene to encode TrpF specialists, HisA specialists, and bifunctional generalists. We found that selection affects enzyme structure and dynamics, and thus substrate preference, simultaneously and sequentially. Bifunctionality is associated with two distinct sets of loop conformations, each essential for one function. We observed two mechanisms for functional specialization: structural stabilization of each loop conformation and substrate-specific adaptation of the active site. Intracellular enzyme performance, calculated as the product of catalytic efficiency and relative expression level, was not linearly related to fitness. Instead, we observed thresholds for each activity above which further improvements in catalytic efficiency had little if any effect on growth rate. Overall, we have shown how beneficial substitutions selected during real-time evolution can lead to manifold changes in enzyme function and bacterial fitness. This work emphasizes the speed at which adaptive evolution can yield enzymes with sufficiently high activities such that they no longer limit the growth of their host organism, and confirms the (βα)8 barrel as an inherently evolvable protein scaffold.
A central question in biology is how new genes, and thus new enzymes, emerge. Experimental evolution and phylogenetic sequence analysis have shown that horizontal gene transfer, de novo evolution, gene fusion/fission, and the duplication of preexisting genes with subsequent divergence all contribute to this process (1–5). One model for duplication and divergence, the IAD (innovation-amplification-divergence) model (6), posits that a candidate for duplication is a gene whose protein product not only performs its primary function, but also carries out a secondary, nonessential function (innovation). Such promiscuous proteins are common in all organisms (7). If conditions arise that make the secondary function important, then selection for increased gene copy number could satisfy the need for more of that protein (amplification). Once a mutation in one extra copy improves the secondary function of the resulting protein, the survival of that duplicate and the probability of its evolution toward specialization increases (divergence).
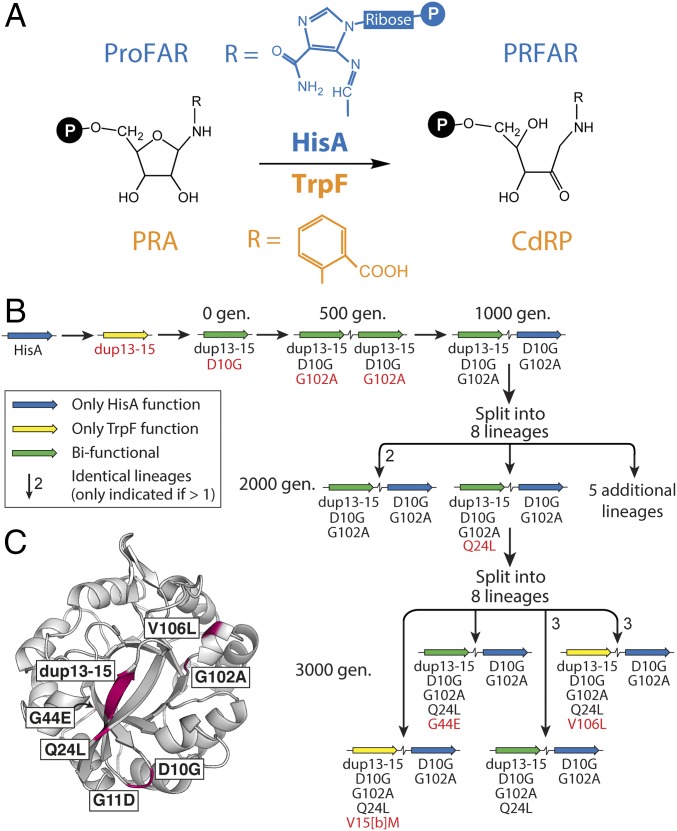
The IAD model was previously tested experimentally in Salmonella enterica, to show that new genes can evolve rapidly in bacteria to perform novel functions (8). The model system involved the N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (ProFAR) isomerase, HisA, which catalyzes an essential step in histidine biosynthesis (Fig. 1A). In an S. enterica strain in which the trpF gene was deleted from the chromosome, a spontaneous hisA mutation was selected that could complement the resulting tryptophan auxotrophy. Growth rate data showed that this mutant, HisA(dup13-15/D10G), catalyzed both the HisA and TrpF (phosphoribosylanthranilate isomerase) reactions (Fig. 1A), albeit suboptimally. Using this bifunctional mutant as a starting point, continuous selection for improved HisA and TrpF activities led to the creation of new genes by duplication and divergence. In fewer than 3,000 generations of growth under selection, the ancestral hisA variant was amplified, and then individual gene copies acquired point mutations, allowing them to diverge to encode either TrpF specialists, HisA specialists, or generalists performing both reactions (Fig. 1B).
Fig. 1.
Real-time evolution of new TrpF and HisA enzymes. (A) The analogous reactions catalyzed by HisA (blue) and TrpF (orange). HisA converts ProFAR into N'-[(5′-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (PRFAR). TrpF converts PRA to CdRP. (B) Mutational trajectory for real-time evolution of HisA into a bifunctional ancestor [HisA(dup13-15/D10G)] and then during continuous selection for improved TrpF and HisA activities (8). Amino acid substitutions arising in each population are shown below the gene symbols, with new mutations identified at each step shown in red. (C) Structure of a HisA variant, HisA(D7N/dup13-15/D10G), with the locations of all mutations from B shown in purple.
To understand how growth under selection honed the bifunctional HisA(dup13-15/D10G) variant during real-time evolution, we have characterized 11 enzymes—3 HisA specialists (including HisA itself), 3 TrpF specialists, and 5 generalists—using various structural and functional approaches. This has allowed us to relate changes in genotype (mutations) to changes in phenotype (enzyme kinetics, expression level, structure, and dynamics) and to organismal fitness (growth rate).
Results
Functional Analyses.
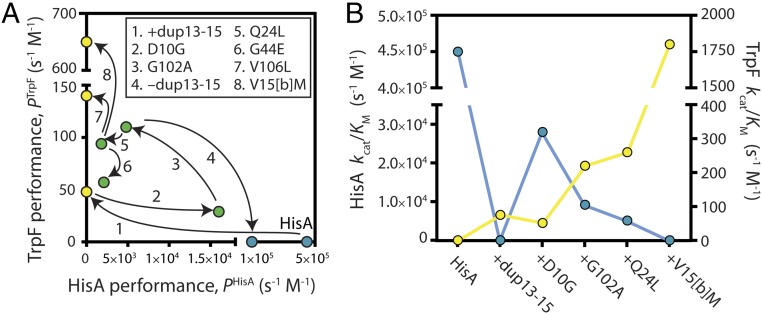
We first characterized each of the nine individual enzymes that make up the evolutionary trajectory in Fig. 1B (mutation sites indicated in Fig. 1C). We purified these enzymes and measured their Michaelis–Menten kinetic parameters in vitro. In parallel, we used a multiplexed tandem mass tagging approach to determine their relative expression levels during exponential-phase growth. This allowed us to determine a performance parameter for each enzyme, P = (kcat/KM) × (relative enzyme level), which determines the relative rate of intracellular product formation. The performances of the enzymes, plotted along the trajectories in which they evolved, are shown in Fig. 2A. Kinetic parameters and relative expression levels of each enzyme are presented in Table 1.
Fig. 2.
Genotype-phenotype landscapes. (A) Mutational effects on HisA and TrpF performance. Points connected by an arrow differ by a single mutation, as shown in the Inset. The intracellular performance of each enzyme, P, is defined as the product of its catalytic efficiency (kcat/KM) and its relative expression level during exponential-phase growth, with the expression level of WT HisA set to 1. Blue, HisA specialists; green, generalists; yellow, TrpF specialists. (B) Activity trade-offs along a trajectory of evolving HisA variants. Blue, HisA activity; yellow, TrpF activity.
Table 1.
Steady-state kinetic parameters and relative expression levels for HisA and evolved enzymes
| Enzyme | TrpF kcat, s−1 | TrpF KM, mM | TrpF kcat/KM, s−1 M−1 | HisA kcat, s−1 | HisA KM, μM | HisA kcat/KM, s−1 M−1 | Relative expression |
| HisA | ND | ND | ND | 7.8 ± 2.4 | 17 ± 0.1 | 4.5 × 105 | 1.00 ± 0.01 |
| HisA(dup13-15) | >0.15 | >2 | 75 ± 2 | ND | ND | ND | 0.64 ± 0.02 |
| HisA(dup13-15/D10G) | 0.09 ± 0.02 | 2.1 ± 1.0 | 51 ± 14 | 0.05 ± 0.01 | 1.7 ± 0.2 | 2.8 × 104 | 0.56 ± 0.02 |
| HisA(dup13-15/D10G/G102A) | >0.44 | >2 | 220 ± 30 | 0.05 ± 0.02 | 5.7 ± 1.6 | 9.2 × 103 | 0.52 ± 0.03 |
| HisA(D10G/G102A) | ND | ND | ND | 3.9 ± 0.1 | 24 ± 3 | 1.6 × 105 | 0.67 ± 0.01 |
| HisA(dup13-15/D10G/G102A/Q24L) | >0.52 | >2 | 260 ± 30 | 0.05 ± 0.01 | 10 ± 2 | 5.1 × 103 | 0.36 ± 0.04 |
| HisA(dup13-15/D10G/G102A/Q24L/V106L) | >0.53 | >2 | 260 ± 70 | ND | ND | ND | 0.53 ± 0.01 |
| HisA(dup13-15/D10G/G102A/Q24L/V15[b]M) | >3.6 | >2 | (1.8 ± 0.1) × 103 | ND | ND | ND | 0.36 ± 0.02 |
| HisA(dup13-15/D10G/G102A/Q24L/G44E) | >0.29 | >2 | 140 ± 60 | 0.18 ± 0.02 | 35 ± 2 | 5.2 × 103 | 0.41 ± 0.06 |
| HisA(dup13-15/D10G/G102A/G11D/G44E) | >0.26 | >2 | 130 ± 20 | 0.67 ± 0.05 | 100 ± 20 | 6.7 × 103 | 0.65 ± 0.01 |
| HisA(D10G) | ND | ND | ND | 7.6 ± 0.1 | 29 ± 10 | 2.6 × 105 | 0.77 ± 0.03 |
ND, not detected. SEs for steady-state kinetic parameters are from two independent enzyme preparations, each assayed in triplicate. Except for HisA(dup13-15/D10G), the HisA variants could not be saturated with PRA, so it was only possible to estimate kcat/KM, and then infer kcat. Expression levels are reported relative to HisA, with SEs from two independent experiments.
The first innovations in the previous evolution experiment generated a gene encoding the weakly bifunctional ancestor, HisA(dup13-15/D10G). Growth experiments previously showed that duplication of codons 13–15 (encoding amino acids V15[a]-V15[b]-R15[c]) imparted TrpF activity, but destroyed HisA activity (8). Assays of purified HisA(dup13-15) confirmed that its TrpF activity was weak but measurable (kcat/KM = 75 s−1 M−1), whereas it was inactive as a HisA enzyme (Table 1). The introduction of D10G further decreased TrpF activity, to kcat/KM = 51 s−1 M−1, but restored HisA activity to 6% of the wild-type (WT) level. The intracellular performances of HisA(dup13-15) and HisA(dup13-15/D10G) were further diminished by their expression levels, which were reduced to ∼60% of HisA (Table 1).
Starting from HisA(dup13-15/D10G), duplication and divergence yielded a series of new genes encoding enzymes with one or both of the activities under selection. In the first 500 generations of growth under selection, amplification of the gene for HisA(dup13-15/D10G) was accompanied by fixation of the additional mutation, G102A (Fig. 1B). G102A increased TrpF performance fourfold, while diminishing HisA performance by a similar amount (mutation 3 in Fig. 2A). Thus, the ratio of the specificity constants, (kcat/KM)HisA ÷ (kcat/KM)TrpF, dropped from 550 in HisA(dup13-15/D10G) to 42 in HisA(dup13-15/D10G/G102A). As expected, this change in kinetic parameters was accompanied by significant improvements in organismal growth rates under conditions requiring TrpF activity or both TrpF and HisA activity (SI Appendix, Table S1). Despite its reduced HisA activity, HisA(dup13-15/D10G/G102A) also conferred improved growth when only this activity was required (i.e., in tryptophan-supplemented medium). Further studies are needed to explain this observation.
Divergence was first observed after 1,000 generations of evolution, with emergence of the gene encoding HisA(D10G/G102A) (Fig. 1B). Loss of the three-residue VVR duplication ablated TrpF activity in this enzyme. The bacterial cells maintained HisA(D10G/G102A) as their HisA specialist (kcat/KM for the HisA reaction = 1.6 × 105 s−1 M−1) for the remainder of the evolution experiment, whereas a gene duplicate acquired further mutations that modulated TrpF and HisA activities. Among the gene variants, the three mutations that effected the largest improvements in TrpF activity (dup13-15, G102A, and V15[b]M) either significantly decreased or abolished HisA activity (Fig. 2B). The variant with the greatest TrpF activity was HisA(dup13-15/D10G/G102A/Q24L/V15[b]M). This variant reached fixation in one lineage after 3,000 generations of growth under selection (Fig. 1B), and had a kcat/KM for the TrpF reaction of 1.8 × 103 s−1 M−1. Approximately 10% of all enzymes that have been characterized from amino acid, fatty acid, and nucleotide metabolism have specificity constants at or below this level (9). Thus, our study emphasizes the speed with which adaptive evolution can yield new enzymes with kinetic parameters that are comparable to those of existing specialized enzymes. The final mutation that increased TrpF performance was V106L when introduced into HisA(dup13-15/D10G/G102A/Q24L) (mutation 7 in Fig. 2A). This abolished HisA activity and increased PTrpF via an increased expression level (Table 1). No mutation increased both activities simultaneously (Fig. 2).
In total, seven of the enzymes shown in Fig. 1B exhibited TrpF activity (three TrpF specialists and four bifunctional isomerases). Notably, none of these were saturated at the highest assayable concentration (2 mM) of the substrate, phosphoribosylanthranilate (PRA). Instead, kcat/KM was estimated from the linear part of the Michaelis–Menten plot. This finding implies that all of the variants have KMPRA >2 mM. The variant with the lowest Michaelis constant, HisA(dup13-15/D10G), appeared to have a KMPRA close to 2 mM. In contrast, the highest KMProFAR measured for any enzyme with HisA activity was 100 µM (Table 1 and vide infra). In the bifunctional enzymes (and thus the TrpF specialists descended from them), selection acted to hone turnover rather than ground state discrimination of the two competing substrates. In contrast, TrpF-active mutants generated by error-prone PCR from an artificial HisA/HisF chimera (10), as well as PriA, a bifunctional HisA/TrpF enzyme from Mycobacterium tuberculosis (11), exhibit micromolar values for KMPRA. This discrepancy highlights the different biochemical outcomes that can be realized depending on the starting scaffold and the nature of selection (from entirely artificial to adaptive over millions of years).
In addition to the trajectory shown in Fig. 1B, the real-time evolution experiment (8) also yielded a single example in which cells had lost the specialist HisA(D10G/G102A). Instead, the variant HisA(dup13-15/D10G/G102A/G11D/G44E) was solely responsible for flux through both the histidine and tryptophan biosynthetic pathways. Compared with the other bifunctional enzymes, its kinetic parameters were relatively poor for both reactions (Table 1), yet it conferred substantial growth advantages on S. enterica cells grown in the absence of histidine, or in the absence of both histidine and tryptophan (SI Appendix, Table S1). Similarly complex relationships between enzyme activity and organismal fitness were observed when bifunctional variants of ProA were evolved to become responsible for steps in both proline and arginine biosynthesis (12). Perhaps significantly, HisA(dup13-15/D10G/G102A/G11D/G44E) had the highest measured KM for the HisA substrate, ProFAR (KMProFAR = 100 µM; Table 1).
Structural Analyses.
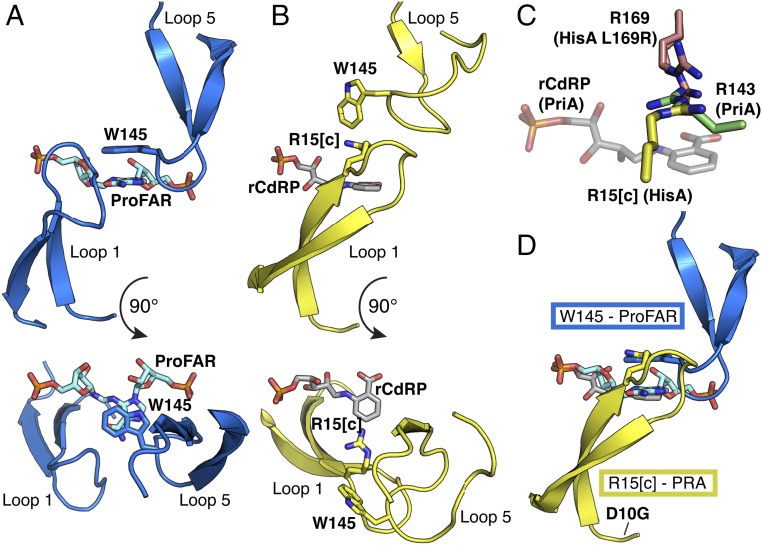
We determined a total of 13 apo and complexed crystal structures of the evolved HisA variants, all of which form similar (βα)8 barrels (SI Appendix, Figs. S1–S3 and Table S2). The long loops on the catalytic face of WT HisA enclose and trigger a structural change in ProFAR before catalysis (Fig. 3A) (13). In the apo structures of the evolved variants, active site loops 1, 5, and 6 are partly disordered or open, and phosphate or sulfate ions from the crystallization buffer often mimic the two phosphate groups of ProFAR in the active site. Selected mutants, including some with disabling mutations of the catalytic base (Asp7), also were subjected to cocrystallization or soaking with either ProFAR or the TrpF product analog, reduced 1′-(2′-carboxyphenylamino)-1′-deoxyribulose 5′-phosphate (rCdRP). This resulted in some structures in which the active site loops are more ordered (SI Appendix, Fig. S1 and Table S2) allowing us to observe closed, presumably active conformations. Owing to the low affinity of rCdRP, its ligand density was always too poor for unambiguous interpretation. Apart from G44E, all mutations are on the substrate-binding, catalytic face of the barrel (Fig. 1C). Several mutations are on loop 1 and thus are only visible in some of the structures.
Fig. 3.
Active site loop structures are critical for activity of HisA mutants. (A) In the HisA-active conformation, W145 forms a critical stacking interaction with the carboxamide aminoimidazole moiety of ProFAR (cyan), as shown in HisA(D7N/D176A)-ProFAR (PDB ID code 5A5W) (13). (B) Dup13-15 enables HisA to adopt an assumed TrpF-active conformation, where R15[c] is available for interaction with PRA, as shown in HisA(D7N/dup13-15/D10G), overlaid with rCdRP from PriA-rCdRP (PDB ID code 2Y85) (11). (C) Convergent positioning of active site arginines in three enzymes with TrpF activity: HisA(D7N/dup13-15/D10G/G102A/Q24L) (yellow), HisA(L169R) (double conformation, salmon), and PriA-rCdRP (PDB 2Y85) (green, ligand in transparent gray). (D) Bifunctionality involves competition between the two substrates ProFAR (cyan) and PRA (product analog rCdRP in gray), as well as between loops 1 and 5 (as positioned in B and A, respectively) in the active site of HisA.
The VVR duplication in loop 1, which initially provided TrpF activity, induces a new, extended conformation of loop 1 toward loop 5 that is observed in structures of two bifunctional enzymes and one TrpF specialist, in three different crystal forms (SI Appendix, Fig. S1). In this presumably TrpF-active conformation, the duplicated arginine, Arg15[c], is positioned in the active site, available for interaction with the TrpF substrate PRA (Fig. 3B). A different arginine, Arg143, has an equivalent interaction with rCdRP in the active site of PriA (11). Another S. enterica mutant with TrpF activity, HisA(L169R) (8), places Arg169 in a similar position (Fig. 3C and SI Appendix, Table S2), as does TrpF itself (14). The appropriate positioning of an arginine appears to be a shared innovation for PRA binding and a catalytically competent active site (11, 14) in the emergence of TrpF activity on the (βα)8 barrel scaffold.
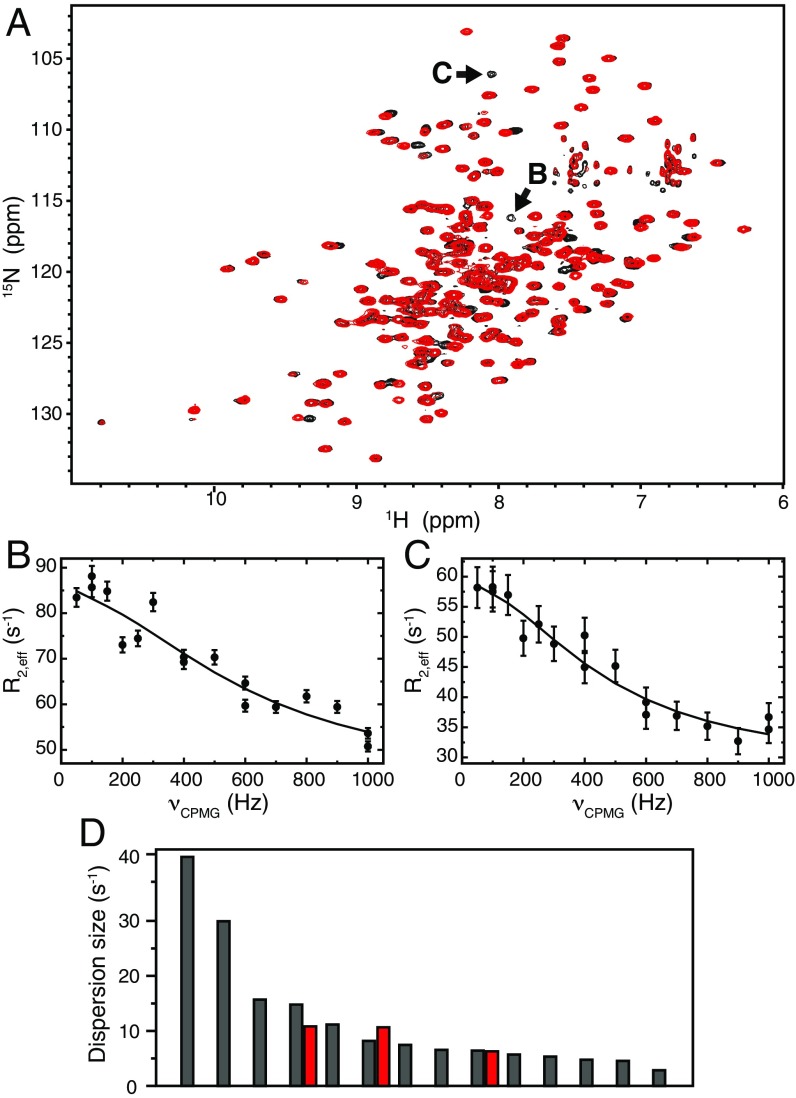
HisA activity is abolished in HisA(dup13-15) because the extended conformation of loop 1 blocks Trp145 (on loop 5) from interacting with ProFAR (Fig. 3D). A HisA(W145A) mutant was unable to rescue the histidine auxotrophy of S. enterica ΔhisA, showing that the Trp145–ProFAR interaction is essential for ProFAR isomerase activity. Mutagenesis of the equivalent tryptophan in PriA also destroys HisA activity (11). The D10G mutation in loop 1, which reintroduced HisA activity into HisA(dup13-15), does not induce any changes in the crystal structure, but is predicted to allow more flexibility of loop 1, so that loop 5 can compete for the active site (Fig. 3D). To test this hypothesis, we performed NMR relaxation dispersion experiments, which demonstrated that D10G leads to increased conformational dynamics (Fig. 4). Significant microsecond-millisecond motions were detected at 14 backbone 15N positions for HisA(dup13-15/D10G), compared with only three positions for HisA(dup13-15) (Fig. 4D; and SI Appendix, Fig. S4). Importantly, the two resonances with the largest dispersions are unique to HisA(dup13-15/D10G) (Fig. 4 A–C). Thus, the D10G mutation, which confers bifunctionality in presence of dup13-15, leads to increased dynamics of HisA, supporting our hypothesis.
Fig. 4.
The solution structure of HisA(dup13-15/D10G) is similar to that of HisA(dup13-15), but more dynamic on the microsecond-millisecond time scale. (A) 15N-1H correlation maps of HisA(dup13-15/D10G) (black) and HisA(dup13-15) (red). (B and C) Relaxation dispersion profiles for the HisA(dup13-15/D10G) 15N resonances indicated with arrows in A. (D) Dispersion sizes for residues with significant 15N dynamics for HisA(dup13-15/D10G) (gray) and HisA(dup13-15) (red), sorted in order of decreasing dispersion size. The two leftmost gray bars correspond to the dispersions in B and C.
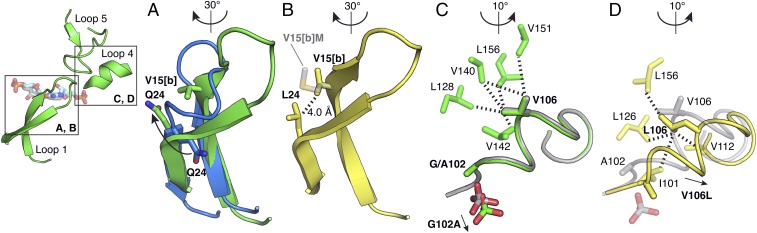
These data suggest that the HisA and TrpF reactions are catalyzed by enzymes adopting mutually exclusive loop conformations, where either loop 1 or loop 5 forms a critical interaction with the substrate (i.e., Arg15[c] with PRA or Trp145 with ProFAR; Fig. 3D). Further support for this model is provided by structures containing the Q24L mutation. In what we assume is the TrpF-active conformation of loop 1, the Gln24 side chain flips from its native position, inducing a backbone turn that brings it close to Val15[b] of the duplication, which is on the other strand of the β-hairpin (Fig. 5A). Q24L thus introduces a new hydrophobic interaction that stabilizes this TrpF-active loop conformation (Fig. 5B). Val15[b] is mutated to methionine in the variant with the highest TrpF activity (Table 1). Although V15[b]M is disordered in the available apo structure of this variant, based on the position of Val15 in the TrpF-active conformation, we can predict that the mutation further stabilizes the interaction with Leu24 (Fig. 5B). This seems to favor the TrpF-active conformation of loop 1 to the extent that the enzyme can no longer adopt its HisA-active conformation, and HisA activity is lost.
Fig. 5.
Structural insights into mutations that favor TrpF activity. (A) Conformational shift of loop 1 from HisA(D7N/D176A) (13) in blue, to HisA(D7N/dup13-15/D10G) in green. (B) In HisA(D7N/dup13-15/D10G/G102A/Q24L) (yellow), the shifted conformation is stabilized by a hydrophobic interaction between L24 and V15[b]. The predicted position of M15[b] is shown in transparent gray. (C) The phosphate-binding site in HisA(dup13-15/D10G)-PO4 (gray) is shifted 1.2 Å in HisA(dup13-15/D10G/G102A)-SO4 (green). Hydrophobic interactions of V106 with residues within 4.5 Å are indicated with dashed lines. (D) The V106L mutation in HisA(dup13-15/D10G/G102A/Q24L/V106L) (yellow) repositions loop 4 and Ala102 compared with HisA(dup13-15/D10G/G102A), gray, preventing binding of phosphate. Hydrophobic interactions of L106 with residues within 4.5 Å are indicated with dashed lines.
When HisA(dup13-15/D10G/G102A) evolved from HisA(dup13-15/D10G), its diminished performance as a HisA (Fig. 2) was related to an increase in KMProFAR (Table 1). The structure of HisA(dup13-15/D10G/G102A) (Fig. 5C) reveals the Ala102 side chain oriented into the binding site for the second phosphate of ProFAR. This phosphate-binding site is not required for PRA binding, as the TrpF substrate is only monophosphorylated (Fig. 1A). Subsequently, fixation of the mutation V106L in one lineage, after 3,000 generations of evolution (Fig. 1B), converted the bifunctional generalist into a TrpF specialist. V106L induces a shift of loop 4 that pushes G102A even further, preventing ionic phosphate from binding to the protein in the crystal structure (Fig. 5D). The blocked phosphate binding site presumably prevents productive binding of ProFAR in the active site and leads to abolished HisA activity.
Phenotype-Fitness Relationship.
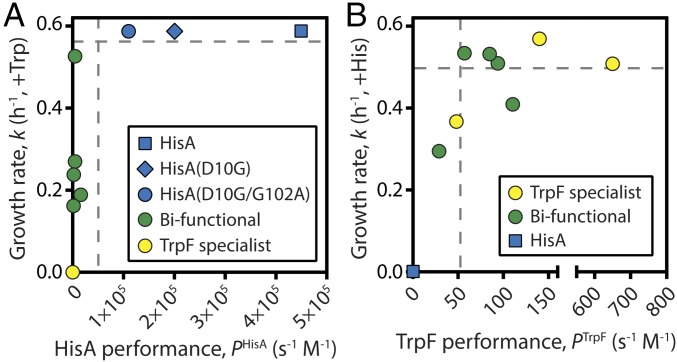
Our data enabled us to relate enzyme phenotype to organismal fitness. We found that the relationship between enzyme performance and growth rate is not linear for either HisA or TrpF activity. Instead, we observed biphasic relationships in which small changes in the activities of low-performance enzymes have large effects on fitness, until a threshold above which large changes in enzyme performance have little effect on fitness. This behavior was marked for growth in the absence of histidine (Fig. 6A). To assess the effects of single mutations, we added HisA(D10G) to our analysis. In vitro, HisA(D10G) was more active (kcat/KM = 2.6 × 105 s−1 M−1) than the dominant HisA specialist from the evolution experiment, HisA(D10G/G102A); however, the two enzymes conferred identical growth rates on strains grown in the absence of histidine (Fig. 6A). A threshold at PHisA ∼105 s−1 M−1 was sufficient to confer maximal growth, and thus the gene for HisA(D10G/G102A) was maintained in the evolving population for 2,000 generations, despite being one point mutation away from the more active HisA(D10G) variant. The threshold for conferring maximal growth in the absence of tryptophan was substantially lower, at PTrpF ∼50 s−1 M−1 (Fig. 6B), consistent with the reduced cellular demand for tryptophan (15).
Fig. 6.
Phenotype-fitness maps for evolved HisA and TrpF enzymes. (A) Biphasic correlation between HisA performance and the cellular growth rate conferred by each enzyme in the absence of exogenous histidine (in presence of tryptophan). Each point represents one of the mutants characterized in this study. Performance and growth data are provided in Table 1 and SI Appendix, Table S1, respectively. (B) Correlation between TrpF performance and growth rate in the absence of tryptophan (in presence of histidine).
Discussion
Taken together, our data provide exquisite detail on the evolution of new enzymes via IAD, and on the evolution of new functions on the (βα)8 barrel scaffold. Unlike other recent studies on the structural mechanisms of transitions in enzyme function (16, 17), here we have focused on a real-time evolution process in which a population of cells was under continuous selection for essential metabolic functions. Our kinetics emphasize the high sensitivity of selection. Even for a core metabolic process such as tryptophan biosynthesis, very poor catalysts (kcat/KM < 102 s−1 M−1) still can provide sufficient activity to be selectable in an evolving population.
In contrast to results from in vitro evolution (18), mutations conferring diminishing returns on enzyme performance were not observed as the TrpF evolutionary trajectory proceeded. Instead, the mutation imparting the single largest improvement in TrpF activity, V15[b]M (Fig. 2B), was among the last to be fixed (Fig. 1B). As reported previously (19–21), this finding suggests epistatic interactions between adaptive mutations. Our structural data (Fig. 5A) suggest that both the VVR duplication and the Q24L substitution are required before the adaptive benefit of V15[b]M can be realized. In addition, the latter part of the evolutionary trajectory might have been influenced by negative selection to lose HisA activity, and thus to prevent potential inhibition of TrpF activity when the HisA substrate ProFAR is also present in the cell. The potential for such inhibitory cross-talk to reduce fitness has been demonstrated in a similar case involving a serendipitous pathway for cofactor biosynthesis (22). Experiments to test the roles of epistasis and inhibitory cross-talk are ongoing in our laboratories.
Although not observed for enzyme performance, a strong trend of diminishing returns was observed with respect to improving organismal fitness. We identified thresholds for each enzymatic activity above which large changes in enzyme performance elicited only small changes in fitness (Fig. 6), suggesting that another enzyme in the pathway was now rate-limiting. Similar observations have been reported previously (23, 24). In our system, this finding has two significant implications. First, selection acted rapidly to take the evolving new activity (TrpF) across its performance threshold. Here 2,000 generations of growth under selection were sufficient to yield HisA(dup13-15/D10G/G102A/Q24L), which conferred near-maximal fitness on S. enterica grown in the absence of exogenous tryptophan (SI Appendix, Table S1). Second, the HisA and TrpF performance thresholds were associated with low enzyme activities. This was especially true for TrpF, where enzymes with catalytic efficiencies on the order of 102-103 s−1 M−1 conferred maximal growth rates. In comparison, the Escherichia coli TrpF enzyme has kcat/KM = 6.8 × 106 s−1 M−1 (25) and the bifunctional M. tuberculosis PriA has kcat/KM = 1.7 × 105 s−1 M−1 for the TrpF reaction (11). In E. coli, TrpF is synthesized at a rate of ∼110 molecules per generation when tryptophan is present in the medium, rising by approximately eightfold when the trp operon is derepressed in minimal medium (26). In our experimental system, the HisA variants were expressed from a constitutive promoter (8). Even if this artificially increased expression by 1–2 orders of magnitude (making our HisA variants among the most highly expressed metabolic enzymes in each strain), the total intracellular TrpF activities of the evolved S. enterica strains would still be lower than those of E. coli and M. tuberculosis. Thus, some bacteria may have TrpF enzymes that exceed their performance thresholds and have “excess capacity” at this node in the metabolic network (27).
The (βα)8 barrel is the most common enzyme fold. Its extraordinary functional diversification is generally ascribed to the mutability of its active site-forming loops. Experimental tests of this hypothesis have focused on engineering by point mutagenesis or loop swaps (28). Our real-time evolution experiment yielded a different functional innovation, the VVR duplication in loop 1, which would have been difficult to discover with currently used design algorithms and engineering methods. The duplication places Arg15[c] in an analogous (but not homologous) position to functionally equivalent arginines in HisA(L169R), PriA and TrpF (Fig. 3C), whereas D10G allows conformational switching between the two mutually exclusive loop orientations that are necessary for TrpF and HisA activities. This emphasizes the important contribution of loop mutability to (βα)8 barrel evolvability (29, 30), as well as the potential of this fold’s overall loop architecture to enable convergent evolutionary solutions.
Our detailed study of a real-time evolution process explains how mutations acquired under continuous selection can result in structural and functional innovations in the encoded enzymes over a relatively small number of generations. The result is a rich—and at present, largely unpredictable—landscape of evolved proteins.
Materials and Methods
The materials and methods used in this study are summarized below. More detailed information is provided in SI Appendix, Materials and Methods.
Molecular Biology.
The cloning of hisA mutants and site-directed mutagenesis were carried out as described previously (13).
Protein and Ligand Preparation.
All proteins were purified as described previously (13). ProFAR was synthesized as reported previously (13), and rCdRP was purchased from Chemir.
Enzyme Kinetics.
HisA assays were conducted as described previously (13). TrpF activities were quantified using a coupled spectrophotometric assay (31).
Mass Spectrometry.
Relative expression levels of each HisA variant were determined using multiplexed tandem mass tagging, as described previously (32).
Structure Determination.
Crystallization conditions for each mutant are listed in SI Appendix, Table S2. Data were collected at the European Synchrotron Radiation Facility (Grenoble, France), Diamond Light Source (Didcot, U.K.), and PETRA (Hamburg, Germany), and structures were solved by molecular replacement, as described for WT HisA (13). Data and refinement statistics are provided in SI Appendix, Table S2.
NMR.
15N Carr–Purcell–Meiboom–Gill relaxation dispersion experiments were recorded for 15N-labeled HisA(dup13-15) and HisA(dup13-15/D10G), as described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility, Diamond Light Source, and PETRA III for access to beamlines for crystallography experiments, and we thank Dr. Per Jemth for his comments on the manuscript. This work was supported by a grant from the Marsden Fund and a Rutherford Discovery Fellowship (to W.M.P.) and grants from the Swedish Research Council (to M.S. and D.I.A.). The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (Grant Agreement 283570).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.H.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5G1T, 5AC7, 5AC8, 5L9F, 5AB3, 5G1Y, 5L6U, 5G4E, 5G4W, 5G2I, 5G2W, 5G2H, and 5G5I).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618552114/-/DCSupplemental.
References
- 1.Nelson-Sathi S, et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvunis A-R, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers RL, Hartl DL. Chimeric genes as a source of rapid evolution in Drosophila melanogaster. Mol Biol Evol. 2012;29:517–529. doi: 10.1093/molbev/msr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 5.Dittmar K, Liberles D. Evolution After Gene Duplication. Wiley; New York: 2010. [Google Scholar]
- 6.Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: Evolution of new genes under continuous selection. Proc Natl Acad Sci USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 8.Näsvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-Even A, et al. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 10.Claren J, Malisi C, Höcker B, Sterner R. Establishing wild-type levels of catalytic activity on natural and artificial (βα)8-barrel protein scaffolds. Proc Natl Acad Sci USA. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Due AV, Kuper J, Geerlof A, von Kries JP, Wilmanns M. Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis. Proc Natl Acad Sci USA. 2011;108:3554–3559. doi: 10.1073/pnas.1015996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanal A, Yu McLoughlin S, Kershner JP, Copley SD. Differential effects of a mutation on the normal and promiscuous activities of orthologs: Implications for natural and directed evolution. Mol Biol Evol. 2015;32:100–108. doi: 10.1093/molbev/msu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Söderholm A, et al. Two-step ligand binding in a (βα)8 barrel enzyme: Substrate-bound structures shed new light on the catalytic cycle of HisA. J Biol Chem. 2015;290:24657–24668. doi: 10.1074/jbc.M115.678086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henn-Sax M, et al. Two (βα)8-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms and common strategies for protecting their labile substrates. Biochemistry. 2002;41:12032–12042. doi: 10.1021/bi026092h. [DOI] [PubMed] [Google Scholar]
- 15.Neidhardt FC, Umbarger HE. In: Chemical Composition of Escherichia coli. Escherichia coli and Salmonella. 2nd Ed. Neidhardt FC, editor. ASM Press; Washington, D.C.: 1996. pp. 13–16. [Google Scholar]
- 16.Campbell E, et al. The role of protein dynamics in the evolution of new enzyme function. Nat Chem Biol. 2016;12:944–950. doi: 10.1038/nchembio.2175. [DOI] [PubMed] [Google Scholar]
- 17.Kaltenbach M, Jackson CJ, Campbell EC, Hollfelder F, Tokuriki N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. eLife. 2015;4:e06492. doi: 10.7554/eLife.06492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuriki N, et al. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- 19.Salverda ML, et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011;7:e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang GI, Desai MM. The spectrum of adaptive mutations in experimental evolution. Genomics. 2014;104:412–416. doi: 10.1016/j.ygeno.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miton CM, Tokuriki N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 2016;25:1260–1272. doi: 10.1002/pro.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Copley SD. Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. Proc Natl Acad Sci USA. 2012;109:E2856–E2864. doi: 10.1073/pnas.1208509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dykhuizen DE, Dean AM, Hartl DL. Metabolic flux and fitness. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 25.Hommel U, Eberhard M, Kirschner K. Phosphoribosyl anthranilate isomerase catalyzes a reversible amadori reaction. Biochemistry. 1995;34:5429–5439. doi: 10.1021/bi00016a014. [DOI] [PubMed] [Google Scholar]
- 26.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton MS, Arcus VL, Patrick WM. Rapid bursts and slow declines: On the possible evolutionary trajectories of enzymes. J R Soc Interface. 2015;12:20150036. doi: 10.1098/rsif.2015.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterner R, Höcker B. Catalytic versatility, stability, and evolution of the (βα)8-barrel enzyme fold. Chem Rev. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- 29.Babbitt PC, Gerlt JA. Understanding enzyme superfamilies: Chemistry as the fundamental determinant in the evolution of new catalytic activities. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 30.Tóth-Petróczy A, Tawfik DS. The robustness and innovability of protein folds. Curr Opin Struct Biol. 2014;26:131–138. doi: 10.1016/j.sbi.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Patrick WM, Matsumura I. A study in molecular contingency: Glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase. J Mol Biol. 2008;377:323–336. doi: 10.1016/j.jmb.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knöppel A, Näsvall J, Andersson DI. Compensating the fitness costs of synonymous mutations. Mol Biol Evol. 2016;33:1461–1477. doi: 10.1093/molbev/msw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.